Abstract
Background.
Patients contracting influenza A(H7N9) infection often developed severe disease causing respiratory failure. Neuraminidase (NA) inhibitors (NAIs) are the primary option for treatment, but information on drugresistance markers for influenza A(H7N9) is limited.
Methods.
Four NA variants of A/Taiwan/1/2013(H7N9) virus containing a single substitution (NA-E119V, NA-I222K, NA-I222R, or NA-R292K) recovered from an oseltamivir-treated patient were tested for NAI susceptibility in vitro; their replicative fitness was evaluated in cell culture, mice, and ferrets.
Results.
NA-R292K led to highly reduced inhibition by oseltamivir and peramivir, while NA-E119V, NA-I222K, and NA-I222R caused reduced inhibition by oseltamivir. Mice infected with any virus showed severe clinical signs with high mortality rates. NA-I222K virus was the most virulent in mice, whereas virus lacking NA change (NA-WT) and NA-R292K virus seemed the least virulent. Sequence analysis suggests that PB2-S714N increased virulence of NA-I222K virus in mice; NS1-K126R, alone or in combination with PB2-V227M, produced contrasting effects in NA-WT and NA-R292K viruses. In ferrets, all viruses replicated to high titers in the upper respiratory tract but produced only mild illness. NA-R292K virus, showed reduced replicative fitness in this animal model.
Conclusions.
Our data highlight challenges in assessment of the replicative fitness of H7N9 NA variants that emerged in NAI-treated patients.
Keywords: influenza virus, H7N9, oseltamivir, peramivir, R292K, E119V, I222K, I222R, mice, ferrets
Recent human infections in China with avian influenza A(H7N9) resulted in substantial morbidity and mortality [1, 2] and provoked global public health concern. The majority of infected patients, most of whom reported direct contact with poultry or having visited live bird markets, were hospitalized and experienced acute respiratory syndrome [3, 4]. Unlike highly pathogenic avian influenza (HPAI) A(H5N1), influenza A(H7N9) lacks molecular markers associated with high pathogenicity in chickens and causes mild or no disease in poultry, hindering control efforts [3, 5].
Neuraminidase (NA) inhibitors (NAIs) have been used for treatment of influenza. Two NAIs, oral oseltamivir and inhaled zanamivir, are approved by the Food and Administration, while intravenous zanamivir is available for compassionate use in severely ill hospitalized patients in the United States [6]. A long-acting inhaled NAI, laninamivir, is available in Japan [7], and intravenous peramivir is marketed in South Korea, Japan [8], and China [9]. Patients who contracted influenza A(H7N9) infection were commonly treated with oseltamivir [3, 10, 11], because the virus is resistant to M2 blockers. The therapeutic effectiveness of NAIs, however, can be compromised by emergence of drug-resistant variants.
Molecular markers for NAI resistance are subtype specific and not well characterized for influenza A(H7N9). It was reported that A/Shanghai/1/2013 (Shanghai/1), collected from the patient soon after initiation of oseltamivir treatment, contained the NA substitution Arg292Lys (R292K in N2 amino acid numbering; R289K in 2013 H7N9 numbering) [3, 12]. This substitution was associated with in vitro resistance to NAIs in the N9 subtype [13] and has been reported in several influenza A(H7N9) viruses [3, 11, 14, 15], at least one of which was recovered from a deceased patient [11]. Furthermore, a mixture of 119Glu/Val (119E/V) was detected in influenza A(H7N9) collected from a patient following oseltamivir treatment; the patient died of acute respiratory distress syndrome [16]. Both NA changes, R292K and E119V, were previously associated with oseltamivir resistance in patients infected with seasonal influenza A(H3N2) [17–19]. While oseltamivir resistance-conferring NA substitutions in H3N2 seasonal influenza viruses were detected predominantly in children and/or immunocompromised patients [17, 19–21], influenza A(H7N9) NA variants have been recovered from otherwise healthy adults [3, 11, 14, 15]. The efficacy of oseltamivir treatment of patients infected with influenza A(H7N9) containing NA substitutions, as well as replicative fitness of the emerged NA variants, remains unknown.
Substitutions in the NA active site are known to impair enzymatic activity and may affect virus fitness. For example, replication of influenza A(H3N2) containing NA-R292K was impaired when tested in ferrets [22]. However, oseltamivir-resistant viruses can overcome reduction in fitness; seasonal influenza A(H1N1) carrying NA-H275Y spread globally during the 2008–2009 influenza seasons [23]. Their replicative fitness and transmissibility were comparable to those of drug-susceptible viruses in mice, guinea pigs, and ferrets [24–27]. At present, information on the fitness of drug-resistant influenza A(H7N9) is limited. The Shanghai/1 NA-R292K virus was shown to replicate efficiently in cell culture [28]. Recombinant influenza A(H7N9) with or without NA-R292K displayed comparable virulence in mice [29, 30] and transmissibility in guinea pigs [30].
Virus population with a 292R/K mixture was reported in the sample collected from a patient in Taiwan on 24 April 2013 [14]. The specimen was propagated once in chicken eggs (E1) and shared with the Centers for Disease Control and Prevention (CDC; Atlanta, GA) for further analysis. In addition to NA-R292K, pyrosequencing identified changes at conserved residues NA-E119V, NA-I222K, or NA-I222R in the E1 isolate. The latter 2 substitutions have not previously been reported in influenza A(H7N9) but were shown to reduce oseltamivir inhibition in both N1 and N2 NA subtypes [31–34]. The NAI susceptibility of the 4 identified NA variants was assessed in an NA inhibition (NI) assay, and their replicative fitness and virulence were determined in mice and ferrets.
METHODS
Detailed information on viruses, sequence analysis, plaque purification, the NA inhibition (NI) assay, and statistical analysis are in the Supplementary Materials.
Ethics Statement
Animals were housed and experiments were conducted in strict compliance with guidelines of the CDC Institutional Animal Care and Use Committee (IACUC), in accordance with Public Health Services Policy, the Animal Welfare Act (US Department of Agriculture), and the Guide for Animal Care and Use of Laboratory Animals. Animal protocols for working with ferrets and mice were approved by the CDC IACUC. All procedures were performed under animal biosafety level 3+ conditions, and animal welfare was observed daily.
Infectivity and Virulence of Influenza A(H7N9) in Mice
Pathogen-free female BALB/c mice (age, 6–8 weeks) were used (Jackson Laboratory, ME) and underwent acclimation for at least 3 days prior to virus inoculation. The mouse median lethal doses (MLD50; n = 4/group) and mouse median infectious doses (MID50; n = 3/group) were determined by intranasally in-oculating animals with 10-fold serial dilutions of virus (101–105 median tissue culture infective doses [TCID50]; 50 μL per mouse). During the course of infection, animals were observed for disease signs and physical abnormalities. For MID50 determination, lungs were collected on day 3 after inoculation, washed thoroughly in phosphate-buffered saline (PBS), and homogenized (Omni Bead Ruptor 24, OMNI International, Kennesaw, GA) in 1 mL of PBS. Cellular debris was cleared by centrifugation at 2000 × g for 10 minutes. Supernatants were serially diluted and used for infection of Madin-Darby canine kidney (MDCK)–SIAT1 cells to determine infectious virus titers, using the TCID50 assay.
Replicative Fitness of Influenza A(H7N9) in Ferrets
Male Fitch ferrets (Mustela putorius furo) aged 3–5 months (Triple F Farms, PA) and serologically negative, by hemagglutination inhibition (HI) assay, for currently circulating 2009 pandemic influenza A(H1N1), influenza A(H3N2), and influenza B virus were used in this study. Ferrets were housed individually and monitored for at least 3 days for acclimation and to establish baseline body temperature prior to starting the study. Clinical signs of illness, such as activity level, sneezing, and nasal and ocular discharge (details for scoring are in the Supplementary Materials), and body weight were recorded daily through-out the 14-day study. Body temperature was measured twice daily by subcutaneous implantable temperature transponders (Bio Medic Data Systems, DE). Intranasal inoculations (3–4 ferrets/group) were performed under anesthesia induced by intramuscular administration of a ketamine/xylazine/atropine mixture (25, 2, and 0.05 mg/kg body weight, respectively), using a 106 TCID50 inoculation dose (0.5 mL total volume; 250 μL per nostril).
Nasal wash specimens were collected daily (under anesthesia) for 10 days after inoculation by flushing both nostrils with 1 mL of PBS and were further processed for determination of infectious virus titers, inflammatory cell counts, and protein concentration. Briefly, nasal washes were centrifuged at 1000 × g for 10 minutes. The cell pellet was resuspended in PBS, and inflammatory cells were counted using the Scepter 2.0 handheld automated cell counter (EMD Millipore, MA). The protein concentration in cell-free nasal wash supernatants was determined using the BCA Protein Assay Kit (Thermo Scientific, IL). Serum samples were collected 14 days after inoculation, treated with receptor-destroying enzyme at 37°C for 18 hours, heat inactivated at 56°C for 30 minutes, and tested by HI assay, using the respective virus with 0.5% packed turkey erythrocytes.
RESULTS
Isolation and Sequencing of NA Variants of A/Taiwan/1/2013 (H7N9) (Taiwan/1)
Multiple swab samples were collected from a patient infected with influenza A(H7N9) in Taiwan who received prolonged oseltamivir treatment, starting on 16 April 2013 [10]. The emergence of virus with R292K was detected in samples collected on 22 April and 25 April [15]; a 292R/K mixture was reported in the sample collected on 24 April [14]. The specimen collected on 24 April was propagated once in chicken eggs (E1) prior to Sanger sequence analysis (Global Initiative on Sharing All Influenza Data [GISAID] accession no. EPI445909–EPI445916; Supplementary Table 2) and was shared with the CDC for further analysis.
Pyrosequencing analysis of the E1 isolate revealed nucleotide polymorphisms at 3 conserved amino acid residues in the NA enzyme active site: E119, I222, and R292. Since each change could affect NAI susceptibility, plaque purification of the E1 isolate was performed. Over 100 individual plaque-purified viruses were screened using NA gene pyrosequencing and Sanger sequencing. The most abundant plaque-purified NA variant was I222R (49%), followed by R292K (23%), E119V (22%), I222K (4%), and wild type (WT; 1%). In all instances, substitutions occurred at the second nucleotide in the respective triplet: for R292K, AGG→AAG; for E119V, GAA→GTA; for I222R, ATA→AGA; and for I222K, ATA→AAA. No analyzed NA variant contained changes at >1 of these substitutions. The 4 NA variants and WT differed by only a single amino acid in the NA and were used in all subsequent experiments (Table 1).
Table 1.
Comparison of Amino Acid Sequences of Plaque-Purified Neuraminidase (NA) Variants of A/Taiwan/1/2013(H7N9) Virus and Mammalian Pathotypes
| Viral Proteins With Amino Acid Differencesa | Amino Acid Substitutions, No. | MID50b | MLD50,b (Fold-Increase) | Versus NAWT Virus | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/Taiwan/1/2013 (H7N9) | PB2 | PB1 | PA | HAc | NAd | NS1 | Virulence in Mice | Replicative Fitness in Ferrets | |||||||||
| 227 | 714 | 632 | 90 | 554 | 556 | 281 | 340 | 119(115) | 222 (219) | 292 (289) | 126 | ||||||
| E1 isolatee | V | S | A | V | V | Q | N | D | E | I | R | K | 4 | … | … | … | … |
| NA-WT | M | … | … | … | I | … | S | … | … | … | … | R | … | 32 | 104.5 | … | … |
| NA-E119V | … | … | … | … | … | R | S | … | V | … | … | … | 5 | <10 | 102.7 (68) | Increased | Comparable |
| NA-I222K | … | N | … | … | I | … | S | … | … | K | … | … | 4 | <10 | 101.5 (1000) | Increased | Comparable |
| NA-I222R | … | … | … | … | … | … | S | … | … | R | … | … | 4 | <10 | 102.3 (150) | Increased | Comparable |
| NA-R292K | … | … | V | M | … | … | … | G | … | … | K | K/R | 8 | <10 | 104.5 (1) | Comparable | Reduced |
Markers of resistance/reduced susceptibility to NA inhibitor(s) are shown in bold.
Abbreviation: WT, wild type.
No amino acid differences were detected in the following genes: PB1-F2, NP, M1/M2, and NS2.
Mouse median infectious dose (MID50) and mouse median lethal dose (MLD50) values are expressed as median tissue culture infective doses.
Mature HA (straight numbering of 2013 influenza [A]H7N9 virus).
Seasonal influenza A(H3N2) NA numbering (straight 2013 influenza A[H7N9] NA virus numbering in parentheses).
The 24 April specimen was propagated once in chicken eggs (E1) at the Taiwan Centers for Disease Control and Prevention; sequences of this virus isolate were deposited to the Global Initiative on Sharing All Influenza Data and used as reference.
Full-genome sequences of the plaque-purified viruses were compared to the original Taiwan/1 E1 isolate consensus sequence. Besides the NA changes, all viruses differed from each other and the E1 consensus sequence by at least 2 additional amino acid substitutions (Table 1). NA-WT sequence differed from the E1 consensus by 4 amino acid changes: PB2-V227M, PA-V554I, HA-N281S, and NS1-K126R. One substitution, PB2-V227M, was unique to this virus and not found in other influenza A(H7N9) sequences available in GISAID. NS1-K126R was rare, with a K/R mixture detected in the NS of NA-R292K only. The NA-E119V virus had arginine (R) at PA-556, while the other viruses had the glutamine (Q) consensus sequence at this position. The NA-I222K virus possessed asparagine (N) in PB2–714; other viruses contained serine (S). The HAs of 4 plaque-purified viruses differed from the E1 consensus sequence by a single amino acid (N281S), whereas NA-R292K contained D340G (Table 1). Notably, the HA sequence of all 5 viruses maintained the presence of proline (P) at HA-226, which was present only in the Taiwan/1 consensus sequence in GISAID.
All plaque-purified viruses contained lysine (K) at PB2–627 and aspartic acid (D) at PB2–701, as seen in the 2 most studied influenza A(H7N9) viruses to date, Anhui/1 and Shanghai/1 [3, 35]. The first substitution has been shown to increase the virulence of influenza A(H7N9) [36] and HPAI A(H5N1) [37] in mice. Sequences of influenza A(H7N9) isolated directly from poultry or environmental samples contain PB2–627E, while both K and E are found in human isolates. The Taiwan/1 viruses contained M2-S31N, a marker of resistance to M2 blockers found in all 2013–2014 influenza A(H7N9) viruses.
In Vitro Susceptibility to NAIs
Susceptibility of plaque-purified viruses to 5 NAIs (oseltamivir, zanamivir, peramivir, laninamivir, and A-315675) was determined in the NI assay. NA-R292K virus had the highest median inhibitory concentration (IC50) values (Table 2), with the >1000-fold increase in oseltamivir and peramivir IC50 values consistent with previous reports [12, 28–30]. NA-E119V virus had the second highest oseltamivir IC50 (an 84-fold increase), together with a 9-fold increase in the zanamivir IC50, with no effect on inhibition by peramivir, laninamivir, or A-315675. NA-I222K and NA-I222R viruses shared a similar resistance profile; oseltamivir IC50 values were elevated by 32–37-fold, while 6–14-fold increases were detected for the remaining 4 NAIs. Applying the WHO Antiviral Working Group criteria [38], NA-R292K virus was characterized as exhibiting highly reduced inhibition by oseltamivir and peramivir and reduced inhibition by zanamivir, laninamivir, and A-315675. NA-E119V virus exhibited reduced inhibition by oseltamivir only. The acquisition of NA-I222R or NA-I222K resulted in reduced inhibition by oseltamivir and laninamivir, while NA-I222R also conferred reduced inhibition by zanamivir.
Table 2.
Susceptibility of Plaque-Purified Neuraminidase (NA) Variants of A/Taiwan/1/2013(H7N9) Virus to NA Inhibitors
| IC50, nM ± SD (Fold-Difference)a | |||||
|---|---|---|---|---|---|
| A/Tai wan/1 | Oseltamivir | Zanamivir | Peramivir | Laninamivir | A-315675 |
| NA-WT | 0.28 ± 0.02 | 0.65 ± 0.13 | 0.08 ± 0.02 | 0.74 ± 0.16 | 0.12 ± 0.01 |
| NA-E119V | 23.56 ± 4.84 (84) | 6.01 ± 2.57 (9) | 0.10 ± 0.01 (1) | 1.31 ± 0.25 (2) | 0.14 ± 0.01 (1) |
| NA-I222K | 8.82 ± 0.27 (32) | 4.91 ± 0.68 (8) | 0.48 ± 0.08 (6) | 9.34 ± 1.17 (13) | 0.82 ± 0.16 (7) |
| NA-I222R | 10.43 ± 0.62 (37) | 7.89 ± 1.82 (12) | 0.94 ± 0.13 (12) | 10.55 ± 1.06 (14) | 0.71 ± 0.08 (6) |
| NA-R292K | 2873 ± 1334 (>10 000) | 35.68 ± 4.28 (55) | 126.96 ± 22.73 (1587) | 16.33 ± 3.19 (22) | 1.60 ± 0.11 (13) |
| Reference viruses A(H3N2) | |||||
| A/Washington/01/2007, NA-WT | 0.07 ± 0.02 | 0.23 ± 0.03 | 0.23 ± 0.03 | 0.29 ± 0.11 | 0.18 ± 0.07 |
| A/Bethesda/956/2006, NA-R292K | 3974 ± 1445 (>10 000) | 6.83 ± 1.01 (30) | 16.27 ± 0.89 (71) | 2.51 ± 0.09 (9) | 2.46 ± 0.33 (14) |
Amino acid substitution in the NA according to N2 amino acid numbering; substitutions correspond to E115V, I219K/R and R289K in the NA of 2013 H7N9 virus. No changes in the NA of wild-type (WT) virus. The World Health Organization Antiviral Working Group criteria are as follows: normal inhibition, <10-fold; reduced inhibition, 10–100-fold; highly reduced inhibition, >100-fold.
Abbreviation: SD, standard deviation.
The median inhibitory concentration (IC50) is defined as the concentration needed to inhibit NA activity by 50%; fold-difference, compared with the IC50 of WT virus.
Virus Replication in Cell Cultures
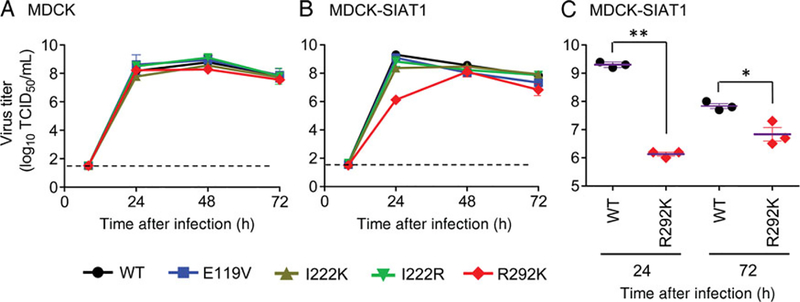
Assessment of the replicative potential of the NA variants of the Taiwan/1 virus was conducted in 2 cell lines, conventional MDCK cells and MDCK-SIAT1 cells that overexpress α−2,6-NeuAc-containing receptors [39]. Cell monolayers were inoculated at a low multiplicity of infection (0.0001), and infectious virus yields were determined 8, 24, 48, and 72 hours after infection. Very low or no virus titers were detected 8 hours after infection in both cell lines. In MDCK cells, all viruses showed efficient replication and produced comparable titers, which peaked 24–48 hours after infection (Figure 1A). In MDCK-SIAT1 cells, NA-WT, NA-E119V, NA-I222K, and NA-I222R viruses replicated as efficiently as in MDCK cells, with titers peaking 24 hours after infection (Figure 1B). In contrast, NA-R292K virus exhibited delayed growth, with infectious titers 2–3 logs lower than those of the other viruses and the highest titer 48 hours after infection. The NA-R292K virus titer was significantly lower than that for WT 24 (P < .0001) and 72 (P = .0175) hours after infection (Figure 1C). Therefore, replication of NA-R292K virus was notably attenuated in MDCK-SIAT1 cells but not in MDCK cells.
Figure 1.
Replication kinetics of influenza A(H7N9) isolated from a patient in Taiwan. Madin-Darby canine kidney (MDCK; A) or MDCK-SIAT1 (B) cells were infected with each plaque-purified virus at a multiplicity of infection of 0.0001. Virus-containing supernatants were collected at the indicated time points, and virus titers were determined in MDCK-SIAT1 cells. The statistically significant difference between wild-type (WT) and R292K viruses are shown (C). Dotted lines indicate the limit of virus titer detection. *P = .0175; **P < .0001. Abbreviation: TCID50, median tissue culture infective doses.
Infectivity and Virulence in Mice
To determine whether the NA variants of Taiwan/1 virus were attenuated in a mouse model, BALB/c mice were inoculated with 10-fold serially diluted viruses (101–105 TCID50 per mouse), and their lungs were harvested on day 3 after inoculation to detect the presence of infectious virus. The mouse MID50 for NA-WT virus was 32 TCID50. Unexpectedly, <10 TCID50 were required to infect 50% of animals with the 4 NA variant viruses, including NA-R292K virus (Table 1). Infections with 104 and 105 TCID50 of any virus resulted in rapid weight loss (Figure 2) and severe signs of illness, including lethargy, ruffled fur, hunched posture, and dyspnea. NA variants with a substitution at I222 caused rapid weight loss and 100% lethality with doses as low as 102 and 103 TCID50 (Figure 2D and 2F), while the NA-WT virus produced a similar effect only at the highest challenge dose, 105 TCID50 (Figure 2J). NA-E119V virus caused 100% lethality in animals infected with 104 TCID50 (Figure 2H). Notable differences in virulence were also observed, based on comparison of the MLD50 values (Table 1). NA-I222K virus was the most virulent (101.5 TCID50), while NA-WT and NA-R292K viruses exhibited 1000-fold lower virulence (104.5 TCID50). NA-E119V and NA-I222R viruses showed intermediate virulence.
Figure 2.
Virulence of Taiwan/1(H7N9) neuraminidase variants in mice. Animals (n = 4/group) were intranasally inoculated with 101, 102, 103, 104, or 105 median tissue culture infective doses (TCID50) of indicated viruses. Body weight change (A, C, E, G, and I) and survival (B, D, F, H, and J) were monitored daily. Mice that lost ≥25% (dotted line) of their baseline weight were humanely euthanized. Each value (A, C, E, G, and I) represents the average percentage loss of body weight (±SD). Abbreviation: SD, standard deviation.
Replicative Fitness in Ferrets
For ferret model experiments, animals were inoculated intranasally with 106 TCID50 of virus, after which nasal wash specimens were collected daily for 10 days to assess infectious virus titers, numbers of inflammatory cells, and protein concentrations. All viruses replicated to comparable titers peaking (approximately 106 TCID50/mL) between days 2 and 4 after inoculation (Figure 3A). Notably, replication of NA-R292K virus was delayed; no virus was detected in 2 of 4 animals on day 1 after inoculation, yet these 2 ferrets shed virus on day 7 after inoculation. NA-R292K virus titers were approximately 1–2 logs lower than that of NA-WT virus on days 1, 2 (P = .004), and 3 (P = .003) after inoculation. Quantitative virus shedding, defined as the area under the curve (AUC) during days 1–6 (days 1–7 for NA-R292K virus), was measured and expressed as log10 TCID50/mL. The AUCs were 29.3 for NA-WT, 29.0 for NA-E119V, 26.6 for NA-I222K, 25.9 for NA-I222R, and 26.6 for NA-R292K viruses, indicating a trend toward lower replication rates of NA variants in the ferret’s upper respiratory tract, with the exception of NA-E119V virus.
Figure 3.
Replicative fitness of Taiwan/1(H7N9) neuraminidase variants in ferrets. Animals (n = 3–4/group) were intranasally inoculated with 106 median tissue culture infective doses (TCID50) of the indicated viruses. Nasal washes were collected daily for 10 days, and virus titers (A) (*P ≤ .004), inflammatory cell counts (B), and protein concentrations (C) were determined. The body weight change (D) and temperature (E) were recorded daily. The statistically significant difference in nasal wash virus titers between wild-type and R292K viruses is shown. Dotted lines indicate limit of virus titer detection. SDs and P values are not shown if <3 animals shed virus. Abbreviation: SD, standard deviation.
Inflammatory cell counts (Figure 3B) and total protein concentrations (Figure 3C) in nasal wash specimens peaked on days 5 and 7 after inoculation with all 5 viruses; values for both parameters were lower (P < .05) in NA-R292K virus–infected animals during days 5–9 after inoculation. NA variants maintained their substitutions during the shedding period (Supplementary Table 1).
Unlike the severe disease observed in mice, all viruses produced only modest clinical symptoms in ferrets. No signs of lethargy were seen in any of the ferret groups, and no infected ferrets had remarkable weight loss over the course of infection (Figure 3D). Notably, ferrets infected with NA-R292K virus did not experience weight loss and gained body weight at a faster rate than those infected with the other influenza A(H7N9) viruses tested. Body temperatures were stable in all infected animals across the course of infection (Figure 3E). Intermittent pyrexia (≥1.5°C above baseline) was detected early and late after inoculation in 1 animal infected with NA-I222R virus early and late after inoculation. Serum specimens collected from inoculated ferrets on day 14 after inoculation showed seroconversion against homologous influenza A(H7N9), with HI titers ranging from 160 to 1280.
DISCUSSION
The presence of viral quasispecies in the NA genes of viruses recovered from patients undergoing antiviral treatment is not unexpected. Analysis of individual plaques of Taiwan/1 (H7N9) virus led to recovery of NA variants without or with 1 change at residues E119, I222, or R292. Laboratory data for virus with NA-R292K, the most commonly detected substitution in the influenza A(H7N9) subtype, indicate clinically relevant resistance to oseltamivir and, possibly, peramivir. Thus, R292K may reduce the effectiveness of treatment and prophylaxis with these drugs.
For risk assessment purposes, it was imperative to investigate the replicative fitness of the identified NA variants. R292 participates in catalysis and interacts directly with the sialic acid–containing receptors, while E119 and I222 are framework residues that support the NA binding pocket [40]. Therefore, NA-R292K is likely to cause a greater functional loss of NA enzymatic activity [28, 41] than the framework substitutions, potentially leading to a greater deficit of virus replicative fitness.
In this study, the NA-R292K virus replicated to similarly high titers as the NA-WT virus in MDCK cells but showed less efficient replication in MDCK-SIAT1 cells modified to overexpress α2,6-NeuAc-receptors [39]. Because the need for functional NA is greater when virus replicates in vivo, we anticipated reduced replicative fitness and virulence of the NA variants in animals. To prove this hypothesis, we used 2 animal models, mice and ferrets. When tested in ferrets, all 4 NA variants and the NA-WT virus replicated efficiently, according to high titers in nasal washes, which peaked on days 2–4 after inoculation. However, all viruses caused only mild disease signs in ferrets, despite the use of a high inoculation dose (106 TCID50 per animal). The NA-R292K virus appeared to be the most attenuated, based on fewer counts of inflammatory cells and lower protein levels in nasal washes and a steady increase in body weights. Notably, nasal wash titers of the NA-R292K virus were significantly lower than those of the NA-WT virus in the first 3 days after inoculation, suggesting impaired replicative fitness. Similarly, Yen et al [42] demonstrate that NA-R292K virus (Shanghai/1 background) showed competitive fitness loss in ferrets; this virus, however, transmitted at comparable efficiency as WT virus to animals via direct or respiratory droplet contact.
Unexpectedly, the NA-WT virus showed approximately 3-fold reduced infectivity in mice (MID50, 32 vs <10), compared with NA variants. Moreover, NA-WT virus required a much greater dose to produce lethality in mice, when compared to the 3 NA variants (E119V, I222R, and, especially, I222K). The NA-R292K virus showed an MLD50 similar to that of the NA-WT virus, consistent with a recent report [30] indicating comparable virulence of recombinant Shanghai/1 viruses, with or without NA-R292K. Of note, use of different mouse strains (eg, BALB/c vs C57BL/6) infected with the same influenza A (H7N9) resulted in substantial differences in pathogenicity and inflammatory responses [43].
One limitation of the present study was a lack of the original clinical sample to characterize a quasispecies composition and to recover NA variants. Another limitation was the use of plaque-purified versus reverse-genetically generated viruses to address the effect of each NA change on virus fitness. Nevertheless, our approach, coupled with full-genome sequence analysis, provided valuable insights into viral phenotypes, which would have not been obtained otherwise. The plaque-purified NA-WT virus showed reduced infectivity and virulence in mice. This virus contained 2 substitutions PB2-V227M and NS1-K126R, not seen in any other influenza A(H7N9), including the published Taiwan/1 sequence. Peculiarly, the NA-R292K virus possessed a mixture of R/K in NS1–126 and it also exhibited reduced virulence in mice. Although the role of NS1-K126R in the virulence of influenza A(H7N9) has not been demonstrated, introduction of NS1-K126R in the A/Puerto Rico/8/34(H1N1) virus led to increased viral yields in cell culture and virulence in mice [44]. In contrast, this substitution was shown to cause a 10-fold decreased replication of a recombinant influenza A(H3N2) in cell culture [45]. In our study, NS1-K126R had no apparent effect on influenza A(H7N9) replication in ferrets, indicating its host-specific nature.
The NA-I222K virus showed the highest virulence in mice and contained a substitution at PB2-S714N, which was not found among influenza A(H7N9) sequences in GISAID. A switch from serine to arginine at PB2–714 was demonstrated to increase viral RNA transcription activities in vitro [46]. The potential role of asparagine at PB2–714 remains unknown. The identified HA substitutions (HA-S281N and HA-D340G) are located in positions not known to affect receptor binding or antigenicity; the role of other identified internal gene mutations is also unknown at this time.
Overall, interpretation of results used for risk assessment is challenging. The NA variants were highly virulent in mice, but the results were complicated by the presence of other substitutions in the viral internal genes. In the ferret model, animals were asymptomatic when infected with any influenza A(H7N9), which is contrary to what was observed in humans. Because Taiwan/1 virus caused severe disease in the patient, these findings in ferrets should be interpreted cautiously.
In summary, we report recovery of 4 NA variants from the Taiwan/1(H7N9) virus collected from an oseltamivir-treated patient. The lack of an apparent compromise in the replicative fitness of these NA variants is concerning. Close monitoring of viruses for changes at these NA residues needs to be conducted, particularly in patients treated with NAIs. Ideally, specimens should be collected systematically from patients infected with influenza A(H7N9), regardless of illness severity or treatment, to better understand the emergence of NAvariants and quasispecies dynamics. This study also highlights the need for development of novel antiinfluenza drugs and drug combination therapies.
Supplementary Material
Acknowledgments.
We thank Dr Margaret Okomo-Adhiambo, for valuable discussion; Marnie Levine, for assisting with pyrosequencing analysis; Peter Eworonsky and Lester Slough (CDC), for excellent assistance with animal care; F. Hoffmann-La Roche (Basel, Switzerland), for kindly providing oseltamivir carboxylate; GlaxoSmithKline (Uxbridge, United Kingdom), for kindly providing zanamivir; BioCryst Pharmaceuticals (Durham, NC), for kindly providing peramivir; Biota (Victoria, Australia), for kindly providing laninamivir; and Abbott Laboratories (Abbott Park, IL), for kindly providing A-315675.
Financial support. This work was supported by the Centers for Disease Control and Prevention (CDC) Influenza Division and by an interagency agreement between Biomedical Advanced Research and Development Authority and the CDC.
Footnotes
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Publisher's Disclaimer: Disclaimer. The findings and conclusions of this report are those of the authors and do not necessarily represent the views of the funding agency or the Centers for Disease Control and Prevention.
References
- 1.WHO. Number of confirmed human cases of avian influenza A(H7N9) reported to WHO. http://www.who.int/influenza/human_animal_interface/influenza_h7n9/14_ReportWebH7N9Number_20140228.pdf?ua=1. Accessed 11 March 2014.
- 2.WHO. Human infection with avian influenza A(H7N9) virus—update 2014. http://www.who.int/csr/don/archive/year/2014/en/index.html. Accessed 11 March 2014.
- 3.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368:1888–97. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Zhou L, Zhou M, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med 2014; 370:520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Liang W, Yang S, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 2013; 381:1916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marty FM, Man CY, van der Horst C, et al. Safety and pharmacokinetics of intravenous zanamivir treatment in hospitalized adults with influenza: an open-label, multicenter, single-arm, phase II study. J Infect Dis 2014; 209:542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugaya N, Ohashi Y. Long-acting neuraminidase inhibitor laninamivir octanoate (CS-8958) versus oseltamivir as treatment for children with influenza virus infection. Antimicrob Agents Chemother 2010; 54:2575–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shetty AK, Peek LA. Peramivir for the treatment of influenza. Expert Rev Anti Infect Ther 2012; 10:123–43. [DOI] [PubMed] [Google Scholar]
- 9.Su CT-T, Ouyang X, Zheng J, Kwoh C-K. Structural analysis of the novel influenza A(H7N9) viral neuraminidase interactions with current approved neuraminidase inhibitors oseltamivir, zanamivir, and peramivir in the presence of mutation R289K. BMC Bioinformatics 2013; 14 (Suppl 16):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang SY, Lin PH, Tsai JC, Hung CC, Chang SC. The first case of H7N9 influenza in Taiwan. Lancet 2013; 381:1621. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Lu S, Song Z, et al. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 2013; 381:2273–9. [DOI] [PubMed] [Google Scholar]
- 12.Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg Infect Dis 2013; 19:1521–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol 1998; 72:2456–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mok CK, Chang SC, Chen GW, et al. Pyrosequencing reveals an oseltamivir-resistant marker in the quasispecies of avian influenza A (H7N9) virus. J Microbiol Immunol Infect 2013; S1684–1182(13) 00201–6. [DOI] [PubMed] [Google Scholar]
- 15.Lin PH, Chao TL, Kuo SW, et al. Virological, serological, and antiviral studies in an imported human case of avian influenza A(H7N9) virus in Taiwan. Clin Infect Dis 2014; 58:242–6. [DOI] [PubMed] [Google Scholar]
- 16.Qi Y, Fan H, Qi X, et al. A novel pyrosequencing assay for the detection of neuraminidase inhibitor resistance-conferring mutations among clinical isolates of avian H7N9 influenza virus. Virus Res 2014; 179:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 2004; 364:759–65. [DOI] [PubMed] [Google Scholar]
- 18.Okomo-Adhiambo M, Demmler-Harrison GJ, Deyde VM, et al. Detection of E119V and E119I mutations in influenza A (H3N2) viruses isolated from an immunocompromised patient: challenges in diagnosis of oseltamivir resistance. Antimicrob Agents Chemother 2010; 54: 1834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piralla A, Gozalo-Marguello M, Fiorina L, et al. Different drug-resistant influenza A(H3N2) variants in two immunocompromised patients treated with oseltamivir during the 2011–2012 influenza season in Italy. J Clin Virol 2013; 58:132–7. [DOI] [PubMed] [Google Scholar]
- 20.Hurt AC, Leang SK, Tiedemann K, et al. Progressive emergence of an oseltamivir-resistant A(H3N2) virus over two courses of oseltamivir treatment in an immunocompromised paediatric patient. Influenza Other Respir Viruses 2013; 7:904–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura D, Sugaya N, Ozawa M, et al. Frequency of drug-resistant viruses and virus shedding in pediatric influenza patients treated with neuraminidase inhibitors. Clin Infect Dis 2011; 52:432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herlocher ML, Carr J, Ives J, et al. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res 2002; 54:99–111. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen HT, Fry AM, Gubareva LV. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther 2012; 17:159–73. [DOI] [PubMed] [Google Scholar]
- 24.Abed Y, Pizzorno A, Bouhy X, Rheaume C, Boivin G. Impact of potential permissive neuraminidase mutations on viral fitness of the H275Y oseltamivir-resistant influenza A(H1N1)pdm09 virus in vitro, in mice and in ferrets. J Virol 2014; 88:1652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baz M, Abed Y, Simon P, Hamelin ME, Boivin G. Effect of the neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J Infect Dis 2010; 201:740–5. [DOI] [PubMed] [Google Scholar]
- 26.Memoli MJ, Davis AS, Proudfoot K, et al. Multidrug-resistant 2009 pandemic influenza A(H1N1) viruses maintain fitness and transmissibility in ferrets. J Infect Dis 2011; 203:348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seibert CW, Kaminski M, Philipp J, et al. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol 2010; 84:11219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yen HL, McKimm-Breschkin JL, Choy KT, et al. Resistance to neuraminidase inhibitors conferred by an R292K mutation in a human influenza virus H7N9 isolate can be masked by a mixed R/K viral population. mBio 2013; 4 pii: e00396–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe T, Kiso M, Fukuyama S, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 2013; 501:551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hai R, Schmolke M, Leyva-Grado VH, et al. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun 2013; 4:2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baz M, Abed Y, McDonald J, Boivin G. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis 2006; 43:1555–61. [DOI] [PubMed] [Google Scholar]
- 32.Hurt AC, Holien JK, Barr IG. In vitro generation of neuraminidase inhibitor resistance in A(H5N1) influenza viruses. Antimicrob Agents Chemother 2009; 53:4433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard M, Ferraris O, Erny A, et al. Combinatorial effect of two framework mutations (E119V and I222L) in the neuraminidase active site of H3N2 influenza virus on resistance to oseltamivir. Antimicrob Agents Chemother 2011; 55:2942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon P, Holder BP, Bouhy X, Abed Y, Beauchemin CA, Boivin G. The I222V neuraminidase mutation has a compensatory role in replication of an oseltamivir-resistant influenza virus A/H3N2 E119V mutant. J Clin Microbiol 2011; 49:715–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kageyama T, Fujisaki S, Takashita E, et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill 2013; 18:20453. [PMC free article] [PubMed] [Google Scholar]
- 36.Mok CK, Lee HH, Lestra M, et al. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol 2014; 88:3568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001; 293:1840–2. [DOI] [PubMed] [Google Scholar]
- 38.Meetings of the WHO working group on surveillance of influenza antiviral susceptibility - Geneva, November 2011 and June 2012. Wkly Epidemiol Rec 2012; 87:369–74. [PubMed] [Google Scholar]
- 39.Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol 2003; 77:8418–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colman PM, Varghese JN, Laver WG. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 1983; 303:41–4. [DOI] [PubMed] [Google Scholar]
- 41.Wu Y, Bi Y, Vavricka CJ, et al. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res 2013; 23:1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen HL, Zhou J, Choy KT, et al. The R292K mutation that confers resistance to neuraminidase inhibitors leads to competitive fitness loss of A/Shanghai/1/2013 (H7N9) influenza virus in ferrets. J Infect Dis 2014; 210:1900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao G, Liu C, Kou Z, et al. Differences in the pathogenicity and inflammatory responses induced by avian influenza A/H7N9 virus infection in BALB/c and C57BL/6 mouse models. PLoS One 2014; 9:e92987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Y, Zhong G, Zhu L, et al. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J Immunol 2010; 184: 5777–90. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C, Hsiang TY, Kuo RL, Krug RM. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc Natl Acad Sci U S A 2010; 107:2253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Wang Q, Wang J, Mizumoto K, Toyoda T. Two mutations in the C-terminal domain of influenza virus RNA polymerase PB2 enhance transcription by enhancing cap-1 RNA binding activity. Biochim Biophys Acta 2012; 1819:78–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.