Abstract
Introduction
Nephrotic syndrome (NS) is a characterized by massive proteinuria, edema, hypoalbuminemia, and dyslipidemia. Glucocorticoids (GCs), the primary therapy for >60 years, are ineffective in approximately 50% of adults and approximately 20% of children. Unfortunately, there are no validated biomarkers able to predict steroid-resistant NS (SRNS) or to define the pathways regulating SRNS.
Methods
We performed proteomic analyses on paired pediatric NS patient plasma samples obtained both at disease presentation before glucocorticoid initiation and after approximately 7 weeks of GC therapy to identify candidate biomarkers able to either predict steroid resistance before treatment or define critical molecular pathways/targets regulating steroid resistance.
Results
Proteomic analyses of 15 paired NS patient samples identified 215 prevalent proteins, including 13 candidate biomarkers that predicted SRNS before GC treatment, and 66 candidate biomarkers that mechanistically differentiated steroid-sensitive NS (SSNS) from SRNS. Ingenuity Pathway Analyses and protein networking pathways approaches further identified proteins and pathways associated with SRNS. Validation using 37 NS patient samples (24 SSNS/13 SRNS) confirmed vitamin D binding protein (VDB) and APOL1 as strong predictive candidate biomarkers for SRNS, and VDB, hemopexin (HPX), adiponectin (ADIPOQ), sex hormone–binding globulin (SHBG), and APOL1 as strong candidate biomarkers to mechanistically distinguish SRNS from SSNS. Logistic regression analysis identified a candidate biomarker panel (VDB, ADIPOQ, and matrix metalloproteinase 2 [MMP-2]) with significant ability to predict SRNS at disease presentation (P = 0.003; area under the receiver operating characteristic curve = 0.78).
Conclusion
Plasma proteomic analyses and immunoblotting of serial samples in childhood NS identified a candidate biomarker panel able to predict SRNS at disease presentation, as well as candidate molecular targets/pathways associated with clinical steroid resistance.
Keywords: biomarkers, nephrotic syndrome, proteomics, steroid resistance
See Commentary on Page 1
Steroid resistance is a major clinical challenge for both physicians and patients with a wide array of diseases, including NS, asthma, rheumatoid arthritis, and other inflammatory conditions primarily treated with steroids. NS is one of the most common forms of glomerular disease and one of the leading causes of end-stage kidney disease in both children and adults. Although GCs have been the primary therapy for NS for >60 years, they unfortunately induce remission of NS in only approximately 50% of adults and approximately 80% of children,1,2 with unresponsive patients being labeled as having SRNS. Unfortunately, no validated biomarkers have yet been identified that are able to predict steroid resistance, leaving patients at high risk for both toxic side effects of GC treatment, as well as disease progression. Thus, the identification and validation of biomarkers able to predict the clinical response to GC before the initiation of treatment could enable avoidance of GC-induced drug toxicity, and rapid initiation of alternative treatments more likely to induce remission and delay or prevent disease progression. In addition, the identification of candidate biomarkers able to determine specific molecular pathways and targets associated with clinical steroid resistance could enable the development of more effective and less toxic targeted future therapies for NS.
Mass spectrometric-based discovery proteomic methods have contributed significantly to our understanding of idiopathic renal disease by identification of proteins integral to disease processes.3,4 State-of-the-art methods have the ability to characterize both high- and low-abundant proteins and develop both qualitative data to identify proteins and to suggest relative abundance or absolute concentration differences.5, 6, 7, 8 These studies and other “omics” studies are providing valuable pilot and confirmatory information for the identification of disease biomarkers.9, 10, 11, 12, 13, 14, 15 Recently, a few studies have approached urinary biomarker identification using proteomics in patients with NS either classified by histology or by clinical response.16,17
The present studies were designed to test the primary hypothesis that proteomic analyses with subsequent validation of paired plasma samples from children with SSNS and SRNS can be used to identify biomarkers predictive of steroid resistance at the time of disease presentation before therapy (Figure 1a). The secondary aim of the present study was to identify mechanistic molecular pathways or targets associated with clinical steroid resistance in NS (Figure 1b). To test this hypothesis, we analyzed paired plasma biosamples collected from 2008 to 2014 through the Midwest Pediatric Nephrology Consortium from children with NS that were obtained both at the time of disease presentation (before initiation of steroid therapy) and an average of approximately 7 weeks post-GC treatment, when the clinical determination of SRNS versus SSNS was made by the treating nephrologist.
Figure 1.
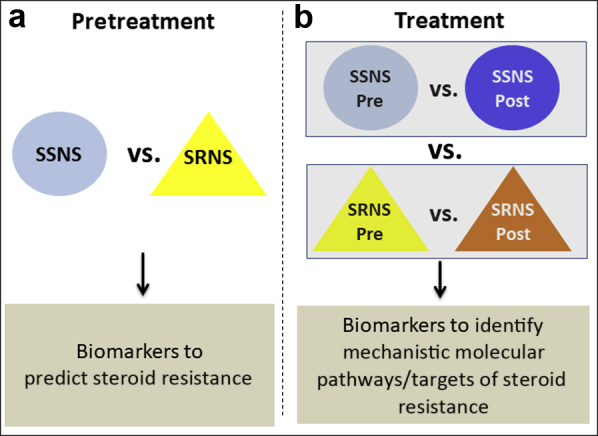
Study hypothesis and design. The present studies were designed to test the hypothesis that proteomic analyses with subsequent validation in paired plasma samples from children with steroid-sensitive nephrotic syndrome (SSNS) and steroid-resistant nephrotic syndrome (SRNS) can be used to identify biomarkers able to (a) predict clinical steroid resistance, and (b) mechanistically define specific molecular pathways or targets associated with clinical steroid resistance.
Methods
Study Approval, Ethics Statement, Patients, and Plasma Collections
All research protocols and consent documents were approved by the institutional review board of Nationwide Children's Hospital as the coordinating center (approval numbers IRB07–00400, IRB12–00039, and IRB05–00544), as well as by each of the other participating centers of the Midwest Pediatric Nephrology Consortium. Paired plasma samples were collected for each patient, with the first sample “pretreatment” at the time of disease presentation before even a single dose of GC, and the second sample “post-treatment” after 6 to 10 weeks of GC therapy when the clinical determination of SSNS versus SRNS had been determined by the treating nephrologist. See Table 1 for patient demographics and Supplementary Methods for clinical data and sample collection details.
Table 1.
Patient demographics for proteomic discovery and Western blotting validation study cohorts
| n | Discovery (proteomics) |
Validation (immunoblotting) |
||||||
|---|---|---|---|---|---|---|---|---|
| Total |
SSNS |
SRNS |
P value | Total |
SSNS |
SRNS |
P value | |
| 15 | 7 | 8 | 37 | 24 | 13 | |||
| Weeks between pre- and post-treatment samples | 6.0 ± 0.5, n = 15 | 5.8 ± 0.7, n = 7 | 6.3 ± 0.7, n = 8 | ns | 6.9 ± 0.4, n = 37 | 6.9 ± 0.6, n = 24 | 6.7 ± 0.5, n = 13 | ns |
| Disease onset samples verified to be pretreatment | 15 (100%) | 7 (100%) | 8 (100%) | 37 (100%) | 24 (100%) | 13 (100%) | ||
| Age | 8.7 ± 1.0, n = 15 | 6.5 ± 1.7, n = 7 | 10.6 ± 0.9, n = 8 | 0.04a | 7.0 ± 0.7, n = 35 | 5.6 ± 0.8, n = 22 | 9.5 ± 1.1, n = 13 | 0.006b |
| Sex, n (%) | ||||||||
| Male | 0 (0) | 0 (0) | 0 (0) | 15 (42) | 11 (48) | 4 (31)c | 0.026c (2-tailed) | |
| Female | 15 (100) | 7 (100) | 8 (100) | 21 (58) | 12 (52) | 9 (69)c | ||
| Not reported | 1 | 1 | ||||||
| Height | 135.1 ± 6.4, n = 15 | 118.8 ± 9.2, n = 7 | 149.3 ± 5.2, n = 8 | 0.01a | 124.5 ± 4.4, n = 36 | 114 ± 4.5, n = 23 | 143.1 ± 6.8, n = 13 | 0.0008d |
| Weight | 43.9 ± 6.0, n = 15 | 28.1 ± 5.0, n = 7 | 57.6 ± 7.5, n = 8 | 0.007b | 35.9 ± 3.9, n = 36 | 25.9 ± 2.6, n = 23 | 53.5 ± 7.6, n = 13 | 0.0002d |
| Race, n (%) | ||||||||
| White | 9 (60) | 4 (57) | 5 (62.5) | 17 (46) | 11 (45.8) | 6 (46) | ||
| Asian | 1 (7) | 1 (14.3) | 0 (0) | 4 (10.8) | 4 (16.7) | 0 (0) | ||
| African American | 4 (27) | 1 (14.3) | 3 (37.5) | 10 (27) | 4 (16.7) | 6 (46)e | <0.0001e | |
| Biracial | 1 (7) | 1 (14.3) | 0 (0) | 2 (5.4) | 1 (4.2) | 1 (7.7) | ||
| Native American | 0 (0) | 0 (0) | 0 (0) | 1 (2.7) | 1 (4.2) | 0 (0) | ||
| Not reported | 0 (0) | 0 (0) | 0 (0) | 3 (8.1) | 3 (12.5) | 0 (0) | ||
ns, not significant; SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome.
Significance determined by at test.
binomial test.
χ2 test.
P < 0.05.
P < 0.01.
P < 0.001.
Proteomics Discovery Workflow
The proteomics workflow (Supplementary Figure S1) addressed the label-free comparison of high mass accuracy data sets developed from 15 paired patient plasma samples (n = 30) using a fast protein liquid chromatography antibody-based method to immune-deplete the 20 most common plasma proteins before trypsinization. To address the limitation of small sample numbers and potential gender variability, the discovery proteomics dataset was developed with only female patient plasma samples, whereas the confirmation cohort was expanded to include both male and female patients. These data were then filtered to identify predictive and mechanistic biomarkers (Figure 1) and used for various other analyses described in Supplementary Table S1. The data filtering approaches were directed by absolute or relative differences in the protein abundance, unbiased statistical or pathways approaches, and, last, expert review of the proteomic data. These procedures of “Immuno-Depletion of Highly Abundant Proteins from Plasma, Sample Proteolysis and Liquid Chromatography–Mass Spectrometry (LCMS) Data Acquisition” are detailed in the Supplementary Material.
Informatics Analysis of LCMS Datasets
Proteome Discoverer (Thermo Fisher Scientific, Waltham, MA) v2.0.0.802 was used to analyze the data collected by the mass spectrometer with SequestHT searches performed using the July 7, 2015 version of the UniprotKB. See Supplementary Methods for details.
Immunoblot Analyses for Differential Protein Abundance
Plasma proteins were resolved on 6% to 20% gradient gels by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and immunoblotted as described in detail in the Supplementary Material. X-ray films were scanned using a calibrated ArtixScan M1 transillumination scanner (Microtek Lab, Cerritos, CA) controlled by the ScanWizard Pro program (version 7.042, Microtek International Inc., Hsinchu, Taiwan; Microtek Lab Inc., Carson, CA) using standard settings. Densitometry analyses of the integrated band densities were performed using ImageJ (version 1.39, National Institutes of Health, Bethesda, MD, standard settings; http://rsb.info.nih.gov/ij/) and values plotted using GraphPad (LaJolla, CA) Prism software version 6.00 for Windows.
Statistical Evaluation of Trends in Protein Abundance
Discovery Proteomic Studies
To determine trends in protein abundance, all censored or missing values were replaced by a minimal global protein abundance value divided by the square root of 2 (minimal observed label-free signal ÷ √2) or 2280.3.18 The estimations for significance of the between-group protein abundance differences were calculated using medians of protein abundance and the Mann-Whitney 1-way analysis of variance. Protein name, gene name, accession numbers, and associated protein abundance values estimated as intensity-based absolute quantification values were exported into an Excel file from Scaffold.19 See Supplementary Methods for details.
Targeted Validation Studies
Statistical significance was determined by unpaired or paired t tests using the GraphPad Prism software version 6.00 for Windows. P values were considered significant at P < 0.05. Data shown include representative blots as well as quantitation of all the samples tested, and are displayed as means ± SEM. The ability of the quantified immunoblot data and patient sex to classify patient samples as SSNS and SRNS was determined using backward stepwise logistic regression analysis with −2log likelihood, Cox & Snell R2, and Nagelkerke R2 to assess goodness of fit for the reduced terms. The sensitivity and specificity for the reduced terms to classify patient samples was determined by the area under the curve from a receiver operator characteristic. Multivariate statistical tests were performed using SAS version 9.4 (SAS Inc., Cary, NC) and SPSS version 24 (IBM Corp., Armonk, NY).
Cluster Analysis With Heatmaps
To evaluate and illustrate group trends in protein relative abundance, the intensity-based absolute quantification data were analyzed by cluster analysis with heatmap illustration. To address the dynamic range differences across the proteomic data set, the protein intensity-based absolute quantification scores were normalized on a per protein level by conversion to a mean abundance fractional value across patient samples and rescaled to absolute values of 0 to 1 per row. These normalized, fractional abundance values were used for hierarchical clustering and heatmap generation using MatLab (2016b) (MathWorks, Natick, MA) and the function “clustergram.”
Pathways Analyses
The proteomic data (differentially regulated gene products with the Log2 fold change of SRNS to SSNS for both the pretreatment sample set and the post-treatment sample sets) were qualitatively assessed to provide approaches to functional annotation of data by submitting lists of identified proteins and expression patterns for pathways analysis using Ingenuity Pathways Analysis software (http://ingenuity.com). See Supplementary Methods.
Results
Patients and Demographics
Eighty-eight pediatric patients were enrolled over a 10-year period, although only approximately 50% of these patients (n = 45) were able to be enrolled before they received even a single dose of steroids. Of these, 37 patients with paired samples verified to include pretreatment samples and detailed clinical data were included in this study (Table 1). Twenty-four of these were clinically phenotyped as SSNS, because they achieved complete remission of proteinuria within an average of approximately 7 weeks of steroid therapy, whereas 13 patients did not achieve remission and were thus phenotyped as SRNS. Approximately equal numbers of children with SSNS (n = 7) and SRNS (n = 8) were used for the proteomics discovery studies, and only female patients were analyzed to compensate for potential gender variability. The validation cohort was expanded to include all available male and female patients. Children with SSNS were found to be different from those with SRNS in age, height, and weight, with SRNS patients presenting at a later age than those with SSNS (9.5 vs. 5.6 years; P = 0.006), consistent with known mean ages of presentation for these different forms of NS. To account for differences in pharmacodynamics of steroids in children with SSNS versus SRNS due to differences in weight, height, and body mass index, we calculated the average prescribed steroid dosage in the discovery cohort of these 2 groups. The differences in steroid dosage (SSNS, 1.81 ± 0.23 mg/kg per day vs. SRNS, 1.24 ± 0.14 mg/kg per day) were found to be not significantly different (nonparametric Mann-Whitney test; P > 0.05). Moreover, African American patients comprised a greater percentage of SRNS patients than SSNS patients (46% vs. 17%; P < 0.05).
Proteomic Profiling
Paired pre- and post-treatment plasma samples (n = 30 samples; SSNS, n = 7 pairs; SRNS n = 8 pairs) were depleted of high abundant proteins, achieving a 95% high and moderate abundance protein depletion. A total of 226 proteins were identified by high-resolution 1D-liquid chromatography–mass spectrometry in both pre- and post-treatment SSNS and SRNS samples. Of the 20 immunodepletion targets, 9 (IgG, Transferrin, IgA, IgM, α1-acid glycoprotein, IgD, ceruloplasmin, plasminogen, and prealbumin) were sufficiently depleted so as to not be observed within the LCMS results. Three (haptoglobin, complement C1q, and α1-antitrypsin) were observed at low intensity-based absolute quantification scores across less than 30% of samples. Eight (alpha-2 macroglobulin; albumin; apolipoproteins-A1, -AII, and -B/; complement 3; complement C4; and fibrinogen) were observed in a large fraction (≥75%) of samples, although none achieved statistical difference between sample groups.
Identification of Candidate Biomarkers Able to Predict and Mechanistically Define Steroid Resistance
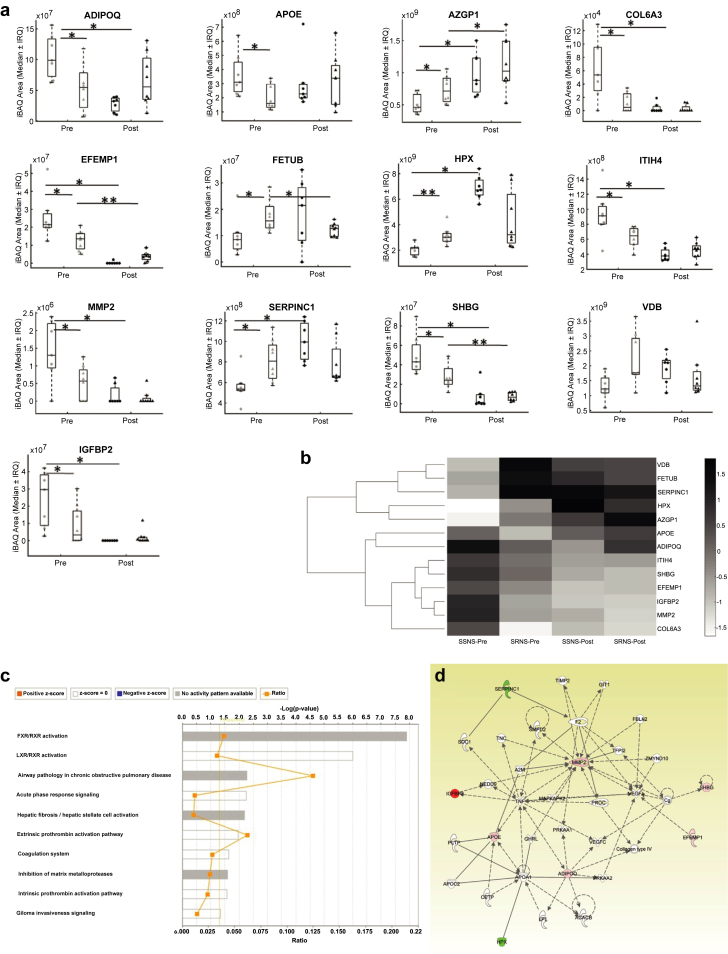
Protein lists were curated by requiring observation in at least 6 of 7 SSNS or 6 of 8 SRNS samples. This requirement resulted in 119 and 122 proteins, respectively, considered for subsequent statistical analysis. Following Wilcoxon and Mann-Whitney testing, 13 predictive (Table 2 and Figure 1b) and 66 mechanistic (Table 3 and Figure 1b) candidate biomarkers were retained for pathways analysis. The effects of steroid response on the relative abundance of the 13 predictive proteins is also illustrated in Figure 2a.20
Table 2.
Candidate biomarkers with potential to predict steroid resistance before therapy
| Name | Protein | Detection rate (SSNS) | Detection rate (SRNS) | P value (rank sum) | Area ratio (SSNS:SRNS) | Log2 (SSNS:SRNS) | Protein function |
|---|---|---|---|---|---|---|---|
| Collagen alpha-3(VI) chain | COL6A3 | 86 | 50 | 0.02 | 9.1 | 3.2 | Alpha chain of type VI Collagen that acts as a cell-binding protein and is implicated in muscle and connective tissue related diseases |
| Insulin-like growth factor-binding protein 2 | IGFBP2 | 100 | 63 | 0.04 | 9 | 3.2 | Inhibits IGF-mediated growth and developmental rates. IGF-binding proteins prolong the half-life of the IGFs and either inhibit or stimulate the growth promoting effects of the IGFs by altering the interaction of IGFs with their cell surface receptors |
| 72 kDa type IV collagenase | MMP2 | 86 | 63 | 0.03 | 2.3 | 1.2 | Ubiquitous metalloproteinase that is involved in diverse functions such as remodeling of the vasculature, angiogenesis, tissue repair, tumor invasion, inflammation, atherosclerotic plaque rupture, and degradation of extracellular matrix proteins |
| Apolipoprotein Ea | APOE | 100 | 100 | 0.04 | 1.9 | 0.9 | Mediates the binding, internalization, and catabolism of lipoprotein particles |
| Adiponectin | ADIPOQ | 100 | 100 | 0.01 | 1.8 | 0.8 | Important adipokine involved in the control of fat metabolism and insulin sensitivity, with direct antidiabetic, anti-atherogenic and anti-inflammatory activities |
| Sex hormone–binding globulin | SHBG | 100 | 100 | 0.03 | 1.8 | 0.8 | Functions as an androgen transport protein and regulates the plasma metabolic clearance rate of steroid hormones |
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 100 | 100 | 0.01 | 1.6 | 0.7 | Binds EGFR, induces EGFR autophosphorylation and activation of downstream signaling pathways |
| Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | 100 | 100 | 0.01 | 1.4 | 0.5 | Also known as type II APP; it is involved in inflammatory response to trauma |
| Hemopexin | HPX | 100 | 100 | 0 | 0.7 | −0.5 | Plasma protein with high affinity for heme and associates with HDL and influences its inflammatory properties |
| Vitamin D binding protein | VDB | 100 | 100 | 0.03 | 0.7 | −0.5 | Belongs to albumin gene family and major role is transport of various forms of Vitamin D metabolites; enhancement of the chemotactic activity of C5 alpha for neutrophils in inflammation and macrophage activation |
| Antithrombin-III | SERPINC1 | 100 | 100 | 0.01 | 0.7 | −0.5 | Serine protease inhibitor in plasma |
| Zinc-alpha-2-glycoprotein | AZGP1 | 100 | 100 | 0.03 | 0.6 | −0.7 | Stimulates lipid degradation in adipocytes |
| Fetuin-B | FETUB | 100 | 100 | 0.02 | 0.5 | −1 | Protease inhibitor |
APP, acute-phase protein; EGFR, epidermal growth factor receptor; HDL, high density lipoprotein; IGF, insulin-like growth factor; SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome.
Entry not observed in “mechanistic analysis.”
Table 3.
Candidate biomarkers with potential to identify mechanistic molecular pathways/targets of steroid resistance
| No. | Name | Gene name | Difference pretreatment |
SSNS post − prea |
SRNS post − prea |
Change with GC therapy |
Group trend | |||
|---|---|---|---|---|---|---|---|---|---|---|
| [SSNS/SRNS] | Median proteomic (iBAQ) signal area | P value signrank | Median proteomic (iBAQ) signal area | P value signrank | SSNS | SRNS | ||||
| 1 | Afamin (vitamin E binding protein) | AFM | 1.21E+08 | 0.016 | 1.80E+07 | 0.547 | ↑b | ns | Increase in both | |
| 2 | Angiotensinogen | AGT | 2.62E+08 | 0.031 | 1.45E+08 | 0.203 | ↑b | ns | ||
| 3 | Apolipoprotein D | APOD | 1.35E+07 | 0.813 | 1.07E+08 | 0.016 | ns | ↑b | ||
| 4 | Apolipoprotein L1 | APOL1 | 7.98E+06 | 0.016 | 4.26E+06 | 0.055 | ↑b | ns | ||
| 5 | Alpha-2-glycoprotein 1, zinc-binding | AZGP1 | 0.6 | 5.02E+08 | 0.031 | 3.63E+08 | 0.039 | ↑b | ↑b | |
| 6 | Carboxypeptidase B2 | CPB2 | 1.09E+07 | 0.016 | 2.40E+06 | 0.148 | ↑b | ns | ||
| 7 | Gelsolin | GSN | 8.40E+07 | 0.016 | 2.50E+06 | 0.945 | ↑b | ns | ||
| 8 | Hyaluronan-binding protein 2 | HABP2 | 1.21E+07 | 0.016 | 2.85E+06 | 0.461 | ↑b | ns | ||
| 9 | Hemopexin | HPX | 0.7 | 4.85E+09 | 0.016 | 1.05E+08 | 0.461 | ↑b | ns | |
| 10 | Insulin-like growth factor-binding protein complex acid labile subunit | IGFALS | 7.09E+07 | 0.016 | 3.95E+06 | 0.742 | ↑b | ns | ||
| 11 | Alpha-1-antichymotrypsin | SERPINA3 | 3.65E+08 | 0.031 | 1.23E+08 | 0.383 | ↑b | ns | ||
| 12 | Kallistatin | SERPINA4 | 4.54E+07 | 0.016 | 7.25E+06 | 0.25 | ↑b | ns | ||
| 13 | Plasma serine protease inhibitor | SERPINA5 | 4.60E+06 | 0.016 | 5.58E+05 | 0.844 | ↑b | ns | ||
| 14 | Alpha-2-macroglobulin | A2M | −3.20E+09 | 0.016 | −4.84E+08 | 0.383 | ↓b | ns | Decrease in both | |
| 15 | Alpha-1 microglycoprotein (bikunin) | AMBP | −1.00E+09 | 0.016 | −3.25E+08 | 0.039 | ↓b | ↓b | ||
| 16 | Apolipoprotein M | APOM | −2.25E+07 | 0.016 | −1.50E+06 | 0.641 | ↓b | ns | ||
| 17 | Attractin | ATRN | −3.18E+07 | 0.016 | −1.94E+07 | 0.109 | ↓b | ns | ||
| 18 | Cholinesterase (Butyrylcholine esterase) | BCHE | −1.94E+07 | 0.016 | −8.72E+06 | 0.063 | ↓b | ns | ||
| 19 | Complement C1r subcomponent | C1R | −1.11E+07 | 0.219 | −5.45E+06 | 0.008 | ns | ↓b | ||
| 20 | C4b-binding protein alpha chain | C4BPA | −1.80E+08 | 0.016 | −2.77E+07 | 0.383 | ↓b | ns | ||
| 21 | Monocyte differentiation antigen CD14 | CD14 | −2.90E+06 | 0.813 | −1.18E+07 | 0.039 | ns | ↓b | ||
| 22 | Complement Factor H | CFH | −3.90E+08 | 0.031 | −2.20E+07 | 0.641 | ↓b | ns | ||
| 23 | Clusterin | CLU | −5.50E+07 | 0.047 | −6.80E+07 | 0.195 | ↓b | ns | ||
| 24 | Collagen alpha-3(VI) | COL6A3 | 9.1 | −4.14E+04 | 0.031 | −3.63E+03 | 0.188 | ↓b | ns | |
| 25 | Carboxypeptidase N catalytic chain | CBPN1 | −1.00E+07 | 0.016 | −6.39E+06 | 0.313 | ↓b | ns | ||
| 26 | Carboxypeptidase N subunit 2 | CPN2 | −6.16E+07 | 0.016 | −9.10E+07 | 0.016 | ↓b | ↓b | ||
| 27 | Fibulin-2 (EGF-containing fibulin-like extracellular matrix protein 1) | EFEMP1 | 1.6 | −2.14E+07 | 0.016 | −8.66E+06 | 0.008 | ↓b | ↓b | |
| 28 | Fibulin-1 | FBLN1 | −3.73E+07 | 0.016 | −2.41E+07 | 0.039 | ↓b | ↓b | ||
| 29 | Fibrinogen alpha chain | FGA | −4.39E+06 | 0.047 | −4.69E+06 | 0.461 | ↓b | ns | ||
| 30 | Insulin-like growth factor-binding protein 2 | IGFBP2 | 9 | −2.96E+07 | 0.016 | −1.71E+06 | 0.094 | ↓b | ns | |
| 31 | Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | −4.56E+08 | 0.047 | −3.70E+07 | 0.461 | ↓b | ns | ||
| 32 | Inter-alpha-trypsin inhibitor heavy chain H3 | ITIH3 | −4.17E+07 | 0.016 | −8.63E+06 | 0.078 | ↓b | ns | ||
| 33 | Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | 1.4 | −4.80E+08 | 0.016 | −2.27E+08 | 0.055 | ↓b | ns | |
| 34 | Phosphatidylcholine-sterol acyltransferase | LCAT | −1.52E+07 | 0.016 | −8.65E+06 | 0.148 | ↓b | ns | ||
| 35 | Galectin-3-binding protein | LGALS3BP | −3.73E+07 | 0.047 | −3.84E+07 | 0.008 | ↓b | ↓b | ||
| 36 | Lumican | LUM | −1.87E+08 | 0.016 | −9.11E+07 | 0.008 | ↓b | ↓b | ||
| 37 | 72 kDa type IV collagenase | MMP2 | 2.3 | −1.30E+06 | 0.031 | −5.69E+05 | 0.063 | ↓b | ns | |
| 38 | Prostaglandin-H2 D-isomerase | PTGDS | −1.54E+07 | 0.031 | −7.10E+06 | 0.156 | ↓b | ns | ||
| 39 | Sulfhydryl oxidase 1 | QSOX1 | −4.66E+05 | 0.297 | −6.43E+05 | 0.039 | ns | ↓b | ||
| 40 | Heparin cofactor 2 | SERPIND1 | −3.60E+07 | 0.078 | −7.90E+07 | 0.039 | ns | ↓b | ||
| 41 | Plasma protease C1 inhibitor | SERPING1 | −4.37E+08 | 0.016 | −5.66E+08 | 0.016 | ↓b | ↓b | ||
| 42 | Sex hormone-binding globulin | SHBG | 1.8 | −3.82E+08 | 0.016 | −1.68E+07 | 0.008 | ↓b | ↓b | |
| 43 | Alpha-2-HS-glycoprotein | AHSG | 4.30E+08 | 0.297 | −1.06E+09 | 0.023 | ns | ↓b | Increase in SSNS Decrease in SRNS | |
| 44 | Complement Factor I | CFI | 1.99E+07 | 0.016 | −1.75E+07 | 0.313 | ↑b | ns | ||
| 45 | Tetranectin | CLEC3B | 4.08E+07 | 0.016 | −2.39E+07 | 0.25 | ↑b | ns | ||
| 46 | Coagulation factor XII | F12 | 5.75E+07 | 0.016 | −2.40E+06 | 0.945 | ↑b | ns | ||
| 47 | Prothrombin | F2 | 3.60E+08 | 0.047 | −2.65E+07 | 0.742 | ↑b | ns | ||
| 48 | Fetuin B | FETUB | 0.5 | 8.27E+06 | 0.109 | −3.32E+06 | 0.039 | ns | ↓b | |
| 49 | Vitamin D-binding protein | VDB | 0.7 | 4.70E+08 | 0.031 | −4.10E+08 | 0.055 | ↑b | ns | |
| 50 | Insulin-like growth factor-binding protein 3 | IGFBP3 | 3.50E+06 | 0.047 | −4.75E+05 | 1 | ↑b | ns | ||
| 51 | Corticosteroid binding globulin | SERPINA6 | 1.05E+08 | 0.016 | −1.69E+07 | 0.461 | ↑b | ns | ||
| 52 | Thyroxine-binding globulin | SERPINA7 | 5.20E+06 | 0.734 | −1.05E+07 | 0.008 | ns | ↓b | ||
| 53 | Antithrombin-III | SERPINC1 | 0.7 | 3.85E+08 | 0.016 | −4.75E+07 | 0.641 | ↑b | ns | |
| 54 | Pigment epithelium-derived factor | SERPINF1 | 4.96E+07 | 0.031 | −9.90E+06 | 0.945 | ↑b | ns | ||
| 55 | Alpha-2-antiplasmin | SERPINF2 | 1.50E+08 | 0.031 | −1.09E+08 | 0.188 | ↑b | ns | ||
| 56 | Vitronectin | VTN | 1.85E+08 | 0.156 | −1.53E+08 | 0.016 | ns | ↓b | ||
| 57 | Adiponectin | ADIPOQ | 1.8 | −8.86E+07 | 0.016 | 1.99E+07 | 0.383 | ↓b | ns | Decrease in SSNS Increase in SRNS |
| 58 | Apolipoprotein A1 | APOA1 | −9.00E+08 | 0.375 | 2.04E+09 | 0.047 | ns | ↑b | ||
| 59 | Apolipoprotein B | APOB | −2.50E+07 | 0.016 | 4.21E+06 | 0.844 | ↓b | ns | ||
| 60 | Apolipoproten C1 | APOC1 | −5.57E+08 | 0.031 | 1.17E+08 | 0.133 | ↓b | ns | ||
| 61 | Apolipoprotein C2 | APOC2 | −3.62E+08 | 0.016 | 6.50E+06 | 0.844 | ↓b | ns | ||
| 62 | Complement factor H-related protein 1 | CFHR1 | −9.10E+07 | 0.016 | 2.35E+07 | 1 | ↓b | ns | ||
| 63 | Properdin | CFP | −1.36E+07 | 0.016 | 6.01E+06 | 0.195 | ↓b | ns | ||
| 64 | Hepatocyte growth factor activator | HGFAC | −9.69E+06 | 0.031 | 0.00E+00 | 0.25 | ↓b | ns | ||
| 65 | Perlecan | HSPG2 | −1.49E+05 | 0.031 | 0.00E+00 | 0.5 | ↓b | ns | ||
| 66 | Vasorin | VASN | −2.77E+06 | 0.031 | 4.55E+05 | 0.844 | ↓b | ns | ||
EGF, epidermal growth factor; GC, glucocorticoid; iBAQ, intensity-based absolute quantification; ns, not significant; SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome.
Negative (−) values indicate a decrease in relative plasma abundance. Positive (+) values indicate an increase in relative plasma abundance.
Bolded proteins are found in Table 2.
Italicized proteins known to be responsive to GCs.
Values represent difference in median values of post-treatment sample − pretreatment sample.
P < 0.05.
Figure 2.
Candidate biomarkers able to predict steroid resistance and their informatics analysis to examine emergent properties. (a) Median intensity-based absolute quantification (iBAQ) areas (middle hash), interquartile range (IQR); boxed area and whisker for maximum and minimum values for candidate biomarkers able to predict steroid resistance were plotted for pre- and post-treatment samples for children with steroid-sensitive nephrotic syndrome (SSNS) and steroid-resistant nephrotic syndrome (SRNS) (SSNS Pre, light circle; SSNS Post, dark circle; SRNS Pre, light triangle; SRNS Post, dark triangle). All the pretreatment samples were significantly different between the SSNS versus SRNS groups (Table 2). Post-treatment time point comparator is added for illustration purposes (Table 3). *P < 0.05; **P < 0.01. (b) Candidate proteins (n = 13) significantly differentiating pre-steroid exposure patient samples were analyzed by hierarchical clustering. Protein abundance (iBAQ scores) were normalized and scaled by the clustergram function in MatLab (MathWorks, Natick, MA). Values are expressed as a fractional value around the median. Gene names and fold-changes (SSNS to SRNS) for significantly regulated pretreatment plasma proteins were submitted for (c) canonical molecular pathways analysis and (d) network analysis by Ingenuity Pathways Analysis (IPA) to consider implications of abundance difference trends within the proteomic dataset. (c) The top 10 canonical molecular pathways illustrated show significant enrichment, including 2 highly enriched pathways (Farnesoid X receptor FXR/retinoid X receptor [RXR] activation and liver X receptor [LXR]/RXR activation). Ratio data demonstrate the fraction of the submitted gene names to the gene names contained within the canonical pathways. (d) The top canonical network included 2 downregulated (SSNS < SRNS) and 6 upregulated (SSNS > SRNS) proteins, of which 3 upregulated proteins (matrix metalloproteinase 2 [MMP-2], APOE, and adiponectin [ADIPOQ]) occupied network node space. Tumor necrosis factor (TNF) is a central node within this network and inference based on its known regulation of ADIPOQ, APOE, IFGBP2, and MMP-2 expression (Activation Z-score 0.152; overlap P < 0.0001). ADIPOQ, adiponectin; HPX, hemopexin; SHBG, sex hormone–binding globulin; VDB, vitamin D binding protein. (c,d) Copyright © 2000–2017 Qiagen. The authors acknowledge that the networks and functional analyses were generated through the use of IPA (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/).20
Plasma Protein Abundance as a Function of Steroid Resistance
A hierarchical clustering method with heatmap visualization was used to guide analysis of the predictor candidate biomarkers (Figure 2b). The visualization of individual patients on heatmap is shown in Supplementary Figure S2 and analysis of the entire proteomic dataset is also included in Supplementary Figures S3 and S4 as supplemental results.
Pathway Mapping and Interaction Networking of Candidate Biomarkers
Ingenuity Pathway Analysis of predictive proteins (see Table 2) suggested 10 canonical pathways (P < 0.01) and 1 significant protein-protein network (Figure 2c and d). The top 6 canonical pathways were as follows: (i) Farnesoid X receptor/retinoid X receptor activation, (ii) liver X receptor/retinoid X receptor activation, (iii) airway pathology in chronic obstructive pulmonary disease, (iv) acute-phase response signaling, (v) hepatic fibrosis/hepatic stellate cell activation, and (vi) extrinsic prothrombin activation pathway. The principal network for the predictive biomarkers was built using 8 of 13 submitted proteins, including ADIPOQ, APOE, EFEMP1, HPX, IGFBP2, MMP-2, antithrombin-III (SERPINC1), and SHBG. As shown in Figure 2d, 5 proteins (SHBG, EFEMP1, HPX, IGFBP2, and antithrombin-III [SERPINC1]) were positioned at network edges, whereas MMP-2, ADIPOQ, APOE, and VEGF-A were positioned as network nodes, with MMP-2 and APOE nodes under the indirect regulation (dashed lines) of the cytokine tumor necrosis factor.
The top 5 canonical pathways identified by Ingenuity Pathway Analysis of defining/mechanistic candidate marker gene names were (i) liver X receptor/retinoid X receptor activation, (ii) Farnesoid X receptor/retinoid X receptor activation, (iii) acute-phase response signaling, (iv) coagulation system, and (v) atherosclerosis signaling. The top 3 networks assembled by Ingenuity Pathway Analysis overlapped and had common disease/functional attributions that included metabolic disease, hematological system development and function, inflammatory response, cell death, and survival. Network 1, but not networks 2 and 3, contained candidate biomarkers as nodal components (Supplementary Figure S5; MMP-2, IGFBP3, ADIPOQ, and F2).
Biomarker Validation
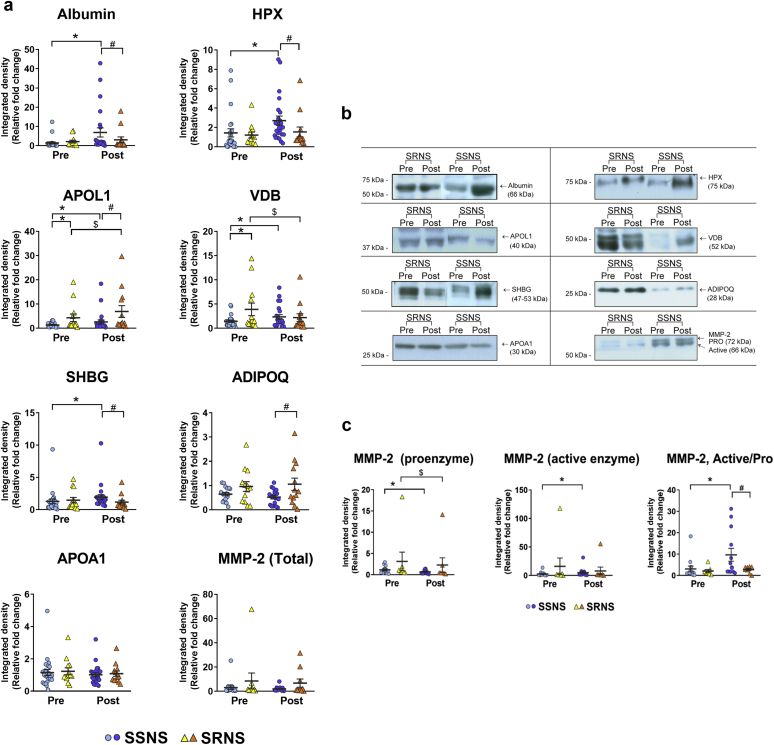
Thirty-seven patients (n = 74 samples) comprising 24 SSNS and 13 SRNS patients were analyzed by immunoblotting with specific antibodies for the validation of several of the predictive and mechanistic candidate biomarkers discovered previously in the proteomics analyses (Figure 3). Albumin levels were analyzed as a reference protein and, as expected, it confirmed a significant difference between SSNS and SRNS post-treatment samples (Figure 3a and b). HPX was significantly increased in the SSNS post-treatment group, but not in the SRNS group (Figure 3a and b), thus corroborating the proteomic results (Table 3 and Figure 2). APOL1 was increased in both the SSNS and SRNS groups post-treatment, while maintaining the differences in its levels both pre- and post-treatment between the 2 groups (Figure 3a and b). Although it corroborated some of the proteomics results (significant increase in SSNS and moderate increase in SRNS, Table 3), its confirmation also underscored its relevance as a predictive biomarker, as well as a biomarker of disease remission (difference between SSNS and SRNS both pre- and post-treatment, Figure 3). VDB was a very strong predictive marker of steroid resistance on validation, and it followed the same pattern between pre- and post-treatment as observed in the proteomics results (Table 3, Figure 2, and Figure 3a and b). SHBG, apolipoprotein A1 (APOA1), and ADIPOQ did not show differences in pretreatment between SSNS and SRNS, although both SHBG and ADIPOQ levels were altered differently in SSNS and SRNS. MMP-2 showed 2 bands, representing the active (lower band, 64 kDa) and proenzyme (upper band, 72 kDa) forms. The relative active form of MMP-2 was significantly increased in SSNS post-treatment, and thus appears to be a potential marker to differentiate steroid sensitivity from resistance (Figure 3a–c).
Figure 3.
Biomarker validation studies of selected candidate biomarkers to predict or define steroid resistance in childhood nephrotic syndrome. (a) Validation graphs and (b) representative blots are shown from the analyses of 37 patients (n = 74 samples) comprising 24 steroid-sensitive nephrotic syndrome (SSNS) and 13 steroid-resistant nephrotic syndrome (SRNS) patients by immunoblotting with specified antibodies for the validation of selected predictive and defining biomarkers outlined in Tables 2 and 3. A control sample was run on every gel, and test patient samples were normalized to control by densitometry. (c) Western blot semiquantitative comparisons of the candidate biomarker matrix metalloproteinase 2 (MMP-2). MMP-2 immunoblotting of 54 patient samples (16 SSNS and 10 SRNS patients) showed 2 bands, representing the active (lower band, 64 kDa) and proenzyme (upper band, 72 kDa) forms of the enzyme. These were individually semiquantitated by densitometry and the active versus proenzyme ratios measured. Statistical significance was determined by unpaired or paired t tests using the GraphPad Prism software version 6.00 (LaJolla, CA) for Windows. P values were considered significant at P < 0.05 (*P < 0.05 vs. SSNS pretreatment; #P < 0.05 vs. SSNS post-treatment; $P < 0.05 vs. SRNS pretreatment). ADIPOQ, adiponectin; HPX, hemopexin; SHBG, sex hormone–binding globulin; VDB, vitamin D binding protein.
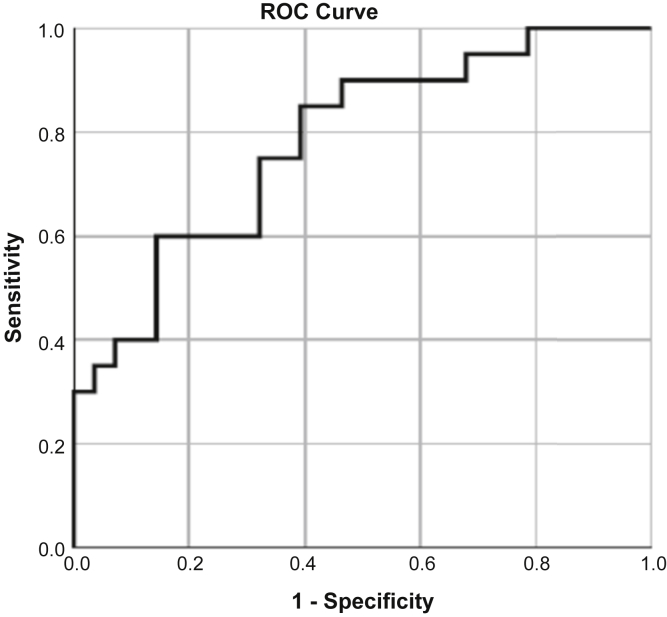
The immunoblot data for the variables sex (male/female), SHBG, VDB, HPX, APOAI, ApoL1, albumin, ADIPOQm and MMP2 were analyzed by logistic regression using a backward stepwise regression. The minimal features set remaining in the SSNS/SRNS classification model included ADIPOQ, VDB, and MMP2, but not sex. The ability of the immunoblot densitometry data for these 3 plasma proteins to classify patient samples was significant (P = 0.003), with a net improvement by –2log likelihood = 51.591 (Cox & Snell R2 value = 0.247; Nagelkerke R2 value = 0.332). Last, to evaluate the sensitivity and specificity for these proteins identified by logistic regression, a receiver operator characteristic curve was constructed (Figure 4) and the area under the curve was calculated to be 0.78.
Figure 4.
Receiver operating characteristic (ROC) curve. Logistic regression analysis of confirmatory immunoblot studies identified vitamin D binding protein (VDB), adiponectin (ADIPOQ), and matrix metalloproteinase 2 (MMP-2) as a minimal, significant set of plasma proteins predicting steroid response. An ROC analysis for these 3 proteins to classify steroid response in patients with NS (n = 37 paired samples) returned an area under the curve of 0.78.
Proteomic Landscape With Different Approaches
Additional analyses approaches outlined in Supplementary Table S1 were applied to the proteins identified in the SSNS and SRNS pre- and post-treatment groups to develop candidate biomarker protein lists (Supplementary Table S2).
Conclusion
NS is a kidney disease that affects both children and adults, and 20% to 50% of patients present with or subsequently develop clinical steroid resistance, which is associated with greatly increased risks for treatment side effects and disease progression. The current study tested the hypothesis that paired plasma proteomic sample analyses could identify candidate biomarkers able to either predict steroid resistance before initial treatment or mechanistically define critical molecular pathways/targets that regulate clinical steroid resistance. We used a pilot proteomic discovery approach in 30 paired plasma samples obtained before and after initial steroid treatment and identified a panel of 13 candidate protein biomarkers predictive of steroid resistance and several candidate biomarkers that mechanistically define specific molecular pathways/targets of clinical steroid resistance in NS. Candidate biomarkers found to predict steroid resistance included VDB, HPX, Fetuin-B (FETUB), and ADIPOQ (Table 4). Candidate protein biomarkers found to mechanistically define specific molecular pathways/targets associated with steroid resistance included VDB, HPX, SHBG, antithrombin-III (SERPINC1), Fetuin-B (FETUB), ADIPOQ, MMP-2, and APOA1. Subsequent confirmatory studies in 74 patient samples of several of these candidate biomarkers by immunoblotting confirmed a few auspicious biomarkers with high potential to be able to either predict steroid resistance or to mechanistically define molecular pathways that regulate steroid resistance in childhood NS (Table 4).
Table 4.
Candidate protein biomarkers to predict or define molecular pathways/targets of steroid resistance in pediatric nephrotic syndrome
| Protein | Name | Predictive biomarkera |
Defining biomarkerb |
||
|---|---|---|---|---|---|
| Proteomics | IBc | Proteomics | IBc | ||
| COL6A3 | Collagen alpha-3(VI) chain | X | X | ||
| IGFBP2 | Insulin-like growth factor-binding protein 2 | X | X | ||
| MMP2 | 72 kDa type IV collagenase | X | X | X | |
| APOE | Apolipoprotein E | X | |||
| ADIPOQ | Adiponectin | X | X | X | |
| SHBG | Sex hormone–binding globulin | X | X | X | |
| EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | X | X | ||
| ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 | X | X | ||
| HPX | Hemopexin | X | X | X | |
| VDB | Vitamin D binding protein | X | X | X | X |
| SERPINC1 | Antithrombin-III | X | X | ||
| AZGP1 | Zinc-alpha-2-glycoprotein | X | X | ||
| FETUB | Fetuin-B | X | X | ||
| APOL1 | Apolipoprotein L1 | X | X | X | |
Approaches defined in Figure 1.
Immunoblotting.
Proteome profiling of urine, and in some cases serum or plasma, has been analyzed over the past decade in patients with NS using various approaches.17,21, 22, 23, 24, 25, 26 More recently, proteomic profiling of patients with NS based on their histology and clinical phenotype has yielded a panel of proteins able to distinguish between these different groups.16,17 These studies had important differences from the current study. First, both of these studies analyzed urine samples. Second, neither of these studies included analyses of pretreatment samples that could enable identification of truly predictive biomarkers before initiation of any therapy. Third, the NS classification of Choi et al.17 was based on histology (minimal change disease, focal segmental glomerulosclerosis, membranous nephropathy) and included only adult patients, whereas the classification of Bennett et al.16 was similarly based on SRNS versus SSNS in children. From the discovery set, a few proteins, such as VDB, antithrombin-III (SERPINC1), and HPX, were common between our predictive markers of SRNS versus SSNS, and the markers of Choi et al.,17 which were able to distinguish among minimal change disease, focal segmental glomerulosclerosis, and membranous nephropathy cases. However, none of these markers were validated in the study by Choi et al.17 In the Bennett et al.16 study (which had more commonalties in study design), of their 13 identified proteins, VDB, HPX, APOA1, TBG (thyroxine-binding globulin or SERPINA7), a closely related protein to Fetuin-B (Fetuin-A) and zinc-alpha 2 glycoprotein (AZGP1) were commonly identified in our discovery sets of either predictive or mechanistic biomarkers (Tables 2 and 3). Of these, VDB, HPX, APOA1, Fetuin-B, and AZGP1 were also identified in our predictive set, of which VDB was validated in both studies. The current study extends these previous findings by combining the use of a state-of-the-art proteomics approach with analyses of paired plasma samples from highly phenotyped children presenting with NS in whom the pretreatment samples were fully verified to represent a “disease-only” state before the administration of even a single dose of steroids. Such careful phenotyping of samples is critical, as we have previously found that even a 30-minute exposure of podocytes to steroids can significantly alter their proteomic profile.27 Our use of paired samples obtained both pretreatment and post-treatment, when patients had been clinically declared to have either SSNS or SRNS, enabled us to search for potential biomarkers able to both predict subsequent clinical steroid resistance, as well as to search for molecular pathways and targets associated with steroid resistance versus steroid responsiveness.
HPX is a plasma protein with high affinity for heme. A variety of biological activities have been attributed to hemopexin, including both pro- and anti-inflammatory activities.28 Regarding NS, hemopexin has been shown to induce nephrin-dependent reorganization of the podocyte actin cytoskeleton,29 and to distinguish SRNS via a urine proteomics approach, although it was not identified as a candidate on validation by enzyme-linked immunosorbent assay (ELISA).16 Our studies showed that HPX could discriminate patients with SSNS versus SRNS pretreatment, and that steroid treatment significantly increased plasma HPX levels in SSNS (but not SRNS) patients.
APOA1 is an important component of HDL and it has been shown to be present in more heterogeneous forms in the plasma of patients with NS.30 Dyslipidemia is also a prominent feature of NS, and our study identified APOL1 and other apolipoproteins that could distinguish children with SSNS versus SRNS, both before and after steroid treatment.7,8,31, 32, 33, 34, 35
Adiponectin (ADIPOQ) levels have also been shown to be increased in patients with NS.36 Our studies found that although adiponectin levels started lower and decreased with steroid treatment in children with SSNS, adiponectin levels started higher and increased further with steroid treatment in those with SRNS.
A few studies have implicated matrix metalloproteinases in the pathogenesis of NS.37,38 We found a trend toward higher MMP-2 levels in children with SRNS versus SSNS, both before and following steroid treatment. MMP-2 comprises both an active and a proenzyme form, and our studies underscored a potential role for the relative ratios of these forms, rather than absolute levels, in distinguishing SSNS versus SRNS.39 Molecular weight forms consistent with higher active/proenzyme ratios in SSNS versus SRNS post-treatment, suggests that increases in this ratio may play a beneficial role in the clinical response to steroids.
Nephrotic rats exhibit urinary loss of SHBG-bound testosterone, and steroid-binding proteins such as SHBG act as gatekeepers of steroid hormone action in the plasma.40,41 Our studies showed that SHBG levels increased with steroid treatment in SSNS but decreased in SRNS. These findings may simply reflect reduced urinary losses of SHBG in children with SSNS as they enter remission. However, given the reported role of SHBG in regulating steroid action, these findings may also identify a novel potential opportunity to enhance steroid responsiveness by supplementing or pharmacologically enhancing its production during steroid treatment.
VDB has recently emerged as a urinary marker of steroid resistance in NS, whereas vitamin D has also been reported to have a role in the protection of podocytes against NS-related injury.16,42,43 Our studies in addition identified a 3-protein predictive candidate biomarker panel (VDB, ADIPOQ, and MMP-2) with a significant ability to differentiate at disease presentation patients who will develop SRNS from those who will have SSNS (P = 0.003; area under the receiver operating characteristic curve = 0.78; Table 4).
This study had several limitations and strengths. Unlike some previous studies in which ELISA was used for confirmation of identified biomarkers, we used an immunoblotting confirmatory strategy. Although more labor intensive, this strategy enabled significantly improved specificity for the detection of proteins of interest. Many antibodies used in ELISAs and immunoblotting bind nonspecifically to other proteins, as we also observed in our blots. Immunoblotting enabled us to successfully circumvent this potential lack of specificity by performing quantitative densitometric analyses of bands, and identify only the proteins of interest. This was specifically relevant for VDB, which belongs to the same gene family and shares significant homology with albumin, and thus could confound confirmatory results using ELISA.44 This approach also allowed us to evaluate the active versus proenzyme fractions of MMP-2, which is a better marker of its activity than total MMP-2 levels.45 In addition, immunoblotting enabled the use of small sample volumes compared with ELISAs. To further enhance accuracy, we also used the same control sample in every gel and blot, enabling us to directly normalize all patient samples tested. A larger cohort will be needed in the future to validate the other biomarkers identified in the discovery analysis, as we were not able to validate them all in the present study. Our validation studies were targeted just toward the most relevant/significant markers known in NS or differentially expressed markers, and were also limited by commercial reagent availability (specific antibodies). The predictive and defining power of the identified proteins is likely to benefit from expanding the validation list in future studies.
Although most similar biomarker studies have been performed using urine, our studies used plasma samples. It would thus be of interest to attempt to further validate our identified biomarkers in urine samples from a larger and separate cohort in the future, and such studies are under way. However, identification of biomarkers in plasma is highly relevant, because it directly reflects the true concentrations of these proteins in blood, without perturbation by renal tubular secretion or reabsorption. Furthermore, SRNS typically presents clinically at older ages with higher average weight than children with SSNS. Because this is a typical generalized difference between the 2 groups, accounting for these confounders was not found to be essential and clinically relevant. Last, we are aware that some of the children with SRNS may have had an underlying genetic cause of disease. However, because these samples were collected over a decade, there were no provisions to screen them for monogenetic causes at that time. Despite this limitation, this study emphasizes differences between children with SRNS and SSNS that are clearly detectable at the time of clinical presentation, regardless of underlying genetic causes, and that would likely be more rapidly available and thus more clinically useful to treating physicians than genetic studies.
A major strength of this study was the evaluation of paired samples from each patient, both before any steroid treatment and following an average of approximately 7 weeks of daily oral steroids. This enabled us to use multiple approaches to analyze the dataset, including identification of potential biomarkers predictive of the subsequent clinical response to steroids, as well as to identify specific mechanistic molecular pathways and/or targets associated with both steroid response and steroid resistance. Because the discovery studies were limited to an all-female subgroup, we expanded our validation cohort to include male and more female patients and using an orthogonal (antibody-based) method, as has been done previously in similar studies.16 Logistic regression analysis for our candidate predictor biomarker classification of male and female patient samples did not identify sex as a significant classification variable. These logistic regression findings suggest additional studies on the mechanistic candidate biomarkers are justified. Notably, this same group of patient samples has also undergone plasma metabolomics analyses,46 and future studies will benefit from identification of additional relevant molecular pathways and biomarkers of SRNS using approaches that integrate the proteomics and metabolomics datasets.
In summary, the current studies used paired plasma samples from children with SSNS and SRNS obtained both before and after approximately 7 weeks of daily oral steroids to identify several candidate proteomic biomarkers with the potential to predict subsequent treatment response, and to define specific molecular pathways or targets associated with both steroid sensitivity and steroid resistance. In addition to NS, steroids are also a primary therapy for many other diseases, such as asthma, rheumatoid arthritis, autoimmune hepatitis, and ulcerative colitis. Interestingly, approximately 10% to 30% of these patients also present with or develop steroid resistance during their disease course, leading to increased risks for both drug toxicity and disease progression. Thus, further validation of these results could greatly improve our ability to predict the risk of clinical steroid resistance at disease onset for a large and diverse group of patients. These findings also could improve our understanding of the molecular mechanisms that regulate the clinical response to GCs and help identify potential future molecular targets to improve the treatment of NS as well as other conditions treated with steroids.
Appendix
Members of The Midwest Pediatric Nephrology Consortium
Drs. John Mahan, Hiren Patel, and Richard F. Ransom (NCH, Columbus, OH, USA); Cynthia Pan (Medical College of Wisconsin, Milwaukee, WI, USA); Denis F. Geary (The Hospital for Sick Children, Toronto, ON, Canada); Myra L. Chang (West Virginia University, Charleston, WV, USA); Keisha L. Gibson (University of North Carolina, Chapel Hill, NC, USA); Franca M. Iorember (Louisiana State University, New Orleans, LA, USA); Patrick D. Brophy (Children’s Hospital, University of Iowa, Iowa City, IA, USA); Tarak Srivastava (Children’s Mercy Hospital, Kansas City, MO, USA); and Larry A. Greenbaum (Emory University School of Medicine, Atlanta, GA, USA).
Disclosure
All the authors declared no competing interests. An international application under the patent cooperation treaty was published on May 24, 2018 (publication no. WO 2018/094021) by WES, SA, MLM, and JBK.
Acknowledgments
We thank the Midwest Nephrology Consortium, its participating centers, physicians, study and nurse coordinators for their contributions toward the collection of the plasma samples used in this study. These include Drs. John Mahan, Hiren Patel, and Richard F. Ransom (NCH, Columbus, OH), Cynthia Pan (Medical College of Wisconsin, Milwaukee, WI), Denis F. Geary (The Hospital for Sick Children, Toronto, ON, Canada), Myra L. Chang (West Virginia University, Charleston, WV), Keisha L. Gibson (University of North Carolina, Chapel Hill, NC), Franca M. Iorember (Louisiana State University, New Orleans, LA), Patrick D. Brophy (Children’s Hospital, University of Iowa, Iowa City, IA), Tarak Srivastava (Children’s Mercy Hospital, Kansas City, MO) and Larry A. Greenbaum (Emory University School of Medicine, Atlanta, GA). We also thank the Biopathology Core at NCH for storing and maintaining the sample biorepository.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases–National Institutes of Health (DK110077 to JBK, WES; DK091584 to DWW, JBK, MLM), National Institute of General Medical Sciences (P20GM113226 to MLM), internal funds from NCH (to WES and to SA); and clinical research funding from the University of Louisville Kidney Disease Program (to JBK, MLM, MEB, and AEG).
Author Contributions
SA and MLM conceptualized and designed the studies, performed experiments, analyzed and interpreted the data, prepared the figures and tables, and drafted and edited the manuscript. JK, MAC, ML, DWW, AEG, and MEB performed experiments, analyzed and interpreted the data, and prepared the figures and tables. JRG and SS interpreted the data and edited the manuscript. WES and JBK conceptualized and designed the study, analyzed and interpreted the data, and edited the manuscript. All the authors approved of the final version of the manuscript.
Data Sharing
Data files for acquired liquid chromatography–mass spectrometry data (.RAW), search engine files (.mgf), peak list files (.mzML) files, and search results aggregated into a Scaffold3 (.sf3, ProteomeSoftware.com) have been deposited with MassIVE (http://massive.ucsd.edu/) data repository with the Center for Computational Mass Spectrometry at the University of California, San Diego (MSV000082114) and shared with the ProteomeXchange (PXD009058; www.proteomexchange.org).
Footnotes
Supplementary Materials and Methods.
Figure S1. Proteomics workflow for candidate biomarker selection.
Figure S2. Heat map with hierarchical clustering portraying individual patients.
Figure S3. Hierarchical clustering method with heatmap visualization of the entire proteomic data set.
Figure S4. Protein-protein interaction networks derived from unique 2-member HC heatmap cluster.
Figure S5. Ingenuity Pathway Analysis–identified molecular networks derived from defining candidate biomarkers.
Table S1. Various approaches used to identify additional proteins/molecular pathways that distinguish SSNS versus SRNS and pre- versus posttreatment samples.
Table S2. Proteins identified using various analyses (A–F from Supplementary Table S1) that distinguish SSNS versus SRNS and pre- versus posttreatment samples.
Contributor Information
Shipra Agrawal, Email: Shipra.agrawal@nationwidechildrens.org.
Michael L. Merchant, Email: michael.merchant@louisville.edu.
Jon B. Klein, Email: jon.klein@louisville.edu.
William E. Smoyer, Email: William.smoyer@nationwidechildrens.org.
The Midwest Pediatric Nephrology Consortium:
John Mahan, Hiren Patel, Richard F. Ransom, Cynthia Pan, Denis F. Geary, Myra L. Chang, Keisha L. Gibson, Franca M. Iorember, Patrick D. Brophy, Tarak Srivastava, and Larry A. Greenbaum
Supplementary Material
References
- 1.Nourbakhsh N., Mak R.H. Steroid-resistant nephrotic syndrome:past and current perspectives. Pediatr Health Med Ther. 2017;8:29–37. doi: 10.2147/PHMT.S100803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canetta P.A., Radhakrishnan J. The evidence-based approach to adult-onset idiopathic nephrotic syndrome. Front Pediatr. 2015;3:78. doi: 10.3389/fped.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck L.H., Jr., Bonegio R.G., Lambeau G. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med. 2009;361:11–21. doi: 10.1056/NEJMoa0810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomas N.M., Beck L.H., Jr., Meyer-Schwesinger C. Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med. 2014;371:2277–2287. doi: 10.1056/NEJMoa1409354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheelock K.M., Cai J., Looker H.C. Plasma bradykinin and early diabetic nephropathy lesions in type 1 diabetes mellitus. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keshishian H., Burgess M.W., Gillette M.A. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol Cell Proteomics. 2015;14:2375–2393. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H., Hoek M., Yi P. Rapid detection and quantification of apolipoprotein L1 genetic variants and total levels in plasma by ultra-performance liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2013;27:2639–2647. doi: 10.1002/rcm.6734. [DOI] [PubMed] [Google Scholar]
- 8.Kozlitina J., Zhou H., Brown P.N. Plasma levels of risk-variant APOL1 do not associate with renal disease in a population-based cohort. J Am Soc Nephrol. 2016;27:3204–3219. doi: 10.1681/ASN.2015101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birmingham D.J., Merchant M., Waikar S.S. Biomarkers of lupus nephritis histology and flare:deciphering the relevant amidst the noise. Nephrol Dial Transplant. 2017;32:i71–i79. doi: 10.1093/ndt/gfw300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rovin B.H., Klein J.B. Proteomics and autoimmune kidney disease. Clin Immunol. 2015;161:23–30. doi: 10.1016/j.clim.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caster D.J., Korte E.A., Merchant M.L. Autoantibodies targeting glomerular annexin A2 identify patients with proliferative lupus nephritis. Proteomics Clin Appl. 2015;9:1012–1020. doi: 10.1002/prca.201400175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fufaa G.D., Weil E.J., Nelson R.G. Urinary monocyte chemoattractant protein-1 and hepcidin and early diabetic nephropathy lesions in type 1 diabetes mellitus. Nephrol Dial Transplant. 2015;30:599–606. doi: 10.1093/ndt/gfv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merchant M.L., Gaweda A.E., Dailey A.J. Oncostatin M receptor beta and cysteine/histidine-rich 1 are biomarkers of the response to erythropoietin in hemodialysis patients. Kidney Int. 2011;79:546–554. doi: 10.1038/ki.2010.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merchant M.L., Perkins B.A., Boratyn G.M. Urinary peptidome may predict renal function decline in type 1 diabetes and microalbuminuria. J Am Soc Nephrol. 2009;20:2065–2074. doi: 10.1681/ASN.2008121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thongboonkerd V., Barati M.T., McLeish K.R. Alterations in the renal elastin-elastase system in type 1 diabetic nephropathy identified by proteomic analysis. J Am Soc Nephrol. 2004;15:650–662. doi: 10.1097/01.asn.0000115334.65095.9b. [DOI] [PubMed] [Google Scholar]
- 16.Bennett M.R., Pleasant L., Haffner C. A Novel biomarker panel to identify steroid resistance in childhood idiopathic nephrotic syndrome. Biomarker Insights. 2017;12 doi: 10.1177/1177271917695832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi Y.W., Kim Y.G., Song M.Y. Potential urine proteomics biomarkers for primary nephrotic syndrome. Clin Proteomics. 2017;14:18. doi: 10.1186/s12014-017-9153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Succop P.A., Clark S., Chen M. Imputation of data values that are less than a detection limit. J Occup Environ Hyg. 2004;1:436–441. doi: 10.1080/15459620490462797. [DOI] [PubMed] [Google Scholar]
- 19.Schwanhausser B., Busse D., Li N. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 20.Krämer A., Green J., Pollard J., Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y.J., Huang S.M., Zhang A.H. [Comparison of urinary proteomics between steroid-sensitive and steroid-resistant minimal change nephrotic syndrome in children] Nan Fang Yi Ke Da Xue Xue Bao. 2007;27:1507–1510. [PubMed] [Google Scholar]
- 22.Candiano G., Musante L., Petretto A. Proteomics of plasma and urine in primary nephrotic syndrome in children. Contrib Nephrol. 2008;160:17–28. doi: 10.1159/000125897. [DOI] [PubMed] [Google Scholar]
- 23.Huang A.W., He Q.N., Zhou P. [Preliminary analysis of urinary proteomics in children with steroid-resistant and steroid-sensitive nephrotic syndrome] Zhongguo Dang Dai Er Ke Za Zhi. 2009;11:341–345. [PubMed] [Google Scholar]
- 24.Wang Q.X., Wang J.H., Cao L. [Pretreatment methods of urine proteomics in children with primary nephrotic syndrome] Zhongguo Dang Dai Er Ke Za Zhi. 2011;13:157–160. [PubMed] [Google Scholar]
- 25.Bhowmik D., Agarwal S.K. Serum proteomics for the diagnosis of nephrotic syndrome:is there a ray of hope? Indian J Med Res. 2012;135:273–275. [PMC free article] [PubMed] [Google Scholar]
- 26.Kuribayashi-Okuma E., Shibata S., Arai S. Proteomics approach identifies factors associated with the response to low-density lipoprotein apheresis therapy in patients with steroid-resistant nephrotic syndrome. Ther Apher Dial. 2016;20:174–182. doi: 10.1111/1744-9987.12356. [DOI] [PubMed] [Google Scholar]
- 27.Ransom R.F., Vega-Warner V., Smoyer W.E. Differential proteomic analysis of proteins induced by glucocorticoids in cultured murine podocytes. Kidney Int. 2005;67:1275–1285. doi: 10.1111/j.1523-1755.2005.00205.x. [DOI] [PubMed] [Google Scholar]
- 28.Mauk M.R., Smith A., Mauk A.G. An alternative view of the proposed alternative activities of hemopexin. Protein Sci. 2011;20:791–805. doi: 10.1002/pro.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennon R., Singh A., Welsh G.I. Hemopexin induces nephrin-dependent reorganization of the actin cytoskeleton in podocytes. J Am Soc Nephrol. 2008;19:2140–2149. doi: 10.1681/ASN.2007080940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santucci L., Candiano G., Petretto A. Protein-protein interaction heterogeneity of plasma apolipoprotein A1 in nephrotic syndrome. Mol Biosyst. 2011;7:659–666. doi: 10.1039/c0mb00127a. [DOI] [PubMed] [Google Scholar]
- 31.Adeyemo A., Esezobor C., Solarin A. HLA-DQA1 and APOL1 as risk loci for childhood-onset steroid-sensitive and steroid-resistant nephrotic syndrome. Am J Kidney Dis. 2018;71:399–406. doi: 10.1053/j.ajkd.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freedman B.I., Kopp J.B., Langefeld C.D. The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. J Am Soc Nephrol. 2010;21:1422–1426. doi: 10.1681/ASN.2010070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genovese G., Friedman D.J., Ross M.D. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopp J.B., Nelson G.W., Sampath K. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal S., Zaritsky J.J., Fornoni A. Dyslipidaemia in nephrotic syndrome: mechanisms and treatment. Nat Rev Nephrol. 2017;14:70. doi: 10.1038/nrneph.2017.175. [DOI] [PubMed] [Google Scholar]
- 36.Zoccali C., Mallamaci F., Panuccio V. Adiponectin is markedly increased in patients with nephrotic syndrome and is related to metabolic risk factors. Kidney Int Suppl. 2003;(84):S98–S102. doi: 10.1046/j.1523-1755.63.s84.49.x. [DOI] [PubMed] [Google Scholar]
- 37.Wasilewska A.M., Zoch-Zwierz W.M. Urinary levels of matrix metalloproteinases and their tissue inhibitors in nephrotic children. Pediatr Nephrol. 2008;23:1795–1802. doi: 10.1007/s00467-008-0881-3. [DOI] [PubMed] [Google Scholar]
- 38.Korzeniecka-Kozerska A., Wasilewska A., Tenderenda E. Urinary MMP-9/NGAL ratio as a potential marker of FSGS in nephrotic children. Dis Markers. 2013;34:357–362. doi: 10.3233/DMA-130980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai S.H., Huang P.H., Hsu Y.J. Inhibition of hypoxia inducible factor-1alpha attenuates abdominal aortic aneurysm progression through the down-regulation of matrix metalloproteinases. Sci Rep. 2016;6:28612. doi: 10.1038/srep28612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond G.L. Plasma steroid-binding proteins:primary gatekeepers of steroid hormone action. J Endocrinol. 2016;230:R13–R25. doi: 10.1530/JOE-16-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elias A.N., Carreon G., Vaziri N.D. The pituitary-gonadal axis in experimental nephrotic syndrome in male rats. J Lab Clin Med. 1992;120:949–954. [PubMed] [Google Scholar]
- 42.Cheng X., Zhao X., Khurana S. Microarray analyses of glucocorticoid and vitamin D3 target genes in differentiating cultured human podocytes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett M.R., Pordal A., Haffner C. Urinary vitamin D-binding protein as a biomarker of steroid-resistant nephrotic syndrome. Biomark Insights. 2016;11:1–6. doi: 10.4137/BMI.S31633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoentgen F., Metz-Boutigue M.H., Jolles J. Complete amino acid sequence of human vitamin D-binding protein (group-specific component):evidence of a three-fold internal homology as in serum albumin and alpha-fetoprotein. Biochim Biophys Acta. 1986;871:189–198. doi: 10.1016/0167-4838(86)90173-1. [DOI] [PubMed] [Google Scholar]
- 45.Klein T., Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41:271–290. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gooding J.R., Agrawal S., McRitchie S. Predicting and defining steroid resistance in pediatric nephrotic syndrome using plasma metabolomics. Kidney Int Rep. 2020;5:81–93. doi: 10.1016/j.ekir.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.