Abstract
Sensory gene families are of special interest for both what they can tell us about molecular evolution and what they imply as mediators of social communication. The vomeronasal type-1 receptors (V1Rs) have often been hypothesized as playing a fundamental role in driving or maintaining species boundaries given their likely function as mediators of intraspecific mate choice, particularly in nocturnal mammals. Here, we employ a comparative genomic approach for revealing patterns of V1R evolution within primates, with a special focus on the small-bodied nocturnal mouse and dwarf lemurs of Madagascar (genera Microcebus and Cheirogaleus, respectively). By doubling the existing genomic resources for strepsirrhine primates (i.e. the lemurs and lorises), we find that the highly speciose and morphologically cryptic mouse lemurs have experienced an elaborate proliferation of V1Rs that we argue is functionally related to their capacity for rapid lineage diversification. Contrary to a previous study that found equivalent degrees of V1R diversity in diurnal and nocturnal lemurs, our study finds a strong correlation between nocturnality and V1R elaboration, with nocturnal lemurs showing elaborate V1R repertoires and diurnal lemurs showing less diverse repertoires. Recognized subfamilies among V1Rs show unique signatures of diversifying positive selection, as might be expected if they have each evolved to respond to specific stimuli. Furthermore, a detailed syntenic comparison of mouse lemurs with mouse (genus Mus) and other mammalian outgroups shows that orthologous mammalian subfamilies, predicted to be of ancient origin, tend to cluster in a densely populated region across syntenic chromosomes that we refer to as a V1R “hotspot.”
Keywords: V1R, vomeronasal system, pheromone, Lemuriformes, synteny, gene family evolution
Introduction
The evolutionary dynamics of sensory gene families are of fundamental interest as a model for how molecular evolutionary processes can shape the content and structure of genomes and for their ability to characterize the life history and ecological traits of organisms. Vomeronasal type-1 receptor (V1R) genes comprise one such gene family and have been the subject of increasing interest in both the molecular genetics (Adipietro et al. 2012) and evolutionary genetics (Yohe and Brand 2018) communities. Vomerolfaction is a form of chemosensation that mediates semiochemical detection and occurs in the vomeronasal organ (VNO) of mammals (Leinders-Zufall et al. 2000). V1Rs are expressed on vomeronasal sensory neurons in the VNO and have been demonstrated to detect pheromones in mice (Boschat et al. 2002; Haga-Yamanaka et al. 2014). For example, impaired vomeronasal function in mice, either through a knockout of V1Rs or removal of the VNO, alters appropriate chemosensory behaviors such as conspecific avoidance of sick animals, interspecies defensive cues, male sexual behavior, and maternal aggression (Del Punta et al. 2002; Papes et al. 2010; Boillat 2015). Thus, the evolution of V1Rs can have direct consequences for both the emitter and the receiver of pheromone signals, with ample evidence indicating that molecular evolution of V1Rs is associated with the speciation process (Lane et al. 2002; Kurzweil et al. 2009; Hohenbrink et al. 2012; Nikaido et al. 2014).
V1Rs are ideally suited for study within the context of “sensory drive” wherein mate preferences in communication systems diverge in the face of novel environmental opportunity (Boughman 2002). Communication mechanisms for mate recognition have been recognized as an important component for driving rapid reproductive isolation (Mendelson 2003; Dopman et al. 2009; Servedio and Boughman 2017; Brand et al. 2019) and with the reinforcement of species boundaries (Servedio and Noor 2003). Sensory drive can affect diverging populations in two ways, by targeting the pheromone receptors or their signaling molecules. As examples, adaptation of a likely V1R signal in mice, androgen binding protein, is associated with assortative mating between Mus musculus subspecies (Karn et al. 2010; Chung et al. 2017; Hurst et al. 2017) just as fixation of nonsynonymous polymorphisms among V1Rs is associated with the speciation of Mus spretus and M.musculus (Kurzweil et al. 2009). Moreover, it has been shown that differential expression of vomeronasal and olfactory receptor genes, including V1Rs, is associated with assortative mating in a pair of house mouse subspecies (Loire et al. 2017) and is likely reinforcing the subspecies along their hybrid zone.
The V1R gene family has experienced many duplications and losses in the evolutionary history of mammals, and the availability of duplicate copies can allow for divergence among sequences, gene expression, and ultimately function (Lynch and Conery 2000; Des Marais and Rausher 2008). Though not directly addressed in this study, it is worth noting that changes in gene expression often occur rapidly after gene duplication events (Makova and Li 2003; Keller and Yi 2014; Guschanski et al. 2017) and are often accompanied by shifts in rates of molecular evolution (Chen et al. 2010; Yang and Gaut 2011). Although the mechanisms that explain variable rates of molecular evolution, specifically the nonsynonymous to synonymous substitution rate ratio (dN/dS), are complex, there is some interdependence on expression levels (O’Toole et al. 2018) and genome architecture (Dai et al. 2014; Xie et al. 2016). The V1R gene family demonstrates structural complexity (Ohara et al. 2009; Yohe et al. 2019), and gene family expansions and directional selection acting on duplicate copies may be important for the maintenance of species boundaries where vomerolfaction is linked with assortative mating (Luo et al. 2003; Isogai et al. 2011; Fu et al. 2015).
Here, we present a comparative genomic study of V1R evolution within the lemuriform primates, primarily focusing on the mouse lemurs of Madagascar (genus Microcebus). Mouse lemurs are perhaps the most species-rich clade of living primates (Hotaling et al. 2016), and are well-known for high levels of interspecific genetic divergence though with nearly uniform morphological phenotypes. They have thus come to be regarded as a classic example of a cryptic species radiation, perhaps related to their nocturnal lifestyle (Yoder et al. 2016). Mouse lemurs, and the closely related dwarf lemurs, have elaborate olfactory communication behaviors that are associated with adaptive strategies such as predator recognition (Sündermann et al. 2008), fecundity (Drea 2015), and even biased sex ratios (Perret 1996; Perret and Colas 1997). V1Rs take on particular interest in mouse lemurs as we hypothesize that their observed role in both speciation and in the maintenance of species boundaries within Mus may also apply to this speciose clade of primates (Smadja et al. 2015; Loire et al. 2017). We hypothesize that among primates, mouse lemurs will show signatures of sensory drive via genomic elaboration of the V1R complex and evidence of positive selection acting on V1R genes.
There are numerous lines of evidence to lead us to this hypothesis: 1) Previous studies have indicated that V1Rs within the lemuriform clade have evolved under pervasive positive selection (Hohenbrink et al. 2012; Yoder et al. 2014), 2) that the majority of gene copies are intact (Young et al. 2010; Larsen et al. 2014), and 3) that the differential expression of a large number of vomeronasal receptors in both the VNO and main olfactory epithelium of mouse lemurs are associated with different behaviors and chemical signals (Hohenbrink et al. 2014). In fact, along with murids, opossums, and platypus, mouse lemurs have been reported to have among the largest V1R repertoires found in mammals (Young et al. 2010). Even so, numerous obstacles such as complexities of chemical background, chemical signals, and the genetic basis of chemosensation complicate both ecological and experimental approaches for differentiating between cause and effect in the speciation process (Yohe and Brand 2018). This is particularly problematic for studies of mouse lemurs given their remote geographic distribution, nocturnal life history, and endangered status. Thus, we take a comparative genomic approach for reconstructing the evolutionary dynamics of the V1R gene family within the small-bodied and nocturnal mouse and dwarf lemurs (family, Cheirogaleidae).
A Comparative Genomic Approach
V1R loci are highly repetitive and they, along with their surrounding regions, are notoriously challenging for genome assembly. Though previous studies have used targeted sequencing or short-read sequencing to examine the evolutionary dynamics of V1R expansions in a limited number of species (Young et al. 2010; Hohenbrink et al. 2012; Yoder et al. 2014), strepsirrhine primates have until recently remained woefully underrepresented in genomic databases (Perry et al. 2012; Meyer et al. 2015; Larsen et al. 2017; Hawkins et al. 2018). Here, we take advantage of the chromosome-level assembly of the gray mouse lemur, Microcebus murinus, along with short-read sequencing in related species, to characterize the V1R repertoires for lemuriform primates. Recent improvements using long-read sequencing of the mouse lemur genome (Larsen et al. 2017) improve our ability to characterize the V1R repertoire and allow for comparisons of the genomic architecture of V1R-containing regions in expanded and contracted V1R repertoires across mammals.
In this study, we have sequenced and assembled six new cheirogaleid genomes, with a particular focus on the mouse lemurs. Furthermore, to explore intraspecific copy number variation (CNV) and evaluate the effects of assembly error on V1R repertoire counts, we resequenced and de novo assembled genomes from eight M. murinus individuals from a captive breeding colony. Our study thus serves as a timely companion to two recent overviews of comparative genomic studies for illuminating the evolutionary and life-history dynamics of chemosensory gene family evolution in vertebrates (Bear et al. 2016; Hughes et al. 2018). A comparative genomic approach allows us to explore classic predictions of gene family evolution, namely, that genomic drift can operate at very fine scales to produce high intraspecific CNV (Nozawa et al. 2007) and that gene family evolution is often marked by a birth–death process over phylogenetic time scales (Nei et al. 1997; Csűrös and Miklos 2009; Hughes et al. 2018). The latter question is of particular interest for V1R evolution given that adaptive pressures on these genes make them highly vulnerable to pseudogenization in cases of relaxed selection, thus yielding the observed correlations between levels of V1R ornamentation and diverse adaptive regimes. An overview of primates shows that those with elaborate representation of subfamilies have a strong reliance on chemosensory communication whereas those with depauperate V1R representation rely on alternative mechanisms for inter- and intraspecific communication (Yoder and Larsen 2014).
These new genomic resources have also allowed us to address a number of questions regarding rates of molecular evolution in V1Rs. Divergent gene function following gene duplication predicts that some signature of positive selection should be evident in the gene sequences (Zhang et al. 1998), but it remains unknown if selection has acted pervasively over time or has occurred in episodic bursts prior to the diversification of mouse lemurs. We might anticipate selection was largely episodic before repertoire expansions if purifying selection has been operating across species at more recent time scales to preserve gene function among duplicate copies. For strepsirrhine primates (i.e. the lemurs and lorises), pervasive positive selection has been detected at the interspecific level (Hohenbrink et al. 2012; Yoder et al. 2014), whereas strong purifying selection has been found within populations (Hohenbrink et al. 2017). Here, we disentangle pervasive versus episodic positive selection among V1Rs and show that both gene duplication and diversifying selection acting on different V1R subfamilies have shaped the repertoires of dwarf and mouse lemurs. Moreover, through comparison with Mus and other mammals, we show that orthologous subfamilies tend to cluster in a densely populated region on syntenic chromosomes including one V1R “hotspot.”
Materials and Methods
Sampling and DNA Extraction
To improve the resolution of the V1R repertoire expansion in lemurs, we sequenced the genomes of Microcebus griseorufus, M. mittermeieri, M. ravelobensis, M. tavaratra, and Mirza zaza. Tissue biopsies were taken from wild individuals in Madagascar from 1997 to 2015 and from captive individuals at the Duke Lemur Center (supplementary table S13, Supplementary Material online). To investigate within species variation in V1R repertoires, we also resequenced eight individuals from the Duke Lemur Center M.murinus colony. Blood and tissue samples were collected in 2016 in accordance with IACUC guidelines. For the novel strepsirrhine genomes, DNA was extracted following manufacturer instructions using the Qiagen DNeasy Blood and Tissue kit, whereas DNA from the resequenced M.murinus was extracted using the Qiagen MagAttract Kit (Qiagen, Germantown, MD).
Genome Sequencing and Assembly
The genomes of M.griseorufus, M. mittermeieri, M. tavaratra, and M.zaza were sequenced at the Baylor College of Medicine as approximately 400-bp insert libraries on a single lane of an Illumina HiSeq 3000 with paired-end 150 bp reads. We sequenced the M.ravelobensis genome from two libraries, one with an average insert size of 570 bp on an Illumina HiSeq 2000 at Florida State University and the other with a 500 bp insert library on 5.5% of both lanes of an Illumina NovaSeq at the Duke University GCB Sequencing Core. We also generated two additional cheirogaleid assemblies for Cheirogaleus sibreei and C. medius (Williams et al. 2019). C. sibreei. was sequenced from a 300 bp insert library on the Illumina HiSeq 4000 at the Duke University GCB Sequencing Core with paired-end 150 bp reads. A reference genome was generated and assembled for C.medius using Dovetail Genomics. All other genomes were assembled using MaSuRCA v3.2.2 (Zimin et al. 2013). We assumed an insert size standard deviation of 15% and used automatic kmer selection. However, we did not use MaSuRCA’s scaffolds for annotation and downstream analyses. Scaffolds were obtained from SSPACE (Boetzer et al. 2011), which also attempted to correct assembly errors and extend contigs from MaSuRCA. De novo assembly statistics are available in the supplementary material (supplementary table S1, Supplementary Material online) as well as annotation details (supplementary table S14, Supplementary Material online) and SRA identifiers (supplementary table S15, Supplementary Material online).
The eight M.murinus individuals were resequenced from high-molecular weight DNA prepared using the 10× Genomics Chromium platform. Briefly, high-molecular weight molecules of DNA are partitioned into gel beads with unique barcodes then prepared for Illumina sequencing (Weisenfeld et al. 2017). The resulting short-read libraries are barcoded such that individual “linked reads” can be traced back to their molecule of origin assisting the genomic scaffolding process. The libraries were size selected to approximately 550 bp and sequenced on the Illumina HiSeq 4000 system at the Duke University GCB Sequencing Core. We then used the 10× Genomics Supernova assembly software to de novo assemble the resequenced genomes (version 2.0.1, 10× Genomics, San Francisco, CA). One replicate individual was sequenced twice, MMUR_DLC7033, and genomes were assembled de novo from each individual library.
BUSCO version 3.0.2 and Assemblathon2 scripts were used to assess genome quality statistics (supplementary fig. S1 and table S16, Supplementary Material online; Simão et al. 2015). Additional genomes analyzed in this study were downloaded from the NCBI genome database and include all available Strepsirrhine genomes and additional high-quality primate and mammalian genomes for phylogenetic coverage (supplementary table S16, Supplementary Material online).
V1R Repertoire Estimation and Ancestral Count Reconstruction
To assess total V1R gene repertoires in each species, tBlastN searches (e value cut-off = 0.001) were conducted with the BLAST+ software suite (version ncbi-BLAST-2.6.0+; Altschul et al. 1990) using available mouse and mouse lemur V1R query protein sequences downloaded from NCBI GenBank against the genomes analyzed in this study (Camacho et al. 2009). We assumed that allelic diversity was collapsed by the assembly algorithm and treat all V1R counts estimated here as a reflection of gene diversity. Duplicate protein sequences were removed from the query database using CD-HIT version 4.6 (Li and Godzik 2006). Bedtools merge (version 2.27.1) was used to merge overlapping hits within a genome. V1R genes are G-protein-coupled receptors that consist of a single exon typically around 1,000 bp long (Mundy 2003). To remove potential pseudogenes outside of the expected size range for V1Rs, bedtools slop and getFasta were used to extract receptor candidate regions longer than 600 bp with 50 bp of upstream and downstream surrounding sequence (Quinlan and Hall 2010). To remove additional potential pseudogenes from further analyses, we used Geneious version 9.0.5 to predict open reading frames (ORFs) and considered sequences intact if they contained an ORF longer than 801 bp. The number of intact V1Rs, percentage of intact V1Rs, and the total V1R count were calculated for each species. For a full list of V1R containing regions analyzed and included in the repertoire size estimates see supplementary file 1, Supplementary Material online.
To determine subfamily membership for each V1R repertoire, we first generated a maximum likelihood (ML) tree using RAxML of mouse lemur sequences mined from the mmur3.0 genome and previously published M. murinus V1R sequences (Hohenbrink et al. 2012). We used this tree to assign mmur3.0 V1R subfamily designations. We then used MAFFT version 7.187 with the E-INS-i algorithm to align intact sequences from all species using the iterative approach described in Yoder et al. (2014) and Katoh and Standley (2013). The MAFFT algorithm is recommended for approaches analyzing ancestral sequence reconstruction (Vialle et al. 2018). A gene phylogeny was constructed using RAxML version 7.2.8 using the GTRGAMMAI nucleotide model with the rapid bootstrapping and search for best ML scoring tree algorithm with 500 bootstraps (Stamatakis 2014). We assigned primate sequences to the subfamilies Strep/I-IX based off the subfamily designation of the previously analyzed mmur3.0 V1R sequences. A subfamily was defined as a monophyletic group encompassing all mmur3.0 V1R sequences from one subfamily, which was expanded to include any closely related primate sequences; sequences were added to the subfamily until the expansion would require the inclusion of a sequence from a member of Laurasiatheria. We used Count version 10.04 with the Wagner parsimony algorithm and a gain penalty of 2 to infer total ancestral vomeronasal repertoire size and ancestral subfamily membership (Csűrös and Miklos 2009; Csűrös 2010).
Establishing Synteny of V1Rs across Mammalian Species
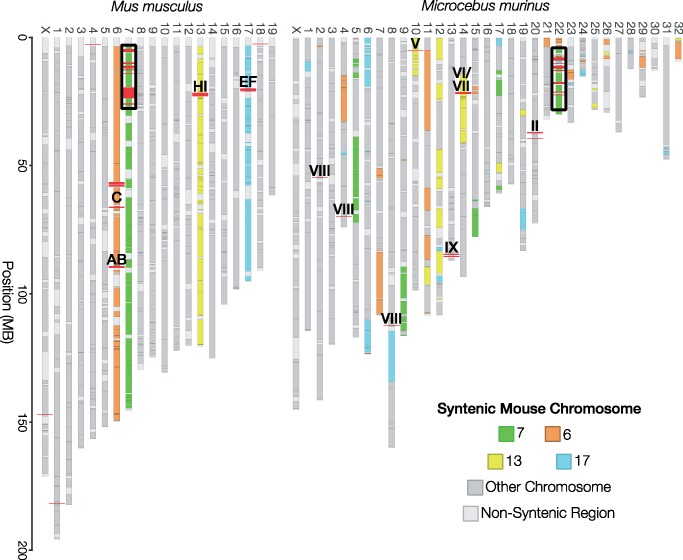
Genomes with chromosome-level scaffolding information (M.musculus, M.murinus, Homo sapiens, Equus caballus, Bos taurus, and Canis familiaris) were used to assess chromosomal synteny of vomeronasal subfamilies among mammalian species. SynChro (Drillon et al. 2014) version SynChro_osx (January 2015) was used to reconstruct synteny blocks between each genome with M.musculus as reference with a delta parameter of 2 using GenBank annotation files from Ensembl release 93 (fig. 1; supplementary figs. S8–S11, Supplementary Material online; Drillon et al. 2014). Orthologous block information was compared with vomeronasal receptor location for each species (fig. 2A and B).
Fig. 1.
—Chromosomal synteny between mouse and mouse lemur V1R-containing regions. Synteny between Mus musculus and Microcebus murinus was estimated using the SynChro software. Chromosomes are colored relative to V1R-containing mouse chromosomes. V1R loci are indicated with red lines and are labeled by subfamily identity. Regions outlined in black are enriched for V1R loci in subfamilies I, III, IV, and V, and are examined in further detail in figure 2B.
Fig. 2.
—Highly orthologous loci on “hotspot” V1R chromosome. (A) RAxML tree of Mus musculus V1R cDNA sequences with intact V1R sequences from the Microcebus murinus genome. Mouse subfamilies are encircled in black and labeled by chromosomal location. Mouse lemur subfamilies are labeled in black and encircled in the color corresponding to the syntenic mouse chromosomal location. (B) Chromosomal “hotspot” regions enriched in V1R loci from several mammalian taxa. Orthologous regions are shaded by syntenic mouse chromosome. V1Rs loci are labeled by phylogenetic relationship to mouse/lemur subfamilies. Starting and end genomic positions are given for each species, and all regions are 23 Mb long with tick marks representing 5-Mb intervals.
Detecting Evidence of Positive Selection
Evidence for positive selection in V1R repertoires was evaluated with PAML 4.8e (Yang 2007). We used two different tests to detect both individual sites under pervasive positive selection throughout the tree (sites models) and individual branches that show an episodic burst of positive selection (branch-site models). For sites models, we applied two tests to each of the nine subfamily trees and alignments: 1) Comparison of the null hypothesis that all sites are a mixture of purifying and neutral rates of molecular evolution (M1a) and the alternative that allows for a third class of sites under positive selection (M2a; Zhang et al. 2005) and 2) a null hypothesis that allows for a mixture of ten discretized beta-distributed site classes (M7), whereas the alternative hypothesis allows an extra component under positive selection (M8; Yang et al. 2000). Each of the recognized lemur subfamilies was analyzed separately. The ggtree R package (Yu et al. 2017) was used to extract subtrees for each subfamily and alignments were parsed with Perl scripts. Because signatures of positive selection may be time-dependent (Peterson and Masel 2009; Pegueroles et al. 2013), we explored variation in sites under positive selection using six different taxonomic filters: 1) Microcebus, 2) Cheirogaleidae, 3) Lemuriformes, 4) Strepsirrhini, 5) Primates, and 6) Euarchontoglires. For each site test, we assumed the likelihood ratio test (LRT) was ∼χ21 and individual sites were detected using the Bayes empirical Bayes procedure where the posterior probability of selection for each site was determined using the MLE dN/dS for the positive selection rate class (Yang et al. 2005). Individual sites were considered to have sufficient evidence for positive selection if the posterior probability was greater than 0.95. Enrichment of sites under selection in transmembrane domains (TMs) used simple chi-square tests and Fisher exact tests in R (R Core Team 2018) for individual transmembrane and loop domains. TMs were predicted using M. murinus sequences from subfamilies Strep/I through IX with TMHMM (Krogh et al. 2001) through the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/; last accessed January 29, 2019). Because V1R genes are expected to have seven TMs (Dulac and Axel 1995), only predicted structures with seven TMs were used to determine TM boundaries in our alignment of the entire V1R repertoire. Predictions that had fewer or more than seven TMs are assumed to be due to inaccuracies of TMHMM (Melén et al. 2003) and not real domain losses or gains.
Of important note, the entire V1R repertoire was prohibitively large for ML optimization over the entire tree; we applied tests for selection to individual subfamilies to circumvent this limitation. This strategy also provided a way to evaluate contributions of alignment and topological errors to evidence of positive selection. First, we evaluated if the ML topology estimated from the entire repertoire was a plausible hypothesis using approximately unbiased (AU) tests (Shimodaira 2004). Second, we estimated the ML topology and branch lengths for each subfamily using the parsed alignments (i.e. the data was not realigned) using the same RAxML model and search strategy as the first analysis. We then realigned translated amino acid data with MAFFT and estimated the phylogeny once more. Site log-likelihoods were then optimized for the three topologies with RAxML and AU P values computed with CONSEL using the default multiscale bootstrapping strategy (Shimodaira and Hasegawa 2001). Bootstrap trees were also collected for the realigned data, but bipartitions were drawn onto the topologies parsed from the entire V1R repertoire tree. The ratio of bootstrap support values was used to identify potential topological errors; bipartitions in the original topology that are absent when the sequences for each subfamily were realigned. Site tests were run for both the parsed and realigned data to check for consistency in the inference of sites under positive selection across alignments.
Branch-site tests (Zhang et al. 2005) were performed for each branch for each subfamily, except subfamily III, which was still computationally limiting. Each test assumed the LRT was ∼χ21, but we applied the Benjamini–Hochberg multiple testing correction (Benjamini and Hochberg 1995). With this correction, we do expect some false positives, but the family-wise error rate should be below 5% (Anisimova and Yang 2007) while not underpowering tests toward the tips of the trees (dos Reis and Yang 2011). Tests were only performed on the parsed topology without removing any species, but branches with evidence of episodic positive selection and the bootstrap ratios with realigned data were mapped to nodes, that those branches were subtending, of the subtree topologies using ggtree to help identify cases where topological errors might lead to false signatures of positive selection (Mendes and Hahn 2016). Individual sites with evidence of episodic positive selection were evaluated using the Bayes empirical Bayes procedure (Yang et al. 2005).
Results and Discussion
Novel Genome Assemblies of Several Strepsirrhine Primates
We de novo assembled seven novel strepsirrhine genomes: M. griseorufus, M. ravelobensis, M. mittermeieri, M. tavaratra, M. zaza, C. sibreei, and C. medius. These efforts have doubled the number of publicly available genomes for the Strepsirrhini with a specific focus on the dwarf and mouse lemur clade. Excluding C. medius, the seven genomes were sequenced to an average depth of coverage between 26× and 45× with scaffold N50s of 17–76 kb (supplementary table S1, Supplementary Material online). The C. medius reference genome was assembled using Dovetail Genomics to an average depth of coverage of 110× and a scaffold N50 of approximately 50 Mb (Williams et al. 2019). We evaluated assembly completeness using the Benchmarking Universal Single-Copy Orthologs tool, BUSCO (Simão et al. 2015), which assesses genomes for the presence of complete near-universal single-copy orthologs (supplementary fig. S1, Supplementary Material online). The seven strepsirrhine assemblies recovered between 77.2% and 92.3% of the mammalian BUSCO gene set. We also resequenced eight M. murinus individuals, with one duplicate individual and here have de novo assembled genomes for each individual with 21×–29× effective coverage using the 10× Genomics Supernova pipeline. The additional scaffolding information provided by the 10× Genomics linked-reads resulted in scaffold N50s of 0.6–1.2 Mb. BUSCO analyses of the resequenced M.murinus individuals revealed that the genome assemblies recovered between 89.9% and 95.5% of the mammalian gene set. A denser sampling of genomes within Cheirogaleidae not only provides an opportunity for illuminating patterns of V1R gene family evolution but also promotes greater understanding of the molecular evolution of primate and strepsirrhine genomes. Genome resequencing of M. murinus individuals allows investigation of intraspecific V1R CNV in a captive population and reveals potential variation in repertoire estimation. Genome resequencing of M. murinus individuals revealed generally consistent estimates of V1R copy numbers. Variation among individuals likely reflects technical artifacts such as assembly errors, which can be observed in the twice-sequenced individual DLC7033, though biological causes of CNV—perhaps relating to the unintentionally outbred nature of the colony (Larsen et al. 2017)—cannot be excluded.
The monophyletic genus Microcebus contains 24 named species (Hotaling et al. 2016), and our results clearly demonstrate that the clade has a uniquely complex V1R repertoire compared with other primates thus far characterized (fig. 3A and B). Contrary to a previous study suggesting that V1R expansion is ubiquitous across the lemuriform clade (Yoder et al. 2014), increased sampling reveals that expansion has been profound in the nocturnal dwarf and mouse lemurs. This is consistent with the original hypothesis that local V1R expansions may play a role in forming or maintaining speciation boundaries within Cheirogaleus and Microcebus as might be predicted given their nocturnal lifestyle. Phylogenetic analyses revealed that expanded V1R repertoires in mouse lemurs demonstrate a remarkably higher rate of duplicate gene retention in comparison to other primates (fig. 3A and B; table 1). The common ancestor of mouse lemurs is not associated with novel subfamily birth though the diversity and number of V1R gene copies is striking (fig. 3A;table 1). Although our methodological strategy does not differentiate between allelic diversity and gene diversity, and genomes generated exclusively from short-read data are vulnerable to collapsing loci in assemblies (Larsen et al. 2014), our inference of increased V1R retention in M. murinus relative to non-cheirogaleid primates was robust to assembly strategies and data sources (supplementary table S2, Supplementary Material online). Furthermore, the resequenced M. murinus individuals reveal low intraspecific variation in copy number (fig. 4), suggesting that the observed differences in repertoire size between mouse lemurs and other non-nocturnal lemurs is not an artifact of individual sampling or assembly error (fig. 5).
Fig. 3.
—V1R subfamily membership across the primate phylogeny. Membership was assessed for available strepsirrhine genomes (A) and for select primate outgroups (B) and estimated using the Count software for ancestral lineages. Bar graphs show absolute gene count for each subfamily. Predicted gene subfamily origins are annotated with arrows. Tree adapted from dos Reis et al. (2018). Circled node numbers correspond with table 1.
Table 1.
Inference of V1R Birth–Death Process within Primates
| Node/Lineage | Clade | Gains–Losses | Subfamilies Retained | Subfamilies Gained | Subfamilies Lost |
|---|---|---|---|---|---|
| Node 1 | Euarchonta | NA | I, II, III, IV, VIII | NA | NA |
| Node 2 | Primates plus Dermoptera | 0–1 | I, II, III, VIII | — | IV |
| Node 3 | Euprimates | 1–0 | I, II, III, VIII | V | — |
| Node 4 | Haplorrhini | 0–2 | I, III, V | — | II, VIII |
| Node 5 | Anthropoidea | 0–1 | III, V | — | I |
| Node 6 | Hominoidea | 0–1 | III | — | V |
| Nomascus leucogenys | White-cheeked gibbon | 1–1 | — | VII | V |
| Node 7 | Strepsirrhini | 1–0 | I, II, III, V, VIII | VI | — |
| Node 8 | Lemuriformes | 1–0 | I, II, III, V, VI, VIII | IV | — |
| Daubentonia madagascariensis | Aye-aye | 1–3 | III, VI, VIII | IX | I, II, V |
| Node 9 | Lemuridae | 0–1 | I, II, III, V, VI | — | VIII |
| Node 10 | Eulemur | 1–0 | I, II, III, V, VI | VII | — |
| Node 11 | Cheirogaleidae | 3–0 | I, II, III, V, VI, VIII | IV, VII, IX | — |
Note.—Node numbers correspond to figure 3. Gains and losses cannot be evaluated for Node 1 because it is the root node of the species tree.
Fig. 4.
—Intraspecific variation in V1R repertoire size estimates across eight closely related M. murinus individuals. Genomes were de novo assembled and mined for loci with significant V1R homology and an ORF longer than 801 bp. Individual DLC7033 was sequenced twice and repertoire size estimates are reported for both assemblies. Squares represent males and circles represent females. Horizontal lines indicate mate pairs (mother and father) and vertical or slanted lines indicate parent to offspring relationship. Numbers inside the symbols represent repertoire size estimates. Individuals represented by gray symbols were not sequenced.
Fig. 5.
—V1R repertoire size estimates across the strepsirrhine phylogeny. Sequences with V1R homology were mined from available strepsirrhine and select outgroup genomes. Total V1Rs consist of all genomic regions with V1R homology that are ≥600 bp in length. Intact genes are defined by vomeronasal homology and a ≥801-bp ORF. Nocturnal species are highlighted in gray. Tree adapted from dos Reis et al. (2018).
The expansion dynamics of V1Rs within Cheirogaleidae do not support a simple linear correlation between species richness and repertoire size. Dwarf lemurs, genus Cheirogaleus, are hypothesized to have as many as 18 species (Lei et al. 2014), and despite having larger repertoire sizes than all diurnal lemurs, they have the smallest V1R repertoires within the cheirogaleids examined here. Conversely, the genus Mirza, with only two recognized species, has a repertoire size that is nearly equal to that of M.murinus. It is notable, however, that the Mirza genome’s expanded repertoire is primarily enriched for subfamily III (fig. 3A). The differential subfamily enrichment among species suggests that despite the similarity in size to Microcebus repertoires, the V1R repertoire of Mirza has experienced independent selective pressures on gene retention and may ultimately fulfill a different functional role compared with Microcebus.
V1R Repertoire Estimation across Primates
We estimated V1R repertoire size evolution across strepsirrhine primates as well as for several well-annotated primate and mammalian genomes for outgroup comparison. Notably, repertoire estimates of extant primates are comparable with previous studies that used trace archive fragments and earlier draft genome versions (fig. 5; supplementary table S2, Supplementary Material online; Young et al. 2010; Moriya-Ito et al. 2018). Previous research suggests that genomes generated exclusively from short-read data may be collapsing loci in assemblies, especially V1R gene sequences (Larsen et al. 2014). Collapsing of repetitive regions of the genome during genome assembly may result in underestimates of total V1R repertoire size, but we assume that this bias should affect all short-read based assemblies equally.
Our analyses reveal that the expanded V1R repertoire within the gray mouse lemur is not ubiquitous across the Strepsirrhini. Rather, repertoire size expanded gradually from a reduced set in the strepsirrhine common ancestor to its peak in the mouse lemur clade. This expansion is characterized by a reduced repertoire in the early diverging aye-aye lineage (genus Daubentonia), moderate repertoires among diurnal lemurs, and an expansion that likely occurred in the common ancestor of Cheirogaleidae (fig. 5). If the origins of many V1R copies in mouse lemur date to the Cheirogaleidae common ancestor, this means that at least some of those duplicates have remained functional and intact since their origins 30 Ma, as would be consistent with divergence time estimates for the cheirogaleid radiation (Yang and Yoder 2003; dos Reis et al. 2018).
Within Cheirogaleidae, repertoire sizes ranged from a low of 58 intact V1Rs in C. medius to highs between 102 and 146 intact V1Rs in the genus Microcebus. The mouse lemurs have universally large V1R repertoires (102–146 intact genes) with notable intragenus variation. Prior to this study, M. murinus had been identified as having one of the largest V1R repertoires within mammals (Young et al. 2010). Additional sampling from Microcebus reveals, however, that among the five mouse lemur species here characterized, M. murinus actually has the smallest repertoire with only 102 intact V1Rs. We also estimated the percent of intact V1Rs contained within the total repertoire for each species. Most haplorrhine primates (Anthropoidea plus Tarsius) species have repertoires with low percentages of intact receptors (<23% intact). Within Lemuroidae, the diurnal lemurs have also small and pseudogenized repertoires (26–49% intact) containing only 22–27 intact V1Rs. In contrast, among nocturnal strepsirrhines excluding aye-aye, we observe intact repertoires between 58% and 66% within Cheirogaleidae, and 61% for the nocturnal lorisiform Otolemur garnettii.
These comparisons do not, however, provide definitive evidence that expanded V1R repertoires in primates are strictly associated with nocturnal life history (Wang et al. 2010; Moriya-Ito et al. 2018). Although O.garnettii shows a proportion of intact V1R copies similar to dwarf and mouse lemurs (fig. 5), subfamilies VII and IX are absent from O. garnettii (fig. 3B). In addition, the genomes of both the aye-aye and the tarsier (Schmitz et al. 2016) contain low numbers of intact V1R gene copies, which appears to contradict the hypothesis that a nocturnal life history alone is sufficient for explaining V1R elaboration in mouse lemurs. Though it is true that both aye-aye and tarsier have more V1R copies than the diurnal primates compared here, they also show a high proportion of putative pseudogenes and an absence of some V1R subfamilies found in Cheirogaleidae (fig. 3A and B).
Our phylogenetic approach revealed multiple gene gain and loss events that are compatible with a birth–death model (Nei et al. 1997; Csűrös and Miklos 2009; Hughes et al. 2018) with an independent V1R expansion in the Cheirogaleidae and three subfamily gains rather than a single more ancient expansion followed by losses in diurnal lineages (table 1). Although the gain and loss dynamics of V1Rs over time is complex with uncertainty in the origins of specific subfamilies, variation in subfamily membership among species suggests that nocturnal primates possess more diverse repertoires than their diurnal counterparts (fig. 3A and B). These results are suggestive of an association between nocturnal life histories and V1R repertoire evolution, as well as the general importance of chemosensation for nocturnal primates. Our findings are not conclusive, however, because the repertoire of aye-aye is depauperate and less diverse relative to the dwarf and mouse lemurs. But, the aye-aye genome assembly is considerably more fragmented than others compared here, with the lowest contig N50 and the lowest complete BUSCO results (supplementary fig. S1 and table S1, Supplementary Material online). An improved genome for aye-aye, a notably solitary primate (Sterling and Richard 1995), as well as genomes for species within the diurnal nest-dwelling genus, Varecia, will allow for more rigorous tests of how life history traits are correlated with V1R copy number.
Subfamily Membership and Ancestral Repertoire Reconstruction
After estimating total repertoire size for each species, we classified repertoire subfamily membership based on homology inferred from a ML tree (fig. 6) and previously described subfamily designations (Materials and Methods; Hohenbrink et al. 2012). During alignment, sequences that introduced excessive gaps to transmembrane regions were iteratively removed, resulting in alignments of increasing conservatism (see Materials and Methods “V1R Repertoire Estimation and Ancestral Count Reconstruction”). We tested whether these varying alignments affected our estimates of subfamily composition and found little impact. Regardless of the number of sequences removed from the alignment, the relative proportions of subfamily membership within each species remained constant (supplementary fig. S2, Supplementary Material online). Although topological errors may contribute to uncertainty in gene count reconstructions, the ML tree shows 70% or greater bootstrap support for 63% of nodes (fig. 6), with little additional improvement possible due to the limitations of a single-exon gene family (supplementary table S3, Supplementary Material online).
Fig. 6.
—ML topology of V1R repertoire. V1R subfamilies in primates are highlighted and circumscribed based on Hohenbrink et al. (2012). There are nine described subfamilies in lemurs, L Strep/I through L IX, although not all lemur sequences fall into these subfamilies. Clades of V1R subfamilies with known function in mice are shown in burnt orange (M AB through M JK). Circles at nodes represent bootstrap support. Black nodes have 100% bootstrap support, dark gray nodes are supported with 70% or more bipartitions from bootstrap trees, and light gray nodes are weak or unsupported with less than 70% of bipartitions across bootstrap replicates. The topology is arbitrarily rooted for visualization. Solid lines represent dwarf and mouse lemur V1Rs (or branches subtending clades of dwarf and mouse lemur V1Rs). Dashed lines represent V1R lineages that are not within Cheirogaleidae.
Subfamily membership varies among strepsirrhines (fig. 3A and B). Although O.garnettii contains a very diverse repertoire, it lacks subfamily VII and IX membership. The diurnal lemurs lack receptors belonging to a few subfamilies, most consistently IV, VIII, and IX. The basal lineage within the lemuriform radiation, Daubentonia madagascariensis, lacks membership for most subfamilies, including Strep/I, II, IV, V, and VII. Subfamily Strep/I, referred to as “V1Rstrep” in Yoder et al. (2014), is used synonymously here for distinction from the mouse subfamily “I.” The repertoires of cheirogaleids are highly enriched for subfamily III, V, and IX membership, whereas the diurnal lemurs are enriched for subfamilies Strep/I, II, and III. In haplorrhine primates, repertoires contain only one or a few subfamilies. Ancestral state reconstruction with asymmetric parsimony (Csűrös and Miklos 2009; Csűrös 2010) revealed that the stem primate possessed only a subset of now extant V1R subfamilies, Strep/I, II, III, IV, and VIII (fig. 3B). Subfamily IX has undergone a notable expansion in Cheirogaleidae, but the aye-aye repertoire also contains members from subfamily IX; thus, subfamily IX is the only subfamily exclusive to nocturnal strepsirrhines, despite its absence in O.garnettii (fig. 3A and B). Our results suggest that both the ancestral primate and the ancestral lemur had repertoires more limited in size and diversity than many living strepsirrhine primates, further supporting the controversial hypothesis that the ancestral primate was diurnal rather than nocturnal (Tan et al. 2005; Borges et al. 2018).
Copy Number Variation in Intraspecific M. murinus Repertoires
We resequenced eight M. murinus individuals of known pedigree from the colony at the Duke Lemur Center in Durham, NC. Using these genomes, we estimated intraspecific variation in V1R repertoire size (fig. 4). For the eight resequenced M. murinus individuals, we observed low levels of intraspecific V1R repertoire size variation relative to size variation between taxonomic families with individual repertoires ranging from 86 to 105 intact V1R loci. Though one might expect that levels of intraspecific variation in V1R repertoire size in a captive population may be reduced relative to wild populations of M. murinus, the colony at the Duke Lemur Center is known from the historical records of its parent colony (Muséum National d'Histoire Naturelle, UMR 7179 CNRS/MNHN, France; Agreement DDPP No. D91-114-1; Brunoy, France) to be comprised descendants from interbreeding of two distinct evolutionary lineages, M. murinus and Microcebusganzhorni (Larsen et al. 2017), presently recognized as distinct species (Hotaling et al. 2016). Therefore, the intraspecific variation observed here may actually be exaggerated, rather than reduced, which increases our confidence in the robustness of repertoire size estimates among species through sampling of single individuals. To test for the potentially confounding effects of sequencing and assembly error, one individual, DLC7033, was sequenced twice as a technical replicate. The duplicate genome assemblies respectively contained 92 or 96 intact loci indicating that sequencing and assembly error likely play a measurable role in generating variation among observed repertoire counts, though the effect appears to be modest. Thus, taking the results of the pedigree analysis as largely accurate, this emphasizes the highly dynamic nature of V1R repertoire size evolution, even over generational timescales.
Complex History of Diversifying Positive Selection in the Dwarf and Mouse Lemurs
Our results agree with previous studies in finding that selection has acted pervasively across the V1R gene family over time (Hohenbrink et al. 2012). Pervasive positive selection was revealed for all subfamilies identified in this study, even when analyzing the genus Microcebus alone (supplementary tables S4 and S5, Supplementary Material online) and additional genome sequences for dwarf and mouse lemurs have likely increased the power of the sites tests. For example, positive selection was not evident for subfamily VII in a previous study limited to only M.murinus (Hohenbrink et al. 2012). Furthermore, some subfamilies have unique profiles of sites under selection (fig. 7). Although lineage-specific rate variation is a confounding factor in V1R gene family evolution (Yoder et al. 2014), our analyses, spanning a range of taxon sampling schemes, show that our ability to characterize the V1R selection profiles are robust to such rate variation (fig. 7). We caution, however, against interpreting detected signatures of positive selection as being specific to any particular lineage produced by speciation or duplication. Given the complex duplication and loss history of the V1R gene family, we have used a combination of branch and branch-site tests to differentiate how selection has acted, and for which domains of V1R genes, at the subfamily level.
Fig. 7.
—Sites under selection across the V1R alignment. Subfamilies are given at the top along with numbers that correspond to taxonomic filters. The first aligned codon starts at the top and the aligned codon position 588 at the bottom, with boundaries of TMs to the right. Sites under selection are colored. Missing columns means that the filter was redundant. Numbers along the bottom are counts of sites under selection detected by both model comparisons and their overlap. The boundaries of loop (L) domains and TMs are shown along the aligned V1R repertoire. A PCA of sites under selection treated all codons as a binary character, determined by whether the site was under selection or not. Circles are 95% CIs for centroids of subfamily variation by taxonomic filters.
We performed two different model comparisons to differentiate between hypotheses of neutrality versus selection, and for the latter, for differentiating between the effects of rate constancy versus heterogeneity among sites. The M7 and M8 model comparisons always recovered more sites under selection than the more conserved M1a and M2a comparisons, but individual sites under selection detected by Bayes empirical Bayes with M2a were subsets of those detected by M8. Both model comparisons use LRTs to detect positive selection and assume dN/dS is constant across branches, but the M2a and M1a comparison (Zhang et al. 2005) uses three finite mixtures of dN/dS whereas the M8 and M7 comparison (Yang et al. 2000) accounts for heterogeneity in dN/dS among nearly neutral sites with a beta distribution. Tests of pervasive positive selection were also performed on data realigned by subfamily, and similar estimates of proportions of sites under positive selection suggested that our site models were not misled by alignment errors (supplementary tables S6 and S7, Supplementary Material online). Most individual sites under positive selection are unique to different subfamilies (supplementary fig. S3, Supplementary Material online) and reflect biases in selective pressures across different loop and TMs (supplementary fig. S4, Supplementary Material online). However, some selection profiles were more differentiated than others, such as Strep/I, II, V, and IX (fig. 7). The divergent selection profiles among subfamilies lead us to interpret positive selection acting on V1R genes in primates to be largely diversifying. Differentiated selection profiles among subfamilies are largely explained by biases among loop domains, and to a lesser extent, TMs (supplementary material; supplementary fig. S5 and tables S8–S10, Supplementary Material online).
Previous studies have indicated that extracellular loops have been primary targets of positive selection in V1Rs (Hohenbrink et al. 2012), and our results agree with these findings. Positive selection acting on extracellular loops two and three from Hohenbrink et al. (2012), identified here as loops three and five, is evident (fig. 7) and enriched in subfamilies II and VI, and subfamilies II, IV, V, and VI, respectively (supplementary fig. S5, Supplementary Material online). These specific domains are probable regions where V1Rs bind to semiochemicals (Hohenbrink et al. 2012). The exposed loop domains have been the primary targets of diversifying selection, and our results show limited evidence for selection acting on the TMs (supplementary material; supplementary fig. S5 and table S8, Supplementary Material online). For example, there are an enriched number of sites under positive selection in TM four, but an enrichment of sites was sensitive to the taxa included in analyses. TMs have likely been subject to strong purifying selection. Notably TM three, which has been previously predicted to form the ligand binding pocket of V1Rs (Kobilka et al. 1988; Pilpel and Lancet 1999; Palczewski et al. 2000; Yoder et al. 2014), shows a depletion of sites under positive selection in subfamilies I and IX (supplementary fig. S5 and table S8, Supplementary Material online). We hypothesize that pervasive diversifying positive selection has accompanied selection for divergent function among V1R subfamilies by affecting substrate affinity of the ligand binding pocket among V1R paralogs, although experimental evidence is needed to associate patterns of molecular evolution with function.
Branch-site models detected evidence of episodic positive selection in the evolution of all evaluated V1R subfamilies except for lemur VIII (supplementary fig. S6 and table S11, Supplementary Material online). Tests of episodic positive selection across the V1R subfamilies in the house mouse have been of little interest (Karn et al. 2010) and our tests of episodic selection here are generally not associated with the expansion of V1Rs in dwarf and mouse lemurs. Instead, signatures of episodic positive selection are detectable only on a few terminal branches with the exception of subfamilies VII and IX. Subfamily IX would be a candidate for further investigation, given that it showed notable levels of episodic positive selection on a deep internal branch and is the only subfamily specific to nocturnal strepsirrhines. Sites identified to be under positive selection by empirical Bayes largely corresponded to TM seven and loop eight, and also mapped to the previously identified ligand binding domains (supplementary fig. S6 and table S12, Supplementary Material online). Exploration of alternative topologies revealed that branches showing episodic positive selection were likely not due to topological errors (supplementary material; supplementary fig. S6, Supplementary Material online).
Comparative Evolution of V1R Repertoires and Genome Architecture across Mammalia
Here, we take advantage of the recently published chromosome-level assembly for M. murinus and other chromosome-level mammalian assemblies in an effort to identify genomic features that are generally associated with V1R repertoire expansion and to investigate the genetic architecture of V1R genes. The molecular environment of V1Rs is predicted to play a role in their regulation and has previously been studied only in mouse, rat, and pig (Lane et al. 2002; Stewart and Lane 2007; Kambere and Lane 2009; Michaloski et al. 2011; Dinka and Le 2018).
Kambere and Lane (2009) hypothesized that the double-stranded DNA breaks created when LINE elements insert into novel locations in the genome promote segmental duplication of repetitive regions of the genome during the DNA repair process. This provides an opportunity for repetitive gene families, such as V1Rs, to increase in copy number with increased LINE activity. Previous studies have found mouse V1R-containing regions to be enriched for LINE elements, and they predict that enrichment of LINE elements is associated with expansion of V1Rs in mammals (Kambere and Lane 2007). We compared the expanded V1R repertoires of mouse and mouse lemur with the putatively contracted V1R repertoires of horse, cow, dog, and human, and find that the expanded V1R repertoires contain more LINE elements than the contracted repertoires, supporting the hypothesis that increased LINE element dynamics may have supported V1R repertoire expansion in mammals (supplementary fig. S7, Supplementary Material online).
We also utilized chromosome-level mammalian genome assemblies to assess the genomic architecture of V1R genes. We find that mouse lemur V1Rs primarily cluster by subfamily at chromosomal locations across the genome as is also characteristic of the V1R repertoire in mouse (figs. 1 and 2B; Kambere and Lane 2007). Only mouse lemur subfamily VIII does not form a cluster but is instead uniquely dispersed across three different chromosomes (figs. 1 and 2B). We also analyzed the locations of all regions demonstrating V1R homology to determine if there are any potential pseudogenized subfamilies or clusters in the genome and found no evidence for pseudogenized clusters of V1Rs in mouse lemur (supplementary fig. S12, Supplementary Material online). Both LINE enrichment and physical clustering of V1R loci have been predicted to be associated with proper regulation of V1Rs (Lane et al. 2002; Kambere and Lane 2007) and may be characteristic of expanded V1R repertoires in general.
To investigate whether homologous subfamilies have retained chromosomal synteny in species with expanded repertoires and across mammals broadly, we evaluated chromosomal synteny for each species relative to mouse using the SynChro software (Drillon et al. 2014; figs. 1 and 2A and B, supplementary figs. S8–S11, Supplementary Material online). In mouse and mouse lemur, most homologous V1R subfamilies retain chromosomal synteny (figs. 6 and 7A and B). Mouse subfamily D is most closely related to mouse lemur subfamily IV on mouse lemur chromosome 22, and both subfamilies share mouse chromosome 7 synteny. Mouse subfamilies G and J/K are most closely related to mouse lemur subfamily V on mouse lemur chromosome 10 and mouse lemur family Strep/I on chromosome 22; these subfamilies all share mouse chromosome 7 synteny. Lemur subfamily III on chromosome 22 is syntenic with mouse E and F on mouse chromosomes 7 and 17. Lemur subfamilies VI and VII on mouse lemur chromosome 14 are syntenic with mouse subfamilies H and I on chromosome 13. Lemur subfamilies not sharing synteny with any mouse subfamily include subfamilies II, VIII, and IX. The expanded subfamilies in Cheirogaleidae, IV, VII, and IX, all map to different chromosomal regions of the M. murinus genome and were not linked on an ancestral syntenic block based on comparisons between M. murinus and mouse. Therefore, subfamily expansions have occurred independently and not as tandem duplications of a single genomic region.
When comparing all mammalian species examined, our results reveal that in each species, one chromosome contains a very dense block of highly homologous subfamilies on a backbone of mouse chromosome 7 synteny, referred to here as a “V1R hotspot” (fig. 2B). The V1R hotspot usually contains receptors of the EF/III, D/IV, JK/V, and G/Strep subfamilies, and cluster order is maintained with a few species-specific subfamily deletions. The chromosomal synteny of the hotspot is rarely interrupted, and if interrupted, it is almost exclusively interrupted by a stretch of synteny from another mouse chromosome containing V1Rs. These interleaving regions across mammalian hotspots are usually from chromosome 13 or 17, indicating that genomic regions where V1Rs cluster are also subject to increased gene duplication rates or survival of duplicate genes. Interestingly, the putatively intact member of the contracted human V1R repertoire, VN1R4, is also contained within this “hotspot” location and shares homology with hotspot subfamilies, specifically Lemur III and Mus E/F.
Previous studies of Laurasiatheria have predicted that the V1R repertoires of cow, horse, and dog consist mostly of highly orthologous loci with evolutionarily conserved functions (Yohe et al. 2019). Although conserved function remains to be shown experimentally, retained synteny of these Laurasiatherian V1Rs within hotspots across Mammalia supports the hypothesized ancient origin of these subfamilies and reinforces the idea that V1Rs in these subfamilies are orthologous in function (Ohara et al. 2009; Yohe et al. 2019). Mouse lemur V1Rs show striking structural similarities to the functionally diverse repertoire of mouse. Considering the independent gains in copy number and novel subfamily evolution, coupled with variable rates of molecular evolution and selective pressures, V1Rs in mouse lemurs may serve as an ideal system for elucidating pheromone evolution in primates. Similar patterns of deep synteny have been described for ∼80 Myr of odorant receptor evolution in bees (Brand and Ramírez 2017). Considered in this context, our results suggest that chemosensory gene family evolution may follow similar molecular “rules” in organisms with vastly different natural histories, even when evolved independently from different ancestral gene families, as would be the case comparing mammals to insects.
Conclusions
We revealed that an expansion of the V1R gene family is shared across the dwarf and mouse lemurs, and that duplicate V1R gene copies have been evolving under strong selective pressures. Divergent patterns of molecular evolution among V1R subfamilies and diversity in subfamily membership and abundance suggests that V1Rs may serve as a test case for studying the evolution of sensory drive in primates. Pheromone detection among nocturnal primates, especially the morphologically cryptic mouse lemurs, may be more important for driving and maintaining species boundaries than previously appreciated. Syntenic analyses with improved genomic resources revealed strikingly similar genetic architecture between the expanded V1R repertoires of mouse and mouse lemur, and that some V1R subfamilies have been maintained in a V1R “hotspot” across ∼184 Myr of mammalian evolution (dos Reis et al. 2012). Characterizing additional features of V1R hotspots across species will be important for future studies translating experimental genetic studies in mice to primates such as mouse lemur.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Malagasy authorities for permission to conduct this research and Duke Lemur Center staff, especially Erin Ehmke, Bobby Schopler, and Cathy Williams, for providing the Microcebus murinus and Mirza zaza tissue samples. We are grateful to our colleagues at Baylor College of Medicine, Jeff Rogers and Kim Worley, for many insightful discussions of mouse lemur genomics. Phillip Brand, Jeff Thorne, and two anonymous reviewers provided critical reviews of the manuscript leading to its significant improvement. We thank Simon Gregory’s lab for preparing the 10× Genomics libraries. We are grateful for the support of Duke Research Computing and the Duke Data Commons (NIH 1S10OD018164-01) and appreciate the donation of complimentary sequencing for Microcebus ravelobensis provided by the Duke GCB Sequencing Core. A.D.Y. gratefully acknowledges support from the John Simon Guggenheim Foundation and the Alexander von Humboldt Foundation during the writing phase of this project. The study was funded by the DTRI pilot program, NIH (R01 DC014423 to H.M.), NIH NRSA (5F31DC017394 to K.Z.), NSF (DEB-1354610 to A.D.Y. and D.W.W.), and Duke University startup funds (to A.D.Y.). This is Duke Lemur Center publication no. 1441.
Data deposition: This project has been deposited at NCBI BioProject under the accession number PRJNA512515. Newly sequenced genome data have been made available through NCBI BioProject PRJNA512515. Complete record information is given in supplementary material (supplementary table S15, Supplementary Material online). Genome assemblies and annotations along with ML trees and alignments are available at the Dryad digital repository (https://doi.org/10.5061/dryad.bzkh1894d).
Literature Cited
- Adipietro KA, Matsunami H, Zhuang H. 2012. Functional evolution of primate odorant receptors In: Hirai H, Imai H, Go Y, editors. Post-genome biology of primates. Tokyo: Springer Tokyo; p. 63–78. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Yang Z. 2007. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol Biol Evol. 24(5):1219–1228. [DOI] [PubMed] [Google Scholar]
- Bear DM, Lassance J-M, Hoekstra HE, Datta SR. 2016. The evolving neural and genetic architecture of vertebrate olfaction. Curr Biol. 26(20):R1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300. [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Boillat M. 2015. The vomeronasal system mediates sick conspecific avoidance. Curr Biol. 25(2):251–255. [DOI] [PubMed] [Google Scholar]
- Borges R, et al. 2018. Adaptive genomic evolution of opsins reveals that early mammals flourished in nocturnal environments. BMC Genomics 19(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschat C, et al. 2002. Pheromone detection mediated by a V1R vomeronasal receptor. Nat Neurosci. 5(12):1261–1262. [DOI] [PubMed] [Google Scholar]
- Boughman JW. 2002. How sensory drive can promote speciation. Trends Ecol Evol. 17(12):571–577. [Google Scholar]
- Brand P, Ramírez SR. 2017. The evolutionary dynamics of the odorant receptor gene family in corbiculate bees. Genome Biol Evol. 9(8):2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, et al. 2019. An olfactory receptor gene underlies reproductive isolation in perfume-collecting orchid bees. bioRxiv, doi: 10.1101/537423. [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinform. 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F-C, Chen C-J, Chuang T-J, Li W-H. 2010. Gene family size conservation is a good indicator of evolutionary rates. Mol Biol Evol. 27(8):1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung AG, Belone PM, Bímová BV, Karn RC, Laukaitis CM. 2017. Studies of an Androgen-binding protein knockout corroborate a role for salivary ABP in mouse communication. Genetics 205(4):1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csűrös M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. [DOI] [PubMed] [Google Scholar]
- Csűrös M, Miklos I. 2009. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 26:2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Xiong Y, Dai X. 2014. Neighboring genes show interchromosomal colocalization after their separation. Mol Biol Evol. 31(5):1166–1172. [DOI] [PubMed] [Google Scholar]
- Del Punta K, et al. 2002. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature 419(6902):70–74. [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454(7205):762–765. [DOI] [PubMed] [Google Scholar]
- Dinka H, Le MT. 2018. Analysis of pig vomeronasal receptor type 1 (V1R) promoter region reveals a common promoter motif but poor CpG islands. Anim Biotechnol. 29(4):293–300. [DOI] [PubMed] [Google Scholar]
- Dopman EB, Robbins PS, Seaman A. 2009. Components of reproductive isolation between North American pheromone strains of the European corn borer. Evolution 64(4):881–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, et al. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc R Soc B 279(1742):3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, et al. 2018. Using phylogenomic data to explore the effects of relaxed clocks and calibration strategies on divergence time estimation: primates as a test case. Syst Biol. 67(4):594–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea CM. 2015. D’scent of man: a comparative survey of primate chemosignaling in relation to sex. Horm Behav. 68:117–133. [DOI] [PubMed] [Google Scholar]
- Drillon G, Carbone A, Fischer G. 2014. SynChro: a fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One 9(3):e92621.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Axel R. 1995. A novel family of genes encoding putative pheromone receptors in mammals. Cell 83(2):195–206. [DOI] [PubMed] [Google Scholar]
- Fu X, et al. 2015. A molecular code for identity in the vomeronasal system. Cell 163(2):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschanski K, Warnefors M, Kaessmann H. 2017. The evolution of duplicate gene expression in mammalian organs. Genome Res. 27(9):1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga-Yamanaka S, et al. 2014. Integrated action of pheromone signals in promoting courtship behavior in male mice. eLife 3:e03025.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins MTR, et al. 2018. Genome sequence and population declines in the critically endangered greater bamboo lemur (Prolemur simus) and implications for conservation. BMC Genomics 19(1):445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenbrink P, Dempewolf S, Zimmermann E, Mundy NI, Radespiel U. 2014. Functional promiscuity in a mammalian chemosensory system: extensive expression of vomeronasal receptors in the main olfactory epithelium of mouse lemurs. Front Neuroanat. 8:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenbrink P, Mundy NI, Radespiel U. 2017. Population genetics of mouse lemur vomeronasal receptors: current versus past selection and demographic inference. BMC Evol Biol. 17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenbrink P, Radespiel U, Mundy NI. 2012. Pervasive and ongoing positive selection in the vomeronasal-1 receptor (V1R) repertoire of mouse lemurs. Mol Biol Evol. 29(12):3807–3816. [DOI] [PubMed] [Google Scholar]
- Hotaling S, et al. 2016. Species discovery and validation in a cryptic radiation of endangered primates: coalescent-based species delimitation in Madagascar’s mouse lemurs. Mol Ecol. 25(9):2029–2045. [DOI] [PubMed] [Google Scholar]
- Hughes GM, et al. 2018. The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol Biol Evol. 35(6):1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst JL, et al. 2017. Molecular heterogeneity in major urinary proteins of Mus musculus subspecies: potential candidates involved in speciation. Sci Rep. 7(1):44992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y, et al. 2011. Molecular organization of vomeronasal chemoreception. Nature 478(7368):241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambere MB, Lane RP. 2007. Co-regulation of a large and rapidly evolving repertoire of odorant receptor genes. BMC Neurosci. 8(S3):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambere MB, Lane RP. 2009. Exceptional LINE density at V1R loci: the Lyon repeat hypothesis revisited on autosomes. J Mol Evol. 68(2):145–159. [DOI] [PubMed] [Google Scholar]
- Karn RC, Young JM, Laukaitis CM. 2010. A candidate subspecies discrimination system involving a vomeronasal receptor gene with different alleles fixed in M. m. domesticus and M. m. musculus. PLoS One 5(9):e12638.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller TE, Yi SV. 2014. DNA methylation and evolution of duplicate genes. Proc Natl Acad Sci U S A. 111(16):5932–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobilka BK, et al. 1988. Chimeric alpha 2-, beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science 240(4857):1310–1316. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer E. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kurzweil VC, Getman M, Comparative Sequencing Program N, Green ED, Lane RP. 2009. Dynamic evolution of V1R putative pheromone receptors between Mus musculus and Mus spretus. BMC Genomics 10(1):74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane RP, Cutforth T, Axel R, Hood L, Trask BJ. 2002. Sequence analysis of mouse vomeronasal receptor gene clusters reveals common promoter motifs and a history of recent expansion. Proc Natl Acad Sci U S A. 99(1):291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PA, et al. 2017. Hybrid de novo genome assembly and centromere characterization of the gray mouse lemur (Microcebus murinus). BMC Biol. 15(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PA, Heilman AM, Yoder AD. 2014. The utility of PacBio circular consensus sequencing for characterizing complex gene families in non-model organisms. BMC Genomics 15(1):720.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei R, et al. 2014. Revision of Madagascar’s dwarf lemurs (Cheirogaleidae: Cheirogaleus): designation of species, candidate species status and geographic boundaries based on molecular and morphological data. Primate Conserv. 28(1):9–35. [Google Scholar]
- Leinders-Zufall T, et al. 2000. Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature 405(6788):792–796. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. CD-HIT: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Loire E, et al. 2017. Do changes in gene expression contribute to sexual isolation and reinforcement in the house mouse? Mol Ecol. 26(19):5189–5202. [DOI] [PubMed] [Google Scholar]
- Luo M, Fee MS, Katz LC. 2003. Encoding pheromonal signals in the accessory olfactory bulb of behaving mice. Science 299(5610):1196–1201. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155. [DOI] [PubMed] [Google Scholar]
- Makova KD, Li W-H. 2003. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 13(7):1638–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melén K, Krogh A, von Heijne G. 2003. Reliability measures for membrane protein topology prediction algorithms. J Mol Biol. 327(3):735–744. [DOI] [PubMed] [Google Scholar]
- Mendelson TC. 2003. Evidence of intermediate and asymmetrical behavioral isolation between orangethroat and orangebelly darters (Teleostei: Percidae). Am Midl Nat. 150(2):343–348. [Google Scholar]
- Mendes FK, Hahn MW. 2016. Gene tree discordance causes apparent substitution rate variation. Syst Biol. 65(4):711–721. [DOI] [PubMed] [Google Scholar]
- Meyer WK, et al. 2015. Evolutionary history inferred from the de novo assembly of a nonmodel organism, the blue-eyed black lemur. Mol Ecol. 24(17):4392–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski JS, et al. 2011. Common promoter elements in odorant and vomeronasal receptor genes. PLoS One 6(12):e29065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya-Ito K, Hayakawa T, Suzuki H, Hagino-Yamagishi K, Nikaido M. 2018. Evolution of vomeronasal receptor 1 (V1R) genes in the common marmoset (Callithrix jacchus). Gene 642:343–353. [DOI] [PubMed] [Google Scholar]
- Mundy NI. 2003. Positive selection during the diversification of class I vomeronasal receptor-like (V1RL) genes, putative pheromone receptor genes, in human and primate evolution. Mol Biol Evol. 20(11):1805–1810. [DOI] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T. 1997. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci U S A. 94(15):7799.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido M, et al. 2014. Multiple episodic evolution events in V1R receptor genes of East-African cichlids. Genome Biol Evol. 6(5):1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Kawahara Y, Nei M. 2007. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci U S A. 104(51):20421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara H, et al. 2009. Conserved repertoire of orthologous vomeronasal type 1 receptor genes in ruminant species. BMC Evol Biol. 9(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ÁN, Hurst LD, McLysaght A. 2018. Faster evolving primate genes are more likely to duplicate. Mol Biol Evol. 35:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, et al. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289(5480):739–745. [DOI] [PubMed] [Google Scholar]
- Papes F, Logan DW, Stowers L. 2010. The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell 141(4):692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Albà MM. 2013. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol. 30(8):1830–1842. [DOI] [PubMed] [Google Scholar]
- Perret M. 1996. Manipulation of sex ratio at birth by urinary cues in a prosimian primate. Behav Ecol Sociobiol. 38(4):259–266. [Google Scholar]
- Perret M, Colas S. 1997. Manipulation of sex ratio at birth and maternal investment in female mouse lemurs (Microcebus murinus, Primates). Appl Anim Behav Sci. 51(3–4):275–283. [Google Scholar]
- Perry GH, et al. 2012. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 4(2):126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson GI, Masel J. 2009. Quantitative prediction of molecular clock and Ka/Ks at short timescales. Mol Biol Evol. 26(11):2595–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Lancet D. 1999. The variable and conserved interfaces of modeled olfactory receptor proteins. Protein Sci. 8(5):969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis M. D, Yang Z. 2011. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 28(7):2161–2172. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/, last accessed November 13, 2018. [Google Scholar]
- Schmitz J, et al. 2016. Genome sequence of the basal haplorrhine primate Tarsius syrichta reveals unusual insertions. Nat Commun. 7(1):12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servedio MR, Boughman JW. 2017. The role of sexual selection in local adaptation and speciation. Annu Rev Ecol Evol Syst. 48(1):85–109. [Google Scholar]
- Servedio MR, Noor M. 2003. The role of reinforcement in speciation: theory and data. Annu Rev Ecol Evol Syst. 34(1):339–364. [Google Scholar]
- Shimodaira H. 2004. Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann Stat. 32(6):2616–2641. [Google Scholar]
- Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smadja CM, et al. 2015. Seeking signatures of reinforcement at the genetic level: a hitchhiking mapping and candidate gene approach in the house mouse. Mol Ecol. 24(16):4222–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling EJ, Richard AF. 1995. Social organization in the aye-aye (Daubentonia madagascariensis) and the perceived distinctiveness of nocturnal primates In: Alterman L, Doyle GA, Izard MK, editors. Creatures of the dark: the nocturnal prosimians. Boston (MA: ): Springer US; p. 439–451. [Google Scholar]
- Stewart R, Lane RP. 2007. V1R promoters are well conserved and exhibit common putative regulatory motifs. BMC Genomics 8(1):253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sündermann D, Scheumann M, Zimmermann E. 2008. Olfactory predator recognition in predator-naïve gray mouse lemurs (Microcebus murinus). J Comp Psychol. 122(2):146–155. [DOI] [PubMed] [Google Scholar]
- Tan Y, Yoder AD, Yamashita N, Li W-H. 2005. Evidence from opsin genes rejects nocturnality in ancestral primates. Proc Natl Acad Sci U S A. 102(41):14712–14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialle RA, Tamuri AU, Goldman N. 2018. Alignment modulates ancestral sequence reconstruction accuracy Thorne J, editor. Mol Biol Evol. 35(7):1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Shi P, Zhu Z, Zhang Y.-P. 2010. More functional V1R genes occur in nest-living and nocturnal terricolous mammals. Genome Biol Evol. 2(0):277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. 2017. Direct determination of diploid genome sequences. Genome Res. 27(5):757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R, et al. 2019. Conservation genomic analysis reveals ancient introgression and declining levels of genetic diversity in Madagascar’s hibernating dwarf lemurs. Heredity, doi: 10.1038/s41437-019-0260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Yang Q-Y, Wang X-T, McLysaght A, Zhang H-Y. 2016. Spatial colocalization of human ohnolog pairs acts to maintain dosage-balance. Mol Biol Evol. 33(9):2368–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Gaut BS. 2011. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol. 28(8):2359–2369. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen A-M. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155(1):431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22(4):1107–1118. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yoder AD. 2003. Comparison of likelihood and Bayesian methods for estimating divergence times using multiple gene loci and calibration points, with application to a radiation of cute-looking mouse lemur species. Syst Biol. 52(5):705–716. [DOI] [PubMed] [Google Scholar]
- Yoder AD, et al. 2014. Molecular evolutionary characterization of a V1R subfamily unique to strepsirrhine primates. Genome Biol Evol. 6(1):213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Larsen PA. 2014. The molecular evolutionary dynamics of the vomeronasal receptor (class 1) genes in primates: a gene family on the verge of a functional breakdown. Front Neuroanat. 8:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Weisrock DW, Rasoloarison RM, Kappeler PM. 2016. Cheirogaleid diversity and evolution: big questions about small primates In: Lehman SM, Radespiel U, Zimmermann E, editors. The dwarf and mouse lemurs of Madagascar. Cambridge: Cambridge University Press; p. 3–20. [Google Scholar]
- Yohe LR, Brand P. 2018. Evolutionary ecology of chemosensation and its role in sensory drive. Curr Zool. 64(4):525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe LR, Davies KT, Rossiter SJ, Davalos L. 2019. Expressed vomeronasal type-1 receptors (V1Rs) in bats uncover conserved mechanisms of social chemical signaling. Genome Biol Evol. 11(10):2741–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Massa HF, Hsu L, Trask BJ. 2010. Extreme variability among mammalian V1R gene families. Genome Res. 20(1):10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 8(1):28–36. [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 95(7):3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, et al. 2013. The MaSuRCA genome assembler. Bioinformatics 29(21):2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.