The citrullinome of P. gingivalis outer membrane vesicles (OMV) has been explored by a novel two-dimensional separation system combined with high resolution mass spectrometry and in-house build software. Analysis of OMVs from wild-type and two PPAD mutants resulted in confident discrimination based on citrullinated peptides. In the wild-type citrullinome 78 proteins were identified having a total of 161 validated citrullination sites. A single citrullination was identified in the C351A mutant and none in the ΔPPAD mutant.
Keywords: Chromatography, protein identification, protein modification, post-translational modifications, omics, bacteria, bioinformatics software, HPLC, HFBA, orthogonal, two-dimensional
Graphical Abstract
Highlights
Novel two-dimensional separation system for identification of citrullinated peptides.
Dedicated software developed for confident validation of citrullination.
P. gingivalis citrullinome: 78 proteins with a total of 161 citrullinated peptides.
Confident discrimination of P. gingivalis OMVs from wild-type and PPAD mutants.
Abstract
Porphyromonas gingivalis is a key pathogen in chronic periodontitis and has recently been mechanistically linked to the development of rheumatoid arthritis via the activity of peptidyl arginine deiminase generating citrullinated epitopes in the periodontium. In this project the outer membrane vesicles (OMV) from P. gingivalis W83 wild-type (WT), a W83 knock-out mutant of peptidyl arginine deiminase (ΔPPAD), and a mutant strain expressing PPAD with the active site cysteine mutated to alanine (C351A), have been analyzed using a two-dimensional HFBA-based separation system combined with LC-MS. For optimal and positive identification and validation of citrullinated peptides and proteins, high resolution mass spectrometers and strict MS search criteria were utilized. This may have compromised the total number of identified citrullinations but increased the confidence of the validation. A new two-dimensional separation system proved to increase the strength of validation, and along with the use of an in-house build program, Citrullia, we establish a fast and easy semi-automatic (manual) validation of citrullinated peptides. For the WT OMV we identified 78 citrullinated proteins having a total of 161 citrullination sites. Notably, in keeping with the mechanism of OMV formation, the majority (51 out of 78) of citrullinated proteins were predicted to be exported via the inner membrane and to reside in the periplasm or being translocated to the bacterial surface. Citrullinated surface proteins may contribute to the pathogenesis of rheumatoid arthritis. For the C351A-OMV a single citrullination site was found and no citrullinations were identified for the ΔPPAD-OMV, thus validating the unbiased character of our method of citrullinated peptide identification.
Citrullination is a deimination of arginine, which results in the loss of a single nitrogen and hydrogen along with the addition of an oxygen, resulting in a mass shift of 0.984 Da and loss of a single charge. Citrullination is a post-translational modification that can only occur on arginine residues, either on the N- or C-terminal of the peptides or internally.
Citrullination occurs in physiological and pathological conditions and is thought to play a range of different functions. The human peptidyl arginine deiminases (PAD)1 1, 2, 3, 4, and 6 exert different roles as a result of expression in different cellular environments. PAD1 citrullinates keratin and filaggrin, which is important for terminal differentiation of keratinocytes (1, 2). PAD2 citrullinates myelin basic protein, which is involved in the myelin sheath formation (3, 4). PAD3 is involved in hair growth by citrullination of trichohyalin (5, 6). PAD4 has been assigned more adverse functions, from regulation of gene-expression (7–11), through immune modulation (12), to auto-citrullination for regulation of citrullinated protein (13). Finally, the PAD6 function is not completely understood because of lack of substrate, but it is thought to have a role in reproduction (14).
Only few species of bacteria belonging to genus Porphyromonas have been found to express peptidyl arginine citrullinating enzyme closely related on the amino acid sequence level, if not identical. (15) The best characterized is Porphyromonas gingivalis peptidyl arginine deiminase (PPAD). The sequence identity between PPAD and the human PADs is low, ∼30% (16), however, similar activity is observed. There are some differences, however, and although human PADs activity is dependent on calcium and targets internal arginine residues, PPAD preferentially citrullinates C-terminal arginine in a calcium-independent manner (17). Apart from the preference for C-terminal arginine citrullination, specificity with respect to preceding residues has not been identified for PPAD. Conversely a study on PAD2 and PAD4 showed very broad specificity with PAD2 favoring Tyr in the +3 position (Assohou-Luy et al. (18)).
Citrullinations have been found to contribute to the pathogenicity of various diseases including rheumatoid arthritis (RA) and periodontitis. P. gingivalis, although absent or at the low level in dental biofilm of periodontally healthy subjects, occurs in high numbers in the mouth of periodontitis patients and is thought to be one of the primary causes of periodontitis (19–21). Apart from PPAD P. gingivalis expresses Arg-specific gingipains, RgpA and RgpB (16), which are important enzymes for citrullination, as they generate peptides and protein fragments with C-terminal Arg, instant substrates for modification by PPAD (22). In line with concerted action of Rgps and PPAD the Rgp-null mutant shows very little citrullination compared with the parental P. gingivalis W83 strain. Nevertheless, PPAD can modify internal arginine residues (23, 24) but this reaction occurs at a rate thousand times slower than citrullination of C-terminal arginine (17). This is in accordance with the topology of the substrate binding site perfectly shaped to accommodate arginine at the C terminus with no room for an extended peptide chain (25).
Several findings implicate PPAD as an important virulence factor of P. gingivalis. Through citrullination of C-terminal residues in epidermal growth factor (24) and C5a anaphylatoxin (26) the enzyme can contribute to the periodontal tissue damage and attenuation of innate immune responses, respectively. Furthermore, P. gingivalis' citrullinome modulates neutrophil activity (27), constrains P. gingivalis biofilm development (28), affects epithelial cells transcriptome (29), contributes to formation of dual-species biofilm with the opportunistic fungus Candidia (30), and is responsible for stimulation of prostaglandin E2 (PGE2) secretion by gingival fibroblasts. (31) The latter activity can be directly linked to periodontitis and RA pathogenicity as PGE2 promotes bone resorption. Apart from that, C-terminally citrullinated peptides generated by concerted action of Rgp and PPAD are considered pivotal in breaking the immunotolerance leading to production of specific anti-citrullinated protein antibodies (ACPA) directly responsible for development of RA. (32) This theory fits well, with the findings of heightened levels of ACPAs in periodontitis and RA patients. (20, 21, 33–36) Likewise, it is supported by data from animal models of RA and P. gingivalis infection in which RA severity is dependent on PPAD expression. (32) In this context, determination of P. gingivalis citrullinome is very important but a challenging task.
The detection of citrullinations started with a color development reagent assay (COLDER assay) according to Clancy et al. (37), which depends on chemical derivatization of the urea group of citrulline (38). This method is mostly used in in vitro assays, because of its poor sensitivity and need of large amounts of citrullinations (37). Antibody based methods also depend on chemical derivatization, the first anti-citrulline assay was developed by Senshu et al. (39). These antibody-based assays have a major disadvantage as they have been found to cross-react with carbamylation, which is a chemical modification of lysine into homocitrulline (40). Currently the most promising technique for the identification of citrullinations regarding sensitivity and specificity is mass spectrometry (MS).
The development during the last decades within the MS field of protein research has made the investigation of clinical samples and whole-cell lysates possible, particularly when fractionated prior to injection. Further, less complex samples can be run directly, leading to significant reduction of manual work prior to analysis. The major problem with MS-based methods for the single Dalton mass shift is the possibility of misinterpretation of a deamination of asparagine or glutamine, picking a wrong isotope on the MS1 level, the loss of a single charge, giving rise to poor ionization and fragmentation, and small retention time shift. We addressed these problems in the present paper using heptafluorobutyric acid (HFBA) as the ion-pairing reagent during two-dimensional fractionation, as well as optimized mass spectrometric data acquisition, and development of specific software, Citrullia, designed for identification and validation of citrullinations.
EXPERIMENTAL PROCEDURES
Bacterial Fraction Preparation
Cultures of P. gingivalis strain W83 and its isogenic mutants; C351A (with a point mutation of the catalytic cysteine residue, C351A, in PPAD rendering the enzyme catalytically inactive) and ΔPPAD (with the ppad gene deleted) were maintained on TSB agar plates with 5% defibrinated sheep blood and supplements: yeast extract (5 mg/ml), l-cysteine (0.5 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml). Liquid cultures were inoculated from 5 to 6 days old plates to liquid TSB medium with supplements and cultured for 18–20 h at 37 °C in an anaerobic chamber. Cultures were then diluted to OD600 = 0.1 in fresh medium and cultured as before for 20–22 h. Aldrithiol-4 (1.5 mm) was added to cultures immediately after incubation. Cultures were then centrifuged (7500 rcf, 15 min, 4 °C), supernatant collected and filtered through 0.45 μm membrane filter. The filtrate was ultracentrifuged at 70,000 rcf for 2 h at 4 °C. The collected sediment encompassing OMV was washed and then suspended in PBS with 1 mm TLCK by gentle sonication. The concentration of protein was determined using the Bradford method with bovine albumin as a standard.
Sample Preparation
OMV were reduced by the addition of dithiothreitol (DTT) to a final concentration of 10 mm. The samples were then incubated for 30 min at 50–57 °C, followed by alkylation with iodoacetamide (IAA) added to a final concentration of 24 mm and incubation in the dark for 20 min. Excess IAA was removed by treatment with DTT and proteins in the sample were digested by overnight incubation at 37 °C with 2% w/w in-house methylated trypsin (41).
For complete analysis without fractionation, the samples were micro purified essentially as described, (42) dried down and resuspended in 0.1% formic acid (FA). For analysis by high performance liquid chromatography (HPLC) fractionation, each sample was dried down and resuspended in 0.05% heptafluorobutyric acid (HFBA) prior to off-line separation.
Off-line HPLC Separation
Samples of 100 μg in a volume of 50 μl 0.05% HFBA were injected into an Agilent 1260 Infinity HPLC system (Agilent Technologies, Santa Clara, CA) equipped with a Phenomenex Aeris™ 2.6 μm PEPTIDE XB-C18 100 Å, LC Column 150 × 2.1 mm (00F-4505-AN)(Phenomenex, Torrance, CA), running at a flow of 0.200 ml/min. Buffer A = 0.05% HFBA, buffer B = 0.05% HFBA and 90% acetonitrile.
The method used a loading time of 2 min with 5% phase B and separated with a gradient of 12.5% B/5 min; 50.0% B/30 min, 60% B/35 min, 90%B/35.5 min, 90% B/36.5 min, 5% B/37 min, followed by equilibration at 5% B for 13 min.
Mass Spectrometry
Samples were run on an EASY- nLC1000 Liquid Chromatography system (Thermo Fisher Scientific, Waltham, MA), using a 3 μm trap column (100 μm inner diameter, 5 μm Reprosilpur 120 C18, Dr. Maisch GmbH, Germany) and an 18 cm analytical column (75 μm inner diameter, 3 μm Reprosilpur 120 C18) coupled online to a Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). The methods applied on the mass spectrometer had the following settings in common: positive mode, an MS1 resolution of 120,000, AGC target of 3e6, maximum injection time of 100 ms, and a scan range of 300–1400 m/z. The common MS2 settings were: resolution of 30,000, AGC target of 1e6, isolation window of 0.8 m/z, and a fixed first mass of 110.0 m/z. Furthermore, peptides with charges ranging from +1 to +6 were included, whereas +7 and above were excluded along with isotopes.
For the duplicate experiment (30-min gradient) the following specific parameters were used: dynamic exclusion of 5 s, maximum injection time of 200 ms, loop count of 5. For the triplicate experiment (30-min gradient) the parameters were changed to: dynamic exclusion of 5 s, maximum injection time of 100 ms, loop count of 10. For the triplicate experiment with long 120-min gradient the parameters were: dynamic exclusion of 15 s, maximum injection time of 100 ms, loop count of 10.
Amino Acid Analysis
To determine protein amounts and composition, amino acid analysis (AAA) was applied essentially as described by Højrup 2015 (43). Samples of 2–4 μg protein were dried in small polypropylene tubes, lids were punctured, and they were placed in 25 ml glass vials along with 200 μl of 6N HCl, 0.1% phenol, 0.1% 2-thioglycolic acid and closed with a MinInert valve (VICI) after being covered by argon and evacuated to <1 mBar pressure. After overnight hydrolysis at 110 °C, samples were dried, re-dissolved, and analyzed on a BioChrom 30+ amino acid analyzer (BioChrom Ltd, Cambridge, UK) using recommended conditions.
Data Handling
Citrullination is the exchange of an amino group with oxygen resulting in a mass increase of 0.984 Da. This difference is readily identified by modern MS search engines, but as the mass difference is identical to the commonly occurring deamidation of asparagine and glutamine and can further be mistaken by wrongly picked isotope in the MS1 spectrum, we developed a program for extraction and display of spectra identified as potentially containing citrullinated residues by the search engine.
In order to improve the validation of citrullinations and enable manual validation within a reasonable timeframe, we developed a program called Citrullia in C# version 7.2 for Windows. In addition, the .NET framework v. 4.7.2 was used in Visual Studio 2019 integrated development environment and the Metro Modern UI (Dennis Magno, https://github.com/dennismagno/metroframework-modern-ui) was used for visual user interface elements. As search engine we chose the publicly available X! Tandem program (version 2017.2.1.4, www.thegpm.org) (44).
As we deemed it necessary also to validate that the correct parent ion isotope had been picked, we used a new mass file format based on the Mascot Generic File format (mgf) but extended with MS1 information. The new format, named mgx (Mascot Generic eXtended), has been developed by MassAI Bioinformatics (www.massai.dk) and extends the common mgf format with a series of new tags, most relating to MS1 level information. In contrast to mgf, mgx is a loss-less format where all peaks are retained on every MS-level. Like mgf, the new mgx format is an open, text-based file format, and the mgx conversion program MGF Filter can be freely downloaded from www.massai.dk/download.html. The mgx files can easily be separated in MS1 and MS2 information for generating the standard mgf format files suitable for standard search engines.
X! Tandem was called with the following parameters: Parent ion precision: 10 ppm, MS2 precision: 0.02 Da, enzyme specificity: Cleavage after Lys and Arg, no cleavage before Pro, number of missed cleavages: < = 2. Fixed modifications: carbamidomethyl [C], variable modifications: deamidation [N, Q], oxidation [M], and citrullination [R]. The database searched was downloaded on 01.10.2018 from UniProt (www.uniprot.org) and contained all sequences from the P. gingivalis strains W83 and ATCC. Both strains were included in the search, as neither strain contained a complete list of proteins. This resulted in a file containing a total of 4417 protein sequences. The inclusion of both strains may have lowered the score slightly because of the increased in size, but this was considered neglible. The identified sequences were mapped to the PG locus numbers by comparing to the P. gingivalis W83 protein database from the Comprehensive Microbial Resource Web site (cmr.jcvi.org, June 2008).
Experimental Design and Statistical Rationale
For the experiment involving identification of citrullinations from three similar strains, one HPLC fractionation was performed for each, followed by two technical replicates of each fraction. The three strains were: P. gingivalis strain W83 WT, its isogenic mutants; C351A, and ΔPPAD (with the ppad gene deleted). Two technical replicates were used for increased depth of analysis for the identification of the citrullinome of the three strains. Based on this analysis, for the detailed analysis of the citrullinome of the WT strain, a single HPLC fractionation and three technical triplicates were performed along with triplicate analysis of the unfractionated sample. The two citrullination negative strain (C351A and ΔPPAD) acted as negative controls and have been subjected to similar conditions throughout the experiment. No statistical method was employed as the current paper is focused on qualitative analysis of the citrullinome of P. gingivalis OMVs.
RESULTS
Citrullia
The citrullination of an arginine residue results in a mass change of +0.9840 Da. This change can be readily identified by modern mass spectrometers, but as pointed out by Küster and co-workers (45) identification is fraught with danger of misinterpretation. As we thus anticipated that all potential citrullinated peptides had to be manually validated, we developed a program, Citrullia, for fast identification and easy manual validation. The data needed is thus presented within a single window containing filename, retention time (RT), sequence, charge-state, e-value from the X! Tandem mass search, parent mass, accession number, table of determined ions, MS1, and MS2 spectra. It further provides information on the isotopic distribution of the MS1 peak, neutral losses, immonium ions, and the elution position for both the first- (HFBA) and the second-dimension (FA) separation. Furthermore, within this single window, each spectrum can be marked as validated. All the validated spectra are saved into a list, which can be extracted as one single list. This makes the validation a relatively straightforward process, as a researcher can quickly scroll through all potential candidates and mark them as validated or non-validated. Citrullia thereby ensures identification and semi-automatic validation of citrullinated peptides.
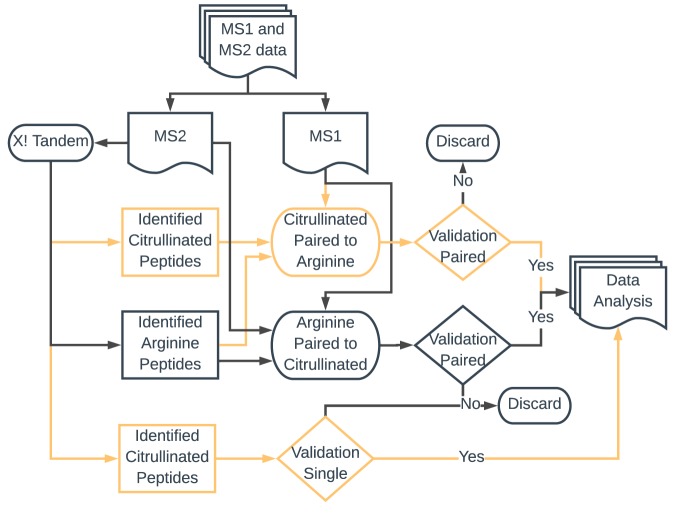
Raw files were first converted into mzXML using MS converter (part of the ProteoWizard package), peak picking was the only filter used, prior to conversion into the MGX file format. When loaded into Citrullia, the MS1 and MS2 data were separated and the MS2 data saved in a standard mgf file format. Each MS2 data file was then searched individually using the X! Tandem search engine, with all parameters set by Citrullia. The 26–30 result files from a given experiment were loaded into Citrullia, and a multiple path search was performed (Fig. 1). Initially peptides identified as citrullinated were extracted, and the entire X! Tandem run was searched for matching peptides with an MS1 mass difference of −0.984. If paired peptides were found, they were forwarded for validation. A second run was then performed using arginine-containing peptides, which were searched for potential citrulline-containing peptides using a mass difference of +0.984. For validation, matching citrullinated and non-citrullinated peptides were displayed with one spectrum mirrored. This enables an easy comparison and visualization of mass shifts and differences in the fragmentation pattern. Validated peptides were then marked as such. Finally, non-matched citrullinated peptides were also validated individually but marked as singles. The two main criteria for validation were:
To establish that citrullinated arginine residues were delineated by fragment ions in the MS2 spectra in order to unambiguously distinguish it from potential deamidated asparagine and glutamine residues.
Verify that the correct monoisotopic ion was picked for parent ion fragmentation.
Fig. 1.
Flowchart of data handling in Citrullia. MS2 data are initially analyzed by the X! Tandem search engine. Peptides identified as citrullinated are then paired with all potential arginine-containing peptides for validation. All not-identified arginine peptides are then compared with potential citrullinated peptides for validation. Finally, all citrullinated peptides that have not found a match to an arginine peptide are forwarded for validation. All validated citrullinations are then saved in a table with relevant information.
Three additional criteria were used for the validation
The fragmentation pattern in agreement with the charge localization in the peptide, e.g. a C-terminally citrullinated tryptic peptide will not have a C-terminally located positive charge which results in a subsequent change in the fragmentation pattern (supplemental Fig. S4).
In reversed phase FA-based chromatography a citrullinated peptides will show delayed retention time relative to the non-citrullinated peptide. (46)
In reversed phase HFBA-based chromatography a citrullinated peptide will show the reversed (leading) retention time behavior. (47)
Citrullinations were thus evaluated both on MS1 specificity, fragment ions, fragmentation pattern, and retention time behavior in one or two dimensions. The user interface of Citrullia is presented in supplemental Fig. S1.
Identification of Citrullination in the OMV of P. gingivalis W83 Strains (WT, C351A, and ΔPPAD)
In order to improve the detection of citrullinated peptides, we decided to evaluate a two-dimensional chromatographic system. This was based on off-line separation of peptides using HFBA as modifier in the first dimension and an on-line FA-based system, both using C18 reversed phase column material. In a publication by Mant et al. (47) it was reported that HFBA as a modifier bound strongly to positively charged residues, in particular arginines, causing arginine-containing peptides to be retained longer in a reversed phase system compared with same peptides with citrulline(s). This was corroborated using synthetic peptides (results not shown). We therefore decided to evaluate whether the HFBA modifier could be used as the first dimension in a standard two-dimensional proteomics setup and introduce a validation step for identification of citrullinated peptides. The first-dimension separation of P. gingivalis OMV tryptic peptides resulted in 26–30 fractions, each of which was subsequently analyzed using a 30-min gradient in FA.
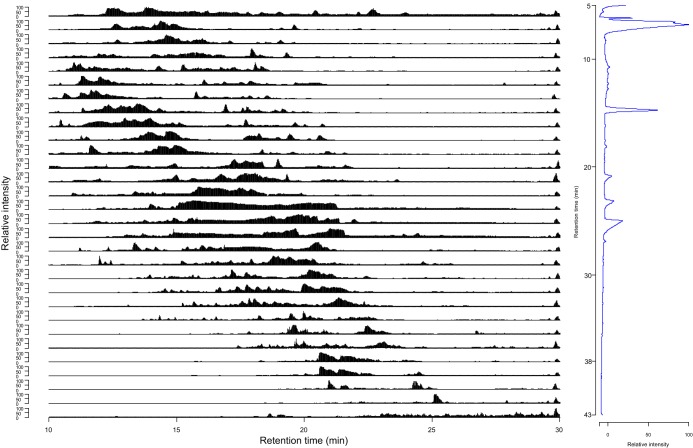
The isolated OMV from three different P. gingivalis strains were analyzed for citrullinations in technical duplicates. The preparations were from wild-type P. gingivalis W83 (WT), the W83 strain with a mutated PPAD, where the active cysteine was replaced with alanine (C351A), and a knockout of PPAD (ΔPPAD) in the W83 strain. Based on previous results on the P. gingivalis OMV secretome (48) we expected WT to contain the majority of citrullinations, whereas fewer were expected for C351A and ΔPPAD. All samples were analyzed using our two-dimensional method. The chromatogram and the total ion count (TIC) for all 30 fractions of the separation of WT replicate 1 are shown in Fig. 2. The orthogonality of the HFBA separation can be observed in the FA-based second dimension, as a broad elution from 10 to 22 min was typically observed for the first 20 fractions of each first-dimension separation. The last 10 first-dimension fractions are shifted toward later elution in the second dimension (Fig. 2). Separation of WT replicate 2, C351A replicate 1 and 2, and ΔPPAD replicate 1 and 2 revealed almost identical separation patterns (supplemental Fig. S2) indicating that the difference in content of citrullinated peptides was not sufficient for a measurable difference in the TIC.
Fig. 2.
Two-dimensional separation of a tryptic digest of P. gingivalis OMV proteins. Left: Second-dimension total ion count traces of the 30 fractions resulting from the first dimension HFBA fractionation. The first set of fractions was obtained from 5 to 10 min elution time. From 10 to 38 min, fractions were collected in 1 min intervals. The last set was collected from 38 to 43 min elution time. Each first-dimension fraction is shown from 10 to 30 min elution. Right: First dimension UV trace (214 nm) of HFBA/acetonitrile separation of 100 μg WT P. gingivalis OMV.
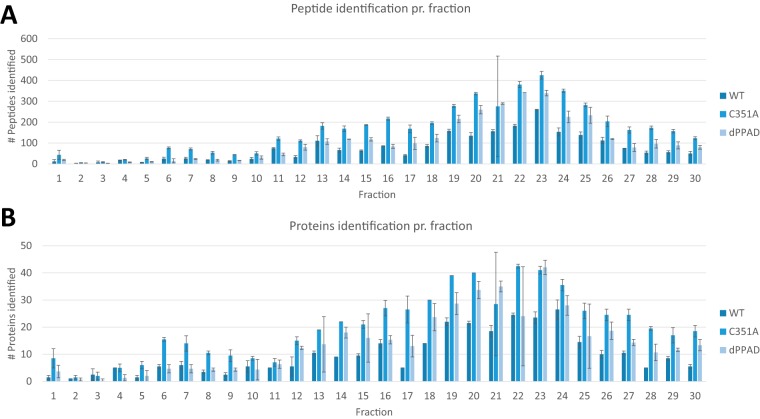
Most of the identified proteins and peptides were found in fractions collected between 12- and 30-min elution from the first-dimension separation. The profile of identified peptides in each fraction was reproducible, and each fraction showed similar identification numbers across the different samples and replicates. Citrullinated peptides were found across the entire fractionation, but the majority of citrullinations were identified in fractions collected between 18 and 26 min (supplemental Fig. S3). Only a few fractions showed large standard deviations in peptide or protein identification, which could be explained by lack of material or too low resuspension buffer volume (Fig. 3).
Fig. 3.
Distribution of identified peptides and proteins from the first-dimension separation of the three P. gingivalis strains. A) Number of peptides identified per fraction. B) Number of proteins identified per fraction. Fraction 1 and fraction 30 have been collected for 5 min, whereas fractions from 2 through 29 were collected for 1 min each during the HPLC gradient development. The error bars indicate the variation between duplicates.
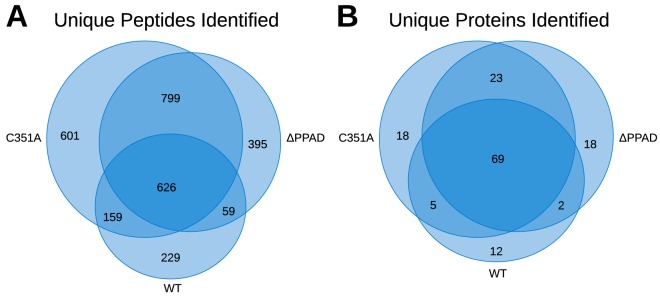
For the majority of fractions, the number of peptides and proteins identified in each OMV fraction was C531A >ΔPPAD> WT (Table I). The number of unique peptides identified in C351A was ∼15% higher than in ΔPPAD and twice that of WT. The number of identified unique proteins was almost identical for C351A and ΔPPAD, but a quarter less for WT (Fig. 4A). This may indicate that the 115 proteins determined in C351A are close to the total number measurable with the current dynamic range and MS settings. However, although the overlap between technical replicates was high, the overlap of peptides between OMV derived from different strains was low, resulting in only 626 common peptides for all three strains (Fig. 4B). On the other hand, the overlap between identified proteins was relatively high (i.e. only 12–18 additional unique proteins identified per strain-fraction OMV), indicating that many of the unique identified proteins are of low abundance.
Table I. Replicates of outer membrane vesicles (OMV) were analyzed for wild-type (WT) P. gingivalis the isogenic mutant in the same strain expressing inactive PPAD (PPADC351A), and in the knockout (ΔPPAD) W83 strain. Each replicate is represented with the combined results of unique proteins and the total number of peptides. For each replicate and sample, number of proteins, peptides, proteins with citrullination, identified citrullinated peptides, paired citrullinations (peptide identified with both arginine and citrulline), and single citrullinations (citrullinated only) are presented. For samples including paired citrullinations, fraction shift and retention time shift are shown.
| Sample | OMV W83 WT |
OMV W83 C351A |
OMV W83 ΔPPAD |
Found in all |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rep. 1 | Rep. 2 | Comb. | Rep. 1 | Rep. 2 | Comb. | Rep. 1 | Rep. 2 | Comb. | 3/3 | |
| Identified unique proteins | 77 | 79 | 88 | 113 | 107 | 115 | 93 | 101 | 112 | 69 |
| Identified unique peptides | 912 | 886 | 1073 | 1969 | 1760 | 2185 | 1549 | 1514 | 1879 | 626 |
| Citrullinated proteins | 30 | 24 | 34 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Identified citrullinated peptides | 43 | 34 | 52 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Paired citrullinations | 9 | 8 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Single citrullinations | 34 | 26 | 39 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ±Cit earlier fraction | 8 | 8 | 13 | - | - | - | - | - | - | - |
| ±Cit same fraction | 0 | 0 | 0 | - | - | - | - | - | - | - |
| ±Cit later fraction | 1 | 0 | 1 | - | - | - | - | - | - | - |
| RT shift from Arg to Cit (Sec) | 49.1 | 50.3 | 49.7 | - | - | - | - | - | - | - |
Fig. 4.
Venn diagrams of the overlap of peptides and proteins between the different OMV isolates. A, Overlap of the combined unique peptides and B, overlap of the combined unique proteins (Table I).
When searching for citrullinations, the vast majority was identified in the WT replicates, as only a single citrullination was identified in replicate 1, fraction 15 of the P. gingivalis PPADC351A strain and none in ΔPPAD. Of the total of 52 unique citrullinations, 13 were found in pairs with the non-citrullinated peptide and the remainders were found as singles, i.e. the corresponding arginine-containing peptide could not be found. Based on the paired citrullinations, fraction shifts in the first dimension HFBA separation can be calculated. Of the 13 paired citrullinations, 12 were found to elute in an earlier fraction and one was found to elute both earlier as well as later, depending on the replicate. The RT shift was calculated for the second dimension FA separation, where the average RT shift has been calculated to 50 s of delayed elution for a citrullinated peptide. This is in accordance with previous observations.
Large differences in the number of observed proteins were found between the WT OMV and the mutant strains when analyzed by the two-dimensional method. In total 115 proteins were identified for C351A and 112 for ΔPPAD, whereas only 88 were identified for the WT. This is likely caused by many WT citrullinated peptides being singly charged and thus more difficult to detect because of less fragmentation relative to multiply charged ions. As protein content and composition in the samples, except for the presence of PPAD, should be identical, the number of lysine terminated peptides should thus remain constant. However, the ratio of arginine/citrulline-terminated peptides may vary depending on the level of citrullination, hereby shifting the ratio. Although the number of identified peptides varied between the samples, the most striking difference is that the arginine/lysine terminated peptide ratio was very low in WT OMV (0.18) whereas much higher and almost identical for C351A (0.82) and ΔPPAD (0.79) (Table II).
Table II. The number of peptides including a C-terminal arginine or lysine residue found in the different replicates, along with the ratio of the two. Clear difference is seen from WT towards Mutant (C351A) and ΔPPAD.
| Sample | #Arginines | #Lysines | R/K |
|---|---|---|---|
| WT_2 | 314 | 1672 | 0.188 |
| WT_3 | 287 | 1625 | 0.177 |
| C351A_2 | 2140 | 2598 | 0.824 |
| C351A_3 | 1881 | 2314 | 0.813 |
| dPPAD_1 | 1368 | 1679 | 0.815 |
| dPPAD_2 | 1321 | 1707 | 0.774 |
Citrullination of OMV Derived from WT P. gingivalis
Based on the above presented results, we decided to analyze the wild type P. gingivalis W83 in more detail, in order to obtain the best characterization of the citrullinome and to evaluate our two-dimensional separation strategy. A single off-line HFBA separation of 100 μg tryptic digest of the WT OMV was separated into 26 fractions. These were analyzed by mass spectrometry as technical triplicate injections using a 30 min gradient and were compared with triplicate injections of the entire tryptic digest using a two-hour gradient.
For the detailed analysis of the WT OMV, mass spectrometric data acquisition was optimized by decreasing the loading time (from 200 to 100 ms) and using a top 10 instead of a top 5 method, thus doubling the number of ions being fragmented and analyzed. For the long 120 min gradient, 2 μg of WT OMV tryptic digest was micro-purified for each injection, as this was the estimated amount injected from the two-dimensional separation method over the central 20 fractions.
As C-terminally citrullinated peptides were expected mainly to be singly charged, these ion species were included for all sample runs. This inclusion had a greater impact for the long gradients where 15 out of 95 identified citrullinated peptides were singly charged. For the two-dimensional system the same occurred in only 6 out of 206 peptides (supplemental Table S3). The reason for the lower number of singly charged peptides in the two-dimensional system is likely that the same peptide has a higher probability of being observed multiple times. If a peptide occurs as a doubly charged species, the resulting improved fragmentation would result in a lower e-value, which would be more favorable, and thereby be selected for further analysis. This can also be seen from the observation that in the long gradient 44% of identified citrullinated peptides were observed without a positively charged side-chain residue (primarily histidine, as tryptic digest takes place after lysine and arginine), which was only the case for 29% of the two-dimensional method.
For the triplicate HFBA fractionated samples, between 1312 and 1452 unique peptides were identified (Table III), approximately two-thirds more than identified in the first duplicate analysis. Although the number of unique proteins identified rose 50% to 133, the number of identified citrullinated peptides almost doubled to 99, an increase of 90%. Of the citrullinated peptides, approximately half were identified along with the corresponding arginine-containing peptide (e.g. paired) whereas the rest were found as singles (e.g. arginine-containing not found). The use of HFBA as modifier in the first-dimension also showed most of the paired peptides (85) eluting in leading fractions whereas eleven peptides eluted in the same and 2 in lagging fractions. The average separation for all paired peptides was 1.7 fractions. As fractions were collected for either 5 (first and last fractions) or 1 min, an even better separation could likely be obtained by collecting smaller fraction (for shorter time windows) in the first-dimension, at the expense of additional MS runs and less material per run. A single citrullinated peptide (AGNHTVQGATR) was found in both lagging and leading fractions, and twice in the same fraction, indicating that it may be sticky and elute over a large part of the chromatogram. In the second dimension, the opposite retention behavior of the citrulline/arginine peptides was observed, with an average separation time of approximately 1 min using a 30-min gradient and 4 mins for the long gradient. This shows that both the first- and second-dimension RT can be used for validation, if both the citrullinated and non-citrullinated peptide are present and have been matched.
Table III. Number of proteins and peptides of the WT-OMV identified in a standard two-hour LC-MS separation and an off-line HFBA separation into 30 fractions combined with 30-minute LC-MS separations of each fraction. Each LC-MS separation was performed in triplicates. In addition, the number of citrullinated proteins and peptides, as well as the number of paired (both native and citrullinated peptide identified) and single (only citrullinated peptide) are shown. For the HFBA fractionated sample, the number of citrullinated peptides found in a leading, in the same, and in lagging HPLC fraction is noted along with the fraction shift.
| Sample | Long Gradient |
Fractionated |
Common for both methods | ||||||
|---|---|---|---|---|---|---|---|---|---|
| WT_T1 | WT_T2 | WT_T3 | Unique | WT_T1 | WT_T2 | WT_T3 | Unique | ||
| # Unique proteins identified | 61 | 61 | 62 | 68 | 119 | 119 | 121 | 133 | 62 |
| # Unique peptides identified | 693 | 668 | 634 | 849 | 1452 | 1312 | 1371 | 1757 | 522 |
| # Citrullinated proteins | 38 | 36 | 39 | 48 | 44 | 39 | 44 | 56 | 31 |
| # Citrullinations | 69 | 73 | 65 | 95 | 74 | 60 | 72 | 99 | 46 |
| # Paired | 3 | 2 | 2 | 4 | 36 | 27 | 35 | 45 | 0 |
| # Single | 66 | 71 | 63 | 92 | 38 | 33 | 37 | 59 | 26 |
| ±Cit leading fraction | - | - | - | - | 31 | 24 | 30 | - | - |
| ±Cit same fraction | - | - | - | - | 4 | 3 | 4 | - | - |
| ±Cit lagging fraction | - | - | - | - | 1 | 0 | 1 | - | - |
| Fraction shift | - | - | - | - | −1.67 | −1.81 | −1.69 | - | - |
| RT shift (sec) | 254 | 230 | 288 | 257.3 | 75 | 56.6 | 63.2 | 64.9 | - |
In addition to serve as a validation step, the HFBA-based separation turned out to be quite efficient as an orthogonal first-dimension chromatography when followed by FA-based separation. For our 30 min gradient the peptides eluted over a period of ≈12 min. For the first 20 one-minute fractions, the elution started at about 10 min., which increased to 17 min. for the last fractions (Fig. 2). Thereby the pooling of fractions, which is often performed when using HILIC (49) or High pH (50) is not possible. However, by using our HFBA-based separation method, increased separation and depth of analysis can be obtained, with a cost of increased MS run time. Further, pooling of fractions would decrease the obtained resolution and obscure information on the exact fraction in which the peptide elutes in the first dimension.
For the experiment using a long gradient, the total number of identified peptides and proteins was half of that observed in the two-dimensional separation (Table II). However, the number of identified citrullinated peptides and proteins were very similar. A major difference was that only a small number of paired citrulline/arginine peptides were found using the long gradient. These were found with an average delayed elution of the citrulline peptide of 4 min in the 2 h FA-based gradient. The difference in retention time compared with the two-dimensional separation is due to the four times longer gradient.
Increased identification of arginine/citrulline terminated peptides where observed using the two-dimensional method compared with the long gradient. The ratio of arginine/lysine terminated peptides for the two-dimensional method was increased by 57% compared with the long gradient (Table IV). This shows that the optimization of data acquisition and the two-dimensional separation had improved the depth of analysis.
Table IV. The number of peptides including a C-terminal arginine or lysine along with their ratios as determined in each replicate using the long gradient or the two-dimensional HFBA-based method.
| Sample | #Arginines | #Lysines | R/K |
|---|---|---|---|
| WT_1_Long | 129 | 817 | 0.158 |
| WT_2_Long | 125 | 786 | 0.159 |
| WT_3_Long | 118 | 721 | 0.164 |
| WT_1_Two-dimensional_method | 834 | 2971 | 0.281 |
| WT_2_Two-dimensional_method | 716 | 2578 | 0.278 |
| WT_3_Two-dimensional_method | 782 | 2707 | 0.289 |
DISCUSSION
P. gingivalis secretes a very active peptidyl arginine deiminase (PPAD) along with Arg-specific gingipains (Rgps) using type IX secretion system (T9SS). During translocation across the outer membrane conserved C-terminal domains are cleaved off by sortase (PorU), and an anionic lipopolysaccharide is attached, anchoring the enzymes to the P. gingivalis cell surface (22), (51). In this way PPAD and gingipains are in very close proximity because they are major components of the surface electron dense layer composed of circa 30 proteins secreted via T9SS (52). Apparently, in this environment Rgps generate C-terminal arginine residues on peptides and protein fragments, which are efficiently converted into citrulline by PPAD (22). By budding of the outer membrane, the OMV coated with the surface electron dense layer are released into the environment carrying inside some periplasmic proteins (53). Formation of the OMV is not a random process and is driven by a mechanism selectively sorting virulence factors into OMVs, excluding at the same time abundant outer membrane proteins (54). In this way OMVs are very important for host-pathogen interactions extending the outreach of P. gingivalis virulence factors, including gingipains, PPAD, and citrullinated proteins into periodontal tissues. Therefore, considering pathogenic potency of citrullinated proteins it was important to develop a technique allowing unbiased identification of citrullination sites and delineation of the citrullinome of the P. gingivalis OMV.
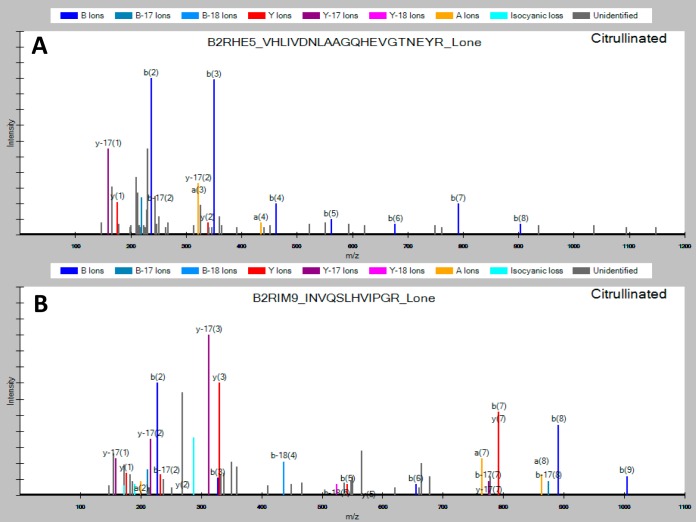
Because PPAD mainly citrullinate C-terminal arginines (55), a large part of the citrullinated peptides in a tryptic digest were expected to be singly charged, because of the peptide size and citrullination. Therefore, we included singly-charged ions in our MS method and database searches. Such an approach increases the number of possible citrullination sites but is usually neglected because of potential false-positive issues and inclusion of many non-peptide ions. In the most comprehensive study to date of the human citrullinome performed by Küster and coworkers (45), the authors excluded all potential C-terminal citrullinations and thereby ignored an estimated 10–48% of their data, which could potentially contain true positives. Furthermore, the authors estimated that the majority of citrullinated species can be identified by a neutral loss of −43 Da. However, careful analysis of our data revealed that approximately one in four was identified without a neutral loss (see Fig. 5). This could be caused by the majority of the citrullinations in the current study being located C-terminally. Therefore, the neutral loss was not included as a selection criterion but may be included into other studies focused on internal citrullinations.
Fig. 5.
Representative spectra of C-terminally citrullinated tryptic peptides from the P. gingivalis OMV. A, Spectrum observed without a neutral loss of isocyanic acid (−43 Da). B, Spectrum observed with the presence of neutral loss (cyan color).
In order to characterize the citrullinome at the P. gingivalis surface and the importance of PPAD, we analyzed proteins located in the OMV of the wild type W83 strain (WT), the mutant with the active cysteine mutated to alanine (C351A), and the knockout mutant (ΔPPAD). The proteomic analysis of the OMV derived from WT and mutants of the W83 using a two-dimensional strategy, identified a combined set of 88 to 115 proteins displaying a common core of 69 proteins (Table I, Fig. 4). The P. gingivalis OMV proteome has previously been determined by Reynolds and co-workers (52), where they established a set of 151 proteins. Comparing our set of identified proteins to theirs, revealed only an ∼50% overlap (supplemental Table S1). The reason for this can either be differences in the purification of the OMVs such as a different bacterial growth phase, gel separation and slicing versus 2D chromatography, or different settings during the database search. Setting our database search parameters like the ones presented in Veith et al. (52) increased our identifications to >650 proteins, but only increased the identification overlap to ∼60%. In a more recent paper (56) the same group using the same strain and preparation, but a slightly different gel and MS strategy, identified 181 proteins. Here 27 (18%) proteins from the earlier paper were not found, showing that the exact details for preparation and analysis are essential for comparison.
We have mapped the identified citrullinated proteins to their predicted subcellular location within the bacteria using the presence of signal peptide and the PG locus database. Three main groups were identified: lipoproteins (24%), T9SS secreted (28%), and others including cytoplasmic and inner membrane proteins (48%), (supplemental Table S4). Comparing our distribution of the citrullinated proteins to the complete OMV proteome of Veith et al., (56) revealed large differences particularly regarding the cytoplasmic and inner membrane proteins. Whereas these proteins constituted only a few percent of the recognized protein in the Veith et al. study, they were found in much larger numbers in our experiment. Also, in contrast to abundance of the T9SS secreted proteins (almost 2/3 of identified proteins) and rarity of lipoproteins identified by Veith et al., in our study, we found an equal number of lipoproteins and T9SS cargo proteins. As outlined above, the differences may be caused by several analytical differences and is likely further skewed by comparing all found proteins against citrullinated proteins. The T9SS secreted proteins are likely more resistant toward proteolysis by Rgps and subsequent citrullination by PPAD, whereas lipoproteins, cytoplasmic, and inner membrane proteins are more sensitive.
In another publication by Stobernack et al. (48), the authors aimed at determining the secreted citrullinome of P. gingivalis. This study showed a similar number of identified proteins, 64, as a core set, with a relatively large difference in the secretome among five different strains of P. gingivalis. Note that the procedure used in this work did not exclusively purify the OMV, and apparently both soluble proteins and those associated with OMV were analyzed, which probably contributed to the observed differences among analyzed strains.
A striking difference between the data presented here and the data of Stobernack et al. (48) is the number of citrullinated proteins identified with high confidence, 34 versus only 2, respectively. Further, they found 11 citrullinated proteins in the ΔPPAD W83 strain, which originated in our laboratory. Although none of these peptides was identified with high confidence, finding such peptides in the PPAD-null strain undermines their approach of unbiased identification of citrullination. Apparently, the van Dijl's group (Stobernack et al. (48)) either could not distinguish Gln/Asn deamidations from Arg deamination, picked up a wrong isotope, or had other technical problems precluding confident identification of citrullinated peptides. For the C351A PPAD mutant we only identified a single citrullination (supplemental Fig. S4, third spectrum [AGRIPK]), showing that the replacement of the cysteine in the active site may not completely remove all activity, but severely diminish it.
As the analysis of the various mutants revealed that the WT OMV was the only sample-type producing a significant number of citrullinations, we used this for setting up a two-dimensional gradient and an MS data acquisition method. Experiments comparing the HFBA-based separation system with a standard high pH chromatography showed similar separation power (data not shown). The HFBA system further has the advantages of being very robust, not needing high-pH stable columns, and HFBA can easily be evaporated. As it additionally provides validation of citrullination when used in the first dimension, we decided to compare it to a triplicate analysis of the WT OMV in a standard 2-hour one-dimensional FA gradient. The total number of validated citrullinated peptides was 99 and 95 for the two- and one-dimensional method, respectively, resulting in the identification of 39 and 35 citrullinated proteins (Table II). Although the total number of identified citrullinations does not vary much between the two methods, the confidence in identification of the citrullinations differed. However, although the number of citrullinations was similar, the two-dimensional method managed to identify twice as many proteins as the simple gradient. This difference in identification efficiency of citrullinated and normal peptides may be caused by citrullinated peptides ionizing as singly charged species, which need a higher ion count in order to fragment sufficiently for identification. Although the vast majority of identified citrullinated peptides were found as C-terminal citrullinations, we did identify a few peptides having internal citrullinations (all spectra presented in supplemental Fig. S4.1 and S4.2). As internally citrullinated peptides generally ionizes as doubly charged ions with resulting higher detection efficiency, it supports the contention that PPAD has a strong to almost exclusive preference for C-terminal arginine residues.
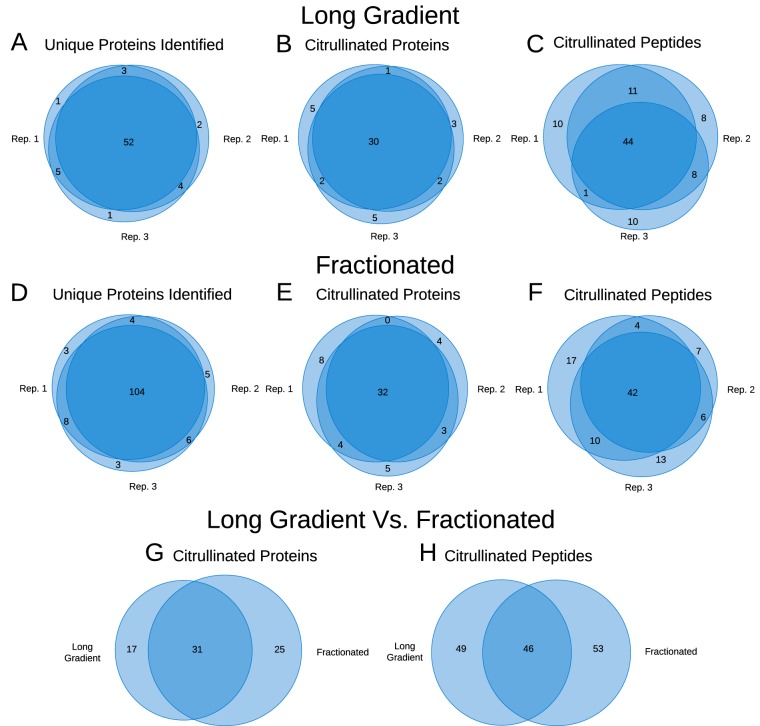
Although identified citrullinated peptides varied somewhat between technical duplicates, the number of identified unique proteins varied surprisingly little (Fig. 6A to 6F). In contrast to this, the difference between both identified citrullinated peptides as well as citrullinated proteins varied more (Fig. 6G and 6H). The variation could be because of the ion pairing reagent utilized (HFBA) in the first-dimension, where it enriches for specific peptides. However, the two-dimensional method doubled the number of identified peptides and proteins. Whether this increase was related to this two-dimensional system and could be reproduced by a high/low pH system (50) or similar was not tested. Even though the number of identified citrullinations was similar between our two systems, our two-dimensional separation clearly shows potential advantages regarding validation of citrullinations and depth of analysis.
Fig. 6.
Venn diagrams, showing overlap between three replicates for the long gradient: A, unique proteins identified, B, unique citrullinated proteins, and C, unique citrullinated peptides. Three replicates for the fractionated samples: D, unique proteins identified, E, unique citrullinated proteins, and F, unique citrullinated peptides. Further, comparison between the long gradient and the fractionated sample G, unique citrullinated proteins and H, unique citrullinated peptides.
On the other hand, the number of identified peptides and proteins was much lower for WT OMV than for C351A and ΔPPAD. As the amount of sample was identical for all samples, the difference is likely caused by citrullination of the WT peptides resulting in lower detection efficiency. This assumption was verified by measuring of the ratio between lysine terminated and arginine terminated peptides in the various samples as shown in Table III and IV. Taken together these results indicate that numerous P. gingivalis proteins were cleaved by RgpA/B, but almost all generated C-terminal arginines were modified by PPAD to citrulline. As expected, however, OMV-associated proteins were not degraded but rather a relatively small fraction of molecules was nicked at some sites, as SDS-gel electrophoresis of the various samples showed clear bands for intact proteins (results not shown).
Based on a sequence logo of the citrullinated peptides (supplemental Fig. S6) no clear consensus sequence for citrullination by the RgpA/B-PPAD system in the P1 to P5 position can be identified. Any RgpA/B preference in the P1′ position was not analyzed, as this will be obscured by the following tryptic cleavage.
The present results clearly show that limitations in the detection of C-terminally citrullinated peptides lie in the detection by mass spectrometry, as most of these peptides are ionized as singly charged species. Several ways to alleviate this can be suggested. The most straightforward way would be to increase the net charge of the peptides, either at the N terminus (e.g. by TMT labeling (57)), or at the C terminus (e.g. using techniques from C-terminomics (58)). A purely MS-based method could be to increase the general charge by supercharging (59) or use a faster MS instrument in combination with a decision tree for optimal data acquisition of singly charged species. A biochemical way of increasing the charge could be to digest with endopeptidase Lys-N that will generate lysine N-terminals at the expense of larger peptides, which may be more difficult to identify. An additional complexity arises when the sample has a low level of citrullination, which is often the case for mammalian citrullination analysis. For these analyses the sample must be enriched. Here a reaction with diols (60) is a possibility, particularly when the diol is coupled to a biotin group, which enables isolation with streptavidin (61). This method has not been generally used, probably because of difficult synthesis of the reagent or problems with the identification. In this way there is ample room for improvements on different levels of the analysis, from MS method optimization to sample preparation.
CONCLUSION
The citrullinations identified and validated by our method are characterized by high overall confidence because of the manual validation steps and no exclusion of data prior to analysis. One drawback is the large amount of data that must be handled manually. This clear downside of the method was mitigated by use of the in-house developed program Citrullia, allowing analysis of hundreds to thousands of spectra in a fair amount of time. Taken together, our results show that the citrullinome of P. gingivalis is much larger than the 6–25 proteins estimated earlier (48). Here we have identified 78 proteins as being citrullinated and having a total of 161 citrullination sites. Nevertheless, our data also show that we most likely identified only a fraction of the actual number of citrullinated peptides present in P. gingivalis OMV. This under-detection of modified peptides is because of the removal of a positive charge by citrullination resulting in lower ionization potential and insufficient fragmentation. Although our results are obtained without chemical derivatization, enrichment or other techniques, in order to obtain the full citrullinome of P. gingivalis new methods need to be implemented, where either a better way of analyzing singly charged peptide ions are applied or citrullinated peptides are derivatized resulting in multiply charged species. Despite these limitations this study reveals that at least 78 proteins of P. gingivalis OMV are citrullinated. Notably the clear majority (51 proteins) possessed a signal peptide targeting proteins to periplasm. Among these 17 are predicted to be lipoproteins and are 9 proteins secreted via T9SS and known to be enriched in the OMV. Apart from a few exceptions of internal or N-terminal citrulline, all identified citrullination sites were at the C terminus. Interestingly however, only one of them was mapped to the native arginine occurring at the C terminus of circa 200 proteins encoded in the P. gingivalis genome (supplemental Table S3). This finding confirms our contention that Rgps and PPAD work in concert in generation of C-terminally citrullinated peptides derived from both bacteria and host proteins. Some of the modified peptides/proteins apparently contribute to the autoimmune response leading to RA (32), others stimulate the PGE2 synthesis pathway (31), affect inter- and intra-microbial interactions in biofilm (27) (29) and finally modify responses of neutrophils (27) and gingival epithelial cells. (29) Recognizing which citrullinated proteins/peptides are responsible for which activity is a challenge and the method described in this report is a first step in the direction of understanding the pathobiological meaning of the P. gingivalis citrullinome.
DATA AVAILABILITY
The current version of Citrullia can be downloaded from https://github.com/Citrullia-SDU/Citrullia.
The mgx conversion program MGF Filter can be downloaded from http://massai.dk/download.html.
All the ms raw files used for searching are available via ProteomeXchange (PRIDE) with the accession number PXD015701.
Supplementary Material
Acknowledgments
We thank Morten I Rasmussen for supplying software for the conversion of raw files to mgx format. Dr. Paul Veith is appreciated for providing the PG locus database. Magnus G. Jespersen is acknowledged for support in handling data and drawing figures.
Footnotes
* The VILLUM Foundation (grant no. 7292) and the Danish Ministry of Higher Education and Science (grant no. 5072-00007B) are gratefully acknowledged for mass spectrometric infrastructure support. J.P. and G.B. were supported by NIH (NIDCR DE 022597 to J.P.) and National Science Center (Krakow, Poland): UMO-2018/30/A/NZ5/00650 (to J.P.) and 2018/29/N/NZ1/00992 (to G.P.B.). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- PAD
- humane peptidyl arginine deiminase
- OMV
- outer membrane vesicles
- WT
- P. gingivalis W83 wild-type
- ΔPPAD
- W83 knock-out mutant of peptidyl arginine deiminase
- C351A
- W83 expressing PPAD with the active site cysteine mutated to alanine
- 2D
- two-dimensional HFBA-based
- LC-MS
- liquid chromatography mass spectrometry
- PPAD
- bacterial P. gingivalis peptidyl arginine deiminase
- RA
- rheumatoid arthritis
- PGE2
- prostaglandin E2
- ACPA
- anti-citrullinated protein antibodies
- RgpA/B
- Arg-specific gingipains
- MS
- mass spectrometry
- HFBA
- heptafluorobutyric acid
- FA
- formic acid
- HPLC
- high performance liquid chromatography
- AAA
- amino acid analysis
- mgf
- Mascot Generic File format
- mgx
- Mascot Generic eXtended
- RT
- retention time
- TIC
- total ion count
- T9SS
- type IX secretion system.
REFERENCES
- 1. Nachat R., Méchin M. C., Takahara H., Chavanas S., Charveron M., Serre G., and Simon M. (2005) Peptidylarginine deiminase isoforms 1–3 are expressed in the epidermis and involved in the deimination of K1 and filaggrin. J. Invest. Dermatol. 124, 384–393 [DOI] [PubMed] [Google Scholar]
- 2. Ishida-Yamamoto A., Senshu T., Eady R. A. J., Takahashi H., Shimizu H., Akiyama M., and Iizuka H. (2002) Sequential reorganization of cornified cell keratin filaments involving filaggrin-mediated compaction and keratin 1 deimination. J. Invest. Dermatol. 118, 282–287 [DOI] [PubMed] [Google Scholar]
- 3. Lamensa J. W. E., and Moscarello M. A. (1993) Deimination of Human Myelin Basic Protein by a Peptidylarginine Deiminase from Bovine Brain. J. Neurochem. 61, 987–996 [DOI] [PubMed] [Google Scholar]
- 4. Finch P. R., Wood D. D., and Moscarello M. A. (1971) The presence of citrulline in a myelin protein fraction. FEBS Lett. 15, 145–148 [DOI] [PubMed] [Google Scholar]
- 5. Steinert P. M., Parry D. A. D., and Marekov L. N. (2003) Trichohyalin mechanically strengthens the hair follicle: Multiple cross-bridging roles in the inner root sheath. J. Biol. Chem. 278, 41409–41419 [DOI] [PubMed] [Google Scholar]
- 6. Tarcsa E., Marekov L. N., Mei G., Melino G., Lee S.-C., and Steinert P. M. (1996) Protein Unfolding by Peptidylarginine Deiminase. J. Biol. Chem. 271, 30709–30716 [DOI] [PubMed] [Google Scholar]
- 7. Cuthbert G. L., Daujat S., Snowden A. W., Erdjument-Bromage H., Hagiwara T., Yamada M., Schneider R., Gregory P. D., Tempst P., Bannister A. J., and Kouzarides T. (2004) Histone deimination antagonizes arginine methylation. Cell 118, 545–553 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y., Wysocka J., Sayegh J., Lee Y.-H., Perlin J. R., Leonelli L., Sonbuchner L. S., McDonald C. H., Cook R. G., Dou Y., Roeder R. G., Clarke S., Stallcup M. R., Allis C. D., and Coonrod S. A. (2004) Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306, 279–283 [DOI] [PubMed] [Google Scholar]
- 9. Li P., Yao H., Zhang Z., Li M., Luo Y., Thompson P. R., Gilmour D. S., and Wang Y. (2008) Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol. Cell. Biol. 28, 4745–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao H., Li P., Venters B. J., Zheng S., Thompson P. R., Pugh B. F., and Wang Y. (2008) Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J. Biol. Chem. 283, 20060–20068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X., Gamble M. J., Stadler S., Cherrington B. D., Causey C. P., Thompson P. R., Roberson M. S., Kraus W. L., and Coonrod S. A. (2011) Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate c-Fos expression in breast cancer cells. PLOS Genet. 7, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Proost P., Loos T., Mortier A., Schutyser E., Gouwy M., Noppen S., Dillen C., Ronsse I., Conings R., Struyf S., Opdenakker G., Maudgal P. C., and Van Damme J. (2008) Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J. Exp. Med. 205, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andrade F., Darrah E., Gucek M., Cole R. N., Rosen A., and Zhu X. (2010) Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 62, 1630–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wright P. W., Bolling L. C., Calvert M. E., Sarmento O. F., Berkeley E. V., Shea M. C., Hao Z., Jayes F. C., Bush L. A., Shetty J., Shore A. N., Reddi P. P., Tung K. S., Samy E., Allietta M. M., Sherman N. E., Herr J. C., and Coonrod S. A. (2003) ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev. Biol. 256, 73–88 [DOI] [PubMed] [Google Scholar]
- 15. Gabarrini G., Chlebowicz M. A., Vega Quiroz M. E., Veloo A. C. M., Rossen J. W. A., Harmsen H. J. M., Laine M. L., van Dijl J. M., and van Winkelhoff A. J. (2017) Conserved citrullinating exoenzymes in Porphyromonas species. J. Dent. Res. 97, 56–562 [DOI] [PubMed] [Google Scholar]
- 16. Mangat P., Wegner N., Venables P. J., and Potempa J. (2010) Bacterial and human peptidylarginine deiminases: targets for inhibiting the autoimmune response in rheumatoid arthritis? Arthritis Res Ther. 12, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goulas T., Mizgalska D., Garcia-ferrer I., Kantyka T., Guevara T., Szmigielski B., Sroka A., Millán C., and Usón I. (2015) Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Sci. Rep. 5, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Assohou-Luty C., Raijmakers R., Benckhuijsen W. E., Stammen-Vogelzangs J., De Ru A., Van Veelen P. A., Franken K. L. M. C., Drijfhout J. W., and Pruijn G. J. M. (2014) The human peptidylarginine deiminases type 2 and type 4 have distinct substrate specificities. Biochim. Biophys. Acta - Proteins Proteomics 1844, 829–836 [DOI] [PubMed] [Google Scholar]
- 19. Montgomery A. B., Kopec J., Shrestha L., Thezenas M. L., Burgess-Brown N. A., Fischer R., Yue W. W., and Venables P. J. (2016) Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase: Implications for autoimmunity in rheumatoid arthritis. Ann. Rheum. Dis. 75, 1255–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reichert S., Schlumberger W., Dähnrich C., Hornig N., Altermann W., Schaller H. G., and Schulz S. (2015) Association of levels of antibodies against citrullinated cyclic peptides and citrullinated α - enolase in chronic and aggressive periodontitis as a risk factor of Rheumatoid arthritis : a case control study. J. Transl. Med. 18, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quirke A., Lugli E. B., Wegner N., Hamilton B. C., Charles P., Chowdhury M., Ytterberg A. J., Zubarev R. A., Potempa J., Culshaw S., Guo Y., Fisher B. A., Thiele G., Mikuls T. R., and Venables P. J. W. (2014) Heightened immune response to autocitrullinated Porphyromonas gingivalis peptidylarginine deiminase : a potential mechanism for breaching immunologic tolerance in rheumatoid arthritis. Ann. Rheum. Dis. 73, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wegner N., Wait R., Sroka A., Eick S., Nguyen K.-A., Lundberg K., Kinloch A., Culshaw S., Potempa J., and Venables P. J. (2010) Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 62, 2662–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abdullah S. N., Farmer E. A., Spargo L., Logan R., and Gully N. (2013) Porphyromonas gingivalis peptidylarginine deiminase substrate specificity. Anaerobe 23, 102–108 [DOI] [PubMed] [Google Scholar]
- 24. Pyrc K., Milewska A., Kantyka T., Sroka A., Maresz K., Kozieł J., and Nguyen K. (2013) Inactivation of epidermal growth factor by Porphyromonas gingivalis. Infect Immun. 81, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bereta G., Goulas T., Madej M., Bielecka E., Solà M., Potempa J., and Gomis-rüth F. X. (2019) Structure, function, and inhibition of a genomic/ clinical variant of Porphyromonas gingivalis peptidylarginine deiminase. Protein Sci. 28, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bielecka E., Scavenius C., Kantyka T., Jusko M., Mizgalska D., Szmigielski B., Potempa B., Enghild J. J., Prossnitz E. R., Blom A. M., and Potempa J. (2014) Peptidyl arginine deiminase from Porphyromonas gingivalis abolishes anaphylatoxin C5a activity. J. Biol. Chem. 289, 32481–32487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kriebel K., Hieke C., Engelmann R., Potempa J., Muller-Hilke B., Lang H., and Kreikemeyer B. (2018) Porphyromonas gingivalis peptidyl arginine deiminase can modulate neutrophil activity via infection of human dental stem cells. J. Innate Immun. 10, 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vermilyea D. M., Ottenberg G. K., and Davey M. E. (2019) Citrullination mediated by PPAD constrains biofilm formation in P. gingivalis strain 381. NPJ biofilms microbiomes 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aliko A., Kaminska M., Bergum B., Gawron K., Benedyk M., Lamont R. J., Malicki S., Delaleu N., Potempa J., and Mydel P. (2018) Impact of Porphyromonas gingivalis peptidylarginine deiminase on bacterial biofilm formation, epithelial cell invasion, and epithelial cell transcriptional landscape. Sci. Rep. 8, 14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karkowska-Kuleta J., Bartnicka D., Zawrotniak M., Zielinska G., Kieronska A., Bochenska O., Ciaston I., Koziel J., Potempa J., Baster Z., Rajfur Z., and Rapala-Kozik M. (2018) The activity of bacterial peptidylarginine deiminase is important during formation of dual-species biofilm by periodontal pathogen Porphyromonas gingivalis and opportunistic fungus Candida albicans. Pathog. Dis. 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gawron K., Bereta G., Nowakowska Z., Lazarz-Bartyzel K., Lazarz M., Szmigielski B., Mizgalska D., Buda A., Koziel J., Oruba Z., Chomyszyn-Gajewska M., and Potempa J. (2014) Peptidylarginine deiminase from Porphyromonas gingivalis contributes to infection of gingival fibroblasts and induction of prostaglandin E2 -signaling pathway. Mol. Oral Microbiol. 29, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Potempa J., Mydel P., and Koziel J. (2017) The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat. Rev. Rheumatol. 13, 606–620 [DOI] [PubMed] [Google Scholar]
- 33. Lundberg K., Wegner N., Yucel-lindberg T., and Venables P. J. (2010) Periodontitis in RA—the citrullinated enolase connection. Nat. Publ. Gr. 6, 727–730 [DOI] [PubMed] [Google Scholar]
- 34. Laugisch O., Wong A., Sroka A., Kantyka T., Koziel J., Neuhaus K., Sculean A., Venables P. J., Potempa J., Möller B., and Eick S. (2016) Citrullination in the periodontium - a possible link between periodontitis and rheumatoid arthritis. Clin. Oral Investig. 20, 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haag S., Schneider N., Mason D. E., Tuncel J., Andersson I. E., Peters E. C., Burkhardt H., and Holmdahl R. (2014) Identification of new citrulline-specific autoantibodies, which bind to human arthritic cartilage, by mass spectrometric analysis of citrullinated type ii collagen. Arthritis Rheumatol. 66, 1440–1449 [DOI] [PubMed] [Google Scholar]
- 36. Pratesi F., Panza F., Paolini I., Petrelli F., Puxeddu I., Casigliani-Rabl S., Ancillotti D., Alcaro C., Rovero P., and Migliorini P. (2015) Fingerprinting of anti-citrullinated protein antibodies (ACPA): Specificity, isotypes and subclasses. Lupus 24, 433–441 [DOI] [PubMed] [Google Scholar]
- 37. Clancy K. W., Weerapana E., and Thompson P. R. (2016) ScienceDirect Detection and identification of protein citrullination in complex biological systems. Curr. Opin. Chem. Biol. 30, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Knipp M., and Vasák M. (2000) A Colorimetric 96-Well Microtiter Plate Assay for the Determination of Enzymatically Formed Citrulline. Anal. Biochem. 264, 257–264 [DOI] [PubMed] [Google Scholar]
- 39. Senshu T., Sato T., Inoue T., Akiyama K., and Asaga H. (1992) Detection of citrulline residues in deiminated proteins on polyvinylidene difluoride membrane. Anal. Biochem. 203, 94–100 [DOI] [PubMed] [Google Scholar]
- 40. Verheul M. K., van Veelen P. A., van Delft M. A. M., de Ru A., Janssen G. M. C., Rispens T., Toes R. E. M., and Trouw L. A. (2017) Pitfalls in the detection of citrullination and carbamylation. Autoimmun. Rev. 17, 136–141 [DOI] [PubMed] [Google Scholar]
- 41. Heissel S., Frederiksen S. J., Id J. B., and Peter H. (2019) Enhanced trypsin on a budget : Stabilization, purification and high-temperature application of inexpensive commercial trypsin for proteomics applications. PLoS One. 14, e0218374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896. [DOI] [PubMed] [Google Scholar]
- 43. Højrup P. (2015) in Houen G. (eds) Peptide Antibodies. Methods in Molecular Biology, 1348th Ed., Humana Press, New York, NY [Google Scholar]
- 44. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 45. Lee C., Wang D., Wilhelm M., Zolg D. P., Schmidt T., Schnatbaum K., Reimer U., Ponte F., Uhle M., Hahne H., and Kuster B. (2018) Mining the human tissue proteome for protein. Mol. Cell. Proteomics, 17, 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fert-bober J., Venkatraman V., Hunter C. L., Liu R., Crowgey E. L., Pandey R., Holewinski R. J., Stotland A., Berman B. P., and van Eyk J. E. (2019) Mapping citrullinated sites in multiple organs of mice using hypercitrullinated library. J. Proteome Res. 18, 2270–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mant C. T., and Hodges R. S. (2006) Context-dependent effects on the hydrophilicity/hydrophobicity of side-chains during reversed-phase high-performance liquid chromatography: Implications for prediction of peptide retention behaviour. J. Chromatogr. A 1125, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stobernack T., Glasner C., Junker S., Gabarrini G., Smit M. De, Jong A. De, Otto A., Becher D., Jan A., Winkelho V., and Dijl J. M. Van (2016) Extracellular Proteome and Citrullinome of the Oral Pathogen Porphyromonas gingivalis. J. Proteome Res. 15, 4532–4543 [DOI] [PubMed] [Google Scholar]
- 49. Alpert A. J. (1990) Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J. Chromatogr. 499, 177–196 [DOI] [PubMed] [Google Scholar]
- 50. Batth T. S., and Olsen J. V (2016) in Phospho-Proteomics: Methods and Protocols, ed von Stechow L. (Springer New York, New York, NY: ), pp 179–192 [Google Scholar]
- 51. Gorasia D. G., Veith P. D., Chen D., Seers C. A., Mitchell H. A., Chen Y.-Y., Glew M. D., Dashper S. G., and Reynolds E. C. (2015) Porphyromonas gingivalis Type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog. 11, e1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Veith P. D., Chen Y., Gorasia D. G., Chen D., Glew M. D., Brien-simpson N. M. O., Cecil J. D., Holden J. A., and Reynolds E. C. (2014) Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 2; 13, 2420–2432 [DOI] [PubMed] [Google Scholar]
- 53. Chen Y.-Y., Peng B., Yang Q., Glew M. D., Veith P. D., Cross K. J., Goldie K. N., Chen D., O'Brien-Simpson N., Dashper S. G., and Reynolds E. C. (2011) The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol. Microbiol. 79, 1380–1401 [DOI] [PubMed] [Google Scholar]
- 54. Haurat M. F., Aduse-Opoku J., Rangarajan M., Dorobantu L., Gray M. R., Curtis M. A., and Feldman M. F. (2011) Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286, 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goulas T., Mizgalska D., Garcia-ferrer I., Kantyka T., Guevara T., Szmigielski B., Sroka A., Millán C., and Usón I. (2015) Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Nat. Publ. Gr., 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Veith P. D., Luong C., Tan K. H., Dashper S. G., and Reynolds E. C. (2018) Outer membrane vesicle proteome of Porphyromonas gingivalis is differentially modulated relative to the outer membrane in response to heme availability. J. Proteome Res. 17, 2377–2389 [DOI] [PubMed] [Google Scholar]
- 57. Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D. F., Burkhard P. R., and Sanchez J. (2008) Relative quantification of proteins in human cerebrospinal fluids by MS / MS using 6-plex isobaric tags. Anal. Chem. 80, 2921–2931 [DOI] [PubMed] [Google Scholar]
- 58. Tanco S., Gevaert K., and Van Damme P. (2015) C-terminomics: Targeted analysis of natural and posttranslationally modified protein and peptide C-termini. Proteomics 15, 903–914 [DOI] [PubMed] [Google Scholar]
- 59. Kjeldsen F., Giessing A. M. B., Ingrell C. R., and Jensen O. N. (2007) Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Anal. Chem. 79, 9243–9252 [DOI] [PubMed] [Google Scholar]
- 60. Stensland M., Holm A., Kiehne A., and Fleckenstein B. (2009) Targeted analysis of protein citrullination using chemical modification and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 23, 2754–2762 [DOI] [PubMed] [Google Scholar]
- 61. Tutturen A. E. V., Holm A., and Fleckenstein B. (2013) Specific biotinylation and sensitive enrichment of citrullinated peptides. Anal. Bioanal. Chem. 405, 9321–9331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The current version of Citrullia can be downloaded from https://github.com/Citrullia-SDU/Citrullia.
The mgx conversion program MGF Filter can be downloaded from http://massai.dk/download.html.
All the ms raw files used for searching are available via ProteomeXchange (PRIDE) with the accession number PXD015701.