Abstract
BACKGROUND:
Loss or mutations of the BRCA1 gene are associated with increased risk of breast and ovarian cancers and with prostate cancer (PCa) aggressiveness. Previously, we identified GADD153 as a target of BRCA1 protein, which increases doxorubicin sensitivity in human p53 −/− PCa cells (PC3). Considering that p53 is a crucial target in cancer therapy, in this work we investigated p53 role in the regulation of transcription of GADD153.
METHODS:
We performed reverse transcription quantitative PCR (RT-qPCR), western blot and luciferase assays to analyze GADD153 and/or BRCA1 expression in response to ultraviolet or doxorubicin exposure in PC3 p53 stable-transfected cells and LNCaP (p53 +/+) cells. BRCA1 protein recruitment to GADD153 promoter was studied by chromatin immunoprecipitation-qPCR. To assess expression of BRCA1 and/or p53 target genes, we used a panel of stable-transfected PCa cell lines. We finally analyzed these genes in vivo using BRCA1-depleted PCa xenograft models.
RESULTS:
We found that GADD153 was highly induced by doxorubicin in PC3 cells; however, this response was totally abolished in LNCaP (p53wt) and in p53-restituted PC3 cells. Furthermore, BRCA1 protein associates to GADD153 promoter after DNA damage in the presence of p53. Additionally, we demonstrated that BRCA1 and/or p53 modulate genes involved in DNA damage and cell cycle regulation (cyclin D1, BLM, BRCA2, DDB2, p21WAF1/CIP1, H3F3B, GADD153, GADD45A, FEN1, CCNB2), EMT (E-cadherin, β-catenin, vimentin, fibronectin, slug, snail) and Hedgehog pathways (SHH, IHH, DHH, Gli1, PATCH1). Furthermore, xenograft studies demonstrated that BRCA1 knockdown in PC3 cells increased tumor growth and modulated these genes in vivo.
CONCLUSIONS:
Although BRCA1 induces GADD153 in a p53 independent manner, p53 abolished GADD153 induction in response to DNA damage. In addition, several important PCa targets are modulated by BRCA1 and p53. Altogether, these data might be important to understand the therapy response of PCa patients.
INTRODUCTION
The efficacy of radiotherapy and many currently used anticancer drugs relies on the presence of p53 wild-type (wt) in tumor cells.1 This is presumably due to the induction of p53 by these DNA-damaging agents and elimination of tumor cells by p53-dependent apoptosis or senescence. The absence of p53 or expression of mutant p53 has been associated with increased resistance to chemotherapy and radiotherapy.2 Thus, p53 mutant promotes cancer by stimulating cell growth, inhibiting cell death and increasing drug resistance. Furthermore, there is a high frequency of p53 mutations in human tumors and p53 mutants associate with poor prognosis in several tumor types.3
Germline mutations in the tumor suppressor gene BRCA1 (breast cancer susceptibility gene 1) are responsible for a large percentage of the inherited predisposition to breast and ovarian cancers4 and are associated with a more aggressive clinical course for prostate cancer (PCa).5–7 BRCA1 loss induces genomic instability,8 a general feature in human cancers. Cells with dysfunctional BRCA1 show defects in survival and proliferation, increased radiosensitivity, chromosomal abnormalities, G2/M checkpoint loss and impaired homologous recombination repair.9
We previously reported that BRCA1 has a central role modulating doxorubicin resistance in PCa10,11 and we identified GADD153 (DDIT3) as an important mediator of the increased sensitivity to genotoxic stress in BRCA1-expressing cells.10 Particularly, we determined that genotoxic stress induces BRCA1 binding to GADD153 promoter, activating its transcription in human prostate tumor PC3 cells (p53 −/−). Thus, GADD153 induction decreases viability and increases apoptosis and cell cycle arrest in the BRCA1 overexpressing cells which in turn induces doxorubicin sensitivity.10 In addition, GADD153 depletion abrogates BRCA1 role on cell cycle progression and cell death in response to doxorubicin treatment in PC3 cells, providing a novel mechanism for BRCA1 functions in p53-mutated PCa.10
In this work, we investigated the effect of p53 over GADD153 regulation by BRCA1 during DNA damage response using human PCa LNCaP cells that express p53 wild-type. Furthermore, in order to increase the knowledge about BRCA1 and p53 regulation in PCa, we used a panel of stable-transfected PCa cell lines to examine BRCA1/p53 modulation of 21 genes involved in cell cycle, DNA damage response, epithelial to mesenchymal transition (EMT) and Hedgehog pathways. We finally analyzed these target genes in vivo using BRCA1-depleted PCa xenograft models.
MATERIALS AND METHODS
Cell lines, reagents and plasmids
PC3 (American Type Culture Collection (ATCC, Mananssas, VA, USA): CRL-1435) and LNCaP (ATCC: CRL-1740) cells were grown in RPMI with 10% fetal bovine serum. PC3 and LNCaP stable-transfected cell lines (pcDNA3, pcDNA3 BRCA1, small hairpin RNA (shRNA) scramble and shRNA BRCA1) were previously described.10,11 PC3 stable cell lines expressing p53 (pcDNA3 p53) and control (pcDNA3) were generated by transfection with 10 μg of plasmids using lipofectamine 2000 (Invitrogen, Sao Pablo, Brazil). Cells were selected with G418 (200 μg ml−1) during 10 days, single clones were amplified and p53 expression was determined by western blot (WB). Doxorubicin (Rontag S.A, Buenos Aires, Argentina.) was prepared in dimethyl sulfoxide. For ultraviolet (UV) treatment, cells were exposed 20 s to UV 80 J m−2, 254 nm (range 240–280) in a Philips ultraviolet lamp (TUV15WG15T8) calibrated to deliver 2.5 J m−2 s−1.10 After UV-irradiation, medium was replaced and cells were maintained at 37 °C in a 5% CO2 humidified incubator for 1 h. GADD15312 and BRCA113 promoter luciferase plasmids were kindly provided by Dr Nikky J. Holbrook (NIA, USA) and Dr Solomon (UMDS, UK), respectively. BRCA1 expression vector (pcDNA3 BRCA1) was previously described.11 shRNA scramble control and shRNA BRCA1 were from Upstate (Billerica, MA, USA). P53 expression vector (pcDNA3 p53) was kindly provided by Dr Norberto Zwirner (IBYME, Argentina).
Western blot
PC3 and LNCaP cells were exposed to the indicated treatments, lysed and immunobloted as previously described10 using antibodies against GADD153 (R20), p53 (DO-1), Lamin A/C (636) and Actin B (I19) proteins from Santa Cruz Biotechnology (Heidelberg, Germany) or using an affinity-purified polyclonal antibody against BRCA1 protein gently provided by Dr Kevin Gardner (NCI—NIH).11 Protein quantitation was determined using Image J 1.41 software (Bethesda, MD, USA).
RNA isolation, complementary DNA synthesis and qPCR (RT-qPCR)
Cells were exposed to the indicated treatments and total RNA was isolated using Tri Reagent (Genbiotech, Buenos Aires, Argentina). Complementary DNA was synthesized from RNA (2 μg) using RevertAid First Strand (Fermentas, Vilnius, Lithuania). qPCR was performed as previously described10,11 using Taq Polymerase (Fermentas) in a DNA Engine Opticon (MJ Research, BioRad, Hercules, CA, USA). Three biological independent replicates were performed. RT-qPCR array figure was generated by average of the selected expression gene from three biological independent experiments for cell lines or five mice for xenografts. Student’s t-test was used to establish statistical significance for each gene with a threshold of P < 0.05. Data were normalized to Actin B and control. Primers sequences are shown in Table 1.
Table 1.
Reverse transcription quantitative PCR primer sequences.
| Gene | Forward | Reverse |
|---|---|---|
| BRCA1 | 5′-TGAAATCAGTTTGGATTCTGC-3′ | 5′-CATGCAAGTTTGAAACAGAAC-3′ |
| Cyclin D1 | 5′-GCGGAGGAGAACAAACAGAT-3′ | 5′-TGAGGCGGTAGTAGGACAGG-3′ |
| BLM | 5′-GAATGGTTAAGCAGCGATG-3′ | 5′-TCAATACATGGAACTTTCTCAG-3′ |
| BRCA2 | 5′-AAGCATTGGAGGAATATCGTAGG-3′ | 5′-CAGGTTCAGAATTATAGGGTGGAG-3′ |
| DDB2 | 5′-TCACTTCCAGCACCTCACAC-3′ | 5′-ACGTCGATCGTCCTCAATTC-3′ |
| CCNB2 | 5′-TTCTGATGCCTTGCTCTGC-3′ | 5′-ATGCGTCCATTTATATCTCTTCC-3′ |
| p21WAF1/CIP1 | 5′-CCCTTGTGCCTCGCTCAG-3′ | 5′-CGTTTGGAGTGGTAGAAATCTGTC-3′ |
| GADD45A | 5′-GAGAGCAGAAGACCGAAAGGATG-3′ | 5′-CCAGCAGGCACAACACCAC-3′ |
| FEN1 | 5′-GGCAACCCCGAACCAAGC-3′ | 5′-GCTCATAGAGGCATCAATGGC-3′ |
| GADD153 | 5′-AAGGCACTGAGCGTATCATGTTAAA-3′ | 5′-TTTCAGGTGTGGTGATGTATGAAGA-3′ |
| H3F3B | 5′-AAAGCCGCCAGGAAAAGC-3′ | 5′-CAGACCCACCAGGTACGC-3′ |
| E-Cadherin | 5′-AAGGTTCACCCAGCACCTTGCA-3′ | 5′-GGCAGAGGGACACACCAGTGTAGTAA-3′ |
| β-Catenin | 5′-CATAACCTTTCCCATCATCGT-3′ | 5′-TGTGGAGAGTTGTAATGGCA-3′ |
| Vimentin | 5′-CACTCCCTCTGGTTGATAC-3′ | 5′-GTGATGCTGAGAAGTTTCG-3′ |
| Slug | 5′-TCGGACCCACACATTACC-3′ | 5′-CAGATGAGCCCTCAGATTTG-3′ |
| Snail | 5′-CCTGCGTCTGCGGAACCTG-3′ | 5′-GTTGGAGCGGTCAGCGAAGG-3′ |
| Fibronectin | 5′-GCTCATCCGTGGTTGTATCAGG-3′ | 5′-TGGTCTGCTTGTCAAAGTGTCC-3′ |
| SHH | 5′-AGTTTCACTCCTGGCCACTG-3′ | 5′-GATGAAGAAAACACCGGAGC-3′ |
| IHH | 5′-AGATAGCCAGCGAGTTCAGG-3′ | 5′-GCTCACCCCCAATTACAATC-3′ |
| DHH | 5′-ACATGTTCATCACGGCAATG-3′ | 5′-AACCCCGACATCATCTTCAA-3′ |
| GLI1 | 5′-CACTGGTCTGTCCACTCTTCG-3′ | 5′-GCTGCTGCGGCGTTCAAG-3′ |
| PATCH1 | 5′-TCTCCAATCTTCTGGCGAGT-3′ | 5′-TGGGATTAAAAGCAGCGAAC-3′ |
| Actin B | 5′-AAGATCATTGCTCCTCCTGAGC-3′ | 5′-CATACTCCTGCTTGCTGATCCA-3′ |
Reporters
Cells were transfected using 1.5 μl of lipofectamine 2000 (Invitrogen) with 1 μg of GADD153 luciferase or BRCA1 luciferase plasmids or co-transfected with 1 μg of reporter plasmid and 1 μg of shRNA BRCA1 or shRNA scramble plasmids. After 24 h, cells were exposed to genotoxic agents as indicated, harvested and lysed with 40 μl of Steady Glo Luciferase System and 40 μl of RPMI medium without phenol red (Promega, Madison, WI, USA). Luciferase activity was measured using 20 μl of lysates in Luminometer (Glomax Multi Detection System, Promega). Data were normalized to total protein.
Chromatin immunoprecipitation
BRCA1-chromatin immunoprecipitation (ChIP) experiments were performed as previously described11 from LNCaP cells exposed or not to UV (50 J m−2) using BRCA1 antibody and nonspecific antibody control (immunoglobulin G). ChIP-DNA was amplified by qPCR using primers mapping at 0.2 Kb upstream from GADD153 transcription start site. Primer sequences are 5′CCTCCGTGAAGCCTCGTGAC3′ and 5′CTCGCATCCGCCACTCAGG3′. Fold enrichment was calculated normalizing data to input and nonspecific binding as previously described.14
Xenografts
Six-week-old male nu/nu mice were housed under pathogen-free conditions following the University of Buenos Aires animal care guidelines. Mice were randomized into two groups (five mice per group) and shRNA scramble or shRNA BRCA1 stable-transfected cells (4.8 × 106 cells) were inoculated subcutaneously into the flank of the mice. Tumor size was measured three times per week using digital caliper and the volume was calculated using the formula: 0.523 × width2 × length, where width is the smallest side of the tumor. Mice were killed 25 days after inoculation. RNA was isolated from tumors as described.10
Statistical analysis
All results are given as mean±s.d. of ‘n’ separate independent experiments unless stated otherwise. Student’s t-tests were used to ascertain statistical significance with a threshold of P < 0.05. Comparisons for in vivo experiments were made with one-way analysis of variance followed by Dunnett’s test, with P < 0.05 as the criterion for statistical significance.
RESULTS
P53 abolished GADD153 induction by DNA damage
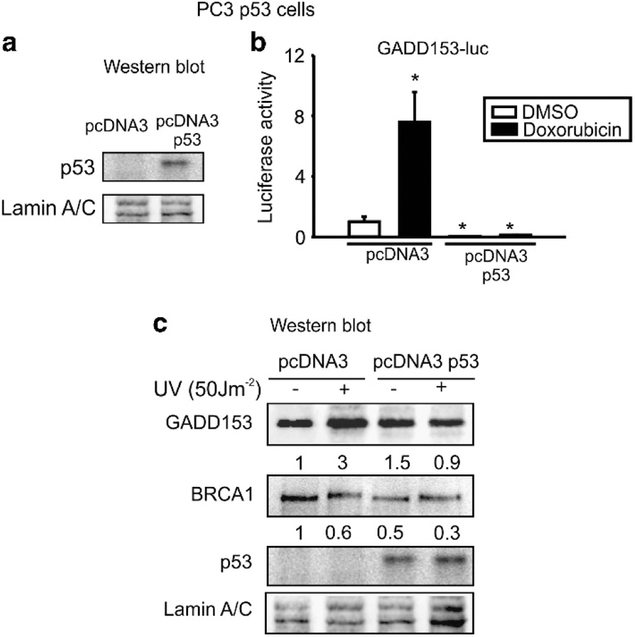
Recently, we reported that BRCA1 induces GADD153 transcription in PC3 (p53 −/−) cells in response to DNA damage.10 It was previously reported that p53 modulates BRCA1.15 To investigate whether GADD153 regulation is dependent on p53, we first generated PC3 cells that stably express p53 protein (pcDNA3 p53) and it’s control (pcDNA3). P53-restored expression in PC3 cells was determined by WB (Figure 1a).
Figure 1.
p53 abolished GADD153 induction by DNA damage. (a) PC3 cells were stable transfected with pcDNA3 p53 or control plasmid and p53 protein expression was analyzed by western blot. (b) PC3 pcDNA3 p53 or pcDNA3 cells were transfected with GADD153-luc vector and exposed to doxorubicin (1 μM, 24 h) or vehicle (dimethyl sulfoxide (DMSO)). Luciferase activity was determined 48 h after transfection. Data were normalized to total protein and control. Media and s.d. values from three independent experiments are shown. *P < 0.05. (c) PC3 pcDNA3 or pcDNA3 p53 stable cell lines were exposed to ultraviolet (UV) (50 J m−2) or not and GADD153, BRCA1, p53 and lamin A/C protein levels were determined by WB. Quantification and normalization to lamin A/C protein levels and control is shown under each band. One representative experiment from three biological replicates is shown.
We exposed pcDNA3 p53 and pcDNA3 stable cell lines to doxorubicin or vehicle after transfection with a GADD153 luciferase plasmid. We found that GADD153 promoter activity was induced by doxorubicin in PC3 pcDNA3 control cells, as we previously reported;10 however, p53-restored expression totally abolished this response (Figure 1b).
In addition, we exposed PC3 cells to UV, a well-known genotoxic agent that induces GADD153 expression.16 UV treatment increased GADD153 protein levels in PC3 pcDNA3 control cells. Nevertheless, p53 restitution abolished GADD153 induction by UV (Figure 1c).
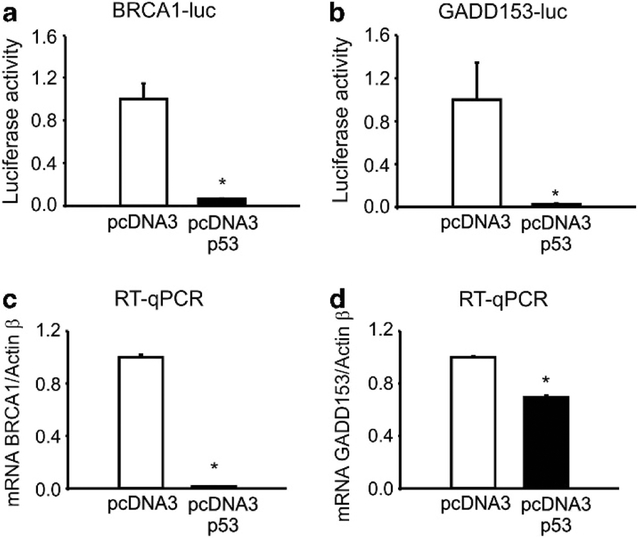
p53 represses BRCA1 and GADD153 transcription
To investigate p53 effect on BRCA1 and/or GADD153 promoter activities, we transfected PC3 pcDNA3 p53 and control cells with BRCA1- or GADD153-luc plasmids and measured luciferase activity. We found that p53 expression dramatically repressed both BRCA1 and GADD153 promoter activities (Figure 2a–b), as was previously reported.15 Accordingly, p53 overexpression significantly decreased BRCA1 and GADD153 mRNA levels (Figure 2c–d).
Figure 2.
p53 represses BRCA1 and GADD153 transcription. PC3 pcDNA3 or pcDNA3 p53 stable cell lines were transfected with BRCA1-luc (a) or GADD153-luc (b) vectors and luciferase activity was determined 48 h post transfection. Data were normalized to total protein. BRCA1 (c) and GADD153 (d) mRNA levels from PC3 pcDNA3 or pcDNA3 p53 stable cell lines were analyzed by real-time quantitative PCR (RT-qPCR). Data were normalized to actin B and control.
Altogether, these results demonstrate that GADD153 induction by genotoxic stress is lost in the presence of p53 wt protein, probably due to p53-mediated BRCA1 transcriptional repression.
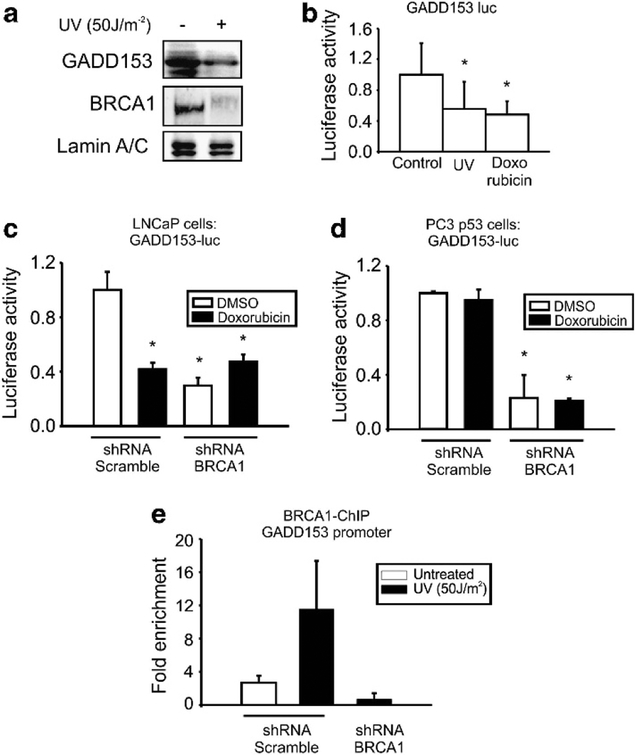
BRCA1 regulates GADD153 transcription in the presence of p53 wt
To determine whether DNA damage modulates GADD153 and BRCA1 expression in the human p53 wt cell line, we exposed LNCaP cells to UV and analyzed the protein levels by WB. We found that UV decreased GADD153 and BRCA1 expression (Figure 3a).
Figure 3.
BRCA1 protein binds and regulates GADD153 expression even in the presence of p53 wt. (a) LNCaP cells were exposed or not to UV (50 J m−2) and BRCA1, GADD153 and lamin A/C levels were analyzed by western blot (WB). LNCaP cells were transfected with GADD153-luc vector (b) or co-transfected with GADD153-luc vector and small hairpin RNA (shRNA) BRCA1 or control (shRNA scramble) (c). Cells were exposed to ultraviolet (UV) (50 J m−2), doxorubicin (1 μM, 24 h) or vehicle (dimethyl sulfoxide (DMSO)) and luciferase activity was determined 48 h post transfection. Data were normalized to total protein and control. (d) PC3 pcDNA3 p53 cells were co-transfected with GADD153-luc vector and small hairpin RNA (shRNA) BRCA1 or shRNA scramble plasmids. Cells were treated with doxorubicin (1 μM, 24 h) or control (DMSO) and luciferase activity was determined 48 h post transfection. Data were normalized to total protein. (e) BRCA1-ChIP experiment was performed in LNCaP control (shRNA scramble) and BRCA1-depleted LNCaP cells (shRNA BRCA1) exposed or not to UV (50 J m−2). BRCA1 binding to GADD153 promoter was analyzed by qPCR. Fold enrichment was determined normalizing data to input and nonspecific antibody (immunoglobulin G). RT-qPCR, real-time quantitative PCR.
Furthermore, GADD153 promoter activity was significantly decreased in LNCaP cells transfected with GADD153-luc plasmid after exposure to either UV or doxorubicin (Figure 3b). These results demonstrated that these genotoxic agents repress GADD153 and BRCA1 expression in p53 wt cells.
Previously, we reported that the transcriptional activity of GADD153 promoter was increased after doxorubicin exposure and this activity was significantly decreased by BRCA1 depletion.10 In order to further understand BRCA1 effect on GADD153 transcription in the presence of p53 wt, we co-transfected LNCaP or PC3 p53-restored cells with GADD153-luc plasmid and shRNA BRCA1 plasmid or shRNA scramble control and determined luciferase activity after doxorubicin exposure. We found that GADD153 promoter activity was not induced by doxorubicin in PC3 pcDNA3 p53 cells (Figure 3d), but was repressed in LNCaP cells (Figure 3c). These results suggest that p53 wt might abolish GADD153 promoter response to doxorubicin.
Nevertheless, BRCA1 depletion markedly decreased GADD153 promoter activity in LNCaP and PC3 p53-restored cells (Figures 3c–d). Considering that we previously reported the same result in p53 −/− cells,10 we speculate that GADD153 regulation by BRCA1 is independent of p53 status.
Furthermore, ChIP-qPCR using a specific antibody for BRCA1 protein11 demonstrated that BRCA1 associates to GADD153 promoter after UV exposure in LNCaP cells (Figure 3e), as we previously reported in PC3 cells.10 This enrichment is specific as BRCA1 recruitment to GADD153 promoter was completely abolished after BRCA1 depletion. Altogether, these results indicate that BRCA1 protein binds and regulates GADD153 promoter probably independently of p53 status.
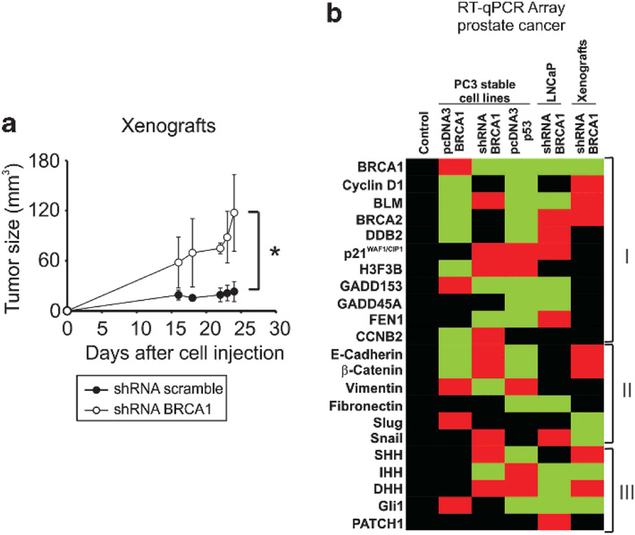
BRCA1 depletion increases PCa tumor growth
We generated a xenograft model by inoculation of BRCA1-depleted (shRNA BRCA1) or control (shRNA scramble) PC3 cells in nude mice. These stable-transfected cells were previously described.10 Tumor volume was measured every 2 days during 25 days. No significant difference was observed in the body weight of mice throughout the experiment (data not shown). Tumor growth was increased threefold (P < 0.05) in shRNA BRCA1 xenografts 16 days post inoculation compared with controls (Figure 4a). Furthermore, the tumor growth rate of the shRNA BRCA1 (5.98 mm3 per day) was 12-fold higher than in control xenografts (0.50 mm3 per day). After mice were killed, tumors were excised, RNA was isolated and RT-qPCR analysis was performed to confirm the BRCA1 knock-down expression in the tumor. BRCA1 expression in the xenografts and cell lines are shown in Figure 4b (first line). We believe this is the first study to demonstrate that BRCA1 suppresses tumor growth in a PCa experimental model.
Figure 4.
BRCA1 regulate several pathways in vivo in prostate cancer. (a) Ten Nu/nu mice were randomly distributed into two groups and inoculated subcutaneously with PC3 small hairpin RNA (shRNA) BRCA1 or shRNA scramble stable cell lines. Tumor size was measured using digital caliper every 2 days during 25 days. Graph indicates the average and s.d. of tumor volumes from five mice. (b) At day 25, mice were killed, tumors extirpated and RT-qPCR array analysis of the indicated genes was carried out. Figure was generated by average of the selected expression gene from three biological independent experiments for cell lines or three mice for xenografts. Student’s t-tests were used to establish statistical significance for each gene with a threshold of P < 0.05. Significantly induced genes were plotted in red, significantly repressed genes were plotted in green and black represents no change for each expression gene. Each data was normalized to Actin B and control.
BRCA1 and p53 regulate several pathways in PCa cells
We have previously identified a range of BRCA1 transcriptional targets in PCa and have linked these genes to cell cycle regulation and DNA damage response.10 In order to extend these studies, we performed an RT-qPCR array to investigate the gene expression profile in PC3 and LNCaP-derived stable cell lines with BRCA1/p53 overexpression or BRCA1 depletion.
BRCA1 expression was previously reported10 and is shown in Figure 4b. Afterwards, we studied three set of genes: (i) DNA damage and cell cycle regulators: cyclin D1, BLM, BRCA2, DDB2, p21WAF1/CIP1, H3F3B, GADD153, GADD45A, FEN1, CCNB2, (ii) EMT markers: E-cadherin, β-catenin, vimentin, fibronectin, slug, snail and (iii) Hedgehog pathway genes: SHH, IHH, DHH, Gli1, PATCH1 (Figure 4b, Supplementary table 1).
In the first set, we identified three clusters of genes: (i) genes that were repressed by BRCA1 and p53: Cyclin D1, BLM, BRCA2 and DDB2. (ii) genes that were modulated in a different manner by BRCA1 and p53: p21WAF1/CIP1, H3F3B, GADD153. (iii) genes that were modulated by BRCA1 or p53: GADD45A, FEN1, CCNB2 (Figure 4b, Supplementary table 1).
In the second set, we observed that p53 and BRCA1 down-regulate epithelial markers (E-cadherin and β-catenin) and induce mesenchymal markers (vimentin and fibronectin). In addition, the EMT effector Snail was induced after BRCA1 knockdown (Figure 4b, Supplementary table 1).
In the third set, we found no consistent modulation of hedgehog pathway’s ligands; however, the effector Gli1 was induced by BRCA1 overexpression and repressed by p53 restitution in PC3 cells (Figure 4b, Supplementary table 1).
BRCA1 regulates several genes in PCa xenografts
To analyze the transcriptional regulation by BRCA1 in vivo, RNA from shRNA BRCA1 PC3 and shRNA scramble xenografts was isolated and a RT-qPCR array was performed (Figure 4b). We found that BRCA1 depletion downregulated target genes involved in DNA damage and cell cycle regulation (cyclin D1, BLM, BRCA2) (Figure 4b). Also, BRCA1 depletion induced the expression of epithelial markers (E-cadherin and β-catenin) and downregulated their repressors (Slug and Snail) (Figure 4b). Mesenchymal markers showed no changes in mRNA levels (vimentin and fibronectin) (Figure 4b). Finally, BRCA1-depleted tumors showed downregulation of the hedgehog pathway target gene Gli1, although no consistent modulation in the hedgehog ligands was observed (Figure 4b).
DISCUSSION
Genetic variations in the tumor suppressor gene p53 contribute to human cancers in different ways.3 First, somatic mutations are frequent in most cancers and the antiproliferative role of p53 protein in response to various stresses makes it a primary target for inactivation in cancer.3 Second, inheritance of a p53 mutation causes predisposition to early-onset cancers.3 Third, p53 is highly polymorphic in coding and noncoding regions and some of these polymorphisms have been shown to increase cancer susceptibility and to modify cancer phenotypes in p53 mutation carriers.17
In addition, BRCA1 mutations confer a more aggressive clinical course for men with localized PCa.5–7 Most of the BRCA1-associated tumors carry p53 mutations.18,19 Owing to p53 gene mutation is a late event in PCa progression and is associated with advanced stage, loss of differentiation and the transition from androgen-dependent to androgen-independent growth,20 testing patients for p53/BRCA1 genes mutations may provide useful prognostic information and could influence the recommended course of treatment.
Previously, we reported multiple regulators of genome stability and cell cycle as transcriptional BRCA1 targets.10,21 Furthermore, we showed that BRCA1 targets GADD153 promoter and increases its transcription during DNA damage response.10 GADD153 depletion significantly abrogates BRCA1 influence on cell cycle progression and cell death in response to doxorubicin treatment.10 In this work, we found that p53 abolished GADD153 induction by DNA damage (Figure 1). These effects might be due to BRCA1 repression by p53 (Figure 2). However, BRCA1 regulates GADD153 transcription in the presence of p53 wt (Figure 3). Genotoxic agents induce GADD153 and BRCA1 expression in p53-deficient cells (PC3); detecting repression of these genes in cells expressing p53 wt (Figure 3). We demonstrate that BRCA1 associates to GADD153 promoter after UV exposure in LNCaP cells by ChIP-qPCR (Figure 3e). These results indicate that BRCA1 protein binds and regulates GADD153 promoter independently of p53 status; however, GADD153 induction by DNA damage is lost in p53-expressing cells probably due to GADD153 and BRCA1 repression by p53. We also found that BRCA1 and p53 regulate several pathways in PCa, including DNA damage and cell cycle regulation, EMT and Hedgehog pathway.
EMT is a critical process for embryogenesis and has a crucial role in carcinogenesis and tumor progression. The molecular alterations involved in this process correlate with a significant downregulation of E-cadherin.22 Thus, E-cadherin is downregulated, silenced, or aberrantly expressed in multiple cancer types.22 However, several recent studies describe E-cadherin reexpression in advanced metastatic tumors,23–25 which was found to correlate with invasion and poor patient prognosis.24,26 In PCa, E-cadherin has been shown to be reexpressed in advanced disease and in metastases.27 The mechanism responsible for the reexpression of E-cadherin in advanced disease and metastases is not clear. To the best of our knowledge, our work is the first to show that BRCA1 depletion in vivo correlates with increased E-cadherin expression suggesting that BRCA1 loss could be involved in EMT process.
It was reported that chronically hyperactive Hedgehog signaling can be oncogenic, especially for the skin or brain.28 It is not clear and controversial whether the Hedgehog signaling has any role in PCa.29 Although Gli knockdown suppresses the in vitro growth of PCa cell lines or xenograft tumor growth in mice,30 the commonly used PCa cell lines show little, if any, evidence for active canonical Hh signaling activity when they are grown in standard culture conditions.31 Although we found in this work that BRCA1 and p53 influence Hedgehog gene expression, the results are not fully understood. As shown in Figure 4b, BRCA1 depletion in mice represses IHH, which in turn represses Gli1. Nevertheless, in the same conditions other ligands (SHH and DHH) are induced. Thus, more specific studies should be performed to clarify BRCA1 effects over Hedgehog pathway.
In summary, BRCA1 and p53 are crucial regulators of transcription in PCa. Moreover, BRCA1 and p53 expression status influence expression of several genes, which in turn could affect tumor aggressiveness, progression and response to therapy.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Argentinean Agency of Science and Technology (ANPCyT PICT 2006-00228, PICT 2006-00367 and PICT 2010-00431). E. Vazquez and A. De Siervi are members of the career of scientific researcher at the National Research Council (CONICET). P De Luca holds postdoctoral fellowship from CONICET. C Moiola and F Zalazar hold PhD scholarships from CONICET. K. Gardner is a principal investigator at the NCI, NIH (USA).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 1993; 74: 957–967. [DOI] [PubMed] [Google Scholar]
- 2.Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 1999; 18: 477–485. [DOI] [PubMed] [Google Scholar]
- 3.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010; 2: a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266: 66–71. [DOI] [PubMed] [Google Scholar]
- 5.Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE. Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 1994; 343: 692–695. [DOI] [PubMed] [Google Scholar]
- 6.Douglas JA, Levin AM, Zuhlke KA, Ray AM, Johnson GR, Lange EM et al. Common variation in the BRCA1 gene and prostate cancer risk. Cancer Epidemiol Biomarkers Prev 2007; 16: 1510–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher DJ, Gaudet MM, Pal P, Kirchhoff T, Balistreri L, Vora K et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res 2010; 16: 2115–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA 2007; 104: 12111–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002; 108: 171–182. [DOI] [PubMed] [Google Scholar]
- 10.De Luca P, Vazquez ES, Moiola CP, Zalazar F, Cotignola J, Gueron G et al. BRCA1 loss induces GADD153-mediated doxorubicin resistance in prostate cancer. Mol Cancer Res 2011; 9: 1078–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Siervi A, De Luca P, Byun JS, Di LJ, Fufa T, Haggerty CM et al. Transcriptional autoregulation by BRCA1. Cancer Res 2010; 70: 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JL, Freebern WJ, Collins I, De Siervi A, Montano I, Haggerty CM et al. Kinetic profiles of p300 occupancy in vivo predict common features of promoter structure and coactivator recruitment. Proc Natl Acad Sci USA 2004; 101: 11554–11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu CF, Chambers JA, Solomon E. Complex regulation of the BRCA1 gene. J Biol Chem 1997; 272: 20994–20997. [DOI] [PubMed] [Google Scholar]
- 14.De Siervi A, De Luca P, Moiola C, Gueron G, Tongbai R, Chandramouli G et al. Identification of new Rel/NFkB regulatory networks by focused genome location analysis. Cell Cycle 2009; 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLachlan TK, Somasundaram K, Sgagias M, Shifman Y, Muschel RJ, Cowan KH et al. BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J Biol Chem 2000; 275: 2777–2785. [DOI] [PubMed] [Google Scholar]
- 16.Anand S, Chakrabarti E, Kawamura H, Taylor CR, Maytin EV. Ultraviolet light (UVB and UVA) induces the damage-responsive transcription factor CHOP/gadd153 in murine and human epidermis: evidence for a mechanism specific to intact skin. J Invest Dermatol 2005; 125: 323–333. [DOI] [PubMed] [Google Scholar]
- 17.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer 2009; 9: 95–107. [DOI] [PubMed] [Google Scholar]
- 18.Holstege H, Joosse SA, van Oostrom CT, Nederlof PM, de Vries A, Jonkers J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res 2009; 69: 3625–3633. [DOI] [PubMed] [Google Scholar]
- 19.Manie E, Vincent-Salomon A, Lehmann-Che J, Pierron G, Turpin E, Warcoin M et al. High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res 2009; 69: 663–671. [DOI] [PubMed] [Google Scholar]
- 20.Navone NM, Troncoso P, Pisters LL, Goodrow TL, Palmer JL, Nichols WW et al. p53 protein accumulation and gene mutation in the progression of human prostate carcinoma. J Natl Cancer Inst 1993; 85: 1657–1669. [DOI] [PubMed] [Google Scholar]
- 21.Moiola C, De Luca P, Cotignola J, Gardner K, Vazquez E, De Siervi A. Dynamic coregulatory complex containing BRCA1, E2F1 and CtIP controls ATM transcription. Cell Physiol Biochem 2012; 30: 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agiostratidou G, Hulit J, Phillips GR, Hazan RB. Differential cadherin expression: potential markers for epithelial to mesenchymal transformation during tumor progression. J Mammary Gland Biol Neoplasia 2007; 12: 127–133. [DOI] [PubMed] [Google Scholar]
- 23.Mitselou A, Batistatou A, Nakanishi Y, Hirohashi S, Vougiouklakis T, Charalabopoulos K. Comparison of the dysadherin and E-cadherin expression in primary lung cancer and metastatic sites. Histol Histopathol 2010; 25: 1257–1267. [DOI] [PubMed] [Google Scholar]
- 24.Chao YL, Shepard CR, Wells A. Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol Cancer 2010; 9: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson LG, Zeineldin R, Stack MS. Phenotypic plasticity of neoplastic ovarian epithelium: unique cadherin profiles in tumor progression. Clin Exp Metastasis 2008; 25: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paredes J, Correia AL, Ribeiro AS, Milanezi F, Cameselle-Teijeiro J, Schmitt FC. Breast carcinomas that co-express E- and P-cadherin are associated with p120-catenin cytoplasmic localisation and poor patient survival. J Clin Pathol 2008; 61: 856–862. [DOI] [PubMed] [Google Scholar]
- 27.Emadi Baygi M, Soheili ZS, Essmann F, Deezagi A, Engers R, Goering W et al. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol 2010; 31: 297–307. [DOI] [PubMed] [Google Scholar]
- 28.Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer 2008; 8: 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen M, Feuerstein MA, Levina E, Baghel PS, Carkner RD, Tanner MJ et al. Hedgehog/Gli supports androgen signaling in androgen deprived and androgen independent prostate cancer cells. Mol Cancer 2010; 9: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez P, Clement V, Ruiz i Altaba A. Therapeutic targeting of the Hedgehog-GLI pathway in prostate cancer. Cancer Res 2005; 65: 2990–2992. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Lipinski R, Shaw A, Gipp J, Bushman W. Lack of demonstrable autocrine hedgehog signaling in human prostate cancer cell lines. J Urol 2007; 177: 1179–1185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.