Abstract
Pericyclic reactions are powerful transformations for the construction of carbon–carbon and carbon–heteroatom bonds in organic synthesis. Their role in biosynthesis is increasingly apparent, and mechanisms by which pericyclases can catalyse reactions are of major interest1. [4+2] cycloadditions (Diels–Alder reactions) have been widely used in organic synthesis2 for the formation of six-membered rings and are now well-established in biosynthesis3–6. [6+4] and other ‘higher-order’ cycloadditions were predicted7 in 1965, and are now increasingly common in the laboratory despite challenges arising from the generation of a highly strained ten-membered ring system8,9. However, although enzyme-catalysed [6+4] cycloadditions have been proposed10–12, they have not been proven to occur. Here we demonstrate a group of enzymes that catalyse a pericyclic [6+4] cycloaddition, which is a crucial step in the biosynthesis of streptoseomycin-type natural products. This type of pericyclase catalyses [6+4] and [4+2] cycloadditions through a single ambimodal transition state, which is consistent with previous proposals11,12. The [6+4] product is transformed to a less stable [4+2] adduct via a facile Cope rearrangement, and the [4+2] adduct is converted into the natural product enzymatically. Crystal structures of three pericyclases, computational simulations of potential energies and molecular dynamics, and site-directed mutagenesis establish the mechanism of this transformation. This work shows how enzymes are able to catalyse concerted pericyclic reactions involving ambimodal transition states.
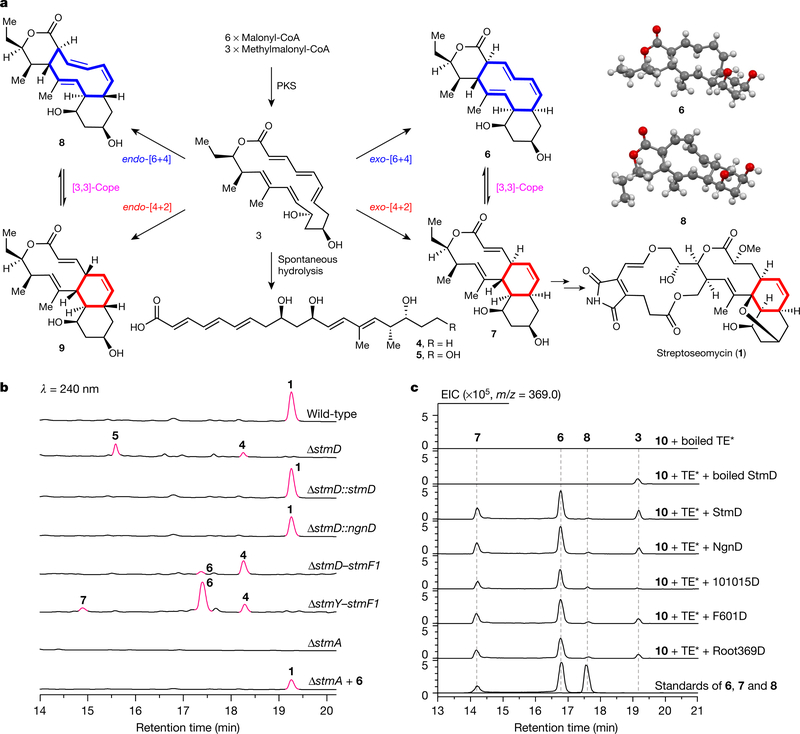
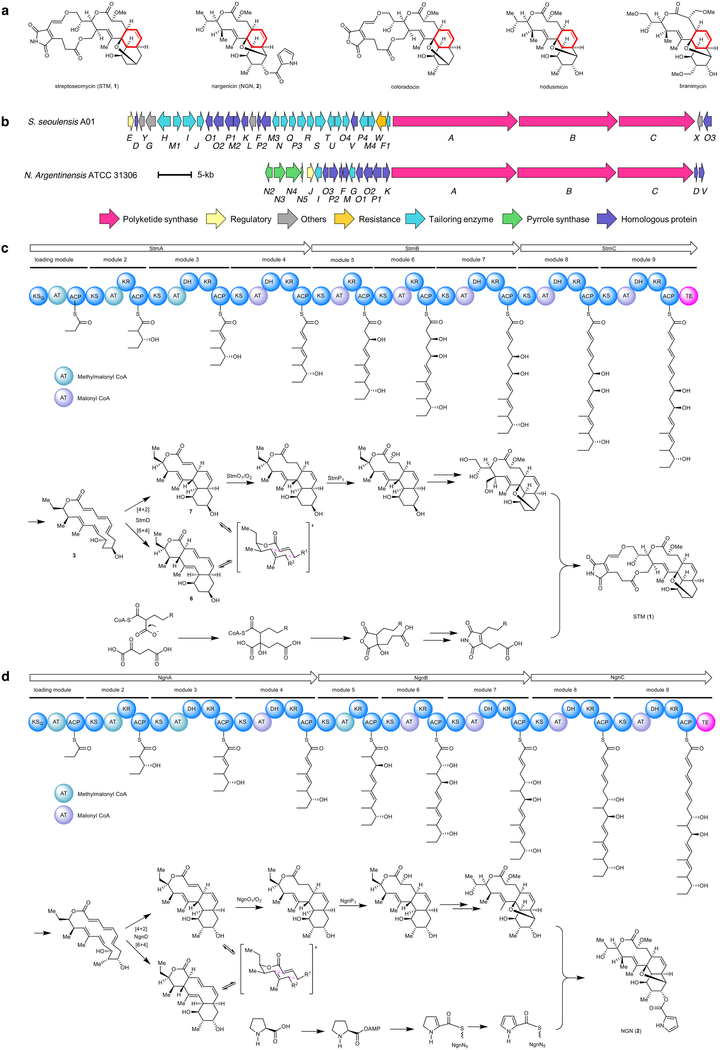
Streptoseomycin (1) is a macrocyclic polyketide that contains a cis-decalin moiety, and was isolated from a marine-derived Streptomyces seoulensis A01 during the screening of antibiotics against pathogenic Helicobacter pylori13. Streptoseomycin is structurally related to nargenicin14 (2), nodusimicin15, coloradocin16 and branimycin17 (Extended Data Fig. 1a), all of which are potent antibiotics and share a 10(9)/6/6-tricyclic ring system. It has been proposed that the cis-decalin ring that is common to these antibiotics is enzymatically formed through an intramolecular [4+2] cycloaddition13,18; however, here we determine that the transformation is more complex and, to our knowledge, unprecedented.
The biosynthetic gene cluster of 1 features a modular type-I polyketide synthase (PKS) system with nine modules13. An 18-membered macrolactone (3) was proposed as the nascent PKS product (Fig. 1a). To facilitate the identification of the cyclase in the streptoseomycin pathway, the nargenicin-producing strain Nocardia argentinensis (ATCC 31306) was sequenced for comparative biosynthetic study. A 69-kb contiguous DNA region consisting of 20 genes was annotated to encode nargenicin biosynthesis (Extended Data Fig. 1b). FigThree homologous PKS proteins (NgnA, NgnB and NgnC) encoded in the ngn gene cluster showed high sequence (>64%) and coverage (>98%) identities when compared with the streptoseomycin-pathway proteins StmA, StmB and StmC, respectively; this is consistent with their almost identical polyketide skeletons. We also located a rather small enzyme, NgnD, with previously unknown function. Its homologue StmD—which shows 85% identity—was found in the streptoseomycin pathway, which suggests that they may have the same roles.
Fig. 1 |. Cycloadditions in natural product biosynthesis.
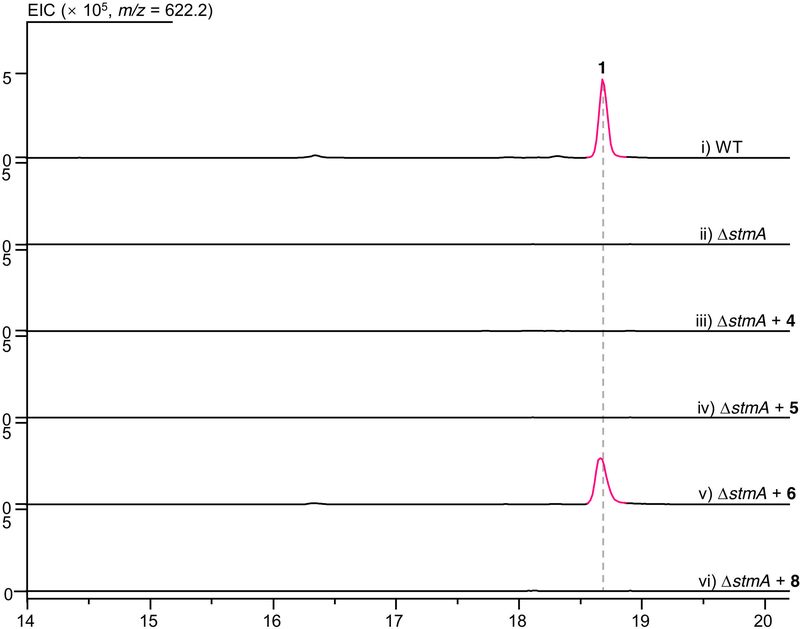
a, Proposed [6+4] and [4+2] cycloadditions involved in the biosynthesis of 1, and crystal structures of 6 and 8. b, HPLC analysis of metabolic extracts from S. seoulensis wild-type, mutant and complementary strains. c, LC–MS analysis of in vitro enzyme-catalysed [6+4]/[4+2] bispericyclic reactions. EIC, extracted ion chromatography. TE* indicates the StmC/ACP-TE. The experiments were independently repeated three times with similar results.
To investigate the function of the unknown protein StmD, we constructed a mutant strain in which the gene encoding it was deleted (ΔstmD). As expected, the production of 1 was abolished in ΔstmD, which confirms the involvement of StmD in the biosynthesis of streptoseomycin. Meanwhile, two new peaks were observed in the high-performance liquid chromatography (HPLC) profile of metabolic extract of ΔstmD (Fig. 1b). This strain was then fermented on a large scale, leading to the characterization of two linear polyketides, 4 and 5, instead of the expected compound 3. The structure of 4 is in agreement with the predicted PKS biosynthetic machinery. Compound 5 was determined to be a C19-hydroxyl derivative of 4, which could be formed by an unknown hydroxylase. The production of 1 was restored when stmD or ngnD was complemented to the ΔstmD mutant through an integrative vector pSET152 (ΔstmD::stmD and ΔstmD::ngnD, respectively; Fig. 1b); this confirms that the two homologous enzymes StmD and NgnD are functionally equivalent. However, neither 4 nor 5 is an on-pathway intermediate, because feeding them into the ΔstmA mutant cannot produce 1 (Extended Data Fig. 2). Therefore, we postulate that PKS product 3 readily undergoes hydrolysis to afford linear polyketides when downstream enzymes are absent.
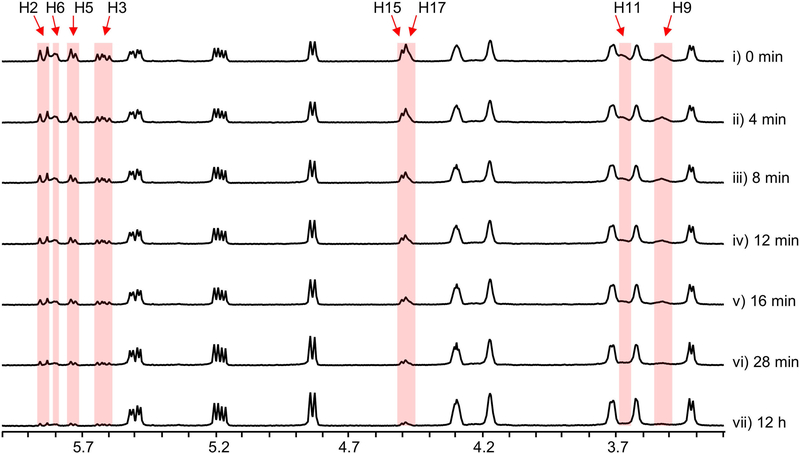
Because we could not obtain the expected intermediate 3 as an enzymatic substrate, we designed a ‘bottom-up’ strategy to establish the activity of StmD in situ. First, to accumulate the substrate of StmD in a clear background, a DNA fragment spanning around 30 kb from stmD to stmF1 was deleted to generate the ΔstmD–stmF1 mutant, leaving three PKS genes and eliminating most of the tailoring gene in the stm cluster. The resultant ΔstmD–stmF1 mutant did not produce 1, but clearly accumulated a major compound 4 (Fig. 1b) along with a minor compound 6. Next, to investigate the function of StmD in situ, we reintroduced the stmD gene into the ΔstmD–stmF1 mutant to afford the ΔstmY−stmF1 strain. Comparison of HPLC traces revealed the appearance of a major product 6 and a minor product 7, along with 4 (Fig. 1b). Large-scale fermentation of the ΔstmY−stmF1 strain led to the isolation of 6 and 8, the latter of which is barely observed in the HPLC profile. Structures of 6 and 8 were unambiguously determined by mass spectrometry, nuclear magnetic resonance (NMR) and X-ray crystallography (Fig. 1a). It is notable that 6 and 8 share a 6/10/6 carbon skeleton, instead of a 10/6/6 tricycle. 6 and 8 are therefore the exo- and endo-[6+4] adducts of 3, and not the Diels–Alder adduct. This is, to our knowledge, the first time that an enzyme-catalysed [6+4] cycloaddition has been observed directly. Attempts to purify 7 were not successful, as it rapidly converted into 6. A mixture of 6 and 7 with a ratio of approximately 3:2 was obtained under dry-ice protection (Extended Data Fig. 3). One- and two-dimensional NMR data for 7 were carefully extracted from the spectra of a mixture of 6 and 7, which enabled us to establish the 10/6/6-tricyclic structure of 7. Compound 7 can convert into 6 spontaneously by a stereoselective [3,3]-Cope rearrangement (Extended Data Fig. 1c), which establishes the stereochemistry of 7. The calculated 13C NMR data of 7 agree well with the experimental spectra, further supporting the structure of 7. Notably, 7 is the expected exo-[4+2] adduct, the skeleton and stereochemistry of which completely match the final product 1.
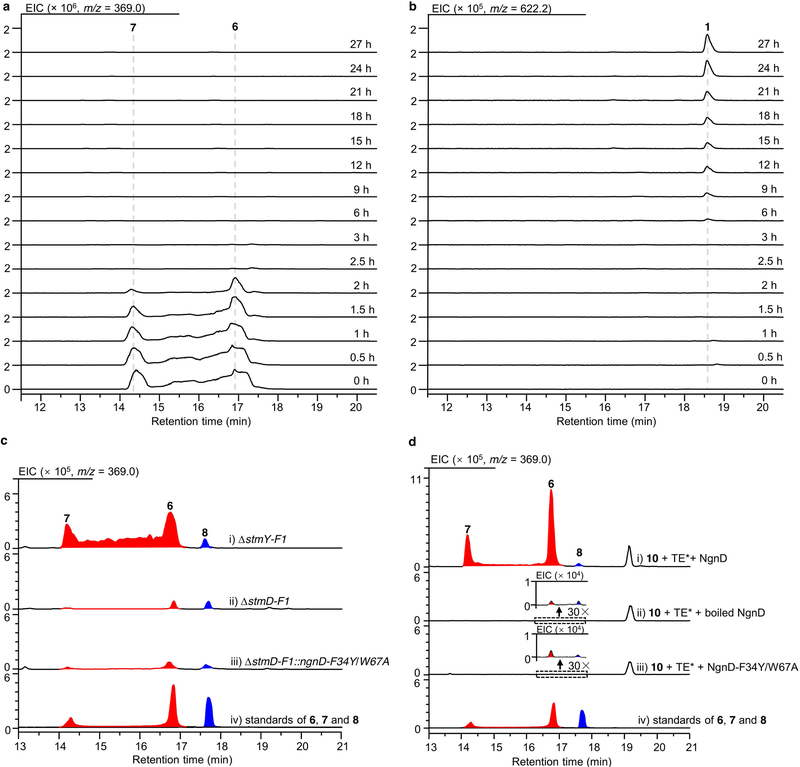
When feeding the exo-[6+4] adduct 6 into the ΔstmA strain, complete conversion of 6 to 1 was observed after 24 h of culture (Fig. 1b, Extended Data Fig. 4a, b), indicating that the thermodynamically more stable [6+4] adduct 6 is converted into the [4+2] adduct 7 through Cope rearrangement when a downstream enzyme—putatively an enoyl reductase-like enzyme (Extended Data Fig. 1c)—is present to reduce the alkene in the α,β-unsaturated ester group of 7. This is a rare example of an enzymatic system that follows the Curtin–Hammett principle19,20. That is, the [6+4] and [4+2] adducts are in equilibrium, but which of these isomers is carried on to the product depends on the transition states of the subsequent reactions, rather than the relative energies of the isomers. By contrast, 8 was determined to be a shunt metabolite, as supplementation of 8 to the ΔstmA strain did not produce 1 (Extended Data Fig. 2). Because cycloadditions can proceed non-enzymatically4,21, we estimated the accelerating effect of StmD on the cycloaddition step. Analysis by liquid chromatography coupled with mass spectrometry (LC–MS) in the extracted ion chromatography mode showed that only tiny amounts of 6–8 are generated in the ΔstmD–stmF1 mutant (Extended Data Fig. 4c), which is consistent with the spontaneous background reaction. By contrast, the combined amount of 6 and 7 increased by around 200-fold in the ΔstmY−stmF1 strain, which indicates that StmD is a pericyclase that accelerates the selective formation of exo-adducts.
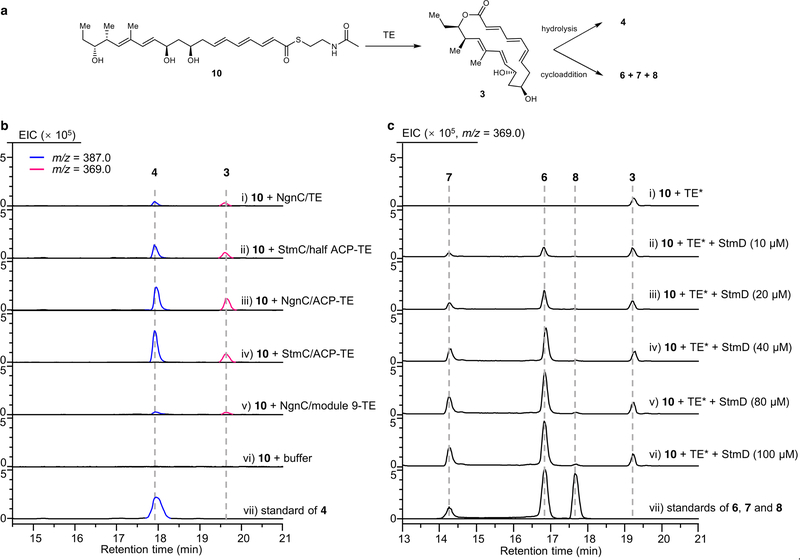
To further confirm the nature of the StmD-catalysed reaction, we reconstructed the cycloaddition step in vitro. As substrate 3 is not available, we used recombinant thioesterases (TE) from the stm or ngn gene clusters to biosynthesize 3. We synthesized the N-acetylcysteamine thioester of 4 (10) to mimic the full-length polyketide intermediate attached to the final acyl carrier protein (ACP) domain, which was then incubated with different thioesterases individually (Extended Data Fig. 5a, b). LC–MS analysis showed that StmC/ACP-TE has the highest efficiency in generating 3 with the expected molecular ion. Therefore, in the presence of StmC/ACP-TE, 10 was incubated with StmD. LC–MS analysis revealed three new peaks that were identical to 6–8 in terms of retention time and mass (Fig. 1c). Moreover, the rate of conversion from 10 to 6 and 7 is notably enhanced as the concentration of StmD increases (Extended Data Fig. 5c). NgnD can also catalyse the same cycloaddition with comparable efficiency (Fig. 1c). These results clearly indicate that StmD and NgnD are functionally equivalent, catalysing [6+4] and [4+2] cycloadditions.
The discovery of StmD and NgnD as [6+4] and [4+2] cyclases prompted us to mine more cyclases in the bacterial genome database. This search retrieved three homologues designated as 101015D, F601D and Root369D from Nocardia tenerifensis NBRC 101015, Streptomyces tsukubaensis F601 and Streptomyces sp. Root369 with 82%, 78% and 48% identities, respectively (Extended Data Fig. 6). Although no streptoseomycin-like compounds have been reported from these strains, we wanted to establish whether these homologues have the ability to catalyse cycloaddition reactions. Notably, all of them were found to catalyse both [6+4] and [4+2] cycloadditions (Fig. 1c), although F601D and Root369D have low activity. These three cyclases are encoded adjacent to several genes that are conserved in the stm and ngn gene clusters, and these strains may be able to produce streptoseomycin-like antibiotics.
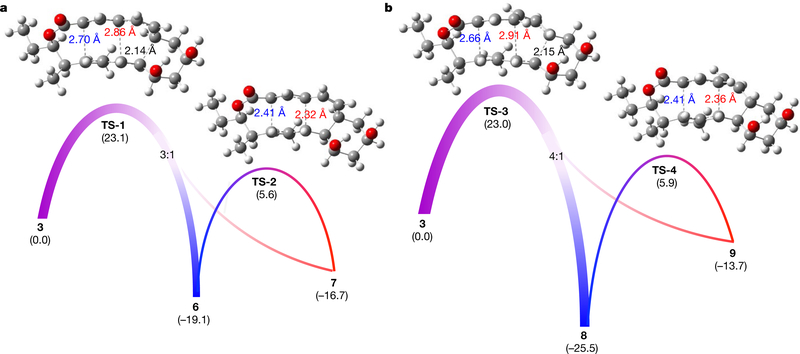
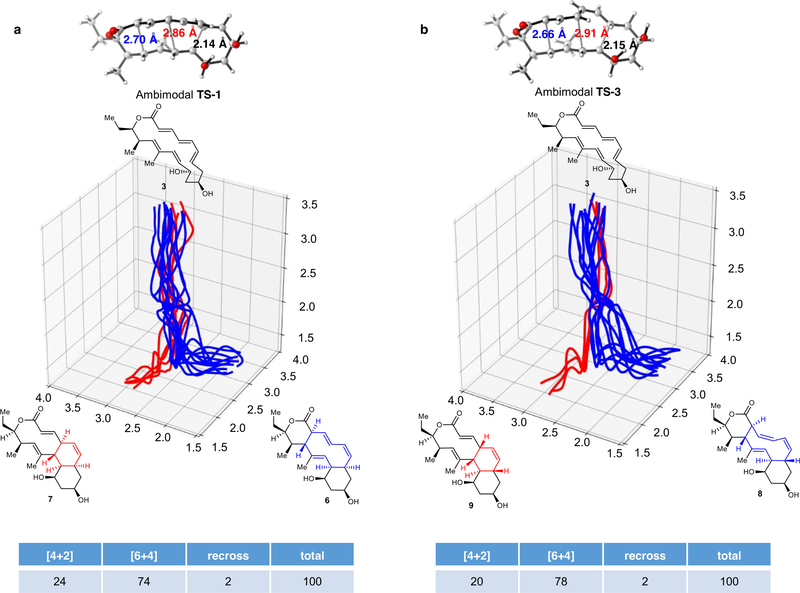
To better understand the aforementioned [6+4] and [4+2] cycloadditions and the [3,3]-Cope reaction, we performed density functional theory (DFT) calculations to evaluate the kinetics and thermodynamics of the reactions. As shown in Fig. 2a, the generation of the exo-[6+4] and exo-[4+2] adducts 6 and 7 involves a single transition state TS-1. To our knowledge, this is a new example of a bispericyclic transition state in biosynthesis1,6. Such a transition state is a normal saddle point on the potential energy surface (PES), but a bifurcation occurs after the transition state to lead to either one product or the other22,23. Here, TS-1 is followed by a bifurcation that leads to the formation of predominantly the [6+4] adduct 6 with a lesser amount of the [4+2] adduct 7 (in an approximately 3:1 ratio of 6:7, Fig. 2a). This conclusion was confirmed by performing quasi-classical molecular dynamics simulations11 (Extended Data Fig. 7a). The preference for [6+4] adduct 6 is consistent with the shorter partial C–C bond (2.70 Å) in TS-1 compared to the longer partial C–C bond (2.86 Å) that leads to [4+2] adduct 724 (Fig. 2a). The calculated activation free energy for this process is 23.1 kcal mol−1, which indicates that the reaction can occur slowly at room temperature without enzyme catalysis. Thermodynamically, the [6+4] adduct 6 is more stable than the [4+2] product 7 by 2.4 kcal mol−1. The two adducts can interconvert via a [3,3]-Cope rearrangement transition state TS-2, with barriers of 22.3 kcal mol−1 (7 to 6) and 24.7 kcal mol−1 (6 to 7) (Fig. 2a).
Fig. 2 |. DFT-computed free energies for the [6+4], [4+2] and [3,3]- Cope reactions.
a, Reaction of 3 via TS-1 to give exo-[6+4] and [4+2] products 6 and 7, and the interconversion between 6 and 7 through the Cope rearrangement transition state TS-2. b, Reaction of 3 via TS-3 to give endo-[6+4] and [4+2] products 8 and 9, and the conversion from 9 to 8 through the Cope rearrangement transition state TS-4. The numbers in parentheses show Gibbs free energies in kcal mol−1, computed with CPCM(water)-M06–2X/6–311+G(d,p)//B3LYP-D3/6–31G(d).
We also calculated the endo pathway for the formation of 8 and 9 (Fig. 1a). The bispericyclic transition state TS-3 has a barrier of 23.0 kcal mol−1 (Fig. 2b), almost identical to that of the exo pathway (23.1 kcal mol−1, Fig. 2a). This accounts for the formation of nearly equal amounts of endo- and exo-adducts under non-enzymatic conditions (Extended Data Fig. 4c, d). The post-transition-state bifurcation was predicted to give an approximately 4:1 ratio of [6+4]:[4+2] products (Extended Data Fig. 7b). However, the endo-[4+2] adduct 9 is much higher in energy than the endo-[6+4] adduct 8—by around 12 kcal mol−1—and the Cope rearrangement from 9 to 8 via TS-4 requires an activation free energy of only 19.6 kcal mol−1 (Fig. 2b). As a result, the [6+4] product 8 should be formed exclusively. All the computational findings are in agreement with experiments (Fig. 1b, c).
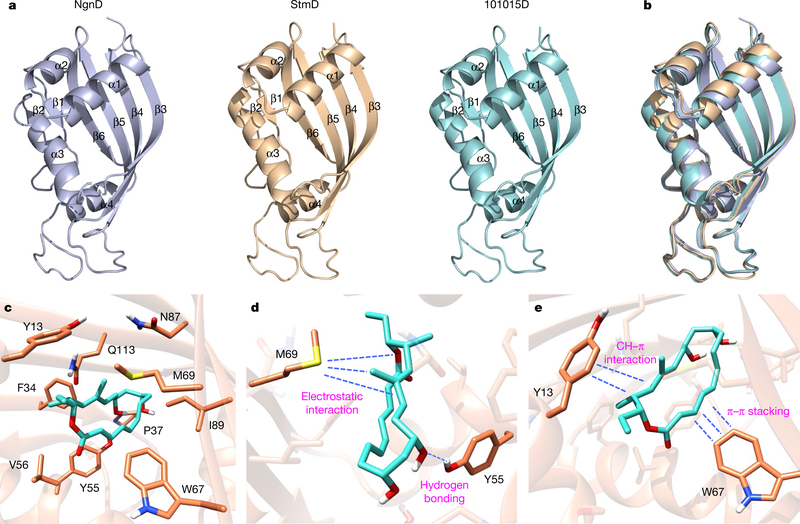
To further understand the structural details that lead to the enzyme-catalysed cycloadditions, we obtained the crystal structures of StmD, NgnD and 101015D at 2.3, 2.1 and 1.6 Å resolution, respectively (Fig. 3a). These proteins are highly similar to each other with root mean square deviation (r.m.s.d.) values of 0.47–0.51 Å (Fig. 3b). The overall structures of StmD, NgnD and 101015D adopt the α+β barrel fold consisting of four α-helices and six β-sheets (Fig. 3a). A structural homologues search25 indicated that these enzymes belong to a dimeric α+β barrel superfamily (Pfam CL0032), enzymes of which are evolutionarily conserved with diverse functions26–30.
Fig. 3 |. Crystal structures of three enzymes and catalytic sites for the bispericyclic reaction.
a, Crystal structures of NgnD, StmD and 101015D. b, The superimposed image of the three homologues. c, Ten residues of NgnD are close to DFT-optimized transition state TS-1. d, Interactions between TS-1 and M69 and Y55 of NgnD. e, Interactions between TS-1 and Y13 and W67 of NgnD.
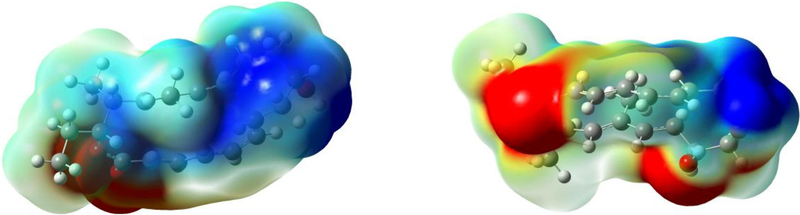
On the basis of the crystal structure of NgnD, we performed molecular dynamics simulations to investigate the active site in NgnD and its mechanism of catalysis. It was found that ten residues are close to the substrate and the transition state (Fig. 3c, Supplementary Information). Three residues—Y55, W67 and M69—have much shorter residue–transition state distances than residue–substrate distances, indicating that there are stronger stabilizing interactions between these residues and the transition state than between these residues and the reactant. These residues are likely to have a crucial role in enzyme catalysis. We also conducted the electrostatic potential analysis of the DFT- optimized transition-state structure TS-1 (Extended Data Fig. 8). There is polarization of the transition state, with electron donation from the diene moiety to the ester-substituted triene part. As shown in Fig. 3d, the electron-rich sulfur atom in residue M69 orientates towards the partially positively charged diene moiety, resulting in an electrostatic interaction that stabilizes TS-1. Meanwhile, the partially negatively charged triene ester in TS-1 is stabilized by the aromatic ring in residue W67 through a T-shaped π–π stacking interaction (Fig. 3e). The residues Y55 and Y13 have hydrogen-bonding and CH–π interactions with TS-1, respectively (Fig. 3d, e).
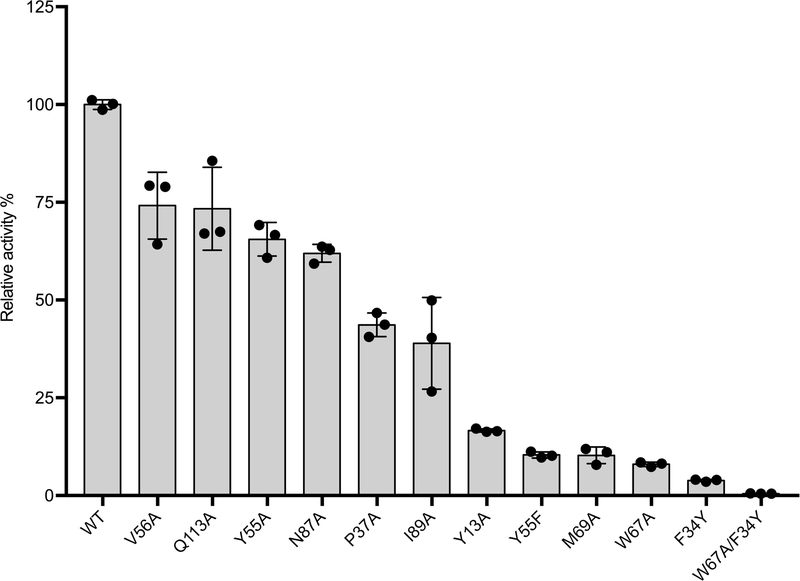
To test our predictions of the putative key residues, we carried out systematic site-directed mutagenesis of NgnD (Extended Data Fig. 9). The activity of each mutant was compared to that of wild-type NgnD by measuring the formation of 6 and 7 by LC–MS. Consistent with molecular dynamics simulations, the activities of the mutants NgnD(M69A), NgnD(W67A) and NgnD(Y55F) decreased substantially, to only 10% that of the wild-type. Moreover, the mutations Y13A, F34Y, P37A, V56A, N87A, I89A and Q113A decreased the activities of the corresponding mutants to 15%, 5%, 45%, 75%, 60%, 40% and 75%, respectively. The F34Y mutation markedly reduces activity, owing to interference with the partially positively charged diene moiety by electrostatic and steric repulsion from the OH group of tyrosine. The double mutation variant NgnD(F34Y/W67A) is nearly devoid of activity. The total amount of 6 and 7 formed by wild-type NgnD is around 500 times that formed by the mutant NgnD(F34Y/W67A), and the introduction of this double-mutant gene into the ΔstmD–stmF1 mutant does not increase the production of 6 or 7 (Extended Data Fig. 4c, d). These results further support the suggestion that NgnD and StmD catalyse the ambimodal [6+4]/[4+2] cycloaddition.
In conclusion, we have fully characterized a group of [6+4]/[4+2] bispericyclases in the biosynthesis of streptoseomycin-type natural products, using in vivo gene deletion, in vitro biochemical experiments and crystallographic structural analysis. DFT calculations reveal that the [6+4] and [4+2] cycloadditions proceed through an ambimodal transition state. The observation of mostly [6+4] adduct 6 with smaller amounts of [4+2] adduct 7 from all the catalytic proteins is strongly supportive of the computational prediction that a single ambimodal transition state leads to both products. The thermodynamically more stable [6+4] adduct 6 can convert into the [4+2] adduct 7 through Cope rearrangement when downstream enzymes are present to further transform 7 to the final product. The crystallographic and computational analyses presented here establish the catalytic mechanism of these pericyclases.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41586–019-1021-x.
METHODS
Data reporting.
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Bacterial strains, plasmids and culture conditions.
Escherichia coli DH5α was used for routine cloning. E. coli ET12567/pUZ8002 was used as the donor strain in intergeneric conjugation. The E. coli-Streptomyces shuttle plasmid pKC1139 was used for gene disruption, and pSET152-KasOp* was used for gene complementation. Ampicillin (100 μg ml−1), apramycin (50 μg ml−1), kanamycin (50 μg ml−1) and chloramphenicol (25 μg ml−1) were used for selection in E. coli. Apramycin (50 μg ml−1) was used for selection of S. seoulensis A01 recombinants. E. coli strains were grown in LB and maintained on LB agar at 37 °C. S. seoulensis A01 and mutant strains were maintained on ISP4 medium at 28 °C, unless otherwise indicated.
DNA isolation, manipulation and PCR.
Genomic DNA was purified from S. seoulensis A01 and N. argentinensis ATCC 31306. Plasmid DNAs were isolated and purified with SanPrep mini-prep kit (Sangon Biotech). Standard procedures, including restriction endonuclease digestion and transformation, were used to manipulate purified DNA. PCRs were carried out using the high-fidelity Q5 DNA polymerase (New England Biolabs) for homologous arm amplification according to the manufacturer’s protocol. The restriction endonuclease was purchased from Invitrogen. 2 × Rapid Taq Master Mix (Vazyme Biotech) was used for identification of the genotype (with additional 5% DMSO).
Gene inactivation and complementation.
To construct a plasmid for inactivation of the stmD gene, the upstream and downstream DNA homologous fragments of the stmD gene were PCR-amplified individually from genomic DNA of S. seoulensis A01. The PCR products with the expected size were cloned into HindIII and EcoRI sites of pKC1139 plasmid by using ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech) to afford pHG02002. Then, pHG02002 was transferred into the E. coli ET12567/pUZ8002 strain, and further introduced into S. seoulensis A01 by conjugation following the standard procedure described previously13. After several rounds of screening, the apramycin-sensitive double-crossover mutants were obtained, and their genotypes were verified by diagnostic PCR and DNA-gel analysis. For gene inactivation of stmA, stmD–stmF1 and stmY–stmF1, similar procedures were carried out. The genotypes of these mutants were verified by diagnostic PCR and DNA-gel analysis.
For gene complementation of the ΔstmD mutant strain, the DNA fragment containing stmD was amplified from genomic DNA of S. seoulensis A01, and then cloned into SpeI and EcoRI sites of pSET152-KasOp*—a Streptomyces integrative plasmid with a strong promoter kasO*—to afford pHG02005 by ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech). Then, pHG02005 was introduced into the ΔstmD mutant by conjugation to afford the complementation strain S. seoulensis ΔstmD::stmD. For gene complementation of the ΔstmD mutant with the ngnD gene, a similar strategy was used to generate a pHG02006 plasmid, which was further introduced into the ΔstmD mutant to afford S. seoulensis ΔstmD::ngnD. For gene complementation of the ΔstmD mutant with the ngnDY34F/W67A gene, the constructed pHG02006 was used as PCR template and NgnD-F34Y/W67A-F/R were used as primers to process two rounds of site mutation to afford the mutated ngnDY34F/W67A complementation plasmid pHG02032.
Fermentation and analysis of streptoseomycin and intermediates.
The fermentation was carried out according to a previously reported method13. In brief, fresh spores of S. seoulensis wild-type or mutant strains were individually inoculated into tryptic soy broth medium at 28 °C for two days. The seed culture was then inoculated into production medium (5 g peptone, 2 g beef extract, 5 g glucose, 1.0 g K2HPO4, 0.5 g FeSO4 in 1 l water, pH 7.0). After cultivation for eight days, the broths were filtered and extracted with ethyl acetate, evaporated under reduced pressure and dissolved in 1.5 ml acetone for HPLC analysis. HPLC was performed using a 23-min solvent gradient system from 20% to 90% methanol in water at a flow rate of 0.5 ml min−1.
Chemical complementation of 4, 5, 6 and 8 in the ΔstmA mutant.
The ΔstmA mutant was cultured on a 50-ml scale at 28 °C in production medium as described above. After 72-h cultivation, compounds 4, 5, 6 and 8 (each 0.5 mg) dissolved in DMSO were separately supplemented into fermentation broth and cultured for another 24 h. After extraction by ethyl acetate, LC–MS was performed on a reverse-phase column (Agilent Proshell 120 EC-C18 (50 × 4.6 mm, 2.7 μm)) with a gradient elution system from 20% methanol to 90% methanol in water (supplemented with 0.1% formic acid) for 23 min with a flow rate of 0.5 ml min−1. The results were analysed by Agilent Qualitative Analysis software. To construct the time course for the feeding of 6, the ΔstmA mutant was cultured on a 300-ml scale at 28 °C in production medium as described above, and 15 ml broth was withdrawn, extracted by ethyl acetate, evaporated and dissolved in 500 μl methanol at each time point.
Large-scale fermentation and secondary-metabolite isolation.
For compound isolation, large-scale fermentation (20 l) of mutant strains (ΔstmD and ΔstmY– stmF1) was carried out individually. The obtained crude extract was loaded onto a medium-pressure liquid chromatography (MPLC) system (Biotage Isolera) equipped with a Biotage SNAP Ultra C18 120 g column, affording 24 fractions by gradient elution with a linear gradient mobile phase from 10% to 90% methanol in water in a total volume of 2 l. The fractions were analysed by analytical HPLC, and those that contained target compounds were combined and concentrated. The resulting residue was loaded onto a Sephadex LH-20 column (GE Biotech) and eluted with methanol to give several fractions, which were analysed by HPLC individually and further isolated by semi-preparative HPLC as follows.
Compound 4 (3 mg, retention time (tR) = 28.1 min) was purified from the ΔstmD mutant strain by semi-preparative HPLC using 35% acetonitrile in H2O containing 0.1% formic acid at a flow rate of 2 ml min−1. Compound 5 (2 mg, tR = 25.1 min) was purified from the same mutant strain by semi-preparative HPLC using 40% acetonitrile in water.
Compound 6 (6.3 mg, tR = 24.8 min) was purified from ΔstmY-F1 by semi-preparative HPLC using 30% acetonitrile in H2O at a flow rate of 2 ml min−1. Compound 7 (1.5 mg, tR = 13.3 min) was obtained from the same mutant strain by semi-preparative HPLC using 25% acetonitrile in H2O at a flow rate of 2 ml min−1 under dry-ice protection. Compound 8 (1.2 mg, tR = 25.9 min) was also purified from the same mutant strain by semi-preparative HPLC using 30% acetonitrile in H2O at a flow rate of 2 ml min−1.
Gene cloning and expression.
All cyclase genes were individually amplified by PCR using genomic DNA or synthesized DNA as templates. After purification, these fragments were cloned into NdeI and HindIII sites of vector pET22b(+) and/or pET28a(+) by using ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech) according to the manufacturer’s protocol, except for fragments from Root369D and F601D, which were cloned only into the same sites of pET28a(+). Similarly, the fragments containing different lengths of thioesterase domains were individually amplified and cloned into pET22b(+) or pET28a(+). For site-directed mutagenesis, mutated fragments were amplified, followed by incubation with DpnI, T4 polynucleotide kinase and T4 DNA ligase according to the standard procedure of Q5 Site-Directed Mutagenesis Kit (NEB Lifescience). Each mutation construct was confirmed by DNA sequencing.
For gene expression, the recombinant plasmids were transferred into E. coli BL21(DE3). In brief, 1 l of LB with appropriate antibiotics was inoculated with overnight seed culture and incubated at 37 °C and 250 r.p.m. until an optical density at 600 nm (OD600) of 0.6 was reached. The culture was then cooled to 4 °C. Gene expression was induced with the addition of isopropyl β-D-1- thiogalactopyranoside (IPTG) to a final concentration of 0.25 mM. The cells were grown overnight at 16 °C. After collecting by centrifugation, the resulting cell pellet was resuspended in lysis buffer (100 mM Tris pH 8.0, 15 mM imidazole, 300 mM NaCl, 10% glycerol), lysed by sonication and centrifuged at 17,000g for 30 min. The supernatant containing the target protein was purified on an ÄKTA fast protein liquid chromatography (FPLC) system equipped with a 5 ml HisTrap HP column (GE Life Sciences) using two column volumes of wash buffer (100 mM Tris pH 8.0, 15 mM imidazole, 300 mM NaCl), and eluted with 50% elution buffer (100 mM Tris pH 8.0, 500 mM imidazole, 300 mM NaCl, 20% glycerol). The resultant pure protein was pooled and desalted using a PD10 column (GE Healthcare) with 100 mM sodium phosphate (pH 7.8) containing 100 mM NaCl, 50 mM KCl and 10% glycerol and concentrated with an Amicon Ultra-15 filter. The concentration of protein was determined from the absorbance at 280 nm using a molar absorptivity constant calculated by the ExPASy/ProtParam tool (https://web.expasy.org/protparam/).
Chemical synthesis of N-acetylcysteamine thioester (SNAC) mimic substrates for in vitro enzymatic assay.
To a solution of compound 4 (8 μmol, 3.1 mg) in dry DMF (300 μl) at 0 °C was added hydroxybenzotriazole (0.6 μmol, 1.3 mg), HSNAC (40 μmol, 4.76 mg) and N,Nʹ-dicyclohexylcarbodiimide (9.6 μmol, 2.0 mg). The reaction was stirred at room temperature overnight. Compound 10 was purified by semi-preparative HPLC using 32% acetonitrile in H2O at a flow rate of 2 ml min−1 to afford 10 (2 mg) at 30.1 min.
Enzymatic activity of thioesterase in vitro.
The thioesterase-catalysed reactions were carried out in a 50 μl reaction system containing 100 mM phosphate buffer (pH 7.0), 200 μM 10 and 10 μM thioesterase. After incubation at 37 °C for 2 h, 50 μl methanol was added to quench the reaction. The reaction was centrifuged at 15,000g for 10 min, and then an aliquot of 10 μl supernatant was subjected to LC–MS analysis. LC–MS was performed on a reverse-phase column (Agilent Proshell 120 EC-C18 (50 × 4.6 mm, 2.7 μm) with a gradient elution system from 20% methanol to 90% methanol in water (supplemented with 0.1% formic acid) for 23 min with a flow rate of 0.5 ml min−1. The results were analysed by Agilent Qualitative Analysis software.
Enzymatic activities of [6+4] and [4+2] cyclase in vitro.
The enzymatic reaction was carried out in a similar way to that described for the thioesterase reaction with the addition of cyclase. In brief, the enzymatic reactions were carried out in a 50 μl reaction system containing 100 mM phosphate buffer (pH 7.0), 200 μM 10, 10 μM StmC/ACP-thioesterase and 80 μM cyclase. The above reaction with boiled cyclase was used as the negative control. The reactions were performed as mentioned above. LC–MS analysis was performed using the same method as described for the thioesterase.
Selenomethionine-labelled NgnD expression and purification.
To prepare the selenomethionine (SeMet)-labelled protein, the cells were cultured in 1 l minimal medium at 37 °C, until the OD600 reached 0.6. After feeding with 0.1 g l-(+)- selenomethionine (J&K Scientific), leucine, isoleucine and valine and 0.2 g lysine, threonine and phenylalanine, the flask was left on the shaker for another 15 min and cooled to 0 °C. Finally, IPTG was added to the flask to a final concentration of 0.25 mM. Cells were incubated at 16 °C overnight. The protein was purified according to the method described above, except that all buffers contained 5 mM β-mercaptoethanol.
Protein crystallization and structure elucidation.
Crystals were grown at 22 °C with the sitting-drop vapour-diffusion method. Drops consisted of a 1:1 ratio of proteins (10 mg ml−1, 50 mM NaCl, 20 mM Tris, pH 8.0) and different crystallization buffers (100 mM sodium citrate, pH 4.2, 40% v/v ethanol, 5% w/v PEG 1000 for NgnD crystallization; 2 M ammonium sulfate, 100 mM MES, pH 6.5, 200 mM NaCl for StmD and 101015D crystallization). Crystals of NgnD and StmD were directly flash-frozen in liquid nitrogen; meanwhile, crystals of 101015D were briefly soaked in the crystallization buffer containing additional 15% glycerol before flash-freezing for protection.
One set of single-wavelength anomalous diffraction data for SeMet-NgnD was collected at BL17U1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF) at wavelengths of 0.97897 Å, whereas data for NgnD, StmD and 101015D were collected at BL18U1 beamline at SSRF at wavelengths of 0.97853 Å, 0.97853 Å and 0.97930 Å, respectively. All diffraction datasets collected at the Shanghai Synchrotron Radiation Facility were processed and scaled using iMosflm31, whereas the dataset of StmD was specially processed by XDS32. The single-wavelength anomalous diffraction phase was determined for the SeMet-derivative dataset and a partial structural model of SeMet-NgnD was traced in AutoSol33. The structural model of SeMet-NgnD was built manually on the basis of the experimental phase and then refined using PHENIX34; subsequently, NgnD (higher resolution), StmD and 101015D were solved with molecular replacement using SeMet-NgnD as the search model and further autobuilt and refined by PHENIX. Coot was used for manual model rebuilding and adjustments35. Finally, additional TLS refinement was performed in PHENIX. The geometries (Ramachandran favoured/outliers) of NgnD, StmD and 101015D are 99.7%/0.3%, 99.7%/0% and 100%/0%, respectively. The final refinement statistics are listed in Extended Data Table 1. Structural diagrams were prepared using the program PyMOL (http://www.pymol.org/) and UCSF Chimera36.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Extended Data
Extended Data Fig. 1 |. Comparative analysis of the stm and ngn gene clusters.
a, Structure of streptoseomycin (1) and structurally related natural products nargenicin (2), coloradocin, nodusmicin and branimycin. b, The stm gene cluster from S. seoulensis A01 and the ngn gene cluster from N. argentinensis ATCC 31306. c, Proposed biosynthetic pathway for 1. d, Proposed biosynthetic pathway for 2.
Extended Data Fig. 2 |. LC–MS analysis of chemical complementation in the ΔstmA mutant strain.
The experiments were independently repeated twice with similar results.
Extended Data Fig. 3 |. Time-course 1H NMR spectra for a mixture of 6 and 7.
Spectra were obtained at room temperature. 1H NMR signals highlighted in red are those from 7.
Extended Data Fig. 4 |. LC–MS analysis of product formation in both in vivo and in vitro enzymatic reactions.
a, b, Time-course analysis of chemical complementation of 6 in the ΔstmA mutant strain. EIC corresponding to 6 and 7 (m/z = 369.0, [M + Na]+) (a); EIC corresponding to 1 (m/z = 622.2, [M + Na]+) (b). c, LC–MS analysis of the production of 6–8 in mutants and in complementary strains. d, LC–MS analysis of the production for 6–8 in enzymatic reactions. TE* indicates the StmC/ACP-TE. The experiments were independently repeated twice with similar results.
Extended Data Fig. 5 |. In vitro biochemical characterization of the efficiency of thioesterase-catalysed macrolactonization and the effects of varying concentrations of StmD in enzymatic cycloaddition.
a, Thioesterase- and cyclase-catalysed tandem reactions. b, LC–MS analysis of different thioesterase-catalysed macrolactonization. c, Effects of varying concentrations of StmD on enzymatic cycloaddition. Each reaction mixture (50 μl 100 mM phosphate buffer at pH 7.0) contained 10 μM StmC/ACP-TE and 200 μM substrate 10, and (i) 0 μM StmD, (ii) 10 μM StmD, (iii) 20 μM StmD, (iv) 40 μM StmD, (v) 80 μM StmD, (vi) 100 μM StmD. (vii) Standard solution of compounds 6, 7 and 8.TE* indicates the StmC/ACP-TE. The experiments were independently repeated three times with similar results.
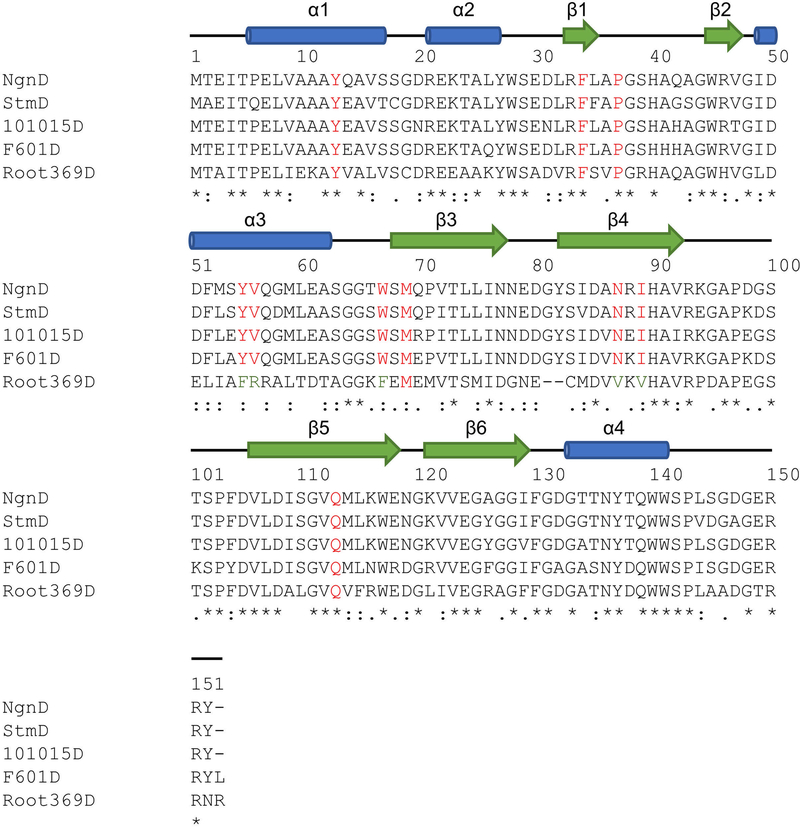
Extended Data Fig. 6 |. Sequence alignment of StmD, NgnD and three homologous proteins 101015D, F601D and Root369D.
The secondary structure elements of NgnD are presented at the top. Blue cylinders and green arrows indicate the α-helices and β-strands, respectively. The proteins 101015D (WP_040742972), F601D (WP_057609890) and Root369D (WP_077974259) are from N. tenerifensis NBRC 101015, S. tsukubaensis F601 and Streptomyces sp. Root369, respectively.
Extended Data Fig. 7 |. Distributions of reactive trajectories initiated from ambimodal transition states TS-1 and TS-3. a, TS-1. b, TS-3.
Fifteen randomly chosen trajectories were plotted in each case. Trajectories leading to a [4+2] adduct are shown in red, and those leading to a [6+4] adduct are shown in blue. The table lists the 100 trajectories that we calculated.
Extended Data Fig. 8 |. The electrostatic potential analysis of the DFT-optimized transition-state structure TS-1.
The blue and red regions represent electrostatic potential regions of positive and negative potential (repulsive and attractive interactions, respectively) with a positive charge, with darker colour representing a ‘more positive’ or ‘more negative’ potential. Two views of TS-1 are shown from front and back.
Extended Data Fig. 9 |. Relative activity of NgnD and its site-specific mutants on enzymatic reactions.
Bars represent mean relative activity averaged over three reactions and error bars indicate the standard deviation. The experiments were independently repeated twice with similar results.
Extended data table 1 |.
Data collection and refinement statistics (molecular replacement) of NgnD, StmD and 101015D
| NgnD | StmD | 101015D | |
|---|---|---|---|
| Data collection | |||
| Space group | P 32 2 1 | P21 221 | P41 2 2 |
| Cell dimensions | |||
| a, b, c (Å) | 117.0, 117.0, 63.2 | 52.1,85.0, 134.8 | 88.6, 85.6, 92.3 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 23.08 −2.05 (2.11–2.05) | 48.6–2.3 (2.38–2.3) | 31.94–1.62(1.68–1.62) |
| Rmerge (%) | 15.2 (91.1) | 6.9(19.9) | 13.4 (29.1) |
| I/σI | 6.6 (1.6) | 20.93 (9.49) | 9.2 (5.0) |
| Completeness (%) | 99.5 (99.2) | 90.7 (98.0) | 99.9 (100) |
| Redundancy | 6.4 (5.1) | 6.4 (6.7) | 7.1 (7.5) |
| Refinement | |||
| Resolution (Å) | 23.08–2.05 (2.12–2.05) | 48.60–2.3 (2.38–2.3) | 31.94–1.62(1.68–1.62) |
| No. reflections | 31334 (3062) | 24828 (2634) | 47324 (4653) |
| Rwork / Rfree | 15.92 (26.21)/18.43 (30.4) | 17.96(19.66)/24.60 (28.73) | 17.44(20.81)/20.06 (22.77) |
| No. atoms | 2543 | 4932 | 2752 |
| Protein | 2316 | 4681 | 2331 |
| Ligand/ion | 0 | 0 | 0 |
| Water | 227 | 251 | 421 |
| B-factors | 36.4 | 28.3 | 18.0 |
| Protein | 35.6 | 28.2 | 15.2 |
| Ligand/ion | 0 | 0 | 0 |
| Water | 45.4 | 29.0 | 33.2 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.009 | 0.006 |
| Bond angles (°) | 0.83 | 0.99 | 0.80 |
Values in parentheses are for highest-resolution shell.
Supplementary Material
Acknowledgements
This work was financially supported by Ministry of Science and Technology (2018YFC1706200), the National Natural Science Foundation of China (21572100, 21803030, 81522042, 81773591, 81530089, 81673333, 21861142005, 21761142001 and 21661140001), the National Thousand Young Talents Program, the Jiangsu Specially-Appointed Professor Plan, the Natural Science Foundation of Jiangsu Province (BK20170631) in China, and the US National Institutes of General Medical Sciences, National Institutes of Health (GM 124480). We are grateful to the High Performance Computing Center of Nanjing University for doing the numerical calculations in this paper on its blade cluster system. We thank the staff at beamlines BL17U1 and BL18U1 of Shanghai Synchrotron Radiation Facility and the National Supercomputing Center in Wuxi for their support.
Footnotes
Data availability
Atomic coordinates of StmD, NgnD and 101015D have been deposited in the Protein Data Bank (PDB) with accession codes 6A5G, 6A5F and 6A5H. The crystallographic data of compounds 6 and 8 have been deposited in the Cambridge Crystallographic Data Centre (https://www.ccdc.cam.ac.uk/) with CCDC numbers 1843311 and 1843314. The DNA sequence of the ngn gene cluster has been deposited in GenBank with the accession number MH544245.
Competing interests The authors declare no competing interests.
Reviewer information Nature thanks Fedor Novikov, Satish Nair and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41586-019-1021-x.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586–019-1021-x.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jamieson CS, Ohashi M, Liu F, Tang Y & Houk KN The expanding world of biosynthetic pericyclases: cooperation of experiment and theory for discovery. Nat. Prod. Rep. 10.1039/C8NP00075A (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takao K, Munakata R & Tadano K Recent advances in natural product synthesis by using intramolecular Diels–Alder reactions. Chem. Rev. 105, 4779–4807 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Jeon BS, Wang S-A, Ruszczycky MW & Liu HW Natural [4 + 2]-cyclases. Chem. Rev. 117, 5367–5388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HJ, Ruszczycky MW, Choi SH, Liu YN & Liu HW Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature 473, 109–112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian Z et al. An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol. 11, 259–265 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Ohashi M et al. SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature 549, 502–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann R & Woodward RB Selection rules for concerted cycloaddition reactions. J. Am. Chem. Soc. 87, 2046–2048 (1965). [Google Scholar]
- 8.Palazzo TA, Mose R & Jørgensen KA Cycloaddition reactions: why is it so challenging to move from six to ten electrons? Angew. Chem. Int. Ed. 56, 10033–10038 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Mose R et al. Organocatalytic stereoselective [8+2] and [6+4] cycloadditions. Nat. Chem. 9, 487–492 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Yu P, Patel A & Houk KN Transannular [6+4] and ambimodal cycloaddition in the biosynthesis of heronamide A. J. Am. Chem. Soc. 137, 13518–13523 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Patel A et al. Dynamically complex [6+4] and [4+2] cycloadditions in the biosynthesis of spinosyn A. J. Am. Chem. Soc. 138, 3631–3634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z et al. Influence of water and enzyme SpnF on the dynamics and energetics of the ambimodal [6+4]/[4+2] cycloaddition. Proc. Natl Acad. Sci. USA 115, E848–E855 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B et al. Discovery, biosynthesis, and heterologous production of streptoseomycin, an anti-microaerophilic bacteria macrodilactone. Org. Lett. 20, 2967–2971 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Snyder WC & Rinehart KL Biosynthesis of nargenicin and nodusmicin. J. Am. Chem. Soc. 106, 787–789 (1984). [Google Scholar]
- 15.Whaley HA, Chidester CG, Mizsak SA & Wnuk RJ Nodusmicin: the structure of a new antibiotic. Tetrahedron Lett. 21, 3659–3662 (1980). [Google Scholar]
- 16.Rasmussen RR, Scherr MH, Whittern DN, Buko AM & McAlpine JB Coloradocin, an antibiotic from a new Actinoplanes. II. Identity with luminamicin and elucidation of structure. J. Antibiot. 40, 1383–1393 (1987). [DOI] [PubMed] [Google Scholar]
- 17.Čikoš A et al. Reinvestigation of the branimycin stereochemistry at position 17-C. Org. Lett. 18, 780–783 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Cane DE & Luo GL Biosynthesis of polyketide antibiotics. Incorporation of a pentaketide chain elongation intermediate into nargenicin. J. Am. Chem. Soc. 117, 6633–6634 (1995). [Google Scholar]
- 19.Seeman JI Effect of conformational change on reactivity in organic chemistry. evaluations, applications, and extensions of Curtin–Hammett/Winstein–Holness kinetics. Chem. Rev. 83, 83–134 (1983). [Google Scholar]
- 20.Medvedev MG et al. Quantifying possible routes for SpnF-catalyzed formal Diels–Alder cycloaddition. J. Am. Chem. Soc. 139, 3942–3945 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Siegel JB et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science 329, 309–313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hare SR & Tantillo DJ Post-transition state bifurcations gain momentum– current state of the field. Pure Appl. Chem. 89, 679–698 (2017). [Google Scholar]
- 23.Hong YJ & Tantillo DJ Biosynthetic consequences of multiple sequential post-transition-state bifurcations. Nat. Chem. 6, 104–111 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Yang Z et al. Relationships between product ratios in ambimodal pericyclic reactions and bond lengths in transition structures. J. Am. Chem. Soc. 140, 3061–3067 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Holm L & Laakso LM Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sciara G et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 22, 205–215 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori T et al. Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis. Nat. Chem. Biol. 13, 1066–1073 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Clarkson WD et al. Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J. Mol. Biol. 272, 716–730 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Sultana A et al. Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J. 23, 1911–1921 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundqvist T et al. Crystal structure of scytalone dehydratase — a disease determinant of the rice pathogen, Magnaporthe grisea. Structure 2, 937–944 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Battye TGG, Kontogiannis L, Johnson O, Powell HR & Leslie AGW iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W XDS. Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger TC et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D 65, 582–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Mustyakimov M, Afonine PV & Langan P Generalized X-ray and neutron crystallographic analysis: more accurate and complete structures for biological macromolecules. Acta Crystallogr. D 65, 567–573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P & Crispin M Structural analysis of glycoproteins: building N-linked glycans with Coot. Acta Crystallogr. D 74, 256–263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.