Chemotherapy resistance and tumor relapse are the major contributors to low patient survival, and both have been largely attributed to cancer stem-like cells (CSCs) or tumor-initiating cells (TICs). Moreover, most conventional therapies are not effective against CSCs, which necessitates the discovery of CSC-specific biomarkers and drug targets.
KEYWORDS: ALDH1A1, SOX9, cancer stem-like cells, chemoresistance
ABSTRACT
Chemotherapy resistance and tumor relapse are the major contributors to low patient survival, and both have been largely attributed to cancer stem-like cells (CSCs) or tumor-initiating cells (TICs). Moreover, most conventional therapies are not effective against CSCs, which necessitates the discovery of CSC-specific biomarkers and drug targets. Here, we demonstrated that the embryonic transcription factor SOX9 is an important regulator of acquired chemoresistance in non-small-cell lung cancer (NSCLC). Our results show that SOX9 expression is elevated in NSCLC cells after treatment with the chemotherapeutic cisplatin and that overexpression of SOX9 correlates with worse overall survival in lung cancer patients. We further demonstrated that SOX9 knockdown increases cellular sensitivity to cisplatin, whereas its overexpression promotes drug resistance. Moreover, this transcription factor promotes the stem-like properties of NSCLC cells and increases their aldehyde dehydrogenase (ALDH) activity, which was identified to be the key mechanism of SOX9-induced chemoresistance. Finally, we showed that ALDH1A1 is a direct transcriptional target of SOX9, based on chromatin immunoprecipitation and luciferase reporter assays. Taken together, our novel findings on the role of the SOX9-ALDH axis support the use of this CSC regulator as a prognostic marker of cancer chemoresistance and as a potential drug target for CSC therapy.
INTRODUCTION
SOX9 is a member of the high-mobility group (HMG)-box class of transcription factors and controls critical stages in embryonic development, stem cell homeostasis, and differentiation. Germ line SOX9 mutations cause severe cartilage and bone malformations and male sex reversal (1). In adults, SOX9 is expressed in stem cells in multiple tissues, including the central nervous system, skin, intestine, liver, and pancreas (1–3). In lungs, SOX9 regulates branching morphogenesis during embryonic development and a response to injuries in adults (4, 5). SOX9 has also been implicated in the formation and progression of multiple cancer types (6). It is overexpressed in various solid tumors, such as breast, prostate, pancreatic, and lung cancers. Furthermore, SOX9 overexpression positively correlates with poor overall and disease-free survival in these malignancies (7, 8), which could be due to its effect on tumor-initiating cells or cancer stem-like cells (CSCs).
CSCs are a small cell population that is highly tumorigenic and generally resistant to apoptosis, chemotherapy, and radiation therapy and are thus thought to be a major source of drug resistance and recurrence in cancer (9). As an imbalance of stem cell homeostasis and differentiation may contribute to tumorigenesis, many stem cell transcription factors, including SOX9, have been linked to CSC regulation. Indeed, SOX9 has been shown to promote self-renewal and inhibit cell differentiation in basal cell carcinoma (10). This transcription factor is also highly expressed in liver CSCs and controls self-renewal and symmetrical cell division, promoting the aggressive behavior of hepatocellular carcinoma cells (11). It cooperates with the master regulator of epithelial-to-mesenchymal transition (EMT) Slug to maintain normal mammary stem cells and breast CSCs and to reprogram differentiated breast cells into stem cells (12). In lungs, a recent study by our group showed that SOX9 expression is associated with the stemness of lung carcinoma, as indicated by enhanced side population and sphere formation properties (13), although the underlying mechanisms are not known.
Consistent with its role in CSCs, increasing evidence suggests that SOX9 may contribute to chemotherapy resistance (14–17). However, a detailed understanding of the process remains to be elucidated. We hypothesized that SOX9 is involved in lung cancer progression and chemoresistance by inducing cancer stem-like properties. We show here that SOX9 is upregulated upon exposure to cisplatin and positively regulates cancer stem-like properties and the chemoresistance of non-small-cell lung cancer (NSCLC) cells. Moreover, we demonstrated for the first time that the stem-cell marker aldehyde dehydrogenase (ALDH) isoform 1A1 (ALDH1A1) is a direct transcriptional target of SOX9 that drives the chemotherapy response in NSCLC cells.
RESULTS
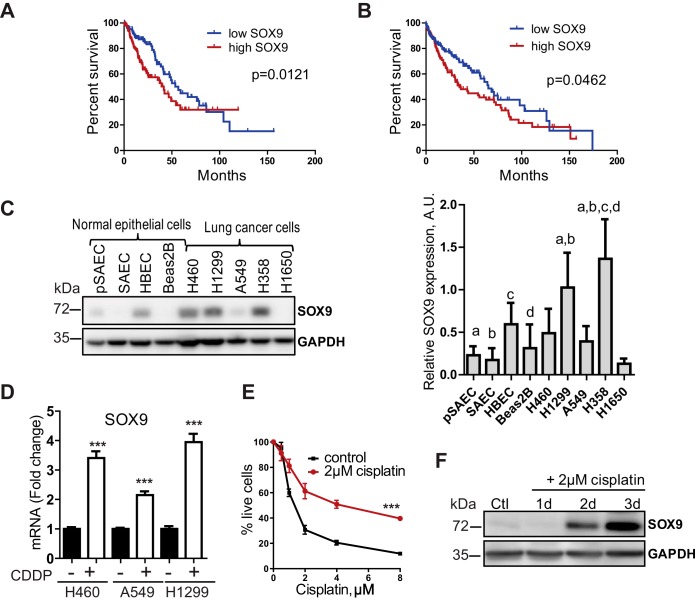
High SOX9 expression correlates with poor survival in NSCLC patients and is induced by cisplatin exposure in human lung cancer cells.
SOX9 is reported to be overexpressed in several cancer types, including lung cancer. We analyzed The Cancer Genome Atlas (TCGA) NSCLC patient cohort to check whether SOX9 overexpression could predict patient survival. Indeed, higher SOX9 expression correlates with poor overall survival in patients with adenocarcinoma and squamous cell carcinoma, two major histological types of NSCLC (Fig. 1A and B). SOX9 is overexpressed in tumors compared to its level of expression in normal tissues in patient samples (18, 19). Consistent with these data, we found that SOX9 expression is highly upregulated in most NSCLC cell lines (H460, H1299, and H358 cells) compared to primary lung epithelial cells (primary small airway epithelial cells [pSAEC]) and several nontumorigenic lung epithelial cell lines (small airway epithelial cells [SAEC] and Beas-2B cells) (Fig. 1C). Together, these observations support the role of SOX9 in lung cancer progression.
FIG 1.
High SOX9 expression correlates with poor survival in NSCLC patients and is induced by cisplatin in lung cancer cells. (A, B) Overall survival in NSCLC patients from the TCGA database. Patients with adenocarcinoma (n = 260) (A) and squamous cell carcinoma (n = 251) (B) with high SOX9 expression (top 25% expression [high SOX9] versus bottom 25% expression [low SOX9]) had lower rates of overall survival. P values were determined by the log-rank (Mantel-Cox) test. (C) (Left) Western blot analysis of SOX9 expression in normal lung epithelial cells and lung cancer cell lines; (right) quantification. P values were determined by one-way ANOVA with Tukey’s multiple-comparison test. a, P < 0.05 versus pSAEC; b, P < 0.05 versus SAEC; c, P < 0.05 versus HBEC; d, P < 0.05 versus Beas-2B cells. A.U., arbitrary units. (D) SOX9 mRNA levels are increased after a 3-day-long treatment with cisplatin (CDDP; 2 μM for H460 cells, 8 μM for A549 and H1299 cells). The data are presented as the mean ± SD. The results are representative of those from 3 independent experiments. ***, P < 0.0001 by one-way ANOVA with Tukey’s multiple-comparison test. (E) H460 cells became resistant to cisplatin after a 3-day exposure to 2 μM cisplatin (IC50, 4.3 μM versus 1.4 μM). Cisplatin-resistant cells were treated with various cisplatin concentrations for 72 h, and cell viability was assessed by the MTT assay. The results are representative of those from 3 independent experiments with 5 technical replicates each. The data are presented as the mean ± SD. ***, P < 0.0001 by two-way repeated-measures ANOVA. (F) Representative Western blot showing SOX9 upregulation in H460 cells in response to cisplatin treatment. The data are from 3 independent experiments. Ctl, control; d, day.
Multiple studies have reported an increased expression of several stem cell markers in drug-resistant cells (20–22). We tested whether SOX9 expression is upregulated in lung cancer cells in response to cisplatin treatment. NSCLC cells (H460, A549, and H1299 cells) were treated with cisplatin at the 50% inhibitory concentrations (IC50) and analyzed for SOX9 expression by reverse transcription (RT)-quantitative PCR (qPCR) and Western blotting. Figure 1D and F show that SOX9 protein and mRNA levels were considerably elevated following a 2- to 3-day exposure to the drug. Furthermore, these cells became highly resistant to cisplatin upon subsequent exposures (Fig. 1E), suggesting a potential role of SOX9 in the resistance to cisplatin.
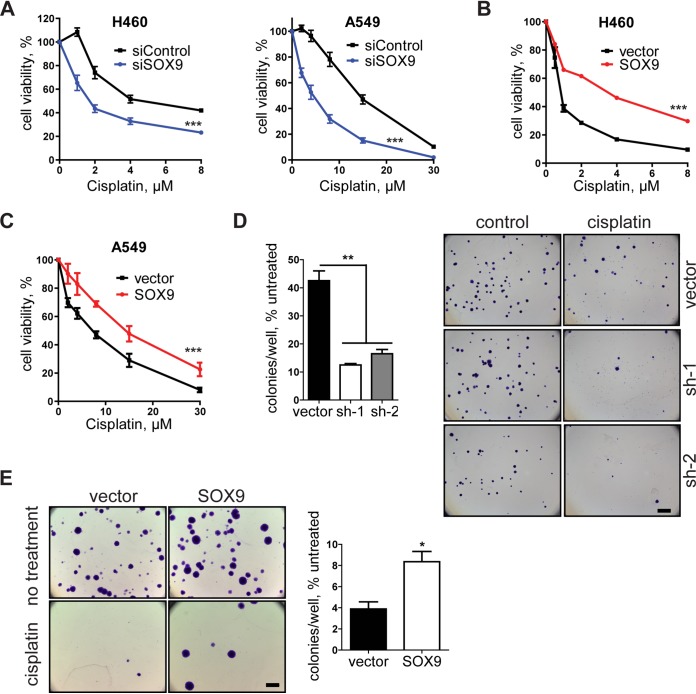
SOX9 overexpression promotes chemoresistance in NSCLC cells.
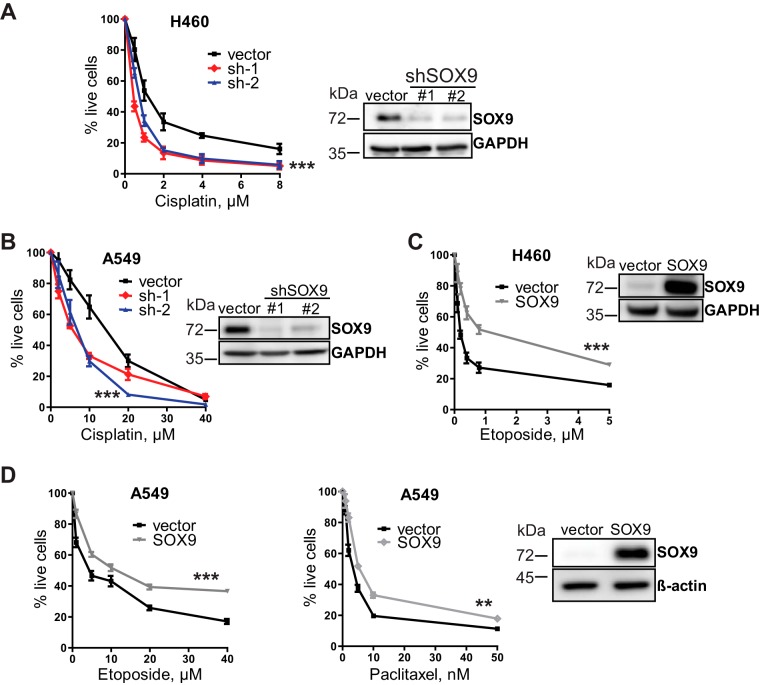
Next, we examined the effects of SOX9 overexpression and knockdown on cell survival upon treatment with chemotherapeutic drugs. Our results showed that SOX9 knockdown in two NSCLC cell lines, H460 and A549 (see Fig. 3 for knockdown and overexpression levels), increased the drug sensitivity of these cells (Fig. 2A and 3A and B), whereas SOX9 overexpression reduced the sensitivity of these cells (Fig. 2B and C). Treatment with other chemotherapeutic drugs, such as paclitaxel and etoposide, yielded similar results (Fig. 3C and D), indicating a broad role of SOX9 in cell survival upon drug exposure. While the standard cell viability assay provides valuable insight into an acute cellular response to drugs, evaluating a continued effect of drug exposure is more clinically relevant with regard to the development of resistance. Thus, a colony formation assay is often used as a surrogate for long-term cell survival after drug treatment. In this study, we preexposed the cells to cisplatin for 2 days, let them recover for 4 days in drug-free medium, and then plated them at a very low density for colony formation. The number of colonies formed by the cisplatin-treated cells was normalized to that of untreated cells to account for any initial differences in colony formation caused by SOX9 knockdown and overexpression. Our results showed that SOX9 knockdown significantly reduced the number of colonies compared to that for the vector control (Fig. 2D), while SOX9 overexpression promoted colony formation (Fig. 2E). These observations are consistent with the cell viability results and indicate a positive regulatory role of SOX9 in the drug resistance of NSCLC cells.
FIG 2.
SOX9 promotes chemoresistance in NSCLC cells. (A) Dose-response curves of H460 and A549 cells transfected with control siRNA (siControl) and SOX9 siRNA (siSOX9). Cell viability was evaluated by the MTT assay following a 3-day cisplatin treatment. The results are representative of those from 3 independent experiments with 4 technical replicates in each. The data are presented as the mean ± SD. (B) Dose-response curve of the empty vector and SOX9-overexpressing H460 cells following a 3-day cisplatin treatment. SOX9 expression was induced by doxycycline treatment 5 days prior to the experiment. The results are representative of those from 5 independent experiments with 5 technical replicates in each. The data are presented as the mean ± SD. ***, P < 0.0001 by two-way ANOVA. (C) Same as panel B but for A549 cells. (D) (Left) Colony formation assay in vector control and SOX9 knockdown H460 cells. Cells were treated with 4 μM cisplatin for 2 days and were then off cisplatin for 4 days and allowed to form colonies for 10 days. The data are presented as the mean ± SD and are for 2 biological replicates. **, P = 0.0021 by one-way ANOVA with Tukey’s multiple-comparison test. (Right) The cells were then fixed, stained with crystal violet, and visualized by light microscopy. Bar = 3 mm. (E) Same as for panel D but with SOX9-overexpressing cells exposed to 2 μM cisplatin. The data are for 3 biological replicates. *, P = 0.0125 by unpaired two-tailed t test.
FIG 3.
SOX9 expression levels correlate with drug sensitivity in NSCLC cells. (A) (Left) SOX9 knockdown renders H460 cells sensitive to cisplatin. The results are shown as the mean ± SD. (Right) A representative Western blot image demonstrates SOX9 knockdown levels generated by 2 shRNAs against SOX9 (sh-1 and sh-2) compared to the levels in the vector control cells. (B) Same as for panel A in A549 cells. (C) (Left) SOX9 overexpression in H460 cells induces resistance to etoposide. The results are shown as the mean ± SD, and the experiment was repeated 3 times. (Right) A representative Western blot image demonstrates the levels of SOX9 overexpression. (D) (Left and middle) SOX9 overexpression renders A549 cells resistant to etoposide and paclitaxel. The results are shown as the mean ± SEM. (Right) Representative Western blot analysis of SOX9 protein levels in SOX9-overexpressing A549 cells and vector control cells. P values were determined by two-way repeated measures ANOVA followed by Bonferroni posttests. **, P = 0.0036; ***, P < 0.0001.
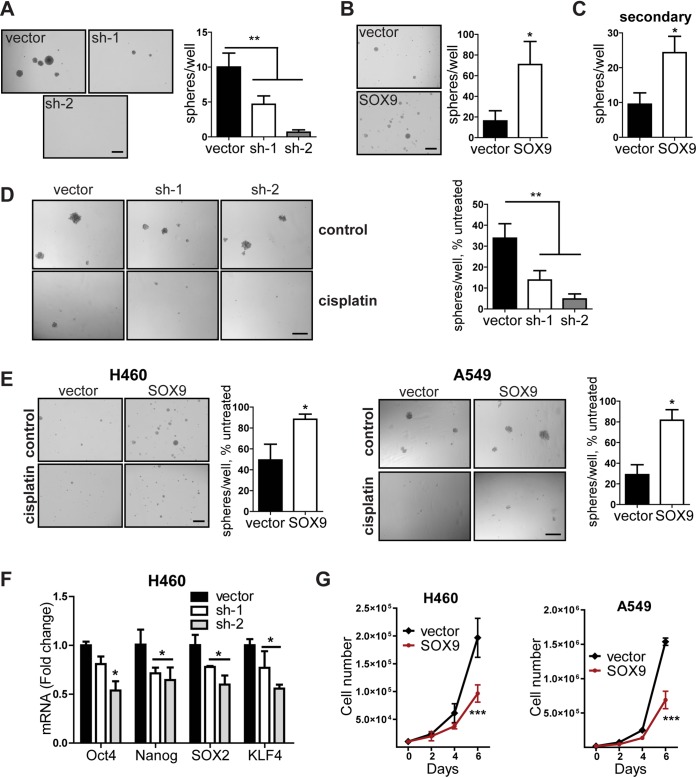
SOX9 positively regulates the cancer stem-like properties of NSCLC cells.
A growing body of evidence suggests that a particular cancer cell population, namely, CSCs, may be responsible for therapeutic resistance and tumor relapses (23). We therefore investigated the role of SOX9 as a CSC regulator in NSCLC cells. First, tumor sphere formation was performed in SOX9 knockdown and SOX9-overexpressing cells. The sphere formation assay is widely used to evaluate multipotency and the self-renewal of stem cells in normal and cancerous tissues and serves as a measure for CSC frequency in vitro (24, 25). We found that tumor sphere formation was substantially reduced in SOX9 knockdown cells (Fig. 4A), whereas it was enhanced in overexpression experiments (Fig. 4B). This effect was also retained during the formation of secondary spheres (Fig. 4C), confirming that SOX9 positively regulates the self-renewal properties of NSCLC cells. In line with these results, the expression of pluripotency-associated transcription factors Oct3/4, Nanog, SOX2, and KLF4 (26) was suppressed in SOX9 knockdown cells (Fig. 4F). Importantly, SOX9 confers cisplatin resistance under stem cell-selective conditions during sphere formation (Fig. 4D and E), suggesting that SOX9 regulates the chemoresistance of CSCs. This result cannot be explained by the initial difference in cell proliferation, since tumor sphere formation in the presence of the drug was normalized to that of untreated cells. In addition, SOX9-overexpressing cells grew slower than the control cells (Fig. 4G). Collectively, these observations indicate that high SOX9 expression is associated with the stem-like properties of NSCLC cells.
FIG 4.
SOX9 promotes cancer stem-like properties. (A) Tumor sphere formation in H460 cells expressing the empty vector or each of the two shRNAs against SOX9. The data are for 3 biological replicates. The data are presented as the mean ± SD. **, P < 0.0001 by one-way ANOVA with Tukey’s multiple-comparison test. Bar = 500 μm. (B, C) Same as for panel A but with the empty vector and SOX9-overexpressing cells (B) and secondary tumor spheres formed by the cells from primary spheres (C). Means ± SD are shown. *, P = 0.005 for primary spheres (B) and 0.012 for secondary spheres (C) by unpaired two-tailed t test. The data are for 3 biological replicates. (D) (Left) Representative pictures of tumor spheres formed by A549 cells expressing the empty vector or shRNAs against SOX9 (sh-1 and sh-2) under standard conditions or with cisplatin (1.5 μM) treatment. Bar = 300 μm. (Right) Quantification of spheroids after treatment with cisplatin. **, P = 0.0042 by one-way ANOVA followed by Tukey’s multiple-comparison test. Bars are the mean ± SD (n = 3). (E) As for panel B, plus the cells were exposed to 0.25 μM cisplatin (H460 cells) and 1.0 μM cisplatin (A549 cells) for the duration of the experiment. *, P = 0.043 and P = 0.0106 by unpaired two-tailed t test for H460 and A549 cells, respectively. The data are for 3 biological replicates. (F) Decreased mRNA expression of stem-cell markers in SOX9 knockdown cells (sh-1 and sh-2) versus vector control cells. The data are presented as the mean ± SD. *, P < 0.05 compared with the vector control cells by two-way ANOVA followed by Bonferroni posttests. (G) SOX9 overexpression slows down cell proliferation in H460 and A549 cells. The proliferation rate of the indicated cells was analyzed by counting the cells every 2 days. The data are for 3 biological replicates. ***, P = 0.011 for H460 cells and P = 0.0068 for A549 cells by two-way repeated-measures ANOVA followed by Bonferroni posttests. Means ± SD are shown.
ALDH1A1 is a downstream target of SOX9 and regulates chemoresistance.
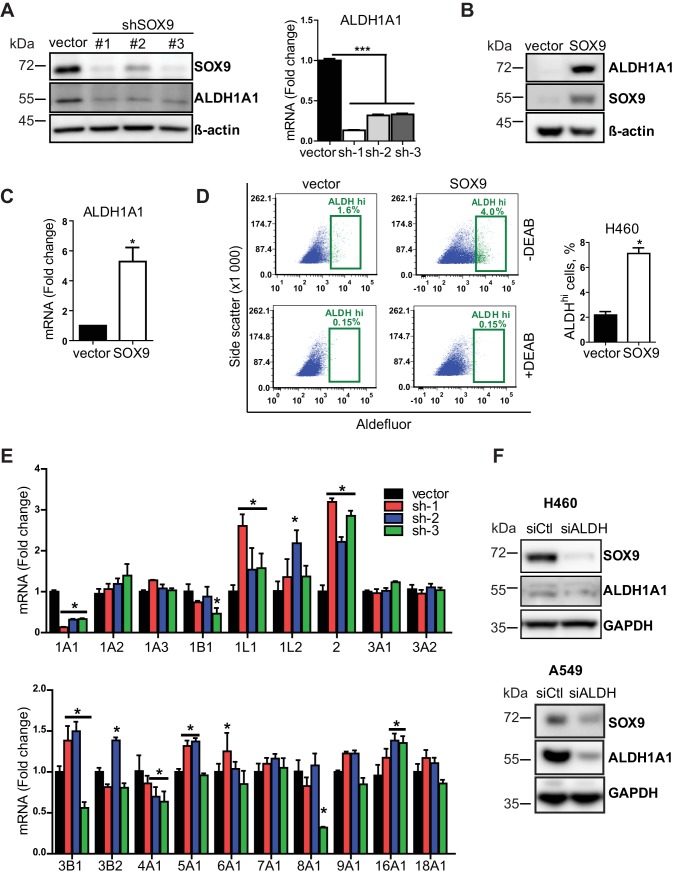
In the search for the underlying mechanisms of CSC regulation by SOX9, we observed a dramatic decrease in ALDH1A1 expression in SOX9 knockdown cells (Fig. 5A). The aldehyde dehydrogenase isoform ALDH1A1 belongs to a group of aldehyde-oxidizing enzymes that includes 19 members in humans (27). While all group members participate in the detoxification of endogenous and exogenous products of metabolism, ALDH1A1 is particularly known as a universal CSC marker (28). Furthermore, ALDH1A1 was shown to regulate cell proliferation and cancer drug resistance (29–31). Thus, we evaluated ALDH1A1 as a potential downstream effector of SOX9. Consistent with the findings of the knockdown experiments, SOX9 overexpression upregulated ALDH1A1 at the protein (Fig. 5B) and mRNA (Fig. 5C) levels. Moreover, ALDH enzymatic activity, measured by the flow cytometry-based Aldefluor assay, was also elevated upon SOX9 overexpression (Fig. 5D). We also profiled the mRNA levels of all 19 human ALDH isoforms in SOX9 knockdown cells since the Aldefluor assay does not appear to be isoform specific (32, 33). Our results showed that only ALDH1A1 expression was suppressed by SOX9 knockdown (Fig. 5E), further suggesting that ALDH1A1 acts downstream of SOX9. Interestingly, we observed a decrease in SOX9 expression in ALDH1A1 knockdown cells (Fig. 5F), suggesting a feedback loop between these proteins. Due to its important role in cell survival, such regulation is likely to be complex and involve indirect mechanisms.
FIG 5.
ALDH1A1 is a downstream target of SOX9. (A) (Left) Representative Western blot analysis of SOX9 and ALDH1A1 expression in SOX9 knockdown (shRNA #1, #2, and #3) and empty vector H460 cells. (Right) ALDH1A1 mRNA levels in the same cells. The data are presented as the mean ± SEM (n = 3). ***, P < 0.0001 by one-way ANOVA with Tukey’s multiple-comparison test. (B) Representative Western blot showing an increased expression of ALDH1A1 in SOX9-overexpressing H460 cells. The data are from 6 independent experiments. (C) SOX9 overexpression-induced ALDH1A1 mRNA levels in H460 cells. The data are presented as the mean ± SD from 7 independent experiments. P was 0.004 by a paired t test. (D) Analysis of ALDH activity by the Aldefluor assay in empty vector and SOX9-overexpressing H460 cells. (Left) A representative flow cytometry gating for the Aldefluor assay; (right) percentage of cells with high ALDH activity. The data are from 3 independent experiments with 2 biological replicates each. The data are presented as the mean ± SD. *, P < 0.001 by a paired two-tailed t test. DEAB, N,N-diethylaminobenzaldehyde, which is an ALDH inhibitor used to set up the background fluorescence level for all flow cytometry experiments. (E) RT-qPCR analysis of 19 ALDH isoforms in SOX9 knockdown (sh-1, -2, and -3) and empty vector H460 cells. The data are presented as the mean ± SD. *, P < 0.05 compared to the vector cells by two-way ANOVA followed by Bonferroni posttests. (F) ALDH1A1 knockdown decreased SOX9 expression. siCtl, control siRNA; siALDH, ALDH siRNA.
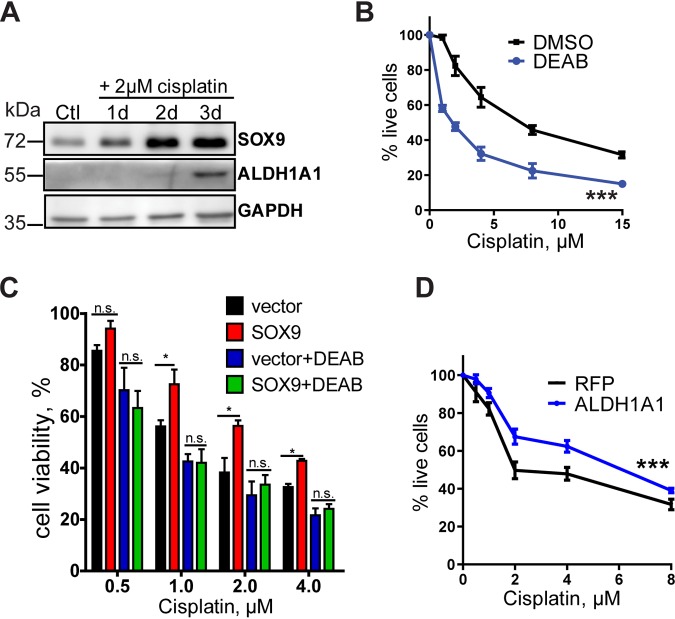
We next evaluated the role of SOX9-mediated ALDH1A1 upregulation in drug resistance. We observed that ALDH1A1 expression is elevated during cisplatin treatment (Fig. 6A) and that inhibition of ALDH activity by a known ALDH blocker, N,N-diethylaminobenzaldehyde (DEAB), reduced cell viability upon exposure to the drug (Fig. 6B). Furthermore, treatment with the ALDH inhibitor reversed the effect of SOX9 overexpression on cisplatin resistance (Fig. 6C), suggesting that SOX9 mediates its chemoresistance effect through ALDH1A1. ALDH1A1 overexpression in H460 cells potentiated drug resistance (Fig. 6D), supporting the results obtained with ALDH inhibition.
FIG 6.
SOX9 promotes chemoresistance through ALDH1A1. (A) Representative Western blot analysis of SOX9 and ALDH1A1 expression in H460 cells after exposure to 2 μM cisplatin. (B) Treatment with the ALDH inhibitor DEAB enhances sensitivity to cisplatin. H460 cells were pretreated with 200 μM DEAB or DMSO for 24 h, followed by cisplatin treatment in the presence of DEAB or DMSO, and were analyzed for cell viability after 2 or 3 days by the MTT assay. DEAB decreased the IC50 of H460 cells 2.5- ± 0.3-fold. The data are from 5 independent experiments. ***, P = 0.0110 by a paired t test. (C) The ALDH inhibitor DEAB reverses the effect of SOX9 overexpression in H460 cells. The cisplatin dose-response curve was evaluated by the MTT assay. The data are presented as the mean ± SD. P values were determined by two-way ANOVA with Bonferroni posttests. All experiments were repeated at least 3 times. *, P < 0.05; n.s., not significant. (D) ALDH1A1 overexpression enhances sensitivity to cisplatin. H460 cells were transfected with pcDNA3-mRFP and pcDNA3-HA-ALDH1A1, followed by cisplatin treatment for 3 days, and were analyzed for cell viability by the MTT assay. The results are presented as the mean ± SD. ***, P < 0.0001 by two-way ANOVA followed by Bonferroni posttests. RFP, red fluorescent protein.
ALDH1A1 is regulated by SOX9 at the transcriptional level.
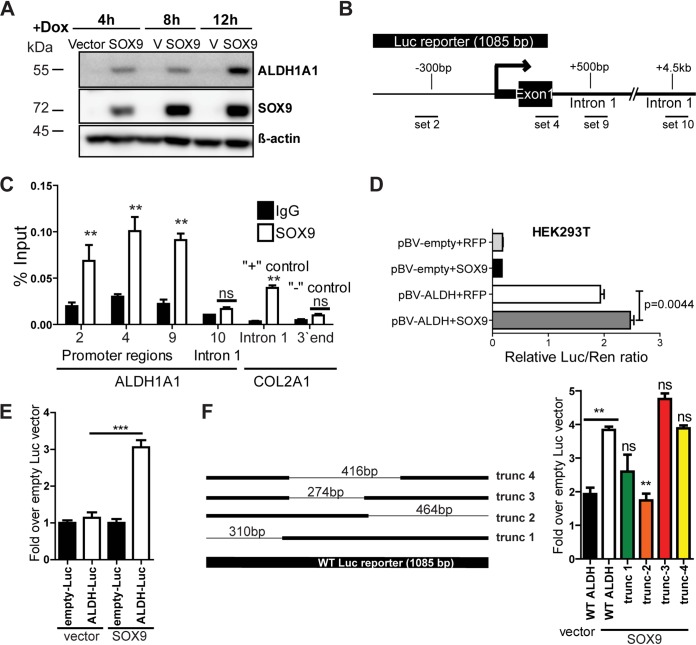
Next, we investigated the underlying mechanism of ALDH1A1 regulation by SOX9. A time course study of ALDH1A1 expression in cells with doxycycline-inducible SOX9 overexpression was conducted first. Interestingly, ALDH1A1 expression was upregulated as early as 4 h after doxycycline was added (Fig. 7A), suggesting that SOX9 may induce ALDH1A1 expression at the transcriptional level. We next performed chromatin immunoprecipitation (ChIP) followed by qPCR analysis. Validated primers for a known SOX9 target, COL2A1 (34), were used as positive and negative controls for SOX9 antibody, while several primer sets were designed for the ALDH1A1 promoter (Fig. 7B). In support of the previous results, SOX9 binding was enriched at several promoter locations, but not in intron 1, 4.5 kb downstream of the transcription start site (Fig. 7C). To further assess whether SOX9 could directly activate transcription from the ALDH1A1 promoter, we performed a luciferase reporter assay. Cells expressing an empty vector or SOX9 were transfected with an empty luciferase reporter plasmid or a reporter plasmid containing the 1-kb-long ALDH1A1 promoter (Fig. 7B). A substantial increase in the luciferase signal was observed in SOX9-overexpressing cells in comparison to that in the vector control cells (Fig. 7E). A similar result was observed in HEK293T cells transfected with both SOX9 and luciferase reporter plasmids (Fig. 7D). Analysis of the truncated promoter constructs identified that SOX9 binds to the ALDH1A1 promoter in the immediate proximity of the transcription start site (Fig. 7F). Collectively, our findings indicate that ALDH1A1 is a direct transcriptional target of SOX9.
FIG 7.
ALDH1A1 is regulated by SOX9 at the transcriptional level. (A) SOX9 overexpression induced by doxycycline (Dox) for the indicated length of time stimulates ALDH1A1 expression. The blots shown are representative of those from two independent experiments. (B) Schematic image of the ALDH1A1 promoter and locations of the primer sets used for the assay whose results are presented in panel C. (C) SOX9 binding is enriched at the ALDH1A1 promoter. SOX9 ChIP results were analyzed by qPCR with primers specific to the indicated DNA regions. IgG was used as a negative immunoprecipitation control. Primers to COL2A1 served as negative and positive controls for ChIP. The data are presented as the mean ± SD. P values were determined by two-way ANOVA with Bonferroni posttests. **, P < 0.01; ns, not significant. (D) SOX9 induces ALDH1A1 luciferase reporter activity in HEK293T cells. Cells were transfected with the indicated constructs, and luciferase activity was measured at 48 h posttransfection. Bars are the mean ± SEM. The indicated P value determined by an unpaired two-tailed t test. Luc/Ren, firefly luciferase/Renilla luciferase ratio. (E) Luciferase reporter assay with the ALDH1A1 promoter. H460 cells stably expressing the empty vector or SOX9 were transfected with the ALDH1A1 luciferase reporter plasmid and the empty luciferase vector. Luciferase activity was analyzed at 48 h posttransfection and normalized to the Renilla luciferase signal. ***, P = 0.028 by an unpaired two-tailed t test and P < 0.0001 by one-way ANOVA with Bonferroni’s multiple-comparison test. Bars are the mean ± SEM. The results are representative of those from two independent experiments. (F) Schematic representation of the truncated (trunc) ALDH1A1 luciferase reporters (left) and luciferase reporter assay with the full-length and truncated ALDH1A1 promoters, performed as described in the legend to panel E (right). Bars are the mean ± SEM. P values were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. **, P < 0.01 versus SOX9 wild-type (WT) ALDH; ns, not significant versus SOX9 wild-type ALDH.
DISCUSSION
Non-small-cell lung cancer (NSCLC) is the second most common cancer in both men and women and a leading cause of cancer-related death worldwide. It claims about 150,000 lives yearly in the United States alone (35). Despite recent advances in cancer therapy, chemotherapy remains a vital component of the overall treatment strategies for lung cancer. However, patient survival continues to be poor due to intrinsic and newly acquired drug resistance, which is an unresolved clinical problem. We identified SOX9 to be a potential regulator of cancer cell survival during chemotherapy, based on its ability to control drug-resistant CSCs. We also observed a dramatic increase in SOX9 levels in cisplatin-treated cells. In contrast, Hong et al. (15) reported that SOX9 is degraded by cisplatin and other DNA-damaging agents, such as doxorubicin. This apparent contradiction may be explained by the fact that the latter study used extremely high drug concentrations (e.g., 30 μM for cisplatin and 7.4 μM for doxorubicin) and a short exposure time that could result in cellular responses distinct from those observed in this study. In another report, SOX9 expression was found to be upregulated in gastric cell lines after a low-dose (10 μM) long-term (5-day) exposure to cisplatin (14). This result is consistent with our findings in lung cancer cells and suggests that SOX9 upregulation may be important for cancer cell survival under a selective therapeutic pressure. This notion is supported by clinical data showing a poor survival outcome in patients with high SOX9 expression (Fig. 1A) (7).
Our major objective was to uncover a mechanism by which SOX9 mediates its effect on chemoresistance. We showed that ALDH1A1 (also identified as ALDH1), which is a common biomarker for normal and cancer stem cells, is a direct transcriptional target of SOX9. According to the cancer stem cell theory, tumors contain a small population of cells that are highly tumorigenic and drug and apoptosis resistant and that possess stem-like properties, such as the ability to self-renew and differentiate (9). The human ALDH family of proteins includes 19 members, but ALDH1A1 is the main isozyme used to identify and/or isolate normal and cancer stem cells in multiple tissues (31). ALDH1A1 is overexpressed in many cancer types and usually correlates with a poor prognosis. For example, high ALDH1 expression correlates with poor overall survival in ovarian cancer (36). Moreover, elevated ALDH1 expression after neoadjuvant chemotherapy is associated with a poor response and early relapse in ovarian, breast, and esophageal cancers (37, 38). While individual reports do not always agree on the prognostic value of ALDH1A1 overexpression, meta-analysis studies demonstrated a strong correlation between ALDH1 expression and overall and disease-free survival in lung cancer (39, 40), colorectal cancer (41), and head and neck squamous cell carcinomas (42). Other studies showed that ALDH1A1 is overexpressed in drug-resistant cell lines (20, 30, 43). Furthermore, high ALDH activity, typical for stem cells, could support CSC survival. For instance, ALDH1A1 can detoxify an active derivate of cyclophosphamide, 4-hydroxycyclophosphamide, and therefore directly protect cancer cells from drugs (44, 45). A recent study also reported that cells with high ALDH activity (ALDHhi) have lower reactive oxygen species levels than their counterparts with low ALDH activity (ALDHlow) (46). Taken together, these results indicate that ALDH1A1 upregulation in response to cytotoxic stress supports CSC survival. Our findings further demonstrate that this process is regulated, at least in part, through SOX9 overexpression. It is worth noting that limited information is available on the direct transcriptional targets of SOX9. The transcription factor can work as a transcriptional activator or repressor, depending on the cellular context. Moreover, several recent studies utilizing a ChIP sequencing assay have identified novel SOX9 binding motifs, indicating that SOX9 binding may be tissue specific or context dependent (10, 34, 47).
While we focused our attention on ALDH1A1, other ALDH isoforms may contribute to CSC properties in general and to chemotherapy resistance in particular. For example, high ALDH3A1 and ALDH1A3 expression is associated with lung cancer progression (48, 49). However, unlike ALDH1A1, these isoforms are not a direct target of SOX9, based on the results of our knockdown studies (Fig. 5E). We also considered other possible roles of SOX9 in the regulation of cell survival and chemoresistance. It has been suggested that the decreased survival of SOX9 knockdown cells in long-term experiments (i.e., in colony formation or tumor sphere formation assays) may be due to a lower rate of cell proliferation (6). However, SOX9low cells were more sensitive to drug treatment, even after taking into account the initial effect of knockdown in untreated cells, and the opposite was true for the overexpression experiments (Fig. 2D and E and 4E). Furthermore, ectopic expression of SOX9 decreased the rate of cell proliferation, while it promoted the survival of drug-treated cells (Fig. 2B and C, 3C and D, and 4G).
In summary, we have demonstrated the role of SOX9 in the CSC regulation and drug resistance of NSCLC cells. Treatment of the cells with cisplatin upregulates SOX9 expression, which is crucial to their long-term survival and resistance to chemotherapeutic drugs. SOX9 mediates its effect on cisplatin resistance through the transcriptional activation of ALDH1A1. This novel finding supports the use of SOX9 as a prognostic marker of chemoresistance and as a potential therapeutic target for advanced and recurrent lung cancers.
MATERIALS AND METHODS
Cell culture and drugs.
H460 (NCI-H460), Beas-2B, HBEC3-KT, H1299, H358, H1650, and A549 cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA) and were passaged less than 15 times. H460 cells were maintained in RPMI medium (Corning, Corning, NY) supplemented with 10% fetal bovine serum (FBS); A549, H1299, H358, and H1650 cells were cultured in Dulbecco modified Eagle medium (Corning) supplemented with 10% FBS. Beas-2B cells and human bronchial epithelial cells (HBEC) were cultured in bronchial epithelial cell growth medium (BEBM) supplemented with Clonetics BEGM BulletKit reagents (Lonza, Walkersville, MD). Primary human small airway epithelial cells (SAECs) immortalized with human telomerase reverse transcriptase were kindly provided by Tom Hei (50). Primary human SAECs were obtained from Lonza. They were cultured in small airway epithelial basal medium (SABM) supplemented with Clonetics SAGM BulletKit reagents (Lonza). All cells were maintained in a humidified atmosphere at 37°C with 5% CO2. Cisplatin, etoposide, doxycycline, and N,N-diethylaminobenzaldehyde (DEAB) were purchased from Sigma-Aldrich Corporation (St. Louis, MO), and paclitaxel was purchased from Acros Organics (Geel, Belgium).
Plasmids and generation of stable cell lines.
SOX9 small interfering RNA (siRNA) (catalog number D-059108-01), ALDH1A1 siRNA (catalog number L-008722-00-0005), nontargeting siRNA (catalog number D-001810-01), and pLKO.1 lentiviral plasmids for SOX9 knockdown (catalog number RHS4533-EG6662) were purchased from Dharmacon (Lafayette, CO). An empty pLKO.1 vector was used as a control, and the virus was produced in HEK293T cells (ATCC) as described previously (51). For SOX9 overexpression, SOX9 cDNA from pCMV-AC-GFP-SOX9 (Origene, Rockville, MD) was amplified by PCR and subcloned into pLUTzeo (a gift from A. Ivanov, West Virginia University, WV). Viral particles were produced as described above, and the empty vector was used as a control. siRNA transfections (50 nM) were performed on 6-well dishes with the Lipofectamine RNAiMAX transfection reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. Cells were lysed or used for subsequent experiments at 48 h posttransfection. The short hairpin RNA (shRNA) and siRNA sequences are listed in Table S3 in the supplemental material. pcDNA3-HA-ALDH1A1 was a gift from Steven Johnson (Addgene plasmid number 11610) with a point mutation fixed by site-directed mutagenesis. pcDNA3-mRFP was a gift from Doug Golenbock (Addgene plasmid number 13032).
Immunoblotting.
Western blot analysis was performed as described previously (51). Immune complexes were detected by enhanced chemiluminescence in an Amersham Imager 600 imager (GE Healthcare Life Sciences, Buckinghamshire, UK) and quantified with Amersham Imager software. The antibodies used in this study are listed in Table S1.
Flow cytometry.
Cells with high aldehyde dehydrogenase activity were identified by staining by use of the Aldefluor kit (Stem Cell Technologies, Cambridge, MA) according to the manufacturer’s instructions. Briefly, for each sample, 3 × 105 cells were incubated in the Aldefluor assay buffer with activated Aldefluor substrate for 45 min at 37°C in the presence or absence of the specific ALDH inhibitor DEAB. Tubes stained with both the Aldefluor substrate and DEAB served as a negative control for each sample. Cells with high ALDH activity (ALDHhi) were detected using a BD Fortessa cell analyzer and analyzed with BD FACSDiva software (BD Biosciences, San Jose, CA). The flow cytometry gates were set to obtain 0.1% ALDHhi cells in tubes with the substrate plus the inhibitor for each sample. All experiments were performed at least 3 times.
Cell proliferation assay.
The rate of cell proliferation was measured by seeding equal numbers of cells onto 6-well plates in duplicate and counting the cells with a Countess automated cell counter (Life Technologies) every 2 days until the growth on the plates become confluent.
Cell viability (MTT) assay.
Cells were plated onto 96-well plates and allowed to recover overnight. On the next day, the cell culture media were replaced with drug-containing media. Ten microliters of 5-mg/ml 1-(4,5-dimethyl-2-thiazolyl)-3,5-diphenylformazan (MTT; Tokyo Chemical Industry, Portland, OR) was added to each well containing 100 μl of cell culture medium following 72 h of drug treatment. After 2 h of incubation under standard cell culture conditions, formazan crystals were dissolved with dimethyl sulfoxide (DMSO), and the absorbance was measured at 570 nm. Cell viability was represented as a percentage of the values for vehicle-treated cells.
Colony formation assay.
Cells were treated with cisplatin for 2 days under standard cell culture conditions and allowed to recover without cisplatin for 4 days. The cells (500 cells/well) were then plated onto 6-well plates and cultured under standard conditions for 10 days. Colonies were fixed in 100% methanol and stained with 0.5% crystal violet solution. Colonies containing over 50 cells were counted using an Olympus MVX10 microscope (Olympus Corporation, Tokyo, Japan) at ×0.63 magnification. The colony numbers were normalized to those of the respective untreated cells. All experiments were performed in triplicate.
Tumor sphere formation assay.
One thousand cells per well were plated onto ultra-low-attachment 24-well plates (Corning) in 0.8% methylcellulose (MC)-based serum-free medium (catalog number H4100; Stem Cell Technologies) supplemented with 20 ng/ml epidermal growth factor (BD Biosciences), 10 ng/ml basic fibroblast growth factor (Sigma-Aldrich), and 5 mg/ml insulin (Sigma-Aldrich). The number of tumor spheres exceeding 50 μm in diameter was quantified under a light microscope after 10 days (H460 cells) or 3 weeks (A549 cells) in culture. All experiments were performed in triplicate and repeated at least twice, with 5 fields of view being analyzed for each replicate. For cisplatin treatments, cisplatin was added to the cultures at the plating time and was replenished 2 times a week thereafter.
ChIP.
Chromatin immunoprecipitation (ChIP) was carried out using a SimpleChIP kit with agarose beads (Cell Signaling Technology, Danvers, MA) according to the manufacturer’s protocol. Briefly, approximately 1 × 107 H460 cells were used for each immunoprecipitation. Cells were cross-linked with 1% formaldehyde, treated with micrococcal nuclease for chromatin digestion, and briefly sonicated to break the nuclear membranes. Two micrograms of each anti-SOX9 antibody (catalog number AB5535; Millipore, Burlington, MA) and negative-control normal rabbit IgG (Cell Technologies) was used for immunoprecipitation. The amount of total and ChIP DNA was quantified by qPCR after the reversal of cross-links and DNA purification. The primer sequences are listed in Table S2.
Luciferase reporter assay.
The ALDH1A1 promoter was obtained from SwitchGear Genomics (Carlsbad, CA) and was moved from the pLightSwitch vector to pBV-luc (Addgene plasmid number 16539; a gift from Bert Vogelstein) by restriction enzyme cloning. The empty pBV-luc vector was used as a control. The Renilla luciferase vector pIS1 (Addgene plasmid number 12179; a gift from David Bartel) served as an internal control for transfection efficiency. SOX9 expression in H460 cells was induced by treating the cells with 0.2 μg/ml doxycycline for 5 days before transfection. Cells were transfected on 12-well plates with pBV-luc or pBV-luc-ALDH1A1 (1 μg/well) and 50 ng of pIS1 using the Viafect reagent (Promega, Madison, WI). Luciferase activities were measured using a dual-luciferase reporter assay (Promega) according to the manufacturer’s instructions. HEK293T cells were transfected with the luciferase plasmids as described above plus pcDNA3.1-RFP or pcDNA3.1-SOX9.
RT-qPCR.
Total RNA was purified using an RNeasy kit (Qiagen, Germantown MD). Reverse transcription was performed with SuperScript III reverse transcriptase (Life Technologies) using oligo(dT) primers. Quantitative real-time PCR was carried out using SYBR green master mix (Applied Biosystems, Foster City, CA) in a Step One Plus PCR cycler (Applied Biosystems). Results were calculated using the 2−ΔΔCT threshold cycle (CT) method; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin served as the internal controls. The primer sequences are listed in Table S2.
Statistical analysis.
The data represent the means ± standard deviations (SD) or standard errors of the means (SEM), as indicated in the figure legends. Statistical comparisons were made using two-tailed Student's t test or analysis of variance (ANOVA) when more than two groups were analyzed. Statistical analysis was performed with Prism software (GraphPad, La Jolla, CA), and a P value of <0.05 was considered statistically significant.
Data availability.
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alexey Ivanov for providing the pLUTz cloning vector.
This work was supported in part by National Institutes of Health grants R01-ES022968 and R01-EB018857. Imaging experiments and image analysis were performed in the West Virginia University Microscope Imaging Facility, which has been supported by the WVU Cancer Institute and National Institutes of Health grants P20RR016440, P20GM103434, and P30RR032138/P30GM103488. Flow cytometry experiments were performed in the West Virginia University Flow Cytometry & Single Cell Core Facility, which is supported by National Institutes of Health equipment grant S10OD016165 and Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P30GM103488 (Cancer CoBRE) and P20GM103434 (INBRE).
We declare that we have no conflict of interest.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, Kang R, Shi LL, Mok J, Lee MJ, Haydon RC. 2014. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis 1:149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. 2011. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 3.Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, Briscoe J. 2010. SOX9 induces and maintains neural stem cells. Nat Neurosci 13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- 4.Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MA, Kopp JL, Sander M, Wellik DM, Spence JR. 2013. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci U S A 110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Zhang H, Min D, Zhang R, Wu J, Qu H, Tang Y. 2015. Sox9 activation is essential for the recovery of lung function after acute lung injury. Cell Physiol Biochem 37:1113–1122. doi: 10.1159/000430236. [DOI] [PubMed] [Google Scholar]
- 6.Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KSE, Skotheim RI, Lothe RA, de Munain AL, Briscoe J, Serrano M, Lovell-Badge R. 2012. Oncogenicity of the developmental transcription factor Sox9. Cancer Res 72:1301–1315. doi: 10.1158/0008-5472.CAN-11-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruan H, Hu S, Zhang H, Du G, Li X, Li X, Li X. 2017. Upregulated SOX9 expression indicates worse prognosis in solid tumors: a systematic review and meta-analysis. Oncotarget 8:113163–113173. doi: 10.18632/oncotarget.22635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei B, Zhang YX, Liu T, Li YW, Pang D. 2016. Sox9 upregulation in breast cancer is correlated with poor prognosis and the CD44+/CD24−/low phenotype. Int J Clin Exp Pathol 9:7345–7351. [Google Scholar]
- 9.Jordan CT, Guzman ML, Noble M. 2006. Cancer stem cells. N Engl J Med 355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 10.Larsimont JC, Youssef KK, Sanchez-Danes A, Sukumaran V, Defrance M, Delatte B, Liagre M, Baatsen P, Marine JC, Lippens S, Guerin C, Del Marmol V, Vanderwinden JM, Fuks F, Blanpain C. 2015. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 17:60–73. doi: 10.1016/j.stem.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Liu L, Chen X, Cheng J, Zhang H, Shen J, Shan J, Xu Y, Yang Z, Lai M, Qian C. 2016. Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 64:117–129. doi: 10.1002/hep.28509. [DOI] [PubMed] [Google Scholar]
- 12.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. 2012. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luanpitpong S, Li J, Manke A, Brundage K, Ellis E, McLaughlin SL, Angsutararux P, Chanthra N, Voronkova M, Chen YC, Wang L, Chanvorachote P, Pei M, Issaragrisil S, Rojanasakul Y. 2016. SLUG is required for SOX9 stabilization and functions to promote cancer stem cells and metastasis in human lung carcinoma. Oncogene 35:2824–2833. doi: 10.1038/onc.2015.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos JC, Carrasco-Garcia E, Garcia-Puga M, Aldaz P, Montes M, Fernandez-Reyes M, de Oliveira CC, Lawrie CH, Araúzo-Bravo MJ, Ribeiro ML, Matheu A. 2016. SOX9 elevation acts with canonical WNT signaling to drive gastric cancer progression. Cancer Res 76:6735–6746. doi: 10.1158/0008-5472.CAN-16-1120. [DOI] [PubMed] [Google Scholar]
- 15.Hong X, Liu W, Song R, Shah JJ, Feng X, Tsang CK, Morgan KM, Bunting SF, Inuzuka H, Zheng XFS, Shen Z, Sabaawy HE, Liu L, Pine SR. 2016. SOX9 is targeted for proteasomal degradation by the E3 ligase FBW7 in response to DNA damage. Nucleic Acids Res 44:8855–8869. doi: 10.1093/nar/gkw748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suryo Rahmanto A, Savov V, Brunner A, Bolin S, Weishaupt H, Malyukova A, Rosén G, Čančer M, Hutter S, Sundström A, Kawauchi D, Jones DT, Spruck C, Taylor MD, Cho Y-J, Pfister SM, Kool M, Korshunov A, Swartling FJ, Sangfelt O. 2016. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J 35:2192–2212. doi: 10.15252/embj.201693889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higashihara T, Yoshitomi H, Nakata Y, Kagawa S, Takano S, Shimizu H, Kato A, Furukawa K, Ohtsuka M, Miyazaki M. 2017. Sex determining region Y box 9 induces chemoresistance in pancreatic cancer cells by induction of putative cancer stem cell characteristics and its high expression predicts poor prognosis. Pancreas 46:1296–1304. doi: 10.1097/MPA.0000000000000945. [DOI] [PubMed] [Google Scholar]
- 18.Zhou CH, Ye LP, Ye SX, Li Y, Zhang XY, Xu XY, Gong LY. 2012. Clinical significance of SOX9 in human non-small cell lung cancer progression and overall patient survival. J Exp Clin Cancer Res 31:18. doi: 10.1186/1756-9966-31-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang SS, Fang WT, Hou YH, Huang SF, Yen BL, Chang JL, Li SM, Liu HP, Liu YL, Huang CT, Li YW, Jang TH, Chan SH, Yang SJ, Hsiung CA, Wu CW, Wang LH, Chang IS. 2010. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin Cancer Res 16:4363–4373. doi: 10.1158/1078-0432.CCR-10-0138. [DOI] [PubMed] [Google Scholar]
- 20.Barr MP, Gray SG, Hoffmann AC, Hilger RA, Thomale J, O'Flaherty JD, Fennell DA, Richard D, O'Leary JJ, O'Byrne KJ. 2013. Generation and characterisation of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PLoS One 8:e54193. doi: 10.1371/journal.pone.0054193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G II, Samuel S, Kim MP, Lim SJ, Ellis LM. 2009. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res 69:1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. 2008. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One 3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean M, Fojo T, Bates S. 2005. Tumour stem cells and drug resistance. Nat Rev Cancer 5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 24.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. 2003. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. 2008. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 26.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. 2007. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 27.Marchitti SA, Brocker C, Stagos D, Vasiliou V. 2008. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol 4:697–720. doi: 10.1517/17425255.4.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pors K, Moreb JS. 2014. Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development? Drug Discov Today 19:1953–1963. doi: 10.1016/j.drudis.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Wei Y, Wu S, Xu W, Liang Y, Li Y, Zhao W, Wu J. 2017. Depleted aldehyde dehydrogenase 1A1 (ALDH1A1) reverses cisplatin resistance of human lung adenocarcinoma cell A549/DDP. Thorac Cancer 8:26–32. doi: 10.1111/1759-7714.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Januchowski R, Wojtowicz K, Sterzyska K, Sosiska P, Andrzejewska M, Zawierucha P, Nowicki M, Zabel M. 2016. Inhibition of ALDH1A1 activity decreases expression of drug transporters and reduces chemotherapy resistance in ovarian cancer cell lines. Int J Biochem Cell Biol 78:248–259. doi: 10.1016/j.biocel.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 31.Tomita H, Tanaka K, Tanaka T, Hara A. 2016. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 7:11018–11032. doi: 10.18632/oncotarget.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan CA, Parajuli B, Buchman CD, Dria K, Hurley TD. 2015. N,N-Diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem Biol Interact 234:18–28. doi: 10.1016/j.cbi.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee P. 2011. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 34.Ma F, Ye H, He HH, Gerrin SJ, Chen S, Tanenbaum BA, Cai C, Sowalsky AG, He L, Wang H, Balk SP, Yuan X. 2016. SOX9 drives WNT pathway activation in prostate cancer. J Clin Invest 126:1745–1758. doi: 10.1172/JCI78815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cronin KA, Lake AJ, Scott S, Sherman RL, Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J, Kohler BA, Jemal A. 2018. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer 124:2785–2800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W, Zang C, Zhang T, Li J, Liu R, Feng F, Lv Q, Zheng L, Tian J, Sun C. 2018. Clinicopathological characteristics and prognostic value of the cancer stem cell marker ALDH1 in ovarian cancer: a meta-analysis. Onco Targets Ther 11:1821–1831. doi: 10.2147/OTT.S160207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Zhang B, Yang YF, Jin J, Liu YH. 2018. Aldehyde dehydrogenase 1 as a predictor of the neoadjuvant chemotherapy response in breast cancer: a meta-analysis. Medicine (Baltimore) 97:e12056. doi: 10.1097/MD.0000000000012056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ajani JA, Wang X, Song S, Suzuki A, Taketa T, Sudo K, Wadhwa R, Hofstetter WL, Komaki R, Maru DM, Lee JH, Bhutani MS, Weston B, Baladandayuthapani V, Yao Y, Honjo S, Scott AW, Skinner HD, Johnson RL, Berry D. 2014. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol Oncol 8:142–149. doi: 10.1016/j.molonc.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huo W, Du M, Pan X, Zhu X, Li Z. 2015. Prognostic value of ALDH1 expression in lung cancer: a meta-analysis. Int J Clin Exp Med 8:2045–2051. [PMC free article] [PubMed] [Google Scholar]
- 40.Wei D, Peng J-J, Gao H, Zhang T, Tan Y, Hu Y-H. 2015. ALDH1 expression and the prognosis of lung cancer: a systematic review and meta-analysis. Heart Lung Circ 24:780–788. doi: 10.1016/j.hlc.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Xia Q, Jiang B, Chang W, Yuan W, Ma Z, Liu Z, Shu X. 2015. Prognostic value of cancer stem cell marker ALDH1 expression in colorectal cancer: a systematic review and meta-analysis. PLoS One 10:e0145164. doi: 10.1371/journal.pone.0145164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou C, Sun B. 2014. The prognostic role of the cancer stem cell marker aldehyde dehydrogenase 1 in head and neck squamous cell carcinomas: a meta-analysis. Oral Oncol 50:1144–1148. doi: 10.1016/j.oraloncology.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 43.Duong HQ, Hwang JS, Kim HJ, Kang HJ, Seong YS, Bae I. 2012. Aldehyde dehydrogenase 1A1 confers intrinsic and acquired resistance to gemcitabine in human pancreatic adenocarcinoma MIA PaCa-2 cells. Int J Oncol 41:855–861. doi: 10.3892/ijo.2012.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Eitzen U, Meier-Tackmann D, Agarwal DP, Goedde HW. 1994. Detoxification of cyclophosphamide by human aldehyde dehydrogenase isozymes. Cancer Lett 76:45–49. doi: 10.1016/0304-3835(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 45.Moreb J, Zucali JR, Zhang Y, Colvin MO, Gross MA. 1992. Role of aldehyde dehydrogenase in the protection of hematopoietic progenitor cells from 4-hydroperoxycyclophosphamide by interleukin 1 beta and tumor necrosis factor. Cancer Res 52:1770–1774. [PubMed] [Google Scholar]
- 46.Mizuno T, Suzuki N, Makino H, Furui T, Morii E, Aoki H, Kunisada T, Yano M, Kuji S, Hirashima Y, Arakawa A, Nishio S, Ushijima K, Ito K, Itani Y, Morishige K. 2015. Cancer stem-like cells of ovarian clear cell carcinoma are enriched in the ALDH-high population associated with an accelerated scavenging system in reactive oxygen species. Gynecol Oncol 137:299–305. doi: 10.1016/j.ygyno.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Shi Z, Chiang CI, Labhart P, Zhao Y, Yang J, Mistretta TA, Henning SJ, Maity SN, Mori-Akiyama Y. 2015. Context-specific role of SOX9 in NF-Y mediated gene regulation in colorectal cancer cells. Nucleic Acids Res 43:6257–6269. doi: 10.1093/nar/gkv568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. 2008. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer 59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 49.Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, Liu H, Behrens C, Shay JW, Wistuba II, Minna JD. 2014. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res 20:4154–4166. doi: 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piao CQ, Liu L, Zhao YL, Balajee AS, Suzuki M, Hei TK. 2005. Immortalization of human small airway epithelial cells by ectopic expression of telomerase. Carcinogenesis 26:725–731. doi: 10.1093/carcin/bgi016. [DOI] [PubMed] [Google Scholar]
- 51.Voronkova MA, Luanpitpong S, Rojanasakul LW, Castranova V, Dinu CZ, Riedel H, Rojanasakul Y. 2017. SOX9 regulates cancer stem-like properties and metastatic potential of single-walled carbon nanotube-exposed cells. Sci Rep 7:11653. doi: 10.1038/s41598-017-12037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.