Abstract
Status epilepticus care and treatment are already being touched by the revolution in data science. New approaches designed to leverage the tremendous potential of “big data” in the clinical sphere are enabling researchers and clinicians to extract information from sources such as administrative claims data, the electronic medical health record, and continuous physiologic monitoring data streams. Algorithmic methods of data extraction also offer potential to fuse multimodal data (including text-based documentation, imaging data, and time-series data) to improve patient assessment and stratification beyond the manual capabilities of individual physicians. Still, the potential of data science to impact the diagnosis, treatment, and minute-to-minute care of patients with status epilepticus is only starting to be appreciated. In this brief review we discuss how data science is impacting the field and draw examples from three main areas: (1) analysis of insurance claims from large administrative datasets to evaluate the impact of continuous EEG monitoring on clinical outcomes, (2) natural language processing of the electronic health record to find, classify, and stratify patients for prognostication and treatment, and (3) real-time systems for data analysis, data reduction, and multimodal data fusion to guide therapy in real time. While early, it is our hope that these examples will stimulate investigators to leverage data science, computer science, and engineering methods to improve the care and outcome of patients with status epilepticus and other neurological disorders.
Keywords: Epilepsy, Status Epilepticus, Big Data, Natural Language Processing, Continuous EEG, Multimodal Data
1. Introduction
Technological advancements in clinical data acquisition and storage are rapidly changing the face of patient care for status epilepticus. In multiple domains, we are now acquiring large-scale data that make it possible to explore previously infeasible clinical and basic science questions[1–3]. For instance, these data come as clinical reports from health-system-wide electronic health records (EHRs) and billing claims from administrative databases, as well as in the form of digital neurophysiologic recordings from continuous EEG or multimodality ICU monitoring. While powerful, each dataset has its own challenges: data can be lacking in clinical detail, noisy, uncurated, or poorly annotated. Claims data is collected for billing purposes and therefore provides little clinical context; however rigorous analysis and careful interpretation allow important inferences from populations of millions of patients. In the case of clinic notes in the EHR, information is not uniformly documented or organized, making automated extraction of relevant data challenging. These issues, combined with the sheer size of these datasets, necessitate the use of new analysis paradigms and technical approaches, such as algorithmic data mining and machine learning, to perform data reduction and glean clinical insights.
The era of “big data” in clinical medicine offers potential to improve care for status epilepticus through several avenues. One goal is to use clinical records to stratify patients, both for prognostication and to identify patient groups that may benefit from particular interventions such as continuous EEG monitoring, surgery, or neuromodulation. In this way, personalized data mining algorithms can be used to optimize and individualize care. Secondly, extensive intracranial EEG datasets recorded using next-generation implantable devices are capable of driving powerful machine learning algorithms for data interpretation, epileptiform event detection, and per-patient optimization of therapeutic stimulation parameters. This streaming data analysis paradigm is now being translated into the neurointensive care unit, where multimodal data inform algorithms for data reduction, automated data monitoring, and notifying caretakers regarding changes in clinical status in near real time. Finally, these data are driving researchers and clinicians to establish data-sharing platforms to propel collaborative research initiatives investigating the mechanisms underlying the pathology of epilepsy and status epilepticus[4–7].
2. Continuous EEG and hospitalization outcomes for patients with status epilepticus
Administrative claims datasets offer a unique opportunity to analyze data from millions of patients at once and to assess trends in healthcare utilization over time. This data is collected by insurers (both private and public insurance datasets are available) and typically contains de-identified information regarding patient demographics, measures of comorbidity, diagnoses, procedures, admission and disposition details, payer information, and hospital characteristics. This type of data is particularly advantageous when studying the care of patients with status epilepticus, a relatively rare occurrence with an annual incidence of 10–41 per 100,000 population[8].
Continuous EEG (cEEG), a tool that provides non-invasive real-time information about brain function, plays an important role in the diagnosis and management of status epilepticus[9,10]. The American Clinical Neurophysiology Society (ACNS) recommends the use of cEEG in seizure diagnosis in critically ill patients as there is a very high incidence of status epilepticus in this population[11] - 8–68% of intensive care patients who are undergoing EEG monitoring are found to have seizures[10]. Continuous EEG is critical to identifying subclinical seizures, which have been associated with neuronal injury[12,13] and neurological decline[14,15]. However, the impact of cEEG utilization on outcomes for patients with critical illness has not been well established. Furthermore, there is great variability in application of cEEG due to resource availability and variation in provider practices.[16,17]. While a randomized controlled trial of cEEG is underway in Europe (“Continuous EEG Randomized Trial in Adults”)[18], this trial will be limited by logistical and ethical considerations, challenges that administrative data analysis does not face.
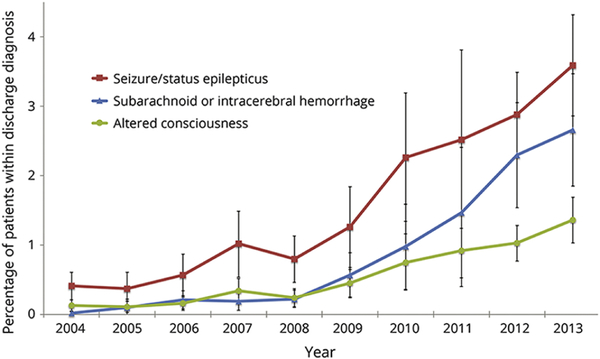
Trends in the utilization of cEEG can be readily characterized with claims data, as many administrative datasets offer a large nation-wide sample of patients and serial years of data. Evaluation of the National Inpatient Sample (NIS), the largest all-payer inpatient national database through the Agency for Healthcare Research and Quality (AHRQ)[19], found an increase in the proportion of critically ill patients receiving cEEG from 0.06% in 2004 to 0.8% in 2013. This increase in utilization was observed across all considered indications: status epilepticus, subarachnoid or intracerebral hemorrhage, and altered consciousness (Figure 1). Yet, cEEG monitoring was only administered for a very small minority of critically ill patients (0.3%) and only employed by 6% of the nationally-sampled hospitals. [20]
Figure 1.
Use of cEEG in critically ill discharges from 2004 to 2013 by diagnostic subcategory. Data derived from National Inpatient Sample database. Numerator is the number of patients who underwent cEEG; denominator is the total discharges with a particular diagnosis. Figure reproduced with permission from Hill, et al., “Continuous EEG is associated with favorable hospitalization outcomes for critically ill patients,” Neurology, Jan 2019, 92 (1) e9–e18[20].
The impact of cEEG monitoring on patient outcomes can also be considered with claims data analysis. Two studies have assessed the association of cEEG monitoring with inpatient mortality within the critically ill population. In a study of 40,945 ventilated patients from the NIS dataset from 2005–2009, undergoing cEEG was associated with a lower risk of in-hospital mortality as compared to patients who received routine EEG only (OR 0.63, 95% CI 0.51–0.76, p<0.001); including epilepsy/convulsion diagnoses as covariates did not alter these findings[21]. A second study of 2004–2013 NIS data identified 7,102,399 ventilated patients, and found a similar association of cEEG with decreased inpatient mortality as compared to patients who did not undergo cEEG (OR 0.83, 95% CI 0.75–0.93, p<0.001); however, this relationship was only significant for patients with subarachnoid/intracerebral hemorrhage or altered mental status, and was not significant for the subgroup with a diagnosis of seizure/status epilepticus[20]. The most plausible explanation for this unexpected finding is that this subanalysis was confounded by disease severity, as the diagnosis of seizure or status epilepticus identifies a highly heterogeneous population with a range of seizure etiologies and related prognoses.
While limited by the absence of detailed clinical information, these administrative claims analyses illustrate the current utilization of cEEG, which is growing at a rapid pace, and should inform the appropriate expansion of cEEG moving forward. Continuous EEG appears beneficial to critically ill patients, however targeting is critical as the observed benefit varies across subgroups of patients.
Claims data offer unique opportunities to find associations between diagnostic approaches, therapies, and outcomes in status epilepticus patients, although it is important to note that such studies are not a substitute for prospective, randomized clinical trials. Rather, studies of this nature can inform the objectives and design of clinical trials such as the Established Status Epilepticus Treatment Trial (ESETT)[22,23]. As health system infrastructures continue to advance across the world, particularly with consolidation of digital medical records[24] and more centralized care delivery, administrative claims research is likely to become increasingly powerful and informative.
3. Natural Language Processing for interpreting epilepsy clinical notes
3.1. Natural Language Processing for clinical data
Large electronic health record (EHR) datasets can reveal patterns in patient populations that are useful for both clinical and research applications. These datasets include clinical notes (such as admission notes, progress notes, discharge summaries, operative notes, and outpatient clinic notes) and diagnostic reports (such as radiology and pathology reports). However, much of EHR data is recorded as unstructured free text [25]. As human review of unstructured text is time intensive and inefficient, natural language processing (NLP) can greatly facilitate analysis of large EHR datasets [26]. NLP is a range of computational techniques used to analyze and process human language[27], critical for information extraction (IE) from unstructured text [28]. In clinical domains, researchers have used NLP IE to aid clinical decision support systems[29,30], identify patient cohorts[31,32], and identify risk factors from clinical notes [33]. While current applications of NLP in epilepsy have so far been limited by algorithm efficacy and generalizability, increasing study sizes in genetic and precision medicine trials are making NLP essential for epilepsy phenotyping and stratification[26].
3.2. Epilepsy specific ontologies
Epilepsy datasets are particularly challenging as they are generated in a wide range of settings using differing terminologies. A seizure with alteration of consciousness may be called a complex partial seizure, a focal dyscognitive seizure, or a dialeptic seizure by different physicians[34]. One way to standardize a vocabulary is to create an ontology, a formal representation of knowledge of a given domain that allows both humans and computers to consistently and accurately interpret terms. Ontologies are now commonly used across biomedical domains to simplify data management, as they enable natural language processing tools to better extract heterogeneous information from clinical free text [35].
A major milestone in epilepsy NLP research was the creation of a comprehensive epilepsy specific ontology by Sahoo et al. in 2012[36]. EpSO (Epilepsy and Seizure Ontology) integrates recommendations from the International League Against Epilepsy (ILAE) Classification and Terminology Commission with elements from existing ontologies like the Neural ElectroMagnetic Ontologies. EpSO includes very specific terms for epilepsy types, seizure types, EEG patterns, symptoms, and brain locations. It uses the World Wide Web Consortium recommended Web Ontology Language (OWL2), a formal knowledge representation language based on description logic that supports software applications[34]. While EpSO has been utilized in several NLP systems, it is not the only ontology currently used in epilepsy NLP research. Several studies from Portugal using text mining of medical records to classify epilepsy in children utilized an ontology based on the Unified Medical Language System and the International Classification of Diseases ICD9 for epilepsy types. This ontology had additional Portuguese words to aid computer analysis of the medical notes [30,37,38]. Other sources of standardized epilepsy terms include the SNOMED-CT (Systematized Nomenclature of Medicine -- Clinical Terms)[39], and HPO (Human Phenotype Ontology)[40]. In 2017 the ILAE released an updated classification of seizure types to allow more flexibility in diagnosis[41]. The ILAE also released an updated classification of epilepsy types with new terminology such as developmental epilepsy and epileptic encephalopathy[42]. These 2017 ILAE classification systems are expected to remain stable for at least a decade to come. As EpSO is now outdated, the field of epilepsy NLP would greatly benefit from a new OWL2 encoded ontology that reflects the updated ILAE recommendations. It is also important to note that these approaches, while efficient for analysis once data are entered, likely sacrifice some of the richness of unstructured data and suffer from the need to revise data entries as terminology, classification, and their scientific underpinnings evolve over time.
3.3. Integrated tools for NLP based epilepsy data management and cohort identification
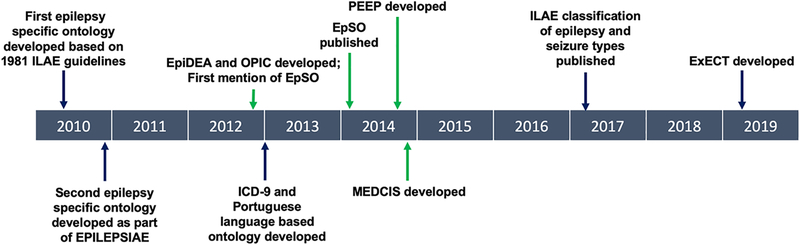
EpSO provided the backbone to a suite of epilepsy specific informatics tools which were developed for a multi-center study funded by the National Institute of Neurological Disorders[31,32,36,43]. The OPIC system, Ontology-driven Patient Information Capturing system for epilepsy, was developed in 2012 to facilitate structured entry of multimodal epilepsy data according to the EpSO format[36]. Another software tool based on EpSO is the Epilepsy Data Extraction and Annotation (EpiDEA) system. The EpiDEA system utilizes Mayo Clinic’s open source NLP software tool cTAKES (clinical Text Analysis and Knowledge Extraction System) to identify and analyze EpSO terminology in Epilepsy Monitoring Unit (EMU) discharge summaries for patient cohort identification[31]. In 2014, the OPIC and EpiDEA system were combined into the Multi-Modality Epilepsy Data Capture and Integration System, MEDCIS, developed to facilitate data integration for large scale multi center EMU studies[43–45]. The robustness of these tools rests in the use of EpSO as the common reference terminology across all software components, with the goal of increasing generalizability across health centers. A timeline of milestones for development of NLP tools for epilepsy is shown in Figure 2.
Figure 2.
Timeline of milestones for NLP in epilepsy. Integrated EpSO-based tools are highlighted with green arrows. EPILEPSIAE = Evolving Platform for Improving the Living Expectations of Patients Suffering from IctAl Events.
3.4. Comparison of recent epilepsy information extraction studies and future directions
Despite the abundance of open source software tools, there are only a few published studies on epilepsy information extraction from the EHR. NLP performance is often measured by three metrics: precision, the fraction of retrieved items relevant to the query, recall, the fraction of relevant items successfully retrieved, and F1 score, the harmonic mean of precision and recall. The EpiDEA system developed by Cui et al. achieved overall precision, recall, and F1 scores of 93.59%, 84.01%, and 88.53% respectively for extraction of EEG patterns and medication information from 104 EMU discharge summaries[31]. Cui et al. also developed the PEEP (Phenotype Extraction in Epilepsy) system for extracting the epileptogenic zone, seizure semiology, lateralizing sign, interictal and ictal EEG pattern from discharge summaries. PEEP achieved overall precision, recall, and F1 scores of 92.4%, 93.1%, and 92.7% respectively when tested over 262 discharge summaries[32]. Most recently, Fonferko-Shadrach et al. developed the ExECT (Extraction of Epilepsy Clinical Text) system which retrieves epilepsy diagnosis, epilepsy type, focal seizures, generalized seizures, seizure frequency, medications, CT, MRI, and EEG data from clinic letters. ExECT achieved overall precision, recall, and F1 scores of 91.4%, 81.4%, and 86.5% respectively [46]. The gold standard in all studies consisted of manual annotations created by clinicians. Though the EpiDEA and PEEP systems have higher scores on the performance metrics than ExECT, this could be because EpiDEA and PEEP utilize EMU discharge summaries while ExECT utilizes clinic letters. Discharge summaries tend to be much more organized than clinic notes, making them easier for an NLP system to classify.
The improvement in classifier performance on pre-structured data compared to free-form notes is clear; however, there are several drawbacks of mandating a structured note format. Predefined categories are not always sufficient to describe every patient and lack the detail and customizability of free-form prose. Clinicians pressed for time may also be less careful with cumbersome, structured data entry systems and may auto-populate forms with default options. Continued work is needed to identify the optimal combination of structured and unstructured data entry to maximize interpretability while offering convenience in clinician workflow. To this end, the field of epilepsy research could benefit greatly from updated data entry tools to balance clinician and researcher interests, an updated ontology to reflect ILAE changes, and increased use of the existing software tools for data organization and natural language processing.
4. Distributed systems for real-time analysis of intracranial EEG
4.1. Next generation implantable devices for iEEG telemetry
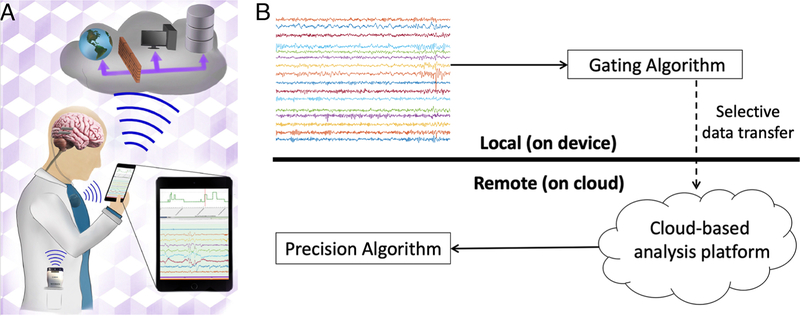
The area with likely the most immediate potential impact of “big data” on the care and science of status epilepticus is the real-time analysis of neurophysiologic data. The next generation of implantable monitoring and therapeutic devices are producing vast amounts of intracranial EEG (iEEG) data capable of being transmitted to analysis systems in real time, and will soon be deployed in the care and prevention of status epilepticus. Traditionally, the computational capabilities of neuroimplantable devices[47,48], such as the NeuroPace RNS system[49], have been limited by device processing power and battery life[50]. Furthermore, such devices typically store only brief segments of iEEG for intermittent review by a physician, precluding evaluation of the continuous record or trending of iEEG features over time. A promising approach for improving the utility of implanted devices is to transmit data from the device to a remote analysis platform[51,52], such as a hand-held personal assist device (PAD) or cloud-based server[53] (Figure 3A). With continuous or intermittent data telemetry, patient data can be archived and annotated in centralized repositories accessible to physicians[54], and can also be exposed to off-board computing resources and custom algorithms.
Figure 3.
Implanted devices with cloud connectivity. (A) Schematic of next-generation epilepsy management system using Medtronic investigational Summit RC+S. The device interfaces with a local PAD and the cloud to create a flexible platform with local and distributed computing, analytics, and data storage. Figure reproduced with permission from Kremen, et al., “Integrating Brain Implants with Local and Distributed Computing Devices: A Next Generation Epilepsy Management System,” IEEE:JTEHM, Sep 2018, Vol 6[53]. (B) Seizure detection paradigm using cloud computing. Potential seizure clips are identified using a hypersensitive, on-board seizure detector. These candidate clips are transmitted to a cloud platform for analysis using a highly accurate, computationally intensive algorithm. Figure reproduced with permission from Baldassano, et al., “Cloud computing for seizure detection in implanted neural devices,” Journal of Neural Engineering, Feb 2019, 16 (2)[88].
A critical benefit of telemetry-capable devices has been the production of high-quality continuous iEEG datasets for both research and clinical use. Automated seizure detection, seizure diary generation, seizure prediction, and behavioral state classification have been demonstrated using the NeuroVista Seizure Advisory System (SAS), an implanted device providing telemetry of iEEG data coupled to off-body analytics, in both humans[55–57] and canines[58–61]. These novel datasets have also been featured in kaggle.com competitions to crowdsource development of seizure detection[56] and prediction[61,62] algorithms, which have produced validated, generalizable, open-source methods at a fraction of the cost of traditional research studies. These datasets have made it possible to investigate questions that require continuous interictal data, such as long-term trends in recording metrics[63], temporal relationships among seizures[64,65], and identification of iEEG biomarkers[66]. Clinically, continuous iEEG data streams offer a means of circumventing reliance on self-reported seizure diaries, which suffer from poor accuracy and hamper assessment of therapy efficacy[67,68].
4.2. Continuous iEEG datasets for optimizing neuromodulation
Neuromodulation (in the form of vagal nerve stimulation (VNS), responsive nerve stimulation (RNS), or deep brain stimulation (DBS)) is a viable treatment option for patients with medication-refractory seizures who are not candidates for surgical intervention[69,70]. While neurostimulation has been shown to reduce seizure frequency by 30–50% on average[71–73], optimizing of stimulation parameters remains a significant challenge. For instance, in the case of the NeuroPace RNS device, the first FDA-approved closed loop stimulator for epilepsy, stimulation parameters such as frequency, pulse width, and burst duration are manually tuned during clinic visits in an effort to decrease seizure frequency[74]. There is also a lack of convincing evidence in the literature as to the optimal location for cortical or subcortical stimulation[75], or whether stimulation should be directed in response to seizure activity[71,76,77] or chronic and subthreshold[78,79].
The next generation of implanted devices with continuous streaming capabilities offers exciting potential to generate comprehensive datasets for algorithmic optimization of neurostimulation. While novel strategies for adjustment of stimulation parameters, such as gradient descent learning[80] and physiologic adaptation[81], have been applied in animal models, they have not yet been translated to human studies[82]. Ongoing work using an investigational implantable neurostimulator (Summit System RC+S, Medtronic Inc.) in a canine model aims to leverage off-body algorithms for optimizing stimulation, in combination with two-way telemetry, to actively probe seizure networks[52,53]. Given the wealth of data produced by continuous iEEG records, it is now possible to dynamically adjust therapeutic stimulation parameters, guided by surrogate interictal markers of seizures such as interictal epileptiform discharges (IEDs), evoked responses, and brain state[52]. Advances in hardware, software, and real-time analysis systems are likely to bring these techniques into the management of status epilepticus patients.
4.3. Cloud computing for seizure detection on intracranial EEG
Effective closed-loop neurostimulation requires highly accurate algorithms to classify brain signals and detect seizures[83–85]. While many machine learning algorithms to detect seizure activity have been published[56,86], such methods are typically far too complex to be implemented in an implanted device. As a result, current responsive stimulation devices, such as the NeuroPace RNS and Medtronic Activa PC+S, are limited to simple feature sets and classifiers[47,48,87] and are consequently plagued by high rates of false detections[72].
Next-generation implantable devices capable of intermittent data telemetry may offer substantial improvements in seizure detection accuracy through off-boarding of computationally-intensive algorithms. Our group recently proposed a novel paradigm for implementing real-time complex seizure detection algorithms on implanted device iEEG data[88]. This method (Figure 3B) relies on two sequential seizure detection algorithms. First, a simple “gating” algorithm running on-board the device is used to control selective transmission of data from the device to a cloud storage and analysis platform. The gating algorithm is designed to be compatible with the computational limitations of an implanted device, and is tuned to be hypersensitive in order to identify every seizure event for transmission. Transmitted data are then analyzed by a “precision” algorithm, a much more accurate and computationally intensive algorithm deployed using cloud computing resources. Evaluated using continuous, human iEEG data collected from the NeuroVista SAS[55], this system achieved seizure detection accuracy far outpacing that of existing neuroimplantable devices with significantly lower false positive rates[72]. This early work is encouraging, as by using telemetry to access cloud computational resources the next generation of implanted devices will be able to employ much more sophisticated algorithms to improve device performance and patient outcomes.
5. Real-time analysis of multimodal ICU data
5.1. Quantitative approaches to continuous EEG
Despite its benefits for detection of non-convulsive status epilepticus (NCSE) [89], a condition with a mortality rate greater than 50%[90], cEEG is underutilized in part due to the significant labor required to initiate recordings and interpret the vast amount of data generated. In many cases, around-the-clock staffing of physicians and EEG technologists to handle the unpredictable needs of cEEG monitoring presents a prohibitive level of cost [91]. Furthermore, cEEG is often read at infrequent intervals which could lead to hours of delay between the onset of a seizure and notification of the event to the patient’s physician. In instances that require rapid clinical action, the therapeutic window may pass before cEEG is interpreted or a patient’s condition may deteriorate and result in an adverse outcome. Automated methods of cEEG interpretation and detection of NCSE hold the potential to solve some of these problems and increase the utilization of cEEG at both large centers and small hospitals.
While there are several approved algorithms for seizure detection in scalp EEG, they suffer from a variety of drawbacks which preclude their widespread clinical use. The most prominent EEG monitoring algorithm is the FDA-approved Reveal algorithm implemented as part of Persyst’s clinical EEG system. Reveal was initially reported to have a sensitivity of 76% with a false positive rate of 0.11/hour [92], though subsequent studies have found a much higher false positive rate [93]. This level of performance leads to a significant proportion of seizures going undetected, as well as a substantial number of false alarms. One accepted method for providing semi-automated ICU EEG analysis is known as quantitative EEG (qEEG) in which signals are evaluated in real time, displaying metrics such as amplitude, rhythmicity, and spectral features in a visual interface [94]. Quantitative EEG can be presented in a temporally compressed format allowing long trends to be quickly visualized, such as in the NeuroTrend algorithm [95]. However, while qEEG may significantly reduce review time by the clinician, sensitivity for seizure identification may be as low as 51–67% [96], limiting its clinical reliability.
Approaches that leverage innovative methods in data science and machine learning may allow future algorithms for automated and semi-automated cEEG analysis to be more impactful than previous attempts. One such method uses quantitative features and a simple classifier to identify patients as belonging to the following groups: normal, isoelectric, slowing, low-voltage, burst suppression, or containing discharges or seizures [97]. While classification accuracy was 85% in the test set, this algorithm is not designed to perform in real time and would not necessarily improve early detection of seizure activity [97]. ICU-ASDA is an algorithm which uses signal amplitude variation and a regularity statistic to detect changes in rhythmicity [98]. This algorithm detected seizures with a 90.4% sensitivity and a false positive rate of 1.6 per 24 hours in a dataset with 24 patients of which 11 had non-convulsive seizures [98]. However, the authors only reported training accuracy and given their small sample size the generalizability of this algorithm is unknown. There remains a clear need for high-performance algorithms designed for real-time deployment, supported by studies which use large, high quality, ICU cEEG datasets.
5.2. Multimodal ICU monitoring: an opportunity for translational clinical data platforms
In addition to cEEG, other modalities of continuous physiologic monitoring are increasingly relied upon for neurointensive care of patients in or recovering from status epilepticus[99,100]. Such modalities include advanced monitoring techniques such as intracranial pressure probes[101], tissue oxygen probes[102], thermal diffusion probes[103], invasive and non-invasive monitors of cardiac output[104], and near-infrared spectroscopy[105], as well as standard physiologic signals such as heart rate, continuous blood pressure, and oxygen saturation. Just as in the case of cEEG, multimodal monitoring data in the ICU must be manually reviewed by physicians, imposing a significant burden to caretakers and increasing cost, latency of response to clinical events, and risk of missing events altogether. In response, development of clinical informatics to fuse outputs from multiple clinical monitors in a meaningful way for monitoring or prediction is flourishing[106]. The ability to detect nonconvulsive seizures after subarachnoid hemorrhage based on cEEG can be improved by incorporating other physiological data such as heart rate, blood pressure, respiratory rate, and cerebral perfusion pressure[107]. Methods incorporating cEEG in combination with metrics such as intracranial pressure, brain tissue PO2, and cerebral microdialysis[108], or demographic and admissions data[109], have been used to detect and monitor cerebral hypoperfusion. Algorithms interpreting multimodal ICU data have also shown efficacy for stratifying patient outcomes after cardiac arrest[110] or severe traumatic brain injury[111].
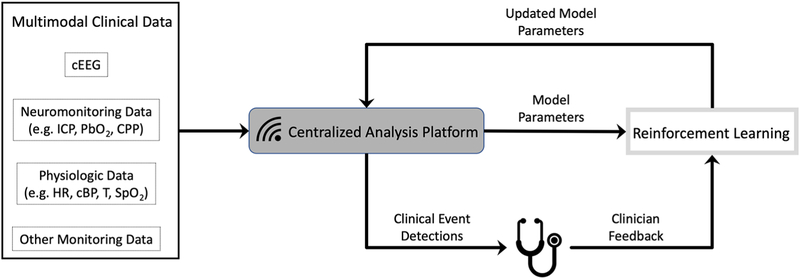
While such methods for inferring patient status and predicting outcomes are increasingly common, there is nevertheless a dearth of tools for practical data reduction and interpretation that integrate with caretaker workflow. Currently, tremendous amounts of data are generated from clinical monitors but are not centralized or archived to allow for remote viewing, annotation, sharing among providers, or retrospective retrieval. In parallel with the advancement of monitoring technology must come an advancement in clinical informatics infrastructure[112]. Specifically, manufacturer-specific data systems must be augmented with customizable, modular data platforms that facilitate data storage, visualization, and analytics (Figure 4).
Figure 4.
ICU data analysis pipeline. The next generation of real-time data analysis platforms will incorporate multimodality data and employ machine learning algorithms to provide custom clinical event detections. By incorporating clinician feedback through reinforcement learning, these algorithms can intelligently adapt over time. cEEG=continuous EEG, ICP=intracranial pressure, PbO2=brain tissue oxygen, CPP=cerebral perfusion pressure, HR=heart rate, cBP = continuous blood pressure, T=continuous temperature, SpO2=blood oxygen saturation.
The need for next-generation data management and analysis platforms is beginning to be addressed by companies such as Blackfynn, Inc.[113,114], a secure cloud-based platform for sharing and analyzing medical data, co-founded by members of our group. Ongoing work in our lab also seeks to meet this need through development of a modular ICU data monitoring platform called IRIS (ICU Real-Time Informatics System) (unpublished). This automated system is capable of streaming ICU data from multimodal clinical monitors in real time, archiving the data on a central server, and carrying out custom analysis modules. Early proof-of-concept trials have included modules to detect faulty cEEG electrodes, automatically trend burst-suppression ratios, and detect sustained elevations in intracranial pressure. IRIS is linked to a custom, HIPAA-compliant API and smartphone application to securely deliver notifications of detected clinical events to caretakers including physicians, nurses, and EEG technicians. Such platforms have great potential to benefit patient care by improving ICU workflow, reducing the cost and caretaker burden associated with continuous monitoring, and exposing existing data streams to state-of-the-art machine learning algorithms. Furthermore, by designing platforms with a modular structure, it is possible to add custom modules based on clinical needs such as methods of seizure detection, spike detection, or vasospasm monitoring. Future work may also integrate therapy in a closed loop system, for applications including titration of medications for treatment of status epilepticus or maintenance of general anesthesia. We hope that our work will encourage the continued implementation of translational medical data systems to optimize care of patients with epilepsy undergoing neurointensive care.
5.3. Economic impact of automated ICU data analysis
Automated analysis of ICU data may have significant ramifications for health care costs in the ICU. As ICU datasets become larger and more descriptive, tools leveraging this data for prognostication will become more effective. In the case of critically ill patients in status epilepticus, improved outcome prediction has potential to significantly influence goals of care and resource allocation at the end of life[115–117]. While automated scoring systems may not yet outperform ICU physicians in predicting mortality[118], their ability to fuse large amounts of multimodal data promises to reveal clinical insights that may affect management decisions. Furthermore, automated analysis of continuous monitoring data will, in itself, change revenue models in critical care. Continuous monitoring studies, such as 24-hour EEG, are costly to patients[119,120], in part due to the need for manual physician review. The implications of this economic impact remain to be seen. In the current, private-insurance driven model, recommendations for continuous monitoring may be monetarily disincentivized by hospitals due to lower billing potential; however, if health care payments shift toward single-payer or bundled payments, automated monitoring studies may be more prevalent due to increased cost effectiveness.
6. Future Directions and Conclusion
The revolution of “big data” in status epilepticus has already begun, and tools for leveraging this data are flourishing. Yet, for extracting data from clinical sources such as claims data and the EHR, we have merely scratched the surface. Part of future progress in these areas is predicated on developing new methods for NLP, integrating clinical notes with imaging and laboratory data, and mining health systems databases. An equally important part will be driven by ambitious pushes to share data and algorithms through collaborative platforms, and to encourage investigators to work together to pose powerful questions. In the realm of physiologic monitoring, implanted devices and ICU systems currently in development are capable of streaming EEG and multimodality data to local computing systems or the cloud. By implementing distributed data reduction and interpretation algorithms, these systems provide real-time analytics for treatment of status epilepticus. As we continue to improve technologies for acquiring, storing, and accessing “big data”, we will be able to rapidly design, test, and employ data-driven methods and classifiers. “Big data” is establishing itself as an agent for change in healthcare that will impact clinical practice, decision making, and care guidelines. In addition, population-based datasets offer opportunities to compare medical management decisions, costs, and outcomes across different health systems, potentially providing new perspectives on health care among countries with a range of payment models. Such information may play an important role in legislative decisions reshaping the US healthcare system. Through data science and engineering, we have potential to improve the quality and efficiency of care for status epilepticus and other neurologic disorders.
Highlights for “Big Data in Status Epilepticus”.
“Big Data” is driving new technologies for treatment of status epilepticus
Administrative claims data provide insight to cEEG usage and efficacy
Natural language processing allows data mining from the electronic health record
Next generation implanted devices are linked to cloud computing resources
Automated analysis of multimodal data is changing continuous monitoring in the ICU
Acknowledgements
Funding for studies described in this review provided by the Mirowski Family Fund, Neil and Barbara Smit, the Family of Johnathan Rothberg, the American Epilepsy Society/Epilepsy Foundation Research and Training Fellowship for Clinicians, the Ashton Fellowship at the University of Pennsylvania, and the National Institutes of Health (NIH) (UH2-NS095495-01, R01NS092882, 1K01ES025436-01).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
Brian Litt is a co-founder of Blackfynn, Inc., an advisor for EpilepsyCo, and has licensed intellectual property to NeuroPace, Inc., through the University of Pennsylvania. No other authors have disclosures to report.
References
- [1].Kleen JK, Lowenstein DH, Progress in Epilepsy: Latest Waves of Discovery, JAMA Neurol. 74 (2017) 139–140. [DOI] [PubMed] [Google Scholar]
- [2].Ben-Menachem E, Epilepsy in 2015: the year of collaborations for big data, Lancet Neurol. 15 (2016) 6–7. [DOI] [PubMed] [Google Scholar]
- [3].Waterhouse EJ, The Epidemiology of Status Epilepticus, in: Drislane FW, Kaplan PW (Eds.), Status Epilepticus: A Clinical Perspective, Springer International Publishing, Cham, 2018: pp. 15–29. [Google Scholar]
- [4].Wagenaar JB, Worrell GA, Ives Z, Matthias D, Litt B, Schulze-Bonhage A, Collaborating and sharing data in epilepsy research, J. Clin. Neurophysiol 32 (2015) 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Duncan D, Vespa P, Pitkänen A, Braimah A, Lapinlampi N, Toga AW, Big data sharing and analysis to advance research in post-traumatic epilepsy, Neurobiol. Dis 123 (2019) 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Istephan S, Siadat M-R, Unstructured medical image query using big data - An epilepsy case study, J. Biomed. Inform 59 (2016) 218–226. [DOI] [PubMed] [Google Scholar]
- [7].Chen Y, Wang Z-Y, Yuan G, Huang L, An overview of online based platforms for sharing and analyzing electrophysiology data from big data perspective, WIREs Data Mining Knowl Discov. 7 (2017) e1206. [Google Scholar]
- [8].Betjemann JP, Lowenstein DH, Status epilepticus in adults, Lancet Neurol. 14 (2015) 615–624. [DOI] [PubMed] [Google Scholar]
- [9].Scheuer ML, Continuous EEG monitoring in the intensive care unit, Epilepsia. 43 Suppl 3 (2002) 114–127. [DOI] [PubMed] [Google Scholar]
- [10].Friedman D, Claassen J, Hirsch LJ, Continuous electroencephalogram monitoring in the intensive care unit, Anesth. Analg 109 (2009) 506–523. [DOI] [PubMed] [Google Scholar]
- [11].Herman ST, Abend NS, Bleck TP, Chapman KE, Drislane FW, Emerson RG, Gerard EE, Hahn CD, Husain AM, Kaplan PW, LaRoche SM, Nuwer MR, Quigg M, Riviello JJ, Schmitt SE, Simmons LA, Tsuchida TN, Hirsch LJ, Critical Care Continuous EEG Task Force of the American Clinical Neurophysiology Society, Consensus Statement on Continuous EEG in Critically Ill Adults and Children, Part I, J. Clin. Neurophysiol 32 (2015) 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vespa PM, McArthur DL, Xu Y, Eliseo M, Etchepare M, Dinov I, Alger J, Glenn TP, Hovda D, Nonconvulsive seizures after traumatic brain injury are associated with hippocampal atrophy, Neurology. 75 (2010) 792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vespa PM, Miller C, McArthur D, Eliseo M, Etchepare M, Hirt D, Glenn TC, Martin N, Hovda D, Nonconvulsive electrographic seizures after traumatic brain injury result in a delayed, prolonged increase in intracranial pressure and metabolic crisis, Crit. Care Med 35 (2007) 2830–2836. [PMC free article] [PubMed] [Google Scholar]
- [14].Payne ET, Zhao XY, Frndova H, McBain K, Sharma R, Hutchison JS, Hahn CD, Seizure burden is independently associated with short term outcome in critically ill children, Brain. 137 (2014) 1429–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Topjian AA, Gutierrez-Colina AM, Sanchez SM, Berg RA, Friess SH, Dlugos DJ, Abend NS, Electrographic Status Epilepticus Is Associated With Mortality and Worse Short-Term Outcome in Critically Ill Children*, Crit. Care Med 41 (2013) 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gavvala J, Abend N, LaRoche S, Hahn C, Herman ST, Claassen J, Macken M, Schuele S, Gerard E, Critical Care EEG Monitoring Research Consortium (CCEMRC), Continuous EEG monitoring: A survey of neurophysiologists and neurointensivists, Epilepsia. 55 (2014) 1864–1871. [DOI] [PubMed] [Google Scholar]
- [17].Abend NS, Dlugos DJ, Hahn CD, Hirsch LJ, Herman ST, Use of EEG Monitoring and Management of Non-Convulsive Seizures in Critically Ill Patients: A Survey of Neurologists, Neurocrit. Care. 12 (2010) 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Continuous EEG Randomized Trial in Adults - Full Text View - ClinicalTrials.gov, (n.d.) https://clinicaltrials.gov/ct2/show/NCT03129438 (accessed June 27, 2019).
- [19].HCUP-US NIS Overview, (n.d.) https://www.hcup-us.ahrq.gov/nisoverview.jsp (accessed June 27, 2019).
- [20].Hill CE, Blank LJ, Thibault D, Davis KA, Dahodwala N, Litt B, Willis AW, Continuous EEG is associated with favorable hospitalization outcomes for critically ill patients, Neurology. 92 (2019) e9–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ney JP, van der Goes DN, Nuwer MR, Nelson L, Eccher MA, Continuous and routine EEG in intensive care: utilization and outcomes, United States 2005–2009, Neurology. 81 (2013) 2002–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cock HR, ESETT Group, Established status epilepticus treatment trial (ESETT), Epilepsia. 52 Suppl 8 (2011) 50–52. [DOI] [PubMed] [Google Scholar]
- [23].Bleck T, Cock H, Chamberlain J, Cloyd J, Connor J, Elm J, Fountain N, Jones E, Lowenstein D, Shinnar S, Silbergleit R, Treiman D, Trinka E, Kapur J, The established status epilepticus trial 2013, Epilepsia. 54 Suppl 6 (2013) 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fritz F, Tilahun B, Dugas M, Success criteria for electronic medical record implementations in low-resource settings: a systematic review, J. Am. Med. Inform. Assoc 22 (2015) 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jensen K, Soguero-Ruiz C, Oyvind Mikalsen K, Lindsetmo R-O, Kouskoumvekaki I, Girolami M, Olav Skrovseth S, Augestad KM, Analysis of free text in electronic health records for identification of cancer patient trajectories, Sci. Rep 7 (2017) 46226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Khankhanian P, Kosaraju N, Pathmanathan J, Ellis C, Helbig I, Litt B, Pollard J, Davis K, On the Feasibility of Natural Language Processing for Standardized Data Extraction from Electronic Medical Records of Epilepsy Patients (P1.283), Neurology. 90 (2018) P1–283.. [Google Scholar]
- [27].Liddy ED, Natural Language Processing, in: Encyclopedia of Library and Information Science, 2nd Ed., Marcel Decker, Inc., 2001. [Google Scholar]
- [28].Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, Liu S, Zeng Y, Mehrabi S, Sohn S, Liu H, Clinical information extraction applications: A literature review, J. Biomed. Inform 77 (2018) 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Demner-Fushman D, Chapman WW, McDonald CJ, What can natural language processing do for clinical decision support?, J. Biomed. Inform 42 (2009) 760–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rijo R, Silva C, Pereira L, Gonçalves D, Agostinho M, Decision Support System to Diagnosis and Classification of Epilepsy in Children, J. Univers. Comput. Sci 20 (2014) 907–923. [Google Scholar]
- [31].Cui L, Bozorgi A, Lhatoo SD, Zhang G-Q, Sahoo SS, EpiDEA: extracting structured epilepsy and seizure information from patient discharge summaries for cohort identification, AMIA Annu. Symp. Proc 2012 (2012) 1191–1200. [PMC free article] [PubMed] [Google Scholar]
- [32].Cui L, Sahoo SS, Lhatoo SD, Garg G, Rai P, Bozorgi A, Zhang G-Q, Complex epilepsy phenotype extraction from narrative clinical discharge summaries, J. Biomed. Inform 51 (2014) 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Khalifa A, Meystre S, Adapting existing natural language processing resources for cardiovascular risk factors identification in clinical notes, J. Biomed. Inform 58 Suppl (2015) S128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sahoo SS, Lhatoo SD, Gupta DK, Cui L, Zhao M, Jayapandian C, Bozorgi A, Zhang G-Q, Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care, J. Am. Med. Inform. Assoc 21 (2014) 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sahoo SS, Zhang G-Q, Lhatoo SD, Epilepsy informatics and an ontology-driven infrastructure for large database research and patient care in epilepsy, Epilepsia. 54 (2013) 1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sahoo SS, Zhao M, Luo L, Bozorgi A, Gupta D, Lhatoo SD, Zhang G-Q, OPIC: Ontology-driven Patient Information Capturing system for epilepsy, AMIA Annu. Symp. Proc 2012 (2012) 799–808. [PMC free article] [PubMed] [Google Scholar]
- [37].Pereira L, Rijo R, Silva C, Agostinho M, ICD9-based Text Mining Approach to Children Epilepsy Classification, Procedia Technology. 9 (2013) 1351–1360. [Google Scholar]
- [38].Pereira L, Rijo R, Silva C, Agostinho M, Using text mining to diagnose and classify epilepsy in children, in: 2013 IEEE 15th International Conference on E-Health Networking, Applications and Services (Healthcom 2013), ieeexplore.ieee.org, 2013: pp. 345–349. [Google Scholar]
- [39].Nivedhitha G, Anandha Mala GS, Enhanced Automatic Classification of Epilepsy Diagnosis Using ICD9 and SNOMED-CT, in: Proceedings of the International Conference on Soft Computing Systems, Springer India, 2016: pp. 259–266. [Google Scholar]
- [40].Son JH, Xie G, Yuan C, Ena L, Li Z, Goldstein A, Huang L, Wang L, Shen F, Liu H, Mehl K, Groopman EE, Marasa M, Kiryluk K, Gharavi AG, Chung WK, Hripcsak G, Friedman C, Weng C, Wang K, Deep Phenotyping on Electronic Health Records Facilitates Genetic Diagnosis by Clinical Exomes, Am. J. Hum. Genet 103 (2018) 58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fisher RS, Cross JH, French JA, Higurashi N, Hirsch E, Jansen FE, Lagae L, Moshé SL, Peltola J, Roulet Perez E, Scheffer IE, Zuberi SM, Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology, Epilepsia. 58 (2017) 522–530. [DOI] [PubMed] [Google Scholar]
- [42].Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, Hirsch E, Jain S, Mathern GW, Moshé SL, Nordli DR, Perucca E, Tomson T, Wiebe S, Zhang Y-H, Zuberi SM, ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology, Epilepsia. 58 (2017) 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang G-Q, Cui L, Lhatoo S, Schuele SU, Sahoo SS, MEDCIS: Multi-Modality Epilepsy Data Capture and Integration System, AMIA Annu. Symp. Proc 2014 (2014) 1248–1257. [PMC free article] [PubMed] [Google Scholar]
- [44].Jayapandian CP, Chen C-H, Bozorgi A, Lhatoo SD, Zhang G-Q, Sahoo SS, Electrophysiological signal analysis and visualization using Cloudwave for epilepsy clinical research, Stud. Health Technol. Inform 192 (2013) 817–821. [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang G-Q, Siegler T, Saxman P, Sandberg N, Mueller R, Johnson N, Hunscher D, Arabandi S, VISAGE: A Query Interface for Clinical Research, Summit Transl Bioinform 2010 (2010) 76–80. [PMC free article] [PubMed] [Google Scholar]
- [46].Fonferko-Shadrach B, Lacey AS, Roberts A, Akbari A, Thompson S, Ford DV, Lyons RA, Rees MI, Pickrell WO, Using natural language processing to extract structured epilepsy data from unstructured clinic letters: development and validation of the ExECT (extraction of epilepsy clinical text) system, BMJ Open. 9 (2019) e023232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Herron J, Denison T, Chizeck HJ, Closed-loop DBS with movement intention, in: 2015 7th International IEEE/EMBS Conference on Neural Engineering (NER), IEEE, 2015: pp. 844–847. [Google Scholar]
- [48].Ryapolova-Webb E, Afshar P, Stanslaski S, Denison T, de Hemptinne C, Bankiewicz K, Starr PA, Chronic cortical and electromyographic recordings from a fully implantable device: preclinical experience in a nonhuman primate, J. Neural Eng. 11 (2014) 016009. [DOI] [PubMed] [Google Scholar]
- [49].Morrell MJ, Responsive cortical stimulation for the treatment of medically intractable partial epilepsy, Neurology. 77 (2011) 1295–1304. [DOI] [PubMed] [Google Scholar]
- [50].Meng E, Sheybani R, Langer R, Langer R, Insight: implantable medical devices, Lab Chip. 14 (2014) 3233. [DOI] [PubMed] [Google Scholar]
- [51].Bourget D, Bink H, Stanslaski S, Linde D, Arnett C, Adamski T, Denison T, An implantable, rechargeable neuromodulation research tool using a distributed interface and algorithm architecture, in: 2015 7th International IEEE/EMBS Conference on Neural Engineering (NER), IEEE, 2015: pp. 61–65. [Google Scholar]
- [52].Kremen V, Brinkmann BH, Kim I, Chang S, Van Gompel JJ, Herron JA, Baldassano S, Patterson EE, Litt B, Denison T, Worrell GA, Continuous active probing and modulation of neural networks with a wireless implantable system, in: 2017 IEEE Biomedical Circuits and Systems Conference (BioCAS), ieeexplore.ieee.org, 2017: pp. 1–4.30406220 [Google Scholar]
- [53].Herron JA, Adamski T, Baldassano S, Integrating Brain Implants With Local and Distributed Computing Devices: A Next Generation Epilepsy Management System, IEEE Journal of Translational Engineering in Health and Medicine. 6 (2018) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wagenaar JB, Brinkmann BH, Ives Z, Worrell GA, Litt B, A multimodal platform for cloud-based collaborative research, in: 2013 6th International IEEE/EMBS Conference on Neural Engineering (NER), IEEE, 2013: pp. 1386–1389. [Google Scholar]
- [55].Cook MJ, O’Brien TJ, Berkovic SF, Murphy M, Morokoff A, Fabinyi G, D’Souza W, Yerra R, Archer J, Litewka L, Hosking S, Lightfoot P, Ruedebusch V, Sheffield WD, Snyder D, Leyde K, Himes D, Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: a first-in-man study, Lancet Neurol. 12 (2013) 563–571. [DOI] [PubMed] [Google Scholar]
- [56].Baldassano SN, Brinkmann BH, Ung H, Blevins T, Conrad EC, Leyde K, Cook MJ, Khambhati AN, Wagenaar JB, Worrell GA, Litt B, Crowdsourcing seizure detection: algorithm development and validation on human implanted device recordings, Brain. 140 (2017) 1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kremen V, Duque JJ, Brinkmann BH, Berry BM, Kucewicz MT, Khadjevand F, Van Gompel J, Stead M, St Louis EK, Worrell GA, Behavioral state classification in epileptic brain using intracranial electrophysiology, J. Neural Eng. 14 (2017) 026001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Davis KA, Sturges BK, Vite CH, Ruedebusch V, Worrell G, Gardner AB, Leyde K, Sheffield WD, Litt B, A novel implanted device to wirelessly record and analyze continuous intracranial canine EEG, Epilepsy Res. 96 (2011) 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Howbert JJ, Patterson EE, Stead SM, Brinkmann B, Vasoli V, Crepeau D, Vite CH, Sturges B, Ruedebusch V, Mavoori J, Leyde K, Sheffield WD, Litt B, Worrell GA, Forecasting seizures in dogs with naturally occurring epilepsy, PLoS One. 9 (2014) e81920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Brinkmann BH, Patterson EE, Vite C, Vasoli VM, Crepeau D, Stead M, Howbert JJ, Cherkassky V, Wagenaar JB, Litt B, Worrell GA, Forecasting Seizures Using Intracranial EEG Measures and SVM in Naturally Occurring Canine Epilepsy, PLoS One. 10 (2015) e0133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Brinkmann BH, Wagenaar J, Abbot D, Adkins P, Bosshard SC, Chen M, Tieng QM, He J, Muñoz-Almaraz FJ, Botella-Rocamora P, Pardo J, Zamora-Martinez F, Hills M, Wu W, Korshunova I, Cukierski W, Vite C, Patterson EE, Litt B, Worrell GA, Crowdsourcing reproducible seizure forecasting in human and canine epilepsy, Brain. 139 (2016) 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kuhlmann L, Karoly P, Freestone DR, Brinkmann BH, Temko A, Barachant A, Li F, Titericz G Jr, Lang BW, Lavery D, Roman K, Broadhead D, Dobson S, Jones G, Tang Q, Ivanenko I, Panichev O, Proix T, Náhlík M, Grunberg DB, Reuben C, Worrell G, Litt B, Liley DTJ, Grayden DB, Cook MJ, Epilepsyecosystem.org: crowd-sourcing reproducible seizure prediction with long-term human intracranial EEG, Brain. 141 (2018) 2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ung H, Baldassano SN, Bink H, Krieger AM, Williams S, Vitale F, Wu C, Freestone D, Nurse E, Leyde K, Davis KA, Cook M, Litt B, Intracranial EEG fluctuates over months after implanting electrodes in human brain, J. Neural Eng. 14 (2017) 056011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Karoly PJ, Nurse ES, Freestone DR, Ung H, Cook MJ, Boston R, Bursts of seizures in long-term recordings of human focal epilepsy, Epilepsia. 58 (2017) 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Karoly PJ, Ung H, Grayden DB, Kuhlmann L, Leyde K, Cook MJ, Freestone DR, The circadian profile of epilepsy improves seizure forecasting, Brain. 140 (2017) 2169–2182. [DOI] [PubMed] [Google Scholar]
- [66].Roehri N, Bartolomei F, Are high-frequency oscillations better biomarkers of the epileptogenic zone than spikes?, Curr. Opin. Neurol 32 (2019) 213–219. [DOI] [PubMed] [Google Scholar]
- [67].Elger CE, Mormann F, Seizure prediction and documentation—two important problems, Lancet Neurol. 12 (2013) 531–532. [DOI] [PubMed] [Google Scholar]
- [68].Hoppe C, Poepel A, Elger CE, Epilepsy, Arch. Neurol 64 (2007) 1595. [DOI] [PubMed] [Google Scholar]
- [69].Nune G, DeGiorgio C, Heck C, Neuromodulation in the Treatment of Epilepsy, Curr. Treat. Options Neurol. 17 (2015) 375. [DOI] [PubMed] [Google Scholar]
- [70].Dalkilic EB, Neurostimulation Devices Used in Treatment of Epilepsy, Curr. Treat. Options Neurol. 19 (2017) 7. [DOI] [PubMed] [Google Scholar]
- [71].Heck CN, King-Stephens D, Massey AD, Nair DR, Jobst BC, Barkley GL, Salanova V, Cole AJ, Smith MC, Gwinn RP, Skidmore C, Van Ness PC, Bergey GK, Park YD, Miller I, Geller E, Rutecki PA, Zimmerman R, Spencer DC, Goldman A, Edwards JC, Leiphart JW, Wharen RE, Fessler J, Fountain NB, Worrell GA, Gross RE, Eisenschenk S, Duckrow RB, Hirsch LJ, Bazil C, O’Donovan CA, Sun FT, Courtney TA, Seale CG, Morrell MJ, Two-year seizure reduction in adults with medically intractable partial onset epilepsy treated with responsive neurostimulation: Final results of the RNS System Pivotal trial, Epilepsia. 55 (2014) 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sun FT, Morrell MJ, Closed-loop Neurostimulation: The Clinical Experience, Neurotherapeutics. 11 (2014) 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Fisher R, Salanova V, Witt T, Worth R, Henry T, Gross R, Oommen K, Osorio I, Nazzaro J, Labar D, Kaplitt M, Sperling M, Sandok E, Neal J, Handforth A, Stern J, DeSalles A, Chung S, Shetter A, Bergen D, Bakay R, Henderson J, French J, Baltuch G, Rosenfeld W, Youkilis A, Marks W, Garcia P, Barbaro N, Fountain N, Bazil C, Goodman R, McKhann G, Babu Krishnamurthy K, Papavassiliou S, Epstein C, Pollard J, Tonder L, Grebin J, Coffey R, Graves N, SANTE Study Group, Electrical stimulation of the anterior nucleus of thalamus for treatment of refractory epilepsy, Epilepsia. 51 (2010) 899–908. [DOI] [PubMed] [Google Scholar]
- [74].Sun FT, Morrell MJ, The RNS System: responsive cortical stimulation for the treatment of refractory partial epilepsy, Expert Rev. Med. Devices 11 (2014) 563–572. [DOI] [PubMed] [Google Scholar]
- [75].Schulze-Bonhage A, Brain stimulation as a neuromodulatory epilepsy therapy, Seizure. 44 (2017) 169–175. [DOI] [PubMed] [Google Scholar]
- [76].Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I, On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy, Nat. Commun 4 (2013) 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Armstrong C, Krook-Magnuson E, Oijala M, Soltesz I, Closed-loop optogenetic intervention in mice, Nat. Protoc 8 (2013) 1475–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lundstrom BN, Worrell GA, Stead M, Van Gompel JJ, Chronic subthreshold cortical stimulation: a therapeutic and potentially restorative therapy for focal epilepsy, Expert Rev. Neurother 17 (2017) 661–666. [DOI] [PubMed] [Google Scholar]
- [79].Lundstrom BN, Van Gompel J, Britton J, Nickels K, Wetjen N, Worrell G, Stead M, Chronic Subthreshold Cortical Stimulation to Treat Focal Epilepsy, JAMA Neurol. 73 (2016) 1370. [DOI] [PubMed] [Google Scholar]
- [80].Panuccio G, Guez A, Vincent R, Avoli M, Pineau J, Adaptive control of epileptiform excitability in an in vitro model of limbic seizures, Exp. Neurol 241 (2013) 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ward MP, Qing KY, Otto KJ, Worth RM, John SWM, Irazoqui PP, A flexible platform for biofeedback-driven control and personalization of electrical nerve stimulation therapy, IEEE Trans. Neural Syst. Rehabil. Eng 23 (2015) 475–484. [DOI] [PubMed] [Google Scholar]
- [82].Nagaraj V, Lee ST, Krook-Magnuson E, Soltesz I, Benquet P, Irazoqui PP, Netoff TI, Future of seizure prediction and intervention: closing the loop, J. Clin. Neurophysiol 32 (2015) 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ramgopal S, Thome-Souza S, Jackson M, Kadish NE, Sánchez Fernández I, Klehm J, Bosl W, Reinsberger C, Schachter S, Loddenkemper T, Seizure detection, seizure prediction, and closed-loop warning systems in epilepsy, Epilepsy Behav. 37 (2014) 291–307. [DOI] [PubMed] [Google Scholar]
- [84].Fountas KN, Smith JR, Murro AM, Politsky J, Park YD, Jenkins PD, Implantation of a closed-loop stimulation in the management of medically refractory focal epilepsy: a technical note, Stereotact. Funct. Neurosurg 83 (2005) 153–158. [DOI] [PubMed] [Google Scholar]
- [85].Sun FT, Morrell MJ, Wharen RE, Responsive Cortical Stimulation for the Treatment of Epilepsy, Neurotherapeutics. 5 (2008) 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tzallas AT, Tsalikakis DG, Karvounis EC, Astrakas L, Tzaphlidou M, Tsipouras MG, Konitsiotis S, Automated epileptic seizure detection methods: a review study, INTECH Open Access Publisher; (2012). [Google Scholar]
- [87].Echauz J, Esteller R, Tcheng T, Pless B, Gibb B, Kishawi E, Litt B, Long Term Validation of Detection Algorithms Suitable for an Implantable Device, Epilepsia, Vol. 42, No Suppl. 7 (2001). [Google Scholar]
- [88].Baldassano S, Zhao X, Brinkmann B, Kremen V, Bernabei J, Cook M, Denison T, Worrell G, Litt B, Cloud computing for seizure detection in implanted neural devices, J. Neural Eng. 16 (2019) 026016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kubota Y, Nakamoto H, Egawa S, Kawamata T, Continuous EEG monitoring in ICU, J. Intensive Care Med. 6 (2018) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Young GB, Jordan KG, Doig GS, An assessment of nonconvulsive seizures in the intensive care unit using continuous EEG monitoring: an investigation of variables associated with mortality, Neurology. 47 (1996) 83–89. [DOI] [PubMed] [Google Scholar]
- [91].Young GB, Continuous EEG monitoring in the ICU: challenges and opportunities, Can. J. Neurol. Sci 36 Suppl 2 (2009) S89–91. [PubMed] [Google Scholar]
- [92].Wilson SB, Scheuer ML, Emerson RG, Gabor AJ, Seizure detection: evaluation of the Reveal algorithm, Clin. Neurophysiol 115 (2004) 2280–2291. [DOI] [PubMed] [Google Scholar]
- [93].Kelly KM, Shiau DS, Kern RT, Chien JH, Yang MCK, Yandora KA, Valeriano JP, Halford JJ, Sackellares JC, Assessment of a scalp EEG-based automated seizure detection system, Clin. Neurophysiol 121 (2010) 1832–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Foreman B, Claassen J, Quantitative EEG for the detection of brain ischemia, Crit. Care 16 (2012) 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Herta J, Koren J, Fürbass F, Hartmann M, Kluge T, Baumgartner C, Gruber A, Prospective assessment and validation of rhythmic and periodic pattern detection in NeuroTrend: A new approach for screening continuous EEG in the intensive care unit, Epilepsy Behav. 49 (2015) 273–279. [DOI] [PubMed] [Google Scholar]
- [96].Haider HA, Esteller R, Hahn CD, Westover MB, Halford JJ, Lee JW, Shafi MM, Gaspard N, Herman ST, Gerard EE, Hirsch LJ, Ehrenberg JA, LaRoche SM, Critical Care EEG Monitoring Research Consortium, Sensitivity of quantitative EEG for seizure identification in the intensive care unit, Neurology. 87 (2016) 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Cloostermans MC, de Vos CC, van Putten MJAM, A novel approach for computer assisted EEG monitoring in the adult ICU, Clin. Neurophysiol 122 (2011) 2100–2109. [DOI] [PubMed] [Google Scholar]
- [98].Sackellares JC, Shiau D-S, Halford JJ, LaRoche SM, Kelly KM, Quantitative EEG analysis for automated detection of nonconvulsive seizures in intensive care units, Epilepsy Behav. 22 Suppl 1 (2011) S69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Suarez JI, Outcome in neurocritical care: Advances in monitoring and treatment and effect of a specialized neurocritical care team, Crit. Care Med. 34 (2006) S232–S238. [DOI] [PubMed] [Google Scholar]
- [100].Busl KM, Bleck TP, Varelas PN, Neurocritical Care Outcomes, Research, and Technology, JAMA Neurol. (2019). doi: 10.1001/jamaneurol.2018.4407. [DOI] [PubMed] [Google Scholar]
- [101].Czosnyka M, Pickard JD, Monitoring and interpretation of intracranial pressure, J. Neurol. Neurosurg. Psychiatry 75 (2004) 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Okonkwo DO, Shutter LA, Moore C, Temkin NR, Puccio AM, Madden CJ, Andaluz N, Chesnut RM, Bullock MR, Grant GA, McGregor J, Weaver M, Jallo J, LeRoux PD, Moberg D, Barber J, Lazaridis C, Diaz-Arrastia RR, Brain Oxygen Optimization in Severe Traumatic Brain Injury Phase-II, Crit. Care Med 45 (2017) 1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Korbakis G, Vespa PM, Multimodal neurologic monitoring, in: Handbook of Clinical Neurology, 2017: pp. 91–105. [DOI] [PubMed] [Google Scholar]
- [104].Saugel B, Cecconi M, Wagner JY, Reuter DA, Noninvasive continuous cardiac output monitoring in perioperative and intensive care medicine, Br. J. Anaesth 114 (2015) 562–575. [DOI] [PubMed] [Google Scholar]
- [105].Green MS, Sehgal S, Tariq R, Near-Infrared Spectroscopy, Semin. Cardiothorac. Vasc. Anesth 20 (2016) 213–224. [DOI] [PubMed] [Google Scholar]
- [106].Hemphill JC, Andrews P, De Georgia M, Multimodal monitoring and neurocritical care bioinformatics, Nat. Rev. Neurol 7 (2011) 451–460. [DOI] [PubMed] [Google Scholar]
- [107].Claassen J, Perotte A, Albers D, Kleinberg S, Schmidt JM, Tu B, Badjatia N, Lantigua H, Hirsch LJ, Mayer SA, Connolly ES, Hripcsak G, Nonconvulsive seizures after subarachnoid hemorrhage: Multimodal detection and outcomes, Ann. Neurol 74 (2013) 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Bouzat P, Marques-Vidal P, Zerlauth J-B, Sala N, Suys T, Schoettker P, Bloch J, Daniel RT, Levivier M, Meuli R, Oddo M, Accuracy of brain multimodal monitoring to detect cerebral hypoperfusion after traumatic brain injury*, Crit. Care Med 43 (2015) 445–452. [DOI] [PubMed] [Google Scholar]
- [109].Amorim E, Rittenberger JC, Zheng JJ, Westover MB, Baldwin ME, Callaway CW, Popescu A, Continuous EEG monitoring enhances multimodal outcome prediction in hypoxic–ischemic brain injury, Resuscitation. 109 (2016) 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Oddo M, Rossetti AO, Early multimodal outcome prediction after cardiac arrest in patients treated with hypothermia, Crit. Care Med. 42 (2014) 1340–1347. [DOI] [PubMed] [Google Scholar]
- [111].Sykora M, Czosnyka M, Liu X, Donnelly J, Nasr N, Diedler J, Okoroafor F, Hutchinson P, Menon D, Smielewski P, Autonomic Impairment in Severe Traumatic Brain Injury: A Multimodal Neuromonitoring Study, Crit. Care Med 44 (2016) 1173–1181. [DOI] [PubMed] [Google Scholar]
- [112].Schmidt JM, De Georgia M, Participants in the International Multidisciplinary Consensus Conference on Multimodality Monitoring, Multimodality monitoring: informatics, integration data display and analysis, Neurocrit. Care 21 Suppl 2 (2014) S229–38. [DOI] [PubMed] [Google Scholar]
- [113].Johnson C, Christini A, Felmeister A, Baglieri C, Kambhampati M, Waanders A, Mason J, Packer R, Resnick A, Nazarian J, DIPG-51. BLACKFYNN: A SECURE, CLOUD-BASED PLATFORM FOR SHARING AND ANALYZING RESEARCH READY DATA FOR PEDIATRIC CNS CANCERS, Neuro. Oncol 20 (2018) i59–i59. [Google Scholar]
- [114].Christini A, Waanders AJ, Wagenaar JB, Felmeister AS, Santi M, Wadhwani NR, Mason JL, Koptyra MP, Lilly JV, Pennington JW, Lulla RR, Resnick AC, Abstract 2593: Accelerating pediatric brain tumor research through team science solutions, Cancer Res. 77 (2017) 2593–2593. [Google Scholar]
- [115].Barnato AE, Angus DC, Value and role of intensive care unit outcome prediction models in end-of-life decision making, Crit. Care Clin 20 (2004) 345–62, vii–viii. [DOI] [PubMed] [Google Scholar]
- [116].Campbell ML, Guzman JA, Impact of a proactive approach to improve end-of-life care in a medical ICU, Chest. 123 (2003) 266–271. [DOI] [PubMed] [Google Scholar]
- [117].Rocker G, Cook D, Sjokvist P, Weaver B, Finfer S, McDonald E, Marshall J, Kirby A, Levy M, Dodek P, Heyland D, Guyatt G, Level of Care Study Investigators, Canadian Critical Care Trials Group, Clinician predictions of intensive care unit mortality, Crit. Care Med 32 (2004) 1149–1154. [DOI] [PubMed] [Google Scholar]
- [118].Sinuff T, Adhikari NKJ, Cook DJ, Schünemann HJ, Griffith LE, Rocker G, Walter SD, Mortality predictions in the intensive care unit: comparing physicians with scoring systems, Crit. Care Med. 34 (2006) 878–885. [DOI] [PubMed] [Google Scholar]
- [119].Vespa PM, Nenov V, Nuwer MR, Continuous EEG monitoring in the intensive care unit: early findings and clinical efficacy, J. Clin. Neurophysiol 16 (1999) 1–13. [DOI] [PubMed] [Google Scholar]
- [120].Jordan KG, Continuous EEG monitoring in the neuroscience intensive care unit and emergency department, J. Clin. Neurophysiol 16 (1999) 14–39. [DOI] [PubMed] [Google Scholar]