Abstract
During the chronic stage of Schistosoma infection, the female lays fertile eggs, triggering a strong anti‐parasitic type 2 helper T‐cell (Th2) immune response. It is unclear how this Th2 response gradually declines even though the worms live for years and continue to produce eggs. Here, we show that Schistosoma mansoni downregulates Th2 differentiation in an antigen‐presenting cell‐independent manner, by modulating the Th2‐specific transcriptional program. Adult schistosomes secrete miRNA‐harboring extracellular vesicles that are internalized by Th cells in vitro. Schistosomal miRNAs are found also in T helper cells isolated from Peyer's patches and mesenteric lymph nodes of infected mice. In T helper cells, the schistosomal miR‐10 targets MAP3K7 and consequently downmodulates NF‐κB activity, a critical transcription factor for Th2 differentiation and function. Our results explain, at least partially, how schistosomes tune down the Th2 response, and provide further insight into the reciprocal geographic distribution between high prevalence of parasitic infections and immune disorders such as allergy. Furthermore, this worm‐host crosstalk mechanism can be harnessed to develop diagnostic and therapeutic approaches for human schistosomiasis and Th2‐associated diseases.
Keywords: extracellular vesicle, miRNA, Schistosoma, Th cells
Subject Categories: Immunology; Membrane & Intracellular Transport; Microbiology, Virology & Host Pathogen Interaction
During chronic Schistosoma infection a strong anti‐parasitic Th2‐type immune response is triggered. The parasite counteracts this by releasing extracellular vesicles that contain miRNAs that modulate Th2 differentiation.
Introduction
Schistosomiasis (Bilharziasis) is caused by infection with the trematode helminth of the genus Schistosoma. It is a common parasite, which affects more than 200 million people, mostly in Africa. The three main species that cause this human disease are Schistosoma mansoni (found mainly in Africa, South America, Caribbean, and the Middle East), Schistosoma haematobium (Africa and the Middle East), and Schistosoma japonicum (China and South East Asia). Schistosome infections have also been diagnosed in non‐endemic areas, often imported by either immigrants or travelers 1, 2.
The life cycle of schistosomes involves snails and humans. Infections of humans take place in freshwater bodies, where the schistosome cercariae penetrate human skin. In the skin, the cercariae transform into the juvenile forms of the helminth, the schistosomula, which migrate from the skin to the lungs, and then to the liver. In the liver, the males and females copulate and mature into adult worms that further migrate to their final locations—urogenital venules for S. haematobium or mesenteric venules for the other species. Pathologically, the acute stage starts 1–2 weeks after skin penetration and continues with the development of the schistosomula until reaching maturation and localization as adult parasites in the blood vessels. With the beginning of egg deposition, the chronic stage starts and can last for many years (there are reports on infected immigrants for even 20–38 years after departing from the endemic areas 3, 4). During the chronic stage, the female produces each day many eggs that reach the water through urine or feces. In the water, the larvae known as miracidia hatch from the eggs and penetrate specific aquatic snail hosts, in which asexual reproduction produces thousands of infective cercariae 5. The eggs which entrapped within the human tissue (intestinal or urogenital tracts) are those which elicit an immunological response with granuloma formation.
The acute and chronic infection stages of schistosomiasis induce different immune responses. The penetration of the cercariae initiates a strong Th1 reaction, which in mice lasts for ~5 weeks 6, 7, whereas the egg production skews the immune response toward the Th2 pathway 8, 9. T helper (Th; CD4+) cells have a fundamental role in shaping the immune response. The first interaction of naïve Th cell with specific antigen on the antigen‐presenting cell (APC, mostly dendritic cell; DC) promotes its differentiation, depending on the context, into either effector—mainly Th1, Th2, and Th17—or regulatory (Treg) lineage 10, 11, 12, 13. Each lineage is specified by a distinct network of transcriptional regulators; the lineage‐specifying transcription factors of Th1, Th2, Th17, and Treg cells are T‐bet, Gata3, Rorγt, and Foxp3, respectively. Consequently, each lineage is characterized by the expression of distinct cytokine profiles with the hallmark cytokines IFN‐γ in Th1 cells, IL‐4, IL‐5, and IL‐13 (the “Th2 cytokines”) in Th2 cells, IL‐17 in Th17 cells, and TGF‐β and IL‐10 in Treg cells (IL‐10 is also expressed by other immune cells such as Th2) 14. These cytokines powerfully promote diverse immune responses: IFN‐γ exerts protective functions, mostly in infection with intracellular parasites, IL‐17 contributes to the host defense against fungi and extracellular bacteria, and Treg cytokines are involved in preventing potential self‐reactivity and dampening hyper‐immune response. The Th2 cytokines mostly play a role in response to extracellular parasites.
The canonical response of Th2 cells is associated with the isotypes IgG1, IgG4, and IgE, and expanding populations of eosinophils, basophils, mast cells, and alternatively activated macrophages 15. IgE isotype facilitates antibody‐dependent cell‐mediated cytotoxicity (ADCC), in which the antibodies cover the parasite while their Fc portion binds receptors on eosinophils, basophils, mast cells, and neutrophils, which, in turn, release granules containing toxic or oxidizing molecules contributing to helminth eradication 15, 16, 17. Th2 pathway may also display a host protective response that reduces immunopathologic damage. Eggs, which penetrate the intestinal wall, carried into the liver via the portal vasculature. These trapped eggs could cause serious damage. Th2 induce granuloma formation around these eggs, constituted by a variety of infiltrating immune cells and fibrosis that sequesters the eggs from the surrounding tissue 8, 9, 17. In fact, knockout mice for Il4 develop fatal hepatic inflammation after the schistosome female starts depositing eggs 18, 19.
A poorly understood aspect of schistosomiasis is the decline in the Th2 responsiveness following its initial peak at approximately week 8 of infection in humans 5, 20. This loss is intriguing because the decline occurs even though the parasitic worms live for years and continue to produce eggs 8, 9, 21. Current theories suggest that an expansion in regulatory cell populations (mostly Treg cells, but also macrophages and B cells) and an increase in the secretion of IL‐10 and TGF‐β, which are observed in both schistosome‐infected humans and mice, play a role in dampening the Th2 reaction. The tegumental schistosome‐specific phosphatidylserine, and S. japonicum conserved peptide of an egg protein, for example, were found to activate the TLR2 on DCs, facilitating their ability to induce the development of IL‐10‐producing Treg cells 22, 23. However, evidence suggests a more complex, multi‐factorial, and systemic effect regulating the long‐lasting Th2 suppression during chronic schistosomiasis 5, 8, 9, 20, 21.
The release of extracellular vesicles (EVs) has recently attracted attention as a means for intercellular communication 24, 25. EVs are small membrane‐bound vesicles that are generally classified into two major types, exosomes and microvesicles, based on their size, biogenesis, and composition. Exosomes are 50–100 nm vesicles originating from the endosome membrane and released into the extracellular space 26. The secretion of EVs was demonstrated in several parasites such as the protozoa Plasmodium falciparum 27 and Trypanosoma brucei 28, and helminths 29 including schistosomes 30, 31, 32. We hypothesized that schistosomes use this long‐distance route to systemically manipulate the immune system of their host. Here, we demonstrate that adult schistosomes deliver EVs containing microRNAs (miRNAs) which preferentially interfere with the differentiation toward the Th2 pathway in an APC‐independent manner by, at least partially, modulating the activation of NF‐κB.
Results
Schistosomes downregulate the differentiation toward the Th2 lineage
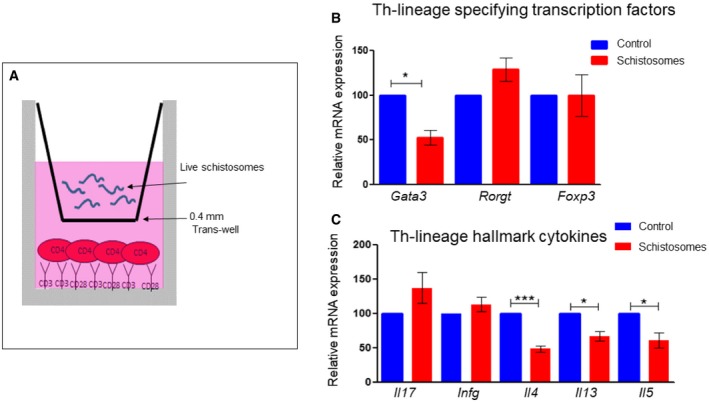
Since Th cells influence the strategy of the immune response, first we wanted to assess whether schistosomes affect Th‐cell differentiation. For that purpose, naive Th cells were exposed to live adult S. mansoni, derived from infected mice, using a trans‐well system 27; freshly isolated Th cells from spleen and lymph nodes of young mice were stimulated with anti‐CD3 and anti‐CD28 antibodies (mimicking stimulation through the T‐cell receptor) in a tissue culture well, while 25–30 adult worms were placed above the cells, in a trans‐well insert with 0.4‐μm pores (Fig 1A). A trans‐well insert with only an unused schistosomal medium was utilized as a control. After 72 h, the inserts were removed, and cells were re‐stimulated for 2 h with phorbol 12‐myristate 13‐acetate and calcium ionophore (P+I) for the induction of cytokine expression. Remarkably, the presence of the schistosomes specifically reduced the differentiation toward the Th2 pathway as reflected by a decrease in the mRNA expression levels of the Th2‐lineage‐specifying transcription factor Gata3 (Fig 1B), and the Th2 hallmark cytokines Il4, Il5, and Il13 (Fig 1C). The mRNA expression levels of other Th‐lineage‐specifying transcription factors and characteristic cytokines have not been changed significantly. Altogether, these results demonstrate that schistosomes, without any direct contact with the cells, restrictedly target the differentiation toward the Th2 pathway.
Figure 1. Schistosomes target the differentiation toward the Th2 pathway.
-
ATh cells (CD4+) were purified using magnetic beads from spleen and lymph nodes of 4‐ to 5‐week‐old mice. The cells were stimulated with anti‐CD3 and anti‐CD28 antibodies, beneath a trans‐well with 0.4‐μm pore containing 25–30 worms. The control wells contained only unused schistosomal medium.
-
B, C72 h after stimulation with anti‐CD3 and anti‐CD28, the cells were re‐stimulated with P+I for 2 h, and the mRNA was subjected to qRT–PCR using the indicated primers for the lineage‐specifying transcription factors (B) or cytokines (C). The graphs represent an average of at least four independent experiments. In each graph, the expression level in the control was set as 100%. P‐value was calculated by t‐test. *P < 0.05, ***P < 0.005.
Schistosomes modulate the Th2 transcriptional program
To reveal genome‐wide schistosomal‐induced alterations in the transcriptional patterns of differentiating Th cells, we repeated the segregated co‐culturing of the schistosomes and differentiating Th cells as in Fig 1, and 72 h later performed an RNA‐seq. We considered only changes in the levels of gene expression by at least 1.5‐fold on average in comparison with the control, in three independent experiments, and with P < 0.05. This analysis revealed that the exposure of differentiating Th cells to the schistosomes resulted in increased expression levels of 275 genes and a decrease in 526 genes (Dataset EV1A and B normalized data; Dataset EV1C and D normalized read count data).
The mRNA sequencing results reinforced the idea that the schistosomes targeted preferentially the Th2 pathway; many of the genes whose expression was decreased significantly in the presence of the parasites are associated with Th2 differentiation (Dataset EV1A and C).
Clustering by the bioinformatic DAVID program 33, 34, indicated that the expression of cytokines, chemokines, and signaling molecules was changed, among other pathways. For example, as expected from the qPCR, the expression levels of the Il4 and Il13 were decreased by 1.53‐ and 1.7‐fold, respectively, whereas the expression levels of other Th‐signature cytokines were unchanged. The expression of Il2, an important cytokine for T‐cell stimulation, was reduced by 1.8‐fold; Il10 by twofold, probably reflecting the decrease in the Th2 population; and Il33, which promotes the Th2 immune response in infected mice with S. japonicum 35, was reduced by 4.6‐fold. IL‐33 plays, in general, a role in the induction and maintenance of the Th2 pathway, e.g., by enhancing the activity of Tnfsf4 (encoding OX40L) 36. Tnfsf4, which was shown to be critical for Th2 development in leishmaniasis 37, was decreased by twofold as well. The expression level of several chemokines and chemokine receptors was also decreased: Ccl22, a chemokine that is specifically involve in the recruitment and polarization of Th2 cells 38, was reduced by twofold. The expression of additional receptors associated with the Th2 differentiation was reduced, such as Tim‐2 (Timd2), which is expressed preferentially in Th2 cells and plays a role in Th2 regulation and cytokine expression 39, was reduced by 1.5‐fold. On the other hand, the expression level of Tim‐3 (Havcr2), another member of this family, which is Th1‐specific 40, was increased by twofold. In addition, the mRNA of cd80 (B7‐1) that interacts with CD28 and stimulates T cells during antigen presentation 41 was decreased by 1.61‐fold. CD80 is required for the activation of the Th2 immune response in mice treated with the S. mansoni egg antigen (SEA) 42.
The idea that schistosomes differentially restrict the Th2 polarization was further supported by analyses of genes whose expression was increased significantly in Th cells in the presence of the schistosome worms. For example, Anxa1, whose overexpression reduces GATA3 and increases T‐bet levels 43, was increased by 1.86‐fold. Schistosomes also elevated by 1.6‐fold the mRNA expression level of CD274 (PD‐L1), an immune checkpoint inhibitor for proliferating T cells. The expression of dynamin (Dnm1) was also increased by 1.79‐fold. Dynamin is required for both membrane budding and exosomal uptake by targeted cells 44, 45. Altogether, the RNA‐seq results indicate that schistosomes preferentially modulate the Th2 response by affecting the transcriptional profile necessary for Th2 lineage differentiation and activation.
Adult schistosomes secrete characteristic EVs
The effect of schistosomes on Th‐cell differentiation across the trans‐well suggested the involvement of long‐distance communication ability such as of EVs. First, we wanted to investigate whether these adult schistosomes secrete typical EVs. The characterization of EVs is based on the vesicle size and protein content. The schistosome worms were cultured in EV‐free medium for up to 1 month, the medium was collected every 2–3 days, and the EVs were extracted by differential centrifugations 46. The EV pellets were analyzed by transmission electron microscopy (TEM) and by atomic force microscopy (AFM) 47. EVs were detected by both microscopes in the schistosome‐derived EV pellet, but not in the control‐derived pellet (Fig 2A and B). Applying the NanoSight, nanoparticle tracking analysis (NTA) revealed that in the fraction extracted from the schistosomal‐growing medium, most of the particles were in size of ~100 nm, in contrast to the control‐derived pellet where almost no particles were found (Fig 2C).
Figure 2. Size characterization of schistosomal EVs.
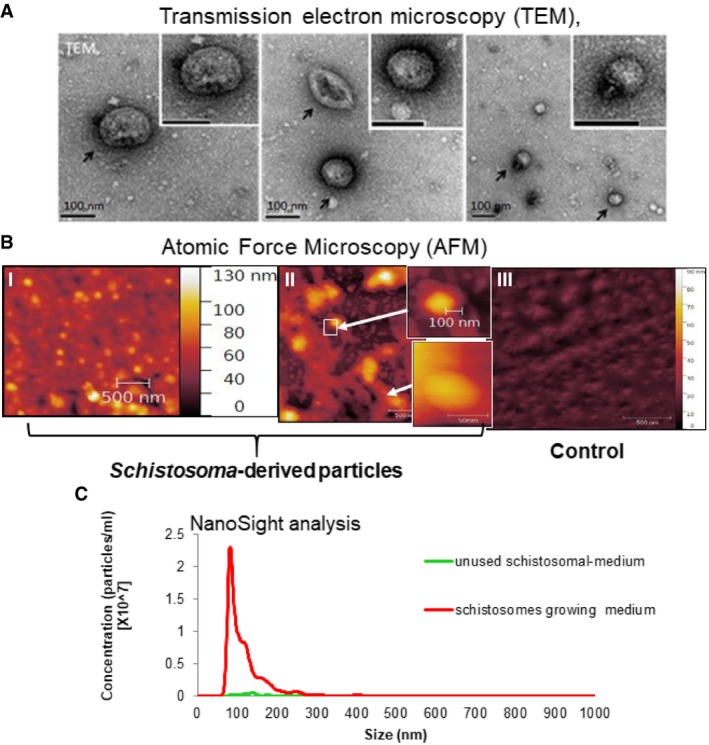
- EVs were extracted from the schistosomal‐growing medium and were subjected to cryo‐TEM analysis: Images were taken as described previously 90. The black arrows indicate EVs at the size of ˜100 nm.
- EVs were extracted from the schistosomal‐growing medium and were subjected AFM analysis: Images were taken from the schistosomal‐growing medium (I) or from 1/10 dilution of this medium (II). The small images magnify the arrow‐indicated EVs. (III) Image of the extract deriving from the unused schistosomal medium.
- EVs were extracted from the schistosomal‐growing medium and were subjected NTA size fractionation of particles analysis; the graph represents average of 5 different measurements.
For further characterization of these EVs, we performed a proteomic analysis of EVs extracted from the schistosomal‐growing medium. The schistosomal‐protein analysis was based on the whole‐genome sequencing containing 11,723 putative proteins 48. The two separated EV proteomic analyses obtained similar results (Dataset EV2A‐1, A‐2, and EV2B). Most of the identified peptides (~96%) were of bovine serum origin (Dataset EV2A‐1 and A‐2). However, none of the detected bovine peptides was of a known EV hallmark protein, such as of the tetraspanin proteins CD9 and CD63, annexin, aldolase, elongation factor 1‐alpha, heat shock protein 70, or programmed cell death protein (Table 1). However, peptides mapped to the schistosomal homologs of the above‐mentioned proteins were identified in at least one of the proteomic analyses. In the first analysis, 423 unique schistosomal peptides were mapped to 97 specific S. mansoni proteins. In the second analysis, 602 unique schistosomal peptides were mapped to 100 S. mansoni proteins (Dataset EVA‐1, A‐2, and EV2B).
Table 1.
The most frequently known EV‐associated proteins are detected in the schistosomal‐EV proteomic analysis
| Mammalian gene symbol | Number of times a protein was identified in other EV studies according to Vesiclepedia 51 | Protein name | Schistosome unique accession number protein from FASTA database | Total number of reads of the identified peptides mapped to the specific protein | The cover of the aligned protein, in percentage by the identified peptides. | Percentage of identity between human and the schistosomal protein | |
|---|---|---|---|---|---|---|---|
| 1 | PDCD6IP | 399 | Programmed cell death 6‐interacting protein | 353233402: putative programmed cell death protein | 3 | 13.22 | 45% |
| 2 | GAPDH | 377 | Glyceraldehyde‐3‐phosphate dehydrogenase | 353229455;353229454;353229456;353229457; glyceraldehyde‐3‐phosphate dehydrogenase | 15 | 51.35 | 73% |
| 3 | HSPA8 | 363 | Heat shock 70 kDa protein 8 | 552242;10168;353229993;353230065;Q27965;Q0VCX2;P0CB32;P19120;Q27975;P34933: putative heat shock protein 70 | 32 | 34.54 | 83% |
| 4 | ACTB | 350 | Beta actin | 678545;924603;353233031;353233035;353233111;360045418;360045419;P62739;P60712;Q3ZC07;P63258;Q5E9B5;P68138;F1MRD0;G8JKX4;F1MKC4; actin | 39 | 46.06 | 97% |
| 5 | ANXA2 | 337 | Annexin A2 | 353232899: putative annexin | 3 | 9.22 | 33% |
| 6 | CD9 | 328 | SM23 molecule | 161030;11890;350646174: integral membrane protein 23 | 6 | 25.23 | 27% |
| 7 | PKM | 327 | Pyruvate kinase | 353230308;353230309 putative pyruvate kinase [Schistosoma mansoni] | 37 | 56.74 | 62% |
| 8 | ENO1 | 327 | Enolase 1, (alpha) | 1002610;360044945;Q3ZC09;A6QR19; enolase | 14 | 23.27 | 75% |
| 9 | HSP90AA1 | 327 | Heat shock protein 90 kDa alpha | 7673568;353230104;353230105;360043333;360043335;Q95M18;Q76LV1;G3N2V5;G5E507; putative heat shock protein | 27 | 30.97 | 66% |
| 10 | CD63 | 306 | CD63 molecule | 353233656;575403085;23305772;353232346;353231401; putative tetraspanin‐CD63 receptor | 45 | 29.29 | 21% |
| 11 | EEF1A1 | 295 | Translation elongation factor 1‐alpha | 353230261;P68103;Q32PH8;G3N2F0;E1BPF4;E1B7J1;E1B9F6;E1BED8 putative elongation factor 1‐alpha (ef‐1‐alpha) | 6 | 11.83 | 78% |
| 12 | PGK1 | 291 | Phosphoglycerate kinase | 556413;360043465 phosphoglycerate kinase [Schistosoma mansoni] | 2 | 13.37 | 69% |
| 13 | ALDOA | 275 | Fructose‐bisphosphate aldolase | 605647;353232336 fructose 1,6 bisphosphate aldolase [Schistosoma mansoni] | 36 | 66.94 | 67% |
Analyzing the ontological characteristic of these proteins by the two bioinformatic tools FunRich and GO Ontology 49, 50 revealed significant enrichment in typical EV‐associated proteins (Appendix Fig S1 and Appendix Table S1). The most abundant known proteins of EVs are listed in the Vesiclepedia (http://microvesicles.org/extracellular_vesicle_markers), the manually updated online database of EV proteins, RNAs, and lipids 51. In our EV proteomics, 13 out of the 16 most characteristic EV proteins were identified (Table 1). In conclusion, and in compliance with the MISEV2018 52, the size and shape of the EVs (in three different analyses; Fig 2), and their protein content (Table 1), strongly suggest that S. mansoni secret typical EVs.
Schistosomal EVs are internalized by primary Th cells
To assess whether schistosomal EVs can enter Th cells, EVs were purified from a schistosomal‐growing medium, fluorescently labeled, and placed together with freshly isolated Th cells simultaneously with their stimulation. As a control, the Th cells were incubated with pellets, which were extracted with the same procedure as the schistosomal EV (ultracentrifugation and labeling), from an unused schistosomal medium. Images were taken by inverted confocal microscopy continually from the same slide after 3, 10, and 25 min of incubation (Fig 3A and Appendix Fig S2). After 10 min, the cytoplasm of 50% of the schistosomal‐EV‐incubated Th cells was stained, but not of the control Th cells. 15 min later, this pattern of staining was maintained, as shown in the graph of Fig 3B. Overall, these results demonstrate uptake of the schistosomal‐derived EVs by primary Th cells.
Figure 3. Schistosomal EVs are internalized by Th cells.
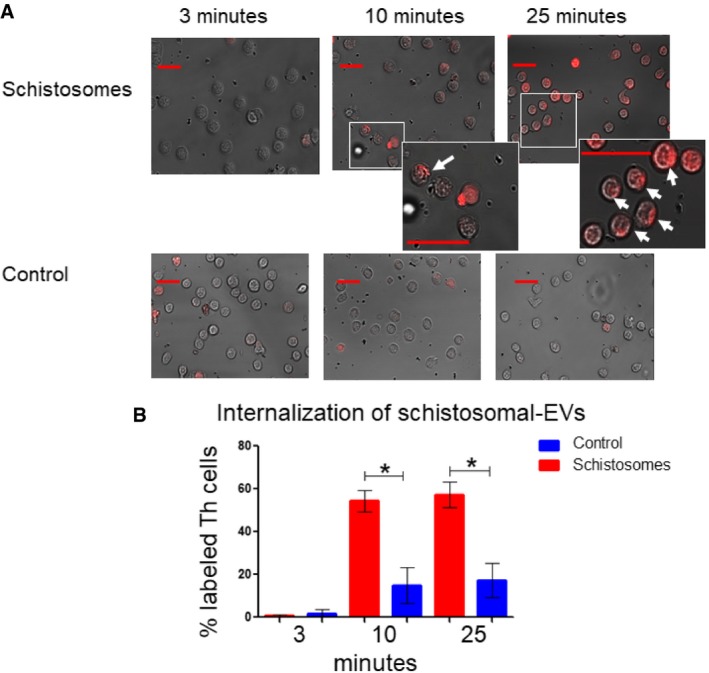
- EVs were purified from either schistosomal‐growing medium (supernatant) or unused schistosomal medium as control. Both pellets were stained using Thiazole Orange. Purified labeled ˜5 × 106 EVs were incubated with 5 × 105 freshly purified Th cells that were stimulated with anti‐CD3 and anti‐CD28 antibodies for 10 min. Images from the same slide were taken by inverted confocal microscopy at the indicated time points. The arrows in the enlarged images point toward cells labeled with EVs. The scale bars in all images represent 20 μm.
- The percentage of labeled cells was calculated by dividing the number of labeled cells by the total numbers of cells in the same image. The mean ± SEM was calculated from three independent images (all are presented in Appendix Fig S2). P‐value was calculated by t‐test. *P < 0.05. The scale bars are representing 20 μm length.
Schistosomal EVs harbor miRNAs
miRNAs are small non‐coding regulatory RNAs that can direct post‐transcriptional repression of mRNAs 53, 54, and were reported to be the main functional executive cargo of EVs 55, 56. To date, the genomes of the Schistosoma genus deposited in the miRBase database contain 225 S. mansoni miRNAs and 79 S. japonicum miRNAs 57, 58, 59, 60. In addition, we identified 65 miRNAs of S. haematobium [unpublished data]. However, so far, very few studies have been conducted to investigate their functions 61, 62. To assess whether selected schistosomal miRNAs are packed within the EVs, we extracted RNA from the schistosomal‐growing medium‐derived EVs, and as control from an unused schistosomal‐medium‐derived pellet. Equal volumes of extracts were subjected to qRT–PCR using specific TaqMan primers to the schistosomal miR‐10, schistosomal Bantam, and schistosomal miR‐125. These three miRNAs were detected only in the EVs extracted from the schistosomal‐growing medium (Fig 4).
Figure 4. Schistosomal EVs harbor miRNAs.
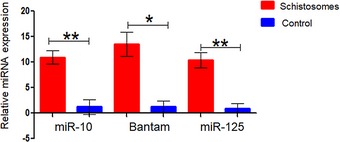
Schistosomal miRNA was extracted from either EVs that were isolated from schistosomal‐growing medium or control pellets that were isolated from unused schistosomal medium as described in Materials and Methods (schistosomal‐EV purification section). Equal aliquots were subjected to qRT–PCR with specific primers to the schistosomal miR‐10, Bantam, and miR‐125; the expression is presented as ΔCt from background Ct that was set as 40 cycles. The mean ± SEM was calculated from 3 independent experiments. P‐value was calculated by t‐test. *P < 0.05, **P < 0.01.
Schistosomal‐EV‐enclosed miRNAs penetrate schistosomal‐exposed Th cells
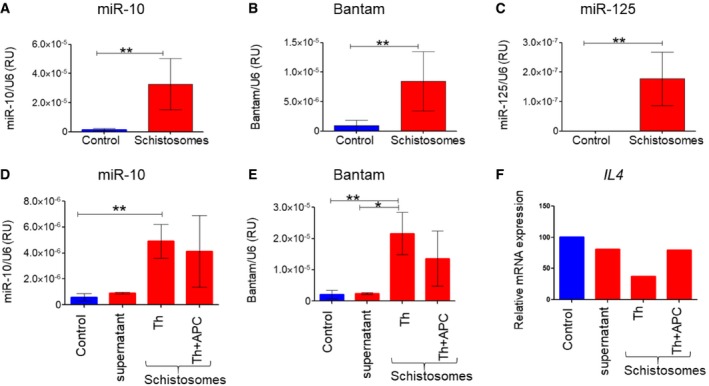
To assess whether schistosomal‐derived miRNAs are uptake by Th cells, we performed qRT–PCR with selected schistosomal‐miRNA primers on the RNA extracted from the schistosomal‐exposed Th cells (using the trans‐well system). Indeed, schistosomal‐exposed Th cells, but not control Th cells, contained the assessed schistosomal miRNAs: miR‐10 (Fig 5A), Bantam (Fig 5B), and miR‐125b (Fig 5C).
Figure 5. Schistosomal‐EV‐enclosed miRNAs penetrate Th cells.
-
A–CThe trans‐well system was performed as in Fig 1. Schistosome‐exposed Th cells or control Th cells were subjected to qRT–PCR with specific schistosomal‐TaqMan primers to (A) miR‐10‐5p, (B) Bantam, and (C) miR‐125b.
-
D–FThe trans‐well system was performed as in Fig 1. The control wells contained only unused schistosomal medium; supernatant wells contained 0.1 μm of the filtered schistosomal‐growing medium; Th wells contained sorted CD4+CD11c− from spleens; Th+APCs contained sorted CD4+CD11c− and CD4−CD11c+ in 5:1 ratio, respectively.
To further confirm that the effect of schistosomes on Th2 cell differentiation was APC‐independent, we sorted cells isolated from spleens into two highly purified groups: CD4+CD11c− (Th cells) and CD4−CD11c+ (APCs). Highly purified Th cells or a mix of Th cells with APCs at ratio of 5:1, respectively, were exposed to the worms using the trans‐well system. In both cases, the schistosomal miRNAs miR‐10 (Fig 5D) and Bantam (Fig 5E) were detected. Moreover, the presence of the schistosomes reduces the expression of Il4 more in the highly purified Th cells than in the Th‐APC mix (Fig 5F). Altogether, these results demonstrate APC‐independent internalization of the schistosomal miRNAs in Th cells.
To verify that the schistosomal miRNAs are packed in EVs rather than in free protein–RNA complexes, we performed the following two experiments: (i) We loaded isolated EVs from the schistosomal‐growing medium on OptiPrep™ density sucrose gradient (Appendix Fig S3). There was almost a complete overlap between the fractions containing the schistosomal miRNAs miR‐10 and Bantam and the HSP70 protein, which is a well‐known marker for EVs (Table 1). These results demonstrate an association between the schistosomal miRNAs and the EV fraction. (ii) Live worms were placed in a trans‐well system as described above. In parallel, schistosomal‐growth medium (used supernatant) was filtered through 0.1‐μm membrane to deplete schistosomal EVs and placed on top of a trans‐well above Th cells. As expected, the schistosomal miRNAs were detected in the Th cells that were exposed to the live schistosoma across the trans‐wells, but were undetected in Th cells that were exposed to the filtrated schistosomal supernatant (supernatant column; Fig 5D–F), excluding the effect of small protein/RNA aggregates. These two experiments strongly suggest that the schistosomal miRNAs are delivered in particles larger than 0.1 μm, which are most likely EVs. These results are consistent with our previous data demonstrating schistosomal miRNAs in the fraction of EVs that were isolated from the sera of Schistosoma‐infected patients 63.
Schistosomal miRNAs are found selectively in Th cells derived from the gut‐associated lymph nodes
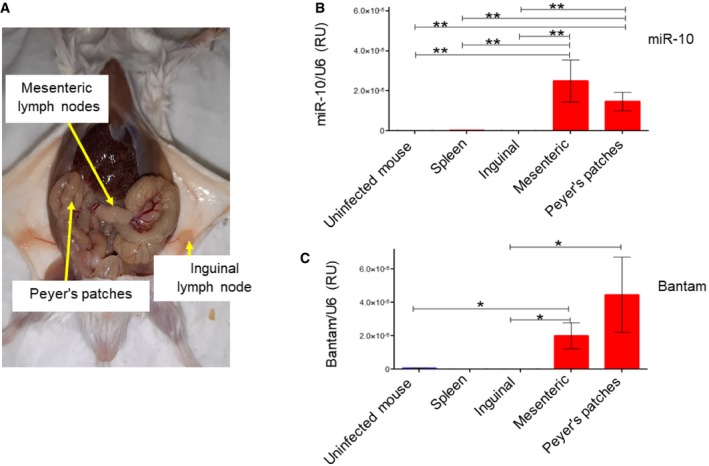
To confirm that the schistosomal miRNAs access Th cells in vivo, we purified Th cells from Peyer's patches, mesenteric lymph node (both are associated with the gastrointestinal tract), inguinal lymph node, and spleen of schistosome‐infected mice (Fig 6A). As a control, we used a mix of Th cells collected from different lymph nodes of uninfected mice. The schistosomal miRNAs miR‐10 and Bantam were detected in Th cells derived from both the mesenteric lymph nodes and Peyer's patches of the infected mice, but not from the inguinal lymph nodes or spleen (even though the spleen was enlarged) (Fig 6B and C). As expected, the schistosomal miRNAs miR‐10 and Bantam were undetected in Th cells derived from lymph nodes of uninfected mice. These results may suggest selective delivery of the schistosomal EVs through the lymphatic system, rather than systemic distribution through the blood.
Figure 6. Schistosomal miRNAs are found in lymph node‐derived Th cells.
-
AImage of a schistosome‐infected mouse. The lymph nodes, from which the Th cells were isolated, are indicated.
-
B, CTh cells were isolated from the indicated lymph nodes and spleen. In the case of uninfected mice, the Th cells from different lymph nodes and the spleen were mixed. qRT–PCR was performed with specific TaqMan primers to either schistosomal miR‐10 or Bantam. The presence of the miRNAs is presented as arbitrary units calculated as 2ΔΔCT of the miRNA divided by snRNA U6. The mean ± SEM was calculated from 4 independent experiments. P‐value was calculated by Mann–Whitney t‐test. *P < 0.05, **P < 0.01.
MAP3K7 (TAK1) is a target of schistosomal miR‐10
To study the biochemical effects of schistosomal miRNAs, we employed the human T‐cell line Jurkat. First, to assess whether schistosomal‐derived EVs can enter these cells, the EVs were fluorescently labeled and were placed together with the Jurkat cells for 10 min. Analysis by Amnis® imaging flow cytometer demonstrated that 40% of the Jurkat cells were labeled if they were incubated at 37°C with the schistosomal EVs, but not with the control‐labeled pellets (Appendix Fig S4). To further assess whether miRNAs were transferred by these EVs, the schistosomal EVs or the control pellets were added to the Jurkat cells. Forty‐eight hours later, the RNA was extracted and subjected to qRT–PCR using the one‐step real‐time RT–PCR protocol that allows detection of very low amount of miRNAs 63. The schistosomal miRNAs miR‐125 and Bantam were detected in schistosomal‐EV‐exposed Jurkat cells but not in the control (Appendix Fig S4).
Then, we looked for putative mRNA targets in Th cells. For this purpose, we decided to further investigate the targets of miR‐10, based on several reasons: (i) In previous analyses of EVs, it appeared as the most abundant miRNA in S. japonicum 31, and one of the most abundant in S. mansoni 32, 64. (ii) miR‐10 was found in sera of rabbits and mice infected with S. japonicum 65. (iii) Most importantly, miR‐10 was the most abundant schistosomal miRNAs among the selected miRNAs we examined in primary Th cells (Fig 5; about 100 times more than miR‐125 and 4 times more than Bantam). (iv) When we looked for predicted targets of selected schistosomal miRNAs, among the genes that were downregulated in schistosomal‐exposed Th cells, with a known Th2 function, using the bioinformatics tool TargetRank (http://hollywood.mit.edu/targetrank/) 66, we realized that miR‐10 has the highest number of putative targets.
Among the predicted targets of miR‐10, whose mRNA expression was decreased in the schistosomal‐exposed Th cells, were Gata3, Ccl22, and Tnfsf4 (OX40L). To assess their relevance, the human 3′UTR of these three genes was cloned into a psiCHECK‐II plasmid downstream to a synthetic Renilla luciferase gene (hRluc), and the plasmids were transfected into Jurkat cells that stably expressed the schistosomal miR‐10. The Renilla activities in both plasmids that included the 3′UTR of Ccl22 or Tnfsf4 were inhibited by miR‐10 (Appendix Fig S5). However, in both Ccl22 and Tnfsf4 the putative binding site of miR‐10 exists only in the human genes, while it is not conserved in mice. In addition, it seems that Gata3 3′UTR is not a biochemical target of miR‐10 (Appendix Fig S5).
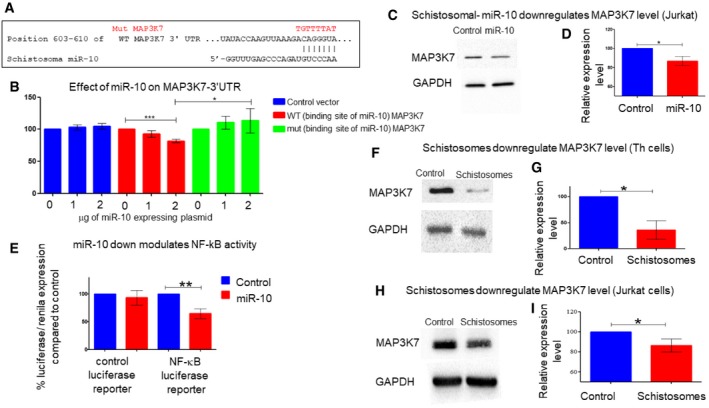
However, screening for miRNA targets by only looking for alterations in the mRNA expression levels is limited, since miRNAs can also regulate expression at the translational levels. Hence, we decided to increase the screen beyond those target genes whose expression was altered in our RNA‐seq analysis of schistosome‐exposed Th cells (Dataset EV1 Appendix Table S1A and C). The RNA‐seq results indicated also a general effect of the parasite on NF‐κB pathway; 29 genes whose expression was decreased in differentiating Th cells in the presence of schistosomes are known NF‐κB targets, Il4, Il10, Il2, Il13, Mmp7, Mmp10, Cx3cr1, Ccl22, Cxcl2, Arg2, and Cd80 (Appendix Table S2). In addition, the expression of 16 known NF‐κB activators was also decreased (Appendix Table S3). NF‐κB is essential for Th2 differentiation; deficiency in NF‐κB in mice results in a defect in the Th2 response, without affecting the Th1 response 67, 68. This is in accordance with the fact that the expression of both Gata3 and Il4 genes is regulated by NF‐κB 69, 70. Therefore, we looked whether any protein in the activation of NF‐κB signaling pathway might be a target of miR‐10. Bioinformatic analysis indicated MAP3K7, a serine/threonine kinase, which is involved in the activation of NF‐κB, as a potential target of miR‐10 (Fig 7A) 71, although its mRNA levels did not change in response to the schistosomes. Active phosphorylated MAP3K7 activates the IKK complex, leading to the phosphorylation, ubiquitination, and degradation of NFKB1A (IkBa). NFKB1A degradation unleashes NF‐κB, which is then free to translocate into the nucleus to regulate gene expression 72. We, therefore, wondered whether the exposure of Th cells to the schistosomal miR‐10 downregulates MAP3K7 expression and consequently the NF‐κB activity. To find whether MAP3K7 is a bona fide molecular target of schistosomal miR‐10, a segment of the 3′UTR containing the putative miR‐10 binding site was cloned to a psiCHECK‐II reporter plasmid 73. HEK‐293 cells were co‐transfected with plasmid expressing miR‐10 and with either control psiCHECK‐II vector, psiCHECK‐WT‐MAP3K7‐3′UTR or psiCHECK‐mut‐MAP3K7‐3′UTR. Renilla activities were significantly inhibited in cells transfected with miR‐10 expressing plasmid, but only if the reporter plasmid had the 3′UTR of MAP3K7 (Fig 7B). Mutation in the MAP3K7 3′UTR binding site of miR‐10 abolished the ability of miR‐10 to decrease luciferase expression, confirming that miR‐10 directly targets MAP3K7 3′UTR (Fig 7B).
Figure 7. Live schistosoma and its miR‐10 affect the expression of MAP3K7.
-
AThe putative binding sites of miR‐10 in the MAP3K7 3′UTR (taken from http://www.targetscan.org/vert_72/). Above the wild‐type seed sequence, in red, the sequences were placed in the mutated plasmid.
-
BHEK‐293 cells were transfected with a plasmid expressing schistosomal miR‐10 at the indicated concentration together with either psiCHECK‐II vector, psiCHECK‐WT‐MAP3K7‐3′UTR or psiCHECK‐mut‐MAP3K7. After 24 h, the cell lysates were subjected to luciferase assay. The results are presented as the ratio of Renilla/luciferase expression that was normalized relative to cells transfected only with psiCHECK‐II. Values are expressed as the mean ± SD of at least three independent experiments. Statistics were performed using t‐test *P < 0.05, ***P < 0.005.
-
CA representative experiment in which protein extracted from Jurkat‐T cells stably over expressing stably control vector or miR‐10 expressing vector was subjected to WB analysis with anti‐MAP3K7 or anti‐GAPDH antibodies.
-
DThe graph presents densitometry analysis of 3 WB analyses performed as in (C). The mean ± SD was calculated from four independent experiments. Statistics were performed using t‐test (*P < 0.05).
-
EJurkat T cells stably expressing either control vector or a vector containing schistosomal miR‐10 were co‐transfected with a control luciferase plasmid or NF‐κB luciferase reporter vector. In all transfections, a plasmid expressing Renilla vector as an internal control was added. 48 h post‐transfection, cells were harvested and were analyzed by luciferase reporter assay. Values were normalized to the Renilla activity. Data are represented as mean ± SEM from five independent experiments. Statistics were performed using t‐test, **P < 0.01.
-
F–IRepresentative WB experiment demonstrating that MAP3K7 is downregulated in schistosomal‐exposed Th cells (F) or to Jurkat (H) in comparison with the control unexposed cells. (G, I) The graphs present densitometry analysis of 3 WB analyses performed as in (F and H), respectively. The mean ± SD was calculated from three independent experiments. Statistics were performed using paired one‐tailed t‐test (*P < 0.05).
To verify whether miR‐10 can also affect the expression of the endogenous MAP3K7, a Western blot (WB) analysis was performed using anti‐MAP3K7 antibodies on protein extract from Jurkat cells stably expressing either the schistosomal miR‐10 or the control plasmid. A 15% decrease was observed in the expression level of MAP3K7 in miR‐10‐expressing cells (Fig 7C and D). To study whether schistosomal miR‐10 affects the expression of NF‐κB‐regulated genes, we used a reporter plasmid containing two NF‐κB DNA‐binding sites linked to a luciferase reporter 74. The overexpression of miR‐10 decreased the reporter luciferase activity by 35% (Fig 7E), indicating that miR‐10 decreases the activity of NF‐κB. These results confirm that schistosomal miR‐10 can decrease the expression of MAP3K7 and consequently the activity of NF‐κB.
Live schistosomes downregulate the expression of MAP3K7 in Th cells
To assess whether the live schistosomes also affect MAP3K7 expression in primary Th cells, we incubated freshly isolated Th cells in the presence of live schistosomes in trans‐well, as illustrated in Fig 1A. After 72 h, the cells were collected, and the protein extracts were subjected to Western blot analysis using anti‐MAP3K7 antibodies. Indeed, the expression of MAP3K7 was reduced by ~50% in the schistosomal‐exposed Th cells (Fig 7F and G), even more profoundly than in Jurkat cells (Fig 7H and I). Altogether, these results show that the schistosomal miR‐10 downregulates the expression of MAP3K7 and consequently the NF‐κB signaling pathway in differentiating Th2 cells.
Discussion
Recently, we found that schistosomal miRNAs can be detected in EVs isolated from sera of Schistosoma‐infected patients 63. The presence of these miRNAs is diminished after treatment, suggesting that these schistosomal miRNAs were released by live worms. Since miRNAs are reported to be the main cargo of EVs, and their main functional executives 55, 56, we hypothesized that these worm‐secreted‐miRNAs are playing an important role in the communication between the parasite and the host immune system. Our current study demonstrates indeed that S. mansoni‐derived EVs modulate host Th‐cell differentiation in an APC‐independent manner; the presence of the worms decreases the expression of the Th2‐lineage‐specifying transcription factor Gata3, and of the Th2 hallmark cytokines. This decrease is not accompanied by a reduction in the expression levels of other cytokines and transcription factors characterizing the other Th lineages—Th1, Th17, or Treg. The idea of preferential modulation of the Th2 response by the parasite was reinforced by the RNA‐seq results in which many known Th2‐associated factors were decreased in cells exposed to the schistosomes such as Il33, Tnfsf4, Tim‐2, and Anxa1, whereas the expression of genes known as polarizing factors toward the Th1 pathway, like Tim‐3, was increased.
Our data strongly suggest that the effect of S. mansoni on Th cells is mediated, at least partially, by miRNA‐loaded EVs: (i) adult S. mansoni secrete EVs, in accordance with previous studies 30, 31, 32; (ii) labeled isolated EVs are internalized into primary Th cells; (iii) schistosomal miR‐10, miR‐125, and Bantam could be detected inside the EVs, and more importantly, within schistosomal‐exposed primary Th cells, as well as schistosomal‐exposed Jurkat cells; (iv) the schistosomal miRNAs could not be detected in Th cells, if the schistosomal‐growing medium was filtrated through 0.1‐μm membrane, countering the idea that transferring of the miRNAs by schistosomes is mediated by free RNA; and (vi) the schistosomal miRNAs miR‐10 and Bantam could be detected in Th cells isolated from lymph nodes of schistosome‐infected mice. Interestingly, these miRNAs were found only in the gut‐associated lymph nodes, Peyer's patches, and mesenteric lymph nodes, and neither in the inguinal lymph node nor in the spleen. It should be noted that in our mice model, the adult S. mansoni are clearly located at the mesenteric and small intestine venules, sites which are drained by the gut‐associated lymph nodes. The restricted delivery of the miRNAs by the Schistosoma strengthens the hypothesis that it mediated through EVs rather than free RNA, but other mechanisms cannot be excluded.
Since the Th2 pathway, which is a major player of the host against the helminth parasites 9, 15, 75, 76, was downmodulated by the schistosomes, we looked for potential Th2 master regulators as targets of the EV cargo. Schistosome miR‐10, one of the most abundant miRNAs in the schistosomal EVs, was chosen for further study. The survey of our RNA‐seq results indicated that NF‐κB is a strong candidate for modulating Th2 differentiation. Indeed, we found a decrease in the expression levels of many genes that are directly regulated by NF‐κB in schistosome‐exposed Th cells. Moreover, NF‐κB regulates both Il4 transcription and Gata3 transcription 67, 68, 70. The regulation of NF‐κB activity is mostly post‐transcriptional 77; therefore, we did not expect to find differences in its mRNA level. Rather, we hypothesized that schistosomes modulate an NF‐κB upstream regulator. Indeed, we found that MAP3K7 is a genuine biochemical target of miR‐10 and that MAP3K7 expression is reduced in primary Th cells in the presence of the parasites. These results can explain, at least partially, the preferential modulation of the Th2 response by adult schistosomes during the chronic stage, which may facilitate the chronic infection. Our findings might be similar in other helminth infections, pinpointing and manipulating the Th2 differentiation process from a distance using EVs as delivering vehicles 15.
Our results are in accordance with the “hygiene hypothesis” 78, 79, 80, 81, associating the dramatic increase in autoimmune and allergic diseases observed in recent decades in Western countries with the reduced exposure to diverse infectious agents. Indeed, epidemiological evidence supports the fact that schistosomiasis can protect against allergic diseases in an endemic population 82, 83. This phenomenon can be attributed to the inhibition of the allergy‐prone Th2 lineage in the infected host and therefore to constrained allergic reactions. Therefore, adopting the schistosome's tactic and harnessing the schistosome EV functions may be a useful approach to treat allergies and other Th2‐associated diseases. However, further studies are necessary to reveal the full scope of schistosomal‐EV manifestations and their communication with the immune system.
Materials and Methods
Maintenance of the schistosome life cycle
The life cycle of S. mansoni was maintained in ICR mice and Biomphalaria glabrata snails. Six‐week‐old male ICR mice were purchased from Harlan Laboratories (Rehovot, Israel) and infected individually by subcutaneous injection with the parasite's cercariae obtained from infected snails which were raised and kept at 26°C in aeriated aquaria. Mice were routinely infected by injection of about 200 cercariae each. Seven to eight weeks post‐infection, schistosome eggs were extracted and purified from the granuloma‐containing livers, hatched, and snails were infected individually by exposure to 7–8 emerging miracidia each. Details of upkeep of the life cycle relied on description by Gold et al 84 and performed at Tel Aviv University (Tel Aviv University ethical committee number 01‐13‐076).
In vitro cultivation of adult schistosome worms
Adult schistosome worms, isolated from infected mice, were cultured in EV‐free medium containing RPMI, 1% penicillin–streptomycin (P/S), 1% l‐glutamine, and 10% EV‐free fetal bovine serum (FBS). The added FBS was ultracentrifuge twice in 150,000 g overnight to remove bovine EVs and filtered twice 0.2 μm aPES membranes (after each centrifugation, the liquid above the precipitated vesicles was collected).
Purification and differentiation of Th cells
Th‐cell differentiation was carried out essentially as described 85, 86. Briefly, CD4+ T cells were purified with magnetic beads from the spleen and lymph nodes of 4‐ to 5‐week‐old mice, using EasySep™ Mouse CD4+ T‐Cell Isolation Kit (Stemcell Technologies, Cat. No. 19852A). Cells were stimulated with 1 μg/ml of anti‐CD3ε (145.2C11, hybridoma supernatant) and 1 μg/ml of anti‐CD28 (Hamster Anti‐Mouse Clone 37.51, catalog no. 553297; Pharmingen San Diego, CA) antibodies. The cells were stimulated in a dish coated with 0.1 mg/ml of goat anti‐hamster antibodies (Whole Molecule Polyclonal Secondary Antibody, MP Biomedicals, catalog no. ICN56984; MP Biomedicals, Inc, Supplier Diversity Partner 0856984) for 3 days. Mice experiments were under the Bar Ilan University ethical committee number 15‐03‐2015 and number 34‐04.
Jurkat‐T cells were cultured in RPMI medium supplemented with 10% FBS, 1% (P/S) antibiotics, and 1% l‐glutamine (purchased from Biological Industries, Kibbutz Beit HaEmek, Israel).
Splenocytes’ sorting protocol
Total splenocytes were stained for CD4 and CD11c, resuspended in cold PBS, and sorted in Astrius (Beckman Coulter) for CD4+ CD11c− and CD4− CD11c+. CD4+‐sorted cells were seeded in 6‐well coated with anti‐hamster in the presence of anti CD3/CD28− stimulating antibodies with or without the presence of dendritic cells (5:1 ratio CD4:CD11c). The cells were then either exposed to S. mansoni or to unused schistosome medium, in 0.4 μm Trans‐well system.
Trans‐well system
Trans‐wells were purchased from Greiner Bio‐One 6‐Well ThinCert™ Cell Culture Inserts for Multiwell Plates, catalog number 657640, pore size 0.4 μm. In each well, cells were seeded in 3 ml of a medium, and 2 ml of medium in the trans‐well.
Purification of schistosomal EVs
25–30 worms were cultured in 5 ml of tissue culture media (in vitro cultivation of schistosomes), using the pre‐centrifuged fetal bovine serum to remove bovine EVs. Under these conditions, the worms can be maintained for 4–5 weeks. The media were collected every other day and frozen at −80°C. For EV purification, a series of centrifugations were performed, briefly, from 150 to 500 ml of collected medium, and cellular debris removed by centrifugation at 1,400 g for 10 min and then at 14,489.28 g for 1 h. The supernatant was concentrated using a Vivaflow 100,000 MWCO PES (Sartorius Stedim Biotech.). Next, the concentrated pellet supernatant was diluted in RPMI to the volume of the tubes (~39 ml) and centrifuged overnight in Beckman Ti50.2 rotor in Beckman Quick‐Seal, REF 342414 tubes, κ‐factor 108 at 100,251.93 g 87. Using this procedure, we obtain ~1011 EVs/ml as quantified by NanoSight. In parallel, as a control, the same volume of unused schistosomal‐growing medium (before adding to the worms) was treated in exactly the same way.
NanoSight particle analysis
Vesicle size distribution and concentration were performed using nanoparticle tracking analysis (NTA) (Malvern Instruments, NanoSight NS300). The instrument settings are as follows: laser beam at 405 nm, camera level 10, and gain 8. Five measurements were done for each sample, and each scan was for 60 s. Sample size distributions were calibrated in a liquid suspension by the analysis of Brownian motion via light scattering. NanoSight provides single particle size and concentration measurements.
EV labeling and FACS analysis
During the EV‐purification procedure, after the supernatant was concentrated using the Vivaflow, the concentrated supernatant was stained with Thiazole Orange (Sigma Cat. No. 390062) for 20 min. To avoid a background staining, after the labeling, the EVs were dissolved in ~70 ml RPMI and ultracentrifuge overnight in Beckman Ti50.2 rotor in Beckman Quick‐Seal, REF 342414 tubes, κ‐factor 108 at 100,251.93 g, to pellet the labeled EVs. As a control, the unused schistosomal‐growing medium was also labeled. Cells were imaged using an inverted confocal microscope or multispectral IFC (ImageStreamX Mark II imaging flow cytometer; Amnis Corp, Seattle, WA, part of EMD Millipore) as previously described in Sisquella et al 88.
Sucrose gradient preparation
OptiPrep™ density gradient EVs, miRNA, and HSP‐70 EV protein detection
The discontinuous iodixanol gradient was prepared (by OptiPrep™ Cosmo Bio USA, Inc., density gradient technique, Cat. No. AXS‐1114542) as described 89. 500 μl of schistosoma extracellular vesicle isolated from 150 ml medium the worms grow in as described above was overlaid onto the top of the gradient, and centrifuged for 22 h at 4°C, in polypropylene tubes #331374, Beckman Coulter, in SW40Ti rotor, κ‐factor at 58,463.35 g, 280.3. 8 fractions, each contains 1 ml, were collected (from top to bottom). Each of the fractions was diluted with 6 ml PBS and centrifuged overnight at 4°C, in Beckman Ti50.3 rotor in Beckman Quick‐Seal, REF 344619 tubes, κ‐factor 59.2 at 113,679.02 g, and the pellet was resuspended in 100 μl PBS.
In the next step, RNA was extracted from 50 μl of each fraction (as described above) and subjected to TaqMan qRT–PCR to detect miRNAs as described by the manufacturer (Applied Biosystems), using specific primers to either schistosomal miRNA bantam (Applied Biosystems Assay ID 006598_mat) or schistosomal miR‐10‐5p (Applied Biosystems Assay ID 244531_mat).
The rest 50 μl of each fraction was subjected to Western blot analysis using anti‐human HSP‐70 antibodies (Antibody (B‐6): sc‐7298, Santa Cruz Biotechnology, Inc.).
Cryo‐electron microscopy
Cryo‐TEM was performed as described previously 90 with a Tecnai G2 F30 (FEI) transmission electron microscope operating at 300 kV with a defocus between 10 and 16 μm across × 15,000 to × 39,000 magnification.
Proteomic analysis
Proteomics was done at the Smoler Proteomics Center of the Technion‐Israel Institute of Technology, Haifa, Israel.
The data of the proteomics are shown in the supplementary Dataset EV2.
Plasmids
Schistosoma miR‐10 expressing plasmid
The S. mansoni pre‐miR‐10 precursor was cloned into the HindIII + EcoRI cut pcDNA3.1 (+) plasmid. Briefly, both sense and antisense oligos of the pre‐miRNA were synthetically synthesized (Sigma, Israel). Sequences were taken from the miRBase database (Accession Number MI0021817) as follows:
sma‐mir‐10 sense primer:
5′AGCTTCCTCAGTATGAACCCTGTAGACCCGAGTTTGGATGCAGTCAGATGCAAATTCGAGTCTATAAGGAAAAATACTTTGGAAG_3′
sma‐mir‐10 antisense primer:
5′AATTCTTCCAAAGTATTTTTCCTTATAGACTCGAATTTGCATCTGACTGCATCCAAACTCGGGTCTACAGGGTTCATACTGAGGA_3′
The bold underline AGCT (the complementary sequence to HindIII digested) was added to the 5′ end of the sense oligo, and the bold underline AATT (the complementary sequence to EcoRI digested) was added to the antisense oligo. Sense and antisense oligos were annealed and ligated into the pcDNA3.1 vector digested with HindIII and EcoRI.
psiCHECK‐MAP3K7‐3′UTR was previously described by Zehavi et al 73.
psiCHECK‐OX40L‐3′UTR and psiCHECK‐CCL22‐3′UTR and psiCHECK‐GATA3‐3′UTR
The 3′UTR of CCL22or OX4OL or GATA3 was cloned into psiCHECK‐II cut with PmeI (Promega, Madison, WI, USA).
The sequences in capital letters in the primers are homologous to the 15 nucleotides up‐ and downstream of the PmeI cut site on psiCHECK‐II. Primers sequences used for the cloning are shown in Table 2. The PCR fragments were extracted from agarose gel and fused into psiCHECK‐II cut with PmeI using In‐Fusion HD Cloning Kit (Takara Bio USA Inc., Mountain View, CA, USA).
Table 2.
Primers used to amplify the putative targets mRNAs 3′UTR
| Gene | NCBI Reference Sequence | Primers 5′–3′ | PCR length |
|---|---|---|---|
| CCL22 | NM_002990.4 | Forward CCCGGGAATTCGTTTattacaggcgtgagctatca | 1,210 bp |
| Reverse GGCCGCTCTAGGTTTaaaggatttattttccagca | |||
| TNFSF4 (OX40L) | NM_003326.4 | Forward CCCGGGAATTCGTTTcctactaggcacctttgtga | 1,200 bp |
| Reverse GGCCGCTCTAGGTTTtagtgtaagggcgaacagcc | |||
| GATA3 | NM_002051.2 | Forward CCCGGGAATTCGTTTgccctgctcgatgctcacag | 1,069 bp |
| Reverse GGCCGCTCTAGGTTTcagccgggccgattgcagga |
In the psiCHECK‐mut‐MAP3K7, the seed miR‐10 binding site, at position 603—610, was changed from ACAGGGTA to TGTTTTAT. Mutation plasmid was generated by using the Q5 Site‐Directed Mutagenesis Kit (NEB, Cat. No. E0554S).
NF‐κB‐Luciferase assay
pGL4 containing two NF‐κB DNA‐binding sites linked to a luciferase cDNA sequence gene was kindly donated by the laboratory of Yinon Ben‐Neriah 74. As a control, we used pGL4‐CMV‐luciferase plasmid. pRL‐CMV Renilla used as an internal control for transfection efficiency (Promega Corporation vector number E2261).
Luciferase assay
HEK‐293 cells were seeded in 24‐well plates (1 × 105/well) 1 day before transfection. Cells were transfected with the indicated vectors for each experiment using Lipofectamine™ 2000 Transfection Reagent (Invitrogen Life Technologies; Thermo Fisher Scientific, catalog number 11668) according to the manufacturer's instructions. 24 h after transfection, luciferase activity was determined using the dual‐luciferase assay system (Promega Corporation, catalog number E1980) according to the manufacturer's instructions.
RNA purification and RT–PCR
RNA was isolated from 106 Th cells which were either exposed to S. mansoni or introduced to miR‐10 according to manufacturer's instructions (Norgene Biotek Corp., Canada, catalog number 17200; Total RNA Purification Kit). RNA was reverse‐transcribed by PrimeScript™ RT Master Mix (Clontech–Takara™, catalog number 2680). The transcripts were then analyzed for the expression of various genes with the appropriate primers shown in Table 3. The cytokines were Ifng (Th1), Il4, Il5, and Il13 (Th2), and Il17 (Th17) and the lineage‐specifying transcription factors were Gata3 (Th2), Rorγt (Th17), and Foxp3 (Treg) 91. PCR was performed using Fast SYBR™ Green Master Mix (Applied Biosystems, Cat. No. 4385610).
Table 3.
Primers used for qRT–PCR of mouse mRNAs
| Oligo name | Sequence 5′–3′ |
|---|---|
| Il4 forward | CCAAGGTGCTTCGCATATTT |
| Il4 reverse | ATCGAAAAGCCCGAAAGAGT |
| Il13 forward | ACCCAGAGGATATTGCATGGC |
| Il13 reverse | CGTGGCGAAACAGTTGCTTT |
| Il5 forward | CACCAGCTATGCATTGGAGA |
| Il5 reverse | TCCTCGCCACACTTCTCTTT |
| Ifng forward | GCGTCATTGAATCACACCTG |
| Ifng reverse | TGAGCTCATTGAATGCTTGG |
| Il17 forward | CTCCAGAAGGCCCTCAGACTA |
| Il17 reverse | GGGTCTTCATTGCGGTGG |
| Gata3 forward | GAGCGTCAGCAACAGTGAAG |
| Gata3 reverse | CCACACTGCACACTGATTCC |
| RORγt forward | AGCTTTGTGCAGATCTAAGG |
| RORγt reveres | TGTCCTCCTCAGTAGGGTAG |
| Foxp3 forward | CCCATCCCCAGGAGTCTT |
| Foxp3 reverse | ACCATGACTAGGGGCACT |
| Beta‐2 microglobulin forward | TTCTGGTGCTTGTCTCACTGA |
| Beta‐2 microglobulin reverse | TTCTGGTGCTTGTCTCACTGA |
miRNAs qRT‐PCR
Quantification of miRNAs was carried out from total RNA, as previously described by Zehavi et al 73 by TaqMan miRNA assays (Applied Biosystems, catalog number 4440886). Target miRNA expression was normalized between different samples based on the values of mouse U6 sma‐bantam‐5p Assay ID 471847_mat, sma‐miR‐10‐5p Assay ID 244531_mat, sma‐miR‐125b Assay ID 006070_mat, and mouse U6 snRNA Assay ID 001973.
RNA sequencing
Total RNAs were extracted from Th cells exposed to S. mansoni. mRNA was purified using NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biotechnologies, USA, catalog number E7490) and cDNA library constructed with NEBNext® Ultra™ RNA Library Prep Kit for Illumina®. Each sample was indexed, multiplexed, and sequenced in the genomic center of the medical faculty of Bar‐Ilan University at Safed, Israel. The data were then aligned using bowtie and further analyzed using seqmonk (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) for differential expression.
Bioinformatics
Mapping of the reads was done by Bowtie, and differential expression was conducted using R DEseq in Seqmonk platform.
Statistics
All statistics tests were done in GraphPad prism program. Only for analyzing the RNA‐seq, seqmonk for differential expression was used.
The microscope used in the study
Zeiss LSM780 inverted confocal microscope, with multi‐photon Chameleon Laser; in vivo capabilities; Lamda‐PMT—identifying all the wavelength spectrum at once (8.9 nm step).
Western blot
Western blot (WB) was performed as previously described by Zehavi et al 73, with anti‐TAK1 (MAP3K7) antibody (Abcam [EPR5984], Cat. No. ab109526), anti‐GAPDH (Cell Signaling, Cat. No. 2118), anti‐HSP70 (B‐6) (Santa Cruz Biotechnology Inc., Cat. No. sc‐7298).
Author contributions
TM, YB, YO‐B, DG, BB; ED and DA carried out the experiments. TM, YB, YO‐B, YS, ES, NR‐R, OA and DA conceived and planned the experiments. TM, YB, YO‐B, YS, ES, NR‐R, OA and DA analyzed and interpreted the results. ES, NR‐R, OA and DA supervised the project. DA and OA lead the writing of the manuscript. All authors discussed the results and contributed to the final manuscript provided critical feedback and helped shape the research, analysis, and manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Dataset EV1
Dataset EV2
Review Process File
Acknowledgements
E.S. and D.A. were supported by the Office of the Chief Scientist, Israeli Ministry of Industry (Kamin program no. 52698); and the Israel Science Foundation (I‐CORE program 41/11). The Weizmann Institute of Science—Sheba Medical Center Collaboration to E.S., D.A. and N.R.‐R. N.R.‐R. is supported by a research grant from the Benoziyo Endowment Fund for the Advancement of Science, the Jeanne and Joseph Nissim Foundation for Life Sciences Research and the Samuel M. Soref and Helene K. Soref Foundation. N.R.‐R. is the incumbent of the Enid Barden and Aaron J. Jade President's Development Chair for New Scientists in Memory of Cantor John Y. Jade. N.R.‐R. is grateful for the support from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no. 757743), and the Israel Science Foundation (ISF) (619/16 and 119034).
EMBO Reports (2020) 21: e47882
Contributor Information
Eli Schwartz, Email: Eli.schwartz@sheba.health.gov.il.
Neta Regev‐Rudzki, Email: neta.regev-rudzki@weizmann.ac.il.
Orly Avni, Email: orly.avni@biu.ac.il.
Dror Avni, Email: droravni@msn.com.
Data availability
The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium http://www.proteomexchange.org/ via the PRIDE partner repository with the dataset identifier PXD012525.
The RNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (http://www.ebi.ac.uk/arrayexpress) under accession number: E‐MTAB‐7658.
References
- 1. Grobusch MP, Muhlberger N, Jelinek T, Bisoffi Z, Corachan M, Harms G, Matteelli A, Fry G, Hatz C, Gjorup I et al (2003) Imported schistosomiasis in Europe: sentinel surveillance data from TropNetEurop. J Travel Med 10: 164–169 [DOI] [PubMed] [Google Scholar]
- 2. Meltzer E, Artom G, Marva E, Assous MV, Rahav G, Schwartzt E (2006) Schistosomiasis among travelers: new aspects of an old disease. Emerg Infect Dis 12: 1696–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hornstein L, Lederer G, Schechter J, Greenberg Z, Boem R, Bilguray B, Giladi L, Hamburger J (1990) Persistent Schistosoma mansoni infection in Yemeni immigrants to Israel. Isr J Med Sci 26: 386–389 [PubMed] [Google Scholar]
- 4. Warren KS, Mahmoud AA, Cummings P, Murphy DJ, Houser HB (1974) Schistosomiasis mansoni in Yemeni in California: duration of infection, presence of disease, therapeutic management. Am J Trop Med Hyg 23: 902–909 [DOI] [PubMed] [Google Scholar]
- 5. Dunne DW, Cooke A (2005) A worm's eye view of the immune system: consequences for evolution of human autoimmune disease. Nat Rev Immunol 5: 420–426 [DOI] [PubMed] [Google Scholar]
- 6. Fallon PG, Smith P, Dunne DW (1998) Type 1 and type 2 cytokine‐producing mouse CD4+ and CD8+ T cells in acute Schistosoma mansoni infection. Eur J Immunol 28: 1408–1416 [DOI] [PubMed] [Google Scholar]
- 7. Pearce EJ, C MK, Sun J, Taylor J, McKee AS, Cervi L (2004) Th2 response polarization during infection with the helminth parasite Schistosoma mansoni . Immunol Rev 201: 117–126 [DOI] [PubMed] [Google Scholar]
- 8. Fairfax K, Nascimento M, Huang SC, Everts B, Pearce EJ (2012) Th2 responses in schistosomiasis. Semin Immunopathol 34: 863–871 [DOI] [PubMed] [Google Scholar]
- 9. Maizels RM, Hewitson JP, Smith KA (2012) Susceptibility and immunity to helminth parasites. Curr Opin Immunol 24: 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu J, Paul WE (2008) CD4 T cells: fates, functions, and faults. Blood 112: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rao A, Avni O (2000) Molecular aspects of T‐cell differentiation. Br Med Bull 56: 969–984 [DOI] [PubMed] [Google Scholar]
- 12. Avni O, Rao A (2000) T cell differentiation: a mechanistic view. Curr Opin Immunol 12: 654–659 [DOI] [PubMed] [Google Scholar]
- 13. Zhou L, Chong MM, Littman DR (2009) Plasticity of CD4+ T cell lineage differentiation. Immunity 30: 646–655 [DOI] [PubMed] [Google Scholar]
- 14. Yao Y, Simard AR, Shi FD, Hao J (2013) IL‐10‐producing lymphocytes in inflammatory disease. Int Rev Immunol 32: 324–336 [DOI] [PubMed] [Google Scholar]
- 15. Allen JE, Maizels RM (2011) Diversity and dialogue in immunity to helminths. Nat Rev Immunol 11: 375–388 [DOI] [PubMed] [Google Scholar]
- 16. Danilowicz‐Luebert E, O'Regan NL, Steinfelder S, Hartmann S (2011) Modulation of specific and allergy‐related immune responses by helminths. J Biomed Biotechnol 2011: 821578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Motran CC, Silvane L, Chiapello LS, Theumer MG, Ambrosio LF, Volpini X, Celias DP, Cervi L (2018) Helminth infections: recognition and modulation of the immune response by innate immune cells. Front Immunol 9: 664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brunet LR, Finkelman FD, Cheever AW, Kopf MA, Pearce EJ (1997) IL‐4 protects against TNF‐alpha‐mediated cachexia and death during acute schistosomiasis. J Immunol 159: 777–785 [PubMed] [Google Scholar]
- 19. de Jesus AR, Silva A, Santana LB, Magalhaes A, de Jesus AA, de Almeida RP, Rego MA, Burattini MN, Pearce EJ, Carvalho EM (2002) Clinical and immunologic evaluation of 31 patients with acute Schistosomiasis mansoni . J Infect Dis 185: 98–105 [DOI] [PubMed] [Google Scholar]
- 20. Pearce EJ, MacDonald AS (2002) The immunobiology of schistosomiasis. Nat Rev Immunol 2: 499–511 [DOI] [PubMed] [Google Scholar]
- 21. Nutman TB (2015) Looking beyond the induction of Th2 responses to explain immunomodulation by helminths. Parasite Immunol 37: 304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van der Kleij D, Latz E, Brouwers JF, Kruize YC, Schmitz M, Kurt‐Jones EA, Espevik T, de Jong EC, Kapsenberg ML, Golenbock DT et al (2002) A novel host‐parasite lipid cross‐talk. Schistosomal lyso‐phosphatidylserine activates toll‐like receptor 2 and affects immune polarization. J Biol Chem 277: 48122–48129 [DOI] [PubMed] [Google Scholar]
- 23. Wang X, Zhou S, Chi Y, Wen X, Hoellwarth J, He L, Liu F, Wu C, Dhesi S, Zhao J et al (2009) CD4+ CD25+ Treg induction by an HSP60‐derived peptide SJMHE1 from Schistosoma japonicum is TLR2 dependent. Eur J Immunol 39: 3052–3065 [DOI] [PubMed] [Google Scholar]
- 24. Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thery C, Ostrowski M, Segura E (2009) Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9: 581–593 [DOI] [PubMed] [Google Scholar]
- 26. Colombo M, Raposo G, Thery C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30: 255–289 [DOI] [PubMed] [Google Scholar]
- 27. Regev‐Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill AF et al (2013) Cell‐cell communication between malaria‐infected red blood cells via exosome‐like vesicles. Cell 153: 1120–1133 [DOI] [PubMed] [Google Scholar]
- 28. Geiger A, Hirtz C, Becue T, Bellard E, Centeno D, Gargani D, Rossignol M, Cuny G, Peltier JB (2010) Exocytosis and protein secretion in Trypanosoma . BMC Microbiol 10: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marcilla A, Martin‐Jaular L, Trelis M, de Menezes‐Neto A, Osuna A, Bernal D, Fernandez‐Becerra C, Almeida IC, Del Portillo HA (2014) Extracellular vesicles in parasitic diseases. J Extracell Vesicles 3: 25040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sotillo J, Pearson M, Potriquet J, Becker L, Pickering D, Mulvenna J, Loukas A (2016) Extracellular vesicles secreted by Schistosoma mansoni contain protein vaccine candidates. Int J Parasitol 46: 1–5 [DOI] [PubMed] [Google Scholar]
- 31. Zhu L, Liu J, Dao J, Lu K, Li H, Gu H, Liu J, Feng X, Cheng G (2016) Molecular characterization of Schistosoma japonicum exosome‐like vesicles reveals their regulatory roles in parasite‐host interactions. Sci Rep 6: 25885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nowacki FC, Swain MT, Klychnikov OI, Niazi U, Ivens A, Quintana JF, Hensbergen PJ, Hokke CH, Buck AH, Hoffmann KF (2015) Protein and small non‐coding RNA‐enriched extracellular vesicles are released by the pathogenic blood fluke Schistosoma mansoni . J Extracell Vesicles 4: 28665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. da Huang W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- 35. Yu Y, Deng W, Lei J (2015) Interleukin‐33 promotes Th2 immune responses in infected mice with Schistosoma japonicum . Parasitol Res 114: 2911–2918 [DOI] [PubMed] [Google Scholar]
- 36. Murakami‐Satsutani N, Ito T, Nakanishi T, Inagaki N, Tanaka A, Vien PT, Kibata K, Inaba M, Nomura S (2014) IL‐33 promotes the induction and maintenance of Th2 immune responses by enhancing the function of OX40 ligand. Allergol Int 63: 443–455 [DOI] [PubMed] [Google Scholar]
- 37. Akiba H, Miyahira Y, Atsuta M, Takeda K, Nohara C, Futagawa T, Matsuda H, Aoki T, Yagita H, Okumura K (2000) Critical contribution of OX40 ligand to T helper cell type 2 differentiation in experimental leishmaniasis. J Exp Med 191: 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Imai T, Nagira M, Takagi S, Kakizaki M, Nishimura M, Wang J, Gray PW, Matsushima K, Yoshie O (1999) Selective recruitment of CCR4‐bearing Th2 cells toward antigen‐presenting cells by the CC chemokines thymus and activation‐regulated chemokine and macrophage‐derived chemokine. Int Immunol 11: 81–88 [DOI] [PubMed] [Google Scholar]
- 39. Chakravarti S, Sabatos CA, Xiao S, Illes Z, Cha EK, Sobel RA, Zheng XX, Strom TB, Kuchroo VK (2005) Tim‐2 regulates T helper type 2 responses and autoimmunity. J Exp Med 202: 437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, Manning S, Greenfield EA, Coyle AJ, Sobel RA et al (2002) Th1‐specific cell surface protein Tim‐3 regulates macrophage activation and severity of an autoimmune disease. Nature 415: 536–541 [DOI] [PubMed] [Google Scholar]
- 41. Riella LV, Sayegh MH (2013) T‐cell co‐stimulatory blockade in transplantation: two steps forward one step back!. Expert Opin Biol Ther 13: 1557–1568 [DOI] [PubMed] [Google Scholar]
- 42. Okano M, Azuma M, Yoshino T, Hattori H, Nakada M, Satoskar AR, Harn DA Jr, Nakayama E, Akagi T, Nishizaki K (2001) Differential role of CD80 and CD86 molecules in the induction and the effector phases of allergic rhinitis in mice. Am J Respir Crit Care Med 164: 1501–1507 [DOI] [PubMed] [Google Scholar]
- 43. Huang P, Zhou Y, Liu Z, Zhang P (2016) Interaction between ANXA1 and GATA‐3 in immunosuppression of CD4(+) T Cells. Mediators Inflamm 2016: 1701059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barres C, Blanc L, Bette‐Bobillo P, Andre S, Mamoun R, Gabius HJ, Vidal M (2010) Galectin‐5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 115: 696–705 [DOI] [PubMed] [Google Scholar]
- 45. Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T (2006) Dynasore, a cell‐permeable inhibitor of dynamin. Dev Cell 10: 839–850 [DOI] [PubMed] [Google Scholar]
- 46. Momen‐Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, Soria CE, Oquin S, Bonebreak CM, Saracoglu E et al (2013) Current methods for the isolation of extracellular vesicles. Biol Chem 394: 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R (2010) Optical and non‐optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost 8: 2596–2607 [DOI] [PubMed] [Google Scholar]
- 48. Protasio AV, Tsai IJ, Babbage A, Nichol S, Hunt M, Aslett MA, De Silva N, Velarde GS, Anderson TJ, Clark RC et al (2012) A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni . PLoS Negl Trop Dis 6: e1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A et al (2015) FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15: 2597–2601 [DOI] [PubMed] [Google Scholar]
- 51. Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, Hendrix A, Mathivanan S (2019) Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res 47: D516–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin‐Smith GK et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7: 1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- 54. Li SC, Chan WC, Hu LY, Lai CH, Hsu CN, Lin WC (2010) Identification of homologous microRNAs in 56 animal genomes. Genomics 96: 1–9 [DOI] [PubMed] [Google Scholar]
- 55. Rayner KJ, Hennessy EJ (2013) Extracellular communication via microRNA: lipid particles have a new message. J Lipid Res 54: 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu L, Yang BF, Ai J (2013) MicroRNA transport: a new way in cell communication. J Cell Physiol 228: 1713–1719 [DOI] [PubMed] [Google Scholar]
- 57. Copeland CS, Marz M, Rose D, Hertel J, Brindley PJ, Santana CB, Kehr S, Attolini CS, Stadler PF (2009) Homology‐based annotation of non‐coding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum . BMC Genom 10: 464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang J, Hao P, Chen H, Hu W, Yan Q, Liu F, Han ZG (2009) Genome‐wide identification of Schistosoma japonicum microRNAs using a deep‐sequencing approach. PLoS One 4: e8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Q, Tuo W, Gao H, Zhu XQ (2010) MicroRNAs of parasites: current status and future perspectives. Parasitol Res 107: 501–507 [DOI] [PubMed] [Google Scholar]
- 60. Wang Z, Xue X, Sun J, Luo R, Xu X, Jiang Y, Zhang Q, Pan W (2010) An “in‐depth” description of the small non‐coding RNA population of Schistosoma japonicum Schistosomulum . PLoS Negl Trop Dis 4: e596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Souza Gomes M, Muniyappa MK, Carvalho SG, Guerra‐Sa R, Spillane C (2011) Genome‐wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni . Genomics 98: 96–111 [DOI] [PubMed] [Google Scholar]
- 62. Simoes MC, Lee J, Djikeng A, Cerqueira GC, Zerlotini A, da Silva‐Pereira RA, Dalby AR, LoVerde P, El‐Sayed NM, Oliveira G (2011) Identification of Schistosoma mansoni microRNAs. BMC Genom 12: 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Meningher T, Lerman G, Regev‐Rudzki N, Gold D, Ben‐Dov IZ, Sidi Y, Avni D, Schwartz E (2017) Schistosomal MicroRNAs isolated from extracellular vesicles in sera of infected patients: a new tool for diagnosis and follow‐up of human schistosomiasis. J Infect Dis 215: 378–386 [DOI] [PubMed] [Google Scholar]
- 64. Samoil V, Dagenais M, Ganapathy V, Aldridge J, Glebov A, Jardim A, Ribeiro P (2018) Vesicle‐based secretion in schistosomes: analysis of protein and microRNA (miRNA) content of exosome‐like vesicles derived from Schistosoma mansoni . Sci Rep 8: 3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cheng G, Luo R, Hu C, Cao J, Jin Y (2013) Deep sequencing‐based identification of pathogen‐specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum . Parasitology 140: 1751–1761 [DOI] [PubMed] [Google Scholar]
- 66. Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB (2007) Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13: 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Artis D, Kane CM, Fiore J, Zaph C, Shapira S, Joyce K, Macdonald A, Hunter C, Scott P, Pearce EJ (2005) Dendritic cell‐intrinsic expression of NF‐kappa B1 is required to promote optimal Th2 cell differentiation. J Immunol 174: 7154–7159 [DOI] [PubMed] [Google Scholar]
- 68. Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A (2001) A critical role for NF‐kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol 2: 45–50 [DOI] [PubMed] [Google Scholar]
- 69. Scheinman EJ, Avni O (2009) Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin‐4 receptor and T cell receptor. J Biol Chem 284: 3037–3048 [DOI] [PubMed] [Google Scholar]
- 70. Li‐Weber M, Giaisi M, Baumann S, Palfi K, Krammer PH (2004) NF‐kappa B synergizes with NF‐AT and NF‐IL6 in activation of the IL‐4 gene in T cells. Eur J Immunol 34: 1111–1118 [DOI] [PubMed] [Google Scholar]
- 71. Sakurai H, Suzuki S, Kawasaki N, Nakano H, Okazaki T, Chino A, Doi T, Saiki I (2003) Tumor necrosis factor‐alpha‐induced IKK phosphorylation of NF‐kappaB p65 on serine 536 is mediated through the TRAF2, TRAF5, and TAK1 signaling pathway. J Biol Chem 278: 36916–36923 [DOI] [PubMed] [Google Scholar]
- 72. Roh YS, Song J, Seki E (2014) TAK1 regulates hepatic cell survival and carcinogenesis. J Gastroenterol 49: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zehavi L, Schayek H, Jacob‐Hirsch J, Sidi Y, Leibowitz‐Amit R, Avni D (2015) MiR‐377 targets E2F3 and alters the NF‐kB signaling pathway through MAP3K7 in malignant melanoma. Mol Cancer 14: 68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nabel G, Baltimore D (1987) An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326: 711–713 [DOI] [PubMed] [Google Scholar]
- 75. Maizels RM, Pearce EJ, Artis D, Yazdanbakhsh M, Wynn TA (2009) Regulation of pathogenesis and immunity in helminth infections. J Exp Med 206: 2059–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Neill DR, McKenzie AN (2011) Nuocytes and beyond: new insights into helminth expulsion. Trends Parasitol 27: 214–221 [DOI] [PubMed] [Google Scholar]
- 77. Karin M, Ben‐Neriah Y (2000) Phosphorylation meets ubiquitination: the control of NF‐[kappa]B activity. Annu Rev Immunol 18: 621–663 [DOI] [PubMed] [Google Scholar]
- 78. Stiemsma LT, Reynolds LA, Turvey SE, Finlay BB (2015) The hygiene hypothesis: current perspectives and future therapies. Immunotargets Ther 4: 143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Alexandre‐Silva GM, Brito‐Souza PA, Oliveira ACS, Cerni FA, Zottich U, Pucca MB (2018) The hygiene hypothesis at a glance: early exposures, immune mechanism and novel therapies. Acta Trop 188: 16–26 [DOI] [PubMed] [Google Scholar]
- 80. Bach JF (2018) The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat Rev Immunol 18: 105–120 [DOI] [PubMed] [Google Scholar]
- 81. Versini M, Jeandel PY, Bashi T, Bizzaro G, Blank M, Shoenfeld Y (2015) Unraveling the hygiene hypothesis of helminthes and autoimmunity: origins, pathophysiology, and clinical applications. BMC Med 13: 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Medeiros M Jr, Figueiredo JP, Almeida MC, Matos MA, Araujo MI, Cruz AA, Atta AM, Rego MA, de Jesus AR, Taketomi EA et al (2003) Schistosoma mansoni infection is associated with a reduced course of asthma. J Allergy Clin Immunol 111: 947–951 [DOI] [PubMed] [Google Scholar]
- 83. Oliveira SM, Bezerra FS, Carneiro TR, Pinheiro MC, Queiroz JA (2014) Association between allergic responses and Schistosoma mansoni infection in residents in a low‐endemic setting in Brazil. Rev Soc Bras Med Trop 47: 770–774 [DOI] [PubMed] [Google Scholar]
- 84. Gold D, Alian M, Domb A, Karawani Y, Jbarien M, Chollet J, Haynes RK, Wong HN, Buchholz V, Greiner A et al (2017) Elimination of Schistosoma mansoni in infected mice by slow release of artemisone. Int J Parasitol Drugs Drug Resist 7: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A (2002) T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3: 643–651 [DOI] [PubMed] [Google Scholar]
- 86. Jacob E, Hod‐Dvorai R, Ben‐Mordechai OL, Boyko Y, Avni O (2011) Dual function of polycomb group proteins in differentiated murine T helper (CD4+) cells. J Mol Signal 6: 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thery C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol/editorial board, Juan S Bonifacino [et al] Chapter 3: Unit 3 22 [DOI] [PubMed] [Google Scholar]
- 88. Sisquella X, Ofir‐Birin Y, Pimentel MA, Cheng L, Abou Karam P, Sampaio NG, Penington JS, Connolly D, Giladi T, Scicluna BJ et al (2017) Malaria parasite DNA‐harbouring vesicles activate cytosolic immune sensors. Nat Commun 8: 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ (2012) Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863‐derived exosomes. Methods 56: 293–304 [DOI] [PubMed] [Google Scholar]
- 90. Coleman BM, Hanssen E, Lawson VA, Hill AF (2012) Prion‐infected cells regulate the release of exosomes with distinct ultrastructural features. FASEB J 26: 4160–4173 [DOI] [PubMed] [Google Scholar]
- 91. Raphael I, Nalawade S, Eagar TN, Forsthuber TG (2015) T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 74: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Dataset EV1
Dataset EV2
Review Process File
Data Availability Statement
The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium http://www.proteomexchange.org/ via the PRIDE partner repository with the dataset identifier PXD012525.
The RNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (http://www.ebi.ac.uk/arrayexpress) under accession number: E‐MTAB‐7658.


