Abstract
Developments in machine learning in recent years have precipitated a surge in research on the applications of artificial intelligence within medicine. Machine learning algorithms are beginning to impact medicine broadly, and the field of spine surgery is no exception. Electronic medical records are a key source of medical data that can be leveraged for the creation of clinically valuable machine learning algorithms. This review examines the current state of machine learning using electronic medical records as it applies to spine surgery. Studies across the electronic medical record data domains of imaging, text, and structured data are reviewed. Discussed applications include clinical prognostication, preoperative planning, diagnostics, and dynamic clinical assistance, among others. The limitations and future challenges for machine learning research using electronic medical records are also discussed.
Keywords: Machine learning, Deep learning, Artificial intelligence, Electronic medical records, Spine surgery
INTRODUCTION
Electronic medical records (EMRs) have experienced widespread adoption globally since their initial development in the 1970s [1]. Recent surveys indicate that 80.5% of United States hospitals and 58.1% of South Korean hospitals report at least basic EMR usage, and EMR adoption is expected to rise in the coming years [2-5]. EMR systems contain a wide variety of data, including demographics, vitals, labs, imaging studies, medications, diagnoses, and more. The breadth of EMR data is also expected to increase with the integration of new types of information such as patient-reported outcome scores [6]. Data may also come from new sources such as wearable devices and patient-driven mobile applications [7,8]. The rise in EMR adoption and usage has generated a large and expanding collection of data.
The growing wealth of data housed within EMR systems has dovetailed well with concurrent advancements in computer processing power and artificial intelligence (AI) techniques [9]. Powerful graphics processing units (GPUs) that had been developed for video game applications were first adapted to nongraphical machine learning (ML) tasks in 2004 [10-12]. Leveraging the large parallel processing capacity of GPUs allowed for a significant reduction in the time required to train AI algorithms. Furthermore, the recent release of open source computer programming libraries like Google’s TensorFlow and Facebook’s PyTorch has democratized the creation of complicated ML algorithms by simplifying the process and expanding accessibility to those with less domain expertise [13,14]. Broadly accessible ML tools now exist to help create algorithms for clinical practice from EMR data. Reflecting the increase in accessibility, the number of articles on the topic of AI and related subtopics in medical literature has undergone exponential growth in recent years (Fig. 1). One bibliometric study found over 60% of articles on AI and its related subtopics were published between 2014 and 2018 [15].
Fig. 1.
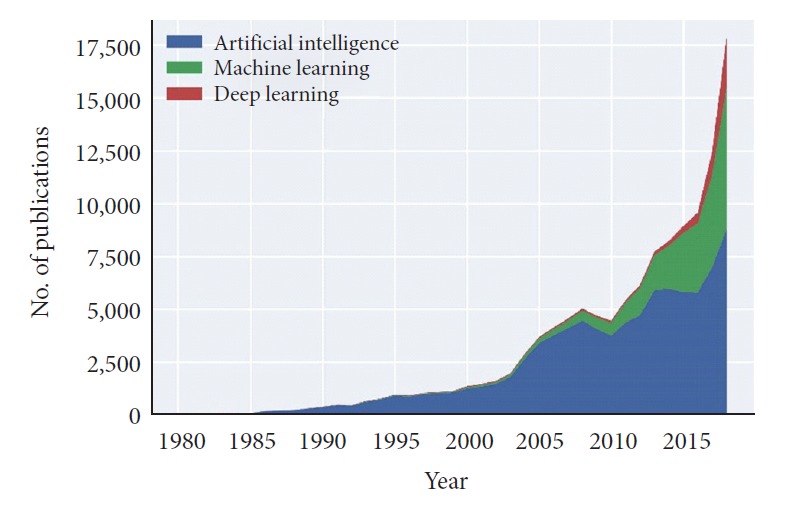
Stacked area chart depicting the number of publications by publication year returned in PubMed searches using the search terms “artificial intelligence,” “machine learning,” or “deep learning.” Results were filtered to include publication dates between 1980 and 2018.
Notable ML studies have used EMR data to create algorithms which optimize neuroradiology workflows, monitor patients for the earliest signs of acute kidney injury, and detect lung cancer with superior accuracy to physicians [16-18]. AI is affecting medicine broadly, and spine surgery is no exception. This narrative review aims to examine the current body of literature to consider the applications of ML using EMRs in the context of spine surgery. ML applications across the EMR data domains of imaging data, text data, and structured data (demographics, vitals, labs, etc.) are discussed. Within these domains, current ML research applied to spine surgery is examined along with adjacent research that may be applicable to spine surgery in the future. These applications span the clinical topics of preoperative risk stratification, preoperative planning, postoperative prognostication, and optimization of the clinical workflow. Finally, the limitations and future challenges for EMR-driven ML are discussed.
ARTIFICIAL INTELLIGENCE, MACHINE LEARNING, AND DEEP LEARNING
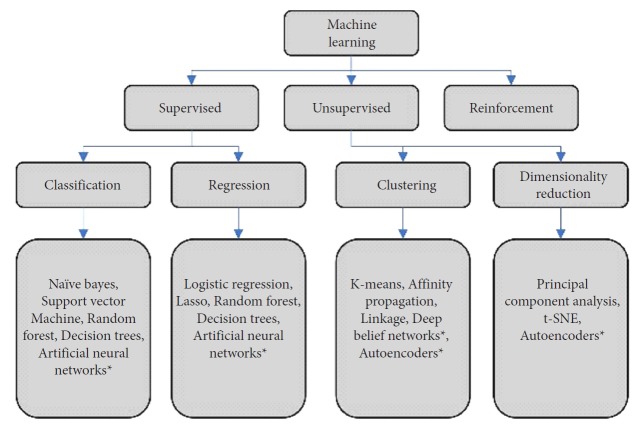
AI refers to a variety of methods which all aim to direct computers to simulate intelligent behavior. ML is a subtype of AI that describes the ability of an algorithm to learn patterns contained in large datasets [19]. Within ML, there are several distinctions (Fig. 2). Within each of these categories, there are wide varieties of algorithm architectures and methods that can be employed for ML problems. Of note, the high impact ML subfield of deep learning (DL) has risen in prominence in recent years (Fig. 1). DL uses neural networks with many layers which allow for complicated, nonlinear processing of input data [20]. This allows DL algorithms to find highly abstracted data representations. In turn, these abstract representations enable strong performance on complex tasks such as imaging classification. Taken together, advances in the subfields of ML have yielded a large variety of algorithm architectures with various strengths and weaknesses. With this large toolkit of ML architectures, much of modern ML research in medicine is aimed at best applying these architectures to clinical tasks.
Fig. 2.
A breakdown of common types of machine learning algorithms used in medical applications. t-SNE, t-Stochastic Neighbor Embedding. *Deep learning algorithms.
MATERIALS AND METHODS
The terms “((electronic medical records OR EMR OR electronic health records OR EHR)) AND (artificial intelligence OR machine learning OR deep learning)”, “(artificial intelligence OR machine learning OR deep learning) AND spine”, and “((electronic medical records OR EMR OR electronic health records OR EHR)) AND spine” were used to search PubMed for relevant articles on the topics of ML, EMR, and spine surgery. The citations from these articles were used to find further relevant research. In addition, other articles known to the authors were pulled for review. The reviewed articles were used to construct this narrative review on applications of ML using EMR in spine surgery.
COMPUTER VISION AND IMAGING DATA
According to estimates by IBM, 90% of all medical data is imaging data [21]. This is especially true in spine surgery as advanced imaging techniques are critical in diagnosis, intraoperative guidance, and postoperative surveillance. Unfortunately, the increasing use of computed tomography (CT) and magnetic resonance imaging (MRI) scans increases the demand for image interpretation [22]. This allows a unique opportunity for the wide variety of powerful ML tools available today to provide vast improvements in the accuracy and speed of automated imaging analysis. This section will discuss some of the most pertinent ML advances in spine imaging and their corresponding impacts in surgical or clinical practice.
1. Automated Visualization and Segmentation
One of the most prominent applications of ML to imaging data in spine surgery is in vertebrae visualization and segmentation. For humans, visualizing and identifying spine landmarks and measuring relevant physical dimensions requires time and effort, which can take up to 15 minutes per patient [23]. In addition, imaging and interpreter variability can lead to skewed interpretations of imaging in EMRs. In turn, this can lead to suboptimal outcomes, particularly in procedures that aim to address spinal deformity, where specific measurements must remain accurate and consistent (e.g., distances between 2 spinous processes of adjacent vertebrae) [24].
Image processing is a difficult task even by the standards of modern computational techniques. To deliver quick and accurate image interpretation, ML in imaging must overcome several challenges. Generally, computational image analysis requires 3 major stages: image classification, object detection, and segmentation. In spine imaging, this might involve first determining the presence of vertebrae (image classification), separating bone from other tissues (detection), and clearly delineating these boundaries (segmentation). The large amount of pixel or voxel information that needs to be evaluated means that exceptionally powerful machines are needed. In addition, algorithms must be flexible enough to address variability in spinal alignment and fusion status. Even carefully constructed algorithms relying on 2-dimensional imaging struggle to properly diagnose spinal imbalances that are important for preoperative diagnostics [25].
Recent advances in computational speed and available medical data have given rise to stronger DL algorithms. In addition, more complex models require large batches of properly annotated data to train, or modify computational parameters to optimize their ability to recognize salient imaging patterns. As CT and MRI scans became available in bulk, they became strong datasets for ML training, yielding stronger ML models [26].
With regards to spinal visualization and segmentation, many groups have applied sophisticated automated analysis of CT and MRI to both normal and pathological spines. One University of Cincinnati group used a “guess-and-revise” ML algorithm on sagittal MRI scans of whole spines. After selecting for the best available slices, the algorithm guesses the location of the center of each vertebra and calculates the location of each vertebral body and intervertebral disk. The initial guesses are revised and optimized based on the locations of other discs until the most probable spine labeling is attained [23]. In this study, automated segmentation and identification were successful, though accuracy was lower on pathological spines. Recently, more advanced DL techniques, such as convolutional neural networks (CNNs), have proven effective in imaging tasks. Such neural nets consist of several computational layers that apply mathematical convolutions in sequence. These networks recognize motifs or patterns in MRI and CT scans that allow the model to see an object and segment it from the rest of the image. One study by Forsberg et al. [27] used these CNNs to attain high accuracy and efficiency in labeling cervical and lumbar vertebrae in MRI images. The authors used a large but minimally-labeled set of MRI data that had only vertebrae positional labels for testing rather than the full vertebral outlines required by previous non-DL models. With one CNN for cervical vertebrae and one for lumbar vertebrae, the authors of the study were able to successfully segment vertebral bodies from IVDs and surrounding tissue with a high degree of precision and label both cervical and lumbar vertebrae with >95% accuracy.
Even these powerful DL tools, however, struggle with boundary problems and incomplete or poor-quality imaging. In the case of the previous study, the model’s most common error was labeling either T12 or S1 as lumbar vertebrae or T1 as cervical vertebrae. A more recent study was able to use an even stronger CNN specialized for processing 3-dimensional images to locate and perform accurate spine segmentation on partial spine CT scans and chest CT scans; the addition of a third dimension to image breakdown and analysis allowed especially efficient localization of the center of vertebral bodies and high accuracy in segmentation, even in images with poor imaging quality or where only parts of some vertebrae are identifiable on the scan [28]. Despite these advances, ML in the near future will likely play a supporting role in spine surgery rather than supply full automation [24].
2. Preoperative Planning and Intraoperative Assistance
ML models in spine imaging analysis have clinical applications beyond examining large visual features. ML can regularize and expedite preoperative planning and intraoperative adjustments, particularly in deformity cases where measurements and corrections rely on estimation and surgeon experience. Any surgical parameter that is manually measured or measured with computer assistance could be made more precise by ML models. For instance, Cobb angles are typically manually measured on a radiograph. Even among experienced surgeons, angle measurements can vary almost 10% [29]. Zhang et al. [29] used a deep neural network (DNN) with 3 neural layers to perform spine segmentation similar to the protocols mentioned above, identify the vertebral end plates, and measure Cobb angles. The DNN was able to attain a high degree of accuracy and consistency, approaching an error of only around 5% from Cobb angles calculated by experienced spine surgeons. Automation could more accurately measure Cobb angles faster than manual measurement and may enable studies of imaging parameters on a massive scale.
Another area of spine surgery that may benefit from ML models is pedicle screw placement. Like Cobb angle determination, pedicle screw placement is an inexact, by-the-eye surgical procedure. While novel navigation and robotics systems for screw placement have increased accuracy, ML can work to continue to optimize the outcomes of screw placement [30]. One DL model from Burström et al. [30] can successfully identify and segment the pedicles on MRI, which may enhance the consistency and accuracy of pedicle screw placement. After utilizing similar techniques and methods as described in the previous section to perform pedicle visualization, the model was able to identify the pedicle midpoint, the optimal site of a pedicle screw; while (like many current algorithms) deficient on spines with severe abnormal curvature, the model was generally able to attain high (>95%) accuracy rates identifying optimal places for pedicle screw placement in normal spines. As a supplement to robotic and navigational aids, an automated pedicle identification model could standardize and improve pedicle screw placement.
3. Diagnostics and Clinical Prognostication
ML techniques are also becoming increasingly useful in clinical diagnosis. ML methods can be extended to check for osteoporosis and fracture using models trained on both MRI and CT images [31,32], which are useful in spine and general orthopedic surgery. Even simple models trained quickly and on relatively scant data, such as regression-based on imaging density of vertebral bodies in a CT scan and demographic information can achieve 90% accuracy in identifying osteoporosis with CT scans [32]. More complex computational models trained with more features, such as topological features of the spine in an MRI scan or projected mechanical qualities of bone, have the capability of yielding even better results [31]. For instance, such models have been used to create an automated system for detecting lytic and blastic bone tumors in spine CT scans [33]: in the corresponding study, extensive image processing on CT scans was performed to isolate out the vertebral bodies, while additional image discernment through a random forest classifier was employed to locate likely lesion centers and the border of the metastases in the bone. These DL models could be used as a fast, low-cost screening for every MRI and CT scan performed. Used in conjunction with traditional radiology, ML techniques can enhance imaging efficiency and open up new pathways for effective diagnosis and treatment.
NATURAL LANGUAGE PROCESSING AND TEXT DATA
EMRs are a significant contributor to stress and burnout among physicians [34,35]. Studies have also shown that EMRs can reduce both the amount of time physicians look at their patients [36] and the number of patients seen per provider [37]. The AI techniques of natural language processing (NLP) can help alleviate these problems by enabling computers to assist clinicians with the arduous process of clinical documentation. NLP transforms unstructured text data into computer-recognizable formats for quantitative analysis. This technique has been utilized in a broad range of surgical and medical disciplines, yet to date there have not been many studies specific to spine surgery. This section will focus on the potential applications of NLP in spine surgery. Although the majority of articles discussed pertain to other fields of medicine, the findings are readily adaptable to spine surgery. Given that EMRs contain a plethora of textual data in the form of provider notes, imaging reports, etc., the applications for NLP are numerous.
1. Data Querying
NLP offers an efficient way to extract clinical data from medical documents, which normally would be labor-intensive and time consuming. For example, groups have used NLP across various medical disciplines to extract key clinical data from text documents such as patient records [38], discharge summaries [38-40], pathology reports [40,41], and radiology reports [40,42]. These techniques can be adapted to assist spine surgeons via data extraction. For example, Tan et al. [43] trained a model to identify information pertaining to low back pain (LBP) from lumbar spine imaging reports. Initially, the group determined 26 distinct findings to extract, such as annular fissure, scoliosis, disc protrusion, or spondylosis. The authors developed both rules-based and ML models for the task. The ML model achieved an average specificity of 0.95 and sensitivity of 0.94. While both models were similar in specificity, the authors found that the ML model achieved greater sensitivity in the detection of “compound findings,” such as “nerve root displaced or compressed.” [43] Another example of NLP is a rules-based approach recently published by Wyles et al. [44] for the analysis of total hip arthroplasty operative notes. Their algorithms captured information pertaining to operative approach, fixation method, and bearing surface. All the models demonstrated an accuracy of >90% and demonstrated external validity [44]. Robust NLP methods may enable unstructured qualitative data to be converted into quantitative statistics for analysis on a massive scale. This can enable the exploration of novel research topics, as well as new methods for quality improvement of clinical operations on a day to day basis.
2. Clinical Assistance
Aside from data extraction, some sophisticated NLP applications have shown promise for more dynamic clinical use, assisting clinicians in real time. One such way is through the automated creation of spine radiological reports, as demonstrated by Han et al. using a weakly supervised DL model [45]. Their model generated reports pertaining to 3 spinal diseases: lumbar vertebrae deformities, neural foraminal stenosis, and intervertebral disc degeneration [45]. The generated text noted any association between the diseases and the location. One example would be “At L3–4, disc degenerative changes are associated with neural foraminal stenosis.” [45] The use of NLP in automated note generation can ease the work burden of clinicians and increase clinical efficiency. These models could be adapted to other text-based documents in spine surgery. In addition to note generation, ML methods can also be employed in speech recognition technologies. A group from Google developed 2 neural network models, connectionist temporal classification (CTC) and listen attend and spell (LAS), to automatically transcribe provider-patient conversations [46]. The latter model produced a word error rate of 18.3% and the former 20.1%. The authors noted that error came in recognizing casual conversations, which may be less clinically relevant [46]. When identifying key medical terms, the CTC model had precision and recall rates ranging from 80%–90%, and the LAS model demonstrated a 98.2% recall in identifying drug names [46]. Another study developed a deep neural network model for medical voice recognition trained on over 270 hours of speech data and compared the performance to professional medical transcriptionists [47]. The model had a 15.4% error rate when applied to a “realistic clinical use case” and performed equally as well as humans [47]. The 2 studies discussed here illustrate the ability of ML to recognize real conversations between patients and providers. With further refinement, this software could be hugely beneficial for clinical and office work, creating less need for providers to manually input notes.
Overall, the use of NLP in spine surgery is still in its early stages. Generally speaking, rules-based approaches have been the dominant approach to NLP tasks. However, as ML libraries and algorithms continue to develop, we may see more sophisticated text-based applications, such as the generation of complex notes or voice recognition software. Given the vast amount of EMR text data available, there is much room for innovation and creativity.
STRUCTURED DATA
The remainder of EMR data is quite comprehensive, including demographic information, labs, vitals, medical history components, social history components, procedures, medications, and other variables. Unlike unstructured imaging and text data, this data benefits from inherent structure. Once extracted from EMR systems, this structured data is primed for integration with ML techniques to create better risk stratification systems, allow for personalized treatment algorithms, improve postoperative clinical prognostication, and refine reimbursement models [26]. As a result, this EMR data constitutes a large proportion of current and potential future research opportunities investigating the applications of ML in spine surgery.
1. Risk Stratification
ML algorithms allow for better risk stratification and the creation of entirely new classification systems that aid spine surgeons in their decision-making process. For example, one recent study by Ames et al. demonstrated the efficacy of ML in patient clustering to better create new classification systems and guide the preoperative decision-making process. While this study used prospective data, as opposed to that from an EMR system, it presents an opportunity to be repeated using retrospective EMR data on larger datasets to establish external validity. This could increase the study’s generalizability and remove any potential bias that may have existed in the original prospective study [26,48]. Also, EMR data can be used to better identify highrisk patients and predict complications and prevent those complications. One recent study by Zhang et al. [49] demonstrated the ability of quantitative CT ML algorithms to assess vertebral strength and predict vertebral fracture risk in elderly patients. This is especially important considering the mortality rate of Medicare patients with vertebral compression fractures is approximately 2 times that in matched cohorts [50].
2. Personalized Treatment Algorithms
The use of ML with EMR data also allows for the creation of more personalized treatment algorithms. One recent study demonstrated the ability of ML to predict a specific patient’s response to functional restoration rehabilitation for chronic LBP. While this study was conducted in a prospective manner, large EMR datasets could be analyzed via this method to increase generalizability. The resultant predictions would allow surgeons and patients to better understand the likelihood of success and make more informed treatment decisions [50,51]. Another study used EMR data to predict the need for intraoperative or postoperative blood transfusion [52]. This allows for the surgical team to better preoperatively optimize these patients and have equipment available to mitigate blood loss and expedite transfusions as needed. Lastly, for certain conditions where comparable treatments exist and overall literature is nonconfirmatory, ML algorithms could help with in-depth analysis of patient information to create personalized treatment algorithms.
3. Clinical Prognostication
The integration of EMR data with ML algorithms could have the greatest potential impact on postoperative clinical prognostication. It would allow surgeons to better predict and prepare for postoperative complications, more efficiently utilize hospital resources, and prioritize patient surveillance on those who are at the greatest risk. In fact, multiple studies have demonstrated the ability to build predictive models using EMR data for major perioperative complications in spine surgery, particularly surgical site infections [53-57]. Other models have also been created using EMR data to predict physical disability, return to work, major complications, readmission rates, walking ability, need for inpatient rehab following spine surgery, discharge, and disposition [58-61]. More specific algorithms have been created to predict preoperative factors impacting survival, discharge, and readmission rates in patients following spine surgery for spinal metastasis [56,62,63]. While some of these studies have used the National Surgical Quality Improvement Program database and insurance databases in the past, similar algorithms could be produced for other indications using large EMR datasets. Then, ML techniques such as ridge linear regression and to a much larger extent nonlinear DL models like ANNs can identify the features most relevant and helpful to predicting each post-surgical outcome [57]. Used in a hospital-wide fashion, these models could identify the most vulnerable postoperative patients and help direct postoperative triage and patient surveillance. Recently, one group even used EMR data to create an application to predict which patients are at higher risk of prolonged postoperative opioid use. This application, which will allow surgeons to better identify patients that may require increased surveillance following surgery, is especially important given the current opioid crisis in the United States [64].
4. Reimbursement
The integration of EMR data with ML algorithms also has huge ramifications on reimbursement processes, particularly in creating new classifications systems for bundled care models [48,65]. Moreover, all of the aforementioned impacts on risk stratification, personalized treatment algorithms, and clinical prognostication will allow for more accurate reimbursement models and financial optimization of clinical practice.
LIMITATIONS AND CHALLENGES
ML applications using EMR data in spine surgery hold immense promise for the future. Despite the impressive advancements of recent years, there are still several challenges that must be addressed to bring robust algorithms into clinical practice. Many of these limitations are not unique to spine surgery. Several limitations of using ML with EMRs are discussed below.
1. Electronic Medical Record Systems
ML applications utilizing EMR data are rapidly advancing, but EMR systems themselves pose a significant challenge to progress. It is often quite difficult to effectively extract data from EMR systems [66]. Notably, many studies reviewed in this article used large publicly available databases rather than attempt to gather the same data from their own institutional EMR systems [52,57,59,63,67]. EMR systems do not follow standardized protocols for data storage and application programming interfaces, making it difficult to interface EMRs with other systems [1]. Not only does this impact data extraction, but it also affects clinical integration. Another major challenge lies in building the necessary infrastructure to clinically integrate ML tools with EMRs for evaluation. Over the coming years, it may be important for EMR systems to adapt to simplify data extraction and to allow integration of ML models into the clinical workflow. Only then will these algorithms begin to benefit clinical practice in a meaningful way.
2. Data Quality
Once data extraction has been completed, the quality of the EMR data is a further challenge. ML techniques are best suited for large amounts of high quality, consistent data with clear labeling. Poor data quality is a common barrier to creating a high performing algorithm for clinical application [68]. Unfortunately, the data found in EMRs is by nature heterogeneous and noisy, and several studies have called the quality of EMR data into question [69-73]. Inaccuracies and missing data points are a common problem. Furthermore, there may be disconnects between the labeling and the reality in EMR data. One common criticism is that International Classification of Diseases codes are often unreliable for diagnosis, as they are primarily used for billing [74]. Training algorithms on such poor-quality data may ultimately be dangerous for patients [75]. Future applications of ML to EMR data will need to employ rigorous data mining techniques to ensure high-quality data for training [76]. Parsing signal from noise in EMR data remains a significant challenge in creating useful ML algorithms.
3. The Black Box Problem
Some studies have created ML algorithms that outperform physicians in clinical tasks [16,77]. These algorithms are exciting, as they point toward the development of tools that could improve the quality of care. However, one drawback of many ML models is that they cannot explain themselves. These models are often referred to as “black boxes,” meaning that although the inputs and outputs are visible, the internal behavior of the model remains hidden [78,79]. Despite impressive validated results, the black box nature of many ML algorithms is a significant barrier to patient and physician trust and ultimate clinical use.
In order to utilize ML tools effectively, ML tools will need to provide explanations for their outputs [80]. This is particularly challenging since ML models may have millions of parameters containing a massive amount of nuanced information. The challenge is to take this high-dimensional model and abstract its relationships into an interpretable form while maintaining adequate fidelity to the model’s inner workings [79]. To this end, some significant progress has already been made in developing methods for dissecting the internal workings of ML models [81]. On imaging data, heatmaps may provide insight by pointing to regions that the algorithm finds important for its ultimate decision. NLP algorithms may highlight particularly important pieces of text. Algorithms integrating labs and vitals might highlight the most influential pieces of data in the decision-making process. Few studies in this review included tools for model interpretation. Future ML developments will need to be accompanied by tools for model interpretation that open up the black box in order to achieve clinical use.
4. Generalizability
ML algorithms work best on tasks with narrow definitions. Unfortunately, slight changes of input data distributions can derail an algorithm altogether. Thus, many ML algorithms fail to generalize across institutions, patient populations, pathologies, and other domains. Multiple studies of ML in spine surgery have discovered issues with the generalizability of proposed models [55,82]. For example, Janssen et al. [55] attempted an external validation of a surgical site infection treatment algorithm on 898 consecutive patients. The results were rather poor compared to the initial study, with a low positive predictive value pointing to problems of overfitting and generalizability [54,55]. One way this limitation can be addressed is through fine-tuning of algorithms with data from the institution to which the algorithm will be applied. Although this is a plausible solution, this route is limited to institutions with large quantities of data. Smaller institutions and private practices might be precluded from employing ML tools if they require fine-tuning to be effective. Therefore, it is essential that ML algorithms are robust and generalize well in order to maximize utility. Unfortunately, few current studies of ML applications to spine surgery pursue external validation [82]. This trend is not limited to spine surgery. One recent review by Kim et al. [83] found that only 6% of AI algorithms designed for diagnostic image analysis included external validation in the results. Fortunately, this appears to be changing, as several calls for “technovigilance” have arisen, advocating careful external validation of ML study results [83,84].
5. Legal
As high-risk decisions begin to be augmented by and delegated to AI systems, questions of legal ramifications will need to be answered. Medical malpractice standards currently govern culpability in care settings. However, it is unclear who is responsible for predictive errors in AI algorithms in current law. Determining responsibility for the outputs of algorithms will be necessary before advancing EMR-based ML to clinical implementation [85].
6. Ethical
One notable ethical challenge that arises frequently in the discussion of ML algorithms in medicine is the potential for bias [85]. Discrepancies in data quality across groups may introduce unwanted bias into algorithms and cause harm to those underrepresented in EMR training data [86]. In some cases, algorithms may provide undue weight to proxy variables or confounders to generate predictions, which may translate to harmful outcomes for patients. Future development of AI systems will need to address these challenges and examine potential biases to avoid unintended consequences before advancing to clinical implementation.
CONCLUSION
EMRs represent a rich source of medical data, and ML algorithms may be able to successfully harness the value of this data to impact the field of spine surgery. Strides have already been made in using ML algorithms to read radiographs, generate reports, and project clinical outcomes for patients with impressive results. However, several challenges still remain to be addressed. Despite these challenges, progress is rapid, and it appears these algorithms will eventually reach a point of meaningful clinical integration with profound impact on the practice of spine surgery.
Footnotes
The authors have nothing to disclose.
REFERENCES
- 1.Evans RS. Electronic Health records: then, now, and in the future. Yearb Med Inform. 2016;Suppl 1:S48–61. doi: 10.15265/IYS-2016-s006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler-Milstein J, Holmgren AJ, Kralovec P, et al. Electronic health record adoption in US hospitals: the emergence of a digital “advanced use” divide. J Am Med Inform Assoc. 2017;24:1142–8. doi: 10.1093/jamia/ocx080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YG, Jung K, Park YT, et al. Rate of electronic health record adoption in South Korea: a nation-wide survey. Int J Med Inform. 2017;101:100–7. doi: 10.1016/j.ijmedinf.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Simborg DW, Detmer DE, Berner ES. The wave has finally broken: now what? J Am Med Inform Assoc. 2013;20:e21–5. doi: 10.1136/amiajnl-2012-001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford EW, Menachemi N, Phillips MT. Predicting the adoption of electronic health records by physicians: when will health care be paperless? J Am Med Inform Assoc. 2006;13:106–12. doi: 10.1197/jamia.M1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azad TD, Kalani M, Wolf T, et al. Building an electronic health record integrated quality of life outcomes registry for spine surgery. J Neurosurg Spine. 2016;24:176–85. doi: 10.3171/2015.3.SPINE141127. [DOI] [PubMed] [Google Scholar]
- 7.Ryu B, Kim N, Heo E, et al. Impact of an electronic health record-integrated personal health record on patient participation in health care: development and randomized controlled trial of MyHealthKeeper. J Med Internet Res. 2017;19:e401. doi: 10.2196/jmir.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SW, Wee JH, Yoo S, et al. Effect of lifestyle modification using a smartphone application on obesity with obstructive sleep apnea: a short-term, randomized controlled study. Clin Exp Otorhinolaryngol. 2018;11:192–8. doi: 10.21053/ceo.2017.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ching T, Himmelstein DS, Beaulieu-Jones BK, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15(141):pii: 20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macedonia M. The GPU enters computing’s mainstream. Computer. 2003;36:106–8. [Google Scholar]
- 11.Das PK, Deka GC. History and Evolution of GPU Architecture. In: Deka G, Siddesh G, Srinivasa K, et al., editors. Emerging research surrounding power consumption and performance issues in utility computing. Hershey (PA): IGI Global; 2016. pp. 109–35. [Google Scholar]
- 12.Oh KS, Jung K. GPU implementation of neural networks. Pattern Recogit. 2004;37:1311–4. [Google Scholar]
- 13.Paszke A, Gross S, Chintala S, et al. Automatic differentiation in PyTorch. 31st Conference on Neural Information Processing Systems (NIPS 2017); Long Beach (CA), USA. [Google Scholar]
- 14.Abadi M, Barham P, Chen J, et al. Tensorflow: a system for large-scale machine learning. 12th USENIX$ symposium on operating systems design and implementation; 2016 Nov 2-4; Savannah (GA), USA. 2016. pp. 265–83. [Google Scholar]
- 15.Tran BX, Vu GT, Ha GH, et al. Global evolution of research in artificial intelligence in health and medicine: a Bibliometric study. J Clin Med. 2019;8(3):pii: E360. doi: 10.3390/jcm8030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–61. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 17.Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572:116–9. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Titano JJ, Badgeley M, Schefflein J, et al. Automated deepneural-network surveillance of cranial images for acute neurologic events. Nat Med. 2018;24:1337–41. doi: 10.1038/s41591-018-0147-y. [DOI] [PubMed] [Google Scholar]
- 19.Rowe M. An introduction to machine learning for clinicians. Acad Med. 2019;94:1433–6. doi: 10.1097/ACM.0000000000002792. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10:257–73. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 21.Papp L, Spielvogel CP, Rausch I, et al. Personalizing medicine through hybrid imaging and medical big data analysis. Front Phys. 2018;6:Article 51. doi: 10.3389/fphy.2018.00051. [DOI] [Google Scholar]
- 22.Hosny A, Parmar C, Quackenbush J, et al. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–10. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Z, Zhong J, Wee W, et al. Automated vertebra detection and segmentation from the whole spine MR images. Conf Proc IEEE Eng Med Biol Soc. 2005;3:2527–30. doi: 10.1109/IEMBS.2005.1616983. [DOI] [PubMed] [Google Scholar]
- 24.Cho BH, Kaji D, Cheung ZB, et al. Automated measurement of lumbar lordosis on radiographs using machine learning and computer vision. Global Spine J. 2019:1–8. doi: 10.1177/2192568219868190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thong W, Parent S, Wu J, et al. Three-dimensional morphology study of surgical adolescent idiopathic scoliosis patient from encoded geometric models. Eur Spine J. 2016;25:3104–13. doi: 10.1007/s00586-016-4426-3. [DOI] [PubMed] [Google Scholar]
- 26.Galbusera F, Casaroli G, Bassani T. Artificial intelligence and machine learning in spine research. JOR Spine. 2019;2:e1044. doi: 10.1002/jsp2.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsberg D, Sjöblom E, Sunshine JL. Detection and labeling of vertebrae in MR images using deep learning with clinical annotations as training data. J Digit Imaging. 2017;30:406–12. doi: 10.1007/s10278-017-9945-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lessmann N, van Ginneken B, de Jong PA, et al. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med Image Anal. 2019;53:142–55. doi: 10.1016/j.media.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Li H, Lv L, et al. Computer-aided Cobb measurement based on automatic detection of vertebral slopes using deep neural network. Int J Biomed Imaging. 2017;2017:9083916. doi: 10.1155/2017/9083916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burström G, Buerger C, Hoppenbrouwers J, et al. Machine learning for automated 3-dimensional segmentation of the spine and suggested placement of pedicle screws based on intraoperative cone-beam computer tomography. J Neurosurg Spine. 2019 Mar 22;:1–8. doi: 10.3171/2018.12.SPINE181397. [Epub]. [DOI] [PubMed] [Google Scholar]
- 31.Ferizi U, Besser H, Hysi P, et al. Artificial intelligence applied to osteoporosis: a performance comparison of machine learning algorithms in predicting fragility fractures from MRI data. J Magn Reson Imaging. 2019;49:1029–38. doi: 10.1002/jmri.26280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nam KH, Seo I, Kim DH, et al. Machine learning model to predict osteoporotic spine with hounsfield units on lumbar computed tomography. J Korean Neurosurg Soc. 2019;62:442–9. doi: 10.3340/jkns.2018.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammon M, Dankerl P, Tsymbal A, et al. Automatic detection of lytic and blastic thoracolumbar spine metastases on computed tomography. Eur Radiol. 2013;23:1862–70. doi: 10.1007/s00330-013-2774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babbott S, Manwell LB, Brown R, et al. Electronic medical records and physician stress in primary care: results from the MEMO Study. J Am Med Inform Assoc. 2014;21:e100–6. doi: 10.1136/amiajnl-2013-001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanford Medicine . White paper: the future of electronic health records. Stanford (CA): Stanford Medicine; 2018. Available from: http://med.stanford.edu/content/dam/sm/ehr/documents/SM-EHR-White-Papers_v12.pdf. [Google Scholar]
- 36.Young RA, Burge SK, Kumar KA, et al. A time-motion study of primary care physicians’ work in the electronic health record era. Fam Med. 2018;50:91–9. doi: 10.22454/FamMed.2018.184803. [DOI] [PubMed] [Google Scholar]
- 37.Hollenbeck SM, Bomar JD, Wenger DR, et al. Electronic medical record adoption: the effect on efficiency, completeness, and accuracy in an academic orthopaedic practice. J Pediatr Orthop. 2017;37:424–8. doi: 10.1097/BPO.0000000000000679. [DOI] [PubMed] [Google Scholar]
- 38.Roberts K, Shooshan SE, Rodriguez L, et al. The role of fine-grained annotations in supervised recognition of risk factors for heart disease from EHRs. J Biomed Inform. 2015;58 Suppl:S111–9. doi: 10.1016/j.jbi.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang M, Chen Y, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc. 2011;18:601–6. doi: 10.1136/amiajnl-2011-000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Zhang Y, Zhang Q, et al. Extracting comprehensive clinical information for breast cancer using deep learning methods. Int J Med Inform. 2019;132:1039. doi: 10.1016/j.ijmedinf.2019.103985. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Huang Y, Boyle B, et al. The utility of including pathology reports in improving the computational identification of patients. J Pathol Inform. 2016;7:46. doi: 10.4103/2153-3539.194838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lou R, Lalevic D, Chambers C, et al. Automated detection of radiology reports that require follow-up imaging using natural language processing feature engineering and machine learning classification. J Digit Imaging. 2019 Sep 3;:1–8. doi: 10.1007/s10278-019-00271-7. [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan WK, Hassanpour S, Heagerty PJ, et al. Comparison of natural language processing rules-based and machine-learning systems to identify lumbar spine imaging findings related to low back pain. Acad Radiol. 2018;25:1422–32. doi: 10.1016/j.acra.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyles CC, Tibbo ME, Fu S, et al. Use of natural language processing algorithms to identify common data elements in operative notes for total hip arthroplasty. J Bone Joint Surg Am. 2019;101:1931–8. doi: 10.2106/JBJS.19.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Z, Wei B, Leung S, et al. Towards automatic report generation in spine radiology using weakly supervised framework. 21st International Conference on Medical Image Computing and Computer Assisted Intervention; 2018 Sep 16-20; Granada, Spain. 2018. pp. 185–93. [Google Scholar]
- 46.Chiu CC, Tripathi A, Chou K, et al. Speech recognition for medical conversations. Interspeech 2018. http://arxiv.org/abs/1711.07274v2.
- 47.Edwards E, Salloum W, Finley GP, et al. Medical Speech recognition: reaching parity with humans. In: Karpov A, Potapova R, Mporas I, et al., editors. Speech and computer. SPECOM 2017. Lecture Notes in Computer Science, vol 10458. Cham (Switzerland): Springer; 2017. pp. 512–24. [Google Scholar]
- 48.Ames CP, Smith JS, Pellisé F, et al. Artificial intelligence based hierarchical clustering of patient types and intervention categories in adult spinal deformity surgery: towards a new classification scheme that predicts quality and value. Spine (Phila Pa 1976) 1976;44:915–26. doi: 10.1097/BRS.0000000000002974. [DOI] [PubMed] [Google Scholar]
- 49.Zhang M, Gong H, Zhang K, et al. Prediction of lumbar vertebral strength of elderly men based on quantitative computed tomography images using machine lear ning. Osteoporos Int. 2019;30:2271–82. doi: 10.1007/s00198-019-05117-0. [DOI] [PubMed] [Google Scholar]
- 50.Lau E, Ong K, Kurtz S, et al. Mortality following the diagnosis of a vertebral compression fracture in the Medicare population. J Bone Joint Surg Am. 2008;90:1479–86. doi: 10.2106/JBJS.G.00675. [DOI] [PubMed] [Google Scholar]
- 51.Jiang N, Luk KD, Hu Y. A Machine Learning-based surface electromyography topography evaluation for prognostic prediction of functional restoration rehabilitation in chronic low back pain. Spine (Phila Pa 1976) 2017;42:1635–42. doi: 10.1097/BRS.0000000000002159. [DOI] [PubMed] [Google Scholar]
- 52.Durand WM, DePasse JM, Daniels AH. Predictive modeling for blood transfusion after adult spinal deformity surgery: a tree-based machine learning approach. Spine (Phila Pa 1976) 2018;43:1058–66. doi: 10.1097/BRS.0000000000002515. [DOI] [PubMed] [Google Scholar]
- 53.Scheer JK, Smith JS, Schwab F, et al. Development of a preoperative predictive model for major complications following adult spinal deformity surgery. J Neurosurg Spine. 2017;26:736–43. doi: 10.3171/2016.10.SPINE16197. [DOI] [PubMed] [Google Scholar]
- 54.Lee MJ, Cizik AM, Hamilton D, et al. Predicting surgical site infection after spine surgery: a validated model using a prospective surgical registry. Spine J. 2014;14:2112–7. doi: 10.1016/j.spinee.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 55.Janssen DMC, van Kuijk SMJ, d’Aumerie BB, et al. External validation of a prediction model for surgical site infection after thoracolumbar spine surgery in a Western European cohort. J Orthop Surg Res. 2018;13:114. doi: 10.1186/s13018-018-0821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han SS, Azad TD, Suarez PA, et al. A machine learning approach for predictive models of adverse events following spine surgery. Spine J. 2019;19:1772–81. doi: 10.1016/j.spinee.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 57.Kim JS, Merrill RK, Arvind V, et al. Examining the ability of artificial neural networks machine learning models to accurately predict complications following posterior lumbar spine fusion. Spine (Phila Pa 1976) 2018;43:853–60. doi: 10.1097/BRS.0000000000002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGirt MJ, Sivaganesan A, Asher AL, et al. Prediction model for outcome after low-back surgery: individualized likelihood of complication, hospital readmission, return to work, and 12-month improvement in functional disability. Neurosurg Focus. 2015;39:E13. doi: 10.3171/2015.8.FOCUS15338. [DOI] [PubMed] [Google Scholar]
- 59.Karhade AV, Ogink P, Thio Q, et al. Development of machine learning algorithms for prediction of discharge disposition after elective inpatient surgery for lumbar degenerative disc disorders. Neurosurg Focus. 2018;45:E6. doi: 10.3171/2018.8.FOCUS18340. [DOI] [PubMed] [Google Scholar]
- 60.DeVries Z, Hoda M, Rivers CS, et al. Development of an unsupervised machine learning algorithm for the prognostication of walking ability in spinal cord injury patients. Spine J. 2019 Sep 13; doi: 10.1016/j.spinee.2019.09.007. [Epub]. pii: S1529-9430(19)30971-4. [DOI] [PubMed] [Google Scholar]
- 61.Papić M, Brdar S, Papić V, et al. Return to work after lumbar microdiscectomy - personalizing approach through predictive modeling. Stud Health Technol Inform. 2016;224:181–3. [PubMed] [Google Scholar]
- 62.Karhade AV, Ahmed AK, Pennington Z, et al. External validation of the SORG 90-day and 1-year machine learning algorithms for survival in spinal metastatic disease. Spine J. 2020;20:14–21. doi: 10.1016/j.spinee.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 63.Karhade AV, Thio QCBS, Ogink PT, et al. Development of machine learning algorithms for prediction of 30-day mortality after surgery for spinal metastasis. Neurosurgery. 2019;85:E83–91. doi: 10.1093/neuros/nyy469. [DOI] [PubMed] [Google Scholar]
- 64.Karhade AV, Ogink PT, Thio QCBS, et al. Development of machine learning algorithms for prediction of prolonged opioid prescription after surgery for lumbar disc herniation. Spine J. 2019;19:1764–71. doi: 10.1016/j.spinee.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Kho AN, Liebovitz D, et al. Learning bundled care opportunities from electronic medical records. J Biomed Inform. 2018;77:1–10. doi: 10.1016/j.jbi.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milinovich A, Kattan MW. Extracting and utilizing electronic health data from Epic for research. Ann Transl Med. 2018;6:42. doi: 10.21037/atm.2018.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goyal A, Ngufor C, Kerezoudis P, et al. Can machine learning algorithms accurately predict discharge to nonhome facility and early unplanned readmissions following spinal fusion? Analysis of a national surgical registry. J Neurosurg Spine. 2019 Jun 7;:1–11. doi: 10.3171/2019.3.SPINE181367. [Epub]. https://doi: 10.3171/2019.3.SPINE181367. [DOI] [PubMed] [Google Scholar]
- 68.Ho LV, Ledbetter D, Aczon M, et al. The dependence of machine learning on electronic medical record quality. AMIA Annu Symp Proc. 2018;2017:883–91. [PMC free article] [PubMed] [Google Scholar]
- 69.Price M, Bowen M, Lau F, et al. Assessing accuracy of an electronic provincial medication repository. BMC Med Inform Decis Mak. 2012;12:42. doi: 10.1186/1472-6947-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greiver M, Barnsley J, Glazier RH, et al. Measuring data reliability for preventive services in electronic medical records. BMC Health Serv Res. 2012;12:116. doi: 10.1186/1472-6963-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feder SL. Data quality in electronic health records research: quality domains and assessment methods. West J Nurs Res. 2018;40:753–66. doi: 10.1177/0193945916689084. [DOI] [PubMed] [Google Scholar]
- 72.Dziadkowiec O, Callahan T, Ozkaynak M, et al. Using a data quality framework to clean data extracted from the electronic health record: a case study. EGEMS (Wash DC) 2016;4:1201. doi: 10.13063/2327-9214.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu CHK, Luk SMH, Holder RL, et al. How do paper and electronic records compare for completeness? A three centre study. Eye (Lond) 2018;32:1232–6. doi: 10.1038/s41433-018-0065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haendel MA, Chute CG, Robinson PN. Classification, ontology, and precision medicine. N Engl J Med. 2018;379:1452–62. doi: 10.1056/NEJMra1615014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Challen R, Denny J, Pitt M, et al. Artificial intelligence, bias and clinical safety. BMJ Qual Saf. 2019;28:231–7. doi: 10.1136/bmjqs-2018-008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cios KJ, Moore GW. Uniqueness of medical data mining. Artif Intell Med. 2002;26:1–24. doi: 10.1016/s0933-3657(02)00049-0. [DOI] [PubMed] [Google Scholar]
- 77.Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29:1836–42. doi: 10.1093/annonc/mdy166. [DOI] [PubMed] [Google Scholar]
- 78.The Lancet Respiratory Medicine Opening the black box of machine learning. Lancet Respir Med. 2018;6:801. doi: 10.1016/S2213-2600(18)30425-9. [DOI] [PubMed] [Google Scholar]
- 79.Ribeiro M, Singh S, Guestrin C, et al. “Why should i trust you?”: Explaining the predictions of any classifier. The 2016 Conference of the North American Chapter of the Association for computational linguistics: human language technologies. Proceedings of the demonstrations session; 2016 Jun 12-17; San Diego (CA), USA. [Google Scholar]
- 80.Cabitza F, Zeitoun JD. The proof of the pudding: in praise of a culture of real-world validation for medical artificial intelligence. Ann Transl Med. 2019;7:161. doi: 10.21037/atm.2019.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sussillo D, Barak O. Opening the black box: low-dimensional dynamics in high-dimensional recurrent neural networks. Neural Comput. 2013;25:626–49. doi: 10.1162/NECO_a_00409. [DOI] [PubMed] [Google Scholar]
- 82.Stopa BM, Robertson FC, Karhade AV, et al. Predicting nonroutine discharge after elective spine surgery: external validation of machine learning algorithms. J Neurosurg Spine. 2019 Jul 26;:1–6. doi: 10.3171/2019.5.SPINE1987. [Epub]. [DOI] [PubMed] [Google Scholar]
- 83.Kim DW, Jang HY, Kim KW, et al. design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20:405–10. doi: 10.3348/kjr.2019.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bates DW. Commentary: the role of “technovigilance” in improving care in hospitals. Milbank Q. 2013;91:455–8. doi: 10.1111/1468-0009.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cath C. Governing artificial intelligence: ethical, legal and technical opportunities and challenges. Philos Trans A Math Phys Eng Sci. 2018;376(2133):pii: 20180080. doi: 10.1098/rsta.2018.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gianfrancesco MA, Tamang S, Yazdany J, et al. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med. 2018;178:1544–7. doi: 10.1001/jamainternmed.2018.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]