Abstract
Recently interest in multi-generational epigenetic phenomena have been fuelled by highly reproducible intergenerational and transgenerational inheritance paradigms in several model organisms. Such paradigms are essential in order to begin to use genetics to unpick the mechanistic bases of how epigenetic information may be transmitted between generations; indeed great strides have been made towards understanding these mechanisms. Far less well understood is the relationship between epigenetic inheritance, ecology and evolution. In this review I focus on potential connections between laboratory studies of transgenerational epigenetic inheritance phenomena and evolutionary processes that occur in natural populations. In the first section, I consider whether transgenerational epigenetic inheritance might provide an advantage to organisms over the short term in adapting to their environment. Second, I consider whether epigenetic changes can contribute to the evolution of species by contributing to stable phenotypic variation within a population. Finally I discuss whether epigenetic changes could influence evolution by either directly or indirectly promoting DNA sequence changes that could impact phenotypic divergence. Additionally, I will discuss how epigenetic changes could influence the evolution of human cancer and thus be directly relevant for the development of this disease.
Keywords: Epigenetics, Evolution, Small RNAs, DNA methylation, Epigenomics, Epimutation
1. Introduction
Epigenetic regulation is a fundamental property of eukaryotic genomes. It enables the development of multicellular organisms by permitting the specification of different cell types starting from identical DNA sequence, and it was in this context that the term was initially proposed [1]. Such a broad definition however leads to confusion whereby almost any process that regulates gene expression might be regarded as epigenetic. A more precise definition states that epigenetic regulation causes a change in gene expression that is heritable through cell division in the absence of any change in the DNA sequence (Holliday 1988). In this article I will follow this definition, recognising that describing a molecular mechanism as epigenetic is a shorthand for stating that it has the capacity to act epigenetically, rather than suggesting that every occurrence is a case of epigenetic regulation. Many molecular pathways have the capacity to act epigenetically, including DNA methylation [2], histone modifications [3], small RNAs [4] and even protein-protein interactions [5].
In addition to specifying cell types through development, there is a growing realisation that epigenetic changes can be propagated across generations in the absence of any sequence differences. Such multi-generational epigenetic phenomena are classified as intergenerational if they pass through one generation, and transgenerational if two or more generations of inheritance occur. Whilst cases of transgenerational epigenetic inheritance have been known about for decades, it is only relatively recently that insight into the mechanisms behind them has been provided, primarily by using carefully controlled studies with isogenic model organisms [6].
The growing appreciation of the molecular mechanisms of transgenerational epigenetic inheritance has led to the recognition of transgenerational epigenetic inheritance as an important phenomenon in mainstream molecular biology. Whether mechanisms of transgenerational epigenetic inheritance contribute to evolutionary processes is much less well understood. Importantly, extrapolating from laboratory studies of transgenerational epigenetic processes to evolution either requires long-term laboratory evolution studies or presents researchers with the difficulty of distinguishing between epigenetic differences and DNA sequence differences in non-isogenic wild populations. Nevertheless, several important insights have been obtained, allowing plausible hypotheses about how evolution and epigenetics impact one another. In the following article, I will explore three areas where important connections between evolution and epigenetics and evolution have been made. In the first section, I will explore whether multi-generational epigenetic inheritance phenomena limited to a few generations could have been selected as adaptive responses to environmental conditions. In the second section I will review studies investigating whether epigenetic changes can underpin evolutionary change in wild populations. Finally, I will explore the possibility that epigenetic differences could lead to evolution through promoting changes in DNA sequence. In the last two sections, the development of cancer in humans is a topic of particular interest, as the rapid mitotic growth of cancer cells subjects them to evolutionary forces in the absence of meiotic changes in epigenetics, potentially making them prime subjects where epigenetic changes could drive evolutionary divergence.
2. Adaptive roles of transgenerational epigenetic inheritance
2.1. Transgenerational epigenetic phenomena in rodents
In rodents several well-controlled research paradigms have been developed where a stimulus applied to an organism leads to phenotypic changes in subsequent generations. Many of these have focused on dietary alterations. One frequently used paradigm is parental undernutrition, in which mothers are provided with 50% of the typical energy content consumed by mice fed ad libitum [7]. F1 Offspring of these mothers showed marked changes in glucose tolerance and importantly F2 offspring derived from F1 males displayed metabolic abnormalities similar to the F1s. The capacity of the phenotype to be passed on through sperm to the F2 generation confirms the transgenerational epigenetic transmission of the phenomenon [8]. Molecular studies performed by Radford et al., showed that DNA methylation patterns in sperm from F1 males derived from undernourished mothers was perturbed [8], although whether these changes are directly responsible for the transmission of the phenotype is unknown. Despite the fact that global DNA demethylation takes place during embryonic development in mammals [9], some regions of the genome escape demethylation, which may provide a route for DNA methylation changes to be passed on to the next generation [10].
Could the heritable epigenetic differences in the offspring of undernourished mothers have an adaptive function? It is tempting to speculate that starved mothers produce offspring that attempt to compensate for an environmental deficient in calories. In a resource-rich environment this may result in overconsumption of food and consequent glucose intolerance/type II diabetes. This possibility was proposed as the “thrifty” phenotype, on the basis of observations made on the association between low birth weight and diabetes later in life [11]. These early observations have subsequently been replicated in a number of studies, though not without controversy (reviewed in [12]). More controversial is a tentative link made on the basis of the offspring of humans exposed to low nutrition during the infamous “Dutch Famine”, where the development of type II diabetes was associated with some changes in DNA methylation [13]. Nevertheless, it is tempting to link these human data with the mouse studies, suggesting conservation across mammals. For this to be valid, two important criteria must be satisfied. First, there must be evidence that, despite the apparent fitness deficit of F1 and F2 individuals when fed ad libitum, they show higher fitness when exposed to starvation conditions. These experiments have yet to be performed. Second, importantly, the idea that the phenotype is adaptive implies that the effect of starvation is under positive selection. For this to be the case, rodents in the wild would have to have experienced similar periods of starvation in order for organisms displaying epigenetic inheritance of the thrifty phenotype to have been subject to positive selection. Whilst it seems reasonable that periods of low food might be part of the natural ecology of a wild mouse (at least if she is a “country” mouse rather than a “town” mouse!), very little is known about their ecology and typical diet making such inferences hard to make. In the absence of this, it would be interesting to test how widespread the phenomenon is across different mouse strains, which have different domestication histories and thus may have evolved subtly different responses to low food.
An alternative well-studied paradigm for transgenerational epigenetic inheritance is to focus on paternally transmitted effects- males contribute less to the offspring than females potentially making it easier to identify the transmitted signal. In this paradigm males are fed with a high-fat [14] or a low-protein diet [15] and the phenotype of the F1 and F2 generation assessed. Metabolic abnormalities persisting up to the F2 generation include altered hepatic cholesterol metabolism [15] and glucose intolerance [14]. The transmission of these effects was shown to be through RNA and, intriguingly, linked to a poorly understood class of small RNAs derived from tRNA cleavage [14,16]. The tRNA fragments are actively transported into developing sperm and the process as a whole is required for normal embryonic development [17,18]. However, exactly how the sperm tRNA fragments result in transgenerational epigenetic inheritance is unclear. Data from RNAseq of embryos derived from low-protein fathers was interpreted to suggest that endogenous retroviral transcripts were targeted by tRNA fragments [16]. Contrastingly, the tRNA fragments in sperm derived from high-fat diet fed mice were suggested to result directly in gene expression changes through promoter-RNA interactions [14]. However there is little direct mechanistic evidence for either of these proposals. Nevertheless, alterations in sperm tRNA fragments and their transmission into the next generation may have a general role in transgenerational epigenetic regulation in mammals.
Despite the existence of a defined mechanism to enable environmental perturbations to be transmitted through sperm, whether these represent adaptive responses is far from clear. Whilst undernutrition, including low protein supply, might be plausible in the natural environment, the overfeeding paradigm seems unlikely to have occurred naturally during mouse evolution. In this regard it is curious that despite an ostensibly opposite stimulus the phenotypic effect on the offspring of overfeeding are similar to the effect of underfeeding, whether transmitted maternally or paternally. This argues against an adaptive role for transgenerational epigenetic inheritance of dietary perturbations. Instead, one could argue that both undernutrition and overnutrition represent examples of pathological stresses that perturb metabolic networks in subsequent generations in a non-adaptive manner. Potentially, diabetes and glucose intolerance are pleiotropric traits that rely on many different inputs thus are highly sensitive to any perturbations in the metabolic network.
In addition to dietary interventions, some of the most spectacular reports of transgenerational epigenetic inheritance in rodents have come from behavioural studies. In one study, Gapp et al., treated newborn mice with a regime involving periods of unnatural separation from maternal care [19]. F1 and F2 offspring from these mice showed several abnormal behavioural traits including in tests supposed to assess depression-like phenotypes. Importantly, Gapp et al., were able to provide mechanistic details of how this was transmitted, demonstrating that small RNAs in the sperm were responsible for transmitting the phenotype. They also observed changes in the RNA profile of sperm, including miRNAs and piRNAs, although whether these differences were responsible for the epigenetic effects in the next generation is unclear; the group did not test for tRNA fragments, which may also have an important role in this phenomenon [19]. Interestingly recent data suggests that long RNA as well as short RNA may have a role in the transmission of this phenotype [20].
Transgenerational effects on behaviour have also been observed resulting from stresses applied to male mice. A recent example studied exposure of male mice to high doses of nicotine, which was shown to lead to learning defects in F1 offspring, some of which persisted into F2 offspring [21]. In this case the phenotype was linked to alterations in DNA methylation of the dopamine receptor in sperm, although as whether alterations in DNA methylation are themselves responsible for transgenerational transmission is unknown.
Taking both behavioural and metabolic paradigms together, it is far from obvious that there is any adaptive role to transgenerational epigenetic inheritance paradigms so far described in rodents. Indeed, the phenotypes so far described seem to be maladaptive. This probably results from the fact that the stresses applied to the P0 generation in order to observe these effects are severe and non-physiological. The question of whether more realistic stresses can give rise to transgenerational effects remains to be answered.
2.2. Transgenerational epigenetic inheritance in C. elegans
Mechanistic information about how transgenerational epigenetic inheritance works in animals is most advanced in the nematode C. elegans, which, due to its short lifecycle has become the model of choice for many researchers interested in these phenomena. The molecular basis of transgenerational epigenetic inheritance in C. elegans was first discovered by examining transgene silencing. Transgenes containing tracts of foreign sequence, such as GFP, can be silenced by supply of exogenous double strand RNA, or piRNAs, a type of endogenously produced small RNA that recognises sequences through imperfect sense-antisense base pairing within the 3’UTR or coding region [[22], [23], [24]]. Importantly both exogenous dsRNA and piRNA-mediated silencing trigger the generation of a further type of small RNA known as 22G-RNAs by RNA dependent RNA polymerases using the target RNA as a template [25,26]. 22G-RNAs are transmitted transgenerationally in complex with an argonaute (HRDE-1) and can trigger further 22G-RNA biogenesis independently of piRNAs, thus acting as the mediators of epigenetic silencing [[22], [23], [24],27]. In the case of piRNA-mediated silencing of transgenes the duration of inheritance can be extremely long [24,28]. Interestingly, mechanisms acting against transgenerational epigenetic silencing have also been proposed. The initiation of 22G-RNA mediated silencing has been proposed to be antagonised by an alternative set of 22G-RNAs bound to the argonaute CSR-1 [29,30], and the duration of small RNA mediated inheritance may be curtailed by feedback mechanisms at the level of small RNA processing genes and at the chromatin level [[31], [32], [33]].
The presence of the HRDE-1 gene, a mechanism to perpetuate transmitted 22G-RNAs and balancing mechanisms that limit the duration of the response all suggest that a dedicated transgenerational epigenetic inheritance machinery exists in C. elegans, which supports an adaptive role for the process. So far however this role is yet to be clearly defined. An important series of studies showed that mutants lacking key components of the transgenerational small RNA inheritance machinery have a so-called “mortal germline” phenotype such that the fertility of the animals gradually declines over several generations [23,34], suggesting that transgenerational inheritance is required for long-term maintenance of fertility. One hypothesis is that this represents the build up of toxic repetitive RNA, a kind of epigenetic anticipation analogous to the gradual worsening of the phenotype of trinucleotide expansion disorders such as Huntingdon’s disease over several generations. In this view, transgenerational inheritance of silencing signals might be an important part of homeostasis due to the short lifecycle of C. elegans relative to the half-life of individual RNA molecules [34]. A similar concept may apply to the role of the alternative CSR-1 pathway of 22G-RNAs, where CSR-1 activity is proposed to have an anti-silencing function [30,35]. Reduced CSR-1 levels leads to progressive accumulation of germline defects, which were linked to decreased expression levels of CSR-1 targets, although the atrophy of the germline in CSR-1 mutants complicates this analysis somewhat [29].
An alternative possibility is that small RNA-mediated epigenetic inheritance exists in order to provide an adaptive response to environmental conditions. For this to be the case, there must exist stimuli that lead to changes in small RNA population in the germline that could be transmitted into the next generation. In support of this possibility, starvation of C. elegans larvae results in gene expression changes, some of which are retained after 3 generations and can be linked to differences in 22G-RNAs [36]. However, the only phenotypic effect linked to this phenomenon was a shortened lifespan, making it difficult to argue that this is an adaptive phenomenon.
Since the piRNA pathway acts upstream of transgenerational epigenetic inheritance in C. elegans, one approach to identify potential adaptive consequences is to search for stimuli that lead to alterations in the strength of the piRNA pathway. An alteration in the strength of piRNA-mediated silencing would lead to either initiation of new silencing or loss of existing silencing which could establish transgenerational epigenetic gene expression changes. A screen for factors affecting piRNA biogenesis showed that a moderate increase in temperature weakened the piRNA pathway leading to desilencing of some targets, which could be inherited transgenerationally. Interestingly however, this could be completely supressed by simultaneous exposure to pathogenic bacteria [37]. Moreover, bacterial infection, whilst compromising fitness in infected individuals, led to enhanced fitness of offspring in subsequent generations when returned to 20C in the absence of infection. Whilst the adaptive function of this in the environment is still unclear, these experiments demonstrate that investigating how environmental triggers impact piRNA biogenesis may be an instructive avenue for future research.
3. Evolution driven by epigenetic changes
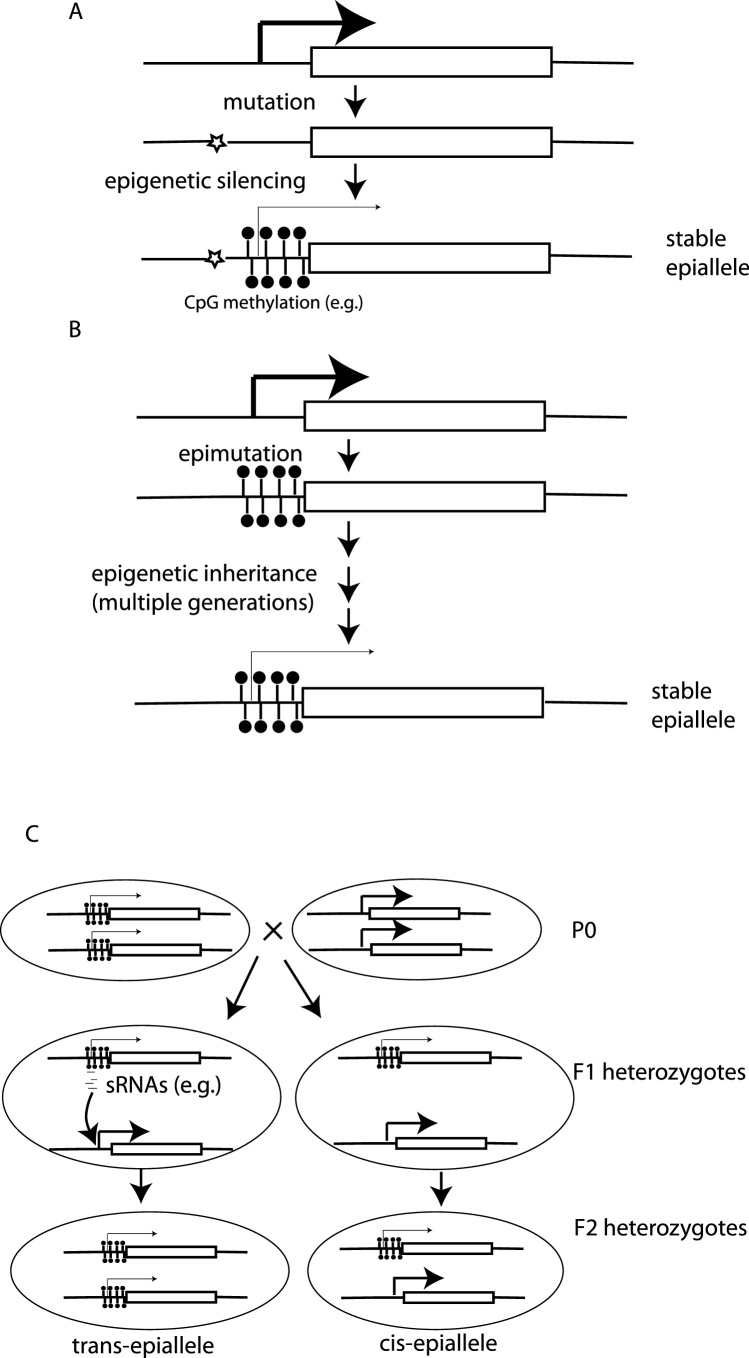
Over long periods of time it is possible that the ability of transgenerational epigenetic inheritance to establish relatively stable gene expression states in the absence of DNA sequence changes could contribute to evolutionary change by providing a source of variation within the population. By analogy to neoDarwinian terminology, gene expression differences within population can be caused by stable “epialleles” in which epigenetic differences account for differential expression. Similarly, different epialleles can be formed or converted into one another by “epimutation”. Epialleles could arise naturally within a population, analogous to a DNA sequence polymorphism; as explained in more detail below, epialleles can also be induced experimentally at endogenous loci through genetic manipulation. In theory, selection and drift could act on epialleles that occur in wild populations, just as they act on DNA sequence polymorphisms, with the result that epiallele frequencies change and different epialleles may even go to fixation within a population. Whilst apparently simple enough, these concepts have historically been controversial [38]. As explained in more detail below part of this controversy relates to whether an epiallele really does behave as a classical allele. The issue is further complicated by the fact that the effect of epigenetic differences on evolutionary processes is studied within the context of two fundamentally different paradigms in which epigenetics plays different roles (Fig. 1). In the first model epigenetic mechanisms are the effectors of DNA sequence variation differences. In this model epigenetic mechanisms act downstream of classical DNA sequence polymorphisms to bring about phenotypic differences. The epigenetic effector model is fully consistent with the modern synthesis as it does not require epigenetic changes to be inherited transgenerationally. In the second model, epigenetic mechanisms alone drive phenotypic changes in the absence of DNA sequence differences. Distinguishing between the effector and driver scenarios can often be difficult as in any natural population both DNA sequence and epigenetic variation are likely to be intermingled. Nevertheless, studies in natural populations and model organisms have provided some examples where epigenetic mechanisms can be shown to act as either drivers or effectors of phenotypic differences.
Fig. 1.
Characteristics of epialleles.
A&B Epialleles in natural populations could either be effectors of DNA sequence change (A) or directly drive phenotypic differences without DNA sequence differences (B).
C- Epialleles can either function in trans, where they convert other epialleles at the same locus on different chromosomes to their own state (left) or in cis where their epigenetic state is transmitted longitudinally between generations without affecting the other allele.
3.1. Epigenetic inheritance of the centromere
In monocentric organisms the correct segregation of chromosomes during cell division depends on the centromere. The centromere is a specialised chromatin structure, with a high degree of compaction of the DNA around modified nucleosome core particles in which histone H3 is replaced by a centromere-specific histone CenH3/CenpA [39]. The presence of CENH3 is essential for the attachment of the centromere to a multiprotein complex known as the kinetochore which links the chromosome to the spindle, enabling chromosome segregation in mitosis or meiosis.
The centromere has been proposed to be an example where epigenetic changes alone could drive evolution. In most organisms studied thus far the DNA sequence of the centromere consists of a number of repeats of varying complexity [40]. For example the human centromere is made up of varying numbers of repeats of the AT-rich alpha satellite sequence [41]. However, early observations showed that some cancer cells have formed an alternative centromere, known as a neocentromere, stably inherited through mitosis apparently without the presence of alpha satellite sequences [42]. Subsequently, many neocentromeres have been described either in naturally occurring or artificial manipulations in model organisms [43]. The formation of a neocentromere in many cases appears to be completely independent of DNA sequence changes. Indeed, ectopic loading of CENH3 artificially into a specific region of the DNA is sufficient to generate a neocentromere in Drosophila [44] Once formed, neocentromeres can persist for many generations, thus fulfilling the definition of an epigenetic driver [45]. Formation of a new centromere might have strong implications for evolution because it could lead to reproductive incompatibility and promote speciation [46].
Whilst these demonstrations show clearly that short term formation of a new centromere can be achieved epigenetically in the laboratory, it is less clear whether these processes explain how centromeres move on an evolutionary timescale. As pointed out above, natural centromeres have unusual and specific DNA sequences. Alpha-satellite repetitive DNA might accumulate at the centromere after its formation through epigenetic processes- indeed evolutionarily recent centromere repositioning is accompanied by accumulation of repetitive sequences [47]. Nevertheless, the DNA sequence itself may determine the initial site for centromere repositioning. Indeed, comparative analysis across primates indicates that new centromeres have formed repeatedly in similar chromosomal locations [48]. A recent study suggests that evolutionarily stable centromeres require formation of non B-DNA sequence, which can be caused by intrinsic sequence features such as possessed by the alpha satellite, or through sequence specific DNA binding proteins such as CENP-B [49]. Thus, whilst transient neocentromeres may be inherited epigenetically, these may be evolutionarily unstable and long-term centromere positioning may be determined predominantly by DNA sequence variation.
3.2. Epialleles in plants
Stable alleles with alternative epigenetic states have a rich history in plants. One well-studied case is paramutation, where one epiallele can convert another epiallele into the same state in trans (Fig. 1C). A good example of paramutation is the B1 locus in maize, discovered to exhibit non-Mendelian inheritance in 1959 [50]. Maize plants can exist as either darkly pigmented (B–I) or lightly pigmented (B’). F1 organisms from a cross between B–I and B’ maize are all light in colour (the B’ phenotype) as are F2 offspring from F1 crosses. Thus exposure of B–I alleles to B’ alleles results in permanent conversion of B–I to B’, a phenomenon known as paramutation [51]. Molecular characterisation of maize paramutation showed that B’ and B–I alleles have the same DNA sequence, but B’ alleles have higher methylation and much lower expression levels. The differences in methylation rely on small RNAs that map to seven tandem repeats upstream of the gene. The small RNAs are able to transmit the B’ phenotype to B–I alleles in trans [52]. The paramutation of the b1 locus in maize thus represents a classic example of an epigenetic driver allele, working independently of sequence variation to bring about phenotypic differences. Nevertheless, it is important to note that alleles of the b1 locus in maize that have only one copy of the tandem repeats are resistant to paramutation [53]. Thus even in this clear-cut case of an epigenetic driver there is a contribution of pre-existing DNA sequence polymorphisms to the paramutation phenomenon.
The evolutionary significance of b1 paramutation in maize is puzzling. The B’ allele is extremely stable. Moreover, B–I alleles are converted into new B’ alleles when in the heterozygous state and even plants homozygous for the B–I alleles spontaneously convert into B’ [54]. Although mathematical analysis indicates that there are parameter ranges within which paramutation processes can be stable [55], the relatively high rate of spontaneous B’ formation means it is somewhat surprising that any B–I alleles exist in the wild. It is possible that the B–I were formed relatively recently, for example through tandem repeat expansion and the B’ allele is currently sweeping through the population; alternatively loss of tandem repeats through unequal recombination might introduce a source of unsilenced alleles into the population [55]. Alternatively, the B–I allele might be formed under an unknown stress situation or promote an advantage under certain environmental conditions, maintaining it at low frequency within the population.
The fact that alleles subject to paramutation are unstable within natural populations perhaps explains why examples in plants are still relatively rare. An alternative type of epiallele is one that maintains its state independent of other epialleles (Fig. 1). These alleles show Mendelian inheritance and thus are easier to maintain in a polymorphic state within a population. Arabidopsis represents a good model to search for such epialleles due to the existence of high quality whole genome methylation maps and fully sequenced natural ecotypes from around the world (known as accessions). Moreover, there is clear experimental evidence in Arabidopsis that newly formed epialleles can be stably inherited through recombination even between divergent genotypes [56]. Recently, large-scale studies have characterised epigenome and genome variation across accessions [57,58]. These studies have revealed abundant epialleles- approximately 80% of cytosines in CpG context show differences in methylation status in at least one accession [57]. Interestingly, some epigenetic differences can be clearly linked to different environmental conditions suggesting that epigenetic variation may have a role in adaptation to the environment although there is no direct evidence for this as yet [59]. A conservative estimate of the fraction of effector epialleles can be obtained by searching for nearby sequence variation that is strongly associated with the presence of each specific epiallele. In a focused study of 150 accessions from Sweden, 45% of epigenetic variation could be clearly associated to cis-acting sequence differences and 21% associated to sequence differences in trans [59]. Much of these sequence differences are likely to be due to transposable element polymorphisms [57]. Given there are likely to be further effector alleles where the sequence differences are complex or difficult to map, this implies that driver epialleles that act independently of DNA sequence variation are rare.
Global studies estimate the overall contribution of DNA sequence variation to epigenomic variation but demonstrating that any particular epiallele is cis-acting requires more careful analysis. One approach is to generate recombinant inbred lines derived from a cross between two strains with different methylation status at the epiallele. With sufficient recombinants it is possible to rule out long-distance and trans effects of DNA sequence. Although driver epialleles may be the minority, a few cases have been robustly mapped. One particularly interesting example where this may have some significance for evolutionary processes was discovered by the Pikaard lab [60]. They discovered that crosses between two accessions led to lethality and mapped the cause of this to a genetic difference in a gene called HIS6A [60]. HIS6A is a developmental gene, which was absent in one but present in the other. Intriguingly, the expression of a paralogue, HIS6B was shown to compensate for the absence of HIS6A; however, HIS6B was epigenetically silenced in the accession in which HIS6A was present. The difference in silencing of his6B behaved as an epiallele, with expression status maintained in cis in the heterozygous state. As a result, recombinants carrying the silent HIS6B epiallele and the HIS6A deletion were inviable, explaining the hybrid incompatibility [60]. This example shows that some true driver epialleles may be evolutionarily and ecologically significant; however, it also demonstrates very clearly the close interplay between sequence and epigenetic information in determining phenotypes in evolution.
Epialleles in vertebrates.
The challenges of identifying naturally occurring epialleles that behave as drivers in vertebrate populations are even larger than in Arabidopsis due to the large size of the genome and the practical difficulty (or impossibility in most wild populations including human) of generating large numbers of recombinant inbred lines. Systematic approaches have been performed in mouse using a panel of inbred mouse strains. Bisulfite sequencing to map DNA methylation patterns across mouse strains concluded that the majority of epialleles are likely to be effectors of DNA sequence change as there was clear evidence for cis and trans acting sequence variation linked to the most epialleles [61]. A more direct approach to map epialleles in near-isogenic animals was recently performed using a single laboratory strain. Here, the authors identified regions of the genome where the epigenetic was particularly variable between individuals and then used breeding experiments to assess the heritability of these differences at a subset of loci. In five out of the six loci investigated there was no correlation between the level of methylation at these loci in the parents and the level in the offspring, with the variability thus being re-established each generation [62]. Together these studies suggest that driver epimutations are likely to be very rare in mouse.
Outside of model organisms there have been few genome-wide methylation studies with sufficient resolution to find potential epialleles with roles in adaptation to the environment. However, with the decreased cost of high-throughput sequencing these studies may become more frequent in future. One intriguing recent example concerns cavefish, which have recently transitioned into a light-limited environment accompanied by degeneration of their eyes. The degeneration is correlated to increased DNA methylation of several genes involved in eye development in cavefish relative to surface fish. Interestingly, treating cavefish with 5-Azacytidine to inhibit DNA methylation led to increased eye size [63]. Most of the genes involved show nearby sequence variation between cavefish and surface fish, which makes it difficult to argue that the epigenetic change is a driver for the phenotypic alteration rather than an effector [63]. Nevertheless, the example does show that gene expression changes with evolutionary significance can be linked to epigenetic alterations.
In human populations, there is much excitement in the possibility that naturally occurring epialleles could be important in human disease. It has even been suggested that epigenetic variation might be a solution to the famous “missing heritability” problem as it could provide heritable variation independent of sequence differences [98]. So far genome-wide studies of epigenetic variation do not support this possibility; studies attempting to disentangle the effects of genetic and epigenetic variation have shown that genetic variation mostly explains epigenetic variation and adding epigenetic variation as an independent factor does not improve estimates of heritability [64]. However the studies so far performed have been on a much smaller scale than studies focusing on genetic variants and it is therefore likely that more epigenetic variation will be uncovered. Another intriguing potential resource so far untapped for DNA methylation studies is trio studies, which could identify de novo silencing events with potential influences on phenotype.
Taken altogether, the results from genome-wide studies across wild populations across eukaryotes have yet to give clear evidence in favour of the driver model where epigenetic changes drive phenotypic changes (Fig. 1). Instead, the interplay of DNA sequence and epigenetic changes common to all these examples rather suggests that epigenetic differences are best considered to be effectors of DNA sequence variation. Studying epigenetic differences in natural populations offers a realistic perspective on the epigenetic and sequence changes that have occurred. However, the limitation of studies of existing variation within natural populations is that they do not give an indication of the dynamics of epiallele formation in relation to DNA sequence changes. As indicated from the examples above, if both sequence variation and epigenetic variation occur at a given locus, it is safest to assume that this indicates that the epigenetic changes are effectors of the DNA sequence variation. However, it is possible that the DNA sequence changes seen are either independent of the phenotype or, even more interestingly, occurred after the epigenetic change. For this reason, the clearest mechanistic insight into the contribution that epigenetic changes can make to evolution has come from the study of evolution in controlled laboratory settings.
3.3. Insights into epigenetic inheritance from evolution in the laboratory
Tremendous insight can come from sampling populations over several generations giving insight into evolutionary processes in real time. Such experiments are most conveniently performed using controlled populations in the laboratory using model organisms. Laboratory evolution experiments can be used to study how populations change in the absence of direct selection, and in response to a specific perturbation in their environment such as nutrient limitation. Several parallel experiments can be performed allowing estimation of the repeatability of epigenetic phenomena to be made [65]. Potentially, this approach offers a great advantage for studies of how epigenetics contributes to evolution. By starting from isogenic populations DNA sequence changes and epigenetic changes can be observed as they appear, allowing the sequence of molecular events (their “dynamics”) to be unravelled.
Studies of the dynamics of epigenetic changes during evolution experiments in the laboratory have been pioneered in Arabidopsis. Arabidopsis has a short lifecycle, making it feasible to follow multiple generations in the laboratory, and a small, fully assembled genome making it straightforward to obtain high coverage bisulfite sequencing data. Taking advantage of these features, a number of laboratories have adapted the classical mutation accumulation assay in order to study the rate and spectrum of epigenetic changes (“epimutations”). Mutation accumulation assays are performed by propagating several independent lines, each descended from an isogenic startic population, for multiple generations [66]. Crucially, the population is passed through a bottleneck every generation in which the population size is kept as low as possible by randomly selecting a small number of individuals. This means that there is virtually no purifying selection because unless a mutation is lethal or results in sterility it has an equal chance be propagated to the next generation [66]. These studies therefore allow a good estimate of the actual mutation rate to be made. Similarly, studying the DNA methylation changes in isogenic lines propagated under conditions of minima selection enables assessment of the actual epimutation rate in the absence of the effect of selection.
Several different epimutation accumulation experiments have been performed in Arabidopsis, each producing quantitatively similar estimates for the rate at which epimutation arises in the absence of selection [[67], [68], [69]]. The interpretation of these studies, however, varies perhaps due to whether one adopts a glass half full or half empty perspective. At the simplest level, an epimutation occurs when methylation is either gained or lost at a single cytosine in the CG, CHH or CHG context. Heritable epimutations were found largely in the CG context and were estimated to occur at a rate of around 1e-4 per base per generation [68,69]. Clearly this is much faster than the rate at which mutations arise, estimated at around 1e-8 per generation [70]. The epimutations observed are highly unlikely to be all linked to underlying DNA sequence changes. However, DNA sequence changes are extremely stable because the rate of back mutation is at usually at least 16-fold lower than the rate of a forward mutation and are usually ignored in modelling MA experiments [66]. Contrastingly, epimutations revert with a high frequency, and indeed were estimated to occur at a 3-fold higher frequency than forward epimuations [68]. The half-life of epimutations is therefore quite short. Thus, although epimutation leads to divergence within lines, this does not increase linearly over time but instead saturates, so the potential for epigenetic changes alone to contribute to long-term evolutionary divergence is limited [71]. Thus, whilst a positive interpretation of these studies suggests epimutations contribute to a dynamic epigenome the more negative perspective is that over the long term DNA sequence variation is likely to be the dominant source of evolutionary change. Consistent with this interpretation, studies comparing a carefully propagated North American Arabidopsis accession to its preserved ancestor confirmed largely stable methylation patterns over 100 years [72].
An important counterargument could be raised against the interpretation that epimutations at single CG sites are unlikely to be important for evolutionary processes. The assessment of stability of an individual epimutation at a CG locus assumes that all CGs act independently. However, DNA methylation status is tightly coupled to the surrounding chromatin environment. The global fast reversion rate of an epimutation at a CG therefore might reflect the fact that if any one CG loses or gains methylation it will be quite likely to be converted back due to the surrounding CG sequences that still carry the original methylation status. Epimutations therefore might act cooperatively, with the forward rate increasing as more surrounding CGs become methylated. Interestingly, a faster on-rate and slower off-rate were shown for methylation at individual CGs within TEs compared to those within genes [68]. It is therefore possible that larger regions exhibiting concerted epigenetic switching could be more stable [56] and therefore have a more imposing effect on long-term divergence.
MA lines are not ideal to investigate concerted epigenetic changes affecting large numbers of CG sites simultaneously because such regions are likely to form very rarely under MA conditions. However, as discussed above, epialleles that are found in natural populations are likely to be linked to nearby sequence changes. An alternative approach to study epialleles in isogenic backgrounds borrows another idea from classical laboratory evolution experiments. To generate sufficient diversity for laboratory evolution, “mutator” lines, which have an elevated mutation rate due to DNA repair defects, are often employed to introduce a lot of variation into the population very rapidly [73]. The variation can then be partitioned amongst different lines known as “Recombinant Inbred Lines” or RILs by crossing the mutator to the wild type and then inbreeding the offspring for multiple generations to make each line homozygous for a different set of mutations [74]. Similar approaches were employed in Arabidopsis to introduce epigenetic variation through disrupting key enzymes in the DNA methylation pathway [75]. The resulting lines show perturbation of 1000s of regions of the genome, forming “epialleles” where several CG sequences in the same region lose methylation. The "epimutator” lines are then crossed back to the wild type and inbred these lines for several generations to generate a set of epiRILs. epiRILs can then be propagated to study how stable the epialleles are [75].
In Arabidopsis, the most fruitful insights into the stability of epialleles came through using ddm1 (decrease in DNA methylation 1), a chromatin-binding protein required for maintenance of CG methylation [76], as an epimutator [77]. Whole-genome bisulfite sequencing of a subset of epiRILs was performed after one generation and 8 generations of selfing [78]. This demonstrated that while many regions of the genome converted back to the wild type epigenetic state, several regions of the genome remained differentially methylated in comparison to the wild type parent line after 8 generations [78]. Regions targeted by small RNAs tended to revert to a greater extent than those without small RNAs, thus suggesting a potential mechanism determining the stability of different epialleles [78,79]. This suggests strongly that some regions of the genome can indeed act cooperatively to preserve an alternative methylation state [80]. Interestingly, epiRILs were shown to have phenotypic differences, and these could even be mapped to different regions which were shown to have DNA methylation differences [77,80].
3.4. Epigenetic changes as drivers for cancer evolution
The capacity of blocks of CG sequences to behave cooperatively as an epimutation with relatively high stability has direct relevance to human disease, in particular cancer. Importantly, cancers divide rapidly meaning that any individual cancer may have gone through >100 somatic divisions at the point of diagnosis. Evolutionary processes such as natural selection and drift therefore operate within cancers [81]. Moreover, the somatic origin of most cancer types mean that they are not subject to processes such as genome-wide reprogramming which make transgenerational epigenetic inheritance rare in mammals [6]. Cancer cells show widespread alterations in epigenetic landscapes compared to their cell of origin (e.g. [82]. The complexity of the genomic changes that occur in cancer [83] mean that unravelling the contribution of sequence and epigenetic changes cannot yet be performed with any certainty. Nevertheless, there are a few tentative indications that epigenetic changes could act as drivers for evolutionary processes in cancer.
A classic example of a cancer-specific epimutation which might fulfil the criteria for a driver is biallelic methylation of the promoter of the MLH-1, which inactivates the mismatch repair pathway [84]. Approximately 10–20% of colon cancers with microsatellite instability are caused by this process with the remaining 50% due to more conventional mutational inactivation of the MLH-1 gene [85] Although mutations often occur in the silenced MLH-1 alleles, there is little evidence for common mutations between different MLH-1-silenced patients, suggesting that these mutations are likely passengers whilst the epigenetic silencing event itself is a driver event for tumour progression [85]. However, the silencing is strongly associated with activation of the oncogene BRAF [86], suggesting that although the methylation is a driver mutation, it is secondary to a different sequence change. In order for the MLH-1 epimutation to be defined as an epigenetic driver, it would have to be demonstrated that MLH-1 silencing can persist, once established, in the absence of oncogene activation, which has yet to be tested.
On a more general scale, a recent study used data from 25 different cancer types to uncover the variability in the methylome of different cancers. Intriguingly, the most hypervariable sites were strongly enriched for enhancer regions. The authors interpreted this as suggesting that changes in methylation at enhancers might promote the growth of cancers, and thus that enhancer methylation changes might be drivers for tumour progression in the absence of underlying sequence variation. However, these changes could also result from activation of an oncogene or tumour suppressor. Moreover, the interpretation that these promote tumour growth might have to be reevaluated in the light of recent work suggesting that the majority of mutational sites in cancer evolve neutrally rather than subject to either purifying selection or positive selection [87] although see [99], and thus paradoxically altered methylation status is more likely to be an indication of lack of function of the specific enhancer in the cancer tissue type. Nevertheless, the observation at least suggests that blocks with altered CG methylation can adopt different methylation status in a cooperative manner leading to a relatively stable epiallele formation.
Whilst studies of epigenetic changes in cancer are most advanced for DNA methylation, it is of course also possible that changes in other epigenetic features could occur in tandem or separately from DNA methylation alterations. Excitingly, recent experiments have used the ATAC-Seq method in order to study changes in global chromatin accessibility in fixed cancer samples [88], offering the possibility of identifying similar epimutations in enhancers associated with changes in chromatin structure such as histone modification, point mutations or nucleosome positioning.
As described for Arabidopsis for epimutations to fulfil the criteria of driver mutations it is crucial that there be a process whereby such changes can occur spontaneously in normal cells, without being caused directly through the activation of other oncogenic processes. One possibility is that epimutations arise through the process of cell division. Coordinated progression of the replication fork, histone assembly onto newly synthesised DNA and DNA methylation is required in order to reestablish epigenetic states after cell division and replication stress can interfere with this process [89,90]. Indeed, interrupting the progression of the replication fork by structured DNA or DNA damage can lead to loss of epigenetic stability and aberrant gene expression which alters the epigenetic state for several generations [91]. This is a plausible mechanism whereby an epiallele could arise spontaneously in a cancer cell independently of either sequence changes at the locus or activation of oncogenes elsewhere in the cell [92]. However, the rate at which such changes occur at endogenous loci in isogenic lines has yet to be established and would be a key criteria for testing this model’s applicability to epimutation accumulation in cancer.
4. Epigenetic drivers of mutation
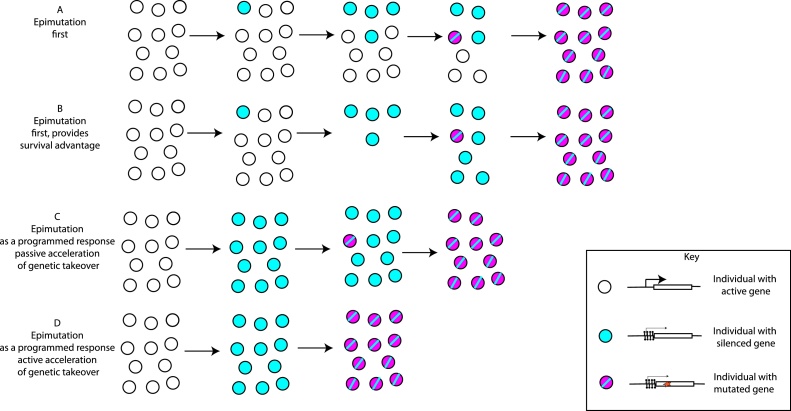
Even if epigenetic changes themselves are too unstable to drive evolutionary processes in the absence of sequence changes, epigenetic inheritance could still play a role in evolution through influencing how sequence variation is formed. This idea comes under the general heading of “genetic takeover” whereby a change that exists in a population as a purely epigenetic phenomenon might be cemented as a genetic change, at which point the epigenetic effect is either no longer required or is stabilised by the genetic alteration [38]. Essentially these ideas are molecular versions of the old concept of canalization proposed by Waddington [1]. Underpinning this broad concept however there are several subtly different scenarios, in which the contribution that the epigenetic change varies (Fig. 2).
Fig. 2.
- An illustration of how epigenetic inheritance could affect DNA sequence evolution. This hypothetical example follows an epimutation caused by acquisition of DNA methylation that leads to silencing of a gene. The phenotypic effect of the inactivation of the gene is beneficial under certain environmental conditions that the population is exposed to. This example is for illustration only; the same process could apply to other types of epimutation with different molecular effects.
A- An epimutation arises in the population due to stochastic fluctuations (i.e. not promoted directly by environmental conditions). The effect of the epimutation is to silence a gene. Although it has a beneficial effect, the epimutation is unstable, because the same stochastic fluctuations can lead to the loss of the epimutation in individuals. However eventually a genetic mutation that inactivates the gene arises in the population. This has the same advantageous effect as the epimutation but is more stable and therefore eventually fixes in the population.
B- An epimutation that silences a gene arises in the population due to stochastic fluctuations as in A, but this time the epimutation provides a survival advantage under adverse conditions. Thus, although the epimutation is still unstable, the survival advantage that it poses gives enough time for the genetic mutation with the same effect to arise by chance and go to fixation within the population.
C- The epimutation that silences a gene occurs as a programmed response to the environment, thus occurs in every individual. Whilst the epimutation is unstable, this has the effect of accelerating the fixation of the inactivating mutation, which has the same effect as the epimutation because the population size remains high.
D- The epimutation that silences a gene occurs as a programmed response to the environment as in C. However in contrast to C, the epimutation has the effect of increasing the mutation rate at the same locus, through the fact that cytosine methylation increases the C–T transition rate (see text for molecular details of this effect). Thus the inactivating DNA sequence mutation occurs even more rapidly within the population than in C and then fixes in the population.
In the simplest model of genetic takeover an epimutation occurs in a population at low frequency in the absence of a sequence change. However, instead of going to fixation as proposed for the “driver epimutation” hypothesis above, the epimutation leads to a transient advantage due to its limited stability. Eventually, a mutation arises in an individual possessing the epimutation which has the same effect as the epimutation. This mutation is more stable and then fixes in the population.
In the simplest version of this hypothesis the epimutation itself has little effect on the dynamics of evolution. This is because it makes no difference whether the mutation arises in an individual with the epimutation or without it. However, a slightly more elaborate version of this model considers a very strong environmental stress, such that all individuals without the epimutation are killed. The epimutation, although transient, then enables the population to survive long enough for the mutation that drives the change to occur.
One further elaboration to the model places the epimutation even more centrally. In this model the epimutation occurs in most individuals as a programmed response to the environmental change. The epimutation allows most individuals in the population to survive thereby speeding up the long-term adaptive process by keeping the population size large so that there is a higher probability that the mutation arises.
A final extension to these scenarios proposes that epimutation directly promotes mutation of the underlying DNA sequence. Acquisition of epigenetic silencing of a gene could lead to an increased rate of mutation of that same gene, hence directly promoting the process of genetic takeover. Such a scenario would be expected to produce the most rapid evolutionary change, but of course requires a clear mechanism linking epigenetic changes to changes in mutation rate.
One very clear way in which an epigenetic change might lead to increased mutation rate is through DNA methylation. Acquisition of DNA methylation in the formation of an epiallele might be expected to increase the rate of DNA sequence polymorphism within the region. Cytosine deamination is one of the most abundant sources of spontaneous DNA damage. Unmethylated cytosine is converted to uracil, which, as uracil is not one of the four canonical DNA bases, is a substrate for the Uracil-DNA glycosylases and targeted for repair [93]. However, methylated cytosine deaminates to a T, which is less easily recognised. Though specialised mechanisms to remove the T-G mismatch do exist [94], they are less efficient and deamination of methylated C can be shown to be the most prominent source of mutations in mammalian genomes [83].
Chromatin environment also affects mutation rate. Studies using cancer genomes showed that the mutation frequency varies by ˜10-fold between different chromatin environments, with repressed chromatin domains highest and active lowest [95]. Clever analyses determined that a significant component of this effect is the mismatch repair system [96], and further analysis suggested a mechanistic explanation through recruitment of mismatch repair to active genes preferentially by the interaction between MSH-2 and H3K36me3 [97]. Thus DNA repair is less efficient in repressed regions of the genome.
Taken together then, formation of an epiallele might lead to an increase in mutation rate. However, whether this process is sufficient to enable it to promote DNA sequence change in wild populations or even in cancer cells is still open to question. The increases in mutation rate although strong, still mean that the rate at which inactivating mutations would arise within a silent epiallele is still lower than the reversion rate of the epimutation. The rapid division and increased genomic instability in cancer might make it a more realistic target for epigenetically driven sequence change. It will be interesting to compare the rate of acquisition of inactivating mutations in epigenetically silenced tumour supressors to test this hypothesis further. At present however, the idea of promotion of DNA sequence change by epigenetic switching must remain speculative.
5. Conclusion
Future work investigating short-term dynamics of adaptation in natural populations may yet implicate an early role for epigenetics in evolutionary change. Nevertheless, so far the molecular studies discussed in this article suggest that transgenerational epigenetic changes in the absence of sequence change may be at best a minor contributor to short term adaptation and long-term evolution.
Based on this conclusion one can turn the question on its head to ask why it is that epigenetic changes do not have more of a significant role in evolution. The existence of epialleles, both in wild and laboratory situations shows that they are mechanistically possible. Yet globally, epigenetic changes are transient. An interesting possibility therefore is that organisms have evolved mechanisms in order to prevent epigenetic changes from becoming fixed in populations. The ability of epigenetic mechanisms to respond directly to the environment might be dangerous if their activities were to result in fixed gene expression differences for subsequent generations. This possibility is supported by the fact that some mutants in C. elegans show improved inheritance of epigenetic information [32,33], albeit so far restricted to artificial situations. Perhaps the existence of specialised mechanisms to restrict transmission of epigenetic changes might indicate that organisms could regulate transgenerational epigenetic memory under particular conditions in order to enable rapid adaptation and fixation of epigenetics within the population. Such a model would also imply that there should be variability across organisms in their reliance on epigenetic inheritance to drive variation.
Even if natural populations turn out not to use epigenetic changes alone, it remains an important and understudied possibility that epigenetic changes could drive the development of cancer. Studying this is extremely difficult given the high degree of genomic instability in cancer and the difficulty in distinguishing driver from passenger mutations. Nevertheless, these studies could be very important as the plastic nature of epigenetic changes make them more easily reversible, for example by drug treatment, compared to DNA mutations. Rapidly growing availability of techniques to assess chromatin structure in small numbers of cells make this an attractive arena for future advances.
Acknowledgements
The author thanks members of the Transgenerational Epigenetic Inheritance and Evolution lab (MRC London Institute of Medical Sciences), Matthias Merkenshlager, Wendy Bickmore, Madan Babu and Ben Lehner for helpful discussion on this topic. The author wishes to apologise for any relevant work that could not be cited for space reasons.
References
- 1.Waddington C.H. Canalization of development and the inheritance of acquired characteristics. Nature. 1942;150:563–565. [Google Scholar]
- 2.Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman P.D., Rando O.J. Chromatin as a potential carrier of heritable information. Curr. Opin. Cell Biol. 2010;22:284–290. doi: 10.1016/j.ceb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bühler M. RNA turnover and chromatin-dependent gene silencing. Chromosoma. 2009;118:141–151. doi: 10.1007/s00412-008-0195-z. [DOI] [PubMed] [Google Scholar]
- 5.Harvey Z.H., Chen Y., Jarosz D.F. Protein-based inheritance: epigenetics beyond the chromosome. Mol. Cell. 2018;69:195–202. doi: 10.1016/j.molcel.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heard E., Martienssen R.A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jimenez-Chillaron J.C., Isganaitis E., Charalambous M., Gesta S., Pentinat-Pelegrin T., Faucette R.R., Otis J.P., Chow A., Diaz R., Ferguson-Smith A. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radford E.J., Ito M., Shi H., Corish J.A., Yamazawa K., Peters A.H.F.M., Patti M., Ferguson-smith A.C. In utero undernourishment perturbs the adult sperm methylome and is linked to metabolic disease transmission. Science. 2015;345:1–26. [Google Scholar]
- 9.Nashun B., Hill P.W.S., Hajkova P. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 2015;34:1296–1308. doi: 10.15252/embj.201490649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hales C.N., Barker D.J.P. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 12.Vaag A.A., Grunnet L.G., Arora G.P., Brøns C. The thrifty phenotype hypothesis revisited. Diabetologia. 2012;55:2085–2088. doi: 10.1007/s00125-012-2589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S., Slagboom P.E., Lumey L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 15.Carone B.R., Fauquier L., Habib N., Shea J.M., Hart C.E., Li R., Bock C., Li C., Gu H., Zamore P.D. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conine C.C., Sun F., Song L., Rivera-Pérez J.A., Rando O.J. Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell. 2018;46:470–480. doi: 10.1016/j.devcel.2018.06.024. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma U., Sun F., Conine C.C., Reichholf B., Kukreja S., Herzog V.A., Ameres S.L., Rando O.J. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell. 2018;46:481–494. doi: 10.1016/j.devcel.2018.06.023. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J., Farinelli L., Miska E., Mansuy I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gapp K., van Steenwyk G., Germain P.L., Matsushima W., Rudolph K.L.M., Manuella F., Roszkowski M., Vernaz G., Ghosh T., Pelczar P. Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol. Psychiatry. 2018:1–13. doi: 10.1038/s41380-018-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy D.M., Morgan T.J., Lowe S.E., Williamson M.J., Spencer T.J., Biederman J., Bhide P.G. Nicotine exposure of male mice produces behavioral impairment in multiple generations of descendants. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C., Doebley A.L., Goldstein L.D., Lehrbach N.J., Le Pen J. PiRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirayama M., Seth M., Lee H.-C., Gu W., Ishidate T., Conte D., Mello C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagijn M.P., Goldstein L.D., Sapetschnig A., Weick E.M., Bouasker S., Lehrbach N.J., Simard M.J., Miska E.A. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gent J.I., Lamm A.T., Pavelec D.M., Maniar J.M., Parameswaran P., Tao L., Kennedy S., Fire A.Z. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sapetschnig A., Sarkies P., Lehrbach N.J., Miska E.A. Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luteijn M.J., van Bergeijk P., Kaaij L.J.T., Almeida M.V., Roovers E.F., Berezikov E., Ketting R.F. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conine C.C., Moresco J.J., Gu W., Shirayama M., Conte D., Yates J.R., Mello C.C. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell. 2013;155:1532–1544. doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wedeles C., Wu M., Claycomb J. Protection of germline gene expression by the C. elegans argonaute CSR-1. Dev. Cell. 2013;27:664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Houri-Ze’evi L., Korem Y., Sheftel H., Faigenbloom L., Toker I.A., Dagan Y., Awad L., Degani L., Alon U., Rechavi O. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 32.Lev I., Seroussi U., Gingold H., Bril R., Anava S., Rechavi O. MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 2017;27:1138–1147. doi: 10.1016/j.cub.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Perales R., Pagano D., Wan G., Fields B.D., Saltzman A.L., Kennedy S.G. Transgenerational epigenetic inheritance is negatively regulated by the HERI-1 chromodomain protein. Genetics. 2018;301456 doi: 10.1534/genetics.118.301456. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon M., Sarkies P., Ikegami K., Doebley A.L., Goldstein L.D., Mitchell J., Sakaguchi A., Miska E.A., Ahmed S. Reduced Insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans piwi mutants. Cell Rep. 2014;7:762–773. doi: 10.1016/j.celrep.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seth M., Shirayama M., Gu W., Ishidate T., Conte D., Mello C. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rechavi O., Houri-Ze’Evi L., Anava S., Goh W.S.S., Kerk S.Y., Hannon G.J., Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belicard T., Jareosettasin P., Sarkies P. The piRNA pathway responds to environmental signals to establish intergenerational adaptation to stress. BMC Biol. 2018;16:1–14. doi: 10.1186/s12915-018-0571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burggren W. Epigenetic inheritance and its role in evolutionary biology: re-evaluation and new perspectives. Biology (Basel) 2016;5:24. doi: 10.3390/biology5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner F.A., Henikoff S. Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev. 2015;31:28–35. doi: 10.1016/j.gde.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Henikoff S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 41.Wevrick R., Willard H.F. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc. Natl. Acad. Sci. U. S. A. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sart Ddu, Cancilla M.R., Earle E., Mao J., Saffery R., Tainton K.M., Kalitsis P., Martyn J., Barry A.E., Choo K.H.A. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 43.Burrack L.S., Berman J. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res. 2012;20:607–619. doi: 10.1007/s10577-012-9296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fulop S., Schepers A., Mendiburo M.J., Heun P., Padeken J. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 45.Tyler-Smith C., Gimelli G., Giglio S., Floridia G., Pandya A., Terzoli G., Warburton P.E., Earnshaw W.C., Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kursel L.E., Malik H.S. The cellular mechanisms and consequences of centromere drive. Curr. Opin. Cell Biol. 2018;52:58–65. doi: 10.1016/j.ceb.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eder V., Ventura M., Ianigro M., Teti M., Rocchi M., Archidiacono N. Chromosome 6 phylogeny in primates and centromere repositioning. Mol. Biol. Evol. 2003;20:1506–1512. doi: 10.1093/molbev/msg165. [DOI] [PubMed] [Google Scholar]
- 48.Ventura M., Weigl S., Carbone L., Cardone M.F., Misceo D., Teti M., D’Addabbo P., Wandall A., Björck E., de Jong P.J. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasinathan S., Henikoff S. Non-B-Form DNA is enriched at centromeres. Mol. Biol. Evol. 2018;35:949–962. doi: 10.1093/molbev/msy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coe E.H. A regular and continuing conversion-type phenomenon at the B locus in maize. Proc. Natl. Acad. Sci. U. S. A. 1959;45:828–832. doi: 10.1073/pnas.45.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arteaga-Vazquez M.A., Chandler V.L. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr. Opin. Genet. Dev. 2010;20:156–163. doi: 10.1016/j.gde.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arteaga-Vazquez M., Sidorenko L., Rabanal F.A., Shrivistava R., Nobuta K., Green P.J., Meyers B.C., Chandler V.L. RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12986–12991. doi: 10.1073/pnas.1007972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belele C.L., Sidorenko L., Stam M., Bader R., Arteaga-Vazquez M.A., Chandler V.L. Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandler V.L. Paramutation’s properties and puzzles. Science. 2010;330:628–629. doi: 10.1126/science.1191044. [DOI] [PubMed] [Google Scholar]
- 55.Geoghegan J.L., Spencer H.G. The evolutionary potential of paramutation: a population-epigenetic model. Theor. Popul. Biol. 2013;88:9–19. doi: 10.1016/j.tpb.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Hofmeister B.T., Lee K., Rohr N.A., Hall D.W., Schmitz R.J. Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation. Genome Biol. 2017;18:155. doi: 10.1186/s13059-017-1288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawakatsu T., Huang S.-S.C., Jupe F., Sasaki E., Schmitz R.J., Urich M.A., Castanon R., Nery J.R., Barragan C., He Y. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166:492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitz R.J., Schultz M.D., Urich M.A., Nery J.R., Pelizzola M., Libiger O., Alix A., McCosh R.B., Chen H., Schork N.J. Patterns of population epigenomic diversity. Nature. 2013;495:193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubin M.J., Zhang P., Meng D., Remigereau M.-S., Osborne E.J., Casale F.P., Drewe P., Kahles A., Voronin V., Song Q. DNA methylation variation in Arabidopsis has a genetic basis and appears to be involved in local adaptation. Elife. 2015;4:1–23. doi: 10.7554/eLife.05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blevins T., Wang J., Pflieger D., Pontvianne F., Pikaard C.S. Hybrid incompatibility caused by an epiallele. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3702–3707. doi: 10.1073/pnas.1700368114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orozco L.D., Morselli M., Rubbi L., Guo W., Go J., Shi H., Lopez D., Furlotte N.A., Bennett B.J., Farber C.R. Epigenome-wide association of liver methylation patterns and complex metabolic traits in mice. Cell Metab. 2015;21:905–917. doi: 10.1016/j.cmet.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kazachenka A., Bertozzi T.M., Sjoberg-Herrera M.K., Walker N., Gardner J., Gunning R., Pahita E., Adams S., Adams D., Ferguson-Smith A.C. Identification, characterization, and heritability of murine metastable epialleles: implications for non-genetic inheritance. Cell. 2018;175:1259–1271. doi: 10.1016/j.cell.2018.09.043. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gore A.V., Tomins K.A., Iben J., Ma L., Castranova D., Davis A.E., Parkhurst A., Jeffery W.R., Weinstein B.M. An epigenetic mechanism for cavefish eye degeneration. Nat. Ecol. Evol. 2018;2:1155–1160. doi: 10.1038/s41559-018-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahsan M., Ek W.E., Rask-Andersen M., Karlsson T., Lind-Thomsen A., Enroth S., Gyllensten U., Johansson Å. The relative contribution of DNA methylation and genetic variants on protein biomarkers for human diseases. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blount Z.D., Lenski R.E., Losos J.B. Contingency and determinism in evolution: replaying life’s tape. Science. 2018;362 doi: 10.1126/science.aam5979. eaam5979. [DOI] [PubMed] [Google Scholar]
- 66.Baer C.F., Miyamoto M.M., Denver D.R. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- 67.Becker C., Hagmann J., Müller J., Koenig D., Stegle O., Borgwardt K., Weigel D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 68.van der Graaf A., Wardenaar R., Neumann D.A., Taudt A., Shaw R.G., Jansen R.C., Schmitz R.J., Colomé-Tatché M., Johannes F. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. 2015;112:6676–6681. doi: 10.1073/pnas.1424254112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmitz R.J., Schultz M.D., Lewsey M.G., O’Malley R.C., Urich M.A., Libiger O., Schork N.J., Ecker J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidalis A., Živković D., Wardenaar R., Roquis D., Tellier A., Johannes F. Methylome evolution in plants. Genome Biol. 2016;17:264. doi: 10.1186/s13059-016-1127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hagmann J., Becker C., Müller J., Stegle O., Meyer R.C., Wang G., Schneeberger K., Fitz J., Altmann T., Bergelson J. Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeyl C., Arjan J., DeVisser G.M. Estimates of the rate and distribution of fitness effects of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 2001;157:53–61. doi: 10.1093/genetics/157.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gray J.C., Cutter A.D. Mainstreaming Caenorhabditis elegans in experimental evolution. Proc. Biol. Sci. 2014;281 doi: 10.1098/rspb.2013.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quadrana L., Colot V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016;50:467–491. doi: 10.1146/annurev-genet-120215-035254. [DOI] [PubMed] [Google Scholar]
- 76.Vongs A., Kakutani T., Martienssen R.A., Richards E.J., Boccara M., Ciaudo C., Cruaud C., Poulain J., Berdasco M., Fraga M.F. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 77.Johannes F., Porcher E., Teixeira F.K., Saliba-Colombani V., Simon M., Agier N., Bulski A., Albuisson J., Heredia F., Audigier P. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ito T., Tarutani Y., To T.K., Kassam M., Duvernois-Berthet E., Cortijo S., Takashima K., Saze H., Toyoda A., Fujiyama A. Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Teixeira F.K., Heredia F., Sarazin A., Roudier F., Boccara M., Ciaudo C., Cruaud C., Poulain J., Berdasco M., Fraga M.F. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 80.Cortijo S., Wardenaar R., Colomé-Tatché M., Gilly A., Etcheverry M., Labadie K., Caillieux E., Hospital F., Aury J.-M., Wincker P. Mapping the epigenetic basis of complex traits. Science. 2014;343:1145–1148. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- 81.Greaves M.F., Maley C.C. Clonal evolution in cancer. Nature. 2012;418:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen B., Wu H., Shinkai Y., Irizarry R.A., Feinberg A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alexandrov L.B., Jones P.H., Wedge D.C., Sale J.E., Campbell P.J., Nik-Zainal S., Stratton M.R. Clock-like mutational processes in human somatic cells. Nat. Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cunningham J.M., Christensen E.R., Tester D.J., Kim C.Y., Roche P.C., Burgart L.J., Thibodeau S.N. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 85.Wong J.J.L., Hawkins N.J., Ward R.L. Colorectal cancer: a model for epigenetic tumorigenesis. Gut. 2007;56:140–148. doi: 10.1136/gut.2005.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oliveira C., Pinto M., Duval A., Brennetot C., Domingo E., Espín E., Armengol M., Yamamoto H., Hamelin R., Seruca R. BRAF mutations characterize colon but not gastric cancer with mismatch repair deficiency. Oncogene. 2003;22:9192–9196. doi: 10.1038/sj.onc.1207061. [DOI] [PubMed] [Google Scholar]
- 87.Williams M.J., Werner B., Barnes C.P., Graham T.A., Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Corces M.R., Granja J.M., Shams S., Louie B.H., Seoane J.A., Zhou W., Silva T.C., Groeneveld C., Wong C.K., Cho S.W. The chromatin accessibility landscape of primary human cancers. Science. 2018;362 doi: 10.1126/science.aav1898. eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Groth A., Corpet A., Cook A.J.L., Roche D., Bartek J., Lukas J., Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 90.Jasencakova Z., Scharf A.N.D., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 91.Sarkies P., Reams C., Simpson L.J., Sale J.E. Epigenetic instability due to defective replication of structured DNA. Mol. Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sarkies P., Sale J.E. Cellular epigenetic stability and cancer. Trends Genet. 2012;28:118–127. doi: 10.1016/j.tig.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 93.Visnes T., Doseth B., Pettersen H.S., Hagen L., Sousa M.M.L., Akbari M., Otterlei M., Kavli B., Slupphaug G., Krokan H.E. Uracil in DNA and its processing by different DNA glycosylases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:563–568. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moréra S., Grin I., Vigouroux A., Couvé S., Henriot V., Saparbaev M., Ishchenko A.A. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 2012;40:9917–9926. doi: 10.1093/nar/gks714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schuster-Böckler B., Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 96.Supek F., Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frigola J., Sabarinathan R., Mularoni L., Muiños F., Gonzalez-Perez A., López-Bigas N. Reduced mutation rate in exons due to differential mismatch repair. Nat. Genet. 2017;49:1684–1692. doi: 10.1038/ng.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Banta J., Richards C. Quantitative epigenetics and evolution. Heriditary. 2018;121:210–224. doi: 10.1038/s41437-018-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tarabichi M., Martincorena I., Gerstung M., Leroi A.M., Markowetz F., The Evolution P.C.A.W.G., Heterogenity Working Group, Spellman P.T., Morris Q.D., Liniaerde O.C., Wedge D.C., Van Loo P. Neutral tumour evolution? Nat. Genet. 2018;50:1630–1633. doi: 10.1038/s41588-018-0258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]