Abstract
Background
Active immunotherapy is an effective, long-lasting, cheap, and safe approach to suppress cancer progression; however, the key issue is to develop appropriate tumour vaccines. Oncoproteins are up-regulated under various stress conditions and promote cell survival. Oncoproteins and their immunogenic domains could serve well as tumour vaccines and prime the hosts’ active anti-tumour immunity.
Methods
Proteomic and bioinformatic analyses were performed to identify potential tumour associated antigens (TAAs). Then, peptides derived from CD151 were designed and synthesized according to the major histocompatibility complex (MHC) I binding and immunogenicity. Cytotoxicity assay, flow cytometry, immunohistochemistry, and in vivo bioluminescence imaging were performed to assess the active anti-tumour immunity triggered by CD151 peptides in H22 primary hepatoma and experimental 4T1 breast cancer lung metastasis models.
Findings
CD151 was identified as an ideal TAA based on proteomic and bioinformatic analyses. CD151 peptides as tumour vaccines triggered active anti-tumour immunity against H22 hepatoma and the lung metastasis of 4T1 breast cancer in two mouse models through the activation of CD8+IFNγ+ lymphocytes and the subsequent targeted cytotoxicity. Further, the peptides suppressed the negative regulators, myeloid-derived suppressor cells. Survival was prolonged for mice with lung metastases from CD151 peptide-immunised groups.
Interpretation
The up-regulated oncoproteins in 8 Gy-irradiated tumour cells are good candidates for designing immunogenic peptides as tumour vaccines. Anti-tumour active immunity primed by peptides from CD151 may be an effective and safe approach to suppress cancer progression.
Keywords: Tetraspanin CD151, Oncoprotein, Irradiation, Peptide vaccine, Active immunotherapy
Abbreviations: TAA, tumour associated antigen; IR, irradiation; TCGA, The Cancer Genome Atlas; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; DLBCL, Diffuse large B-cell lymphoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, brain low-grade glioma; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; THYM, thymoma; CARs, Chimaeric antigen receptors; MHC, major histocompatibility complex; CAR, chimeric antigen receptor; FCM, flow cytometry; IFN, interferon; LDH, lactate dehydrogenase; IHC, immunohistochemistry; MDSC, myeloid-derived suppressor cell; THPA, The Human Protein Atlas
Research in context
Evidence before this study
Active immunity generated by potent vaccines is effective against deadly infectious diseases. Lack of tumour vaccines is related to the difficulties in the discovery of tumour-specific antigens. An alternative is to use tumour-associated antigens, i.e. oncoproteins, to suppress cancer progression.
Added value of this study
We used 8 Gy irradiation to upregulate the expression of a panel of oncoproteins, which were involved in tumour cell survival, anti-apoptosis, metastasis, and angiogenesis. Of these, CD151 expression was significantly higher in tumours than in normal tissues and associated with low differentiation, late stage, and poor prognosis. Two peptides derived from CD151 triggered active immune responses by enhancing CD8+IFNγ+ lymphocytes, which were cytotoxic against both H22 hepatoma cells and 4T1 breast cancer cells in vitro and suppressed the primary tumour or lung metastasis in vivo.
Implications of all the available evidence
Anti-tumour active immunity primed by peptides from oncoproteins/TAAs could serve as an effective, long-lasting, cheap, and safe way to suppress cancer progression.
1. Introduction
Immunotherapy has received tremendous attention for the treatment of cancer [1]. Antibodies and immune checkpoint inhibitors are revolutionising cancer treatment [2]. However, only a few patients exhibit objective tumour responses with long-term survival benefits [3]. One of the mechanisms for treatment failure may involve the downregulation or loss of antigen presentation, which confers tumour cells the ability to become ‘invisible’ and avoid immune attack [4]. Tumour-associated antigens (TAAs) are regarded as foreign antigens, which contribute to immune recognition and trigger anti-tumour immune responses [5]. TAA-derived peptide vaccines are potential tumour immunotherapy regimens which elicit endogenous tumour-specific T-cell responses and have been proved to provide clinical benefits with low toxicity in many phase I and II clinical trials [6], [7], [8], [9], [10]. However, effective TAA-based peptide vaccines are limited. Therefore, a novel and effective cancer peptide vaccine may increase immunotherapy strategies and sensitise patients to the effects of immune checkpoint inhibitors.
Functionally, most TAAs could promote tumour malignant behaviour and hence are described as oncoproteins [11]. At the early stage of cancer, oncoproteins (such as over-expressed Her2, EGFR, Bcl2, etc.) could enhance the proliferation of cells and suppress apoptosis, leading to uncontrolled cell growth [12,13]. At the progression stage, more oncoproteins (such as over-expressed VEGF, HGF, c-MET, ICAM, etc.) are involved in angiogenesis, vascularization, invasion, migration, and metastasis [14], [15], [16]. During chemotherapy and radiotherapy, the stress from drug and radiation stimulates a variety of oncoproteins via the activation of signaling cascades (such as PI3K/AKT/mTOR, NF-κB, etc.); these oncoproteins regulate cell proliferation, survival, and metabolism against the treatment-induced stress [17,18]. Hence, oncoproteins and related pathways are the targets of anti-tumour drugs.
While drug companies are spending billions to develop blockers of oncoproteins and related pathways, we believe that oncoproteins as vaccines is an effective, long-lasting, cheap, and safe strategy to ‘wake up’ the host immune system against cancer progression.
In the present study, we employ proteomic and bioinformatic approaches to show that 8 Gy irradiation (IR) up-regulated a panel of oncoproteins in cancer cells; these oncoproteins may serve as IR-induced endogenous ‘tumour vaccines’ and regulate immune responses. Of these, the tetraspanin oncoprotein CD151 was significantly overexpressed in several tumour cell lines and tumour tissues as compared to the adjacent normal tissues. CD151 is an oncoprotein that plays an important role in cancer progression and is associated with poor prognosis [19,20]. To suppress the malignant functions of CD151, we used the corresponding peptides to activate the host immunity against the progression of both primary 4T1 breast tumour growth and lung metastasis in mouse models. Immunisation with CD151 peptide vaccines activated CD8+ interferon (IFN)-γ+ lymphocytes, killed cancer cells, reduced the negative regulators myeloid-derived suppressor cells (MDSCs), and prolonged the survival of tumour-bearing mice. Our results indicate that CD151 peptides triggered active immune responses against tumour progression.
2. Materials and methods
2.1. Label-free quantitative proteomics for IR-induced differentially expressed proteins
After 8 Gy IR, the lysate from H22 cells was collected at 48 h. Label-free quantitative proteomics was performed to screen differentially expressed proteins using the service of Allied Protein Technology Co. Ltd. (China).
2.2. Clinical database analysis to reveal the association of CD151 with tumour malignant behaviour
The expression data of CD151 in different types of cancers were obtained from Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) [21], and Cancer Cell Line Encyclopedia (CCLE; https://portals.broadinstitute.org/ccle/about) [22].
The gene expression data and clinical information of hepatoma were obtained from The Human Protein Atlas (THPA; https://www.proteinatlas.org) [23], and The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/). In total, 374 tumour tissue and 50 adjacent normal tissues were obtained, and analysed by R software. This project was conducted in accordance with the guidelines provided by the TCGA (http://cancergenome.nih.gov/publications/publicationguidelines).
2.3. Design of CD151 peptides for priming active immunity
To improve the efficacy of CD151 peptide vaccines, two databases (SYFPEITHI: http://www.syfpeithi.com/ and IEDB: http://www.iedb.org/) were used to design CD151 peptides. SYFPEITHI is a database for T-cell epitope prediction [24]. The prediction of T-cell epitopes is based on published motifs derived from pool sequencing and analysis of individual natural ligands, and especially considers amino acids in anchor and auxiliary anchor positions, as well as other frequent amino acids. CD151 peptide1 is designed based on SYFPEITHI database. IEDB is another popular database for peptide vaccine design, containing information on immune epitopes [25]. CD151 peptide2 is designed based on IEDB database. The amino acid sequences of the two peptides were: N’-IYKVEGGCI-C’ and N’-DWQDSEWIRSG-C’. They were synthesised with N-terminal acetylation and C-terminal amidation to prevent degradation by peptidases using the service of Top-peptide Biotech LLC (Shanghai, China).
2.4. Immunisation with CD151 peptides
Female ICR or BABL/c mice (22–25 g, Slaccas Experimental Animal LLC, license# SCXK 2012–0002, Shanghai, China) were randomly divided into two groups as follows: (1) phosphate-buffered saline [PBS]-Freund's adjuvant-immunised group (n = 10); and (2) CD151 peptide-immunised group (n = 10). In brief, 200 μg of each peptide was dissolved in 0.1 mL PBS, mixed, emulsified with an equal volume of Freund's complete adjuvant, and subcutaneously injected into the four feet and eight spots on the back of each mouse [26]. PBS with an equal volume of Freund's adjuvant was used as the control group. On day 10 and 20 after first priming, the mice were boosted with the second and third immunisation with PBS or peptide mixed with Freund's incomplete adjuvant.
Animal experiments were approved by Fujian Medical University Institutional Animal Ethical Committee (FJMU IACUC #2018–075). The applicable institutional guidelines for the care and use of animals were followed.
2.5. Flow cytometry (FCM) analysis for CD8+IFNγ+ t lymphocytes
On day 24 after immunisation, some mice were sacrificed. Splenic lymphocytes from control and CD151 peptide-immunised mice were harvested with Ficoll separation method. After stimulation with CD151 peptides (10 μg/mL), lymphocytes were blocked with 10 μg/mL brefeldin A (Beyotime, Cat #S1536, China) for 2 h and stained with allophycocyanin (APC)-anti-mouse CD8 (Biolegend, Cat #100712, San Diego, CA) for 1 h. The cells were centrifuged at 350 × g for 5 min, fixed with 4% paraformaldehyde-PBS for 15 min, permeabilised with 0.1% Triton-PBS, spin-washed, and stained with phycoerythrin (PE)-anti-mouse IFNγ (Biolegend, Cat #505808, San Diego, CA) for 1 h at 4 °C. The double-stained CD8+IFNγ+ T lymphocytes were analysed with BD Accuri™ C6 Flow Cytometer (BD Inc, Piscataway, NJ).
2.6. Cytotoxicity assay
To test the cytotoxic function of spleen lymphocytes obtained 24 days after immunisation, these cells were co-cultured with H22 or 4T1 tumour cells at different ratios (tumour cell to lymphocyte: 1:1; 1:10; 1:20, or 1:50) in 96-well round bottom plates in triplicates in serum-free RPMI-1640 medium containing 50 ng/mL of CD3 mAb and 20 U/mL of mouse IL-2 (PeproTech, Cat #200–02, US) for 2 days. The cytotoxicity was evaluated from the amount of lactate dehydrogenase (LDH) released in the medium using an LDH Release Assay Kit (Beyotime, Cat #C0016, China). Percentage cytotoxicity was calculated as follows: LDH level in each well/total LDH released from the same number of cells lysed after two freeze-thaw cycles.
2.7. Imaging of co-cultures of 4T1 cancer cells and lymphocytes
The imaging of cell-cell contacts between GFP-positive 4T1 cells and lymphocytes obtained 24 days after immunisation in co-culture was performed under a fluorescence inverted microscope (IX71-F22FL/PH, Olympus, Japan).
2.8. Primary tumour model and measurement of tumour growth
A total of 1 × 106 H22 hepatoma cells in 0.2 mL PBS were subcutaneously injected into the back of H22 mice on day 25 after immunisation. The tumour size in each mouse from either control or CD151 peptide-immunised group was measured with a digital calliper twice per week. Tumour volume was calculated as follows: long diameter × short diameter2/2. Tumour growth curve was obtained by plotting tumour volume against time. At the end of the experiment, the tumours from each group were harvested, imaged, and weighed.
2.9. Experimental metastasis model and live imaging of lungs
Alive 1 × 106 4T1 breast cancer cells transfected with cDNAs of GFP and luciferase in 0.3 ml PBS were intravenously injected into the tail vein of BABL/c mice on day 25 after immunisation. Two weeks later, the tumour growth of 4T1 experimental lung metastases was measured with the IVIS Lumina III living imaging system (PerkinElmer, USA) and the total flux photon (P/S) was measured. At the end of the study, lung metastases of each mouse were counted under dissecting microscope.
2.10. Ficoll seperation and MDSCs staining
Monocytes were isolated using Ficoll density gradient centrifugation (Solarbio, Cat #P8620, China) according to the manufacturer's instruction. Then, 106 monocytes were labeled with fluorochrome-labeled Ab targeting murine CD11b (Biolegend, Cat #101228, San Diego, CA) and GR-1 (Biolegend, Cat #108423, San Diego, CA) for 1 h at 4°C washed, resuspended in 1% formalin, and analyzed via flow cytometry.
2.11. Survival curve analysis
The number of BABL/c mice with lung metastases was counted every day and recorded. The survival percentage in each group was calculated as (the number of survived mice/the number of total studied mice) × 100 and was plotted against time.
2.12. Quantitative real-time PCR
Total mRNA was isolated from tumour tissue using Trizol (Invitrogen, Cat #15596026, UK), and cDNA was synthesised from 100 ng of total RNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Cat #RR047A, Japan). Q-PCR was performed with GoTaq qPCR Master Mix (Promega, Cat #A6001, USA) and Applied Biosystems 7500 Real-Time PCR Systems. All samples were normalized to β-actin mRNA levels. The primer sequences were:
CD151: Forward: 5′-AGCCACGGCCTACATCTTAGT-3′;
Reverse: 5′-TTCCGTCGCTCCTTGAAAGTG-3′;
β-actin: Forward: 5′-GGCTGTATTCCCCTCCATCG-3′;
Reverse: 5′-CCAGTTGGTAACAATGCCATGT-3′.
2.13. Western blot analysis
In brief, 40 µg of protein per sample was analyzed on a 12% gel by SDS-PAGE, and transferred into a nitrocellulose membrane. Then, the bands were blocked with 5% BSA, incubated with anti-mouse CD151 (Abcam, Cat #ab185684, UK) at 4 °C for 12 h, washed 3 times for 15 min each in TBST at room temperature, incubated with horseradish peroxidase-conjugated (HRP) secondary antibody (Abcam, Cat #ab205718, UK) for 2 h at room temperature, and washed 3 times for 15 min each in TBST. The bands were exposed using a FlourChemE system (Protein Simple, US) according to the manufacturer's instructions. The scanned bands were further analyzed by the ImageJ software to obtain densitometry values.
2.14. Immunohistochemistry (IHC) analysis
The H22 and 4T1 tumours harvested from PBS control and CD151 peptide-immunised groups were fixed with 10% formalin overnight and processed into 5-μm-thick paraffin sections. The slides were stained with haematoxylin and eosin (HE) or subjected to IHC with anti-mouse CD8 (Abcam, Cat #ab203035, UK), followed by HRP secondary antibody (Abcam, Cat #ab205718, UK) and DAB treatment. Images were obtained under a microscope (BX43, Olympus, Japan) at a magnification of 200×.
2.15. Hemogram and biochemical indexes
Briefly, a 200 µl blood sample from each mouse was collected and subjected to automatic animal blood cell analyzer (BC-2800 Vet, China) for hemogram detection. Biochemical indexes detection was performed using Roche automatic biochemical analyzer P800.
2.16. Statistical analysis
Student's t-test and Log-rank test were used for statistical analysis. P < 0.05 was considered statistically significant.
3. Results
3.1. Defining potential oncoproteins/TAAs induced by radiation via proteomic analysis
Previous studies have demonstrated that tumour cells could release large amounts of TAAs following high dose and low fractionation radiation (8 Gy dose per fraction), which increased the generation of tumour antigen-specific effector cells [27], [28], [29]. IR could also enhance tumour immunogenicity, such as by increasing expression of MUC-1 and CEA, that ultimately enhances cell killing by cytotoxic T lymphocytes [30,31]. Therefore, proteins upregulated by IR may be ideal TAAs.
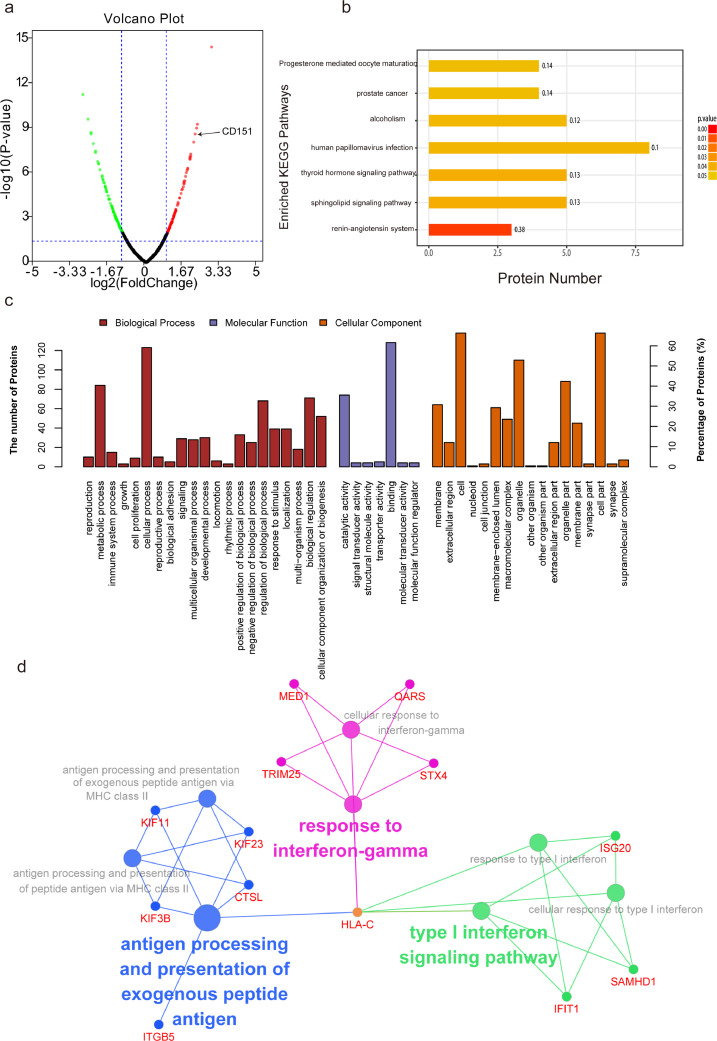
To define the protein alterations of H22 cancer cells, the lysates from non-IR and IR cells were collected and subjected to label-free relative quantitative proteomic analysis. In total, 209 differentially expressed proteins were detected after radiation, including 103 up-regulated proteins and 106 down-regulated proteins (|log2 fold change| > 1 and p < 0.05) (Fig. 1a). The full list of differentially expressed proteins is supplied in supplementary table 1.
Fig. 1.
Defining potential oncoproteins/TAAs induced by radiation via proteomic analysis. Label-free relative quantitative proteomic analysis was performed to identify potential TAAs induced by radiation. (a) Volcano plot showing the distribution of 209 differentially expressed proteins quantified among non-IR and IR cells with the threshold of |log2 Fold change|>1 and P<0.05. Red dots indicate up-regulated proteins and green dots indicate down-regulated proteins. The expression of CD151 is indicated by the arrow. (b) The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed proteins. Y-axis shows KEGG pathway entry, and X-axis shows the number of proteins annotated in the pathway. The intensity of the colour represents P-value. (c) Gene Ontology (GO) function analysis. Y-axis shows the number of proteins annotated in the GO term. X-axis shows the GO pathway, which was mainly categorized as biological process (red), cellular component (orange) and molecular function (purple). (d) The role of differentially expressed proteins induced by IR in immune system process were visualized in ClueGO (ver. 2.5.3) plugin of Cytoscape (ver. 3.5.1).(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that the differentially expressed proteins were mainly enriched in prostate cancer, hormone (thyroid, renin-angiotensin, progesterone, etc.), and stress (such as papillomavirus infection) related signal pathways (Fig. 1b).
Gene Ontology (GO) function analysis revealed the association of these differentially expressed proteins with metabolic process, cellular process, and response to stimuli (Fig. 1c). As shown in Fig. 1d, the IR-induced differentially expressed proteins in immune system process were mainly involved in response to interferon-gamma (such as HLA-C, MED1, QARS, TRIM25 and STX4), type I interferon signaling pathway (such as ISG20, SAMHD1, IFIT1 and HLA-C), antigen processing and presentation of exogenous peptide antigens (HLA-C, KIF3B, KIF11, KIF23 and CTSL). The radiation stress-induced alterations included changes in the expression of oncoproteins involved in several processes which is indicative of their potential as TAAs.
3.2. Identification of CD151 as an ideal target for peptide-based cancer immunotherapy
To achieve effective and safe peptide-based cancer vaccines, it is important to identify appropriate TAAs as targets. The ideal TAAs must have three characteristics to be effective molecular targets: over-expression in tumours; oncogenicity, and immunogenicity. Here, we identified CD151 as an ideal TAA according to proteomics and bioinformatic analysis.
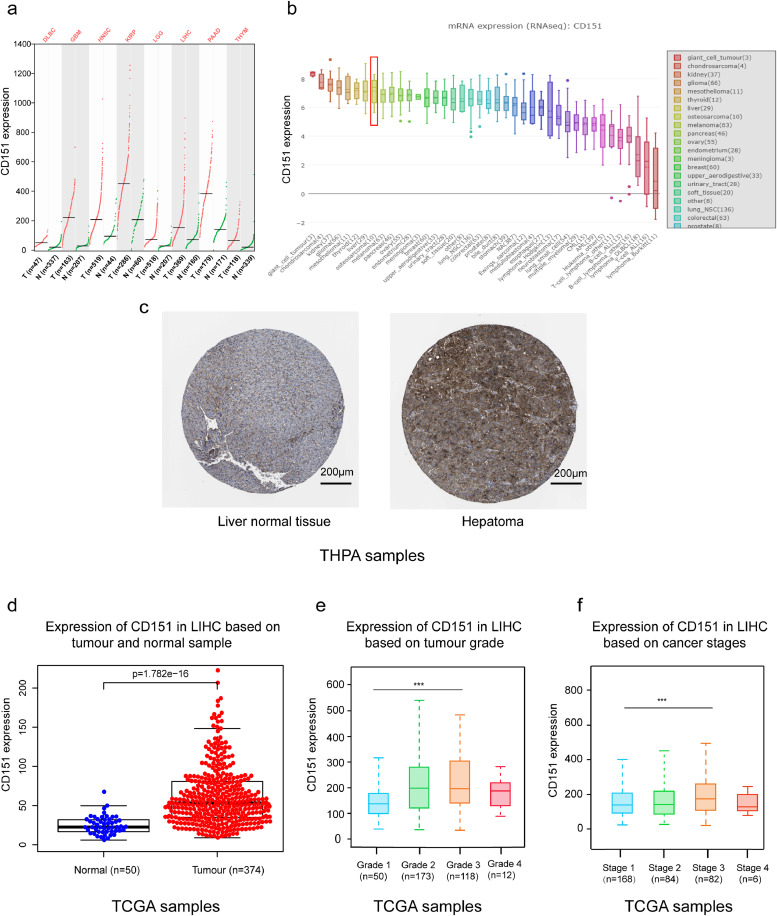
Firstly, CD151 is overexpressed on various tumours, including gastric cancers, hepatocellular carcinoma, and clear cell renal cell carcinoma [32], [33], [34]. Here, we found that more types of cancers express high levels of CD151, including diffuse large B-cell lymphoma (DLBCL), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal papillary cell carcinoma (KIRP), brain low-grade glioma (LGG), liver hepatocellular carcinoma (LIHC), pancreatic adenocarcinoma (PAAD), and thymoma (THYM), through analysis of GEPIA (Fig. 2a).
Fig. 2.
Identification of CD151 as an ideal target for peptide-based cancer immunotherapy. To identify CD151 as an ideal TAA, the Cancer Genome Atlas, GEPIA, the Human Protein Atlas, and CCLE databases were used for bioinformatic analysis. (a) CD151 is overexpressed in 8 types of tumours (figure from http://gepia.cancer-pku.cn/detail.php?gene=cd151). (b) Differential expressions of CD151 in 40 types of cancer cell lines were obtained from CCLE database. Y axis represents CD151 expression, and X axis represents different types of cancer cell lines. Of these, liver cancer cell lines are one of the top hints as the red square indicates (figure from https://portals.broadinstitute.org/ccle/page?gene=CD151). (c) IHC staining of CD151 (antibody HPA011906) in liver normal tissue and hepatocellular carcinoma was obtained from The Human Protein Atlas database (https://www.proteinatlas.org/ENSG00000177697-CD151/pathology/liver+cancer#img). (d) CD151 was over-expressed in LIHC compared to normal samples and was associated with (e) tumour grade and (f) progression stages of LIHC (data from TCGA databases). P-values were determined by two-tailed Student's t-test (***, P <0.001).(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Secondly, as shown in Fig. 2b, the expression of CD151 was differential in various cancer cell lines via using CCLE database. Of these, liver cancer cell lines were one of the top hits.
Thirdly, analysis of liver hepatocellular carcinoma data from THPA (Fig. 2c) and TCGA (Fig. 2d) revealed that CD151 was significantly overexpressed in tumour tissues compared with normal tissues, and over-expression of CD151 was associated with high tumour grade (Fig. 2e), and advanced stage (Fig. 2f).
This data suggest that CD151 is likely to be involved in malignant behaviour and may be an ideal TAA. Considering its location on cell surface [32], [33], [34], CD151 could serve as an ideal target and facilitate the production of TAA-derived vaccines for cancer immunotherapy.
3.3. CD151 peptides trigger active immunity and inhibit primary tumour growth of H22 hepatoma
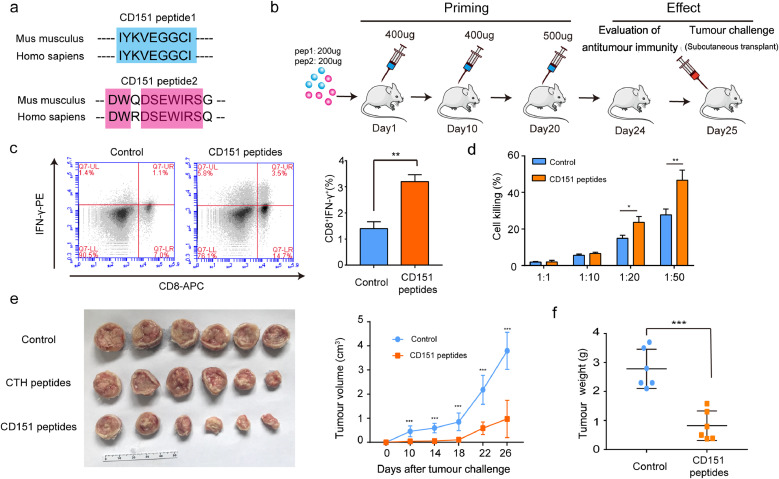
To explore the applicability of CD151 as a target for active immunotherapy, two peptides derived from CD151 were synthesised. The sequences of the two peptide vaccines are highly conserved in human and mice, indicating its potential translatability to human cancer (Fig. 3a).
Fig. 3.
Peptides of CD151-triggered active immunity inhibited primary tumour growth of H22 hepatoma. (a) Two peptides derived from Mus musculus CD151 were synthesized according to MHC I binding and MHC I immunogenicity, and were blasted with sequences of Homo sapiens CD151. (b) Experimental schema depicting vaccination and tumour challenge schedule. Mice were vaccinated with CD151 peptides-Freund's adjuvant and received boost vaccinations 10 and 20 days later, followed by evaluation of anti-tumour immunity and primary tumour challenge. PBS with an equal volume of Freund's adjuvant was used as the control group (n = 10/group). (c) The scatter plots show the differences of spleen CD8+IFNγ+ T lymphocytes between control group and CD151 peptide immunisation group by FCM analysis. The percentage of CD8+IFNγ+ T lymphocytes is presented by the graph in the H22 models (n = 4/group). (d) To test the killing function of spleen lymphocytes, they were co-cultured with H22 tumour cells at different ratios of tumour cells to lymphocytes (1:1; 1:10; 1:20 or 1:50). The killing effect was measured by the LDH released (n = 4/group). (e) Image of primary tumours and growth curve of H22 hepatoma presented by tumour volume (n = 6/group). (f) Tumour weights at the end of experiment (n = 6/group). Results are representative of two independent experiments. P-values were determined by two-tailed Student's t-test (*, P<0.05; **, P < 0.01; ***, P <0.001).
In brief, 200 μg of each peptide was mixed and emulsified with an equal volume of Freund's adjuvant and subcutaneously injected into ICR mouse at day 1, day 10 and day 20. PBS with an equal volume of Freund's adjuvant was used as the control group (Fig. 3b). 24 days after the first immunisation, the elicited immune response against CD151 was evident from the increase in the peptide-stimulated active CD8+IFNγ+ T lymphocytes (Fig. 3c) and the associated cytotoxicity on CD151-overexpressing H22 hepatoma cells (Fig. 3d). 25 days after priming, 106 H22 hepatoma cells were subcutaneously injected into the back of ICR mouse. The H22 tumour growth in the CD151 peptide-immunised mice was greatly suppressed as compared with that in the control mice (immunised with PBS-Freund's adjuvant), as evident from a retarded growth curve, smaller tumour size, and lower tumour weight (Fig. 3e and f). Together, these data demonstrate the capability of CD151 peptides to trigger an active immune response to inhibit growth of primary H22 hepatoma via the activation of T lymphocytes.
As tumours often mutate or downregulate tumour antigens to avoide an immune attack, we further analyzed CD151 expression on the tumour cells after peptide vaccination. As shown in the supplementary Fig. 1a and b, we did not see a difference in CD151 expression between the control and CD151 peptide group after peptide vaccination, indicating that CD151 was not downregulated at the end of experiment.
We also designed another peptide vaccine derived from the screen (CTH peptides). CTH was the top one in the screen, but not overexpressed in hepatoma (supplementary Fig. 2a). However, considering the limited efficacy (supplementary Fig. 2b and 2c) and serious side effects of CTH peptides, such as loss of weight, inflammation at injection site, glomeruloatrophy and renal hypofunction (supplementary Fig. 2d–2 g), we focused on CD151 peptides research.
3.4. CD151 peptides suppress experimental 4T1 lung metastases via triggering active immune responses
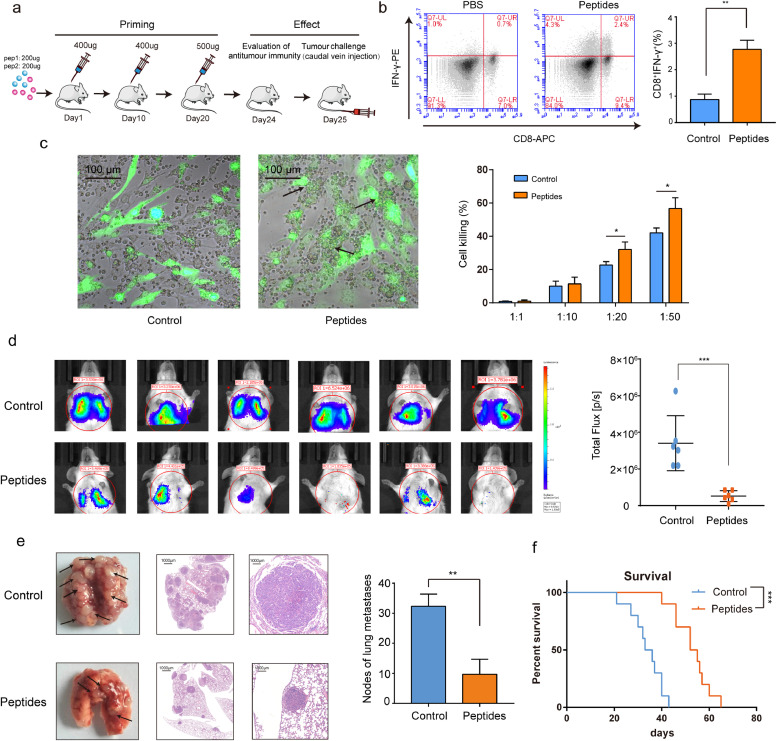
To test whether CD151 peptide-triggered active immune responses were universal against different types of tumours in different models, an experimental metastasis model of 4T1 breast cancer cells (stable and double transfected with GFP and luciferase cDNAs) was used. Similarly, the immunisation with CD151 peptides was performed in BABL/c mice (Fig. 4a). Indeed, the peptide-stimulated active CD8+IFNγ+ T lymphocytes were increased (Fig. 4b). In the co-cultures of 4T1 tumour cells and lymphocytes from peptide-immunised mice, 4T1 tumour cells were closely surrounded by lymphocytes, and this phenomenon was absent in the control mice. Besides, CD8+IFNγ+ T lymphocytes had a strong cytotoxic effect on CD151-overexpressing 4T1 cancer cells (Fig. 4c). The anti-lung metastasis effect triggered by the CD151 peptides was further confirmed by live imaging, demonstrating that the tumour growth in the lung of CD151 peptide-immunised mice was much smaller than that in the control mice which showed less flux photon (Fig. 4d). At the end of the experiment, the harvested lungs from CD151 peptide-immunised mice showed lower metastasis rate and fewer tumour nodules than those from the control mice (Fig. 4e). Importantly, the CD151 peptide-triggered active anti-cancer immunity could result in a prolonged survival (Fig. 4f), confirming the effectiveness of CD151 peptide-activated immunity.
Fig. 4.
Peptides of CD151-triggered active immunity and suppressed the experimental 4T1 lung metastases. (a) The experiment schema for immunisation and experimental lung metastasis challenge. (b) The scatter plots show the differences of spleen CD8+IFNγ+ T lymphocytes between control group and CD151 peptide immunisation group by FCM analysis. The percentage of CD8+IFNγ+ T lymphocytes is presented by the graph in 4T1 lung metastases models (n = 4/group). (c) The imaging of cell-cell contact between GFP-positive 4T1 cells and lymphocytes was taken under fluorescent reverse-microscopy. When co-culture of 4T1 tumour cells with lymphocytes, the 4T1 tumour cells were closely surrounded by lymphocytes from peptide-immunised mice as the arrows indicate, which was not observed in lymphocytes from control mice. The killing effect was measured by the LDH released (n = 4/group). (d)In vivo bioluminescence imaging. GFP-Luc-4T1 cells were intravenously injected into the tail vein of BABL/c mice after immunisation. 14 days later, the tumour growth of 4T1 experimental lung metastases was measured with the IVIS Lumina III living imaging system. Luciferase signals from CD151 peptide group were lower than those from control group (n = 6/group). (e) The gross anatomy and HE staining of lung metastases in the control and CD151 peptide immunised mice, and the statistical analysis of lung metastases nodules per mice (n = 6/group). The arrows indicate lung metastases nodules. (f) The differences in survival between the control and CD151 peptide immunised mice (n = 10/group). Results are representative of two independent experiments. P-values were determined by two-tailed Student's t-test or Log-rank test (*, P<0.05; **, P < 0.01; ***, P <0.001).
3.5. CD151 peptides enhance tumour infiltration of CD8 lymphocytes and reduce MDSCs
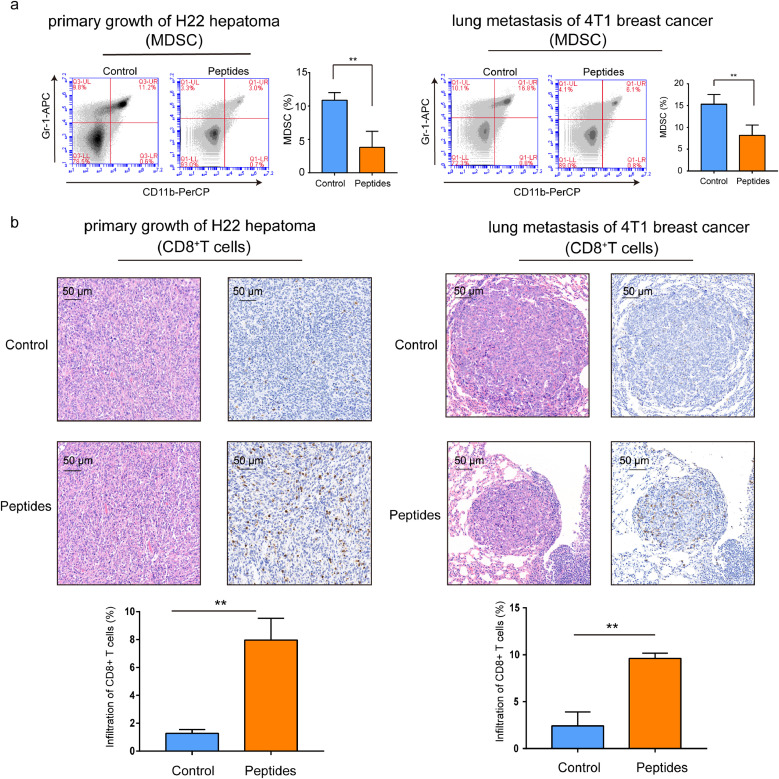
To explore the cellular mechanisms underlying CD151 peptides triggered anti-tumour effects, the alteration of anti-tumour suppression cells, blood MDSCs, were stained with anti-mouse Gr-1-APC and CD11b-PerCP and analyzed with FCM. The result showed that the MDSCs were greatly reduced in CD151 peptides immunised ICR and BABL/c mice (Fig. 5a). In addition, the IHC stain indicated that the tumour infiltrated CD8+ lymphocytes were higher in both CD151 peptides immunised ICR and BABL/c mice than that of controls in two mouse models (Fig. 5b).
Fig. 5.
Peptides of CD151-triggered active immunity enhanced the CD8 cell infiltration and reduced MDSC. (a) MDSCs from ICR and BABL/c mice blood were double stained with anti-mouse Gr-1-APC and CD11b-PerCP and analyzed with FCM (n = 6/group). (b) The H22 primary tumour and 4T1 lung metastases were harvested at the end of experiments and stained with HE and anti-mouse CD8 (pictured at 200 ×) (n = 6/group). Infiltration of CD8+ T cell in tumour microenvironment was calculated. Results are representative of two independent experiments. P-values were determined by two-tailed Student's t-test (*, P<0.05; **, P < 0.01; ***, P <0.001).
3.6. Side-effects test of CD151 peptide vaccines
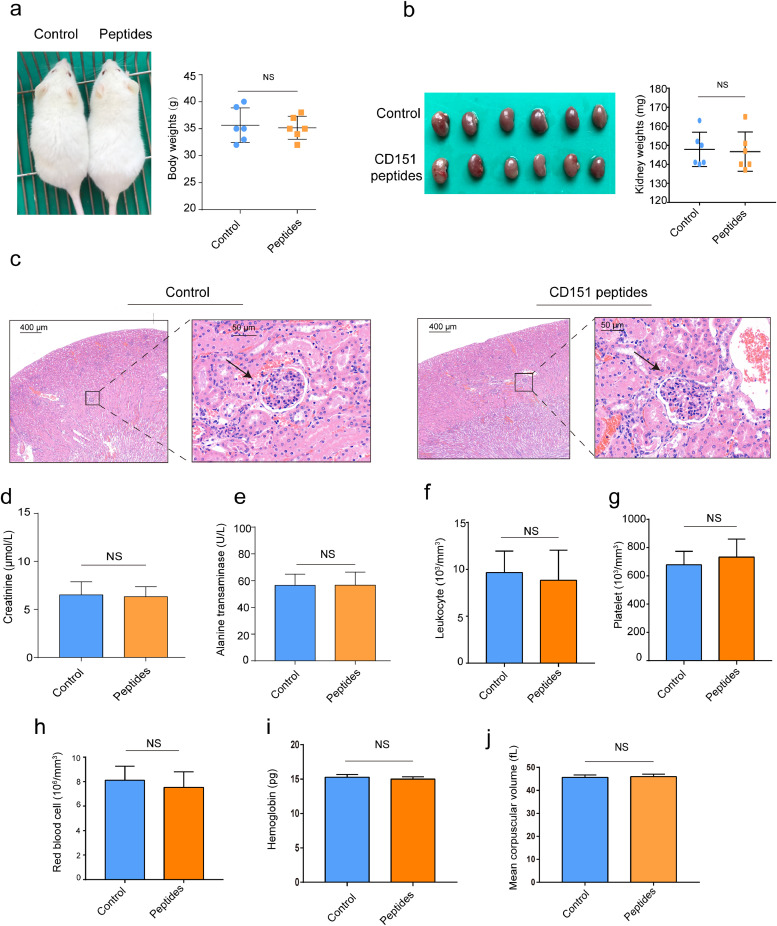
To evaluate the safety of CD151 peptide vaccines, the mouse body weights, kidney weights, glomerular morphology, renal function, liver function, and various indices of blood were detected after immunisation.
As shown in the Fig. 6, there were no differences in the mouse body weights (Fig. 6a), kidney weights (Fig. 6b), glomerular morphology (Fig. 6c), renal function (Fig. 6d), liver function (Fig. 6e), and various indices of blood (Fig. 6f–j) between control group and CD151 peptides group. These results demonstrated that CD151 peptide vaccines were a safe approach to activate anti-tumour immune response.
Fig. 6.
Side-effects test of CD151 peptide vaccines. At the end of the peptides immunisation, side-effects were evaluated by measuring the body weight, blood and biochemical indexes between control and peptides group. (a) Mouse image and body weights; (b) gross anatomy of kidney and weights; (c) glomerular morphology by HE staining and the arrows indicate glomerulus; (d) plasma creatinine; (e) plasma alanine transaminase; (f) number of leukocytes; (g) number of platelets; (h) number of red blood cells; (i) hemoglobin; (j) mean corpuscular volume. Results are representative of two independent experiments (n = 6/group). P-values were determined by two-tailed Student's t-test (NS, P>0.05).
4. Discussion
Using a comparative proteomic approach and a set of bioinformatic analyses, here we demonstrate that IR stress could regulate the expression of several proteins in cancer cells involved in metabolism, biological regulation, cell binding, and biogenesis. As a reaction to IR stress, these altered proteins are likely to rescue the cells from IR-induced apoptosis /death, promote cell survival and proliferation, thereby serving as oncoproteins involved in malignant behaviour. These stress-induced oncoproteins may be responsible for imparting resistance to therapy, relapse, and metastasis. Targeting these stress-induced oncoproteins by various means has been an important goal in medical research [35]. As oncoproteins may serve as TAAs, we chose a unique and specific immune approach using synthetic peptides of CD151 as tumour vaccines. We found that, in both primary and metastasis models, this vaccine-triggered active immune responses, i.e. induction of functional CD8+IFNγ+ T cell infiltration/cytotoxicity and suppression of MDSCs, and could effectively inhibit both tumour growth and metastasis. Two cancer cell lines in two in vivo mouse models showed similar results, confirming the promising therapeutic value of CD151 peptides as tumour vaccines.
The tetraspanin CD151 is overexpressed in various tumours and correlated with tumour progression, metastatic propensity and poor survival of cancer patients [32,33]. The CD151-based antibody has successfully inhibited the tumour growth, angiogenesis and metastasis in different xenograft cancer models [36]. However, passive immunity is limited for its short-term effects and no generation of memory cells [37]. Active immunity is a powerful host weapon against diseases. Various vaccines successfully help the human host to get rid of infectious diseases such as smallpox, measles, cholera, polio, plague, pertussis, typhoid fever, and meningococcal meningitis. In the battle against cancer, active immunity is highly desirable. However, the discovery of tumour-specific antigens and effective utilisation of TAAs are challenging [38]. The successful use of CD151 peptides to reduce tumour growth and metastasis is based on the following principles: (1) as an oncoprotein/TAA, CD151 is induced by IR stress; (2) CD151 is overexpressed in tumours as compared with normal tissues; (3) CD151 expression is associated with cancer stages and progression, which is considered to be barely lost in the process of tumour progression; (4) CD151 is expressed on cell surface [5]. These characteristics suggest that CD151 is always targetable without becoming ‘invisible’ or avoiding an immune attack due to down-regulation or loss of its TAA presentation.
CD151 peptide designed as a tumour vaccine needs to meet MHC restriction [39]. Using the databases IEDB and SYFPEITHI, we focused on the following characteristics: (1) outer surface of cell membrane that may be accessible to immune cells; (2) MHC I-binding capacity; (3) MHC I immunogenicity; (4) solubility for hapten. These properties determined the ability of the peptides to serve as a vaccine and consequently trigger an immune response.
The effectiveness of CD151 peptide as a tumour vaccine requires two stages: priming and function. In priming stage, peptide immunisation primes the host immune system to recognize CD151 as a target. Then, in function stage, the active immunity would be able to kill tumour cells with high CD151 expression. As priming requires about 3 weeks, if CD151+ cancer cells are inoculated before CD151 peptide priming, the fast replication of tumour cells will result in a heavy tumour burden and suppress anti-tumour immunity (data not shown). Based on this time-delay from immunisation priming stage to effectiveness stage, clinically, the best result of active immunotherapy would probably be achieved right after the surgery to remove large tumour burden. This would facilitate the generation of host anti-tumour immunity within 3 weeks with no suppression from tumour burden.
Antibodies that block the interaction between PD-1 and PD-L1 and consequently modulate the host immunity have attracted attention in immunotherapy research [40]. However, only a small portion of cancer patients gain long-term benefits, partly owing to the lack of infiltrated T cells [41], [42], [43]. The use of peptide vaccine generated from TAAs, such as CD151, could increase the infiltrated T cells, which may sensitise patients to immune checkpoint inhibitors.
A multitude of chimaeric antigen receptors (CARs) targeting an array of TSAs/TAAs have been reported for their remarkable anti-tumour effects in vitro or in vivo, including targeting cell surface tumour antigens in hematologic malignancies and solid tumours [44]. Immunotherapies with T cells expressing chimeric antigen receptor (CAR-T) using CD19, CD20, CD30, and CD123 as targets have been very successful in treating hematologic malignancies, while prostate-specific membrane antigen (PSMA) CAR-T or EGFR-vIII CAR-T against solid tumours have been less successful [45], [46], [47], [48]. CD151 as surface oncoprotein/TAA might be a novel tumour target for CAR-T immune treatment.
In summary, the peptide vaccines derived from IR stress-induced oncoprotein/TAA could prime host anti-tumour immunity and effectively reduce tumour growth and metastasis by increasing functional CD8+IFNγ+ T cell infiltration/cytotoxicity and suppressing MDSCs. This approach may expand immunotherapy strategies and sensitise patients to immune checkpoint inhibitors.
Author contributions
Lurong Zhang and Junxin Wu conceived the project and designed the experiments. Wanzun Lin conducted experiments and wrote the manuscript. Jun Liu and Juhui Chen helped to perform FCM analysis. Jiancheng Li and Sufang Qiu helped to perform immunohistochemical analysis. Jiayu Ma and Xiandong Lin helped to write the manuscript. All authors read and provided input on the manuscript.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgments/Funding
The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
The project was funded by the grants of Fujian Province Natural Science Foundation (2017J01260); Science & Technology Program of Fujian Province, China (#2018Y2003). The funders had no role in study design, data collection, data analysis, interpretation, or writing of the report.
Junxin Wu and Lurong Zhang had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.025.
Contributor Information
Lurong Zhang, Email: lz8506@163.com.
Junxin Wu, Email: junxinwufj@aliyun.com.
Appendix. Supplementary materials
References
- 1.Mandal R., Chan T.A. Personalized oncology meets immunology: the path toward precision immunotherapy. Cancer Discov. 2016;6::703–713. doi: 10.1158/2159-8290.CD-16-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Mutational Load The more the merrier for immune checkpoint blockade therapies. EBioMedicine. 2016;13::1–2. doi: 10.1016/j.ebiom.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao X., Subramanian S. Intrinsic resistance of solid tumors to immune checkpoint blockade therapy. Cancer Res. 2017;77::817–822. doi: 10.1158/0008-5472.CAN-16-2379. [DOI] [PubMed] [Google Scholar]
- 4.De Charette M., Marabelle Aurélien, Houot R. Turning tumour cells into antigen presenting cells: the next step to improve cancer immunotherapy? Eur J Cancer. 2016;68::134–147. doi: 10.1016/j.ejca.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Hirayama M., Nishimura Y. The present status and future prospects of peptide-based cancer vaccines. Int Immunol. 2016;28::319–328. doi: 10.1093/intimm/dxw027. [DOI] [PubMed] [Google Scholar]
- 6.Iinuma H., Fukushima R., Inaba T., Tamura J., Inoue T., Ogawa E. Phase I clinical study of multiple epitope peptide vaccine combined with chemoradiation therapy in esophageal cancer patients. J Transl Med. 2014;12::84. doi: 10.1186/1479-5876-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kono K., Iinuma H., Akutsu Y., Tanaka H., Hayashi N., Uchikado Y. Multicenter, phase II clinical trial of cancer vaccination for advanced esophageal cancer with three peptides derived from novel cancer-testis antigens. J Transl Med. 2012;10::141. doi: 10.1186/1479-5876-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki H., Fukuhara M., Yamaura T., Mutoh S., Okabe N., Yaginuma H. Multiple therapeutic peptide vaccines consisting of combined novel cancer testis antigens and anti-angiogenic peptides for patients with non-small cell lung cancer. J Transl Med. 2013;11::97. doi: 10.1186/1479-5876-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzentruber D.J., Lawson D.H., Richards J.M., Conry R.M., Miller D.M., Treisman J. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N Engl J Med. 2011;364::2119–2127. doi: 10.1056/NEJMoa1012863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pol J., Bloy N., Buqué A., Eggermont A., Cremer I., Sautès-Fridman C. Trial watch: peptide-based anticancer vaccines. Oncoimmunology. 2015;4: doi: 10.4161/2162402X.2014.974411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pistoia V., Morandi F., Pezzolo A., Raffaghello L., Prigione I. MYCN: from oncoprotein to tumor-associated antigen. Front Oncol. 2012;2::174. doi: 10.3389/fonc.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf-Yadlin A., Kumar N., Zhang Y., Hautaniemi S., Zaman M., Kim H.D. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol. 2006;2::54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yewale C., Baradia D., Vhora I., Hautaniemi S., Zaman M., Kim H.D. Epidermal growth factor receptor targeting in cancer: a review of trends and strategies. Biomaterials. 2013;34::8690–8707. doi: 10.1016/j.biomaterials.2013.07.100. [DOI] [PubMed] [Google Scholar]
- 14.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69::4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto K., Umitsu M., De Silva D.M., Roy A., Bottaro D.P. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci. 2017;108::296–307. doi: 10.1111/cas.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roland C.L., Harken A.H., Sarr M.G., Barnett C.C. ICAM-1 expression determines malignant potential of cancer. Surgery. 2007;141::705–707. doi: 10.1016/j.surg.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Porta C., Paglino C., Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014;4::64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia Y., Shen S., Verma I.M. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2::823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadej R., Grudowska A., Turczyk L., Kordek R., Romanska H.M. CD151 in cancer progression and metastasis: a complex scenario. Lab Invest. 2014;94::41–51. doi: 10.1038/labinvest.2013.136. [DOI] [PubMed] [Google Scholar]
- 20.Kumari S., Devi G., Badana A., Dasari V.R., Malla R.R. CD151-a striking marker for cancer therapy. Biomark Cancer. 2015;7::7–11. doi: 10.4137/BIC.S21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1) doi: 10.1093/nar/gkx247. W98-w102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barretina J., Caponigro G., Stransky N. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlén M., Fagerberg L., Hallström B.M. Proteomics. tissue-based map of the human proteome. Science (New York, NY) 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 24.Schuler M.M., Nastke M.D., Stevanovikć S. SYFPEITHI: database for searching and T-cell epitope prediction. Method Molecul Biol (Clifton, NJ) 2007;409:75–93. doi: 10.1007/978-1-60327-118-9_5. [DOI] [PubMed] [Google Scholar]
- 25.Vita R., Mahajan S., Overton J.A. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1) doi: 10.1093/nar/gky1006. D339-d43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billiau A., Matthys P. Modes of action of Freund's adjuvants in experimental models of autoimmune diseases. J Leukoc Biol. 2001;70::849–860. [PubMed] [Google Scholar]
- 27.Hodge J.W., Sharp H.J., Gameiro S.R. Abscopal regression of antigen disparate tumors by antigen cascade after systemic tumor vaccination in combination with local tumor radiation. Cancer Biother Radiopharm. 2012;27(1):12–22. doi: 10.1089/cbr.2012.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein M.B., Krishnan S., Hodge J.W., Chang J.Y. Immunotherapy and stereotactic ablative radiotherapy (ISABR): a curative approach. Nat Rev Clin Oncol. 2016;13(8):516–524. doi: 10.1038/nrclinonc.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lugade A.A., Moran J.P., Gerber S.A., Rose R.C., Frelinger J.G., Lord E.M. Local radiation therapy of B16 melanoma tumors increases the generation of tumor antigen-specific effector cells that traffic to the tumor. J Immunol. 2005;174::7516–7523. doi: 10.4049/jimmunol.174.12.7516. [DOI] [PubMed] [Google Scholar]
- 30.Vanpouille-Box C., Alard A., Aryankalayil M.J., Sarfraz Y., Diamond J.M., Schneider R.J. DNA exonuclease trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. 2017;8::15618. doi: 10.1038/ncomms15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo L., Lv M., Zhuang X., Zhang Q., Qiao T. Irradiation increases the immunogenicity of lung cancer cells and irradiation-based tumor cell vaccine elicits tumor-specific T cell responses in vivo. Onco Targets Ther. 2019;12:3805–3815. doi: 10.2147/OTT.S197516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang B.W., Lee D., Chung H.Y., Han J.H., Kim Y.B. Tetraspanin CD151 expression associated with prognosis for patients with advanced gastric cancer. J Cancer Res Clin Oncol. 2013;139::1835–1843. doi: 10.1007/s00432-013-1503-4. [DOI] [PubMed] [Google Scholar]
- 33.Ke A.W., Shi G.M., Zhou J., Wu F.Z., Ding Z.B., Hu M.Y. Role of overexpression of CD151 and/or c-Met in predicting prognosis of hepatocellular carcinoma. Hepatology. 2009;49::491–503. doi: 10.1002/hep.22639. [DOI] [PubMed] [Google Scholar]
- 34.Yoo S.H., Lee K., Chae J.Y., Moon K.C. CD151 expression can predict cancer progression in clear cell renal cell carcinoma. Histopathology. 2011;58::191–197. doi: 10.1111/j.1365-2559.2011.03752.x. [DOI] [PubMed] [Google Scholar]
- 35.Hinrichs C.S. Cell-based molecularly targeted therapy: targeting oncoproteins with T cell receptor gene therapy. J Clin Invest. 2018;128::1261–1263. doi: 10.1172/JCI120386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haeuw J.F., Goetsch L., Bailly C., Corvaia N. Tetraspanin CD151 as a target for antibody-based cancer immunotherapy. Biochem Soc Trans. 2011;39(2):553–558. doi: 10.1042/BST0390553. [DOI] [PubMed] [Google Scholar]
- 37.Baxter D. Active and passive immunization for cancer. Hum Vaccin Immunother. 2014;10(7):2123–2129. doi: 10.4161/hv.29604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo C., Manjili M.H., Subjeck J.R., Sarkar D., Fisher P.B., Wang X.Y. Therapeutic cancer vaccines: past, present, and future. Adv Cancer Res. 2013;119::421–475. doi: 10.1016/B978-0-12-407190-2.00007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneble E., Clifton G.T., Hale D.F., Peoples G.E. Peptide-Based cancer vaccine strategies and clinical results. Methods Mol Biol. 2016;1403::797–817. doi: 10.1007/978-1-4939-3387-7_46. [DOI] [PubMed] [Google Scholar]
- 40.Wilky B.A. Immune checkpoint inhibitors: the linchpins of modern immunotherapy. Immunol Rev. 2019;290::6–23. doi: 10.1111/imr.12766. [DOI] [PubMed] [Google Scholar]
- 41.Sznol M., Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res. 2013;19::1021–1034. doi: 10.1158/1078-0432.CCR-12-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi M., Jiao D., Xu H., Liu Q., Zhao W., Han X. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17::129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Liang L., Dai W., Cai G., Xu Y., Li X. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol Cancer. 2016;15::55. doi: 10.1186/s12943-016-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M.G., Kim D., Suh S.K., Park Z., Choi M.J., Oh Y.K. Current status and regulatory perspective of chimeric antigen receptor-modified T cell therapeutics. Arch Pharm Res. 2016;39::437–452. doi: 10.1007/s12272-016-0719-7. [DOI] [PubMed] [Google Scholar]
- 45.Frigault M.J., Maus M.V. Chimeric antigen receptor-modified T cells strike back. Int Immunol. 2016;28::355–363. doi: 10.1093/intimm/dxw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe K., Terakura S., Martens A.C., van Meerten T., Uchiyama S., Imai M. Target antigen density governs the efficacy of anti-CD20-CD28-CD3 ζ chimeric antigen receptor-modified effector CD8+ T cells. J Immunol. 2015;194::911–920. doi: 10.4049/jimmunol.1402346. [DOI] [PubMed] [Google Scholar]
- 47.Savoldo B., Rooney C.M., Di S.A., Abken H., Hombach A., Foster A.E. Epstein Barr virus specific cytotoxic T lymphocytes expressing the anti-CD30zeta artificial chimeric T-cell receptor for immunotherapy of Hodgkin disease. Blood. 2007;110::2620–2630. doi: 10.1182/blood-2006-11-059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walter R.B. The role of CD33 as therapeutic target in acute myeloid leukemia. Expert Opin Ther Targets. 2014;18::715–718. doi: 10.1517/14728222.2014.909413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.