Abstract
Background
In multiple sclerosis (MS), immune up-regulation is coupled to subnormal immune response to interferon-β (IFN-β) and low serum IFN-β levels. The relationship between the defect in IFN signalling and acute and long-term effects of IFN-β on gene expression in MS is inadequately understood.
Methods
We profiled IFN-β-induced transcriptome shifts, using high-resolution microarrays on 227 mononuclear cell samples from IFN-β-treated MS Complete Responders (CR) stable for five years, and stable and active Partial Responders (PR), stable and active untreated MS, and healthy controls.
Findings
IFN-β injection induced short-term changes in 1,200 genes compared to baseline expression after 4-day IFN washout. Pre-injection after washout, and in response to IFN-β injections, PR more frequently had abnormal gene expression than CR. Surprisingly, short-term IFN-β induced little shift in Th1/Th17/Th2 gene expression, but up-regulated immune-inhibitory genes (ILT, IDO1, PD-L1). Expression of 8,800 genes was dysregulated in therapy-naïve compared to IFN-β-treated patients. These long-term changes in protein-coding and long non-coding RNAs affect immunity, synaptic transmission, and CNS cell survival, and correct the disordered therapy-naïve transcriptome to near-normal. In keeping with its impact on clinical course and brain repair in MS, long-term IFN-β treatment reversed the overexpression of proinflammatory and MMP genes, while enhancing genes involved in the oligodendroglia-protective integrated stress response, neuroprotection, and immunoregulation. In the rectified long-term signature, 277 transcripts differed between stable PR and CR patients.
Interpretation
IFN-β had minimal short-term effects on Th1 and Th2 pathways, but long-term it corrected gene dysregulation and induced immunoregulatory and neuroprotective genes. These data offer new biomarkers for IFN-β responsiveness.
Funding
Unrestricted grants from the US National MS Society, NMSS RG#4509A, and Bayer Pharmaceuticals
Keywords: Gene expression, ILT, Immune regulation, Interferon-beta, Multiple sclerosis, RNA microarray
Research in context
Evidence before this study
The cause of multiple sclerosis is unknown, but hundreds of cytokines, scores of viruses, and over 100 DNA polymorphisms have been implicated. We and others have also defined multiple abnormalities in immune subsets and immune functions in stable and active MS, but many studies used whole blood from clinical mixed groups of patients and less comprehensive expression arrays. We now provide gene expression architecture for immune, metabolic, and neuroprotective interactions in MS, and link this to the abnormal interferon signaling that characterizes untreated MS.
Added value of this study
We expand identified abnormalities in MS immune cell RNA transcription, with 8,800 abnormally expressed genes in untreated MS.
Rigorous kinetics of short-term and long-term in vivo IFN responses provide more comprehensive understanding of, immune regulation, antiviral activity, neuroprotection, and MS complexity.
We define 277 genes, after a 4-day therapy washout that differ between complete and partial clinical responders to IFN-therapy, and identify biomarkers of successful treatment with IFN, world-wide the most frequently used therapy for MS.
We investigate the intrinsically defective interferon signaling pathway in MS and demonstrate that long-term interferon therapy induces multiple discrete immunoregulatory pathways that coincide with decreased inflammatory profiles. These are pathways that could be targeted with agonists or second agents.
This number of identified genes is greater than in prior MS studies. We provide approaches to increase sensitivity of gene expression in autoimmune disease by using RNA microarrays that detect nearly 70,000 gene products, purified mononuclear cells, high quality RNA, careful in vivo stimulation kinetics, and well-defined forms of MS.
We provide a comprehensive public repository of gene expression data for genetic, neuroprotection, and immune studies, and a resource for effective targeting and discovery of new drugs for MS and autoimmune diseases.
Implications of all of the available evidence
MS should be viewed not only as an inflammatory CNS disease, but as one of defective immune regulation and suppression. To an important extent, this defect arises from subnormal interferon responses; the defective immune balance is corrected by long-term interferon treatment.
Many of the most effective therapies for MS cause a shift to immunoregulation (fingolimod, glatiramer, interferon-beta), sometimes after depletion of immune subsets (alemtuzumab, ocrelizumab). Our approach will allow characterization of immune regulation before and after treatment, characterization of responders and non-responders to therapies.
Abbreviations: CR, Complete Responders; DEG, differentially expressed gene; FDR, false discovery rate; GEO, Gene Expression Omnibus; GO, gene ontology; GWAS, genome-wide association studies; HC, Healthy Controls; HTA, Human Transcriptome Arrays; IFN, type I interferon; IPA, Ingenuity Pathway Analysis; ISG, IFN-stimulated gene; lncRNA, long non-coding RNA; PBMNC, peripheral blood mononuclear cells; MS, multiple sclerosis; MU, million unit; NAb, neutralizing antibody; PCA, principal component analysis; PR, Partial Responders; SEM, standard error of the mean; SQ, subcutaneous
1. Introduction
Multiple sclerosis (MS) affects 2 million people world-wide, but its cause is unknown. There is no accepted MS antigen, proven inciting virus, or ubiquitous mutation that catalyzes MS. In untreated MS, excessive monocyte and Th1, Th17, and B cell activity is coupled to impaired regulatory/suppressor cell function in the periphery. These peripheral blood mononuclear cells (PBMNC) penetrate the blood-brain barrier and give rise to MS relapses and to CNS inflammation and demyelination [1], [2], [3]. Later in the course, excessive innate immunity and neurodegeneration predominate. The underlying cause of both forms of inflammation is unknown. We hypothesize that dysregulation of the interferon pathway underlies some of these abnormalities.
Genome-wide association studies (GWAS) implicate polymorphisms in multiple genes that control immunity and regulate IFN signaling [4]. In functional studies, the IFN-α/β pathway is strongly down-regulated in (PBMNC) from most untreated MS patients [5]. IFN-driven genes have low expression during remissions in untreated MS, with further reduction during exacerbations and progression. The disruption in RNA and protein expression in type I IFN-driven pathways outstrips disturbances in Th1/Th17/Th2 cell pathways [6], [7], [8], [9], [10], and could disrupt adaptive and innate immunity and abet neuronal death.
The mechanism of how IFN-β injections favorably affect MS remains incompletely resolved. Here, we sought to identify molecular regulators of the abnormal type I interferon system of MS. We collected 227 PBMNC samples from 54 subjects representing five clinical groups: Complete Clinical Responders to IFN-β therapy (CR) and Partial Clinical Responders to IFN-β therapy (PR) followed over five years, therapy-naïve stable MS, therapy-naive active MS, and healthy controls (HC). IFN control of gene expression was tested at different doses, with short- and long-term kinetics, and during stable and active disease in CR and PR. Gene expression was profiled using high-resolution whole transcriptome microarrays covering 67,000 genes and 285,000 coding and non-coding transcripts [11]. We used injection of IFN-β to examine induction of rapid or long-term expression of immune-regulatory and neuroprotective genes that could affect MS disease activity.
2. Materials and methods
2.1. Study design
We first studied rapid, short-term changes in PBMNC gene expression after injection of different doses of IFN-β (Fig. S1). Clinically stable MS patients who had been on IFN-β therapy for an average of 8 years, had initial study sampling after injections of IFN-β, 500 μg (16 million units [MU]) and then one month later, 250 μg (8 MU). Five years later, patients were split into two groups, CR who had remained attack-free, and PR who had experienced at least one attack over 5 years. Spontaneous (no IFN-β given) gene expression in PBMNCs from therapy-naïve clinically stable MS patients, therapy-naïve clinically active MS patients, and healthy controls was compared to expression in PBMNCs from CR and PR subjected to a 4-day IFN-β-free washout. This duration of washout cleans residual in vivo IFN-β-driven gene expression. To reduce variability in sample collection, gene expression, and data acquisition, patients had clearly-defined clinical activity, and carefully-timed drug administration and phlebotomy. PBMNC were isolated in order to remove background noise from other whole blood RNAs that are minimally relevant to MS immunopathology.
2.2. Subjects
227 samples were collected from 54 subjects (Tables S1 and S2). 27 well-characterized relapsing/remitting MS (RRMS) patients had received IFN-β therapy for 8•14 ± 0•86 years (range 1–19 years) prior to sampling. Eight years of IFN-β therapy was considered likely to reveal chronic effects of IFN-β treatment, since 4–5 years of IFN-β-1b therapy in the pivotal IFN-β-1b trial led to a 46% prolonged survival over 21 years, compared to a placebo group, untreated during the 5 year trial, then placed on therapy [12]. IFN-β-treated patients had an EDSS of 3•4 ± 0•4, and duration of MS since symptom onset was 13•8 ± 1•9 years at time of entry into our study. Patients were followed longitudinally over five years in a paired design. No patients who stopped or changed therapy were included, one PR stable patient was missing the 16 MU, 24-hour data point.
Twelve patients were CR to IFN-β therapy, as defined in [5,8,13] (age 47•3 ± 1•7 [mean ± SEM], 10F/2 M). They were exacerbation-free and progression-free for at least 6 months before sampling (time since last attack averaged 6•4 ± 1•7 years) and for 5 years after sampling. Fifteen PR patients had been stable for 6 months or more when studied (average 1•0 ± 0•3 years) and were also followed for 5 years (age 46•7 ± 3•1, 12F/3 M). PR were additionally sampled during an exacerbation that occurred during this study, indicative of incomplete response to IFN-β. Activity in PR patients required an ongoing severe flare with an at least 1-point increase on the EDSS, or 1 point increase on two of its subscales [14], lasting >24 h, starting within the prior 2 weeks, and without evidence of infection. Gadolinium enhancement on MRI, without clinical worsening, was not accepted as an attack. Age, sex ratio, race, duration of MS, time on therapy, and time after last injection of CR and PR, were not significantly different from each other, student's t-test. CR and PR, demographically close before study start, serve as internal replication cohorts for each other. An independent validation cohort consisted of 10 HC (age 48±5, 6F/4 M) and 8 age- and disability-matched clinically-stable relapsing-remitting MS patients (age 43•3 ± 6•1, 8F/0 M) who were receiving therapy with IFN-β−1a (30 mcg IM weekly), and who injected again after a 6•5-day washout.
Therapy-naïve RRMS patients, who had opted against disease-modifying drug therapy, included 10 with stable MS and 9 during exacerbations (stable: age 45•2 ± 2•6, 8F/2 M; active: age 46•3 ± 3•5, 8F/1 M). They were studied on a single occasion for spontaneous gene expression, without IFN-β exposure. They were matched with the treated patients for disease duration and disability severity. Eight healthy controls were sampled once, without IFN-β exposure (age 42•3 ± 4•8, 5F/3 M).
Inclusion criteria for all patients were clinically definite or laboratory-supported MS, ages 18–65, EDSS of 0–6•5. Exclusion criteria were medical problems such as cardiovascular disease or significant concurrent infections, and glucocorticoid use within six months prior to sampling. Patients who dropped out of the five-year study or who discontinued or changed therapy were not studied. Glucocorticoids were not given before or during on-study exacerbations. All subjects signed University of Chicago IRB-approved informed consents and the study was conducted according to the Declaration of Helsinki principles.
2.3. Interferon treatment in vivo with standard (250 ug) and double-dose (500 ug) IFN-β injections
Clinically stable patients had a planned >60-hour therapy-free interval before injections, to attenuate acute IFN-β-induced gene expression. After the IFN-β washout, pre-injection study blood was drawn between 8 and 10 AM (0 h). Patients then self-injected IFN-β in the clinic. Gene expression during stable disease was induced with an initial 500 ug (16 million units [MU] IFN-β (two injections of the regular dose of 250 ug [8 MU] of IFN-β−1b SQ, N = 26, or of 44 ug IFN-β−1a SQ, N = 1, grouped as 500 ug in text), followed by blood draws at 4 h and 24 h. One month later, the process was repeated, with a single dose of IFN-β (250 ug of IFN-β−1b or 44 ug of IFN-β−1a). CR and PR patients completed the two kinetic studies while clinically stable. All PRs also experienced attacks outside of the stable period of testing. They injected 500 ug of IFN-β in the clinic the morning after reporting the attack but not the evening before, with blood samples obtained as above. In all conditions, blood was drawn at tightly-controlled times, 4•06 ± 0•04 and 23•6 ± 0•14 h after injections (mean ± SEM). After these kinetic studies, patients reverted to their usual program of 250 ug every other day. Blood was drawn from healthy controls and therapy-naïve MS patients between 8 and 10 AM. The validation cohort was measured before and after injections after a 6•5-day therapy washout, with blood samples obtained as above.
PBMNC from 30 ml heparinized blood were purified on Ficoll density gradients. PBMNC were isolated within 1–4 h of phlebotomy and lysates were stored at −80Co in “buffer RLT plus” (Qiagen). Storage times of RNA in lysis buffer over this long-term study ranged from 1 to 63 months. Average storage times were as follows: group A = 54.3 ± 1.8 months, B = 48.6 ± 3.5, C = 35.9 ± 7.4, D = 2.4 ± 0.5, and E = 4.1 ± 1.1. There was no difference in purified RNA quality between long and short storage times; any trend was for better integrity with longer storage times. All RNA was freshly purified one week before the first plate scan. The scan dates were all within three weeks.
The washout time from the last home self-injection to the study injection for single doses was 103•0 ± 6•7 h, and for double doses was 116•7 ± 9•3 h, and was statistically equivalent between clinically stable CR and PR groups. The delay from last injection to phlebotomy was 73•5 ± 9•5 h in the exacerbating PR patients, not statistically different from the delay in the stable phases. These intervals well exceed the immediate gene-inducing effects of injections of IFN-β. IFN-β has largely left the circulation within one hour of injection [15], [16], [17] and new IFN-β-induced RNA expression in PBMNC is minimal beyond 24 h [7].
2.4. RNA preparation and microarray hybridization
Total RNA was extracted from each sample, reverse-transcribed, and amplified using the RNeasy Reagent Kit (QIAGEN, Valencia, CA). Quantitation and integrity of total RNA samples was measured by Agilent 2100 Bioanalyzer. RNA Integrity Number was calculated based on the entire electrophoretic trace of the RNA sample to evaluate the presence or absence of degradation products. RNA median integrity was 10, and average was 9.70±0.08 (RIN of 10 = intact; 0 = severely degraded). For RNA quality control, we evaluated Labeling Controls, Hybridization Controls, and Positive vs. Negative Area Under the Curve (AUC) measures, using TAC 4.0 software of CHP files. All 227 samples passed the Hybridization Controls and AUC, and all but one sample (99.56%) passed the Labeling Controls measure. The one sample that did not pass the Labeling Controls Threshold in TAC 4.0 was SM152 (Table S2). Since it passed the other two thresholds, and did not show as an outlier on PCA map, we decided to still include this one sample in downstream analysis.
Samples were randomized and then 100 ng RNA was hybridized to GeneChipⓇ Human Transcriptome Arrays (HTA) 2•0 according to manufacturer's instructions (Affymetrix, Santa Clara, CA). One sample with only 60 ng of RNA was assayed with no degradation in signal. HTA identify a broad dynamic range of expression in protein-coding RNAs, long non-coding RNAs (lncRNA, >200 bp), and other RNA biotypes, and detect very low-level signals such as lncRNAs and RNAs coding for cytokines and neuroprotective genes.
2.5. Gene expression analysis
Raw CEL files were imported, log2-transformed, and normalized using GC Correction and Space Transformation Robust Multi-array Average (GC-STT-RMA) algorithms in Affymetrix Power Tools (APT, version 1•17•0). Detection p-values <0•05 for each probeset with expression levels significantly higher than background noise were identified. 913,055 probesets designed for expression profiling were mapped to 67,539 coding and non-coding transcripts using HTA 2•0 annotation (Affymetrix version na34, human reference genome assembly hg19). Low-expressed transcripts are defined as those with at least half of the probesets not significant at the 0•05 level. Significant DEGs were identified using limma (Linear Models for Microarray and RNA-Seq Data) (version 3•26•8) [18], filtered by fold change of ≥1•5 (or ≤ −1•5), and FDR-corrected with p-value <0•05.
For detection of differentially expressed genes (DEGs), we excluded transcripts low-expressed in fewer than 5 samples (half sample size of the smallest group). In addition, we excluded from analysis 1 CR and 3 PR subjects with neutralizing antibodies to IFN-β (NAb+). Batch effects were evaluated for age, gender, race, and microarray scan date using pvca (Principal Variance Component Analysis) (version 1•10•0) [19]. These factors carried minimal batch effects in the study cohort except for microarray scan date, which was subsequently included as a covariate in the limma model, so as to adjust for potential batch effects introduced by different dates. DEGs within each group with significant IFN-β-induced short-term effects were detected at 0, 4, and 24 h, with 250 and 500 ug IFN-β doses, and two clinical statuses (stable and active). Both the factor of interest and the subject name were included in the model. Long-term sustained effects were studied by comparing CR and PR samples at baseline (0 h, 250 ug groups, clinically stable, after 4-day washout) to the healthy controls and to therapy-naïve stable and active MS groups. IFN response differences at 0 vs. 4 h between CR and PR were compared using subject name as the blocking factor in the limma model to adjust for individual variance. Canonical pathways significantly enriched in the genes of interest were identified by Ingenuity Pathways Analysis (IPA) (IngenuityⓇ Systems, www.ingenuity.com) based on experimental evidence from the Ingenuity Knowledge Base (release date 2015/06/27).
2.6. QuantiGene assay of RNA expression
Gene expression on microarrays was confirmed with branched DNA technology that relies on highly-specific cooperative hybridization between a target mRNA and multiple target-specific probes (QuantiGene Plex, ThermoFisher). This complex is detected through signal amplification by a branched DNA amplifier and then fluorescence signal generation, with a final result that is proportional to the original target abundance. The assay can reliably detect 10% differences, even in RNA of very low abundance. We tested 10 differentially expressed genes from biologically relevant pathways and three housekeeping genes, GUSB, HPRT, and PPIB, ranging from low to moderate expression. DEG included those induced immediately after IFN-β injection (4 short-term– CXCL10, LILRB4/ILT3, MX1, and TNFSF13B/BAFF) those expressed at higher levels in untreated MS compared to long-term-treated MS and healthy controls (6 long-term– CCL22, CD80, EIF2AK3/PERK, LILRB2/ILT4, SPP1/OPN, and IFNG-AS1/Tmevp1/ NeST, a lncRNA)).
Samples were assayed following manufacturer's protocol and all were run on one plate to avoid plate-to-plate variation. Sample median fluorescence intensity was corrected for background signal, and then was divided by the geometric mean of the three housekeeping genes to correct for amount of input RNA. Dose-response curves from 33 to 900 ng input RNA showed linear signals (Pearson's correlation r = 0•98), using serial dilutions from a pair of 0 and 4 h samples from a 16 MU PR stable patient, and compared to Quantigene kit Universal Standard RNA. For all assayed samples, 100 and 300 ng RNA replicates had close correspondence (coefficient of variation = 0•99).
2.7. Multiplex analysis of serum proteins
We constructed a 29-protein serum multiplex assay (ProcartaPlex™ Immunoassay, Affymetrix/Panomics/eBioscience, Santa Clara, CA), that included cytokines, chemokines, plus neuroendocrine and neurotrophic proteins. Assays were run with duplicate samples according to manufacturer's guidelines, but standard curves with 8 two-fold dilutions were expanded to 9 dilutions. Patient subtypes and kinetics groups were randomized between plates.
2.8. Statistical analysis
If not otherwise indicated, a Welch two sample t-test was used to compare gene expression differences between two groups, and Fisher's exact test was used to compare frequency of categorical factors between two groups. DEGs were identified using linear regression model constructed in limma. p < 0•05 was considered significant, and multiple testing correction was performed using Benjamini-Hochberg-FDR method. Values are given as mean ± SEM. Statistical analysis was performed using R 3•2•2•1 and Bioconductor.
2.9. Data availability
Processed data files are provided as supplementary tables. Raw microarray data files have been deposited into the NCBI Gene Expression Omnibus (GEO) repository, Accession #GSE138064.
3. Results
3.1. Short-term and long-term responses to IFN-β injection in MS
Short-term gene expression in paired, serial PBMNC samples were analyzed before (0 h) and 4 and 24 h after IFN-β injection (250 and 500 ug, or 8 and 16 MU) in CR and PR (Fig. S1). Long-term responses were compared between CR and PR groups after a 4-day IFN-β washout (0 h), and to untreated stable or active MS patients and healthy controls. 1 CR and 3 PR contained anti-IFN-β neutralizing antibodies (NAb+) in serum, hence excluded. 50 subjects were used for comparison of transcriptomes in response to IFN-β injection (Tables S1 and S2).
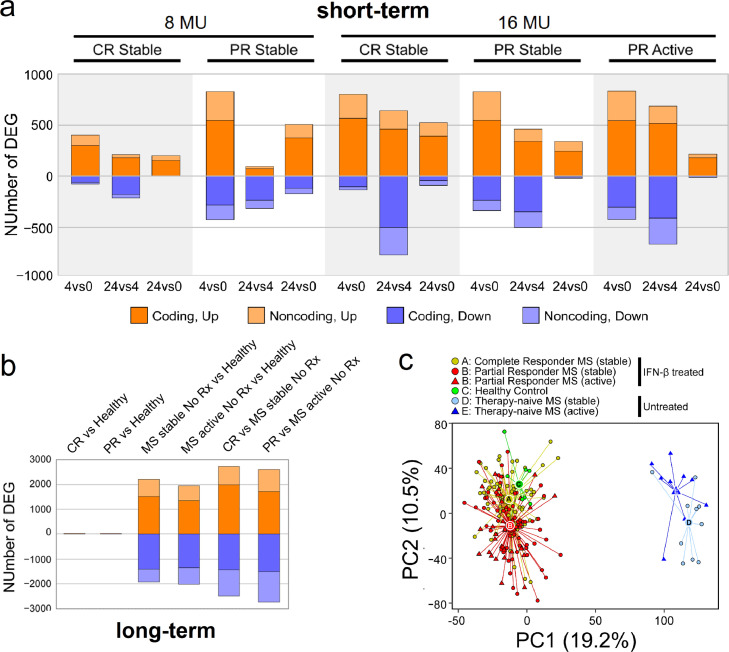
Short-term expression of 1,233 coding and 664 non-coding genes was altered in response to IFN-β injection (FDR-corrected p-value <0•05, fold change ≥ 1•5 or ≤ −1•5) (Fig. 1a). Known IFN-β targets such as CXCL10 (coding for IP-10), CXCL11 (I-TAC), CCL2 (MCP1), and MX1 (MXA) were among genes most strongly induced (Fig. S2a, b, and c). Immune regulation, inflammation, antiviral responses, cell cycle progression, and apoptosis promotion comprised biological processes most significantly affected. 35% of DEGs were lncRNA (Table S3, annotated DEGs), many regulating immune responses [20].
Fig. 1.
Differentially expressed genes (DEGs) after short-term and long-term IFN-β induction. (a) Short-term comparisons after 250 or 500 ug (8 or 16 MU) IFN-β injection. Panels include stable CR and stable and active PR without NAb to IFN-β. Comparisons on the x-axis include time 4 vs. 0 h, 24 vs. 0 h, and 24 vs. 4 h. The number of significant DEGs, with FDR-corrected p-value<0•05 and fold change ≥ 1•5 or ≤ −1•5, is shown on the y-axis. Genes upregulated are in orange; downregulated in blue. Coding DEG are in darker colour; non-coding genes in lighter colour. (b) Long-term compositions among baseline IFN-β-treated CR (stable) and PR (stable or active), therapy-naïve MS (stable or active), and healthy controls (HC), with FDR-corrected p-value<0•05, fold change ≥ 1•5 or ≤ −1•5. Colour coding same as in Fig. 1a. (c) Two-dimensional principal component analysis (PCA) of 227 samples using gene expression in all NAb-negative patients. Each circle or triangle represents one sample, for IFN-β-treated CR (A, yellow) and PR (B, red), 0, 4, and 24 h after injection, HC (C, green), therapy-naïve stable (D, light blue), and therapy-naïve active MS (E, dark blue). The variance of the five IFN-β-treated clinical groups is described by PC1 (19•2%) and PC2 (10•5%). The centroids of each group are labeled by letters A to E in larger circles or triangles. Euclidean distance between: CR and PR = 23•4; CR and HC = 15•4; PR and HC = 37•5; MS stable and MS active = 29•5; CR and MS stable = 132•5; PR and MS active = 124•7.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Long-term effects of IFN-β therapy were identified by comparing baseline gene expression among five groups: 4-day washout (time 0) IFN-β-treated stable CR, IFN-β-treated PR while stable and during a relapse, untreated therapy-naïve stable MS, untreated therapy-naïve MS, and healthy controls (Fig. S1). CR and PR had received standard IFN-β therapy for an average of 8 years before undergoing a 4-day washout to bring short-term therapy-induced gene expression back to baseline. 6,434 coding and 2,362 non-coding genes were significantly altered in one or more of the treated and untreated MS groups compared to healthy controls (Fig. 1b; Table S4). Remarkably, 95% (over 8,000) of altered DEGs in MS were discovered within the therapy-naïve groups. In contrast, during long-term IFN-β treatment, clinically stable and active patients retained an expression profile similar to that of HC.
Principal component analysis (PCA) showed that all 227 samples segregated by long-term treatment (on PC1) and by disease activity (on PC2) (Fig. 1c). The segregation pattern of long-term treated compared to untreated MS retains when using only the post-washout baseline expression profile, without short-term induction (Fig. S3). Thus, there is profound gene dysregulation at all times in untreated MS. Long-term IFN-β therapy induced a sustained shift to a near-normal profile in treated patients, where normalization likely persists beyond 4-day washout. Persistence of therapeutic effect may explain the clinical observation that after IFN-β treatment is stopped, weeks to months can elapse before overt clinical activity resumes [21].
3.2. Partial responders and complete responders differ in short-term and long-term responses to IFN-β
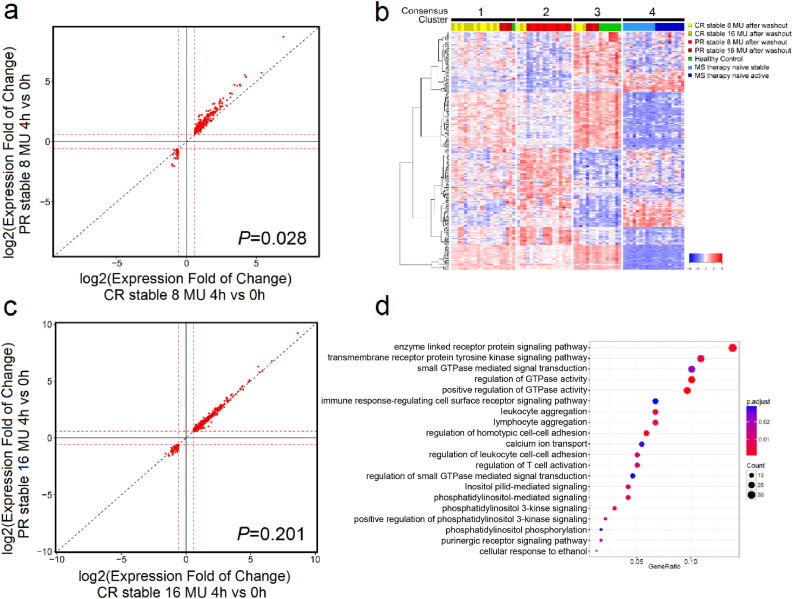
To examine short-term IFN-β-induced effects between stable PR and CR, we compared genes altered at 4 h after an IFN-β injection, relative to 0 h Among DEGs shared by CR and PR, an overall higher fold change of expression was observed in stable PR after 250 ug injection (slope >1•0, p-value = 0•028, Welch's t-test, two-sided) (Fig. 2a), and was also seen in Fig. 1a, with PR > CR after 8 MU injection. In contrast, similar fold changes were observed between stable PR and CR after 500 ug injection (Fig. 2b), suggesting most responses may have reached maxima.
Fig. 2.
Short-term IFN-β-induced gene expression distinguishes clinically stable PR from CR patients. (a) 250 ug (8 MU) or (b) 500 ug (16 MU) log2-transformed fold change (FC) of gene expression at time 4 vs. 0 h is shown for CR stable (x-axis) and PR stable (y-axis). A statistically significant stronger response is observed in PR compared to CR patients after 250 ug IFN-β injection, indicated by upward deviation of data points from the diagonal line (p-value = 0•028, Welch's t-test, two-sided), which is not observed after 500 ug (p-value = 0•201, Welch's t-test, two-sided). Only significant DEGs are shown. The red dashed line represents the 1•5 FC threshold. (c) 277 PR-specific genes in long-term IFN-β-induced gene expression patterns with FDR-corrected p-value<0•05 (gene list provided in Table S5). 50 samples from stable IFN-β-treated NAb-negative CR and PR after a 4-day washout, healthy controls, and therapy-naïve stable and active MS were grouped into 4 consensus clusters (cluster 1 to 4). (d) Gene Ontology (GO) terms significantly enriched in the 277 protein-coding genes from (c). The size of the dots reflects the number of genes; the colour indicates the enrichment p-value after multi-testing correction. The ratio of (number of genes from the 277-gene set) divided by (total number of genes annotated with this GO term) is shown on the x-axis; GO terms are shown on the y-axis.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To probe the long-term effects of IFN-β therapy, we studied PR and CR at their post-IFN-β-washout state (time 0). 277 genes had significant expression shifts in PR alone, 36 in CR alone, and 86 in common, relative to HC. The 277 PR-specific DEGs (Table S5) were used to group 50 subjects into four clusters through bootstrap-based consensus clustering [22] (Fig. 2c). Cluster 1 consisted of 75% CR, cluster 2 of 78% PR, cluster 3 of 50% HC, 25% CR, 25% PR, and cluster 4 of 100% therapy-naïve MS. Similar analysis using the 36 CR-specific DEGs did not distinguish among the 50 subjects. Gene Ontology (GO) analysis indicated that the 277 genes were enriched in regulation of GTPase activity, inflammatory responses, and T cell activation (Fig. 2c). These 277 genes differ from most prior studies in that they are long-term changes, measured after a therapy washout, and do not reflect acute IFN responses.
In PR compared to CR after washout, there were major expression differences in immune response pathways and neurodegenerative processes (IPA z-score >2•0 or <−2•0). We focus on genes relevant to MS immunity and potential CNS damage or repair because of their central role in the immunopathology of MS. For instance, the FoxO1 (forkhead box O1 transcription factor) signaling pathway was activated in PR. FoxO1 is anti-inflammatory as well as pro-inflammatory and neuroprotective [23]. It prevents proliferation of Th1 cells and is necessary for TGF-β-induced differentiation of regulatory CD4 cells (Treg), yet it also induces pro-inflammatory cytokines in adipocytes, dendritic cells, and macrophages [24]. The pro-inflammatory actions of FoxO1 may dominate in untreated MS, but in CR receiving IFN-β therapy, normalization of FoxO1 expression may reduce inflammation.
Pro-inflammatory pathways, GATA1, TGM2, and TP53 [25], were repressed by long-term IFN-β therapy from IPA upstream regulator predictions. The GATA1 transcription factor is vital for DC and mast cell development, antigen presentation and immune regulation. Downregulation of GATA1 has anti-inflammatory effects. Less tissue transglutaminase (TGM2), which activates NF-κB and promotes inflammation, would reduce inflammation. TGM2 also enhances protein cross-linking and amyloid-β deposition in Alzheimer's disease. Inhibition of TGM2 may be relevant to MS, since CSF β-amyloid levels predict disability in MS. Tumor protein p53 enhances IL-1β-induced neuronal death. Inhibition of TP53 expression might be beneficial, because high levels of IL-1β correlate with active neuronal degeneration in MS [26].
3.3. Time-dependent variation in gene expression in short-term responses to IFN-β
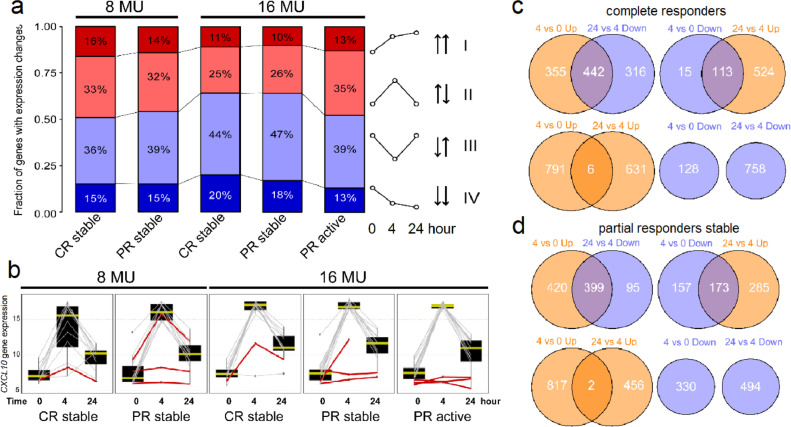
Among short-term gene expression differences, we observed four types of temporal change shared by CR and PR (Fig. 3a). Type I DEGs, composing 15% of transcripts, were upregulated at 4 h and/or at 24 h. Those induced at 4 h include LILRB4 (coding for immunoglobulin-like transcript 3 (ILT3)), is expressed on antigen-presenting cells and potently induces tolerance when binding to CD166 on activated T cells. CD163, a marker for immunosuppressive M2 macrophages, had a similar pattern. C2, rate-limiting in the complement cascade, was induced at both 4 and 24 h. Less frequently, expression increased first at 24 h, as with VDR (vitamin D receptor). Vitamin D synergistically potentiates IFN-β-induced ILT3 synthesis [27].
Fig. 3.
Time-dependent IFN-β effects on gene expression patterns. (a) Proportion of genes in each kinetics category after short-term IFN-β induction in clinically stable CR and stable and active PR, after 250 or 500 ug (8 or 16 MU) IFN-β, excluding NAb+ patients. To make the calculated percentage comparable across groups, we united DEGs that are significant in any of the five groups into one list (short-term category genes shown in Table S3). This is the percentage of genes falling into each of the four types of temporal change from the same total number of DEGs for every group. There was high statistical consistency from uniting DEGs significant in any of the five groups, and provides a common ground to compare the pattern across groups. (b) The CXCL10 gene is up-regulated at 4 vs. 0 h, and down-regulated at 24 vs. 4 h. NAb+ patients show reduced response at 4 h; the average induction of CXCL10 dropped from 8•5-fold in NAb- patients to 1•9-fold in NAb+ patients, a 78% decrease (red lines); also see Fig. S4. (c and d) Venn diagrams of overlapping DEGs, comparing 4 vs. 0 h, and 24 vs. 4 h after 500 ug IFN-β in (c) stable CR and (d) stable PR groups. DEGs were filtered by FDR-corrected p-value<0•05, and fold change ≥ 1•5 or ≥ −1•5. Genes upregulated are in orange; genes downregulated are in blue. Venn diagrams show the number of 1) Genes up at 4 h relative to 0 h, overlapping with genes down at 24 h relative to 4 h (left, top); 2) Genes down at 4 vs. 0 h, overlapping with genes up at 24 vs. 4 h (right, top); 3) Genes up at 4 vs. 0 h, overlapping with genes up at 24 vs. 4 h (left, bottom); and 4) Genes down at 4 vs.0 h, overlapping with genes down at 24 vs. 4 h (right, bottom).(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Type II DEGs (30%) were induced at 4 h but reverted towards baseline by 24 h. An example is CXCL10, codes for a chemoattractant for PBMNC (Fig. 3b). Chemokines are classic IFN-induced genes that have strong effects on immune cell migration and retention in lymphoid and target tissues and cause lymphopenia during IFN-β therapy [28]. To demonstrate the downstream consequences of RNA induction and relevance to MS, we studied chemokine protein expression with a Luminex multiplex assay, and validated the findings in a second independent cohort of 10 MS patients receiving IFN-β therapy (see Methods). I-TAC (IFN-inducible T cell alpha chemoattractant, CXCL11), IP-10 (IFN-induced protein-10, CXCL10), and MCP1 (macrophage chemotactic protein-1, CCL2) were induced at 24 h and returned towards baseline levels at 48 h (Fig. S2b and c). This paralleled the type II pattern of RNA expression, with a 1-day delayed recapitulation of peak levels.
Type III DEGs (40%) were repressed at 4 h but shifted back towards baseline by 24 h. An example is IFNGR1 (IFN-γ receptor α chain). Diminished levels of IFN-γ receptor could lessen IFN-γ-driven inflammation. Type II and III DEGs, unlike Type I and IV DEGs, often showed greater response to 500 ug than to 250 ug of IFN-β (Fig. 3c and d).
Type IV DEGs (15%) were repressed at 4 h and equally or more so at 24 h. Among these, KLRB1 (killer cell lectin-like receptor B1, CD161) codes for a costimulatory molecule that is increased in untreated MS patients [29]. Consistent with that, KLRB1 transcription was down-regulated in all IFN-β-treated groups in this study. In support, IFN-β therapy reduces the number of CD8+ IL-17-secreting cells in MS [30]. CHD7 (chromodomain helicase DNA binding protein 7), not previously linked to MS, was repressed in CR and stable PR, but not in active PR. It is required for lymphocyte development, and has a role in neural crest guidance where its absence causes cranial nerve dysfunction [31]. In Fig. 3a, downregulated genes are increased (blue bars) compared to Figs. 1 and 2A/B, indicating that upregulated genes predominate in response to IFN-β.
Taking the short-term DEGs, we investigated their pattern in the four NAb+ patients. Of note, only 7% of the genes showed >80% reduction at 4 h after injection (Fig. 3b, red lines; Fig. S4a and b, stars; Table S6). Repression was nearly complete at a titre of >500 NU (neutralizing antibody units) for 250 ug injections, but was partially overcome with 500 ug injections (Fig. S4c). Overall, a clear anti-correlation was observed between NAb units and IFN-β induced gene expression fold changes, with higher titres having less than 30% of the short-term DEGs induced in the NAb+ patients (Spearman's correlation ρ = −0•93, p-value = 0•002). A prior study, using pre-selected patients with no in vivo induction of MxA protein, found complete suppression of responses to IFN by NAb [32]. In unselected NAb+ patients, we see a titre-dependent reduction in responses at standard dose IFN-β injection, with a threshold at ∼500 NU.
To examine the spectrum of short-term genes across treated patients, we grouped DEGs into 12 clusters using unsupervised hierarchical clustering (Fig. S5). Gene expression of each cluster rose or fell in a repetitive manner across 0, 4, and 24 h after 250 or 500 ug of IFN-β. Clusters 1–7 contain genes induced by IFN-β at 4 h, with the strongest response in cluster 1 and weakest in cluster 7. In contrast, clusters 8–12 contain genes downregulated at 4 h, with magnitude of inhibition increasing from cluster 8 to 12. These results suggested highly reproducible and consistent time-dependent IFN-β-induced modulation under short-term conditions.
3.4. Acute transcriptional shifts after IFN-β injection are minimal in Th1/Th17/Th2 pathways
IFN-β has been ascribed a prominent role in the direct blunting of CNS-destructive Th1 and Th17 responses, as well as a role in enhancing protective Th2 cell responses. Nonetheless, much of the MS literature is discordant [33]. In this study, short-term shifts at the RNA level were minimal at 4 and 24 h after IFN-β injection for almost all genes in these pathways, in all treated MS groups, for 250 and 500 ug IFN-β (Fig. S6a, short-term).
There were several exceptions with Th1/Th17/Th2-involved genes. IFNGR1 was repressed, potentially diminishing response to IFN-γ (Fig. S2a). IFNG (coding for IFN-γ, a Th1 cytokine) showed a trend for reduction at 4 h that was restricted to clinically active PR patients (p-value=0•03, unadjusted; p-value=0•11, FDR-corrected). Despite minimal short-term induction, profound long-term effects were observed (described below).
3.5. Long-term IFN-β treatment induces expression of neuroprotective protein-coding RNA and lncRNA
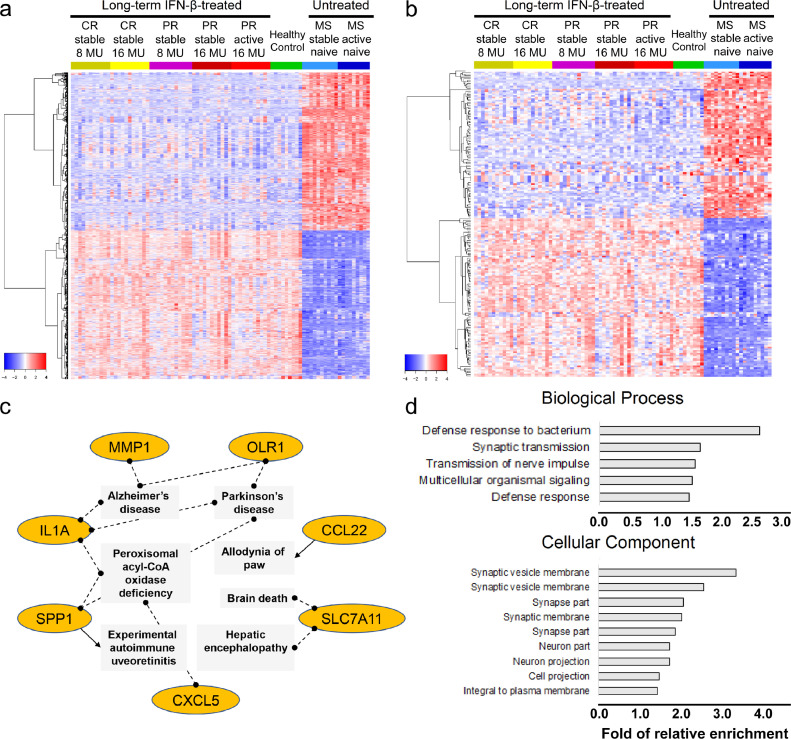
Long-term IFN-β treatment led to a profound reversal of approximately 6,000 protein-coding genes and 2,000 lncRNAs dysregulated in untreated MS. The 50 subjects form two large clusters based on gene expression profiles, the treated, post-washout stable and active MS which shows similar patterns to healthy controls, and the untreated stable and active MS, each equipped with unique sets of DEGs (Fig. 4a and b). Unsupervised hierarchical clustering grouped long-term DEGs into 12 clusters based on variations in magnitude of expression change (Fig. S7). Cluster 1 showed the highest expression in PR, CR, and HC, and the lowest expression in untreated stable and active MS. Cluster 12 showed the lowest expression in PR, CR, and HC, and the highest expression in untreated patients.
Fig. 4.
Long-term IFN-β treatment induction of dramatic transcriptome changes. (a) 6,434 significant protein-coding DEGs and (b) 2,362 significant non-coding DEGs are shown. Clinically distinct groups are labeled in the horizontal annotation bar above the heatmap: IFN-β therapy clinical responders (CR, yellow) and partial responders (PR, red) after therapy washout, healthy controls (HC, green), therapy-naïve stable MS (light blue), and therapy-naïve active MS (dark blue). CR and PR refer to samples collected at 0 h after 4-day washout. Genes are on the row, and samples on the column. Dendrogram of gene clusters is shown at left of heatmap. (c) Neurodegenerative network genes enriched in long-term gene cluster 12 from Fig. S7. Genes upregulated in therapy-naïve MS stable and active groups compared to baseline CR, baseline PR, and HC are shown in orange. (d) Gene Ontology terms significantly enriched in the 80 annotated lncRNA genes (FDR-corrected p<0•05). The bar indicates the relative fold enrichment. 250 ug = 8 MU; 500 ug = 16 MU IFN-β.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We pursued with DEGs from cluster 12, which showed the largest magnitude of expression upregulation in untreated MS and inhibition in long-term IFN-β-treated patients. Interestingly, pathway analysis revealed that those genes are enriched in detrimental inflammation (SPP1, coding for OPN, osteopontin), CCL22, CXCL5, IL1A, and MMP1) and degenerative CNS diseases (OLR1, SLC7A11) (Fig. 4c). Osteopontin binds integrin α4β1 to activate NF-κB, promote Th1 and Th17 cell activation, and enhance secretion of MMP1 that facilitates immune cell passage across the blood-brain barrier. OPN is overexpressed in experimental autoimmune encephalomyelitis (EAE), an antigen-specific animal model of MS, and in MS lesions [34]. In support of our findings, IFN-β inhibits production of OPN protein in CD4+T cells [35].
OLR1 (aka LOX1) and SLC7A11 were overexpressed in untreated MS but downregulated by long-term IFN-β therapy. OLR1 (oxidized low-density lipoprotein receptor 1) codes for a C-type lectin receptor on endothelial cells, monocytes, and microglia that clears myelin breakdown products and oxidized low density lipoproteins. It also promotes humoral responses and atherosclerosis [36]. OLR1 appears at the rims of chronic active MS lesions [37]. In this study, during exacerbations in IFN-β-treated patients (PR active), OLR1 expression gets reduced to normal levels, while in exacerbating untreated MS, it was induced by 365-fold (Table S4). In this unbiased approach, expression of a large number of neurodegenerative genes was altered. SLC7A11 (solute carrier family 7, member 11) codes for a sodium-independent cysteine/glutamate antiporter found in PBMNC. In astrocytes, it regulates synaptic activity; in CNS microglia, it is implicated in neurotoxicity and death of oligodendroglia [38].
Additionally, neuroprotective genes such as HGF and CNTF were upregulated by long-term IFN-β exposure (Table S4). HGF (hepatocyte growth factor) induces Treg, drives dendritic cells to tolerize T cells, and induces IL-10, which inhibits EAE [39]. CNTF (ciliary neurotrophic factor) induces growth of neurons and oligodendrocytes. It counteracts TNF-α-mediated damage to oligodendrocyte precursor cells [40], shows protection in EAE, and is under study in MS clinical trials [41]. In summary, the results suggested that long-term IFN-β therapy reversed overexpression of inflammatory signaling and tissue-destruction, and promoted signals of neuroprotection. lncRNAs modify transcription and translation of coding genes and have an emerging role in autoimmunity [42]. We identified 664 altered lncRNA transcripts in MS PBMNC 4 h after IFN-β injections. 81 had annotated gene descriptions, and were enriched for genes implicated in immune defence, synaptic transmission, and neuronal protection (Fig. 4d). An example is IFNG-AS1 (alias Tmevp1, NeST), which was induced in 60–70% of CR and PR patients at 4 h after injection. In long-term treatment, IFNG-AS1 remained at higher levels in treated patients after IFN-β washout than in untreated patients. Consistent with our findings, IFNG-AS1 enhances IFN-γ secretion by Th1 and CD8 T cells and drives inflammation [43]. Of note, IFNG-AS1 also has regulatory effects, potently inhibiting IL-17 production, augmenting indoleamine 2,3 dioxygenase (IDO) secretion, and suppressing EAE. It remains unknown whether IFNG-AS1 acts likewise on regulatory CD8 cells, which secrete IFN-γ and IL-10 to suppress activated T cells.
Changes in lncRNA expression relative to healthy controls, accounted for a third of the DEGs in long-term IFN-β-treated patients after washout, or in untreated MS groups (Fig. 4b; Table S4). Among those, NRAV (alias DYNLL1-AS1) is a virus-induced lncRNA that suppresses type I IFN responses, inhibiting anti-viral immunity [44]. Our previous work reported its deficient responses to IFN [5], which may be relevant to the high NRAV levels detected in untreated stable and active MS patients. In contrast, low NRAV expression in CR and PR after long-term IFN-β therapy, was comparable to that in healthy controls. Reversal of NRAV levels by IFN-β therapy could potentially enhance anti-viral immunity and eliminate inhibition of type I IFN responses.
3.6. Long-term IFN-β therapy reverses the gene dysregulation of therapy-naïve MS
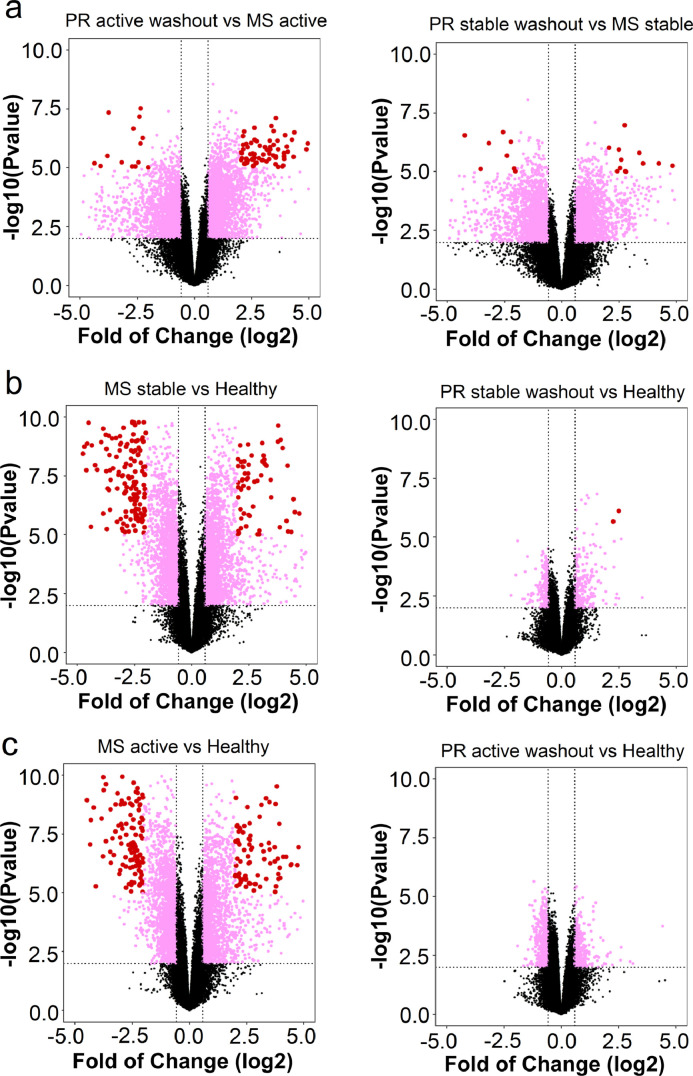
During relapses, untreated patients showed considerably more frequent and more extreme changes in gene expression than IFN-β-treated PR patients (Fig. 5a). 113 genes were highly-affected (log2 fold change ≥ 4•0 or ≤ −4•0) (Fig. 5a, red dots; Table S7). Among these, 33 are associated with immunity, six with immune suppression, 21 with neuronal function or CNS disease, and six with IFN signaling or anti-viral responses. Compared to healthy controls, gene expression was highly dysregulated in untreated clinically stable and relapsing MS (Fig. 5b and c, left). In contrast, IFN-β-treated PR patients after 4-day washout had a transcriptome profile similar to HC (Fig. 5b and c, right).
Fig. 5.
Gene expression changes comparing long-term IFN-β-treated to untreated patients. (a) Left: IFN-β-treated clinically active PR patients after 4-day washout compared to untreated clinically active MS. Right: IFN-β-treated clinically stable PR patients after 4-day washout compared to untreated clinically stable MS. (b) Left: untreated clinically stable MS compared to healthy controls. Right: IFN-β-treated clinically stable PR after 4-day washout compared to healthy controls. (c) Left: untreated clinically active MS compared to healthy controls. Right: IFN-β-treated clinically active PR after 4-day washout compared to healthy controls. Log2-transformed gene expression fold change is shown on the x-axis, and −log10 transformed p-value (unadjusted) on the y-axis. Each data point represents one gene. Genes with p-value <0•01 and fold change ≥ 1•5 or ≤ −1•5 are labeled in pink; those with fold change ≥ 4•0 or ≤ −4•0 are in red.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Compared to healthy controls and long-term treated patients (baseline, after 4-day washout), untreated stable and active MS patients’ PBMNC showed increased expression of many genes coding for components of NF-κB, such as RELA/p65, RELB/p52, REL, NFKB1, and NFKB2, which induce inflammatory cytokine expression. The results indicated that without long-term IFN-β treatment, proinflammatory cytokines and failed immune restraint may create an immune system poised for activation and tissue destruction in MS. As a counterpoise, long-term IFN-β substantially downregulated NF-κB, potentially reducing matrix metalloproteases (MMPs), costimulatory molecules, and inflammatory cytokines and chemokines (Fig. S6, long-term). MMP9 disrupts the basement membrane of brain venules and glia limitans, and thereby permits autoreactive immune cell penetration into the brain parenchyma. Of note, MMP9 was among the most highly affected DEGs, with 17-fold expression reduction during long-term treatment (Table S4).
The costimulatory molecule, CD80, promotes antigen-specific immune activation. We previously found that its over-expression on B cells in MS was reversed by IFN-β therapy [45]. In this study, CD80, highly expressed in untreated patients, was repressed by three- to five-fold in treated CR and PR, to near-normal levels. Long-term IFN-β also induced regulatory genes such as immunosuppressive CD163, a marker for immunosuppressive M2 macrophages, and ILTs (Fig. S8). During long-term therapy, increased expression of immune regulatory genes, coupled to substantially decreased expression of genes linked to inflammation and to neurodegeneration, should quell inflammation in the periphery and possibly in the CNS.
3.7. Short-term and long-term IFN-β treatments have different effects on gene signatures
Contrasting DEG lists identified genes with significant short-term effects only, long-term effects only, or both. For short-term effects, we restricted DEGs to those arising from 250 ug injections, a stimulus comparable to standard long-term IFN-β MS therapy.
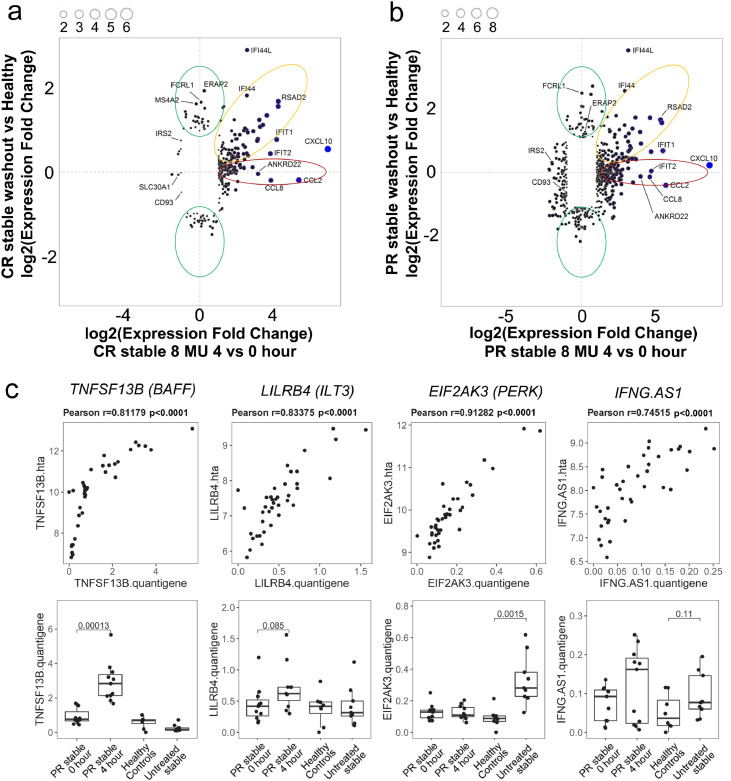
Differences in CR at 4 h after IFN-β injections were correlated with differences in CR baseline (after 4-day-washout) compared to healthy controls (Fig. 6a), and similarly in stable PR (Fig. 6b). QuantiGene Plex validated expression of selected genes, with high correspondence to HTA microarrays (Fig. 6c; Fig. S9).
Fig. 6.
Short-term compared to long-term effects of IFN-β injections in CR and PR patients and validation of DEG expression. (a) Short-term effect in stable CR after 250 ug IFN-β injection at 4 vs. 0 h is displayed on the x-axis, showing log2-transformed gene expression fold changes. Long-term effect in stable CR at baseline (0 h, after 4-day washout) vs. healthy controls is on the y-axis. (b) Short-term effect in stable PR after 250 ug IFN-β at 4 vs. 0 h is compared to long-term effect in stable CR at baseline vs. healthy controls. Of 67,518 genes captured on the array (including coding and non-coding transcripts), 6344 genes significantly altered by short-term and/or long-term IFN-β exposure are shown in (a) and (b). Red ellipse, genes significantly induced by short-term, but not by long-term IFN-β exposure. Yellow ellipse, genes significant after both short- and long-term exposure. Green ellipse, genes significantly altered by long-term, but not by short-term exposure. (c) Correlation of gene expression between microarrays and QuantiGene Plex validation assays, and expression differences between patient groups. Genes shown are highly correlated after short-term IFN induction (TNFSF13B/BAFF and LILRB4/ILT3) and long-term IFN induction (EIF2AK3/PERK, and IFNG-AS1/Tmevp1/ NeST, a lncRNA). Upper panels: gene expression correlation between HTA microarray platform (y-axis) and QuantiGene (x-axis), with Pearson's correlation coefficient r and p-values shown above each panel. Bottom panels: gene expression changes measured by QuantiGene in four patient groups (PR stable patients at 0 and 4 h after IFN-β injection, healthy controls, and untreated MS stable patients). Data shown are from 100 ng input RNA; replication with 300 ng had a very similar profile (data not shown). Four representative genes of 10 tested; the other six are shown in Fig. S9. Pearson's correlation was used in (c) upper panel, and Student's t-test was used in (c) bottom panel.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Three major categories emerged: genes (1) induced by short-term, but not by long-term IFN-β exposure (Fig. 6a and b, red ellipse), (2) altered by long-term, but not by short-term exposure (green), and (3) induced after both short- and long-term exposure (yellow). PR, compared to CR, showed a higher number of genes with significant response to both short- and long-term IFN-β exposure. The greatest relative changes arose acutely 4 h after an injection (red). The majority of the DEGs fell into category 3 (yellow), suggesting that after nearly a decade of therapy, many of the genes most responsive to IFN-β at 4 h remain responsive 4 days post-washout.
CR showed 35 genes that were upregulated by at least 2-fold 4 h after injection, but not during long-term treatment (Fig. 6a, red). Among these were immune response genes, IDO1, CD274, CCL2, CCL8, TAP1, and STAT1. Indoleamine 2,3-dioxygenase (IDO) and PD-L1 (CD274) are immunoregulatory; the others affect immune cell migration, antigen responses, and cytokine production. IDO is strongly induced by type I and type II IFNs in cultured plasmacytoid dendritic cells (pDC), where it promotes its own expression for a month. In concert with TGF-β, IDO-driven kynurenine induces CD4+FoxP3+ Treg [46]. IFN-β induced expression of IDO1 at 4 and 24 h potentially corrects subnormal Treg function in MS. After long-term treatment, IDO1 expression was reduced by at least 7-fold in treated CR and PR, to a near-normal level comparable to that of healthy controls, indicating less immune dysregulation. Conversely, nine other genes with minimal short-term change in CR were induced long-term (Fig, 6a, green) (BCL2, CHRM3-AS2, KIR2DS4, NELL2; AREG, CPA3, FKBP5, MAL, and MS4A2).
PR exhibited 104 genes with short-term induction (Fig. 6b, red), similar to CR, involve many immunoregulatory genes. Eleven genes with only long-term responses showed induction (Fig. 6b, green) (BCL2, CHRM3-AS2, KIR2DS4, NELL2; CD22, CD79A, KLHL14, RAB30, RALGPS2, SNORD51, and TAPT1); the first four overlap with long-term-only induction in CR. BCL2 showed 2•5-fold long-term, but no short-term, induction in PR and CR relative to HC. Persistent long-term elevation of anti-apoptotic BCL2 may protect PBMNC, oligodendroglia, and neurons against inflammation-induced cell demise.
3.8. Interferon affects short- and long-term transcription in multiple pathways implicated in MS immunopathology
Activated macrophages and microglia attack oligodendrocytes in MS plaques. EIF2AK3 codes for eIF2a kinase (alias PERK, protein kinase RNA-like ER kinase). eIF2a kinase is an essential participant in the integrated stress response that protects oligodendroglia from toxic misfolded proteins induced by oxidative stress during viral infection, inflammation, and CNS damage [47]. It is related to EIF2AK2 (PKR), an antiviral gene strongly induced by IFN-β [48]. EIF2AK3 is higher in therapy-naïve patients, but was down-regulated in PBMNC of stable PR at 4 and 24 h after IFN-β injection. Less EIF2AK3 could subvert the brain-protective integrated stress response [47]. DDIT3 (DNA Damage Inducible Transcript 3, alias CHOP) is downstream from eIF2a kinase, but conversely, activates apoptosis once proteostasis has failed. DDIT3 is a potential vector of oligodendrocyte death in MS [47], and was down-regulated by 2-fold at 4 h after IFN-β in stable and active PR. A similar trend was detected in CR. These short-term decreases in DDIT3 could prevent apoptosis of PBMNC and possibly of oligodendrocytes. With long-term IFN-β treatment, EIF2AK3 expression is low in all groups, suggesting that there is less ongoing inflammatory CNS damage during therapy. DDIT3 however was down-regulated by 3-fold in clinically active PR compared to untreated patients during exacerbations. Thus, long-term IFN-β therapy may reduce the neurotoxic integrated stress response.
Some IFN-β-induced genes suppress inflammation directly. IL-15 expression increased 4 and 24 h after injection of IFN-β (Fig. S6, short-term; Table S3). IL-15 induces proliferation of cytolytic CD8 cells, yet also blunts their function [49]. Cytolytic CD8 cells can sever axons in vitro [50]. However, IL-15 also induces regulatory CD4+FoxP3+ and CD8+CD28- T cells that secrete IFN-γ, IL-10, and other cytokines capable of resolving inflammation and enhancing tissue healing [49]. In support, we find that in vitro and in vivo IFN-β therapy enhances regulatory CD8 cell function in MS [51].
BAFF (B-cell activating factor, alias TNFSF13B, BLyS) induces regulatory B cells that produce IL-10, and that help generate CD4+CD25+FoxP3+ regulatory Tregs, the T cell subset with the highest expression of BAFF receptors [52]. Arguing for a protective role, serum BAFF levels are higher in stable than in active disease in untreated patients, and levels rise with IFN-β therapy [53]. Blocking BAFF and APRIL (alias TNFSF13), which share receptors, exacerbates MS. In this study, 4 and 24 h after IFN-β injection, BAFF expression rose in CR and stable and active PR (Fig. S6; Table S3). Expression of 62 genes, including BAFF, was altered in a longitudinal study of IFN-β-induced gene expression, two days to one year after starting therapy [54]. 46 of these overlap with genes having short-term expression change (Table S3, footnote) and 15 do not. Of these 15, 4 are changed long-term (Table S4, footnote). After long-term IFN-β therapy, BAFF and APRIL expression was upregulated relative to untreated MS (Fig. S6, long-term). Elevated BAFF may promote the higher immunoglobulin secretion typical for MS. BAFF also induces regulatory B and T cells and neurite outgrowth, potentially important in CNS repair.
CD8+CD28- regulatory T cells are found in chronic inflammatory MS plaques [55]. Regulatory CD8 T cell contact doubles expression of ILT3 on primate macrophages and dendritic cells. ILT3 levels on monocytes fall 5-fold during MS exacerbations, but return to normal levels during remissions [56]. We and others find that IFN-β induces ILT3 on monocytes in vitro and in vivo, reducing inflammation by binding to CD166 on activated T cells, tolerizing them after cognate antigen exposure [27,56]. In PR during MS exacerbations, IFN-β injection induced ILT3 at 4 and 24 h (Fig. S8a), potentially increasing the otherwise low levels of this anti-inflammatory factor. Long-term IFN-β therapy also increased immune control by upregulating ILT2 (LILRB1), coding for a protein on regulatory NK and T cells; ILT4 (LILRB2), with its protein expressed on endothelial cells and antigen presenting cells; and ILT5 (LILRB1), with ILT5 functioning as an immune-inhibitory MHC class I binding protein (Fig. S8b).
Engagement of Toll-like receptors (TLR) 3, 7, 8, and 9 activates innate anti-viral immune responses and induces type I IFN secretion. IFN-β injection rapidly increased expression of TLR3 and TLR7, coding for proteins that detect viral dsRNA and ssRNA, respectively (Fig. S8c). Long-term therapy increased the low expression of TLR in untreated MS to a level close to healthy controls, including TLR4 (receptor for LPS), TLR5 (for flagellin), TLR7 (for viral ssRNA), TLR8 (for ssRNA), and TLR9 (for CpG DNA) (Fig. S8d). IFN-β-induced upregulation of TLR3 and TLR7 could increase IFN-α and β secretion and thus prolong effects of injected IFN-β [57].
4. Discussion
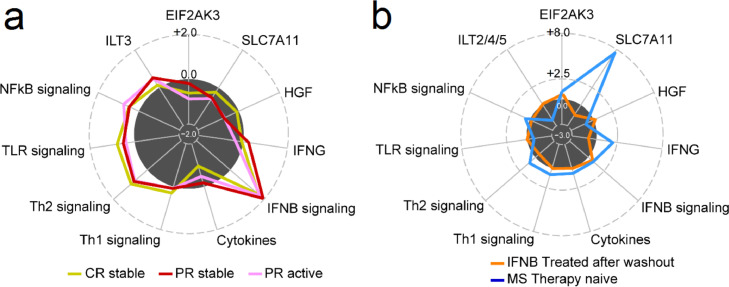
IFN-β induces short-term and long-term gene expression in immunosuppressive pathways and induces long-term reduction of expression for CNS-destructive Th1 and Th17 cell products. Short-term and long-term induction of neuroprotective genes by IFN-β may foster brain repair after inflammatory damage (Fig. 7; Table S8).
Fig. 7.
Transcription in genes and pathways relevant to MS immunopathology. (a) Short-term expression fold change (FC) in three patient groups: stable CR (gold), stable PR (red), and active PR (pink). For each group, an average FC was calculated across doses (250 ug and 500 ug) and clinical status (stable and active), at 4 h relative to 0 h after injection. (b) Long-term expression fold change is shown in baseline treated (orange), and untreated (blue). For each group, an average FC was calculated across disease status (stable CR and stable PR, or stable and active MS), compared to healthy controls. Numbers on the radius represent log2-transformed expression fold change. The edge of dark circle represents no changes (log2FC = 0•0). Genes and pathways include EIF2AK3, essential in unfolded protein response; SLC7A11, neurodegeneration; HGF, neuroprotective; IFN-α and β signaling excluding IFN-γ; NF-κB, cytokine gene activation; and ILT, immune suppression.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
An IFN-β injection changes short-term expression of over 1,200 genes in MS PBMNC. Classic IFN-stimulated genes coding for antiviral proteins and chemokines are upregulated, but the injection does not meaningfully alter expression signals in Th1, Th17, or Th2 pathways. Studies of MS exacerbations in therapy-naïve patients report elevation of inflammatory Th1 and Th17 products and diminution of protective Th2 cell products, which activates CNS-destructive macrophages and microglia [1,2], though evidence is often inconsistent. Our finding parallels the minimal Th1/Th17 response to type I IFNs in EAE, despite a major protective role for IFN-β in this MS model [58]. It raises the prospect that disease-attenuating actions of IFN-β lie elsewhere, such as induction of immune-regulatory or tolerizing genes, plus possible contributions from neuroprotective genes. This would correct the immune milieu in untreated MS, characterised by high levels of immune cytokines and costimulatory molecules, and reduced expression of multiple immunosuppressive pathways that may allow clonal expansion of T and B cells responsive to even low-affinity antigens.
Pre- vs. post-therapy signatures have been extensively studied, although often with mixed patient populations, undefined injection schedules, and less comprehensive arrays (reviewed in [48,59]. We used a different approach to evaluate carefully controlled induction kinetics with two IFN doses in CR and PR to test the abnormal IFN system in MS. Untreated patients had >8000 genes abnormally expressed compared to HC and IFN-β-treated subjects. These included Th1 and Th2 cytokines and type I IFN-stimulated genes, confirming and extending gene expression seen in many prior studies [[6], [7], [8], [9], [10],59]. IFN-stimulated genes (ISG) in the present paper correspond to those in earlier studies but are more numerous, because many evaluated these short-live RNAs at indeterminate times after injections.
Short-term gene induction and inhibition after an IFN-β injection were of greater magnitude and longer duration in clinically stable PR than in CR, and were enriched in inflammatory, immune regulatory, and neuroprotective DEGs. It is counter-intuitive that PR would have greater response to IFN than CR. However, 20% of untreated MS patients have normal or slightly elevated type I IFN levels in blood and strong in vitro responses to IFN-β in monocytes and PBMNC, but do less well after starting IFN-β therapy than those with subnormal IFN levels [8,9,17]. An excessive IFN signature in therapy-naïve MS correlates with more MRI and clinical disease activity reviewed in [59]. We have found more extreme examples of supra-normal IFN signatures in systemic lupus erythematosus (SLE) and demyelinating neuromyelitis optica (NMO), where patients have very high serum type I IFN levels and have strong responses to IFN-β, and will frequently deteriorate with IFN-β therapy [17]. By segregating patients into PR and CR and then evaluating immediate post-injection kinetics, we find that PR have greater and longer responses to IFN-β. CR, with lower levels of abnormal gene expression after washout, had more modest response after injections, perhaps to an optimum level for suppression of relapses.
In contrast, 80% of untreated relapsing/remitting MS patients have low levels of serum type I IFNs and subnormal responses to IFN-β in vitro and in vivo compared to HC [8,13,17,60]. Subnormal IFN response in untreated MS is linked to a worse disease course [5,17]. We find that during ongoing IFN-β therapy, after washout, CR with the lowest resting and IFN-induced IFN signature showed better clinical response to IFN than PR with high resting and induced signature. (Figs. 1a, 2a, 3c, d, 6a, b). A balance between too little IFN-β, and not too much IFN-β, appears relevant to the efficacy of IFN-β therapy in MS and inflammatory CNS diseases.
The effects of IFN-β on RNA expression parallel the dose-dependent IFN-β effects on clinical responses. In formal clinical trials of mildly affected patients with stable MS, higher IFN-β doses did not lessen attack frequency [61,62] or preserve cognition [63], but did enhance repair of MRI brain lesions. In MS patients with moderate disability (Extended Disability Scale Score = 3•5–5•5/10), however, 250 ug of every-other-day IFN-β-1b had more clinical and MRI benefit than 1•6 MU, which was superior to placebo [64]. Similarly, in moderate disability MS, thrice-weekly 44 ug IFN-β-1a slowed accumulation of disability more than weekly injections [64], and had more cognitive benefit than 22 ug injections [63]. Thus, patients with low clinical severity have good clinical responses to both doses of IFN-β, but in more severe MS, higher doses have more benefit. Our data on dosage effects show that it was logical to clinically test even higher IFN-β doses. PR during exacerbations, compared to clinically stable periods, had delayed and reduced gene induction after 500 ug IFN-β injection, but still could generate anti-inflammatory and neuroprotective responses. Our data suggest that doubling the IFN-β dose during attacks might overcome disease activity-associated resistance to IFN-β.
4.1. Long-term IFN-β therapy corrects dysregulated immune and neuroprotective pathways in MS
During remission and attacks, PBMNC of untreated MS patients are in a seriously dysregulated state, aptly termed a “cytokine storm” (52). Long-term IFN-β therapy corrects this cytokine storm, suggested by a near-normal expression profile in stable PR and CR after washout. A state of cytokine harmony appears with long-term IFN-β exposure. The average of 8 years of IFN-β therapy studied here is beyond the 4–5 years treatment known to prolong patient survival [12]. Long-term therapy downregulates expression of pro-inflammatory genes coding for NF-κB, inflammatory cytokines in Th1/2/17 pathways, and costimulatory proteins. This is coupled to upregulated expression of immunoregulatory genes, such as immune-inhibitory cytokines, and IDO, PD-L1, and ILT proteins that enhance regulatory T cell function.
Expression of over 8,000 coding and non-coding RNAs is significantly distorted in untreated MS, compared to HC and to long-term IFN-β-treated MS patients. 162 of these dysregulated genes carry SNPs associated with MS in GWAS studies [4,65]. These DEG are predominantly implicated in immunity, including type I IFN signalling: JAK2, Tyk2, and IRF-8 [66], [67], [68].
CNS-restricted molecules regulating MS pathogenesis and repair can be affected by type I IFNs in PBMNC. Many brain-expressed RNAs are detectable in PBMNC. This may reflect some CNS effects, as gene expression is echoed in tissues throughout the body [69,70]. IFN injections also modify blood immune cells before and after they traffic into the CNS. For instance, IFN-β regulates pDC, the richest cellular source of type I IFN. pDC are tolerogenic and home from blood to the inflamed brain in murine EAE [71]. Plus, type I IFN penetrates into the CNS [72], and more easily crosses the disrupted blood-brain barrier during inflammation, inducing IFN-stimulated genes [73].
DEGs from multiple biological pathways in therapy-naïve and IFN-treated MS patients were identified. Detection of this large number of genes likely is from the use of PBMNC (monocytes and dendritic, NK, Th1, Th17, Th2, Treg, and B cells) instead of whole blood. Purified PBMNC eliminate background noise from granulocytes, nucleated reticulocytes, and megakaryocytes present in whole blood. Up to 95% of whole blood RNA, arising from those lineages, is largely irrelevant to MS pathogenesis [7]. Therefore, analysis of PBMNC considerably improves the RNA signal-to-noise ratio, quality of data, and the measure of MS-relevant transcripts.
Limitations of our study include (1) the lack of a fully Non-Responsive subgroup. This was overcome by evaluating PR during MS exacerbations, a short period of muted responsiveness to IFN-β. (2) The sample size is moderate. We attempted to approach this with a paired experimental design where the same patients were exposed to different IFN-β doses, at precisely controlled serial measures to improve statistical power [74]. (3) There is no public database containing a longitudinal, or PR vs. CR, design comparable to our study to allow independent validation to the best of our knowledge. From that perspective, cross-validation was partially reflected between groups within our study design. For example, untreated stable and untreated active MS are independent groups, yet have high correspondence on the majority of DEGs (Table S4). Post-IFN washout values before 250 and 500 ug injections also closely correlate, and there is minimal remaining short-term gene induction (Fig. S2d). (4) Microarray platforms cannot be used to discover novel transcripts. In this study, 285,000 coding and noncoding transcripts are captured by HTA, presenting a comprehensive collection of transcriptomes. These arrays have high quantification reproducibility over a large dynamic range including low abundance RNAs coding for cytokines and neuroprotective genes. (5) Levels of HGF were below the level of detection of 1,000 transcripts per well on QuantiGene, but were detected with the microarrays. Even though the values from the two assays are highly correlated, not all expression changes of the selected 10 genes can be validated by QuantiGene. This may be due to the relatively low expression level of those transcripts, e.g., the reduced expression of EIF2AK3 at 4 h after injection and low expression of IFNG-AS1 overall (Fig. 6c, on y-axis; most samples have expression <0.5), compared to the other 8 genes. Additionally, fewer samples were used for the QuantiGene assays because of limited amounts of RNA (n = 11, 11, 8, and 9 for PR stable 0 h, PR-stable 4 h, healthy controls, and untreated MS-stable, respectively. The number of samples used for HTA assays were 15, 15, 8, and 10 for the four designated groups shown in Table S1).
Normalized gene expression from long-term IFN-β treatment carries implications for MS therapy. In untreated MS patients, products of dysregulated genes promote inflammatory CNS damage and impede brain repair. Interferon signalling, which regulates these genes, is abnormal in MS. Long-term IFN-β therapy contracts excessive gene expression in MS. It induces multiple modes of immune-regulatory control and enhances expression of neurotrophic genes and pro-survival pathways. Interferons were discovered nearly 60 years ago as anti-viral agents [75], and in 1993 IFN-β-1b was the first approved disease-modifying drug for MS [14,76]. IFNs, however, first appear in cartilaginous and bony fish 400 million years ago [77]. Subsequent gene evolution is likely to have engendered mechanisms beyond anti-viral effects, including regulation of the acute inflammatory response, prevention of cancer cell proliferation, and repair of damaged tissues, including protection of neurons. Some of the genes that change with IFN treatment are likely to reduce the frequency, severity, and duration of breakthrough MS attacks and enhance brain repair [14,76], help preserve cognition [78,79], and perhaps prolong survival in MS [12], and may affect other autoimmune and neurodegenerative diseases.
The large number of dysregulated genes in therapy-naïve MS provides potential new targets for MS treatment. Many are not previously known to be modulated by IFN-β. Pairing IFN-β therapy with rational adjuvants may more fully correct the abnormal Th1/Th17/Th2 cell balance of MS and enhance clinical responsiveness, while maintaining the beneficial effects that induce immune regulation, neuroprotection, and tissue repair.
Author contributions
X.F. and A.T.R. conceived the hypotheses and designed the study. X.F., L.L., and A.T.R. performed the experiments. X.F. and A.T.R. collected and prepared samples and data. F. D performed the NAb titres. R.B. developed the analytical methods and performed the computations. X.F., R.B., and A.T.R. performed data analysis. X.F., R.B., B.G.W.A., and A.T.R. interpreted the results. X.F., R.B., B.G.W.A. and A.T.R. wrote the paper. All authors discussed the results and contributed to the final manuscript.
Declaration of Competing Interest
X.F., R.B., L.L., and B.G.W.A. have no competing interests. A.T.R. has paid consulting relationships with Bayer Pharmaceuticals, Merck-Serono, Biogen, and Novartis, all of whom produce interferons for therapy of MS; these do not include speakers’ bureaus. The authors designed and performed the experiments, independent of commercial interests.
Acknowledgments
This work was supported by unrestricted grants from NMSS (RG#4509A) and Bayer Healthcare Pharmaceuticals. The funders had no role in study design, sample collection, data analysis or interpretation, or writing the report.
We thank S. Causevic and Quentin Howlett-Prieto for assistance with data analysis; A Javed, NP Reder, R Roos, and D White for valuable discussions; M. Jarsulic for technical assistance on the computational infrastructure and software. The Center for Research Informatics is funded by the Biological Sciences Division at the University of Chicago with additional support provided by The Institute for Translational Medicine/Clinical and Translational Award (NIH 5UL1TR002389-02), and The University of Chicago Comprehensive Cancer Center Support Grant (NIH P30CA014599). Bioinformatics analysis was performed on High-Performance Computing clusters at Center for Research Informatics, Biological Sciences Division. Gene analysis was performed at the University of Chicago Genomics Core facility.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.09.059.
Appendix. Supplementary materials
References
- 1.Hemmer B., Kerschensteiner M., Korn T. Role of the innate and adaptive immune responses in the course of multiple sclerosis. Lancet Neurol. 2015;14(4):406–419. doi: 10.1016/S1474-4422(14)70305-9. [DOI] [PubMed] [Google Scholar]
- 2.Antel J.P., Arnason B.G.W., Medof M.E. Suppressor cell function in multiple sclerosis: correlation with clinical disease activity. Ann Neurol. 1979;5:338–342. doi: 10.1002/ana.410050406. [DOI] [PubMed] [Google Scholar]
- 3.Ontaneda D., Thompson A.J., Fox R.J., Cohen J.A. Progressive multiple sclerosis: prospects for disease therapy, repair, and restoration of function. Lancet. 2017;389(10076):1357–1366. doi: 10.1016/S0140-6736(16)31320-4. [DOI] [PubMed] [Google Scholar]
- 4.Beecham A.H., Patsopoulos N.A., Xifara D.K. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng X., Petraglia A.L., Chen M., Byskosh P.V., Boos M.D., Reder A.T. Low expression of interferon-stimulated genes in active multiple sclerosis is linked to subnormal phosphorylation of STAT1. J Neuroimmunol. 2002;129:105–115. doi: 10.1016/s0165-5728(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi K.D., Ruderman D.L., Croze E. IFN-β-regulated genes are abnormally expressed in therapy-naive ms mononuclear cells: unbiased gene expression analysis parallels literature on signaling pathways. J Neuroimmunol. 2008;195:116–120. doi: 10.1016/j.jneuroim.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Reder A.T., Velichko S., Yamaguchi K.D. Interferon-β-1b induces transient and variable gene expression in relapsing-remitting multiple sclerosis patients, independent of neutralizing antibodies or changes in IFN receptor RNA expression. J Interferon Cytokine Res. 2008;28:317–331. doi: 10.1089/jir.2007.0131. [DOI] [PubMed] [Google Scholar]
- 8.Comabella M., Lunemann J.D., Rio J. A type I interferon signature in monocytes is associated with poor response to interferon-β in multiple sclerosis. Brain. 2009;132(Pt 12):3353–3365. doi: 10.1093/brain/awp228. [DOI] [PubMed] [Google Scholar]
- 9.Rudick R.A., Rani M.R., Xu Y. Excessive biologic response to IFNbeta is associated with poor treatment response in patients with multiple sclerosis. PLoS ONE. 2011;6(5):e19262. doi: 10.1371/journal.pone.0019262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturzebecher S., Wandinger K.P., Rosenwald A. Expression profiling identifies responder and non-responder phenotypes to interferon-beta in multiple sclerosis. Brain. 2003;126(Pt 6):1419–1429. doi: 10.1093/brain/awg147. [DOI] [PubMed] [Google Scholar]
- 11.Xu W., Seok J., Mindrinos M.N. Human transcriptome array for high-throughput clinical studies. Proc Natl Acad Sci U S A. 2011;108(9):3707–3712. doi: 10.1073/pnas.1019753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodin D.S., Reder A.T., Ebers G.C. Survival in MS: a randomized cohort study 21 years after the start of the pivotal IFNbeta-1b trial. Neurology. 2012;78(17):1315–1322. doi: 10.1212/WNL.0b013e3182535cf6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Axtell R.C., de Jong B.A., Boniface K. T helper type 1 and 17 cells determine efficacy of interferon-β in multiple sclerosis and experimental encephalomyelitis. Nat Med. 2010;16(4):406–413. doi: 10.1038/nm.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Interferon beta-1b is effective in relapsing-remitting multiple sclerosis. I. Clinical results of a multicenter, randomized, double-blind, placebo-controlled trial. The IFNB multiple sclerosis study group. Neurology. 1993;43(4):655–661. doi: 10.1212/wnl.43.4.655. [DOI] [PubMed] [Google Scholar]
- 15.Chiang J., Gloff C.A., Yoshizawa C.N., Williams G.J. Pharmacokinetics of recombinant human interferon-βser in healthy volunteers and its effect on serum neopterin. Pharm Res. 1993;10:567–572. doi: 10.1023/a:1018902120023. [DOI] [PubMed] [Google Scholar]
- 16.Salmon P., Le Cotonnec J.Y., Galazka A., Abdul-Ahad A., Darragh A. Pharmacokinetics and pharmacodynamics of recombinant human interferon-β in healthy male volunteers. J Interferon Cytokine Res. 1996;16(10):759–764. doi: 10.1089/jir.1996.16.759. [DOI] [PubMed] [Google Scholar]
- 17.Feng X., Han D., Kilaru B.K., Franek B.S., Niewold T.B., Reder A.T. Inhibition of interferon-beta responses in multiple sclerosis immune cells associated with high-dose statins. Arch Neurol. 2012;69(10):1303–1309. doi: 10.1001/archneurol.2012.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie M.E., Phipson B., Wu D. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bushel P. pvca: principal variance component analysis (PVCA) R package version 1120. 2013 [Google Scholar]
- 20.Aune T.M., Crooke P.S., 3rd, Patrick A.E., Tossberg J.T., Olsen N.J., Spurlock C.F., 3rd Expression of long non-coding RNAs in autoimmunity and linkage to enhancer function and autoimmune disease risk genetic variants. J Autoimmun. 2017;81:99–109. doi: 10.1016/j.jaut.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siger M., Durko A., Nicpan A., Konarska M., Grudziecka M., Selmaj K. Discontinuation of interferon beta therapy in multiple sclerosis patients with high pre-treatment disease activity leads to prompt return to previous disease activity. J Neurol Sci. 2011;303(1–2):50–52. doi: 10.1016/j.jns.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumitriu A., Latourelle J.C., Hadzi T.C. Gene expression profiles in Parkinson disease prefrontal cortex implicate FOXO1 and genes under its transcriptional regulation. PLoS Genet. 2012;8(6) doi: 10.1371/journal.pgen.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Zhou Y., Graves D.T. FOXO transcription factors: their clinical significance and regulation. Biomed Res Int. 2014;2014 doi: 10.1155/2014/925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi S., Motta C., Studer V. Interleukin-1beta causes excitotoxic neurodegeneration and multiple sclerosis disease progression by activating the apoptotic protein p53. Mol Neurodegener. 2014;9:56. doi: 10.1186/1750-1326-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waschbisch A., Sanderson N., Krumbholz M. Interferon beta and vitamin D synergize to induce immunoregulatory receptors on peripheral blood monocytes of multiple sclerosis patients. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0115488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y., Majchrzak-Kita B., Fish E.N., Gommerman J.L. Dynamic accumulation of plasmacytoid dendritic cells in lymph nodes is regulated by interferon-β. Blood. 2009;114(13):2623–2631. doi: 10.1182/blood-2008-10-183301. [DOI] [PubMed] [Google Scholar]
- 29.Annibali V., Ristori G., Angelini D.F. CD161(high)CD8+T cells bear pathogenetic potential in multiple sclerosis. Brain. 2011;134(Pt 2):542–554. doi: 10.1093/brain/awq354. [DOI] [PubMed] [Google Scholar]
- 30.Negrotto L., Canto E., Rio J., Tintore M., Montalban X., Comabella M. Peripheral blood non-MAIT CD8+CD161hi cells are decreased in relapsing-remitting multiple sclerosis patients treated with interferon beta. J Neuroimmunol. 2015;288:98–101. doi: 10.1016/j.jneuroim.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Wong M.T., Lambeck A.J., van der Burg M. Immune dysfunction in children with charge syndrome: a cross-sectional study. PLoS ONE. 2015;10(11) doi: 10.1371/journal.pone.0142350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hesse D., Sellebjerg F., Sorensen P.S. Absence of MxA induction by interferon beta in patients with MS reflects complete loss of bioactivity. Neurology. 2009;73(5):372–377. doi: 10.1212/WNL.0b013e3181b04c98. [DOI] [PubMed] [Google Scholar]
- 33.Graber J.J., Dhib-Jalbut S. Biomarkers of interferon Beta therapy in multiple sclerosis. J Interferon Cytokine Res. 2014;34(8):600–604. doi: 10.1089/jir.2013.0144. [DOI] [PubMed] [Google Scholar]
- 34.Lock C., Hermans G., Pedotti R. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–508. doi: 10.1038/nm0502-500. May 2002. [DOI] [PubMed] [Google Scholar]
- 35.Steinman L. Shifting therapeutic attention in MS to osteopontin, type 1 and type 2 IFN. Eur J Immunol. 2009;39(9):2358–2360. doi: 10.1002/eji.200939814. [DOI] [PubMed] [Google Scholar]
- 36.Joo H., Li D., Dullaers M. C-type lectin-like receptor LOX-1 promotes dendritic cell-mediated class-switched b cell responses. Immunity. 2014;41(4):592–604. doi: 10.1016/j.immuni.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendrickx D.A., Koning N., Schuurman K.G. Selective upregulation of scavenger receptors in and around demyelinating areas in multiple sclerosis. J Neuropathol Exp Neurol. 2013;72(2):106–118. doi: 10.1097/NEN.0b013e31827fd9e8. [DOI] [PubMed] [Google Scholar]
- 38.Bridges R., Lutgen V., Lobner D., Baker D.A. Thinking outside the cleft to understand synaptic activity: contribution of the cystine-glutamate antiporter (System xc-) to normal and pathological glutamatergic signaling. Pharmacol Rev. 2012;64(3):780–802. doi: 10.1124/pr.110.003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benkhoucha M., Molnarfi N., Dunand-Sauthier I. Hepatocyte growth factor limits autoimmune neuroinflammation via glucocorticoid-induced leucine zipper expression in dendritic cells. J Immunol. 2014;193(6):2743–2752. doi: 10.4049/jimmunol.1302338. [DOI] [PubMed] [Google Scholar]
- 40.Giess R., Maurer M., Linker R. Association of a null mutation in the CNTF gene with early onset of multiple sclerosis. Arch Neurol. 2002;59:407–409. doi: 10.1001/archneur.59.3.407. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., Kramer E.G., Asp L. Promoting myelin repair and return of function in multiple sclerosis. FEBS Lett. 2011;585(23):3813–3820. doi: 10.1016/j.febslet.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G.C., Pan H.F., Leng R.X. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14(9):798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Gomez J.A., Wapinski O.L., Yang Y.W. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang J., Zhu X., Chen Y. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16(5):616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Genc K., Dona D.L., Reder A.T. Increased CD80+ B cells in active multiple sclerosis, and reversal by IFNβ-1b therapy. J Clin Invest. 1997;99:2664–2671. doi: 10.1172/JCI119455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pallotta M.T., Orabona C., Volpi C. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat Immunol. 2011;12(9):870–878. doi: 10.1038/ni.2077. [DOI] [PubMed] [Google Scholar]
- 47.Way S.W., Popko B. Harnessing the integrated stress response for the treatment of multiple sclerosis. Lancet Neurol. 2016;15(4):434–443. doi: 10.1016/S1474-4422(15)00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hecker M., Hartmann C., Kandulski O. Interferon-beta therapy in multiple sclerosis: the short-term and long-term effects on the patients' individual gene expression in peripheral blood. Mol Neurobiol. 2013;48(3):737–756. doi: 10.1007/s12035-013-8463-1. [DOI] [PubMed] [Google Scholar]
- 49.Correale J., Villa A. Isolation and characterization of CD8+ regulatory t cells in multiple sclerosis. J Neuroimmunol. 2008;195(1–2):121–134. doi: 10.1016/j.jneuroim.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Denic A., Wootla B., Rodriguez M. CD8(+) t cells in multiple sclerosis. Expert Opin Ther Targets. 2013;17(9):1053–1066. doi: 10.1517/14728222.2013.815726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noronha A., Toscas A., Jensen M. Contrasting effects of alpha, beta, and gamma interferons on nonspecific suppressor function in multiple sclerosis. Ann Neurol. 1992;31:103–106. doi: 10.1002/ana.410310119. [DOI] [PubMed] [Google Scholar]
- 52.Saulep-Easton D., Vincent F.B., Quah P.S. The BAFF receptor TACI controls IL-10 production by regulatory B cells and CLL B cells. Leukemia. 2016;30(1):163–172. doi: 10.1038/leu.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kannel K., Alnek K., Vahter L., Gross-Paju K., Uibo R., Kisand K.V. Changes in blood B cell-activating factor (BAFF) levels in multiple sclerosis: a sign of treatment outcome. PLoS ONE. 2015;10(11) doi: 10.1371/journal.pone.0143393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goertsches R.H., Hecker M., Koczan D. Long-term genome-wide blood RNA expression profiles yield novel molecular response candidates for IFN-beta-1b treatment in relapsing remitting MS. Pharmacogenomics. 2010;11(2):147–161. doi: 10.2217/pgs.09.152. [DOI] [PubMed] [Google Scholar]
- 55.Arosa F.A., Esgalhado A.J., Padrao C.A., Cardoso E.M. Divide, conquer, and sense: CD8+CD28- T Cells in perspective. Front Immunol. 2016;7:665. doi: 10.3389/fimmu.2016.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jensen M.A., Yanowitch R.N., Reder A.T., White D.M., Arnason B.G. Immunoglobulin-like transcript 3, an inhibitor of T cell activation, is reduced on blood monocytes during multiple sclerosis relapses and is induced by interferon B-1b. Mult Scler. 2010 doi: 10.1177/1352458509352794. [DOI] [PubMed] [Google Scholar]
- 57.Kono D.H., Baccala R., Theofilopoulos A.N. TLRs and interferons: a central paradigm in autoimmunity. Curr Opin Immunol. 2013;25(6):720–727. doi: 10.1016/j.coi.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prinz M., Schmidt H., Mildner A. Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity. 2008;28(5):675–686. doi: 10.1016/j.immuni.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Goertsches R.H., Zettl U.K., Hecker M. Sieving treatment biomarkers from blood gene-expression profiles: a pharmacogenomic update on two types of multiple sclerosis therapy. Pharmacogenomics. 2011;12(3):423–432. doi: 10.2217/pgs.10.190. [DOI] [PubMed] [Google Scholar]
- 60.Reder A.T., Feng X. Aberrant type I interferon regulation in autoimmunity: opposite directions in MS and SLE, shaped by evolution and body ecology. Front Immunol. 2013;4:1–13. doi: 10.3389/fimmu.2013.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mikol D.D., Barkhof F., Chang P. Comparison of subcutaneous interferon beta-1a with glatiramer acetate in patients with relapsing multiple sclerosis (the REbif vs Glatiramer acetate in relapsing MS disease [REGARD] study): a multicentre, randomised, parallel, open-label trial. Lancet Neurol. 2008;7(10):903–914. doi: 10.1016/S1474-4422(08)70200-X. [DOI] [PubMed] [Google Scholar]
- 62.O'Connor P., Filippi M., Arnason B. 250 ug or 500 ug interferon beta-1b versus 20 mg glatiramer acetate in relapsing-remitting multiple sclerosis: a prospective, randomised, multicentre study. Lancet Neurol. 2009;8:889–897. doi: 10.1016/S1474-4422(09)70226-1. [DOI] [PubMed] [Google Scholar]
- 63.Patti F., Morra V.B., Amato M.P. Subcutaneous interferon beta-1a may protect against cognitive impairment in patients with relapsing-remitting multiple sclerosis: 5-year follow-up of the cogimus study. PLoS ONE. 2013;8(8):e74111. doi: 10.1371/journal.pone.0074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ebers G.C., PRISMS (Prevention of Relapses and Disability by Interferon β-1a Subcutaneously in Multiple Sclerosis) Study Group Randomised double-blind placebo-controlled study of interferon β-1a in relapsing/remitting multiple sclerosis. Lancet. 1998;352:1498–1504. November 7, 1998. [PubMed] [Google Scholar]
- 65.de Jager P.L., Jia X., Wang J. Meta-analysis of genome scans and replication identify CD6, IRF8, and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–785. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Jager P.L., Baecher-Allan C., Maier L.M. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0813310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Doherty C., Favorov A., Heggarty S. Genetic polymorphisms, their allele combinations and IFN-beta treatment response in Irish multiple sclerosis patients. Pharmacogenomics. 2009;10(7):1177–1186. doi: 10.2217/PGS.09.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sawcer S., Hellenthal G., Pirinen M. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dudley J.T., Tibshirani R., Deshpande T., Butte A.J. Disease signatures are robust across tissues and experiments. Mol Syst Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ardlie K.G. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duraes F.V., Lippens C., Steinbach K. pDC therapy induces recovery from EAE by recruiting endogenous pDC to sites of CNS inflammation. J Autoimmun. 2016;67:8–18. doi: 10.1016/j.jaut.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan W., Banks W.A., Kastin A.J. Permeability of the blood-brain and blood-spinal cord barriers to interferons. J Neuroimmunol. 1997;76:105–111. doi: 10.1016/s0165-5728(97)00034-9. [DOI] [PubMed] [Google Scholar]
- 73.Hovanessian A.G., Marcovistz R., Rivière Y., Guillon J.-.C., Tsiang H. Production and action of interferon in rabies virus infection. In: Smith RA, editor. Interferon treatment of neurologic disorders. Marcel Dekker, Inc.; New York: 1988. pp. 157–186. [Google Scholar]
- 74.Ching T., Huang S., Garmire L.X. Power analysis and sample size estimation for RNA-Seq differential expression. RNA. 2014;20(11):1684–1696. doi: 10.1261/rna.046011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borden E.C., Sen G.C., Uze G. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C., Blumhardt L.D. Randomised, double blind, placebo controlled study of interferon beta-1a in relapsing-remitting multiple sclerosis analysed by area under disability/time curves. J Neurol Neurosurg Psychiatry. 1999;67(4):451–456. doi: 10.1136/jnnp.67.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Secombes C.J., Zou J. Evolution of interferons and interferon receptors. Front Immunol. 2017;8:209. doi: 10.3389/fimmu.2017.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pliskin N.H., Hamer D.P., Goldstein D.S. Improved delayed visual reporduction test performance in multiple sclerosis patients receiving interferon β–1β. Neurology. 1996;47:1463–1468. doi: 10.1212/wnl.47.6.1463. [DOI] [PubMed] [Google Scholar]
- 79.Lacy M., Hauser M., Pliskin N., Assuras S., Valentine M.O., Reder A. The effects of long-term interferon- beta-1b treatment on cognitive functioning in multiple sclerosis: a 16-year longitudinal study. Mult Scler. 2013:1765–1772. doi: 10.1177/1352458513485981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed data files are provided as supplementary tables. Raw microarray data files have been deposited into the NCBI Gene Expression Omnibus (GEO) repository, Accession #GSE138064.