Abstract
Purpose
Computed tomography (CT) is an effective method for detecting and characterizing lung nodules in vivo. With the growing use of chest CT, the detection frequency of lung nodules is increasing. Noninvasive methods to distinguish malignant from benign nodules have the potential to decrease the clinical burden, risk, and cost involved in follow‐up procedures on the large number of false‐positive lesions detected. This study examined the benefit of including perinodular parenchymal features in machine learning (ML) tools for pulmonary nodule assessment.
Methods
Lung nodule cases with pathology confirmed diagnosis (74 malignant, 289 benign) were used to extract quantitative imaging characteristics from computed tomography scans of the nodule and perinodular parenchyma tissue. A ML tool development pipeline was employed using k‐medoids clustering and information theory to determine efficient predictor sets for different amounts of parenchyma inclusion and build an artificial neural network classifier. The resulting ML tool was validated using an independent cohort (50 malignant, 50 benign).
Results
The inclusion of parenchymal imaging features improved the performance of the ML tool over exclusively nodular features (P < 0.01). The best performing ML tool included features derived from nodule diameter‐based surrounding parenchyma tissue quartile bands. We demonstrate similar high‐performance values on the independent validation cohort (AUC‐ROC = 0.965). A comparison using the independent validation cohort with the Fleischner pulmonary nodule follow‐up guidelines demonstrated a theoretical reduction in recommended follow‐up imaging and procedures.
Conclusions
Radiomic features extracted from the parenchyma surrounding lung nodules contain valid signals with spatial relevance for the task of lung cancer risk classification. Through standardization of feature extraction regions from the parenchyma, ML tool validation performance of 100% sensitivity and 96% specificity was achieved.
Keywords: artificial intelligence, computed tomography, pulmonary nodule, radiomics, risk assessment
1. Introduction
Lung cancer is the leading cause of cancer‐related deaths in the United States with an estimated 228,150 new diagnoses in 2019.1 Computed tomography (CT) is vital technology used for lung nodule detection via an incidental finding, diagnostic evaluation of symptomatic patients, and most recently as the standard for lung cancer screening in nonsymptomatic patients at high risk for lung cancer.2 This increased lung nodule detection power comes with concerns related to increased cumulative radiation exposure, risks from diagnostic procedures for false‐positive screens, and overdiagnosed cancers.3 Guidelines such as the Lung Imaging Reporting and Data System (Lung‐RADS) criteria for lesions identified with low‐dose CT screening, and the Fleischner society guidelines for incidental nodules use the size (and change in size) of the lung nodule as a key indicator to determine appropriate clinical follow‐up procedures.4, 5 However, this approach does not include much of the information captured in the CT data. Machine learning (ML) tools can utilize quantitative imaging features extracted directly from the CT scans, such as nodule shape and texture, to differentiate between malignant and benign disease states.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 The traditional focus of imaging‐based risk models for lung cancer has been on nodule and border features with a range in area under the receiver operator curve (AUC‐ROC) performance (0.821‐0.962).7, 8, 9, 10, 11, 12, 13
The perinodular parenchyma has biological importance with respect to cell migration, inflammation, and vascularization. Morphological characteristics from this region including spiculation and structural distortion of the parenchyma have been explored in the context of lung nodule malignancy.21 Recently, improvements in lung nodule classification have been demonstrated through the inclusion of perinodular parenchymal features using both machine learning with AUC‐ROC: 0.9386 and 0.91520 and deep learning methods have indirectly examined parenchymal inclusion with AUC‐ROC of 0.993, 0.899, 0.946, 0.984 13, 17, 18, 19 but the degree to which parenchyma has been included has varied. From the reported literature, it is not clear where the optimal parenchymal radiological signal resides, and how these features interplay with features from the nodule and its borders.
In this study, we systematically investigate the optimal regions of perinodular parenchyma to use for feature extraction and classification. With a focus on feature transparency, we explore the trends within and between regions of perinodular parenchyma by nodule‐standardized parenchymal quartile bands. Finally, we compare the value this ML tool could have on follow‐up pathways versus the established Fleischner Society guidelines in an independent validation dataset.
2. Materials and Methods
A systematic processing pipeline was developed to identify the optimal feature set for ML, and independent validation testing, as depicted in Fig. 1. ML tool development involved the selection of a feature set and classifier training, while validation was executed on the top performing candidate tool.
Figure 1.
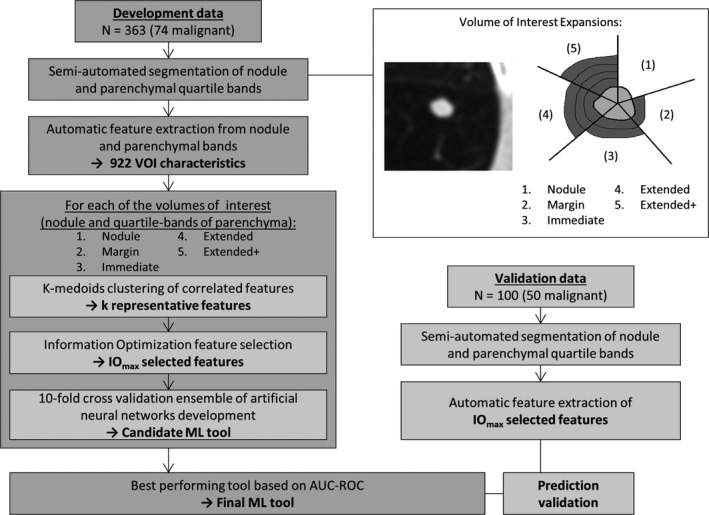
Overview of machine learning tool development and validation pipeline. The depiction of the varying amounts of parenchyma tested through the pipeline (in quartile bands) include; (1) Nodule, (2) Margin [nodule, 25%], (3) Immediate [nodule, 25%, 50%], (4) Extended [nodule, 25%, 50%, 75%], (5) Extended+ [nodule, 25%, 50%, 75%, 100%]. Abbreviations: ML, machine learning; VOI: volume of interest, IOmax, information objective function maximum point; AUC‐ROC, area under the receiver operating characteristic curve.
2.1. Study population
Subjects were retrospectively selected and included 363 pulmonary nodules, ≤30 mm in diameter (74 malignant, 289 benign) from three study data sources: the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease study (COPDGene), the National Lung Screening Trial (NLST), and the SPIE LungX Challenge.2, 22, 23 A subset of 50 subjects from the current 363 cohort was previously used to demonstrate the value of feature extraction from the lung parenchyma.6 The criterion for inclusion in this study was based on the availability of diagnostic information: malignant nodules were confirmed on histopathology, and benign nodules were diagnosed by histopathology or size stability or resolution on more than 24 months follow‐up. An independent validation cohort of 100 (50 malignant, 50 benign) pulmonary nodules from the INHALE study was used to test the top performing ML tool.24 Malignancy was confirmed in this cohort through histological confirmation accessed through the Detroit area SEER registry, and benign cases selected to match size characteristics. Further demographics and scanning parameters for the two cohorts are described in Table 1, pictorial representation of a randomly selected sample of 24 nodules (12 malignant, 12 benign) is included in the supplemental online content (see Figures S1–S4).
Table 1.
Demographics and scanning parameters of study cohorts.
| Malignant | Benign | P‐value | |
|---|---|---|---|
| Development | |||
| Subjects | 74 | 289 | – |
| Study (COPDGene:NLST:LungX) | 30:6:38 | 239:8:42 | <0.001c |
| Age, yrs (mean ± SD) | 65.5 ± 11.3 | 53.2 ± 13. | <0.001 |
| Sex (Female: Male) | 34:40 | 179:110 | <0.001c |
| Pack‐yearsa, yrs (mean ± SD) | 37.7 ± 30.4 | 10.7 ± 15.7 | <0.001 |
| Nodule size, mm Range (mean ± SD) | 5–30 (13.6 ± 6.2) | 4–30 (7.79 ± 13.3) | <0.001 |
| Nodule size ≤ 15mm | 50 | 240 | 0.005c |
| LDCT screening eligiblea (Yes: No) | 33:3 | 69:178 | <0.001c |
| Kilovoltage, kVp (range, mean) | 120–120, 120 | 120–120, 120 | 1.00 |
| Tube current, mA (range, mean) | 60–440,b 339 | 40–500,b 330 | 0.89 |
| Slice thickness, mm (range, mean) | 0.6–1.3, 0.8 | 0.6–1.3, 0.7 | 0.97 |
| CT manufacturer (GE:Philips:Siemens) | 19:35:20 | 86:97:106 | <0.001c |
| Validation | |||
| Subjects | 50 | 50 | – |
| Age (mean ± SD) | 64.0 ± 10.7 | 62.5 ± 10.9 | 0.456 |
| Sex (Female: Male) | 35:15 | 31:19 | 0.339c |
| Pack‐yearsa, yrs (mean ± SD) | 33.5 ± 30.1 | 30.1 ± 23.6 | 0.505 |
| Nodule size, mm Range (mean ± SD) | 5–30 (19.9 ± 7.4) | 9–30 (13.66 ± 4.8) | <0.001 |
| Nodule size ≤ 15mm | 17 | 35 | <0.001c |
| LDCT screening eligible (Yes: No) | 18:32 | 16:34 | 0.674c |
| Kilovoltage, kVp (range, mean) | 120–120 , 120 | 120–120 , 120 | 1.00 |
| Tube Current, mA (range, mean) | 160–351, 237 | 160–386, 265 | 0.62 |
| Slice thickness, mm (range, mean) | 0.6–0.8, 0.7 | 0.6–0.8, 0.7 | 0.98 |
| CT Manufacturer (GE:Philips:Siemens) | 17:19:14 | 16:22:12 | 0.817c |
COPD, chronic obstructive pulmonary disease; NLST, National Lung Screening Trial; SD, standard deviation; LDCT, low‐dose computed tomography; GE, General Electric.
smoking pack‐year data were not available for the LungX Challenge.
low‐dose NLST scans included were reconstructed to higher resolution at time of acquisition.
discrete (categorical) data significance performed with chi‐square test.
2.2. Segmentation of nodule and parenchyma
The nodule and parenchyma were semiautomatically segmented into volumes of interest (VOI) using a modified version of the proposed pipeline by Mukhopadhyay.25 The nodule mask was grown using a binary image dilation to produce parenchyma quartile bands: 25%, 50%, 75%, and 100% of the maximum in‐plane diameter of the nodule (Fig. 1).
2.3. Development of ML tool
Here, we provide a brief summary of our method of selecting feature sets and training artificial neural network (ANN) classifiers which has been previously described on breast masses.26 Quantitative CT features were extracted from the nodule and each of the parenchyma quartile volumes. These included 14 volumetric measures of intensity histogram (V‐IH), 136 volumetric Law's energy measures (V‐LTEM), 13 volumetric gray‐level run length measures (V‐GLRL), 13 volumetric gray‐level size‐zone measures (V‐GLSZ), 5 volumetric neighborhood gray tone difference measures (V‐NGTD), and 17 measures of size and volumetric shape (V‐SzSp) including 11 border measures.6, 27, 28, 29, 30, 31
Features were clustered based on pairwise correlations using the k‐medoids method resulting in k clusters with k representative medoid features.32 Determination of k was done by optimization of the average cluster silhouette with the method adjusted to not penalize for clusters of one feature. From the reduced group of medoids, a set of predicting features was selected using an objective function of information theory measures. The final selected set size was determined from the information objective function maximum point (IOmax). In cases where the IOmax was larger than one predictor for every 5–10 cases the set size was capped at 72 features. The selected feature sets were used to train the ANNs with performance analyzed through tenfold cross validation on a per subject basis. As random initialization of weights in ANN development can affect classifier performance, we further developed an ensemble of ten ANNs for final prediction values. Individual ANNs were trained using an in‐house developed MatLab (Mathworks, Natick MA) script using stochastic gradient descent and hyperbolic tangent activation function following random initialization of weights;33 this method which was adjusted to allow for trials of randomly assigned hyperparameter values (momentum, learning rate, error signal, hidden layer node number).
2.4. Performance and comparison
Tool performance was assessed using area under the receiver operating characteristic curve (AUC‐ROC) and area under the precision‐recall curve (AUC‐PR).34 To determine the statistical advantage of one ML tool over another on a given dataset, we employed Delong's assessment of AUC‐ROC.35 Risk thresholds were determined using Youden's J Statistic with McNemar's test to compare differences in threshold misclassifications between ML tools.36 Statistical difference of features between malignant and benign predictors was assessed with either a two‐sample t‐test (for normally distributed features) or a Wilcoxon rank sum test (for non‐normally distributed features).37, 38
3. Results
3.1. Study population
The development cohort (N = 363) was diverse in subject demographics, scanner parameters, and CT manufacturer; statistical difference in subject demographics existed between malignant and benign nodules (Table 1). As this cohort was established by combination of different parent academic studies, we explored the number of subjects that would have met lung cancer low‐dose CT (LDCT) screening eligibility criteria based on age and smoking pack‐years. Scanning parameters in the development cohort were in accordance with recommended protocols for high‐resolution CT studies,39 with the exception of the NLST cases which was a LDCT protocol (reconstructed thin slice thickness). The demographics and scanning parameters for the validation cohort (N = 100) were more evenly matched between malignant and benign cases (Table 1).
3.2. Perinodular ML tool performance
The best performing ML tool included the nodule and surrounding tissue from the 25%, 50%, and 75% quartile bands (Extended ML). In the development cohort, the Extended ML tool achieved an AUC‐ROC of 1.0 — or complete separation of the classes along the ensemble‐ANN decision boundary (Fig. 1) and achieved the highest AUC‐PR (0.945). The performance of the undivided, inclusive (border‐to‐75%) ring was also calculated and achieved weaker measures (AUC‐ROC = 0.938, AUC‐PR = 0.916). On the independent validation cohort, the Extended tool achieved an AUC‐ROC = 0.965 (accuracy 98%, sensitivity 100%, specificity 96%). Delong's comparison showed the four ML tools incorporating parenchymal signal (Margin, Immediate, Extended, Extended+) were statistically better than the Nodule ML tool (P < 0.01); there was no statistical difference between the ML tools developed with parenchymal features (P > 0.05). AUC‐ROC graphs for the five ML tools are presented in Figure S5. Pairwise McNemar's tests showed binary predictions from Youden threshold in the Nodule ML tool were significantly worse (P < 0.01) than the four ML tools incorporating parenchymal signal; no statistically significant difference in threshold binary predictions among the parenchymal ML tools was found using McNemar's tests (P > 0.05).
For the Extended and Extended+ ML tools the objective function's IOmax of 76 and 85 predictors respectively were adjusted to 72 predictors to prevent overfitting (Table 2). To test if fewer selected features were needed to maintain this boundary plane we built ensemble ANNs for each feature set size between 2 and 72 predictors. Complete class separation was achieved for all ensemble ANNs between 50 and 72 predictors (AUC‐ROC = 1.0, AUC‐PR = 0.945) for the Extended ML tool. The remainder of the results will focus on the Extended ML tool built using the top 50 features (Fig. 2). At the suggestion of the editor, the methodology for development and validation of the ML tool was repeated using the LungX dataset as the blinded validation cohort. This achieved a development AUC‐ROC of 0.957 (COPDGene, NLST, INHALE) and a validation AUC‐ROC of 0.924 (LungX); refer to Supplemental Text and Table S4, Figure S6 for full results of this investigation.
Table 2.
Development cohort performance results from differing amounts of surrounding parenchyma utilized.
| ML tool | IOmax | HLSM | AUC‐ROC | AUC‐PR | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| Nodule | 22 | 8 | 0.919 | 0.891 | 89% | 85% | 92% |
| Margin | 38 | 7 | 0.982 | 0.916 | 95% | 90% | 98% |
| Immediate | 55 | 8 | 0.998 | 0.943 | 97% | 93% | 98% |
| Extended | 76 a (72) | 8 | 1.000 | 0.945 | 100% | 100% | 100% |
| Extended+ | 85a (72) | 6 | 0.998 | 0.944 | 97% | 93% | 98% |
ML, Machine learning; IOmax, feature set size from information optimization; HLSM, hidden layer size maximum; AUC‐ROC, area under curve receiver operating characteristic; AUC‐PR, area under curve precision recall.
Indicates IOmax beyond N/5 limitation, classifier set was curbed at 72 features. This highest performing ML tool is highlighted in bold.
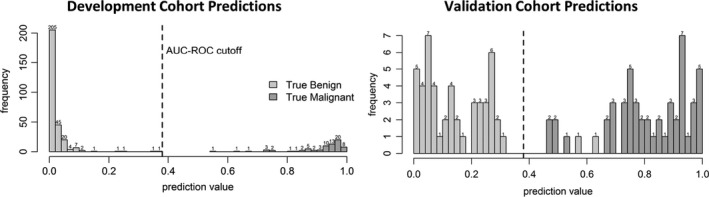
Figure 2.
The final Extended machine learning tool lung cancer risk prediction values result in complete division of malignant and benign lung nodule cases in cross validation testing of the development cohort (363 cases), with a threshold of 0.38 as determined using the Youden's J statistic. Output lung cancer risk prediction values range from 0 (confidently benign) to 1(confidently malignant).
3.3. Extended ML tool feature set
The 50 selected features included 13 V‐IH, 15 V‐GLRL, 12V‐GLSZ, 6 V‐NGTM, 1 B‐ASC, and 3 V‐SzSp. The region from which the included features were extracted included 19 nodular, 2 of band 25%, 12 of band 50%, and 17 of band 75%, such that the final model contained more features extracted from the parenchyma than from the nodule. In the development cohort, cutoff values of 0.38 from the receiver operating characteristic prioritize the correct classification of malignant cases (Fig. 2). Application of the Extended ML tool in the independent validation cohort resulted in an accuracy of 98%, with no misclassification of malignant cases.
A complete list of the 50 selected features for the Extended ML tool with mean, standard deviation, p‐value (from t‐test or Wilcoxon rank sum test as appropriate), and Pearson's correlation with nodule size is included in the online data supplement (see Table S1). The AUC‐ROC values for each of the 50 features as independent predictors is also included in the online data supplement(see Table S2). The individual AUC‐ROC for diameter was 0.594 (95% confidence‐interval: 0.526–0.663) and for volume was 0.542 (95% confidence‐interval: 0.478–0.606); Delong comparisons of these individual predictors to the Extended ML tool indicated statistical difference (P < 0.01).
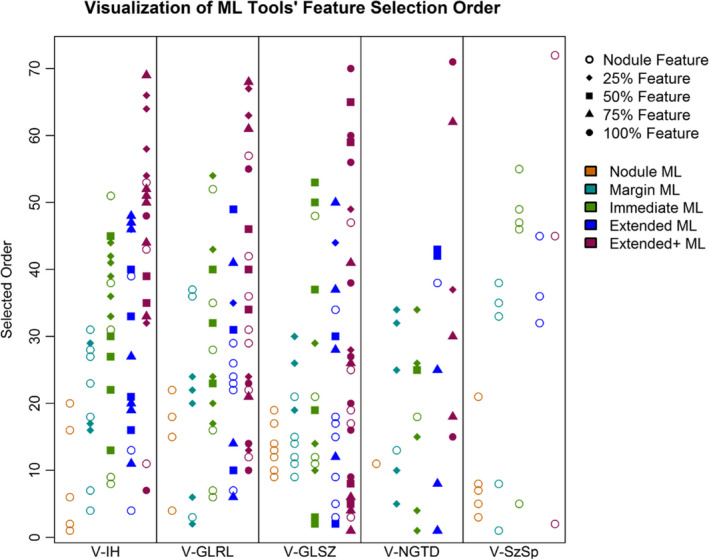
To summarize, the first five features selected included two features from the surrounding parenchyma quartile bands, followed by three nodule features. Selected first was a V‐NGTD feature describing the coarseness of texture in the 75% parenchyma quartile band, which is a high order feature where large values represent areas where the gray tone differences are small, therefore, leading to a high degree of local uniformity in intensity. Malignant cases had lower values (mean ± SD: 0.005 ± 0.013) than benign cases (0.011 ± 0.017) and this was statistically different (P = 0.023). Next, feature selection chose a V‐GLSZ texture feature indicating large zone emphasis in the 50% parenchyma quartile band. This feature multiplies each zone by the size of the zone squared, thus high values imply large zones within the texture. Again, malignant cases had lower values (159.5 ± 386.4), than benign (3087.1 ± 8332.7), this was statistically significant (P = 0.007). The third selected feature was V‐GLSZ extracted from the nodule indicating small zone low gray‐level emphasis with malignant cases having lower values (0.026 ± 00.23) than benign (0.036 ± 0.025) at a significant level (P < 0.001). This feature is larger when there is an emphasis of small zones of low intensity within the texture. The next two selected features were also nodule‐based being the entropy (V‐IH) and high gray‐level zone emphasis (V‐GLSZ). Figure 3 illustrates the order of feature selection for the five ML tools (Nodule, Margin, Immediate, Extended, Extended+), partitioned based on the intensity histogram, texture, or size/shape‐based feature class. This figure demonstrates how the inclusion of features from the parenchyma affect the selection of features from the nodule.
Figure 3.
Visualization of feature selection by category for each of the five candidate ML tools: (1) Nodule, (2) Margin [nodule, 25%], (3) Immediate [nodule, 25%, 50%], (4) Extended [nodule, 25%, 50%, 75%], (5) Extended+ [nodule, 25%, 50%, 75%, 100%. The shape of the point indicates the feature's origin (nodule, 25% band, etc.), color shows which tool it was selected into, and distance from horizontal axis shows the feature selection prioritization (ranging from an IOmax of 22 for the Nodule ML tool to a cap of 72 for the Extended + tool. Abbreviations: ML, Machine learning; V‐IH, intensity histogram features; V‐GLRL, gray‐level run length texture measures; V‐GLSZ, gray‐level size‐zone texture measures; V‐NGTD, neighborhood gray tone difference texture measures; V‐SzSp, size and shape.
Presented in Table 3 are the features used in the Extended ML tool, which were selected from more than one location (i.e., nodule and parenchyma, or different quartile bands of parenchyma). Twenty three out of the 50 total features were selected from more than one location. While these features are extracted in the same manner, the spatial location of the extracted region is effective. Large values in entropy features indicate a large amount of randomness in gray levels of the VOI. Full‐width‐at‐half‐maximum (FWHM) of the histograms of parenchyma bands tended to be smaller in malignant cases indicating a thinner, more peaked histogram shape. The gray‐level nonuniformity demonstrates while the malignant nodule showed increased nonuniformity, the tissue surrounding the malignant nodules tended to be lower than their benign counterparts. Run length variance tended to be lower in malignant cases indicating more homogeneous runs. The small zone low gray‐level emphasis demonstrated a converse effect with malignant nodules tending to have lower values while the tissue surrounding those nodules obtained a mean higher than that of benign nodule's surrounding tissues. Similarly, the contrast in texture showed a converse effect between nodule and parenchyma signal as this feature is high in the surrounding malignant nodules indicating increased amount of local variation in intensity. On the other hand, contrast texture tends to be lower in the nodule proper indicating a smaller amount in local intensity variation. While there was a size bias in the development cohort (malignant: 13.6 mm ± 6.2, benign: 7.8 mm ± 13.3, P < 0.001), the maximum in‐plane diameter was selected later (39/50) in the Extended ML. In addition, on nodules with size ≤ 15 mm, the Extended ML tool maintained high performance in both development (AUC‐ROC = 1.0, AUC‐PR = 0.943) and validation (AUC‐ROC = 0.998, AUC‐PR = 0.877).
Table 3.
Example feature trends in malignant nodules from Extended ML tool. For a complete list of selected features see online supplement (Table S1).
| Group | Feature | Rank | Trend | Malignant | Benign | P | ||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| V‐IH | Nodule Entropy | 4 | ↑ | 7.38 | 0.93 | 7.25 | 1.03 | 0.326 |
| 50% Entropy | 33 | ↑ | 8.58 | 0.72 | 8.03 | 0.74 | <0.001 | |
| 50% Maximum intensity | 21 | ↑ | −220.4 | 56.3 | −236.6 | 68.9 | <0.001 | |
| 75% Maximum Intensity | 47 | ↑ | −235.0 | 97.1 | −265.6 | 120.7 | <0.001 | |
| 50% FWHM | 40 | ↓ | 0.04 | 0.05 | 0.06 | 0.04 | <0.001 | |
| 75% FWHM | 19 | ↓ | 0.05 | 0.05 | 0.06 | 0.04 | <0.001 | |
| V‐GLRL | Nodule GL Nonuniformity | 29 | ↑ | 0.08 | 0.04 | 0.07 | 0.05 | 0.091 |
| 25% GL Nonuniformity | 35 | ↓ | 0.05 | 0.04 | 0.06 | 0.04 | <0.001 | |
| 50% GL Nonuniformity | 49 | ↓ | 0.06 | 0.02 | 0.08 | 0.04 | <0.001 | |
| Nodule Run Length Variance | 17 | ↓ | 1.8E‐04 | 2.1E‐04 | 2.5E‐04 | 3.4E‐04 | 0.035 | |
| 50% Run length Variance | 10 | ↓ | 2.4E‐04 | 1.4E‐04 | 2.8E‐04 | 9.9E‐05 | <0.001 | |
| Nodule GL Variance Runs | 24 | ↑ | 0.08 | 0.08 | 0.07 | 0.06 | 0.349 | |
| 50% GL Variance Run | 31 | ↑ | 0.04 | 0.06 | 0.03 | 0.04 | 0.014 | |
| 75% GL Variance Run | 14 | ↑ | 0.02 | 0.03 | 0.02 | 0.03 | 0.015 | |
| V‐GLSZ | Nodule GL Variance Zones | 9 | ↑ | 0.06 | 0.02 | 0.05 | 0.01 | <0.001 |
| 75% GL Variance Zones | 37 | ↑ | 4.3E‐03 | 1.9E‐02 | 3.6E‐03 | 1.1E‐02 | 0.019 | |
| Nodule Large Zone Emphasis | 34 | ↓ | 443.6 | 1089.9 | 1708.6 | 4862.4 | 0.360 | |
| 50% Large Zone Emphasis | 2 | ↓ | 159.5 | 386.4 | 3087.1 | 8332.7 | 0.007 | |
| Nodule Small Zone Low GL Emphasis | 3 | ↓ | 0.03 | 0.02 | 0.04 | 0.02 | <0.001 | |
| 75% Small Zone Low GL Emphasis | 50 | ↑ | 0.02 | 0.01 | 0.02 | 0.01 | 0.134 | |
| V‐NGTD | Nodule Contrast Texture | 38 | ↓ | 0.41 | 0.87 | 0.50 | 0.61 | 0.018 |
| 50% Contrast Texture | 42 | ↑ | 0.37 | 0.30 | 0.22 | 0.19 | <0.001 | |
| 75% Contrast Texture | 8 | ↑ | 0.26 | 0.25 | 0.12 | 0.14 | <0.001 | |
FWHM, Full‐width‐at‐half‐maximum; GL, gray level; SD, standard deviation; V‐IH, intensity histogram, V‐GLRL, gray‐level run length texture; V‐GLSZ, gray‐level size‐zone texture; V‐NGTD, neighborhood gray tone difference texture.
3.4. Fleischner society guidelines comparison
We analyzed the potential effect the Extended ML tool would have on the follow‐up response compared to the Fleischner Society Pulmonary Nodule Follow‐up Guidelines as the INHALE study used for validation was not a lung cancer screening cohort (see Table S3, which provides the results of applying the guidelines to the validation cohort).24 These guidelines are stratified by size and nodule composition, as all nodules in this study were solid we can separate into three categories: Category 1: CT in 12 months, Category 2: CT in 6–12 months, and Category 3: CT, biopsy, or positron emission tomography in 3 months. No size distribution criterion was enforced on nodule inclusion in this study. As such, 97% of the validation cohort fell into the third size‐based category; this differed from the development cohort where the split was more balanced (Category 1: 23%, Category 2: 35%, Category 3: 41%). The Extended ML tool identified 50 malignancies that would have required follow‐up with a waiting period of 3–12 months.4 The Extended ML tool also recognized 48 benign lesions that would have required a 3‐month follow‐up with imaging or biopsy. We demonstrate the potential acceleration of malignant follow‐ups over the Fleischner guidelines; for three malignant cases, the Fleischner Society guidelines would have recommended a CT in 6–12 months while the Extended ML tool would immediately send these patients to treatment. Similarly, for an additional 45 subjects with malignant nodules, the guidelines would have recommended a follow‐up in 3 months of imaging and/or biopsy.
4. Discussion
We have developed a high performing lung nodule classification approach using radiomic features of the lung and surrounding parenchyma extracted from CT data, and validated the performance in an independent validation cohort. We discovered that features from three separate perinodular parenchymal quartile bands contributed various texture features to improve the model performance, at a level that was not achievable with one inclusive area of comparable size.
Other studies have explored the inclusion of perinodular features from the surrounding parenchyma for classification of lung nodules. A recent study comparing the performance of human observers to a computer algorithm showed observer‐interpreted broader characteristics such as spiculation, and disruption of perinodular parenchymal architecture as significant indicators of malignancy; however, subjective assessment of these characteristics is associated with a high degree of observer variability.21 Dilger et al, demonstrated the potential of quantitative texture features for improved classification in a cohort of 50 subjects, using bounding boxes for capturing parenchymal signal approximately proportional to nodule size and whole lung density measures, with optimal classifier AUC‐ROC of 0.938. Huang et al. more recently demonstrated using a cohort of 186 subjects from the NLST trial that a ML tool system constructed with perinodular features achieved an AUC‐ROC of 0.915. Their published results show a direct agreement with two nodule features selected in the Extended ML: nodule entropy and nodule variance. Also, two of their perinodular features selected from the small parenchyma ring surrounding the nodule appear similar to our selected parenchyma quartile band features: surrounding variance (at 25%, 50%) and surrounding parenchyma maximum intensity (at 25%, 50%, 75%).
The method of feature set selection used in our study is not only independent of classifier performance but also provides separate insight into the connections among imaging features and between characteristics and disease classification. In this study, the top two features were extracted from parenchymal bands distant to the nodule which provides evidence that there are more global changes in the lobe characteristics that imaging can detect. Decoding the spatial relationship between radiomic features from the parenchyma surrounding lung nodules, presents future opportunity to advance ML tool analysis beyond a binary diagnosis. The field of transport oncophysics is relatively new, but holds promise in understanding the mass transport differentials of malignancy.40 With a dataset classified in these differentials, the Extended ML tool could be used as an effective delineator of mass transport.
This study did include some limitations. The malignant tumors in both the development and validation cohorts were larger on average than their benign counterparts creating a size bias between the classes. While RECIST was selected in the final ML tool model (39/50), it was not predominantly ranked, the other selected features were not highly correlated with the nodule size. While this bias exists in both cohorts, there was still a range in nodule size with some small malignant cases and some large benign cases; if size was a driving factor, we would have expected to see a greater disparity in performance in these nodules particularly. The number of malignant and benign nodules are matched in the validation dataset, it is possible given a more population representative validation dataset (more benign nodules) performance could be altered. The CT data quality used in this study is not the current clinical standard (LDCT or clinical chest with contrast) but rather a cohort of high‐resolution multicenter trial CT scans; however, it does demonstrate the performance advantage in using high‐quality scans and incorporating the perinodular signal. Our group has previously demonstrated the effects of LDCT and ultra‐LDCT protocols on quantitative lung and airway measurements.41 The ML tool here purports the diagnostic quality of features extracted from high quality scans, of which most were not LDCT scans or subjects eligible for LDCT screening. Further studies investigating the transference of these high‐resolution features to LDCT is needed to determine the performance of the Extended ML tool on lower resolution scans. If transference of features to LDCT were to decrease the performance of the ML tool, this would show the increased value of high‐dose CT for the characterization of disease and reduction of repeated imaging studies would keep radiation dosage low.
We included only solid nodules in this study. In our validation cohort, only 34% would have met LDCT screening eligibility, making comparisons to the Fleischner guidelines more suitable for comparison than Lung‐RADS. Assuming all follow‐ups complied with the guidelines and patients were seen at the earliest follow‐up point, the Extended ML tool would reduce patient wait time on malignant nodules by a cumulative 165 months, or on average 3.3 months per patient.
This study included a large dataset with histopathology confirmed malignant cases. The dataset we have assembled includes multicenter variability, indicative of generalizability to a wide study population. As the algorithm is based only on radiological features, the approach presents a pipeline integration advantage without the need for separate (and potentially subjective) data extraction and inclusion. The high accuracy of our approach can support clinician's higher confidence in risk assessment output and hence adherence to follow‐up in concordance with the assigned class. This presents the potential to decrease the burden of unnecessary clinical follow‐up of benign lesions and the timely and efficient treatment of those with cancerous lesions.
In conclusion, we demonstrate ML tool accuracy using nodule standardized, perinodular parenchyma features. We quantified the theoretical benefit the Extended ML tool could have on the follow‐up response compared to the Fleischner Society Pulmonary Nodule Follow‐up Guidelines in reducing follow‐up of benign nodules and expediting treatment of malignant nodules.
Funding
This research was funded in part by the Holden Comprehensive Cancer Center (grant NCI P30 CA086862). The COPDGene Study was supported by NHLBI U01 HL089897 and U01 HL089856. The COPDGene study (NCT00608764) is also supported by the COPD Foundation through contributions made to an Industry Advisory Committee comprised of AstraZeneca, Boehringer‐Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens and Sunovion. The INHALE study was supported by Award Number R01CA141769 and P30CA022453 from the National Cancer Institute, Health and Human Services Award HHSN26120130011I and the Herrick Foundation. The SPIE LungX challenge data were obtained from The Cancer Imaging Archive (TCIA) sponsored by the SPIE, NCI/NIH, AAPM, and The University of Chicago.
Conflicts of Interest
The authors have no relevant conflicts of interest to disclose.
Supporting information
Fig. S1: Example centroid slice images of six benign cases. Red arrow indicates location of nodule. See Figs. S2–S4 for additional nodule images.
Fig. S2: Example centroid slice images of six benign cases. Red arrow indicates location of nodule. See Figs. S1; S3–S4 for additional nodule images.
Fig. S3: Example centroid slice images of six malignant cases. Red arrow indicates location of nodule. See Figs. S1–S2; S4 for additional nodule images.
Fig. S4: Example centroid slice images of six malignant cases. Red arrow indicates location of nodule. See Figs. S1–S3 for additional nodule images.
Fig. S5: Development Cohort Receiver‐Operator Characteristic (ROC) Curves. Curves were constructed using the R package “pROC” (Xavier Robin, Natacha Turck, Alexandre Hainard, Natalia Tiberti, Frédérique Lisacek, Jean‐Charles Sanchez and Markus Müller (2011). pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics, 12, p. 77.).
Fig. S6: Receiver‐operator curve for validation cohort (LungX) on Extended ML‐tool. Curves were constructed using the R package “pROC” (Xavier Robin, Natacha Turck, Alexandre Hainard, Natalia Tiberti, Frédérique Lisacek, Jean‐Charles Sanchez and Markus Müller (2011). pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics, 12, p. 77.).
Table S1: List of Extended machine learning tool selected features with mean, standard deviation (SD), and p‐value. Pearson's correlation of the feature with nodule size is also shown as r‐size.
Table S2: List of Extended ML tool selected features with single‐AUC calculated using “pROC” R package.
Table S3: Extended machine learning tool effect on follow‐up response.
Acknowledgments
We thank Dr. Samantha K.N. Dilger, Kimberly Sprenger, Debra O'Connel‐Moore, and Mark Escher for technical assistance and also thank Drs. Surya Bhatt and John Buatti for helpful discussion and revision of the manuscript.
References
- 1. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2. Aberle DR, Adams AM, Berg CD, et al. Reduced lung‐cancer mortality with low‐dose computed tomographic screening. New Engl J Med. 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patz EF, Pinsky P, Gatsonis C, et al. Overdiagnosis in low‐dose computed tomography screening for lung cancer. JAMA Intern Med. 2014;174:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacMahon H, Naidich DP, Goo JM, et al. Guidelines for management of incidental pulmonary nodules detected on ct images: from the fleischner society 2017. Radiology. 2017;284:228–243. [DOI] [PubMed] [Google Scholar]
- 5. Radiology ACo . Lung CT Screening Reporting and Data System (Lung‐RADS). http://wwwacrorg/Quality-Safety/Resources/LungRADS(accessed 9 Feb 2018).
- 6. Dilger SK, Uthoff J, Judisch A, et al. Improved pulmonary nodule classification utilizing quantitative lung parenchyma features. J Med Imaging. 2015;2:041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Way TW, Hadjiiski LM, Sahiner B, et al. Computer‐aided diagnosis of pulmonary nodules on CT scans: segmentation and classification using 3D active contours. Med Phys. 2006;33:2323–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Way TW, Sahiner B, Chan HP, et al. Computer‐aided diagnosis of pulmonary nodules on CT scans: improvement of classification performance with nodule surface features. Med Phys. 2009;36:3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee MC, Boroczky L, Sungur‐Stasik K, et al. Computer‐aided diagnosis of pulmonary nodules using a two‐step approach for feature selection and classifier ensemble construction. Artif Intell Med. 2010;50:43–53. [DOI] [PubMed] [Google Scholar]
- 10. Zhu Y, Tan Y, Hua Y, Wang M, Zhang G, Zhang J. Feature selection and performance evaluation of support vector machine (SVM)‐based classifier for differentiating benign and malignant pulmonary nodules by computed tomography. J Digit Imaging. 2010;23:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferreira JR, Oliveira MC, de Azevedo‐Marques PM. Characterization of pulmonary nodules based on features of margin sharpness and texture. J Digit Imaging. 2018;31:451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Felix A, Oliveira M, Machado A, Raniery J. Using 3D Texture and Margin Sharpness Features on Classification of Small Pulmonary Nodules. Paper presented at: 2016 29th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI); 4–7 Oct. 2016, 2016.
- 13. Causey JL, Zhang J, Ma S, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Scientific Reports. 2018;8:9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun T, Zhang R, Wang J, Li X, Guo X. Computer‐aided diagnosis for early‐stage lung cancer based on longitudinal and balanced data. PLoS ONE. 2013;8:e63559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Amir GJ, Lehmann HP. After detection: the improved accuracy of lung cancer assessment using radiologic computer‐aided diagnosis. Acad Radiol. 2016;23:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hawkins S, Wang H, Liu Y, et al. Predicting malignant nodules from screening CT scans. J Thorac Oncol. 2016;11:2120–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng JZ, Ni D, Chou YH, et al. Diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep. 2016;6:24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nibali A, He Z, Wollersheim D. Pulmonary nodule classification with deep residual networks. Int J Comput Assist Radiol Surg. 2017;12:1799–1808. [DOI] [PubMed] [Google Scholar]
- 19. Sun W, Zheng B, Qian W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med. 2017;89:530–539. [DOI] [PubMed] [Google Scholar]
- 20. Huang P, Park S, Yan R, et al. Added value of computer‐aided ct image features for early lung cancer diagnosis with small pulmonary nodules: a matched case‐control study. Radiology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Riel SJ. Malignancy risk estimation of pulmonary nodules in screening CTs: Comparison between a computer model and human observers. PLoS ONE. 2017;12:e0185032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Regan EA, Hokanson JE, Murphy JR, et al. Genetic epidemiology of COPD (COPDGene) study design. Copd. 2010;7:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Armato SG 3rd, Drukker K, Li F, et al. LUNGx Challenge for computerized lung nodule classification. J Med Imaging. 2016;3:044506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwartz AG, Lusk CM, Wenzlaff AS, et al. Risk of lung cancer associated with copd phenotype based on quantitative image analysis. Cancer Epidemiol Biomark Prev. 2016;25:1341–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mukhopadhyay SA. Segmentation framework of pulmonary nodules in lung CT images. J Digit Imaging. 2016;29:86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uthoff J, Sieren JC. Information theory optimization based feature selection in breast mammography lesion classification. Conf Proc IEEE Eng Med Biol Soc. 2018.
- 27. Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recogn Lett. 1990;11:415–419. [Google Scholar]
- 28. Dasarathy BV, Holder EB. Image characterizations based on joint gray level—run length distributions. Pattern Recogn Lett. 1991;12:497–502. [Google Scholar]
- 29. Galloway MM. Texture analysis using gray level run lengths. Comput Graph Image Process. 1975;4:172–179. [Google Scholar]
- 30. Thibault G, Fertil B, Navarro C, et al. Shape and texture indexes application to cell nuclei classification. Int J Pattern Recognit Artif Intell. 2013;27:1357002. [Google Scholar]
- 31. McCollough C, Bakalyar DM, Bostani M, et al. Use of water equivalent diameter for calculating patient size and size‐specific dose estimates (SSDE) in CT: The Report of AAPM Task Group 220. AAPM report. 2014;2014:6–23. [PMC free article] [PubMed] [Google Scholar]
- 32. Rousseeuw PJ, Kaufman L. Finding Groups in Data . Wiley Online. Library.1990.
- 33. Dilger SKN. Pushing the boundaries: feature extraction from the lung improves pulmonary nodule classification: Biomedical Engineering, Univeristy of. Iowa. 2016.
- 34. Davis J, Goadrich M.The relationship between Precision‐Recall and ROC curves. Proceedings of the 23rd international conference on Machine learning; Pittsburgh, Pennsylvania, USA; 2006.
- 35. DeLong ER, DeLong DM. Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 36. Mc NQ. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12:153–157. [DOI] [PubMed] [Google Scholar]
- 37. Jarque CM, Bera AK. Efficient tests for normality, homoscedasticity and serial independence of regression residuals. Econ Lett. 1980;6:255–259. [Google Scholar]
- 38. Wilcoxon F, Wilcox RA. Some rapid approximate statistical procedures. Lederle Laboratories. 1964.
- 39. Sieren JP, Newell JD Jr, Barr RG, et al. Protocol for multicenter quantitative computed tomography to phenotype the lungs. Am J Respir Crit Care Med. 2016;194:794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eugene JK, Mauro F. Transport Oncophysics in silico, in vitro, and in vivo. Phys Biol. 2014;11:060201. [DOI] [PubMed] [Google Scholar]
- 41. Hammond E, Sloan C, Newell JD Jr, et al. Comparison of low‐ and ultralow‐dose computed tomography protocols for quantitative lung and airway assessment. Med Phys. 2017;44:4747–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Example centroid slice images of six benign cases. Red arrow indicates location of nodule. See Figs. S2–S4 for additional nodule images.
Fig. S2: Example centroid slice images of six benign cases. Red arrow indicates location of nodule. See Figs. S1; S3–S4 for additional nodule images.
Fig. S3: Example centroid slice images of six malignant cases. Red arrow indicates location of nodule. See Figs. S1–S2; S4 for additional nodule images.
Fig. S4: Example centroid slice images of six malignant cases. Red arrow indicates location of nodule. See Figs. S1–S3 for additional nodule images.
Fig. S5: Development Cohort Receiver‐Operator Characteristic (ROC) Curves. Curves were constructed using the R package “pROC” (Xavier Robin, Natacha Turck, Alexandre Hainard, Natalia Tiberti, Frédérique Lisacek, Jean‐Charles Sanchez and Markus Müller (2011). pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics, 12, p. 77.).
Fig. S6: Receiver‐operator curve for validation cohort (LungX) on Extended ML‐tool. Curves were constructed using the R package “pROC” (Xavier Robin, Natacha Turck, Alexandre Hainard, Natalia Tiberti, Frédérique Lisacek, Jean‐Charles Sanchez and Markus Müller (2011). pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics, 12, p. 77.).
Table S1: List of Extended machine learning tool selected features with mean, standard deviation (SD), and p‐value. Pearson's correlation of the feature with nodule size is also shown as r‐size.
Table S2: List of Extended ML tool selected features with single‐AUC calculated using “pROC” R package.
Table S3: Extended machine learning tool effect on follow‐up response.


