Seedlings of salt-tolerant Galapagos tomatoes display a wide variation of combined responses to salinity.
Abstract
Traits of modern crops have been heavily selected in agriculture, leaving commercial lines often more susceptible to harsh conditions compared with their wild relatives. Understanding the mechanisms of stress tolerance in wild relatives can enhance crop performance under stress conditions such as high salinity. In this study, we investigated salinity tolerance of two species of wild tomato endemic to the Galapagos Islands, Solanum cheesmaniae and Solanum galapagense. Since these tomatoes grow well despite being constantly splashed with seawater, they represent a valuable genetic resource for improving salinity tolerance in commercial tomatoes. To explore their potential, we recorded over 20 traits reflecting plant growth, physiology, and ion content in 67 accessions and two commercial tomato lines of Solanum lycopersicum. Salt treatments were applied for 10 d using supported hydroponics. The Galapagos tomatoes displayed greater tolerance to salt stress than the commercial lines and showed substantial natural variation in their responses. The accessions LA0317, LA1449, and LA1403 showed particularly high salinity tolerance based on growth under salinity stress. Therefore, Galapagos tomatoes should be further explored to identify the genes underlying their high tolerance and be used as a resource for increasing the salinity tolerance of commercial tomatoes. The generated data, along with useful analysis tools, have been packaged and made publicly available via an interactive online application (https://mmjulkowska.shinyapps.io/La_isla_de_tomato/) to facilitate trait selection and the use of Galapagos tomatoes for the development of salt-tolerant commercial tomatoes.
High soil salinity is one of the main agricultural challenges in the modern world (Rengasamy, 2016). Salt stress affects the growth and development of plants, thus reducing their yield (Arzani and Ashraf, 2016). Global cultivated lands cover ∼1.5 billion hectares, and an estimated 32 million hectares are damaged by salinity. Irrigated lands, having the highest productivity, comprise just 230 million hectares, of which an estimated 20% have yields reduced by high soil salinity (Munns, 2005). Water availability for agriculture is another major concern, not only in desert regions but at a global level, as freshwater supplies are being depleted (Famiglietti, 2014). Salt-affected areas worldwide are predicted to continue expanding at a rate of ∼10% per year due to low precipitation, high surface evaporation, erosion of rocks, irrigation with saline water, and poor cultural practices (Foolad, 2004).
Wild relatives of modern crops have been used for crop improvement for more than 60 years (Hajjar and Hodgkin, 2007). In their natural habitats, they withstand harsh conditions such as high soil salinity (Muñoz et al., 2017). Adaptation to wide-ranging environments has enriched the genetic pool of these wild relatives (Gruber, 2017), thus representing a rich source of potentially beneficial alleles that can be explored to improve salinity tolerance (Zamani Babgohari et al., 2013). Therefore, performing phenotypic screens to capture the natural variation of the wild germplasm can help uncover new genetic sources for enhancing stress tolerance (Arzani and Ashraf, 2016).
The Galapagos Islands, an isolated environment close to the center of origin of the current domesticated tomato (Blanca et al., 2012), hold a rich genetic diversity of tomato wild relatives, notably the two endemic species, Solanum cheesmaniae (formerly Lycopersicon cheesmanii) and Solanum galapagense (formerly Lolanum cheesmanii forma minor), adapted to thrive in harsh environments and highly saline coastal habitats (Rick, 1956; Rush and Epstein, 1976). Previous studies hypothesized that at least some accessions of Galapagos tomatoes, collected from both coastal and inland regions, are able to survive higher NaCl concentrations than the domesticated tomato (Rush and Epstein, 1976, 1981; Tal and Shannon, 1983). However, little is known about the specific mechanisms by which the Galapagos tomatoes thrive on saline soil.
The known mechanisms involved in plant salinity tolerance can be classified into three types: (1) osmotic tolerance, involving the sensing and signaling modules occurring before shoot Na+ accumulation and causing reductions in growth rate; (2) ion exclusion, the limitation of ion accumulation in the shoot by ion sequestration in the roots; and (3) tissue tolerance, where high Na+ concentrations in the shoot are compartmentalized in the vacuoles to reduce their toxic effects (Munns and Tester, 2008; Roy et al., 2014). These considerations have been developed further by Morton et al. (2018) to include a wider range of other physiological traits, focusing in particular on the ability of plants to maintain processes in saline conditions relative to nonsaline conditions. Traits specific to salinity tolerance in tomato have also been described well by Cuartero et al. (2002). The technical approaches that can be taken to measure these traits are detailed in Negrão et al. (2017). In this work, a range of traits from across these different articles that could be quantified in our greenhouse conditions were measured in control and salt-grown plants, and the ability of plants to maintain those traits in saline conditions relative to control conditions was quantified.
An in-depth characterization of Galapagos tomato accessions would allow us to understand the mechanisms for salinity tolerance used in these species, and to identify the most tolerant accessions for further studies, thus helping to unlock their potential use as genetic resources for improving salinity tolerance of commercial tomato. However, the wild nature of these plants makes them difficult to compare with domesticated commercial lines, as they differ in growth rates, morphology, and plant architecture, all of which affect a plant’s performance under saline conditions. Hence, developing a robust phenotyping method that is suitable for investigating wild germplasm, accounting for all variations in growth, was necessary. The application of salt treatments at the same growth stage for all accessions is important when growth rates vary substantially. To effectively deliver the salt treatment, a hydroponics growth system is preferred, since it allows the precise control of salt concentration in the medium (Genc et al., 2007; Munns et al., 2010; Negrão et al., 2017). Moreover, the effects on the ionic activity of micronutrients, such as calcium, by the interaction between salt and nutrients in the medium, can be calculated and balanced by adding supplemental nutrients (Tester and Davenport, 2003). Also, given that so many tomatoes are grown commercially in hydroponic systems, such results can even be of direct relevance to commercial applications.
In this study, the phenotypic traits related to salinity tolerance of 67 different accessions of wild tomato from the Galapagos Islands were scored, using a flood-and-drain hydroponic growth system. Traits reflecting growth, physiology, and ion content were explored using multivariate analysis, leading to a better and more comprehensive understanding of the role of these traits in salinity tolerance in Galapagos tomatoes. Wide variations in growth, physiology, and ion content were observed across the accessions, demonstrating great natural diversity underlying the three main mechanisms of salinity tolerance within the Galapagos tomatoes. No one trait was found to confer salinity tolerance, but different traits contributed to salinity tolerance in different lines.
RESULTS
Galapagos Tomatoes Are More Salt Tolerant Than the Commercial Tomato Varieties Tested
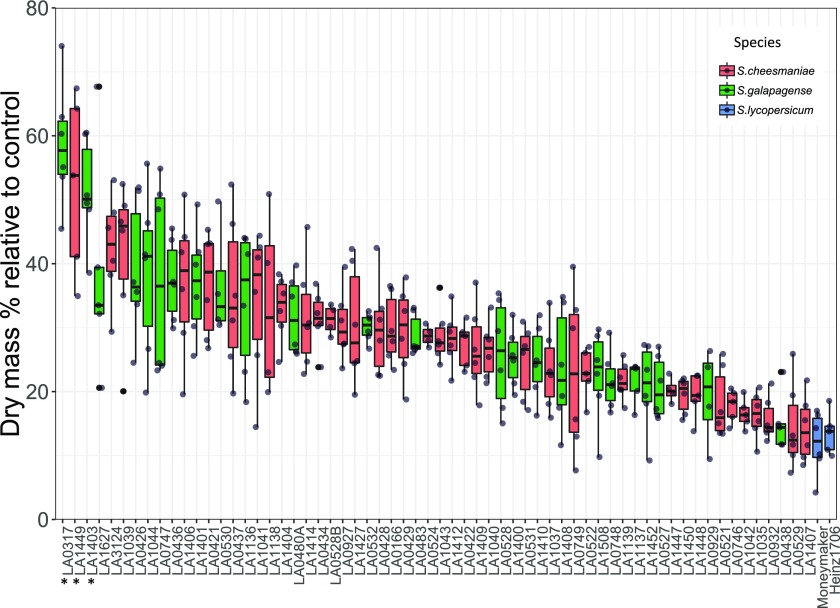
As suggested by Negrão et al. (2017), responses to salinity stress were measured only for the time when the plants were stressed (by taking measurements before and after the stress treatment). S. cheesmaniae and S. galapagense accessions were better able to maintain growth (based on dry mass) during the salt stress period than the Solanum lycopersicum varieties tested (Fig. 1). This effect is less apparent when biomass is determined only at the endpoint of the experiment, since S. lycopersicum plants are bigger than Galapagos tomato plants throughout the developmental stages (Supplemental Fig. S1). There was a large variation in salinity tolerance between accessions, ranging from a difference in dry mass in saline conditions relative to control conditions of 12% to 55% (Fig. 1).
Figure 1.
Salinity tolerance across the studied Galapagos tomato accessions. Salinity tolerance measured as the difference in dry mass between the start and end of the treatment of plants grown in saline conditions relative to plants grown in control conditions (n = 6). Different colors represent the different species S. cheesmaniae (coral), S. galapagense (green), S. lycopersicum (blue). Top performing accessions are marked with an asterisk (*). Boxplot whiskers extend 1.5× the interquartile range from 25th and 75th percentiles.
Correlation Analysis of Different Seedling Traits Revealed Trait Groups
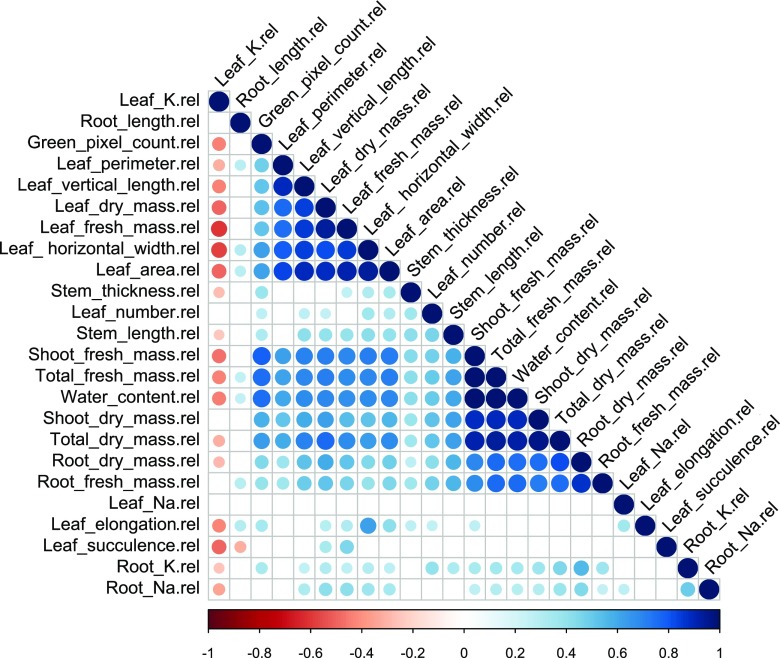
The correlation matrix (Fig. 2) shows that leaf traits such as perimeter, vertical length, dry and fresh mass, horizontal width, and area are all positively and significantly correlated (correlation coefficients 0.79–0.93, P = 0.001; Supplemental Fig. S2). Plant growth-related traits, such as shoot fresh and dry mass, root fresh and dry mass, total fresh and dry mass, and total water content (fresh weight − dry weight), are also positively and significantly correlated (correlation coefficients 0.69–1.00, P = 0.001; Supplemental Fig. S2).
Figure 2.
Pearson correlation matrix of the recorded traits under salinity relative to control conditions (n = 6). Large circles represent strong correlations and smaller circles represent weaker correlations. The color scale indicates the sort of correlation, where 1 denotes completely positive correlation (dark blue) and −1 denotes completely negative correlation (dark red) between two traits. Only significant correlations are shown (P < 0.05).
Interestingly, leaf K concentration in salt-treated plants relative to control plants is negatively correlated with all the leaf traits, some plant growth-related traits, and Na and K in the root. On the other hand, leaf Na concentration in salt-treated plants, relative to control plants, did not have a significant correlation with any of the other measured traits. In most cases, leaf Na concentration in control plants was negligible, so the leaf Na concentration in salt-treated plants, relative to control plants, was very similar to the leaf Na concentration in salt-treated plants. Na and K in the root have a slight positive correlation with leaf and plant growth-related traits.
Plant size can be assessed by counting the green pixels of the seedling image. Green pixel count was significantly correlated with most of the salinity tolerance-related traits (Fig. 2). For most of the accessions tested, the green pixel count had a positive correlation with the shoot fresh mass of salt-treated plants relative to control plants (Supplemental Fig. S3A). However, green pixel count proved a more useful measure of plant growth in S. cheesmaniae (r2 = 0.85), than in S. galapagense (r2 = 0.64; Supplemental Fig. S3, B–D). Based on the correlation analyses, the following set of plant traits representing each of the groups of traits that have been identified as potentially useful predictors of salinity tolerance in plants were selected for further analysis: Na and K concentration in root and leaf, leaf area, leaf elongation, leaf succulence, leaf number, stem and root length, and total fresh mass. To compare the different species, the traits in salt stress relative to control conditions for the same accession were used.
PCA Revealed Tendencies and Contributions of Selected Traits
A principal component analysis (PCA) was performed to reduce data dimensionality and reveal the potential relationships among representative salinity-tolerance traits. In this study, the four main PCA axes had eigenvalues >1 (Supplemental Table S1), which indicates that each principal component (PC) accounts for more variance than is accounted for by one of the original variables in the standardized data. This was used as a cutoff to determine the number of PCs to retain.
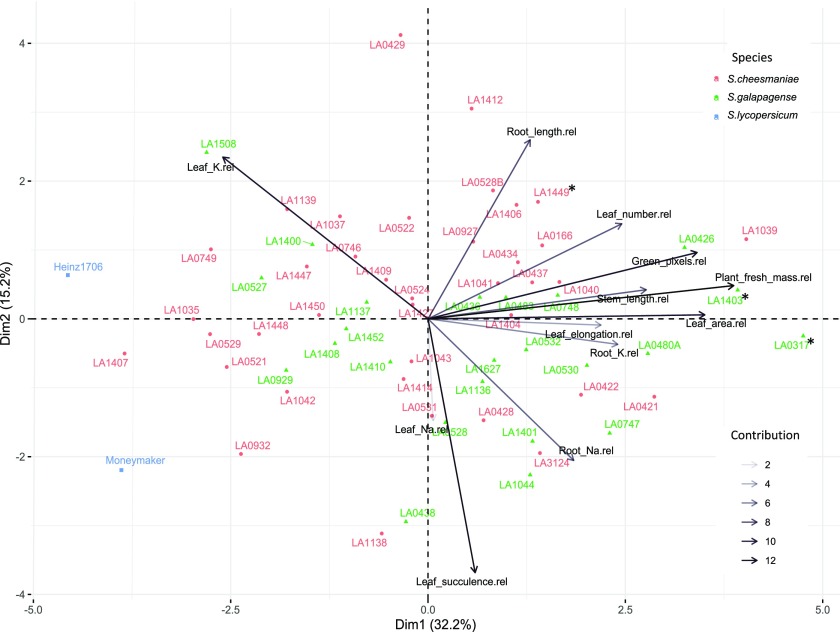
PC1 explained 32.2% of the total variability between traits/individuals and was associated with most traits, except leaf Na concentration and leaf succulence (Fig. 3; Table 1). The most significant trait for PC1 was the total fresh mass (Table 1). The accessions at the lower end of PC1 are those whose growth was most affected by salinity but which were still able to retain high levels of K in the leaf, while those at the higher end are the accessions with higher levels of plant growth, leaf area, leaf number, and stem and root length.
Figure 3.
PCA biplot of 62 Galapagos tomato accessions and two commercial tomato varieties (n = 6), based on the variance in 11 salt stress-related physiological traits, explained by two PC axes (Dim1 and Dim2). The two components explained 32.2% and 15.2% of the variance, respectively. Arrows denote the strength of the trait influence on the first two PCs. The transparency of the arrows indicates the contribution to the variance in the dataset, ranging from 2% (lightest) to 12% (darkest). The direction and length of the arrows indicate how each trait contributes to the first two components in the PCA. Aligned vectors indicate a strong positive correlation between the two traits. Vectors at right angles and opposites indicate no correlation and negative correlation, respectively. The first component shows that leaf K concentration is negatively correlated with the other representative traits. The second component shows that leaf elongation, leaf succulence, leaf and root Na concentrations, and root K concentration are negatively correlated with the other representative traits. Individual accessions are placed on the ordination plane. Different colors and symbols represent the different species, S. cheesmaniae (coral circles), S. galapagense (green triangles), S. lycopersicum (blue squares). Top performing accessions are marked with an asterisk (*).
Table 1. Contributions of plant traits to the four main PCA axes.
Values were obtained from a matrix of 11 traits × 64 Galapagos and commercial tomato accessions and are ranked in order of magnitude in PC1 (eigenvalue >1). Traits significantly correlated to each PCA axis (α = 0.05) are indicated in bold. All the traits represent the trait value under salt stress relative to control conditions. The highest contribution per PC is indicated by an asterisk (*).
| Traits in Salt Stress Relative to Control Conditions | PC1 (32.2%) | PC2 (15.2%) | PC3 (11.5%) | PC4 (10.2%) |
|---|---|---|---|---|
| Plant fresh mass | 19.9* | 0.66 | 1.93 | 0.95 |
| Leaf area | 16.3 | 0.01 | 1.65 | 3.13 |
| Green pixels | 15.4 | 2.60 | 11.7 | 1.15 |
| Stem length | 10.1 | 0.50 | 2.23 | 4.44 |
| Leaf K+ | 8.97 | 15.5 | 8.46 | 5.65 |
| Leaf number | 8.00 | 5.35 | 11.9 | 6.95 |
| Root K+ | 7.67 | 0.38 | 2.27 | 20.8 |
| Leaf elongation | 6.37 | 0.02 | 6.30 | 31.2* |
| Root Na+ | 4.52 | 11.9 | 9.39 | 7.50 |
| Root length | 2.22 | 19.0 | 2.41 | 7.81 |
| Leaf succulence | 0.47 | 38.1* | 1.34 | 0.00 |
| Leaf Na+ | 0.01 | 6.13 | 40.4* | 10.5 |
PC2 accounted for an additional 15.2% of the total variability among seedling traits and appeared to be related to the ion content and some growth traits (Fig. 3; Table 1). The accessions with succulent leaves and higher accumulation of Na in the leaf were located at the lower end of PC2, while those with increased leaf number and K retention in the leaf were located at the higher end of PC2. PC2 also divided the root Na concentration and root length, where those accessions with high Na concentration in the root had the shortest roots.
PC3 accounted for 11.5% of the total variability among salinity tolerance-related traits. It was significantly associated with total fresh mass but had a stronger association with leaf traits, such as elongation factor (length/width) and Na concentration (Table 1). This could suggest that Na concentration in the leaf is independent of the other traits.
PC4 accounted for an additional 10.2% of the total variability and is significantly associated with Na and K accumulation in the leaf and root, but also with leaf number and area, and root length (Table 1).
Overall, the PCA indicates that in this experiment, the primary traits that varied and associated with each other were total fresh mass, ion content, and some leaf traits, such as elongation factor.
Cluster Analysis Suggests That Salinity Tolerance at the Seedling Stage Is Best Defined by the Ability of the Plant to Maintain Growth under Salt Stress Conditions
Cluster analysis is a suitable method to study large datasets involving multiple variables. It allows the grouping of accessions with similar traits and the recognition of hidden patterns or trends in the data. Therefore, we used cluster analysis to examine how the different accessions grouped by traits related to salinity tolerance and to see if any of the traits predominantly explain the overall variation.
The K-means cluster analysis (MacQueen, 1967) of the surviving 38 accessions of S. cheesmaniae and 24 accessions of S. galapagense after treatment was used to classify accessions into different clusters (K), where the accessions within the same cluster are as similar as possible, while accessions from different clusters are as dissimilar as possible. The number of clusters K = 2 was chosen with the aim of separating the most tolerant accessions from the least tolerant while considering some salinity tolerance-related traits. A total of 11 nonredundant traits were selected based on their strong correlation with other traits measured and their lack of correlation with each other (Fig. 1).
Considering the values of the selected traits, the Euclidean distance between each accession and the cluster mean was calculated to assign the accession to the nearest cluster. A new mean value of each cluster was calculated after an accession was assigned to it, and every accession was checked again to see if it was closer to a different cluster. These steps were iteratively repeated until convergence was achieved.
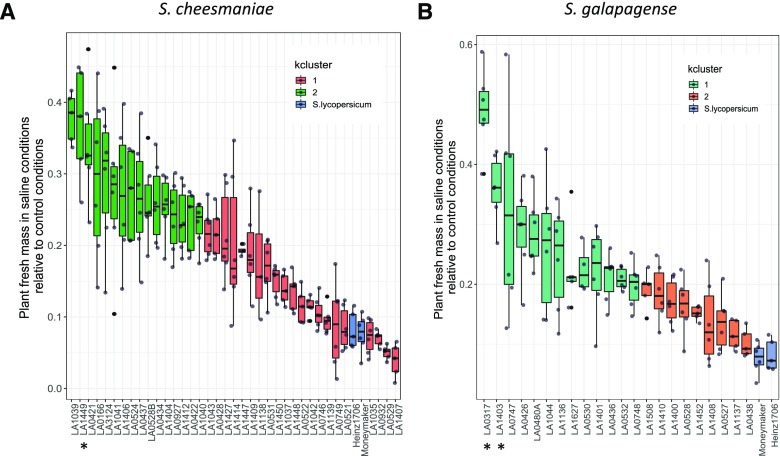
Bar plots were used to visualize the distribution of the accessions by cluster for each specific trait. A similar visualization strategy is commonly used when plotting Q-matrices and identifying K clusters in population structure studies (Pritchard et al., 2000). The accessions were arranged in descending order and the bars are colored by cluster (Supplemental Fig. S4). By visualizing bar plots for all traits, it was easy to identify that the plant fresh mass was predominantly defining the clustering by K = 2. From this, it was observed that the accessions of both species were best grouped by their fresh mass production under salt stress relative to control conditions (Supplemental Fig. S4). Thus, the two clusters divide the accessions of each species of Galapagos tomato into those with high tolerance and those with low tolerance to salinity, in terms of growth maintenance (Fig. 4). Cluster 1 included accessions with higher fresh mass production during salt stress relative to controls, indicative of their ability to better maintain growth under salt stress. Cluster 1 of S. cheesmaniae had 23 members (Fig. 4A) and cluster 1 of S. galapagense had 14 members (Fig. 4B).
Figure 4.
K-means clustering (K = 2) of each of the Galapagos tomato species. A boxplot for each trait was plotted with the accessions (n = 6) in descending order, where different colors represent different cluster assignments. Cluster number was chosen based on the most informative grouping. A, S. cheesmaniae within-cluster sums of squares (SS) by cluster were 217.6 and 136.9, respectively (between SS/total SS = 20.2%). B, S. galapagense within-cluster SS by cluster were 115.9 and 94.1, respectively (between SS/total SS = 23.9%). Both species showed a clean cluster separation when accessions were arranged in descending order by the total plant fresh mass in salt stress relative to control conditions. S. lycopersicum cultivars were added for reference. Top performing accessions are marked with an asterisk (*). Boxplot whiskers extend 1.5× the interquartile range from 25th and 75th percentiles.
Natural Variation in Salinity Tolerance Exists across Galapagos Tomato Accessions
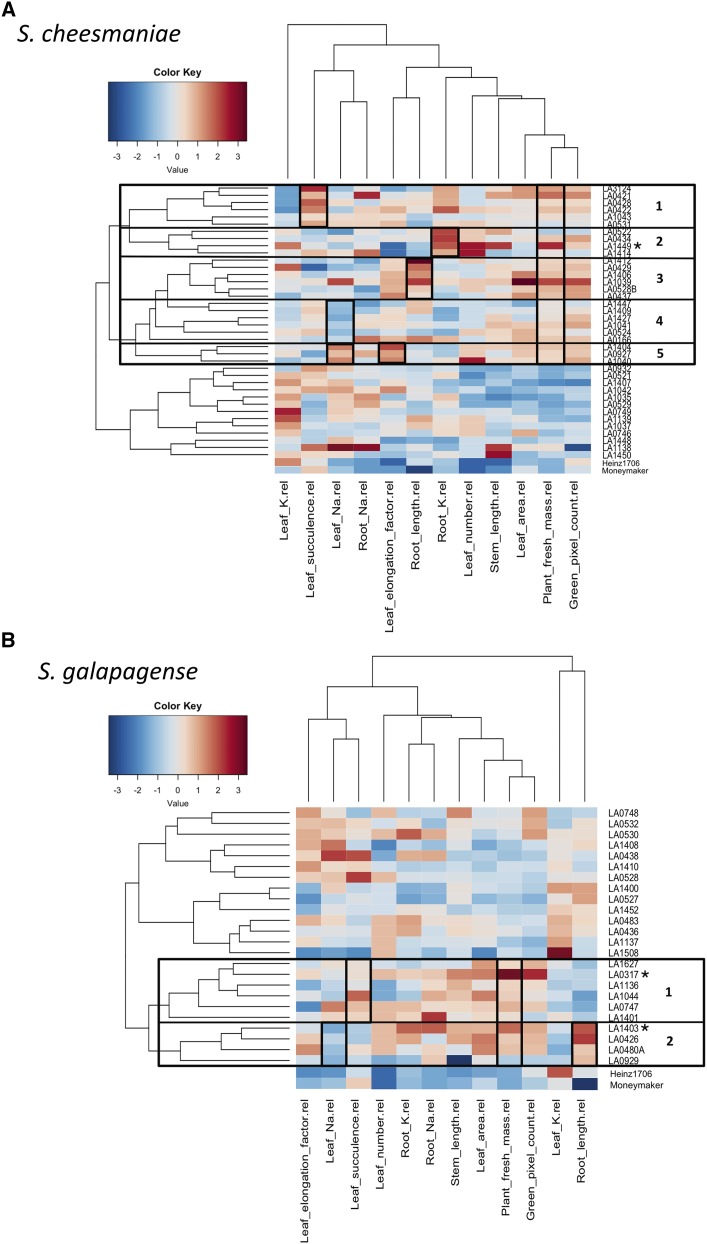
Phenotypic data were also analyzed using a hierarchical clustering approach (Fig. 5), which was found to be more complex, but more informative, than the K-means clustering method. A heat map paired with the dendrogram obtained by hierarchical clustering provides a way to visualize and simplify large datasets. This method is routinely used for gene expression data analysis (Eisen et al., 1998) and metabolomics (Tikunov et al., 2005). More recently, it has proven useful also to analyze genotype and trait interactions (Chen et al., 2014; Awlia et al., 2016; Clark, 2016; Julkowska et al., 2016).
Figure 5.
Hierarchical clustering and heat map of Galapagos tomato accessions and important salinity tolerance-related traits, divided by species. Each column represents a trait and each row represents an accession (n = 6). Accessions with large fresh mass in salt stress relative to control conditions clustered together. However, they differed in other traits. Further clustering by similarity in traits is indicated in the figure, showing diversity in salt stress responses among Galapagos tomato accessions. A, S. cheesmaniae accessions divide into two main clusters. The accessions in the upper cluster share high values of total fresh mass. This cluster is further divided into five other clusters (indicated in the figure), each with a distinctive trait that differentiates it from other clusters (Table 2). B, S. galapagense accessions divide into two main clusters. The accessions in the lower cluster share high values of total fresh mass. This cluster is further divided into two other clusters (indicated in the figure), each with a distinctive trait that differentiates it from other clusters (Table 2). S. lycopersicum cultivars were added for reference. Top performing accessions are marked with an asterisk (*).
The heat map divided the accessions into two clusters, also defined by fresh mass in salt-treated plants relative to control plants. Moreover, the heat map showed clustering of accessions based on other traits within the main mass-related clusters (i.e. K and Na concentrations in both leaf and root differed greatly between groups of accessions, despite them having similar plant fresh mass). This indicates a pronounced natural variation in salinity tolerance mechanisms within the Galapagos tomato collection.
In S. cheesmaniae, the hierarchical clustering method (Fig. 5A) separated the accessions into two main clusters, which display contrasting values of fresh mass, leaf area, stem length, leaf number, root length, leaf elongation, leaf Na and K concentration, and green pixel count. Within the cluster of accessions with high relative fresh mass, five different clusters could be distinguished that differed in leaf succulence, root K concentrations, root length, leaf Na+ concentration, and leaf elongation (Table 2). Based on leaf Na concentration it can be determined whether the plant is using Na+ exclusion or Na+ compartmentalization mechanisms. High leaf Na, paired with growth maintenance (salinity tolerance), means that Na+ is being compartmentalized in the leaf vacuoles and photosynthesis can still take place. Low leaf Na, paired with growth maintenance, means that the plant is preventing Na+ from reaching the leaves, thus protecting the photosynthetic organs.
Table 2. General description of the main mechanisms identified in each of the clusters.
Top performing accessions are marked with an asterisk (*).
| Species | Highly Tolerant Accession Cluster | Accessions in the Cluster | High Maintenance of Particular Feature in Saline Conditions: |
|---|---|---|---|
| S. cheesmaniae | 1 | LA3124, LA0421, LA0428, LA0422, LA1043, LA0531 | Leaf water |
| 2 | LA0522, LA0434, LA1449*, LA1414 | Root K+ | |
| 3 | LA1412, LA0429, LA1406, LA1039, LA0528B, LA0437 | Root length | |
| 4 | LA1447, LA1409, LA1427, LA1041, LA0524, LA0166 | Leaf Na+ exclusion | |
| 5 | LA1404, LA0927, LA1040 | Leaf elongation and Na+ compartmentalization | |
| S. galapagense | 1 | LA1627, LA0317*, LA1136, LA1044, LA0747, LA1401 | Leaf water |
| 2 | LA1403*, LA0426, LA0480A, LA0929 | Leaf Na+ exclusion and root length |
The S. galapagense accessions also separated into two clusters (Fig. 5B), based on their relative fresh mass, leaf area, root Na, root K, leaf K, and green pixel count. The cluster with the high relative fresh mass was divided into two clusters differing in leaf succulence, leaf Na concentration, and root length (Table 2).
The phenotypic data collected were integrated into a Shiny App, Isla_Tomate (https://mmjulkowska.shinyapps.io/La_isla_de_tomato/), allowing interactive clustering of the data. The identified clusters can be validated using the Isla_Tomate App by grouping the accessions into clusters based on the chosen trait and examining the significant differences between the clusters based on each trait. Significance was calculated using Tukey’s honestly significant difference pairwise comparison with P < 0.05. The data for both species showed that clustering by plant fresh mass forms two significant groups (Supplemental Fig. S5), which we can divide into the two groups of high- and low salinity-tolerance accessions.
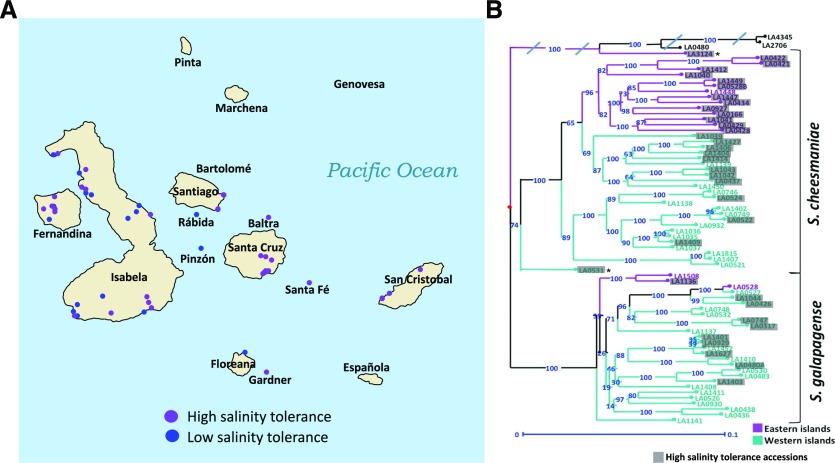
The dendrograms presented in Figure 5 represent how similar individual accessions react to salt, based on the selected traits. When the hierarchical clustering groups of the accessions were compared to their geographical origin or genetic distance between them (Pailles et al., 2017), a connection was observed. Accessions from the high- and low-salinity tolerance clusters were located in the Galapagos Islands map (Fig. 6A); high-tolerance accessions are distributed across the archipelago, while low-tolerance accessions are only present in the western islands, which receive higher precipitation than the eastern islands (Rick and Fobes, 1975) and therefore have abundant access to fresh water. The highly tolerant accessions were also marked in the Galapagos tomatoes’ genetic distance tree (Pailles et al., 2017), from which it was observed that the accessions with similar tolerance are also genetically close together (Fig. 6B). This suggests that geographical origin and evolutionary history have some influence on Galapagos tomatoes’ salinity tolerance mechanisms.
Figure 6.
Geographical and genetic relation with hierarchical clustering. Based on figure 3 from Pailles et al. (2017), hierarchical clustering results were drawn into the Galapagos Islands map and genetic distance tree. A, Geographical origin of Galapagos tomato accessions corresponds with salinity tolerance clustering. Low salinity tolerance accessions are found only in locations with high annual precipitation, as described by Rick and Fobes (1975). B, Genetic distance between Galapagos tomato accessions corresponds with salinity tolerance clustering. Accessions with high salinity tolerance (based on hierarchical clustering) are highlighted in gray. It is observed that accessions with similar salinity tolerance are also genetically close.
The two types of trait association analyses (PCA and clustering analyses) indicated that similar trait influences determine differences between accessions and their responses to salinity.
DISCUSSION
Wild relatives of modern crops possess useful traits that can potentially improve plant performance under salt stress. Hence, there is a need for characterizing and screening the available germplasm. However, their wild nature and great natural variation in many traits make them difficult to study quantitatively with conventional methods. The specific mechanisms through which Galapagos tomatoes are tolerant to salinity are not known. Identification of salinity tolerance mechanisms in Galapagos tomatoes will facilitate the improvement of salinity tolerance in current tomato elite varieties.
In this study, most available accessions of wild tomatoes endemic to the Galapagos Islands were screened for salinity tolerance traits. The first objective was to develop an efficient screening method that allowed the quantitative comparison of salt stress responses of different wild tomato seedlings. For this, a commercial ebb-and-flow supported hydroponics system was used. The use of hydroponics for experimental screenings allows better control of the growth media, stress exposure, and experimental reproducibility (Genc et al., 2007, Munns et al., 2010). In this case, the hydroponics system facilitated even delivery of NaCl to the plants at the root level. The plastic-beads substrate conserved moisture during drainage, supported the roots, and protected them from breaking and allowing uncontrolled Na+ influx into the root system (Miller, 1987). An opaque substrate was preferred to simulate the light-blocking properties of soil and to limit algal growth. The system’s flexibility allowed treatment at the same developmental stage, independent of the growth rate of each of the different genotypes. This is necessary for large-scale experiments with a large number of genotypes from different species that have widely different rates of growth.
The second objective of the study was to determine the most informative traits indicating salinity tolerance in Galapagos tomato seedlings while highlighting the most tolerant accessions and their potential underlying mechanisms for salinity tolerance. Salt stress affects many aspects of plant growth, such as biomass production, yield, photosynthesis, and leaf metabolites (Chinnusamy et al., 2006; Munns and Tester, 2008; Munns and Gilliham, 2015; Negrão et al., 2017). Hence, there are many traits that could be recorded and analyzed to accurately assess the salinity tolerance of a plant. In this study, it was found that many of the traits were highly and significantly correlated, so the salinity tolerance could be assessed by focusing attention on only a few representative traits. From the highly correlated leaf-related traits, leaf area was chosen as it was previously reported to be an important trait for salinity tolerance in tomato (Cuartero and Fernández-Muñoz, 1998; Dogan et al., 2010). From the highly correlated plant growth-related traits, total fresh mass was chosen, since it describes the increase in biomass and water retention at the whole-plant level. Other traits, which did not correlate as strongly with the leaf- or growth-related traits, such as root and stem length, leaf number, leaf elongation, leaf succulence, and Na and K concentrations in root and leaf, were included in further analyses. Leaf K was the only trait that negatively correlated with multiple traits. In general, the two Galapagos tomato species, S. cheesmaniae and S. galapagense, did not differentiate in PCA. This indicated high phenotypic variability within both species that was greater than differences between the two species, which are clear at the genetic level (Pailles et al., 2017).
The PCA revealed that the selected traits contributing most to describe the variation across accessions were total fresh mass, leaf area, leaf succulence, and leaf K concentration. Plants with greater fresh mass and leaf area shared a tendency to have more leaves, longer stems and roots, and higher root K concentration, whereas leaf succulence, leaf elongation, K and Na concentrations in leaf, and root Na concentration tendencies differed greatly. The presence of similar tendencies in the two species suggests that salinity tolerance-related traits are conserved across these two species.
Total plant fresh mass, which is a measure of growth maintenance during salt stress, was the trait driving most of the variation across accessions. Growth maintenance has been widely acknowledged to be a good estimate of salinity tolerance (Genc et al., 2007; Negrão et al., 2017), especially at the seedling stage, since it is not possible in young plants to measure any commercially relevant traits such as yield. Thus, the genotypic variability for salinity tolerance was assessed in this study based on the maintenance of growth under saline conditions relative to control conditions. This assumption was further confirmed by K-means clustering analysis, where the plant fresh mass of salt-treated plants relative to control plants was identified as the main trait driving the accession clustering. The accessions were categorized into two groups with high or low salinity tolerance, based on the relative fresh mass. Within these groups, it was tested whether Galapagos tomato accessions showed covarying groups of tolerance traits that are consistent across accessions, to categorize them as traits of influence in salinity tolerance. However, remarkable trait diversity was found within the highly tolerant group, which suggests the presence of different mechanisms of salinity tolerance between accessions within the species, consistent with previous reports by Rush and Epstein (1981) and Cuartero et al. (1992).
Maintenance of growth, defined by an increase in mass, is one of the most important mechanisms contributing to salinity tolerance. According to Munns and Termaat (1986), leaf growth is more affected by salinity than root growth. In this study, leaf area was also observed to be an important trait contributing to plant mass. However, the physiological mechanisms underlying leaf growth inhibition under salt stress are not fully understood (Neves-Piestun and Bernstein, 2001). The current results showed a strong and significant correlation between leaf area and leaf elongation, followed by leaf area and leaf number. These correlations could provide insights into the mechanisms by which salt stress affects leaf growth.
K concentration was found to be generally lower in all the high-tolerance accessions when compared to the low-tolerance accessions, especially in leaves. However, one group of S. cheesmaniae tolerant accessions showed higher levels of K in the root and leaf samples. Percey et al. (2016) reported that reduction in K+ efflux in halophytes is linked to reduced H+ efflux, which saves energy, allowing more resources to be redirected for plant growth. Therefore, the ability to maintain K+ uptake and a high K+:Na+ ratio under salt stress may be an important mechanism of salinity tolerance (Chen et al., 2007; Shabala and Cuin, 2008). K+ deficiency in plants can impair photosynthesis (Cakmak, 2005), as well as many other aspects of cellular function, such as protein synthesis (Flowers and Dalmond, 1992). The four most salt-tolerant accessions of S. cheesmaniae and the two most salt-tolerant accessions of S. galapagense showed high K in their roots, which could indicate that they are good at maintaining K uptake under salt stress. In some accessions, higher K in the roots seems to go hand in hand with low K in the leaves, which could be explained by higher K+ retranslocation, which assists in NO3− uptake and distribution (Taleisnik and Grunherg 1994), or lower K+ translocation from roots to shoot. In addition, bigger leaves appeared to have a lower K concentration compared to smaller leaves, which could be due to a dilution effect, e.g. having a similar amount of K to that of the smaller leaves but more water content (Jarrell and Beverly, 1981).
Increase in leaf succulence (measured as water per unit leaf area), a strategy for reducing salt concentrations in photosynthetic tissues (Han et al., 2013), is another known mechanism of salinity tolerance in some plants, including tomato (Cuartero and Fernández-Muñoz, 1998). The hierarchical clustering of accessions and traits showed that both S. cheesmaniae and S. galapagense accessions each formed a cluster of accessions with increased leaf succulence and low leaf Na concentrations. This might be caused by the succulence increasing cell size, thereby diluting the salt without increasing the leaf area (Munns et al., 2016).
The accumulation of Na+ relative to biomass may also be an indicator of salinity tolerance. However, Na+ is toxic when it accumulates in the cell cytosol, resulting in ionic disequilibrium (Hanin et al., 2016). Additionally, Na+ reduces the availability of K+ binding sites for important metabolic processes in the cytoplasm (Wei et al., 2017). For the plant to protect itself when exposed to salt stress, it has to either limit the entry of Na+ through the roots or control Na+ concentration and distribution once it has entered (Tester and Davenport, 2003; Hanin et al., 2016). The Na+ that enters the root cells is extruded from the cytoplasm into the apoplastic space and compartmentalized into the vacuole (Maggio et al., 2007). This process is called tissue tolerance (Munns et al., 2016). One cluster from each species included tolerant accessions with high Na concentrations in their leaves, which suggests significant levels of tissue tolerance (Munns et al., 2016). The S. cheesmaniae cluster appeared to respond to high Na accumulation in the leaves by growing more leaves, while the S. galapagense cluster appeared to increase leaf succulence.
In conclusion, our seedling screen study allowed characterization of the responses of 67 Galapagos tomato accessions to salt stress. Individual accessions were classified based on the phenotypic traits contributing to salinity tolerance. Interestingly, it was observed that individual salt-tolerant accessions from the Galapagos Islands use different mechanisms to maintain their growth at the seedling stage under saline conditions. The different combinations of characteristics found across all the studied accessions, while maintaining a good relative fresh mass, indicate that the Galapagos tomatoes are naturally diverse and have different mechanisms to tolerate high salinity. In terms of growth maintenance under stress, the accessions LA0317, LA1449, and LA1403 displayed exceptional salinity tolerance at the seedling stage. However, to assess which mechanisms are the most effective for salinity tolerance, the tolerant accessions should be studied further at later growth stages, such as the reproductive stage, to evaluate the effect of salinity on yield. Additionally, trials to evaluate their performance under field conditions are recommended. Together with our results, dissecting the genetic basis of salinity tolerance mechanisms through a genetic characterization and/or transcriptomic approach would facilitate the selection of useful accessions as genetic sources for breeding salinity tolerance traits into commercial tomatoes.
MATERIALS AND METHODS
Plant Material and Seed Treatments
A collection of 67 Galapagos tomato accessions (Pailles et al., 2017) was characterized and screened for salinity tolerance, of which 39 are Solanum cheesmaniae and 28 are Solanum galapagense. Two commercial Solanum lycopersicum varieties, Heinz 1706 and Moneymaker, were also used for comparison. Heinz 1706 has a published reference genome sequence (Tomato Genome Consortium, 2012), and Moneymaker was previously shown to have mild tolerance to salt stress (Cuartero et al., 1992). The seeds were obtained from the Tomato Genetics Resource Center, University of California Davis, and propagated under controlled conditions (temperature [T] = 27°C day, 23°C night), in the greenhouse of King Abdullah University of Science and Technology (KAUST). Seeds from a single plant were used in the experiment.
To sterilize the surface of the seeds and to break their dormancy, all seeds were treated with 10% (v/v) bleach solution for 10–30 min, until the seed coat was softened and had become transparent, then washed several times in tap water. This treatment was necessary for the germination of most of the Galapagos tomato seeds (Rush and Epstein, 1976). Although the commercial varieties did not require bleaching to germinate, the same treatment was applied to all the seeds.
Experimental Hydroponics Setup
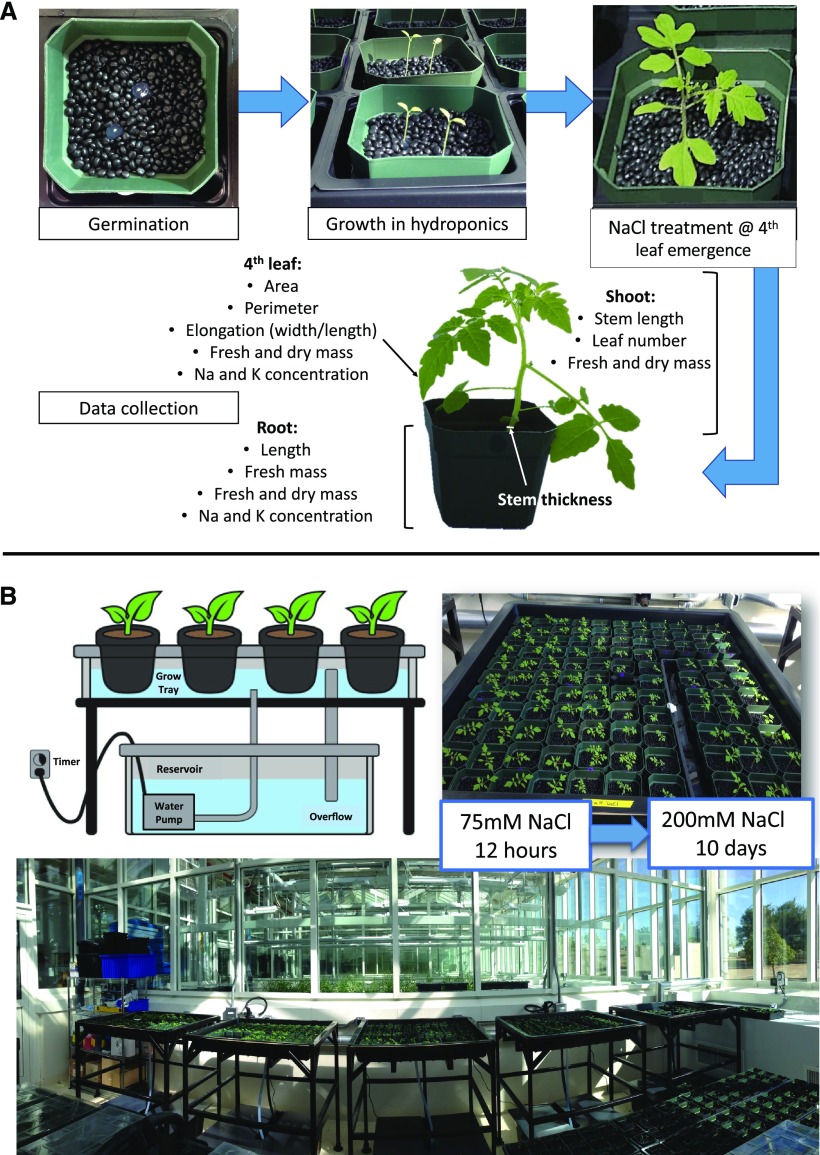
Eight-centimeter square pots were filled with plastic pellets as a substrate to support the roots. The pellets were chosen for their inert quality and dark color to protect the roots from light. The plastic pellets were made of 20% (w/w) talc-filled polypropylene, black, and with a density of 1.05 g/cm3 to sink in water (Edwards Industrial Repair). Treated seeds were germinated directly in the pots, on 0.8% (w/v) agar plugs (8 mm diameter, 12 mm deep) containing one-quarter strength Murashige and Skoog salts inserted in the plastic pellets (Fig. 7A). Two agar plugs, each with one seed, were placed in each pot to increase the chances of germination of at least one seed per pot. Sown pots were placed in nursery trays filled with fresh water and covered with a transparent plastic cover, then kept at 26°C. Germination took between 3 and 8 d. After germination, the two seedlings per pot were thinned to one by choosing plants with even size and healthy appearance. Treatment with 10% (v/v) bleach was repeated for those seeds that did not germinate one week after the first treatment (Darwin, 2009). Six seedlings per genotype were used as individual replicates for each control and salt treatment. Because different species with different growth habits were being compared, another six seedlings were grown, to be harvested before the salt treatment started. Thus, the effects of salinity on growth that occurred only during the time of the salt treatment could be calculated and the calculations used to correct for differences in growth that occurred prior to the salinity treatment.
Figure 7.
Description of the screening system. A, Workflow from germination to final harvest began with one seed on an agar plug and two agar plugs per individual pot filled with plastic beads. After germination and thinning to only one plant per pot, plants were moved to grow in a hydroponics system. After 10 d of salt stress, the plants were harvested to record the effects of salinity on their physiology. Recorded traits are listed. B, Supported hydroponics system for screening plants for salinity tolerance at the seedling stage (diagram adapted from Kruger and Doyle [2016]). Pots are held on top of the grow tray. A 100 L reservoir tank with nutrient or saline solution is located under the grow tray and a pump is used to feed the solution up to the grow tray. The pump is controlled by a timer, programmed for 15 min ON/OFF intervals. Solution floods the grow tray for 15 min and drains back into the reservoir tank when the pump turns off for another 15 min.
When the cotyledons had emerged fully and the radicle was long enough, the pots were transferred from the nursery into a greenhouse room with controlled conditions of T = 27°C day, 23°C night, and photoperiod = natural daylight occurring in Thuwal, Saudi Arabia, from September to November 2014, which were 12 h on average. Pots were placed on supported hydroponics systems (EconoTray, American Hydroponics, Inc.), which use an ebb-and-flow scheme for root aeration (Fig. 7B). Each system consists of a grow tray, a tray frame, a 100 L nutrient reservoir tank, and a submersible aquarium pump (ViaAqua 360, Commodity Axis). The tray frame height was modified, from 0.5 to 1.06 m, to hold the plants above the greenhouse walls and thus avoid shading. Each growth tray was able to hold up to 108 pots (8 cm2) with seedlings at the cotyledon stage and 96 pots (8 cm2) growing tomato plants up to leaf stage 7 or 8. The growth tray rests on the tray frame above the reservoir tank, which contains the nutrient solution. The solution was pumped to the grow tray to deliver nutrients to the plants, then drained back into the reservoir tank, allowing root aeration. Aquarium pumps inside the reservoir tanks were controlled by a programmed timer to be on for 15 min, pumping nutrient solution up to fill the grow tray, and off for 15 min, allowing the solution from the grow tray to drain back into the tank. The nutrient solutions were prepared using 100 L of tap water from the greenhouse and 33 mL of each nutrient stock, FloraGro, FloraMicro, and FloraBloom (General Hydroponics), as suggested by the manufacturer. The tap water was tested for calcium, chloride, potassium, sodium, ammonium, and nitrate ion content using the Multi-Ion Kit (CleanGrow Europe) before preparing the solutions. These measurements were recorded for future normalization. Nutrient depletion was monitored weekly using the Multi-Ion Kit ion-sensitive electrodes (CleanGrow Europe). The nutrient solution was changed at the start of the salt stress treatment and no further substantial nutrient depletion occurred throughout the rest of the experiment (10 d).
Salt Stress Treatment and Considerations
The use of eight units of the hydroponics system allowed the salt treatment of a total of 69 accessions of three different tomato species (S. cheesmaniae, S. galapagense, and S. lycopersicum), with six biological replicates per accession per treatment, plus six seedlings per accession that were harvested as a baseline before treatment. The plants were subjected to salt stress treatment at the same developmental stage, when the fourth leaf started to emerge. Given that S. lycopersicum plants were bigger than Galapagos tomatoes throughout all developmental stages (as leaves were larger and stems were thicker), they were treated when the third leaf started to emerge, to compensate for the size difference (Supplemental Fig. S1), since bigger plants are often better able to tolerate salt stress than smaller plants.
The salt stress treatment was administered gradually to the plants. As each plant reached the desired developmental stage (i.e. emergence of leaf 3 or 4), it was moved from the nutrient-only hydroponics systems to a system supplemented with NaCl. Plants were first moved to a 75 mm NaCl hydroponic system and 12 h later to a 200 mm NaCl hydroponic system. Seedlings remained in the 200 mm NaCl hydroponic system for 10 d.
Supplemental CaCl2 was added to the solutions to compensate for the decrease in Ca2+ activity arising from the addition of NaCl (Tester and Davenport, 2003). The amount of CaCl2 added to the NaCl solutions was calculated using GEOCHEM-EZ software (Shaff et al., 2010) to maintain Ca2+ activity at 0.4 mm, which was the normal Ca2+ activity in the nutrient solution prior to NaCl addition (Supplemental Table S2).
Sample Collection and Recording Traits Related to Salinity Tolerance
The principles of the phenotyping were as described by Negrão et al. (2017). Plants were photographed and tissues harvested to measure traits related to plant growth, leaf area, and ion allocation (Fig. 7A). Photographs of each plant were taken at the start and end of the salt treatment, using a Photosimile 200 light-box and a Nikon D5100 digital single-lens reflex camera. The in-camera white balance calibration function was used with a reference photo of the empty light box white background, taken at the intended light intensity. This ensured a consistent and accurate color capture in all photographs taken. The images were used to test a nondestructive approach to estimate the salinity tolerance of Galapagos tomato seedlings. The photographs were processed using a Matlab script for green pixel count (green_finder_V2), which can be found in the Supplemental Data. All raw images have been uploaded to Figshare and can be accessed via the following link: https://doi.org/10.6084/m9.figshare.c.4658156.v1. For destructive sample harvesting before and after salt treatment, the plant was carefully extracted from the pot, its roots rinsed in 10 mm MgCl2 solution, and the excess solution dried off using tissue paper. Tissues from plant roots, shoots, and leaves 3 or 4 were each weighted separately. Leaves 3 and 4, taken from S. lycopersicum and Galapagos accessions, respectively, were further characterized as they had developed under salt stress conditions. Each leaf was scanned using an EPSON scanner, and leaf area, perimeter, height, and width were calculated from the images using WinFolia software (Regent Instruments). In terms of physical measurements, stem thickness was measured at the base using a caliper, while the stem and root length were measured using a ruler. The different tissues were then stored in paper envelopes and dried at 60°C for 3 d to then measure their dry mass. Dry leaf samples (without petiole) and root samples were digested in 50 mL Falcon tubes with 5 mL of 1% (v/v) nitric acid in a HotBlock (Environmental Express) at 80°C for 4 h. Sodium and potassium contents were measured in leaf and root samples using a flame photometer (model 420, Sherwood Scientific Ltd.). All the traits recorded and the methods used to calculate or measure each trait are listed in Supplemental Table S3.
Data Analysis
A multivariate analysis was performed to assess the effect of the salt-induced changes in the plants under salt stress conditions. The mean of six replicates was calculated for all the measured traits, with the exception of leaf number, for which the mode was calculated instead.
All traits were corrected by subtracting the initial measurement (before salt stress treatment) to measure only the differences that occurred during treatment (Supplemental Fig. S6), except for traits measured in leaves 3 or 4. To facilitate comparison of different species and accessions with large differences in early growth (Supplemental Fig. S7), relative traits calculated as below were used for analysis (Negrão et al., 2017).
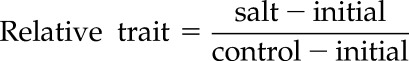 |
To determine the salt stress effects on the different accessions, correlations between all of the traits measured were analyzed using the cor and corrplot functions from the corrplot package (Wei and Simko, 2016) in R (R Core Team, 2017). A total of 11 representative traits were selected for further analysis.
The variability in traits related to salinity tolerance was described using a PCA on a matrix of 11 traits × 64 Galapagos and commercial tomato accessions. Note that of the 67 accessions screened, five had no survivors of the salt stress treatment: LA0526, LA0930, LA1141, LA1411, and LA1815. Since the variables have different units, they were scaled to have a variance of 1 and a mean of 0, by subtracting the mean and dividing by the sd, using the scale function in R (R Core Team, 2017). PCA analysis was carried out using the PCA function from the FactoMine R package (Lê et al., 2008) in R (R Core Team, 2017).
The two Galapagos tomato species were observed to have distinctly different morphologies, so the phenotypic data for each species were analyzed separately. To reduce the data complexity, two clustering approaches were used to group the accessions that showed similar phenotypes. The two clustering methods employ different statistics, enriching data analysis and allowing a cross check of results. The K-means clustering method (MacQueen, 1967) was used to reveal groups within the data. The clustering method was run with different numbers of clusters (2–6) and K = 2 was selected by calculating and plotting the within-groups sum of squares. Two clusters also provided the most interpretable output, in terms of accession clustering by traits. The accessions were grouped based on 11 traits related to salinity tolerance: Na and K concentration in root and leaf, leaf area, leaf elongation (width-to-length ratio), leaf succulence (leaf water content-to-leaf area ratio), leaf number, stem and root length, and total fresh mass. K-means were calculated using the stats package in R (R Core Team, 2017), using the standard kmeans function, with K = 2 as the number of centers, 10 maximum iterations and 20 as the number of starting partitions.
To identify possible tolerance mechanisms, all trait measurements from the accessions of each species were compared using a heat map, drawn by the function heatplot(x, scale = “none”, dualScale = FALSE, method = “ward.D2”) of the made4 R package (Culhane et al., 2005). The dendrograms of the traits and accessions were drawn by the same function, using a correlation similarity metric and average-linkage hierarchical clustering (Eisen et al., 1998).
All the phenotypic data were integrated into an Isla_Tomate App, available at https://mmjulkowska.shinyapps.io/La_isla_de_tomato/. The App allows interactive exploration of the correlations between individual traits, as well as cluster analysis of the accessions based on the chosen traits. The App was developed with the shinyapp package. The code used for the App is available at https://github.com/mmjulkowska/La_isla_de_tomato, and the instructions on how to use the App can be found at https://mmjulkowska.github.io/La_isla_de_tomato/.
Accession Numbers
The accession numbers of the Galapagos tomato collection used for this study are described in detail in Pailles et al. (2017).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Leaf stage 4 in Galapagos tomatoes, and leaf stage 3 in S. lycopersicum.
Supplemental Figure S2. Pearson correlation matrix of the recorded traits in salinity relative to control conditions with statistical significance.
Supplemental Figure S3. Green pixel count.
Supplemental Figure S4. Traits plotted in descending order colored by K-means-assigned clusters.
Supplemental Figure S5. Validation of clustering by plant fresh mass.
Supplemental Figure S6. Correction of data are recommended to find significant results.
Supplemental Figure S7. Variability in growth.
Supplemental Table S1. Eigenvalues, and variance contributed by each of the PCs.
Supplemental Table S2. CaCl2 supplement for a NaCl solution, to maintain a Ca2+ activity coefficient (α) of 0.4.
Supplemental Table S3. Phenotyping methods used for measuring or calculating each trait.
Supplemental Data. Matlab script for green pixel count (green_finder_V2.m).
Acknowledgments
We thank Igor Silva (Center for Desert Agriculture, KAUST) and Derek Burgess (Procurement Department, KAUST) for assisting with logistics throughout the project and Shireen Hammoud (Center for Desert Agriculture, KAUST) for assisting with sample collection and processing. We are also grateful to Muppala Reddy, Marina Khashat, and Gomerito Sagun (KAUST greenhouse) for providing the experimental facilities and technical support. Substantial text and content editing input from Neelima Sinha (University of California Davis) is also gratefully acknowledged.
Footnotes
This work was supported by King Abdullah University of Science and Technology (KAUST).
Articles can be viewed without a subscription.
References
- Arzani A, Ashraf M (2016) Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit Rev Plant Sci 35: 146–189 [Google Scholar]
- Awlia M, Nigro A, Fajkus J, Schmoeckel SM, Negrão S, Santelia D, Trtílek M, Tester M, Julkowska MM, Panzarová K (2016) High-throughput non-destructive phenotyping of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front Plant Sci 7: 1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanca J, Cañizares J, Cordero L, Pascual L, Diez MJ, Nuez F (2012) Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS One 7: e48198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I. (2005) The role of potassium in alleviating detrimental effects of abiotic stresses in plants. J Plant Nutr Soil Sci 168: 521–530 [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26: 4636–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhou M, Newman IA, Mendham NJ, Zhang G, Shabala S (2007) Potassium and sodium relations in salinised barley tissues as a basis of differential salt tolerance. Funct Plant Biol 34: 150–162 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu J-K (2006) Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng (N Y) 27: 141–177 [DOI] [PubMed] [Google Scholar]
- Clark JS. (2016) Why species tell more about traits than traits about species: Predictive analysis. Ecology 97: 1979–1993 [DOI] [PubMed] [Google Scholar]
- Cuartero J, Fernández-Muñoz R (1998) Tomato and salinity. Sci Hortic 78: 83–125 [Google Scholar]
- Cuartero J, Romero-Aranda R, Yeo AR, Flowers TJ (2002) Variability for some physiological characters affecting salt tolerance in tomato. Acta Hortic 573: 435–441 [Google Scholar]
- Cuartero J, Yeo AR, Flowers TJ (1992) Selection of donors for salt‐tolerance in tomato using physiological traits. New Phytol 121: 63–69 [Google Scholar]
- Culhane AC, Thioulouse J, Perrière G, Higgins DG (2005) MADE4: An R package for multivariate analysis of gene expression data. Bioinformatics 21: 2789–2790 [DOI] [PubMed] [Google Scholar]
- Darwin SC. (2009) The systematics and genetics of tomatoes on the Galapagos Islands. PhD thesis. University College London, London
- Dogan M, Tipirdamaz R, Demir Y (2010) Salt resistance of tomato species grown in sand culture. Plant Soil Environ 56: 499–507 [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famiglietti JS. (2014) The global groundwater crisis. Nat Clim Chang 4: 945–948 [Google Scholar]
- Flowers TJ, Dalmond D (1992) Protein synthesis in halophytes: The influence of potassium, sodium and magnesium in vitro. Plant Soil 146: 153–161 [Google Scholar]
- Foolad MR. (2004) Recent advances in genetics of salt tolerance in tomato. Plant Cell Tissue Organ Cult 76: 101–119 [Google Scholar]
- Genc Y, McDonald GK, Tester M (2007) Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ 30: 1486–1498 [DOI] [PubMed] [Google Scholar]
- Gruber K. (2017) Agrobiodiversity: The living library. Nature 544: S8–S10 [DOI] [PubMed] [Google Scholar]
- Hajjar R, Hodgkin T (2007) The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 156: 1–13 [Google Scholar]
- Han Y, Wang W, Sun J, Ding M, Zhao R, Deng S, Wang F, Hu Y, Wang Y, Lu Y, et al. (2013) Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants. J Exp Bot 64: 4225–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci 7: 1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell WM, Beverly RB (1981) The dilution effect in plant nutrition studies. Adv Agron 34: 197–224 [Google Scholar]
- Julkowska MM, Klei K, Fokkens L, Haring MA, Schranz ME, Testerink C (2016) Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J Exp Bot 67: 2127–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger T, Doyle P (2016) Hydroponics 101: For Investors—The Basics. LinkedIn, https://www.linkedin.com/pulse/hydroponics-101-investors-basics-thean-leonard-kruger [Google Scholar]
- Lê S, Josse J, Husson F (2008) FactoMineR : An R package for multivariate analysis. J Stat Softw 25: 253–258 [Google Scholar]
- MacQueen J. (1967) Some methods for classification and analysis of multivariate observations. Proc Fifth Berkeley Symp Math Stat Probab 1: 281–297 [Google Scholar]
- Maggio A, Raimondi G, Martino A, De Pascale S (2007) Salt stress response in tomato beyond the salinity tolerance threshold. Environ Exp Bot 59: 276–282 [Google Scholar]
- Miller DM. (1987) Errors in the measurement of root pressure and exudation volume flow rate caused by damage during the transfer of unsupported roots between solutions. Plant Physiol 85: 164–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton MJL, Awlia M, Al‐Tamimi N, Saade S, Pailles Y, Negrão S, Tester M (2018) Salt stress under the scalpel—dissecting the genetics of salt tolerance. Plant J 97: 148–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. (2005) Genes and salt tolerance: Bringing them together. New Phytol 167: 645–663 [DOI] [PubMed] [Google Scholar]
- Munns R, Gilliham M (2015) Salinity tolerance of crops—what is the cost? New Phytol 208: 668–673 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Gilliham M, Flowers TJ, Colmer TD (2016) Tissue tolerance: An essential but elusive trait for salt-tolerant crops. Funct Plant Biol 43: 1103–1113 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Sirault XRR, Furbank RT, Jones HG (2010) New phenotyping methods for screening wheat and barley for beneficial responses to water deficit. J Exp Bot 61: 3499–3507 [DOI] [PubMed] [Google Scholar]
- Munns R, Termaat A (1986) Whole-plant responses to salinity. Aust J Plant Physiol 13: 143–160 [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Muñoz N, Liu A, Kan L, Li MW, Lam HM (2017) Potential uses of wild germplasms of grain legumes for crop improvement. Int J Mol Sci 18: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrão S, Schmöckel SM, Tester M (2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Piestun BG, Bernstein N (2001) Salinity-induced inhibition of leaf elongation in maize is not mediated by changes in cell wall acidification capacity. Plant Physiol 125: 1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pailles Y, Ho S, Pires IS, Tester M, Negrão S, Schmöckel SM (2017) Genetic diversity and population structure of two tomato species from the Galapagos Islands. Front Plant Sci 8: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percey WJ, Shabala L, Wu Q, Su N, Breadmore MC, Guijt RM, Bose J, Shabala S (2016) Potassium retention in leaf mesophyll as an element of salinity tissue tolerance in halophytes. Plant Physiol Biochem 109: 346–354 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017) R: A Language And Environment For Statistical Computing. R Found Stat Comput, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- Rengasamy P. (2016) Soil Salinization. Oxford Research Encyclopedia of Environmental Science, New York: https://oxfordre.com/environmentalscience/view/10.1093/acrefore/9780199389414.001.0001/acrefore-9780199389414-e-65 [Google Scholar]
- Rick CM. (1956) Genetic and systematic studies on accessions of Lycospersicon from the Galapagos Islands. Am J Bot 43: 687–696 [Google Scholar]
- Rick CM, Fobes JF (1975) Allozymes of Galapagos tomatoes: Polymorphism, geographic distribution, and affinities. Evolution 29: 443–457 [DOI] [PubMed] [Google Scholar]
- Roy SJ, Negrão S, Tester M (2014) Salt resistant crop plants. Curr Opin Biotechnol 26: 115–124 [DOI] [PubMed] [Google Scholar]
- Rush DW, Epstein E (1981) Comparative studies on the sodium, potassium, and chloride relations of a wild halophytic and a domestic salt-sensitive tomato species. Plant Physiol 68: 1308–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush DW, Epstein E (1976) Genotypic responses to salinity: Differences between salt-sensitive and salt-tolerant genotypes of the tomato. Plant Physiol 57: 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiol Plant 133: 651–669 [DOI] [PubMed] [Google Scholar]
- Shaff J, Schultz B, Craft E, Clark R, Kochian L (2010) GEOCHEM-EZ: A chemical speciation program with greater power and flexibility. Plant Soil 330: 207–214 [Google Scholar]
- Tal M, Shannon MC (1983) Salt tolerance in the wild relatives of the cultivated tomato: Responses of Lycopersicon esculentum, L. cheesmanii, L. peruvianum, Solanum pennellii and F1 hybrids to high salinity. Aust J Plant Physiol 10: 109–117 [Google Scholar]
- Taleisnik E, Grunherg K (1994) Ion balance in tomato cultivars differing in salt tolerance. I. Sodium and potassium accumulation and fluxes under moderate salinity. Physiol Plant 92: 528–534 [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y, Lommen A, de Vos CHR, Verhoeven HA, Bino RJ, Hall RD, Bovy AG (2005) A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol 139: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Zhang W, Wang C, Meng Q, Li G, Chen THH, Yang X (2017) Genetic engineering of the biosynthesis of glycinebetaine leads to alleviate salt-induced potassium efflux and enhances salt tolerance in tomato plants. Plant Sci 257: 74–83 [DOI] [PubMed] [Google Scholar]
- Wei T, Simko V (2016) corrplot: Visualization of a Correlation Matrix. https://github.com/taiyun/corrplot
- Zamani Babgohari M, Niazi A, Moghadam AA, Deihimi T, Ebrahimie E (2013) Genome-wide analysis of key salinity-tolerance transporter (HKT1;5) in wheat and wild wheat relatives (A and D genomes). In Vitro Cell Dev Biol Plant 49: 97–106 [Google Scholar]