Under simulated high light, diel growth conditions, the photosynthetic apparatus of Chlamydomonas reinhardtii exhibits an almost constitutive photoprotection capacity through non-photochemical quenching.
Abstract
The photosynthetic apparatus must be able to withstand light conditions that exceed its capacity for carbon fixation. Photosynthetic organisms developed nonphotochemical quenching (NPQ), a process that dissipates excess absorbed light energy as heat and limits the production of reactive oxygen species and cellular damage. In the green alga Chlamydomonas reinhardtii, the LHCSR pigment-binding proteins are essential for NPQ. These complexes are not constitutively present in the thylakoid membranes; however, in laboratory conditions their expression depends on prior high light exposure of cells. To investigate the role of NPQ, we measured cells grown under a day-night cycle with a high light peak at mid-day. LHCSRs are present and NPQ is active consistently throughout the day, likely due to their slow degradation in vivo. This suggests that in physiologically relevant conditions, Chlamydomonas cells are prepared to immediately activate photoprotection, as is the case in vascular plants. We further reveal that state transitions are fully functional under these conditions and that PsbS is highly expressed throughout the day, suggesting it might have a more impactful role than previously thought.
In photosynthesis, pigments absorb light energy which is then used for photochemistry. The light-driven separation of charges occurs within specialized pigment-protein complexes, PSI and PSII. The electrons are in consequence transferred from water to NADP+. Simultaneously, the proton translocation to the lumen takes place, creating an electrochemical gradient of protons across the thylakoid membrane that is used by the ATP synthase to synthesize ATP. Finally, ATP and NADPH are used to reduce CO2 and to produce carbohydrates. However, if the light absorbed is in excess with respect to the capacity of photosynthesis to fix CO2, the dwelling of the excitation on photosynthetic pigments can lead to the production of reactive oxygen species (ROS; Roach and Krieger-Liszkay, 2014). To avoid ROS formation and thus photodamage, photosynthetic organisms developed nonphotochemical quenching (NPQ, also known as energy-dependent quenching; Erickson et al., 2015; Ruban, 2016), a mechanism that depopulates the excited states within the PSII pigment array upon lumen acidification.
In the green alga Chlamydomonas reinhardtii (Chlamydomonas), the proteins Light-Harvesting Complex Stress-Related 1 (LHCSR1) and LHCSR3, which are members of the LHC family (e.g. Croce and Nicol, 2018), are essential for NPQ (Peers et al., 2009). Whereas in vascular plants, the major NPQ-inducing protein, PsbS, is constitutively present in the thylakoid membranes (Li et al., 2000), the expression of LHCSRs in Chlamydomonas is triggered primarily by high light (HL) stress (Allorent et al., 2013; Maruyama et al., 2014; Polukhina et al., 2016). PsbS is also present in Chlamydomonas, but it was described to be only transiently expressed at the onset of HL exposure (Correa-Galvis et al., 2016; Tibiletti et al., 2016; Strenkert et al., 2019) and to provide a limited contribution to NPQ.
Although it was shown that low pH induced quenching in LHCSR3, this complex was suggested to be in a partially quenched state already at pH 7.5 (Bonente et al., 2011). This might explain why they are not expressed in low light (LL), when the presence of quencher would be wasteful (Niyogi and Truong, 2013). If this is indeed the case, it is expected that these proteins are rapidly degraded when not needed, to avoid a negative impact on photosynthetic productivity when the alga is in light-limiting conditions. However, no data regarding the lifespans of the LHCSR proteins are currently available.
Another mechanism of short-term light adaptation, namely state transitions (ST), which are a phosphorylation-dependent movement of the antenna between PSII and PSI (Rochaix, 2007; Nawrocki et al., 2016), is active in Chlamydomonas acclimated to LL. STs were proposed to photoprotect the alga upon a LL-to-HL shift, before the LHCSRs are expressed (Allorent et al., 2013), providing Chlamydomonas with complementary light-response strategies.
In this work, we measured the LHCSR content and NPQ capacity upon a transition from HL to LL to determine the rate of LHCSR degradation. We observed a slow degradation of these proteins, which suggests only a limited loss of the photoprotective capacity overnight. To verify this, we grew Chlamydomonas in a 12-h day/12-h night cycle with a sinusoidal light amplitude profile and an intensity that mimicked field conditions and assessed the resulting photosynthetic and photoprotection parameters, as well as the composition of photosynthetic apparatus. We show that the LHCSRs are present—and NPQ is active—consistently throughout the day, which means that in physiologically relevant conditions, Chlamydomonas cells are prepared to activate photoprotection in seconds, as is the case in vascular plants.
RESULTS AND DISCUSSION
Chlamydomonas Exhibits NPQ Capacity and Retains LHCSR Proteins for 2 d Following HL Acclimation
We studied the loss of NPQ capacity after HL exposure on three reference wild-type strains chosen based on their large genetic diversity (Gallaher et al., 2015). Cells were acclimated to continuous HL and then shifted to LL. The decrease of maximal NPQ was measured as a function of time the cells spent in LL conditions. To obtain insight into the interplay between the two major light-dependent photosynthetic regulation processes, mutants of NPQ (npq4) and ST (stt7-9), were also measured, together with a double mutant impaired in both mechanisms (npq4/stt7-9).
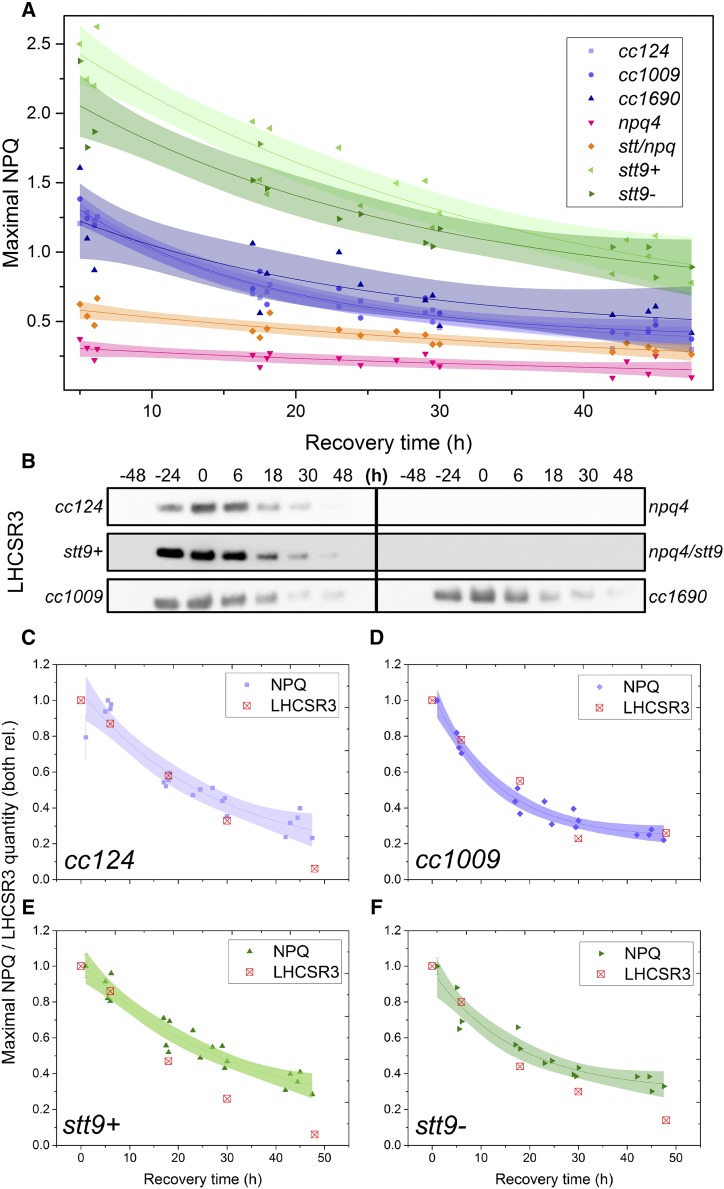
As shown in Figure 1A, the NPQ level of all the strains decreased exponentially upon LL acclimation, showing an average lifespan of 15–40 h, depending on the strain. The decrease of the LHCSR3 protein, the major molecular player in NPQ in Chlamydomonas (Peers et al., 2009), is shown in Figure 1, B–F. The cells exhibited a tight correlation between the levels of NPQ and LHCSR3, in agreement with reports from experiments under continuous light (Allorent et al., 2013; Roach and Na, 2017; Tian et al., 2019). These results confirm the substantial contribution of LHCSR3 to NPQ in Chlamydomonas cells. Although a correlation is also seen between NPQ and LHCSR1 content (Supplemental Fig. S1), the virtual absence of NPQ in the LHCSR3-lacking npq4 strain argues that the contribution of LHCSR1 can only be very limited in our conditions.
Figure 1.
NPQ capacity and LHCSR3 abundance in LL following HL acclimation. The strains were first acclimated to continuous HL (500 μmol photons m−2 s−1) for 2 d, then transferred to continuous LL (15 μmol photons m−2 s−1) at t0. A. Decrease in the maximal NPQ capacity upon recovery from HL acclimation. The points are average values of three biological replicates. The lines are monoexponential decay fits and the shaded areas represent the 95% confidence intervals of the fits. B, Immunoblot analysis with αLHCSR3 antibodies throughout the experiment. Whole-cell extracts were loaded on an equal-protein-content basis. See Supplemental Figure S1 for dynamics of other photosynthetic proteins during the treatment. C to F, Correlations between the loss of NPQ and the decrease in LHCSR3 content in cc124 (C), cc1009 (D), stt9+ (E), and stt9− (F) strains. Both the value of NPQ and the quantity of LHCSR3 were normalized to 1 at their maxima. The lines correspond to a monoexponential-decay fit of the NPQ data, and the shaded areas are 95% confidence intervals of the fits.
The long protein lifespan of the LHCSRs in vivo agrees with their proposed function as pH-dependent switches that can act in both light harvesting and photoprotection depending on the conditions (Liguori et al., 2013; Tokutsu and Minagawa, 2013; Ballottari et al., 2016; Dinc et al., 2016). The remarkably long time (as compared to natural light intensity variations) needed for the LHCSR proteins (and NPQ capacity) to decrease in LL (Fig. 1; Supplemental Fig. S1; see also Supplemental Fig. S2 for behavior of other proteins during the HL-to-LL switch) implies that in natural conditions there is a limited loss of NPQ capacity overnight. To check this hypothesis, we measured the photoprotective capacity of the cells acclimated to 12-h day/12-h night cycles.
NPQ Is Virtually Unchanged during the Day in Cells Acclimated to Day-Night Cycle
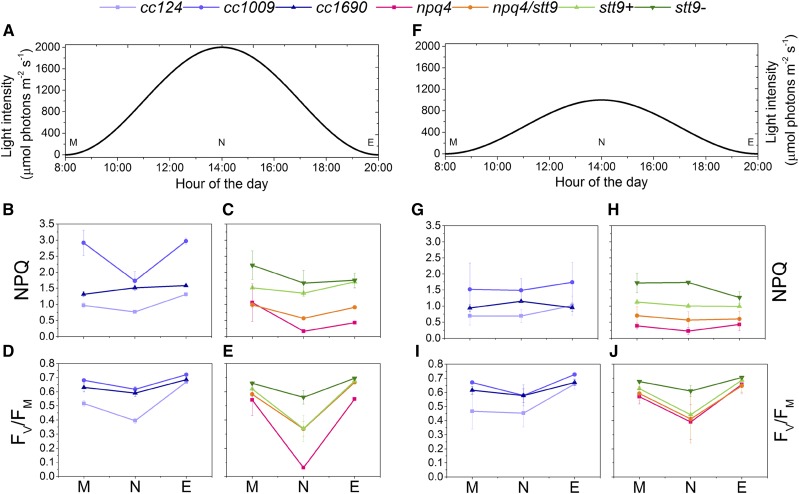
In this experiment, the light intensity was set to follow a sine function to mimic the progressive sunrise and sunset. We investigated two light regimes with maximum light intensities of 1,000 and 2,000 μmol photons m−2 s−1 (Fig. 2, A and F). The samples were assessed in the morning, at noon, and in the evening, corresponding to the beginning, peak, and end of the light period. As shown in Figure 2, B, C, G, and H, the NPQ capacity remained relatively stable throughout the day in each individual strain, whereas the differences between strains were maintained at different time points (see NPQ kinetics in Supplemental Fig. S3). Exceptions to this are cc1009 and npq4 under 2,000 μmol photons m−2 s−1, in which NPQ decreased after HL exposure at noon. As expected, the NPQ amplitude was larger when the cells were grown under the higher light intensity. Similarly, the FV/FM ratio, a parameter corresponding to the maximum efficiency of open PSII reaction centers, decreased in most of the strains at noon and recovered in the evening. FV/FM was similar in the two growth conditions, except in the npq4 strain, which suffered from photodamage to a higher extent under peak 2,000 μmol photons m−2 s−1.
Figure 2.
Photoprotection is active throughout the day-night cycle in Chlamydomonas. Uppercase letters M, N, and E denote samples measured at the onset of illumination (morning), at the peak light intensity (noon), and at the end of the light period (evening), respectively. A, Profile of the daily light intensity used for experiments in B–E and for Figures 3 and 4. B and C, Maximal NPQ value following 3 min of 1,500 μmol photons m−2 s−1 illumination in wild-type strains (B) and photoprotection mutants (C). D and E, Maximal FV/FM value upon 30 min dark adaptation of wild-type strains (D) and photoprotection mutants (E). F, Profile of the daily light intensity used for experiments in G–J. G and H, Maximal NPQ value following 3 min of 1,500 μmol photons m−2 s−1 illumination in wild-type strains (G) and photoprotection mutants (H). I and J, Maximal FV/FM value upon 30 min dark adaptation of wild-type strains (I) and photoprotection mutants (J). Values in B–E and G–J are means ± sd calculated for three biological replicates.
Pigment analysis indicated that the chlorophyll (Chl) a/b ratio slightly increased at noon in all strains, as compared to morning and evening samples (Supplemental Fig. S4). Since Chl b is only present in the outer antenna complexes of both photosystems (Tanaka and Tanaka, 2011; Blankenship, 2014), this result indicates a light-dependent reduction of the LHC proteins or, conversely, an increase in PS core synthesis. The Chl-carotenoids ratio showed the opposite trend, decreasing at noon and increasing back to the morning value during the second part of the day (Supplemental Fig. S4), in agreement with a relative increase of carotenoids under continuous HL (Niyogi et al., 1997; Bonente et al., 2012; Polukhina et al., 2016).
Whereas the npq4 mutant performed poorly in physiologically relevant growth conditions, as expected, the double npq4/stt9 mutant surprisingly appeared to perform better (higher FV/FM and higher NPQ capacity). This is in line with the observation that both stt9 strains exhibit higher NPQ than the genetically close reference strain cc124, as observed before (Bonente et al., 2011).
Taken together, these results indicate that the cellular capacity for photoprotection throughout the day is largely stable. Even though at noon some of the strains experienced photodamage, the changes in NPQ and FV/FM during the day for each strain were smaller than the differences between strains, even among the wild-type strains, indicating that the genetic background plays a primary role in light acclimation.
LHCSR Protein Content in Cells Grown in Day-Night Cycles Remains Stable throughout the Day
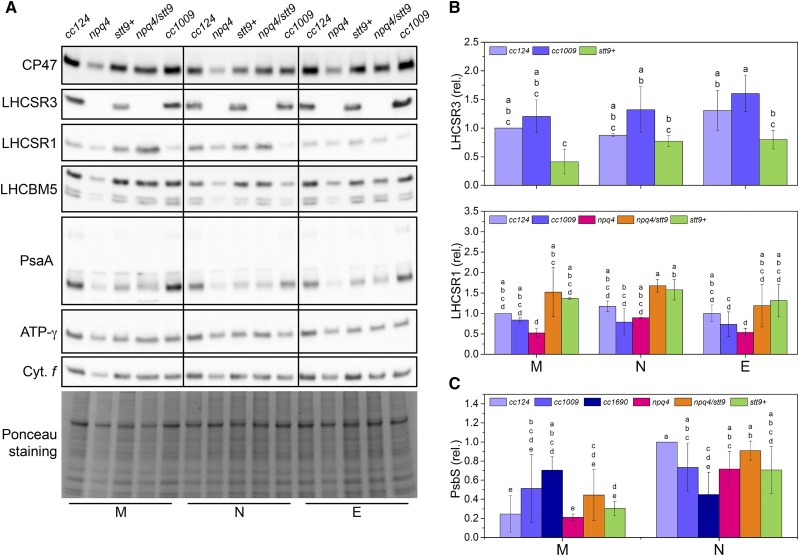
Next, we checked whether the NPQ capacity in cells grown in physiologically relevant conditions stems from the presence of the LHCSR proteins. Figure 3 shows that the amounts of both LHCSR3 and LHCSR1 did not significantly change throughout the day in any of the individual strains, in agreement with stable NPQ capacity. We hypothesize that the combination of slow degradation (Fig. 1), HL-driven expression in the afternoon, and diurnal-cycle-driven expression (Savard et al., 1996; Strenkert et al., 2019) results in a stable amount of LHCSR proteins during the day. This agrees with their steady presence in Chlamydomonas grown under a constant intensity—moderate-light 12-h day/12-h night cycle, as demonstrated recently (Strenkert et al., 2019)—and suggests that this was also the case in earlier studies (Niyogi et al., 1997).
Figure 3.
The main proteins involved in photoprotection are present in Chlamydomonas cells throughout the day. M, N, and E denote samples isolated at the onset of illumination (morning), at the peak light intensity (noon), or at the end of the light period (evening), respectively. Shared lowercase letters between means denote the absence of a statistically significant difference (P < 0.05; see “Materials and Methods” for details). A, Immunoblot analysis with antibodies against the subunits of major photosynthetic complexes assessed throughout the day-night cycle. Whole-cell extracts were loaded on an equal-protein-content basis. Ponceau staining of the membrane as a loading control is shown at the bottom. B, Quantification of the LHCSR content throughout the day-night cycle. The means ± sd were calculated for three biological replicates. C, Quantification of the PsbS content throughout the day-night cycle. The means ± sd were calculated for three biological replicates.
Finally, we verified the presence of PsbS in Chlamydomonas cells. Whereas PsbS is responsible for NPQ induction in vascular plants (Li et al., 2000), its contribution to photoprotection in Chlamydomonas is considered to be limited (Correa-Galvis et al., 2016; Tibiletti et al., 2016). PsbS was recently reported to be expressed in the early stage of HL treatment (Allorent et al., 2013; Tibiletti et al., 2016) and under UV-B treatment (Allorent et al., 2016). During the diurnal cycle, the PSBS mRNA and gene product are present only upon a dark-to-light transition (Strenkert et al., 2019). To check for its presence during a HL day-night cycle, we isolated thylakoids and probed the quantity of PsbS from the strains in the morning and at noon. As shown in Figure 3 (see Supplemental Fig. S5 for individual immunoblots), the protein was present in all the strains, and increases in protein amount correlated with an increase in light intensity, i.e. more PsbS accumulated at noon, at least in cc124, npq4, and npq4/stt9. These results suggest that the role of PsbS in Chlamydomonas in physiologically relevant growth conditions might be more prominent than previously thought.
Interestingly, whereas the amount of LHCSR3 correlated with the level of NPQ in each individual strain when cells were grown under constant light (Fig. 1), we found no clear relationship between the LHCSR content and NPQ throughout the day-night cycle (Supplemental Fig. S6). This indicates that other factors, such as PSII photoinhibition or electron transfer, are limiting factors in the development of NPQ.
State Transitions Amplitude Is Substantial in Cells Grown in a Day-Night Cycle
ST, the mechanism redistributing LHCII between PSII and PSI, is a LL-adaptation process in terms of electron flow regulation. This is because under HL, when photosynthesis is saturated at the level of PQH2 oxidation by the cytochrome b6f, moderate changes in the cross section of either PSII or PSI have no influence on the net electron flow (Sacksteder and Kramer, 2000). Nonetheless, a decrease of PSII antenna size has been shown in Chlamydomonas to be beneficial, as reduced PSII absorption reduces ROS production (Allorent et al., 2013). STs were shown to be active in LL-grown cells and after HL treatment (Allorent et al., 2013; Nawrocki et al., 2016), but no functional data are available for diurnal cycle-grown Chlamydomonas. Since STs were proposed to protect cells only at the beginning of their exposure to HL, when the LHCSRs are not yet present (Allorent et al., 2013), it might be that they are reduced or lost in Chlamydomonas adapted to physiologically relevant conditions, with rapid NPQ prevailing over the phosphorylation-dependent regulation.
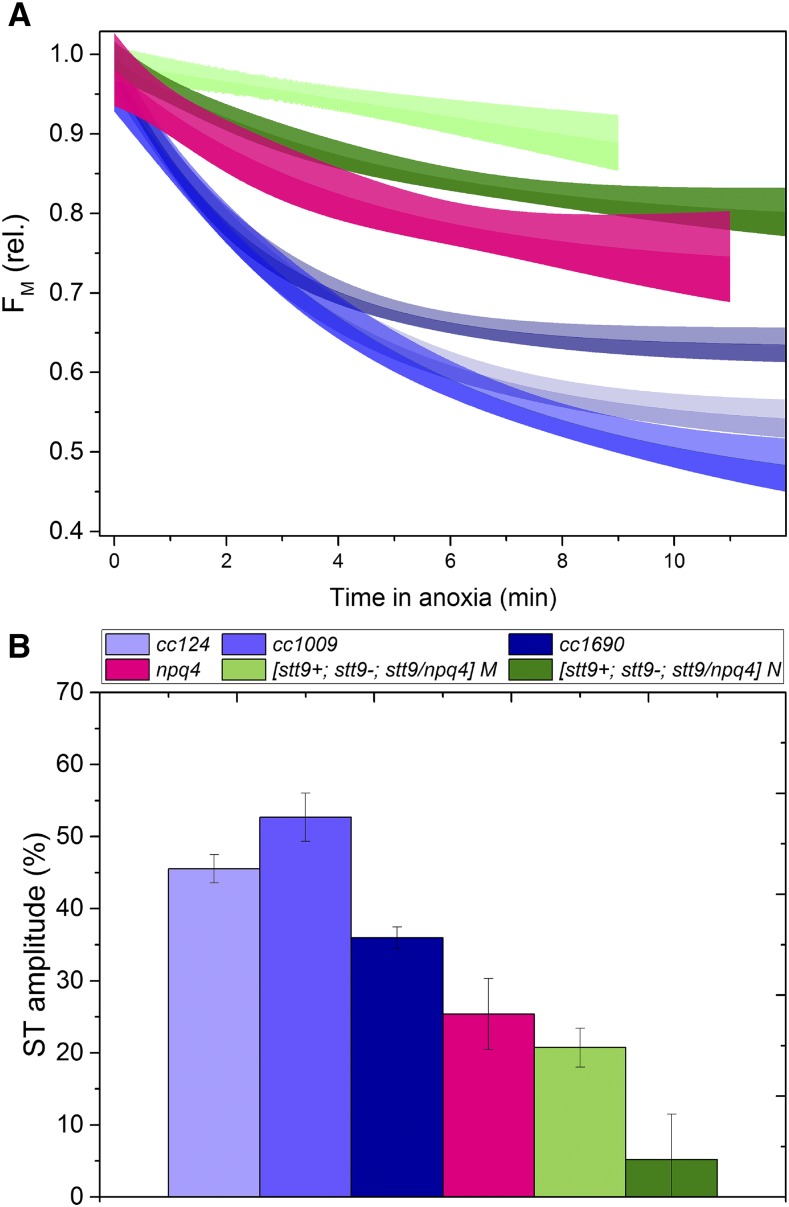
To assess this, we examined the full capacity of STs in cells acclimated to a day-night cycle by first placing them in state I (LL illumination in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea [DCMU]) and then recording the evolution of FM in darkness following the addition of Glc and Glc oxidase. These additions result in immediate establishment of an anoxic condition (Supplemental Fig. S7) and rapid reduction of the photo-quenching pool, inducing a state I-to-state II switch (Nawrocki et al., 2016). The use of anoxia instead of light treatment allows the ST to be distinguished from NPQ.
As shown in Figure 4, each of the wild-type strains exhibited a substantial ability to perform the ST, significantly higher than that for cells grown under constant LL conditions (Nawrocki et al., 2016). The extent of ST did not differ whether assessed in the morning or at noon in the three wild-type strains and the npq4 mutant, whereas in all the STT7-lacking mutants, slight differences were observed depending on the time of the day. Note that, as shown before, the stt9 allele is leaky for phosphorylation (Bergner et al., 2015), which is reflected by the nonnegligible amplitude of STs in the STT7-lacking strains. Furthermore, we observed that in npq4, STs were partially impaired, in agreement with Roach and Na (2017). One could hypothesize that the absence of LHCSR3, which was suggested to be part of the mobile antenna pool (Allorent et al., 2013; Bergner et al., 2015; Roach and Na, 2017), could reduce the amplitude of STs. However, this effect is far too small to account for the ∼25% decrease in ST amplitude in npq4 (Fig. 4), as LHCSR3 is highly substoichiometric with regard to LHCII (Semchonok et al., 2017).
Figure 4.
State transitions in cells acclimated to the day-night cycle. A, Kinetics of the transition from full state I (achieved with illumination in the presence of DCMU) to complete state II upon anoxia. The line represents a monoexponential decay fit of three independent experiments, whereas the shaded areas correspond to 95% confidence intervals of the fit. B, Amplitude of the state I to state II transition. Means ± sd from three independent experiments are shown.
Our data strengthen the view that LHCSR3 is responsible for the majority of the NPQ capacity in Chlamydomonas upon HL treatment, as well as in physiologically relevant day-night conditions, as proven by results in the npq4 background strains. Nonetheless, a small amount of photoprotection can be achieved with LHCSR1 and PsbS. LHCSRs act as triggers for rapid quenching, and the combination of their circadian rhythm-driven synthesis and slow degradation results in a continuously available defense line against HL in physiologically relevant conditions. Cells grown in these conditions also retain the ability to perform STs, which provides them with an additional mechanism to decrease the functional PSII antenna size. We also show that PsbS is highly expressed during the diurnal cycle, likely indicating that its role in Chlamydomonas is more important than previously thought. By unraveling the photoprotective strategy of Chlamydomonas, we highlight, in line with recent work (Sasso et al., 2018; Strenkert et al., 2019), that it is crucial to use conditions that mimic the natural habitat of the experimental organism to fully grasp its adaptation to the environment.
MATERIALS AND METHODS
Strains and Growth Conditions
The Chlamydomonas reinhardtii strains used in this study were cc124, cc1009, and cc1690 (Chlamydomonas Centre; https://www.chlamycollection.org), npq4 (Peers et al., 2009), stt7-9 (Bellafiore et al., 2005), and stt7-9/npq4 (Allorent et al., 2013).
For the experiments described in Figure 1, the cells were pregrown in 500-mL Erlenmeyer flasks in Tris-acetate phosphate medium (heterotrophic growth; Gorman and Levine, 1965), acclimated to high-salt medium (photoautotrophic growth; Sueoka, 1960) in LL (≤15 μmol photons m−2 s−1) for 1 d, and transferred to constant HL for 48 h. HL intensity was set at 500 μmol photons m−2 s−1, because higher intensities proved unsuitable for some of the strains. The HL source was a custom-built white light-emitting diode (LED) setup (see Supplemental Fig. S8 for the light spectrum; BeamBio) illuminating the flasks from the bottom and placed on a shaker (constant 110 rpm) inside a growth chamber (Infors) with the temperature set at 25°C.
The photoperiod (Figs. 2–4) was set to 12-h darkness/12-h light. During the latter part of the period, the light intensity followed a sine function, peaking at midday at 1,000 or 2,000 μmol photons m−2 s−1. The latter intensity, 2,000 μmol photons m−2 s−1, is a conservative estimate for an average daily peak intensity at Massachusetts coordinates where Chlamydomonas reinhardtii zygospores were initially isolated (Gallaher et al., 2015), according to the NASA MERRA-2 database (https://gmao.gsfc.nasa.gov/reanalysis/MERRA-2/). The same setup described above (BeamBio) was used for the illumination. The cells were diluted every 2 d to an optical density of ∼0.1 in order to avoid a shading-induced decrease in effective light intensity. The NPQ capacity of the cells was verified each day until a steady-state level was achieved after at least 10 day-night cycles and approximately four dilutions. Between the preculture (Tris-acetate phosphate medium, ≤15 μmol photons m−2 s−1) and the final diurnal growth conditions, the cells were transferred for 2 d to continuous 500 μmol photons m−2 s−1 light in high salt medium to allow gradual acclimation to HL conditions.
Biochemistry
Total cell extracts were prepared according to previously published methods (Ramundo et al., 2013) in the presence of protease inhibitor (Roche). Total extracts were quantified using a mix of copper (II) sulfate solution and bicinchoninic acid solution (Sigma-Aldrich) in a 1:50 ratio, together with a standard curve referring to bovine serum albumin control. Thylakoid membranes were prepared according to the method of Chua and Bennoun (1975). The membrane proteins were quantified by 80% (v/v) acetone, and the spectra were fitted as in Croce et al. (2002) using the Chl extinction coefficients reported in Porra et al. (1989).
Total extract or thylakoid membrane proteins were separated using Bolt 4% to 12% (w/w) Bis-Tris Plus gels (Invitrogen, catalog no. NW04120BOX). Five micrograms total extracts and 1 μg thylakoids of each sample were loaded.
Immunoblots were performed according to Dinc et al. (2014). Primary antibodies were diluted as follows: PsaA (1:5,000); CP47 (1:1,000); LHCSR1 (1:1,000); LHCSR3 (1:4,000); LHCBM5 (1:5,000), which reacts with the three major LHCBMs (e.g. Nama et al., 2019; https://www.agrisera.com/en/artiklar/lhcbm5-.html); ATPC (1:2,000); and Cyt f (1:3,000). All antibodies were from Agrisera (Sweden). PsbS antibody was a gift from Stefano Caffarri (Tibiletti et al., 2016) and was diluted to 1 μg/mL prior to use. The image was recorded using the Image Quant LAS 4000 system. Densitometry analysis was performed using ImageJ software (https://imagej.nih.gov/ij/), in which the rectangular tool was used to select the bands. For PsbS quantification, the signals from both the monomeric and dimeric bands (the latter persisting even in denaturing conditions) were summed during the analysis.
Statistical Analysis
Statistical analysis of the protein amounts was performed using DPS (Data Processing System) software, version 9.50 (http://www.dpsw.cn/dps_eng/index.html; Sinyosoft; Tang and Zhang, 2013; Sun et al., 2018), with two-way ANOVA test using Fisher's lsd to calculate significance (P < 0.05). Different lowercase letters from the lsd test (P < 0.05) are used to describe multiple comparisons. Letters starting from “a” indicate the largest mean in the group of data, while “b” indicates a statistically significant group with a smaller mean compared to “a”. Sharing of at least one letter between two datasets indicates the absence of a statistically significant difference (P < 0.05) between them.
Pigment Quantification
The pigments were extracted with 80% (v/v) acetone and analyzed as described before (Croce et al., 2002). The values of Chl a/b and Chl/carotenoids ratios were obtained from multiple technical replicates of the two growth conditions (1,000 and 2,000 μmol photons m−2 s−1), as no differences were observed between the latter.
Functional Measurements
Fluorescence kinetics were recorded in diluted cultures with the JTS-10 spectrophotometer (BioLogic) equipped with a custom-made cuvette holder with homogenous, sideways actinic LED illumination (BeamBio). The LEDs peak at 630 nm. All strains were dark-adapted while being vigorously shaken for at least 20 min prior to illumination to ensure that the FV/FM [(FM − F0)/FM] parameter was measured correctly. NPQ was calculated as (FM − FM′)/FM′.
ST capacity was assessed as described in Nawrocki et al. (2016); in brief, the cells treated with DCMU were illuminated to bring them to full state I, after which Glc (10 mm) and Glc oxidase (50 U/mL) were added to induce anoxia (Supplemental Fig. S4, time 0). Every ∼60 s, a short saturating light pulse was used to close PSII and the FM′ was recorded. This kinetics of full state I-to-full state II transition was then fitted with a monoexponential function (Fig. 4A), and the amplitude of the fit is shown in Figure 4B.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: XP_001697526.1 (LHCBM5); AAO63768.1 (Stt7); XP_001696138.1 (LHCSR3); XP_001696125.1 (LHCSR1); XP_001689923.1 (PSBS); XP_001696335.1 (ATPC chloroplast ATP synthase gamma chain); CAA40911.1 (CYT F chloroplast); AAA84154.1 (CP47); and ACJ50108.1 (PSAA).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Correlations between the loss of NPQ and the decrease in LHCSR1 content in cc124, cc1009, stt9+, and stt9− strains.
Supplemental Figure S2. Immunoblots—and their corresponding Ponceau-stained membranes—of proteins from major photosynthetic complexes in studied strains upon HL acclimation and shift to LL.
Supplemental Figure S3. Averaged NPQ traces from cells grown in peak 2,000 μmol photons m−2 s−1.
Supplemental Figure S4. Variations of Chl a/b and the Chl-carotenoid ratios throughout the day.
Supplemental Figure S5. Immunoblots of PsbS on isolated thylakoids and their corresponding Ponceau stains.
Supplemental Figure S6. Changes in NPQ level throughout the day do not correlate with the amounts of LHCSR1 and LHCSR3 in the cells.
Supplemental Figure S7. Kinetics of oxygen concentration changes upon Glc/Glc oxidase treatment.
Supplemental Figure S8. Spectrum of the LEDs used to grow the cells in all conditions (HL and LL acclimation, day-night cycles).
Acknowledgments
We thank Dr. Giovanni Finazzi for the npq4/stt9 double mutant. PsbS antibody was a kind gift from Prof. Stefano Caffarri. The authors declare no conflict of interest.
Footnotes
This work was supported by the European Commission (Marie Curie Actions Individual Fellowship 799083 to W.J.N.; consolidator grant 281341 to R.C.), the China Scholarship Council (201606910042 to X.L.), and the Netherlands Organization for Scientific Research (NWO; Vici grant 86510013 to R.C.).
Articles can be viewed without a subscription.
References
- Allorent G, Lefebvre-Legendre L, Chappuis R, Kuntz M, Truong TB, Niyogi KK, Ulm R, Goldschmidt-Clermont M (2016) UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 113: 14864–14869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allorent G, Tokutsu R, Roach T, Peers G, Cardol P, Girard-Bascou J, Seigneurin-Berny D, Petroutsos D, Kuntz M, Breyton C, et al. (2013) A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballottari M, Truong TB, De Re E, Erickson E, Stella GR, Fleming GR, Bassi R, Niyogi KK (2016) Identification of pH-sensing sites in the light harvesting complex stress-related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii. J Biol Chem 291: 7334–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix J-D (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bergner SV, Scholz M, Trompelt K, Barth J, Gäbelein P, Steinbeck J, Xue H, Clowez S, Fucile G, Goldschmidt-Clermont M, Fufezan C, Hippler M (2015) STATE TRANSITION7-dependent phosphorylation is modulated by changing environmental conditions, and its absence triggers remodeling of photosynthetic protein complexes. Plant Physiol 168: 615–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship RE. (2014) Molecular mechanisms of photosynthesis. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Bonente G, Ballottari M, Truong TB, Morosinotto T, Ahn TK, Fleming GR, Niyogi KK, Bassi R (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol 9: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Pippa S, Castellano S, Bassi R, Ballottari M (2012) Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J Biol Chem 287: 5833–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua N-H, Bennoun P (1975) Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: Wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA 72: 2175–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Galvis V, Redekop P, Guan K, Griess A, Truong TB, Wakao S, Niyogi KK, Jahns P (2016) Photosystem II subunit PsbS is involved in the induction of LHCSR protein-dependent energy dissipation in Chlamydomonas reinhardtii. J Biol Chem 291: 17478–17487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce R, Canino G, Ros F, Bassi R (2002) Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41: 7334–7343 [DOI] [PubMed] [Google Scholar]
- Croce R, Nicol L (2018) Light harvesting in higher plants and green algae In Croce R, van Grondelle R, van Amerongen H, and van Stokkum I, eds, Light Harvesting in Photosynthesis. CRC Press, Boca Raton, FL, pp 73–90 [Google Scholar]
- Dinc E, Ramundo S, Croce R, Rochaix JD (2014) Repressible chloroplast gene expression in Chlamydomonas: A new tool for the study of the photosynthetic apparatus. Biochim Biophys Acta 1837: 1548–1552 [DOI] [PubMed] [Google Scholar]
- Dinc E, Tian L, Roy LM, Roth R, Goodenough U, Croce R (2016) LHCSR1 induces a fast and reversible pH-dependent fluorescence quenching in LHCII in Chlamydomonas reinhardtii cells. Proc Natl Acad Sci USA 113: 7673–7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson E, Wakao S, Niyogi KK (2015) Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J 82: 449–465 [DOI] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Glaesener AG, Pellegrini M, Merchant SS (2015) Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27: 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP (1965) Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA 54: 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-P, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Liguori N, Roy LM, Opacic M, Durand G, Croce R (2013) Regulation of light harvesting in the green alga Chlamydomonas reinhardtii: The C-terminus of LHCSR is the knob of a dimmer switch. J Am Chem Soc 135: 18339–18342 [DOI] [PubMed] [Google Scholar]
- Maruyama S, Tokutsu R, Minagawa J (2014) Transcriptional regulation of the stress-responsive light harvesting complex genes in Chlamydomonas reinhardtii. Plant Cell Physiol 55: 1304–1310 [DOI] [PubMed] [Google Scholar]
- Nama S, Madireddi SK, Yadav RM, Subramanyam R (2019) Non-photochemical quenching-dependent acclimation and thylakoid organization of Chlamydomonas reinhardtii to high light stress. Photosynth Res 139: 387–400 [DOI] [PubMed] [Google Scholar]
- Nawrocki WJ, Santabarbara S, Mosebach L, Wollman FA, Rappaport F (2016) State transitions redistribute rather than dissipate energy between the two photosystems in Chlamydomonas. Nat Plants 2: 16031. [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Björkman O, Grossman AR (1997) The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA 94: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Truong TB (2013) Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr Opin Plant Biol 16: 307–314 [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Polukhina I, Fristedt R, Dinc E, Cardol P, Croce R (2016) Carbon supply and photoacclimation cross talk in the green alga Chlamydomonas reinhardtii. Plant Physiol 172: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R, Thompson W, Kriedemann P (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix J-D (2013) Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas. Plant Cell 25: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach T, Krieger-Liszkay A (2014) Regulation of photosynthetic electron transport and photoinhibition. Curr Protein Pept Sci 15: 351–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach T, Na CS (2017) LHCSR3 affects de-coupling and re-coupling of LHCII to PSII during state transitions in Chlamydomonas reinhardtii. Sci Rep 7: 43145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J-D. (2007) Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Lett 581: 2768–2775 [DOI] [PubMed] [Google Scholar]
- Ruban AV. (2016) Nonphotochemical chlorophyll fluorescence quenching: Mechanism and effectiveness in protecting plants from photodamage. Plant Physiol 170: 1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder CA, Kramer DM (2000) Dark-interval relaxation kinetics (DIRK) of absorbance changes as a quantitative probe of steady-state electron transfer. Photosynth Res 66: 145–158 [DOI] [PubMed] [Google Scholar]
- Sasso S, Stibor H, Mittag M, Grossman AR (2018) From molecular manipulation of domesticated Chlamydomonas reinhardtii to survival in nature. elife 7: e39233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard F, Richard C, Guertin M (1996) The Chlamydomonas reinhardtii LI818 gene represents a distant relative of the cabI/II genes that is regulated during the cell cycle and in response to illumination. Plant Mol Biol 32: 461–473 [DOI] [PubMed] [Google Scholar]
- Semchonok DA, Sathish Yadav KN, Xu P, Drop B, Croce R, Boekema EJ (2017) Interaction between the photoprotective protein LHCSR3 and C2S2 photosystem II supercomplex in Chlamydomonas reinhardtii. Biochim Biophys Acta 1858: 379–385 [DOI] [PubMed] [Google Scholar]
- Strenkert D, Schmollinger S, Gallaher SD, Salomé PA, Purvine SO, Nicora CD, Mettler-Altmann T, Soubeyrand E, Weber APM, Lipton MS, et al. (2019) Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc Natl Acad Sci USA 116: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci USA 46: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Yang L, Zhang L, Han M (2018) An investigation of Panax ginseng Meyer growth promotion and the biocontrol potential of antagonistic bacteria against ginseng black spot. J Ginseng Res 42: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A (2011) Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim Biophys Acta 1807: 968–976 [DOI] [PubMed] [Google Scholar]
- Tang QY, Zhang CX (2013) Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci 20: 254–260 [DOI] [PubMed] [Google Scholar]
- Tian L, Nawrocki WJ, Liu X, Polukhina I, van Stokkum IHM, Croce R (2019) pH dependence, kinetics and light-harvesting regulation of nonphotochemical quenching in Chlamydomonas. Proc Natl Acad Sci USA 116: 8320–8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibiletti T, Auroy P, Peltier G, Caffarri S (2016) Chlamydomonas reinhardtii PsbS protein is functional and accumulates rapidly and transiently under high light. Plant Physiol 171: 2717–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Minagawa J (2013) Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110: 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]