Abstract
Purpose:
We present a method for generating a T2 MR-based probabilistic model of tumor occurrence in the prostate to guide the selection of anatomical sites for targeted biopsies and serve as a diagnostic tool to aid radiological evaluation of prostate cancer.
Materials and methods:
In our study, the prostate and any radiological findings within were segmented retrospectively on 3D T2-weighted MR images of 266 subjects who underwent radical prostatectomy. Subsequent histopathological analysis determined both the ground truth and the Gleason grade of the tumors. A randomly chosen subset of 19 subjects was used to generate a multisubject-derived prostate template. Subsequently, a cascading registration algorithm involving both affine and non-rigid B-spline transforms was used to register the prostate of every subject to the template. Corresponding transformation of radiological findings yielded a population-based probabilistic model of tumor occurrence. The quality of our probabilistic model building approach was statistically evaluated by measuring the proportion of correct placements of tumors in the prostate template, i.e., the number of tumors that maintained their anatomical location within the prostate after their transformation into the prostate template space.
Results:
Probabilistic model built with tumors deemed clinically significant demonstrated a heterogeneous distribution of tumors, with higher likelihood of tumor occurrence at the mid-gland anterior transition zone and the base-to-mid-gland posterior peripheral zones. Of 250 MR lesions analyzed, 248 maintained their original anatomical location with respect to the prostate zones after transformation to the prostate.
Conclusion:
We present a robust method for generating a probabilistic model of tumor occurrence in the prostate that could aid clinical decision making, such as selection of anatomical sites for MR-guided prostate biopsies.
Keywords: Prostate cancer, Multi-parametric MRI, Tumor occurrence probability map, Prostate registration
Prostate cancer (PCa) is one of the most common malignancies in men [1] with an estimated 2016 incidence of 180,890 cases in the United States [2]. PCa detection primarily rests on measurements of prostate-specific antigen (PSA) levels in patients or digital rectal exams (DRE) [3–5]. Although PSA and DRE have poor specificity for diagnosing prostate cancer [6–8], an abnormal PSA or DRE usually results in a systematic trans-rectal ultrasound (TRUS)-guided template biopsy typically obtaining 12 individual cores of tissue distributed around the prostate [9].
However, TRUS biopsy is imperfect, often detecting clinically insignificant PCa (Gleason 6, < 1 cm length) disease while failing to detect clinically significant PCa (Gleason ≥ 7 and > I cm length) especially in the anterior, central, and apical prostate gland [10]. Here, 3T multi-parametric MRI (mp-MRI) has been shown to significantly improve detection, grading, and staging of clinically significant PCa especially in the era of standardized interpretation and reporting criteria via Prostate Imaging and Reporting Data System version 2 (PI-RADS v2) [11, 12]. 3T mp-MRI also enables targeted biopsy of individually detected lesions using in bore MR-guided [10, 13] or MR-US fusion biopsy, increasing detection of clinically significant PCa [14–16] compared to traditional 12 core TRUS template biopsy alone.
Even with such advancements, up to 50% of clinically insignificant and 20% of clinically significant PCa are not detected on conventional mp-MRI [17]. This highlights the need for tools to further guide targeted biopsy and supplement clinical radiological evaluation of mp-MRI in detection and localization of PCa. Pathologically, PCa lesion distribution in the prostate is inhomogeneous and may actually cluster [18, 19]. Thus, spatially selective biopsies can contribute to improved tumor detection [20]. A probabilistic model of tumor occurrence in the prostate may potentially guide both imaging and imageguided targeted biopsy of PCa. Additionally, correlation between tumor location and tumor characteristics (genotypes, molecular profiles, etc.) could further enrich the process of radiological evaluation of clinical findings on multi-parametric MR.
Tumor probability maps in the prostate have been developed previously through analysis of histopathological slides of prostatectomy specimens [21–25] or through combining histology-based disease localization with an MR-based anatomical atlas [26]. In these studies, tumor margins were localized in histology (with sparse coverage of the prostate as a whole) and then transposed into a 3D space (reconstructions or 3D MRI) for building a volumetric probabilistic model. However, establishing tumor margins in the 3D space from 2D whole-mount sections is a challenging task with limited accuracy. Even if a 3D MR space is used, visualization of internal prostate anatomy in whole-mount images is limited, making co-registration with MR a complex task that is quite susceptible to under/over-estimation-related inaccuracies.
In order to address these short-comings, we present a method for building a probability map/probabilistic model of PCa occurrence based solely on tumor localization in mp-MRI. Our methodology calls for using 3T mp-MRI to localize tumors in the prostate and establish their margins. Such an approach takes advantage of the rich spatial resolution offered by 3T mp-MRI and the full coverage of the prostate achieved in the acquired image data for establishing tumor margins directly in a 3D space. Since this approach still allows for cognitive matching with whole-mount imaging data through our an integrated matching process conducted jointly by radiology and pathology for establishing ground truth, we only rely on whole-mount histopathology data for identifying false positives, i.e., clinical findings on mp-MRI that are not subsequently confirmed through pathology. Finally, we demonstrate our methodology on a large mp-MRI dataset of 266 subjects with biopsyproven PCa, as discussed in the following sections.
Materials and methods
Dataset
This study was compliant with the Health Insurance Portability and Accountability Act (HIPAA) of 1996 and was performed under waiver from the institutional review board (IRB). The study cohort was composed of 266 men who underwent mp-MRI within 90 days of robot-assisted radical prostatectomy (RALP), all with TRUS biopsy confirmed PCa. mp-MRI was performed using an endorectal coil and an external phase-array coil on a 3T magnet (Trio, Verio or Skyra, Siemens Healthcare). All prostatectomy specimens in the study cohort were sectioned at 5-mm intervals in the axial plane to match the 3T mp-MRI sections as closely as possible. Of the 266 subjects, 40 were excluded since no lesions were detected on 3T mp-MRI. Of the remaining 226 subjects, a total of 247 lesions (200 were clinically significant, i.e., Gleason 3 + 3 > 1.5 cm or ≥ Gleason 3 + 4 on histopathology) were detected on both 3T mp-MRI and whole-mount histopathology. The index lesion was not visible on 3T mp-MRI scans of 24 subjects while 70 subjects had at least 1 false-positive 3T mp-MRI finding (83 false positives in total) that could not be confirmed by whole-mount histopathology. The distribution of the clinically significant tumors was as follows: Gleason 3 + 3: 28 lesions, Gleason 3 + 4: 104 lesions, Gleason 4 + 3: 48 lesions, and ≥ Gleason 8: 20 lesions.
The mp-MRI protocol included the following acquisition techniques—(1) high-resolution 3D axial TSE T2, (2) axial and coronal T2 TSE (lower through-plane resolution, i.e., 3-mm slice thickness with no gap), (3) echoplanar diffusion-weighted imaging, and (4) dynamic view-sharing time-resolved angiography with stochastic trajectories gradient-echo T1-weighted sequence [performed over 6 min (4.75 s per acquisition) with a 15-s injection delay (TWIST, Siemens Healthcare)]. For the primary objectives of this study, only the high-resolution 3D T2 acquisition was used; this was acquired as small-FOV 3D axial T2-weighted sequence using spatial and chemical-shift encoded excitation (Siemens SPACE™, Siemens Healthcare, Erlangen, Germany) sequence with echo time (TE) 101 ms, echo repetition time (TR) 3.8–5.04 s, and echo train length (ETL) = 13. Using this sequence, images were acquired with 14 cm field of view, 256 9 256 matrix with in-plane resolution 0.66 mm 9 0.66 mm and contiguous slices of 1.5 mm thickness.
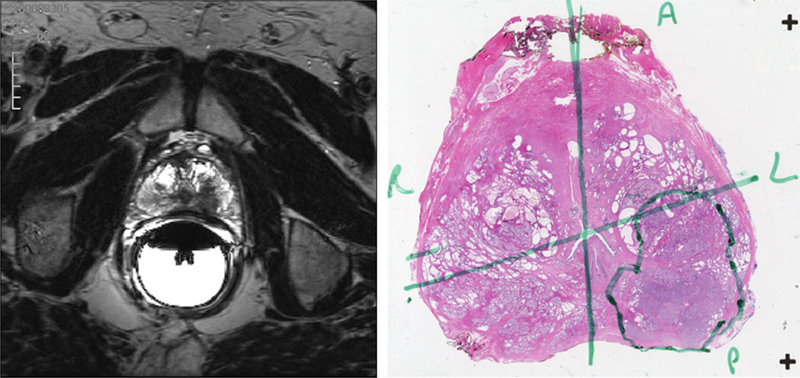
The prostate capsule was segmented using semi-automated tools in ProFuse software (Eigen, Grass Valley, California). Clinically significant findings on MR were described using a published multi-parametric assessment system [15], similar to but predating PI-RADS v1.0; these findings were segmented manually. For a small subset (19 cases), internal prostate anatomy, i.e., the peripheral zone, the transition zone, the central zone, and the anterior fibromuscular stroma, was also manually segmented to assist with the validation of our prostate registration approach, as discussed below. All segmentations were performed by an experienced radiologist (WCL 5 years) on the high-resolution 3D T2 sequence. Subsequently, MR findings were cognitively matched with the whole-mount histopathology ground truth based on cystic, cellular, and stromal features keeping the relative distance offset between MR slices consistent. The matching process was conducted by an experienced radiologist (DJAM 10 years) and pathologist (JH 15 years) in a joint review of MR findings with pathological findings to designate each lesion detected on 3T mp-MRI as true or false positive in comparison to whole-mount histopathology. Representative images of tumors on high-resolution T2 MR and images of whole-mount prostate sections are shown in Fig. 1.
Fig. 1.
One example prostate tumor from the dataset used in this study. A single axial slice of the prostate, as visualized on high-resolution 3D axial TSE (turbo spin echo) T2-weighted MR (left) and the closest matching whole-mount section from histopathology (right). The tumor was localized on the left posterior region of the prostate as seen by the low MR signal intensity areas in the otherwise high MR signal intensity peripheral zone. The MR findings were confirmed through histopathology; the tumor as localized by the pathologist is annotated in green.
With the 3D contours of both the prostate and suspicious lesions on mp-MRI, and the corresponding tumor label information derived from pathology, the probability map that models of tumor occurrence in the prostate was built using the following techniques.
Prostate MR registration
One basic component of probabilistic model building in the prostate is the registration of prostate anatomy across different subjects to the space of a common template. Using the high-resolution 3D T2-weighted images and the corresponding prostate segmentation for every subject, we proposed a registration framework which used an enriched template with the following components—(1) an intensity-based affine prostate template, (2) an intensity-based non-rigid prostate template, and (3) a binary segmentation (mask) of the non-rigid prostate template, and a cascading registration pipeline involving affine registration followed by a non-rigid B-spline registration of candidate prostate images to the enriched template.
First, the candidate prostate and its corresponding segmentation were affinely registered to the prostate template and its prostate segmentation mask. The registration process was initialized by aligning the centroids of the prostate segmentation in the candidate and template to improve the performance of the affine registration step. Multi-resolution registration over four resolutions via a smoothing image pyramid were then performed to obtain the final affine transformation, where the iteration number was fixed to a maximum of 100 iterations per resolution. A multi-metric cost function was used for the registration, which consists of the sum of the following two components:
Image intensity-based mutual information (MI), with a histogram bin size of 32, between the T2 image and the affine template image.
Sum of square difference (SSD) between the distance transform of the subject prostate segmentation and the template prostate mask.
The weights used for both MI and SSD were 1 and 0.01, respectively, which were empirically determined.
The affine registration was subsequently followed by a non-rigid B-spline registration; here, the candidate prostate was deformed to the same shape as the template. The non-rigid registration only included the prostate, other areas of the images were masked by the corresponding prostate segmentation mask. Multi-resolution registration was then performed over four resolutions via a smoothing image pyramid to register the subject to the template, where the intensity-based mutual information of the mask images, with a histogram bin size of 32, were used as cost function. A maximum of 2000 iterations was used as the stopping criteria for each resolution. The B-spline transformation model used 4-mm control point spacing.
The implementations of the affine and B-spline registration modules were taken from the ELASTIX registration toolbox [27, 28].
Prostate template model
We constructed an enriched prostate template model using 19 randomly chosen subjects from our dataset. For this purpose, we adapted a previously described approach for building average brain models [29].
The high-resolution T2-weighted images and prostate segmentation of a randomly chosen subject (from the subset of 19 subjects used to build the template) served as the affine template component for our enriched template prostate model, and as an initial non-rigid template and prostate segmentation mask. As part of the template building process, all other datasets were then affinely registered to this template. Using the parameters of affine registration as an initial transform, the non-rigid template component of our enriched template prostate model was constructed through an iterative process as follows:
Each of the 19 subjects was registered to the template through a non-linear B-spline deformation.
The average deformation field was then computed from all the deformation fields previously computed, and then applied to all 19 subject prostate images.
The non-rigid template intensities were computed as the median of the deformed image intensities.
The average deformation field was also applied to all prostate segmentation masks; the template mask was identified as the regions where at least 10 of the deformed prostate segmentations overlapped.
The final template model was generated over 5 such iterations.
Probabilistic model building
Using the multi-subject-derived prostate template model and the inter-subject prostate registration described above, each of the 240 candidate prostates from the dataset was registered to the template. Volumetric contours of each lesion (both true and false positives with respect to pathology) were then deformed into the prostate template space using the corresponding transformations computed from the registration process. The average processing time for registration, the candidate prostate and the template, and subsequent transformation of the volumetric lesions into the prostate template space was < 330 s, as measured on Windows platform, 16 MB RAM, 8 cores @ 3392 MHz). The final probabilistic model was computed by evaluating the probability of tumor occurrence at each voxel in the prostate template as the sum of all deformed tumor contours overlapping at that voxel divided by the total number of subjects used to build the probabilistic model. Using this approach, models were built for different sub-populations of tumors from the entire dataset. In this study, probabilistic models of tumor occurrence were created for the following tumor sub-populations—index tumors as determined by pathology, all tumors as confirmed by histopathology, false positives, and histopathology-confirmed tumors stratified by Gleason grade.
Validation
In order to validate the accuracy of our probabilistic model building methodology, we evaluated how well the anatomical location of the lesion within the prostate (with respect to the four zones) is maintained when its tumor margins are transformed from the candidate prostate (on which it was originally segmented) to the prostate template, i.e., does a peripheral zone lesion remain in the peripheral zone of the prostate template after registration? For this purpose, we first established anatomical zones in the prostate template as follows.
Each of the 19 subjects with prostate zone segmentations was registered to the template as previously described. The prostate zone segmentations were then transformed to the same physical space as the prostate template. While segmentations of the same zone across different subjects are expected to overlap, they are not perfectly aligned owing to inter-subject differences in anatomical shape and size. Hence, we computed the probability of a prostate template voxel belonging to a specific zone as M/N, where N = 19 and M was the number of subjects whose corresponding zone segmentations overlap at that voxel. For example, all 19 peripheral zone segmentations overlapping at a voxel in the interior of the prostate zone result in a probability of 1.0 that the voxel in question belongs to the peripheral zone. However, closer to the peripheral zone border, 10 peripheral zone segmentations and 9 transition zone segmentations may overlap at a voxel; this voxel thus has a 0.53 probability (10/19) of belonging to the peripheral zone and a 0.47 probability (9/19) of belonging to the transition zone.
Using this approach, the probability of each voxel in the prostate template belonging to the peripheral zone, the transition zone, the central zone, and the anterior stroma was computed and served as a fuzzy zonal probability map for the prostate template. Once the map was created, every lesion was transformed into the same physical space as the prostate template, and the mean zone probability of all voxels belonging to any lesion was computed for the peripheral zone, transition zone, central zone, and the anterior stroma. The highest zone probability of the lesion was used as an estimate of its anatomical location within the prostate template and compared to the anatomical location reported by the reading radiologist.
A correct placement was defined as the event where the highest computed zone probability of a lesion matched the reported anatomical zone for the lesion. A partially correct placement was defined as an instance where the highest zone probability of a lesion did not match the reported anatomical zone for the lesion, but the computed zone probability of the reported anatomical zone was non-zero. Any other event was defined as an incorrect placement. To simplify the validation, only those lesions which were reported as occurring entirely within a single prostate zone were analyzed. Additionally, only those lesions within the template mask (areas where the prostate segmentation of at least 10 candidates overlapped) were included. Using this inclusion criteria, 250 lesions visualized on MR (including index tumors, secondary tumors and false positives) were evaluated. The accuracy proportion and 95% confidence interval estimates for the number of correct lesion placements after applying our registration methodology were computed.
Given that the prostate registration approach involves deforming the prostate surface contour onto the template prostate surface, the effect of this on the internal prostate anatomy was also examined. A discrete (non-fuzzy) zonal atlas for the prostate template was also created by using the fuzzy zonal probability map and assigning each voxel to the zone with the highest zonal probability. The 19 candidate prostates were then registered to the template and the alignment between the internal anatomy of the prostate template and the internal anatomy of the candidate prostates was also evaluated using the Dice similarity coefficient [30].
All analysis was performed with software developed in-house. Registration modules were incorporated into our software from ELASTIX registration toolbox v4.4. Probabilistic model visualizations were rendered using 3D Slicer v4.4 [31]. Stata 13.0 statistical software (StataCorp LP, College Station, TX) was used to estimate accuracy proportion and exact binomial 95% confidence interval estimates.
Results
Probabilistic model visualization
Using our probabilistic model building approach outlined in previous sections, probabilistic models were built for different sub-populations of tumors. One such probability map that was generated with all tumors localized on high-resolution 3D T2 sequence and correlated with WMHP is shown in Fig. 2. A map created from only those clinically significant tumors, i.e., index tumors as determined through histo-pathologic criteria, is shown in Fig. 3. A map generated from all index tumors with Gleason grade 3 + 4 is shown in Fig. 4. Finally, a map created using false positives, i.e., clinical findings on MR that were not correlated with histopathology, is shown in Fig. 5. All probability maps are color coded to indicate varying degrees of tumor occurrence, i.e., varying number of overlapping tumor contours at different regions within the prostate template.
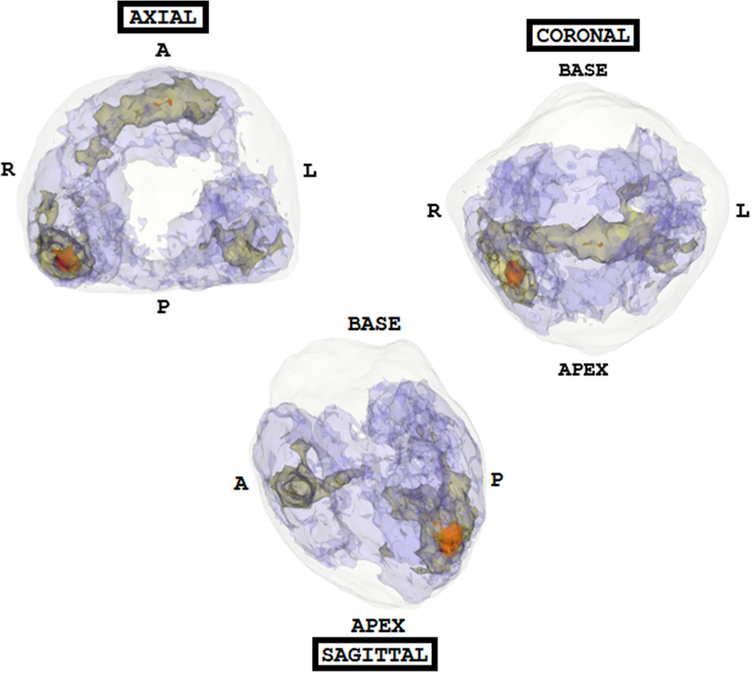
Fig. 2.
A probability map of all tumors (both index and secondary) localized on MR and correlated with whole-mount histopathology. Axial, coronal, and sagittal views allow visualization of tumor hotspots in the prostate template space. The color code is as follows—red indicates the occurrence of more than 15 lesions, yellow indicates the occurrence of 10–15 lesions, and blue indicates the presence of 5–10 lesions. An approximate prostate shell is shown in white; regions with less than 5 lesions are not shown to allow for better visualization of hotspots. The maximum number of tumors overlapping at any voxel within the probabilistic model was 21. As seen here, high-frequency tumor occurrence is noted in the posterior peripheral zone toward the apex and in the mid-anterior and peripheral zone.
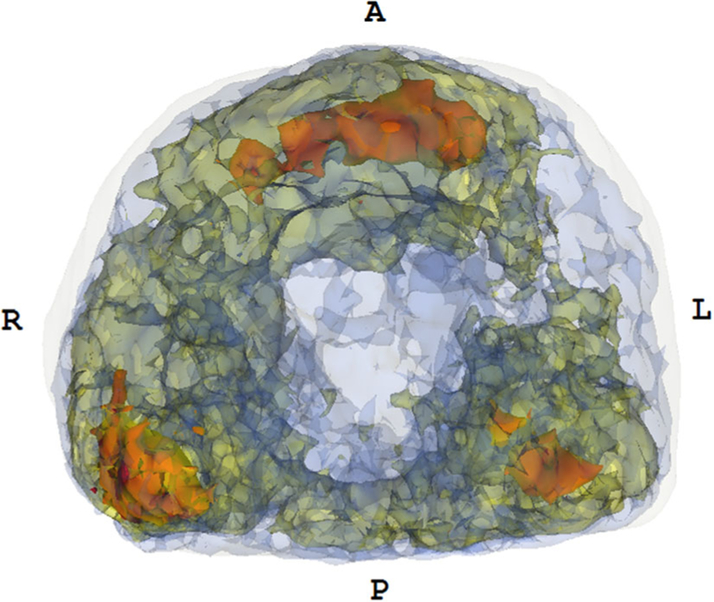
Fig. 3.
A probability map of all index tumors localized on MR (axial view). The color code is as follows—red indicates the occurrence of more than 15 lesions, yellow indicates the occurrence of 10–15 lesions, and blue indicates the presence of 5–10 lesions. An approximate prostate shell is shown in white; regions with less than 5 lesions are not shown to allow for better visualization of hotspots. The maximum number of tumors overlapping at any voxel within the probabilistic model was 19. As seen here, high-frequency tumor occurrence is noted in the posterior peripheral zone toward the apex and in the mid-anterior and peripheral zone.
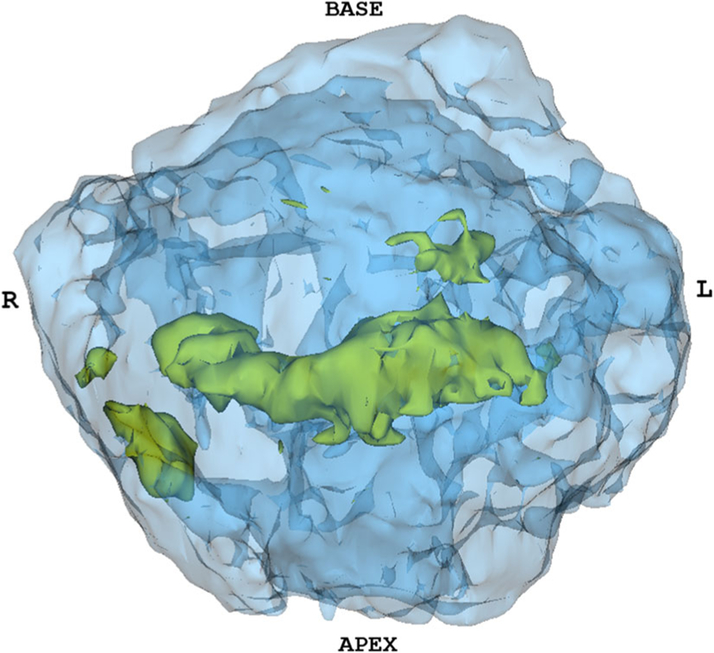
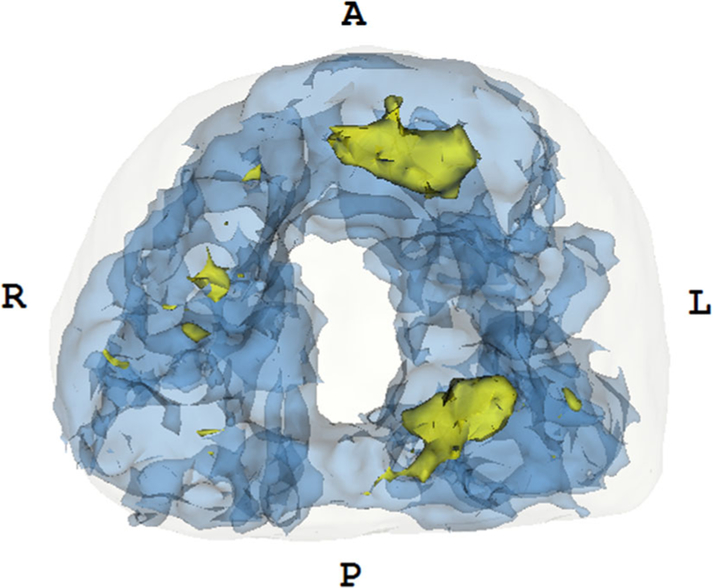
Fig. 4.
Probability map of index tumors localized on MR stratified by Gleason Grade 3 + 4. The color code is as follows—yellow indicates the occurrence of 6–10 lesions, and blue indicates the presence of 1–5 lesions. An approximate prostate shell is shown in white. As seen here, a higher frequency of tumor occurrence is noted in the mid-anterior and peripheral zone.
Fig. 5.
Probability map of all false positives localized on MR. The color code is as follows—yellow indicates the occurrence of 4–7 lesions, and blue indicates the presence of 1–4 lesions. An approximate prostate shell is shown in white.
Validation of prostate registration
As part of the validation of the registration approach used for building the tumor occurrence probability map, the anatomical location of the lesion in the prostate template was compared to its originally reported location in the candidate prostate. As seen in Table 1, our proposed registration approach to building the probability map of prostate tumor occurrence was found to have a high degree of accuracy in transforming lesions from candidate prostates to the corresponding anatomical location in the prostate template. The estimated proportion of correct/partially correct placements was 0.992 with a 95% confidence interval estimate of (0.971, 0.999).
Table 1.
Results of quantitative evaluation of the quality of prostate registration approach through evaluation of whether lesions maintain their anatomical location after being transformed to the prostate template
| Total number of lesions analyzed | 250 |
| Number of correct placements | 233 |
| Number of partially correct placements | 15 |
| Number of incorrect placements | 2 |
Since the prostate registration approach described in this study involved deforming the candidate prostate surface onto the template prostate surface, we examined the effect of such surface deformation on the internal prostate anatomy. Table 2 shows the average DICE similarity coefficient between the prostate template and the 19 candidate prostates used to create it in different regions. The largest overlap was noted in the peripheral and transition zones while the smallest overlap was noted in the central zone and the anterior stroma. Strong overlap was also noted when considering the central zone and transition zone together (central gland).
Table 2.
Average overlap between the internal anatomy of the prostate template created in this study and the 19 candidate prostates used to create the template, as measured with the Dice similarity coefficient
| Prostate region | Dice similarity coefficient |
|---|---|
| Anterior fibromuscular stroma | 0.28 ± 0.13 |
| Central zone | 0.38 ± 0.19 |
| Transition zone | 0.68 ± 0.11 |
| Central gland (central zone + transition zone) | 0.72 ± 0.08 |
| Peripheral zone | 0.76 ± 0.08 |
Discussion
Both MR Imaging and MRI-guided targeted biopsy, while significantly improving PCa detection over traditional TRUS-guided systematic biopsies, still miss an estimated 20% of clinically significant PCa [10, 15]. In this study, we presented a novel method for generating a volumetric T2-weighted MR-based probabilistic model of PCa occurrence in the prostate gland. Such probabilistic models have many potential uses including selection of anatomical sites for maximal yield of MR-guided targeted biopsy specimen collection by increasing sampling in regions of the prostate where tumor occurrence probability is high, highlighting true/false positives hot spots for aiding diagnosis where clinical findings are borderline, and correlating tumor location with clinical and tumor characteristics (genotype, molecular profile, etc.).
Previous approaches at building such probabilistic models have relied solely on images of whole-mount sections for establishing tumor margins, i.e., ground truth while using an artificial or MR 3D space. While this approach has merit, it is also vulnerable to inaccuracies from over-estimated or under-estimated volumetric tumor contours derived from 2D tumor contours on whole-mount sections that do not capture the entire extent of the lesion. Establishing correlations between MR slices and whole-mount sections is not a trivial task; internal prostate anatomy is not distinctly visualized on whole-mount sections and the prostate sectioning plane may not be exactly transversal.
While cascaded algorithms for prostate registration are not new [26, 32], previous attempts have also placed more emphasis on very precise registration. While such registration is desirable, it places an undue burden on manual steps to accurately delineate the margins of different zones or localize different anatomical landmarks for every subject prostate to be registered to the template. We note that such manual steps are time consuming and imprecise, and are not required to achieve a prostate registration that is suitable for building a probabilistic model. A simpler approach involving fewer and easier manual tasks that achieved reasonable overlap of internal prostate anatomy across different subjects may be enough to build a probabilistic model of tumor occurrence.
To this end, we present an approach to building a probabilistic model of tumor occurrence in the prostate based on volumetric contours of tumors visualized on high-resolution T2-weighted MR images of the prostate from a population of subjects. To avoid the inclusion of false positives, i.e., suspicious findings detected on MR but not corroborated by subsequent histopathological analysis, only those MR findings that are cognitively matched to tumors localized in pathology by a matching team of an experienced radiologist and pathologist were used to generate the probabilistic model, as seen in Fig. 3. In this manner, histopathological analysis still served as ground truth but the volumetric contours of the tumor were already in the 3D MR space. Additionally, only the prostate and tumors were segmented for every subject prostate included in the probabilistic model. Our results suggest that the cascaded registration approach used sufficiently aligned internal prostate anatomy to make meaningful region-based tumor probability computations.
An examination of the probabilistic models generated in our study in Figs. 2, 3, 4, and 5 confirmed previous findings that tumors are heterogeneously distributed within the prostate [18, 19]. Specifically, we observed a greater likelihood of tumor occurrence at the mid-gland anterior transition zone and the base-to-mid-gland posterior peripheral zone, which supported findings in other studies [22]. While the probabilistic model that includes all tumors is likely to be most useful for guiding selection of biopsy sites to focus on areas with higher likelihood of tumor occurrence, an examination of tumor occurrence models that focus on specific sub-populations could reveal correlations between specific tumor sub-types and regions where they are likely to occur. For instance, when stratifying tumors by Gleason Grade, we note a higher frequency of Gleason 3 + 4 tumor occurrence in the mid-level anterior transition zone. However, further analysis of the distribution of tumors stratified by Gleason grade was not possible as tumors with Gleason scores of 3 + 3, 4 + 3, and ≥ 8 were under-represented in this dataset. We intend to further investigate the correlation between tumor location and tumor profile through such probabilistic models derived from sub-populations of tumors stratified by different criteria.
Given the variability in the shape, size, and percentage volume of the peripheral zone, transition zone, central zone, and the anterior stroma, and the simplified registration approach which does not explicitly deform the zones of the candidate prostates onto the prostate template, the quality of the probabilistic model of tumor occurrence built must be rigorously evaluated. We proposed a rigorous, externally validated approach to examine how well the anatomical location of lesions with respect to the prostate zones was maintained when the lesions were transformed into the prostate template space. Given that zones are not explicitly registered across two prostates, the anatomical location of lesions with respect to the prostate zones can change during the transformation to the template space where the zone borders are soft, i.e., with fuzzy labels. However, our results suggest that the registration approach utilized in this study maintained a high degree of accuracy in retaining the anatomical location of the lesion relative to the prostate zones when transforming them to the prostate template. We also examined the effect of surfacebased registration components in our approach on the internal prostate anatomy. Our results suggest reasonable alignment between the internal prostate anatomy of different candidate prostates after registration to the prostate template, specifically in the peripheral and transition zones. This is not surprising given that these are the largest regions in the prostate and easiest to visualize and segment accurately.
We acknowledge that our reliance on MR to determine tumor margins is not without drawbacks of its own. Since not all tumors are visualized on MR, our probabilistic model is skewed in favor of tumors that are visible on MR alone. It is not clear if the inability to visualize these tumors on MR is related to the imaging technique or to tumor biology itself, and this will continue to be an area of focus moving forward. Another concern is that MR tends to under-estimate tumor volume, as shown in [33]. The impact of such tumor margin under-estimation on our probabilistic model needs to be investigated further. To this end, we aim to compare and quantify differences in probabilistic models of tumor occurrence in the prostate derived purely from MR to those obtained from transforming tumor margins localized in images of whole-mount sections to the 3D MR space in future studies. Finally, the current approach relies on manual segmentation of the prostate and tumors on MR which can be error prone and subjective in areas where margins are not well-defined. While automated prostate tumor detection and segmentation remains an active area of research [34–37], automated prostate capsule segmentation methods such as [38–40] could be incorporated in our proposed framework.
Conclusion
We presented an approach for building a probabilistic model of tumor occurrence in the prostate using a cascaded prostate registration algorithm and tumors segmented on high-resolution T2-weighted MRI. We validated our model building approach by testing whether the zonal location of the tumor was maintained when the prostate was registered to the template. We demonstrated our approach using a rich dataset of 266 subjects with correlated histo-pathology and visualized the resulting probabilistic models on a multi-subject-derived prostate template. Our results suggest that tumor distribution in the prostate is inhomogeneous; specifically, we observe a greater likelihood of tumor occurrence at the mid-gland anterior transition zone and the base-to-mid-gland posterior peripheral zone. Our longterm goal is to use such probabilistic models in helping to guide the selection of anatomical sites within the prostate when performing MR-US fusion-guided biopsies and in assisting radiological evaluation of prostate MR exams.
Acknowledgments
This work was supported by funds from the Integrated Diagnostics Program, Departments of Radiological Sciences & Pathology, David Geffen School of Medicine at University of California Los Angeles (UCLA). The authors would like to thank Alan Martin Priester from the Department of Bioengineering at UCLA for his assistance with the ProFuse software setup and usage.
Footnotes
Conflict of interest All authors declare no conflict of interest.
Ethics statement All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Since the study involved purely retrospective analysis of previously acquired data, the need for additional informed consent was waived by the Institutional Review Board.
References
- 1.Jernal A, Siegel R, Ward E, et al. (2007) Cancer statistics. CACancer J Clin 57:43–66 [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society (2016) Cancer facts & figures 2016. Atlanta: American Cancer Society [Google Scholar]
- 3.Carter HB, Albertsen PC, Barry MJ, et al. (2013) Early detection of prostate cancer: AUA guideline. J Urol 190(2):419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyer VA (2012) Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 157(2):120–134 [DOI] [PubMed] [Google Scholar]
- 5.Carroll PR, Parsons JK, Andriole G, et al. (2014) Prostate cancer early detection, version 1.2014. Featured updates to the NCCN Guidelines. J Natl Compr Canc Netw 12(9):1211–1219 [DOI] [PubMed] [Google Scholar]
- 6.Schröder FH, Carter HB, Wolters T, et al. (2008) Early detection of prostate cancer in 2007. Part 1: PSA and PSA kinetics. Eur Urol 53(3):468–477 [DOI] [PubMed] [Google Scholar]
- 7.van Leeuwen PJ, van den Bergh RC, Wolters T, et al. (2011) Critical assessment of prebiopsy parameters for predicting prostate cancer metastasis and mortality. Can J Urol 18(6):6018–6024 [PubMed] [Google Scholar]
- 8.Rajinikanth A, Manoharan M, Soloway CT, Civantos FJ, Soloway MS (2008) Trends in Gleason score: concordance between biopsy and prostatectomy over 15 years. Urology 72(1):177–182 [DOI] [PubMed] [Google Scholar]
- 9.Hodge KK, McNeal JE, Terris MK, Stamey TA (1989) Random systematic versus directed ultrasound guided transrectal core biopsies of the prostate. J Urol 142(1):71–74 [DOI] [PubMed] [Google Scholar]
- 10.Tan N, Lin WC, Khoshnoodi P, et al. (2016) In-bore 3-T MR-guided transrectal targeted prostate biopsy: prostate imaging reporting and data system version 2-based diagnostic performance for detection of prostate cancer. Radiology 152827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamie K, Sonn GA, Finley DS, et al. (2014) The role of magnetic resonance imaging in delineating clinically significant prostate cancer. Urology 83(2):369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinreb JC, Barentsz JO, Choyke PL, et al. (2016) PI-RADS prostate imaging—reporting and data system: 2015, version 2. Eur Urol 69(1):16–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felker ER, Lee-Felker SA, Feller J, et al. (2016) In-bore magnetic resonance-guided transrectal biopsy for the detection of clinically significant prostate cancer. Adbom Radiol 41(5):954–962 [DOI] [PubMed] [Google Scholar]
- 14.Sonn GA, Natarajan S, Margolis DJ, et al. (2013) Targeted biopsy in the detection of prostate cancer using an office based magnetic resonance ultrasound fusion device. J Urol 189(1):86–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonn GA, Chang E, Natarajan S, et al. (2014) Value of targeted prostate biopsy using magnetic resonance-ultrasound fusion in men with prior negative biopsy and elevated prostate-specific antigen. Eur Urol 65(4):809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto PA, Chung PH, Rastinehad AR, et al. (2011) Magnetic resonance imaging/ultrasound fusion guided prostate biopsy improves cancer detection following transrectal ultrasound biopsy and correlates with multiparametric magnetic resonance imaging. J Urol 186(4):1281–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan N, Margolis DJ, Lu DY, et al. (2015) Characteristics of detected and missed prostate cancer foci on 3-T multiparametric MRI using an endorectal coil correlated with whole-mount thin-section histopathology. Am J Roentgenol 205(1):W87–W92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen ME, Troncoso P, Tang K, Babaian RJ, Johnston D (1999) Comparison of prostate biopsy schemes by computer simulation. Urology 53(5):951–960 [DOI] [PubMed] [Google Scholar]
- 19.Eskew LA, Bare RL, McCullough DL (1997) Systematic 5 region prostate biopsy is superior to sextant method for diagnosing carcinoma of the prostate. J Urol 157(1):199–202 [PubMed] [Google Scholar]
- 20.Shen F, Shinohara K, Kumar D, et al. (2008) Three-dimensional sonography with needle-tracking. J Ultras Med 27:895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sofer A, Zeng J, Mun SK (2003) Optimal biopsy protocols for prostate cancer. Ann Oper Res 119:63–74 [Google Scholar]
- 22.Opell MB, Zeng J, Bauer JJ, et al. (2002) Investigating the distribution of prostate cancer using three-dimensional computer simulation. Prostate Cancer PD 5(3):204–208 [DOI] [PubMed] [Google Scholar]
- 23.Shen D, Lao Z, Zeng J, et al. (2004) Optimized prostate biopsy via a statistical atlas of cancer spatial distribution. Med Image Anal 8(2):139–150 [DOI] [PubMed] [Google Scholar]
- 24.Zhan Y, Shen D, Zeng J, et al. (2007) Targeted prostate biopsy using statistical image analysis. IEEE Trans Med Imaging 26(6):779–788 [DOI] [PubMed] [Google Scholar]
- 25.Narayanan R, Werahera PN, Barqawi A, et al. (2008) Adaptation of a 3D prostate cancer atlas for transrectal ultrasound guided target-specific biopsy. Phys Med Biol 53(20):N397–N406 [DOI] [PubMed] [Google Scholar]
- 26.Rusu M, Block BN, Jaffe CC, et al. (2014) Prostatome: a combined anatomical and disease based MRI atlas of the prostate. Med Phys 41(7):072301–072312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW (2010) Elastix: a toolbox for intensity based medical image registration. IEEE Trans Med Imaging 29(1):196–205 [DOI] [PubMed] [Google Scholar]
- 28.Shamonin DP, Bron EE, Lelieveldt BPF, et al. (2014) Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front Neuroinform 7(50):1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guimond A, Meunier J, Thirion JP (2000) Average brain models: a convergence study. Comput Vis Image Und 77(2):192–210 [Google Scholar]
- 30.Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302 [Google Scholar]
- 31.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. (2012) 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betrouni N, Iancu A, Puech P, Mordon S, Makni N (2012) ProstAtlas: a digital morphologic atlas of the prostate. Eur J Radiol 81(9):1969–1975 [DOI] [PubMed] [Google Scholar]
- 33.Rud E, Klotz D, Rennesund K, et al. (2014) Detection of the index tumour and tumour volume in prostate cancer using T2-weighted and diffusion-weighted magnetic resonance imaging (MRI) alone. BJU Int 114(6b):E32–E42 [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Yang W, Weinreb J, et al. (2017) Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning. Sci Rep 7(1). 10.1038/s41598-017-15720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalvati F, Wong A, Haider MA (2015) Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med Imaging 15(27). 10.1186/s12880-015-0069-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vos PC, Barentsz JO, Karssemeijer N, Huisman HJ (2012) Automatic computer-aided detection of prostate cancer based on multiparametric magnetic resonance image analysis. Phys Med Biol 57:1527–1542 [DOI] [PubMed] [Google Scholar]
- 37.Groenendaal G, Borren A, Moman MR, et al. (2012) Pathological validation of a model based on diffusion-weighted imaging and dynamic contrast-enhanced MRI for tumor delineation in the prostate peripheral zone. Int J Radiat Oncol Biol Phys 82:537–544 [DOI] [PubMed] [Google Scholar]
- 38.Turkbey N, Fotin SV, Huang RJ, et al. (2013) Fully automated prostate segmentation on MRI: comparison with manual segmentation methods and specimen volumes. Am J Roentgenol 201(5):W720–W729 [DOI] [PubMed] [Google Scholar]
- 39.Martin S, Troccaz J, Daanen V (2010) Automated segmentation of the prostate in 3D MR images using a probabilistic atlas and a spatially constrained deformable model. Med Phys 37(4):1579–1590 [DOI] [PubMed] [Google Scholar]
- 40.Reda I, Shalaby A, Khalifa F, et al. (2016) Computer-aided diagnostic tool for early detection of prostate cancer. In Proceedings of 23rd IEEE international conference on image processing (ICIP), pp 2668–2672 [Google Scholar]