Abstract
Background and Aims
Many monotypic gymnosperm lineages in south-east China paradoxically remain in relict status despite long evolutionary histories and ample opportunities for allopatric speciation, but this paradox has received little attention and has yet to be resolved. Here, we address this issue by investigating the evolutionary history of a relict conifer, Pseudotaxus chienii (Taxaceae).
Methods
DNA sequences from two chloroplast regions and 14 nuclear loci were obtained for 134 samples. The demographic history was inferred and the contribution of isolation by environment (IBE) in patterning genetic divergence was compared with that of isolation by distance (IBD).
Key Results
Three genetic clusters were identified. Approximate Bayesian computation analyses showed that the three clusters diverged in the late Pliocene (~3.68 Ma) and two admixture events were detected. Asymmetric gene flow and similar population divergence times (~ 3.74 Ma) were characterized using the isolation with migration model. Neither IBD nor IBE contributed significantly to genetic divergence, and the contribution of IBE was much smaller than that of IBD.
Conclusions
These results suggest that several monotypic relict gymnosperm lineages like P. chienii in south-east China did not remain in situ and undiversified for millions of years. On the contrary, they have been evolving and the extant populations have become established more recently, having insufficient time to speciate. Our findings provide a new perspective for understanding the formation and evolution of the relict gymnosperm flora of China as well as of the Sino-Japanese Flora.
Keywords: demography, evolutionary history, gene flow, isolation by distance, population divergence, Pseudotaxus chienii
Introduction
Geographical isolation between populations within species (allopatry) is viewed as the default means of speciation (Coyne and Orr, 2004; Harrison, 2012) and has been invoked by biogeographers and ecologists to explain the formation of biodiversity hotspots (Halffter, 1987; Qian and Ricklefs, 2000; Whittaker and Fernández-Palacios, 2007; Steinbauer et al., 2016). Given a sufficient period of strict allopatry, the accumulation of endemic variants through mutation, genetic drift and local adaptation will inevitably result in divergence and subsequently speciation (Won and Hey, 2005). However, strict allopatry is not always encountered in nature (Nosil, 2008; Zhou et al., 2010; Anacker and Strauss, 2014; Stankowski et al., 2015; He et al., 2018); species are composed of multiple populations that are dynamic in size, location, selective pressure and amount of gene exchange. Whether isolated populations speciate or not depends on the competition between unifying (gene flow) and diversifying genetic processes (genetic drift and local adaptation) (Hey and Nielsen, 2004).
China is one of the richest countries in plant diversity and one of the world's 17 mega-diversity countries (Mittermeier et al., 1997). One key reason for the high plant diversity in China is that the extreme physiographical heterogeneity, in conjunction with climate and sea-level changes, has provided abundant opportunities for evolutionary radiation through allopatric speciation (Qian and Ricklefs, 2000). However, China (particularly the central and south-east regions) is home to many relict lineages (Sakai, 1971; Ying et al., 1993; Manchester et al., 2009; Chen et al., 2018) that represent the survivors of clades with a large proportion of extinct members or the remains of a larger distributional area (Grandcolas et al., 2014). These relict lineages always contain only one or very few representatives and are narrowly or extremely disjunctly distributed compared with their larger sisters (Grandcolas et al., 2014). The most famous relicts in China include several monotypic gymnosperm genera (e.g. Ginkgo, Metasequoia, Cathaya) and many woody angiosperm genera (e.g. Davidia, Cercidiphyllum, Cyclocarya) that are monotypic or oligotypic (Manchester et al., 2009). Those evolutionary relicts are referred to as ‘living fossils' and their assembly in China over time is key to understanding the origin of the flora of China and even the evolution of Northern Hemisphere floras (Manchester et al., 2009; Chen et al., 2018). Recently, the demographic and divergence history of these relict lineages has gained renewed interest from plant phylogeographers (e.g. Qi et al., 2012; Tian et al., 2015; Kou et al., 2016). However, except for Cercidiphyllum, the range of which extends to Japan (Qi et al., 2012), clear examples of speciation events in relict tree lineages during the late Cenozoic (late Pliocene to Pleistocene) have never been demonstrated in central and south-east China and many relict tree lineages there remain monotypic. This is particularly true for gymnosperms in central and south-east China which each contain only one species (e.g. Cathaya, Ginkgo, Glyptostrobus, Metasequoia, Pseudotaxus, Pseudolarix; Ying et al., 1993). This phenomenon is perplexing given that those relict gymnosperm lineages have experienced long evolutionary histories and recent topographic changes and climatic shifts have provided ample opportunities for allopatric speciation (Qian and Ricklefs, 2000; Harrison et al., 2001; Qiu et al., 2011).
There are three possible explanations for the relict status of China's monotypic gymnosperms. First, several recent studies have demonstrated that relicts may be more dynamic than previously thought. In other words, the stem lineages are ancient, but the extant species originated or became established in China recently (Chou et al., 2011; Hohmann et al., 2018; Chen et al., 2018). The most striking case is Ginkgo biloba. This is an indisputable living fossil, but the genetic footprint of extant Ginkgo biloba populations in China dates back only 390 000 years (Hohmann et al., 2018). Thus, the relict status does not imply that they have not experienced any evolutionary changes. Instead, those lineages might have diversified recently (e.g. Cycas; Nagalingum et al., 2011) whereas other members went extinct. Alternatively, they might have broader ranges, yet China might have been colonized or re-colonized recently, while populations in other places became extinct. Under either situation, those relicts might have had insufficient time to speciate in China. Second, gene flow among populations, which has been assumed to be extremely limited due to long-term glacial and interglacial isolation (Harrison et al., 2001; Qiu et al., 2011), might be otherwise substantial, especially for plants with wind-mediated pollen and/or effective seed dispersal modes (e.g. Bai et al., 2014; Tian et al., 2015). Third, as most of those relict gymnosperms are geographically restricted, their ranges may span fewer environmental gradients and thus experience weak divergent selection (or local adaptation). This scenario might be more plausible in south-east China compared with south-west China where geological complexity could facilitate speciation (e.g. Li et al., 2013; Liu et al., 2013; reviewed by Favre et al., 2014). While one or two of the above hypotheses have been tested explicitly or implicitly (e.g. Chou et al., 2011; Bai et al., 2014), to our knowledge, no study has explicitly considered all the possibilities simultaneously.
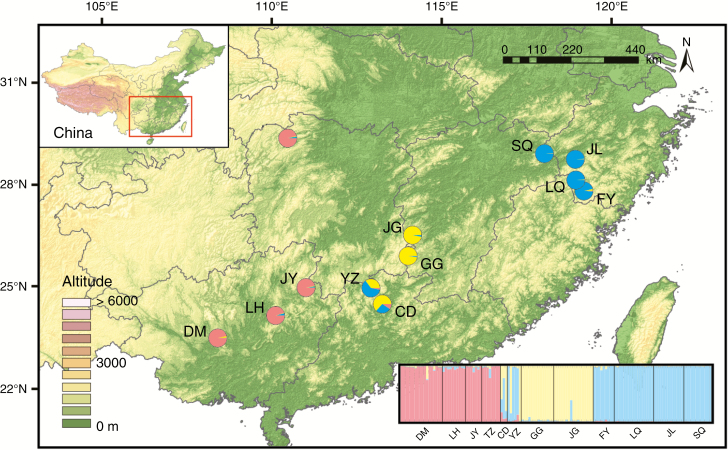
Pseudotaxus is a genus in Taxaceae established by Cheng (1947). Although two species names have been published in this genus [P. chienii (Cheng) Cheng (Cheng, 1947) and P. liana J. Silba (Silba, 1996)], it was considered to be monotypic (Fu et al., 1999; Wang and Yang, 2007) as leaf morphology variation between P. chienii and P. liana individuals is continuous (Wang and Yang, 2007). Pseudotaxus chienii is a dioecious woody shrub or small tree up to 4 m tall, with seeds partly enclosed within a fleshy white aril at maturity (Fu et al., 1999). The species has a restricted distribution, occurring in isolated montane regions of southern Zhejiang, north-eastern and south-western Jiangxi, north-western and southern Hunan, northern Guangxi, and northern Guangdong, China (Fig. 1). Plants primarily grow on scarp slopes, along the sides of valleys or on cliffs at altitudes of 800–1500 m and are adapted to cool, humid climates (Fu and Jin, 1992; Fu et al., 1999; Xu et al., 2008). Although the fossil record of Pseudotaxus is deficient (Wu et al., 2007), the genus can be considered an evolutionary relict according to two of the criteria proposed by Grandcolas et al. (2014): (1) it contains one living representative and diverged from its sister group Taxus very early (Palaeocene or early Eocene, ~54–65 Ma; Leslie et al., 2012; Lu et al., 2014); and (2) it has a much smaller distribution relative to Taxus (Fu et al., 1999). As an officially protected species in China, its conservation status has led to research efforts. The genetic diversity and genetic structure of P. chienii have been investigated using dominant molecular markers such as random amplified polymorphism DNA (RAPD) and inter-simple sequence repeats (ISSRs) (Wang et al., 2006; Su et al., 2009). Low genetic diversity and significant genetic differentiation were reported, and fragmentation and bottleneck events were postulated based on simple summary statistics. To improve conservation efforts, an in-depth understanding of the evolutionary history that accounts for the present distribution of genetic diversity is needed. In particular, the timing of population divergence and admixture, fluctuation in population size, and natural selection experienced by different populations should be addressed with more powerful molecular tools and more sophisticated statistical algorithms (Qiu et al., 2011; Liu et al., 2012).
Fig. 1.
Distribution of three clusters (W, red; C, yellow; E, blue) revealed by STRUCTURE analysis using K = 3, which was determined to be the optimal K value using the ΔK method.
In this study, we sequenced 14 nuclear loci as well as two chloroplast genomic regions from 12 P. chienii populations. Two coalescent-based analyses [isolation-with-migration (IM) and approximate Bayesian computation (ABC)], which are either likelihood-based (IM) or likelihood-free (ABC), were implemented to explicitly test the first and the second hypotheses, by determining population divergence time and historical gene flow among populations. In addition, the importance of isolation by distance (IBD) and isolation by environment (IBE) were estimated to quantify the relative roles of neutral (genetic drift and gene flow) and adaptive processes (local adaptation) on population divergence, which can provide clues to the third hypothesis. By detailing the evolutionary history of a typical relict genus (Pseudotaxus) in south-east China, we attempted to explain a puzzling paradox: Why do many monotypic gymnosperms in south-east China remain in relict status given their long evolutionary histories and ample opportunities for allopatric speciation.
MATERIALS AND METHODS
Plant material, loci sampling, PCR amplification and sequencing
We investigated 34 wild populations that were recorded in herbarium specimens, reported in the literature, listed on websites of nature reserves, and were identified by experienced botanists. Living trees can be found only in 12 populations because some populations have been removed by deforestation and some remain inaccessible. Leaves of 134 individuals were collected and three to 17 individuals per population were sampled according to population size (Fig. 1 and Supplementary Data S1, Table S1). Leaves were dried in silica gel immediately after collection and subsequently used for DNA extraction. Total genomic DNA was extracted from ~20 mg of dried leaf using a modified CTAB procedure (Doyle and Doyle, 1987).
Two chloroplast (cp) DNA regions (rpl16 and psbA-trnH) were sequenced following our previous studies (Kou et al., 2016, 2017). In addition, 14 low-copy nuclear genes (PT23, PT58, PT107, PT122, PT126, PT140, PT154, PT202, PT203, PT222, PT333, PT353, PT373 and PT399) were developed from transcriptome sequences of P. chienii in this study (Supplementary Data Tables S2 and S3). These loci were classified into various functional categories against four protein databases (Nr, GO, KO and Swiss-Prot), with the exception of PT58 and PT222, which had no known function (Table S4). Transcriptome sequencing was performed on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) with 101-base paired-end reads at Novogene Biological Information Technology (Beijing, China). The primer pairs were designed using Primer 3 (Untergasser et al., 2012), with PCR product size ranging from 400 to 700 bp, GC content between 40% and 60%, primer length ranging from 18 to 25 bp, and melting temperature between 55 and 65 °C. The raw sequences of the transcriptome were deposited at figshare (https://doi.org/10.6084/m9.figshare.9788927).
PCR amplification was performed in a 20μL volume containing 20–40 ng genomic DNA, 2× Taq PCR MasterMix (Tiangen, Beijing, China), 5 μM of each primer, and double distilled H2O. The PCR procedure included an initial denaturation for 4 min at 94 °C, followed by 35 cycles of 35 s at 94 °C, 30 s at 55–59 °C and 1 min at 72 °C, ending with a final extension of 7 min at 72 °C. The different annealing temperatures were chosen according to each primer's features (Supplementary Data Table S2). Sequencing reactions were performed with the PCR primers in both directions by the service of Sangon Biotech (Shanghai, China). Sequences of the same locus were aligned using Clustal X 1.81 with default parameters (Thompson et al., 1997) and checked by eye using MEGA 5.05 (Tamura et al., 2011). All obtained DNA sequences have been deposited in the NCBI GenBank database under accession numbers MK331152–MK331582.
Nucleotide diversity, mismatch distribution and neutrality tests
Chloroplast DNA genetic diversity was estimated using the number of haplotypes (Nh), haplotype diversity (Hd) and nucleotide diversity (π; Nei, 1987) in DnaSP 5.10 (Librado and Rozas, 2009). The relationships between haplotypes were constructed using a median-joining network implemented in Network 5.0 (available at http://www.fluxus-engineering.com;Bandelt et al., 1999). The coefficients of differentiation GST (coefficient of genetic variation over all populations) and NST (coefficient of genetic variation influenced by both haplotype frequencies and genetic distances between haplotypes) were evaluated using Permut 1.0 (Pons and Petit, 1996). A mismatch distribution of the observed number of nucleotide differences between pairs of DNA sequences was computed using DnaSP 5.10, and values of Tajima's D (Tajima, 1989) and Fu's FS (Fu, 1997) were calculated using Arlequin 3.01 (Excoffier et al., 2005) to infer demographic history.
All nuclear sequences were assigned to coding and non-coding regions by aligning genomic sequences against their corresponding mRNA sequences. For each nuclear locus, basic population genetic parameters were estimated after phasing sequences using the phase algorithm in DnaSP 5.10. We calculated the number of segregating sites (S), the number of haplotypes (Nh), haplotype diversity (Hd), nucleotide diversity (π), Watterson's parameter (θ w; Watterson, 1975) and the minimum number of recombinant events (Rm). We also tested whether each locus fitted neutral evolution expectations using Tajima's D and Fu and Li's D* and F* statistics (Fu and Li, 1993). These parameters are expected to approach zero under neutrality by comparing the observed value of the summary statistics with their expected distribution.
Population genetic structure
STRUCTURE 2.3.4 (Hubisz et al., 2009) was used to assess population structure with the admixture model and the assumption of correlated allele frequencies using the dataset of 14 nuclear loci. Segregating sites in significant linkage disequilibrium after Bonferroni correction were excluded from this analysis. The number of clusters, K, corresponding to the number of populations, was explored using 20 independent runs per K. Burn-in was set to 20 000 and Markov chain Monte Carlo (MCMC) run length to 200 000. We ran STRUCTURE with K varying from 1 to 12, with 10 runs for each K value. The most likely number of clusters was estimated using LnP(D) (Pritchard et al., 2000) and ΔK statistics (Evanno et al., 2005). The population clusters were visualized using the program DISTRUCT 1.1 (Rosenberg, 2004).
We also used Wright's fixation index (FST; Wright, 1949) to assess population genetic differentiation between geographical groups. FST values for each nuclear locus and across all nuclear loci were calculated using AMOVA in Arlequin 3.01. The significance of FST was tested based on 10 000 permutations as described by Excoffier et al. (1992).
Population divergence and demography
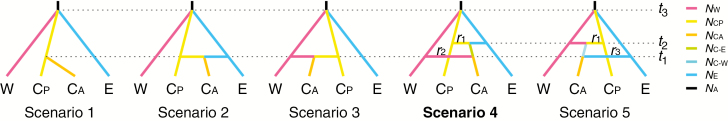
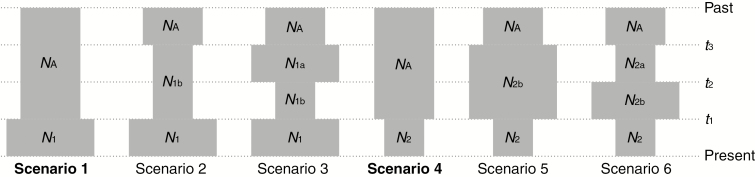
Because cpDNA variation in P. chienii is extremely low (only four substitutions were identified, and nearly half of the sampled populations were invariable; see details in Results), we did not include cpDNA in subsequent analyses. The population divergence models were tested using an ABC approach implemented in DIYABC 2.1.0 (Cornuet et al., 2014). Because two populations in the central cluster contain mixed genetic components in the STRUCTURE analysis (see details in Results), we adopted a three-step ABC analysis. First, we excluded populations with admixed components in the central cluster (CD and YZ, hereafter named CA), and five possible population divergence scenarios were modelled and tested for the west, central (excluding CA) and east clusters (W, CP and E). These scenarios are visualized and described in detail in Supplementary Data Figure S2. Second, after adding genetically mixed populations (CA) into the most likely model inferred in the first step, five scenarios were formulated (Fig. 2): (1) W, CP and E diverged from an ancestral population at the same time, t3, then CA diverged from CP; (2) CA was born from an admixture event between CP and E, or (3) CA formed via admixture between W and CP at time t1; (4) CA was then born from an admixture event between W and the descendant of CP and E; or (5) CA formed from an admixture event between E and the descendant of W and CP. Third, six scenarios (Fig. 3) of population demography were tested using ABC for each genetic cluster [W, CP and E; CA was excluded in this analysis as well as in IMa2 and LAMARC analyses (see below) due to their admixed origin that probably induces underrated calculation; e.g. Li et al. (2013)]: (1) an expansion at time t1, (2) a contraction at time t3 followed by an expansion at time t1, (3) an expansion at time t3 followed by a contraction at time t2, and then followed by an expansion at time t1, (4) a contraction at time t1, (5) an expansion at time t3 followed by a contraction at time t1, and (6) a contraction at time t3 followed by an expansion at time t2, and then followed by a contraction at time t1. The priors of all parameters were set with a uniform distribution (Supplementary Data Tables S5–S7). All one-sample and two-sample summary statistics were chosen to compare observed and simulated datasets. To ensure statistically robust results, at least 2 000 000 simulated datasets were generated for each scenario. We used 1% of the simulated datasets closest to the observed data to estimate the relative posterior probability [with 95% confidence intervals (CIs)] determined for each scenario via logistic regression and posterior parameter distributions (Cornuet et al., 2014). The generation time of 25 years were assumed for P. chienii as this estimate was applied to Taxus wallichiana in the same family Taxaceae (Liu et al., 2013).
Fig. 2.
Five assumed scenarios for population divergence of clusters W, CP, CA and E tested in DIYABC. NW, NCP, NCA and NE represent the population sizes of clusters W (TZ, JY, LH and DM), CP (JG and GG), CA (YZ and CD) and E (SQ, JL, LQ, and FY), respectively. NC-E and NC-W represent the population sizes of the admixed populations. NA represents the ancestral population size. t1, t2 and t3 represent times of population divergence and genetic admixture between clusters W, CP, CA and E. The best model is shown in bold type.
Fig. 3.
Six assumed demographic scenarios tested for clusters W, CP and E in DIYABC. N1 and N2 represent current population sizes, and NA represents the ancestral population size. N1a, N1b, N2a and N2b represent population sizes between the ancestral population and the current population. t1, t2 and t3 represent times of population changes. The best model is shown in bold type.
The IM model (Hey, 2010a, b) was also used to estimate gene flow and divergence time between genetic clusters. The model assumed neutrality, no selection, no recombination within loci, and random mating in ancestral and descendant populations (Hey and Nielsen, 2004, 2007). After extracting the longest non-recombining region of each locus using the Imgc program (Woerner et al., 2007), the demographic parameters, migration rate (m), effective population size (θ) and divergence time (t), were simulated and estimated using MCMC implemented in the IMa2 software package (Hey, 2010a, b). We ran the simulations using a burn-in of 5 000 000 steps and retaining 1 000 000 steps under the HKY mutation model. The demographic parameters from the IM model are scaled by a mean mutation rate. The mutation rate was estimated according to μ = KS/2T, where KS is the average divergence at silent sites between P. chienii and its closest extant relative T. chinensis, and T is the divergence time between the two species (~65 Ma; Leslie et al., 2012). The resulting geometric average mutation rate, 3.44 × 10−10 per site per year, was used to scale the effective population size and divergence time. Although the mutation rate was estimated over seven nuclear loci because PCR amplification of other loci failed for T. chinensis (represented by one individual), it is comparable with estimates in other conifers, such as 2.23–3.42 × 10−10 per site per year for Picea (Bouillé and Bousquet, 2005), 7.0–13.1 × 10−10 for Pinus (Willyard et al., 2007) and 1.94 × 10−10 for Juniperus (Li et al., 2012).
A Bayesian method, which accounts for the genealogical relationships among alleles and allows for population size changes, was implemented to estimate the exponential population growth rate (g) in LAMARC 2.1.8 (Kuhner, 2006). This analysis was performed using two runs of LAMARC, which included 10 initial chains and two final chains. The initial chains were run with 5000 samples and a sampling interval of 20 and using a burn-in of 10 000 samples for each chain. The final chains were carried out with the same burn-in and interval sampling as initial chains, but with 50 000 samples.
Isolation by distance and isolation by environment
To understand the effects of geography and environment on spatial patterns of genetic variation, we quantified the contributions of IBD and IBE in determining patterns of population genetic divergence using multiple matrix regression with randomization (MMRR) with all populations included (Wang, 2013). First, 19 bioclimatic variables were extracted from current climatic data with 30 arc-second resolution (http://www.worldclim.org) and used as proxies of environmental gradients. A climatic distance matrix was constructed using population pairwise Euclidean distances across all 19 bioclimatic variables. A geographical distance matrix was also calculated using the GPS coordinates of populations. Genetic distances (FST) among each pair of populations were calculated using Arlequin 3.01. Next, after standardizing the three distance matrices by Z scores, we analysed the contributions of geographical distance and environmental distance (explanatory variables) to genetic distance (response variable), using the 'MMRR' function in R (Wang, 2013). Additionally, principal component analysis (PCA) and multivariate statistical analysis were performed on the standardized 19 bioclimatic variables using IBM SPSS Statistics 22 (IBM, New York, USA) to evaluate the climatic divergence of three clusters.
RESULTS
Patterns of variation in cpDNA
One inversion, two indels and four substitutions were detected from concatenated sequences of two cpDNA fragments, rpl16 and psbA-trnH, which had a total length of 1452 bp (Supplementary Data Table S8). Seven haplotypes (H1–H7) were recovered from these sequence variations (Table S1). One haplotype (H1) occurred in all populations except for DM. Three haplotypes (H2–H4) were each shared between a population pair (LQ–JL, TZ–JL and JG–DM) (Fig. S1). The other three haplotypes (H5–H7) occurred in a single population each (YZ, JY and DM, respectively). Network analysis revealed that H2 and H4–H7 derived from H1 with one mutation and H3 was further derived from H6 with two mutations (Fig. S1).
Haplotype diversity (Hd) was 0.5933 at the species level and ranged from 0 to 0.6154 within populations. Species-level nucleotide diversity (π) was 0.0009, ranging from 0 to 0.00138 within populations (Supplementary Data Table S1). Five populations did not have cpDNA variation (SQ, FY, GG, CD and LH). Genetic differentiation was moderate among populations (GST = 0.519, NST = 0.598), and no significant difference was detected between GST and NST. The multimodal mismatch distribution for cpDNA (Fig. S3) indicated that populations might have experienced demographic equilibrium or contraction, which was also supported by a non-significant negative value of Tajima's D (−0.74552, P = 0.254) and positive Fu's FS (0.93509, P = 0.673).
Patterns of variation and neutrality tests for nuclear DNA
Fourteen low-copy nuclear genes were sequenced for all 134 individuals from 12 populations. The total aligned length was 6473 bp, with loci ranging from 334 to 702 bp. Three indels, located in non-coding regions of PT122, PT353 and PT399 with lengths of 18, 1 and 2 bp, respectively, were detected and excluded from the data analyses because they have a different mutation mechanism from substitutions.
A total of 109 segregating sites, included 10 singleton sites, were found from 14 nuclear genes. Haplotype diversity (Hd) varied from 0.2520 (PT58) to 0.9000 (PT353) and nucleotide diversity (π t) ranged from 0.00067 (PT58) to 0.00974 (PT122) with an average of 0.00265. Silent nucleotide diversity (π s) varied from 0 (PT23 and PT222) to 0.01192 (PT202) with an average of 0.00358, which was higher than the average non-synonymous diversity (π a = 0.00212). The minimum number of recombination events (Rm) detected across all nuclear loci ranged between zero and five. Average values of Tajima's D and Fu and Li's D* and F* were all positive, and Tajima's D was close to zero, although these values did not significantly deviate from the expectations of a standard neutral model for all nuclear loci (Supplementary Data Table S9).
Genetic structure and genetic differentiation
The Bayesian clustering algorithm (STRUCTURE) revealed that the most likely number of clusters for the entire dataset was K = 3 based on ΔK statistics (Supplementary Data Fig. S4). The three clusters clearly matched the geographical distribution: the west cluster (W) consisted of populations TZ, JY, LH and DM, the east cluster (E) included SQ, JL, LQ and FY, and the central cluster (C) contained populations JG, GG, YZ and CD (Fig. 1). YZ and CD showed signals of genetic admixture and we therefore defined them as CA, and the other two populations in the central cluster (JG and GG) were defined as CP.
We further explored the genetic substructure within each genetic cluster (Supplementary Data Fig. S5). Cluster W was divided into four distinct units when K = 4. While populations in cluster C were grouped into three units when K = 3, populations YZ and CD were not separated when K = 4. For cluster E, STRUCTURE analysis was not able to differentiate the four populations regardless of the K value.
F ST values were highly significant for each pair of genetic clusters (Table 1). Within cluster C, genetic differentiation between CP and CA was non-significant. When mixed populations (CA) were excluded, the FST value between E and CP increased, whereas it decreased between W and CP, indicating that cluster E contributed more to the genetic composition of CA compared with W.
Table 1.
Population differentiation (FST) at 14 nuclear loci among clusters W, C (CP and CA) and E based on pairwise comparisons.
| Locus | W vs. E | E vs. C | W vs. C | E vs. CP | W vs. CP | CP vs. CA |
|---|---|---|---|---|---|---|
| PT23 | −0.08964 | 0.29237 | −0.00048 | 0.40364 | 0.05304 | −0.27831 |
| PT58 | 0.02059 | 0.02934 | −0.05446 | −0.02606 | −0.17540 | 0 |
| PT107 | 0.07616 | 0.17826** | −0.07264 | 0.20517** | −0.11832 | 0.01705 |
| PT122 | 0.21718*** | −0.02168 | 0.17286*** | −0.01229 | 0.10909* | −0.37503 |
| PT126 | 0.12006* | 0.21293*** | 0.10458* | 0.32433* | 0.11656 | −0.01298 |
| PT140 | 0.54957*** | 0.18721* | 0.13424 | 0.21929* | 0.07369 | −0.08641 |
| PT154 | 0.79650* | 0.10853 | 0.42214 | 0.16007 | 0.32832 | −0.12108 |
| PT202 | −0.08902 | −0.00700 | −0.08521 | 0.04835 | −0.07607 | 0.03947 |
| PT203 | 0.23256*** | 0.11600* | −0.04306 | 0.24987** | −0.07565 | 0.48263 |
| PT222 | −0.00540 | 0.12427** | 0.08535 | 0.09315* | 0.04920 | 0.00004 |
| PT333 | 0.24574*** | 0.11564* | 0.43376*** | 0.14059 | 0.40948** | 0.22827 |
| PT353 | 0.31210*** | 0.12404*** | 0.33513*** | 0.17002*** | 0.35686*** | 0.06306 |
| PT373 | 0.00454 | 0.10006* | 0.11531 | 0.09470 | 0.11458 | −0.30421 |
| PT399 | 0.10195*** | 0.10905* | 0.00759 | 0.12134* | −0.05415 | 0.00942 |
| Total | 0.22659*** | 0.11358*** | 0.17654*** | 0.15774*** | 0.13901*** | −0.08293 |
W, western populations (TZ, JY, LH and DM); E, eastern populations (SQ, JL, LQ and FY); C, central populations (JG, GG, YZ and CD); CP, central populations dominated by one genetic component (JG and GG); CA, central populations with admixed genetic components (YZ and CD). *P < 0.05; **P < 0.01; ***P < 0.001.
Population divergence and demographic history
To yield more robust results, ABC modelling of the population divergence history of P. chienii was performed in two steps. First, two mixed populations (YZ and CD), detected from genetic assignment analysis, were excluded from the modelling. Scenario 1, which hypothesized that three clusters (W, CP, and E) diverged from an ancestral population at the same time, received the highest support (59.56 % posterior probability; Supplementary Data Fig. S6, Table S10) among five potential population divergence models (Fig. S2). Second, based on the best model (scenario 1) in the first step, we simulated five scenarios that included two mixed populations (YZ and CD) (Fig. 2). The results suggested that scenario 4 had a high posterior probability (49.05 %) (Fig. S7, Table S11). Posterior parameter estimates for scenario 4 suggested that three clusters (W, CP and E) diverged from a common ancestor around 3.68 Ma (95 % CI: 2.60–4.63 Ma). The origin of CA was quite complex; it was derived from two admixture events: one between CP and E at around 0.68 Ma, and the other between the derivative of CP-E and W at ~0.40 Ma (Table 2; Fig. S8).
Table 2.
Posterior estimates of demographic parameters for the best model (scenario 4) of population divergence in Fig. 2 revealed by Approximate Bayesian Computation.
| Parameter | Mean | Median | Mode | 95 % CI |
|---|---|---|---|---|
| N W | 2.80 × 105 | 2.89 × 105 | 3.03 × 105 | 1.98–3.35 × 105 |
| N CP | 1.80 × 105 | 1.83 × 105 | 1.89 × 105 | 1.11–2.37 × 105 |
| N CA | 4.86 × 104 | 4.69 × 104 | 4.72 × 104 | 1.16–9.19 × 104 |
| N E | 3.62 × 105 | 3.68 × 105 | 3.81 × 105 | 2.31–4.79 × 105 |
| N C-E | 5.20 × 104 | 5.20 × 104 | 3.77 × 104 | 0.81–9.54 × 104 |
| N A | 3.85 × 105 | 3.87 × 105 | 3.48 × 105 | 0.19–5.69 × 105 |
| T 1 (years) | 4.00 × 105 | 3.73 × 105 | 2.19 × 105 | 0.56–8.58 × 105 |
| T 1 (generations) | 1.60 × 104 | 1.49 × 104 | 8.76 × 103 | 0.22–3.43 × 104 |
| T 2 (years) | 6.75 × 105 | 5.65 × 105 | 3.63 × 105 | 0.16–1.58 × 106 |
| T 2 (generations) | 2.70 × 104 | 2.26 × 104 | 1.45 × 104 | 0.64–6.32 × 104 |
| T 3 (years) | 3.68 × 106 | 3.73 × 106 | 3.85 × 106 | 2.60–4.63 × 106 |
| T 3 (generations) | 1.47 × 105 | 1.49 × 105 | 1.54 × 105 | 1.04–1.85 × 105 |
| r 1 | 0.665 | 0.716 | 0.917 | 0.185–0.966 |
| r 2 | 0.274 | 0.233 | 0.136 | 0.051–0.638 |
N W, population size of cluster W (TZ, JY, LH and DM); NCP, population size of cluster CP (JG and GG); NCA, population size of cluster CA (YZ and CD); NE, population size of cluster E (SQ, JL, LQ and FY); NC-E, population size of admixed population; NA, ancestral population size; T, divergence time of clusters W, C and E from the ancestral population; r1 and r2, admixture rates.
Among six potential demographic scenarios in ABC modelling (Fig. 3), cluster E conformed to scenario 1, which indicated a population expansion at 0.62 Ma, while clusters W and CP conformed to scenario 4, which indicated population contractions at 0.48 and 0.49 Ma, respectively (Table 3 and Supplementary Data Table S12, Figs S9 and S10). The LAMARC analysis also suggested that the three clusters have undergone changes in population size. For clusters W and CP, the estimated g values were −151.75 and −59.27, respectively, suggesting population declines. In contrast, the g value of cluster E was 219.77, indicating population growth.
Table 3.
Posterior estimates of demographic parameters for the best models of population demography in Fig. 3 revealed by Approximate Bayesian Computation.
| Cluster (best model) | Parameter | N A | N 1/N2 | T (years) | T (generations) |
|---|---|---|---|---|---|
| W (scenario 4) | Mean | 1.79 × 105 | 9.13 × 104 | 4.78 × 105 | 1.91 × 104 |
| Median | 1.77 × 105 | 8.75 × 104 | 4.63 × 105 | 1.85 × 104 | |
| Mode | 1.52 × 105 | 8.45 × 104 | 2.23 × 105 | 8.92 × 103 | |
| 95 % CI | 0.76–2.87 × 105 | 0.38–1.59 × 105 | 0.68–9.33 × 105 | 0.27–3.73 × 104 | |
| CP (scenario 4) | Mean | 1.79 × 105 | 5.70 × 104 | 4.90 × 105 | 1.96 × 104 |
| Median | 1.79 × 105 | 5.19 × 104 | 4.70 × 105 | 1.88 × 104 | |
| Mode | 1.62 × 105 | 3.86 × 104 | 3.18 × 105 | 1.27 × 104 | |
| 95 % CI | 0.70–2.87 × 105 | 0.17–1.14 × 105 | 0.93–9.38 × 105 | 0.37–3.75 × 104 | |
| E (scenario 1) | Mean | 1.25 × 105 | 2.21 × 105 | 6.20 × 105 | 2.48 × 104 |
| Median | 1.11 × 105 | 2.23 × 105 | 6.93 × 105 | 2.77 × 104 | |
| Mode | 4.39 × 104 | 2.49 × 105 | 9.93 × 105 | 3.97 × 104 | |
| 95 % CI | 0.33–2.59 × 105 | 0.60–3.79 × 105 | 0.79–9.83 × 105 | 0.32–3.93 × 104 |
N A, ancestral population size; N1/N2, current population sizes; T, population expansion/contraction time.
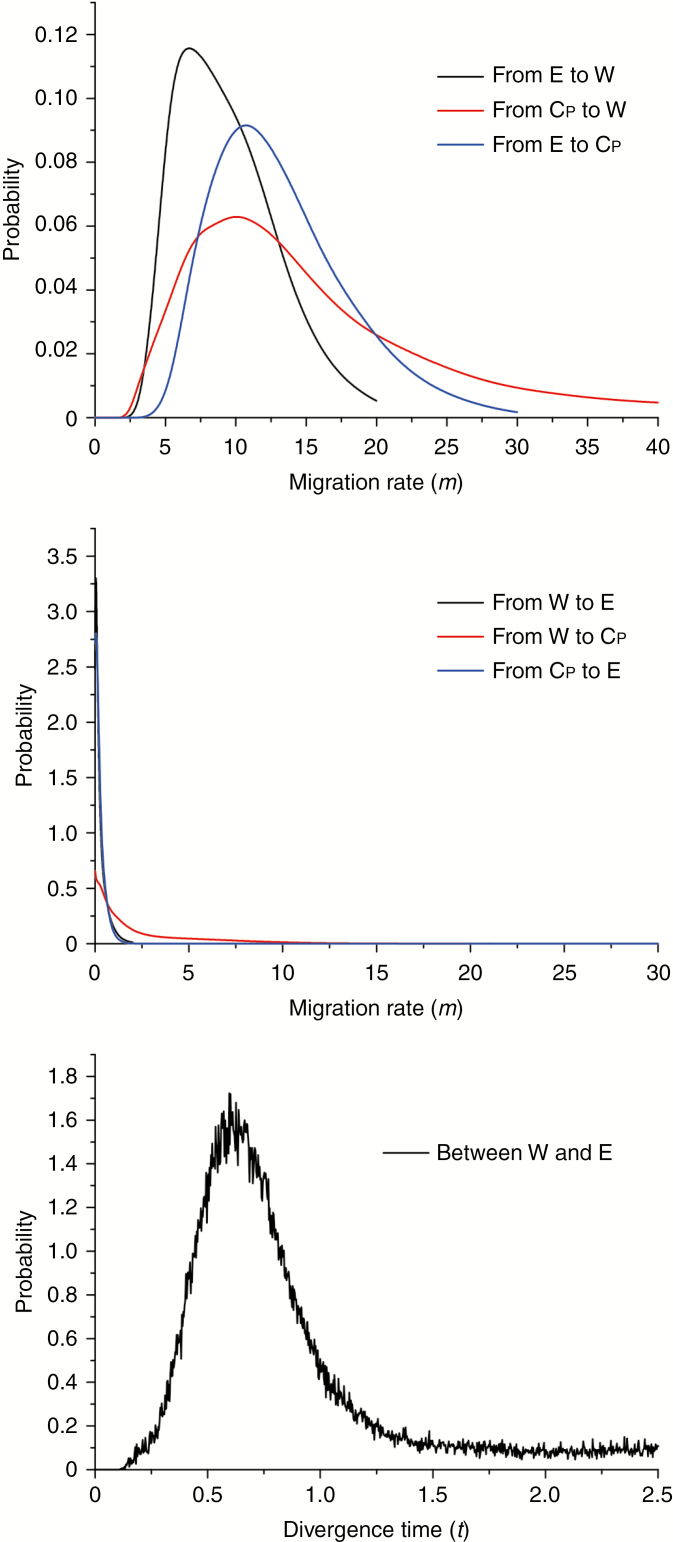
Migration rates (m) estimated under the IM model using IMa2 were asymmetric, with much higher gene flow from E to W (6.69), E to CP (10.73), and CP to W (10.06) than in the reverse directions (approaching zero) (Fig. 4 and Supplementary Data Fig. S11, Table S13). The divergence time estimates using IMa2 between clusters CP and W and between E and CP were unreliable due to a lack of convergence in their marginal posterior densities. However, we obtained a robust time estimate for the divergence between E and W [3.74 Ma; 95% highest posterior density (HPD) interval: 1.31–11.80 Ma) (Fig. 4; Fig. S11, Table S13), which is almost identical to the divergence time of three clusters in ABC modelling.
Fig. 4.
Posterior probability distributions of the migration rate (m) between each pair of clusters (W, E and CP) from both directions and the divergence time (t) between clusters W and E estimated by the IM model.
Contributions of IBD and IBE to population divergence
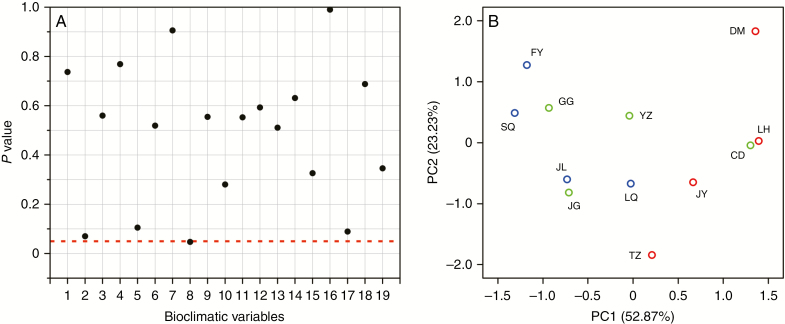
MMRR analysis revealed non-significant effects of both IBD (β IBD = 0.121, P = 0.425) and IBE on population genetic divergence (FST) (β IBE = 0.062, P = 0.758). The impact of IBD on spatial patterns of genetic variation was about twice as much as IBE. Almost none of the 19 bioclimatic variables had a significant contribution to genetic divergence, except for BIO 8 (mean temperature of wettest quarter), which was marginally significant (P = 0.047; Fig. 5A). PCA did not find clear climatic differentiation among three genetic clusters (W, C and E) in either of the first two principal components (PC1: 52.87%, PC2: 23.23%) (Fig. 5B); multivariate statistical analysis also showed no significant differentiation among them (P = 0.207).
Fig. 5.
(A) The effects of 19 bioclimatic variables on population genetic divergence analysed using the MMRR function in R. The red dotted line represents a significance level of 0.05. (B) Principal component analysis of 19 bioclimatic variables among three genetic clusters using IBM SPSS Statistics 22. The red, green and blue circles represent population clusters W, C and E, respectively.
Discussion
Recent establishment of Pseudotaxus chienii populations in south-east China
The divergence time of three genetic clusters (W, CP and E) of P. chienii estimated by ABC was about 3.68 Ma (95 % CI: 2.60–4.63 Ma), indicating that the first hypothesis is most plausible. Given the stem lineage of Pseudotaxus branched very early (~54–65 Ma; Leslie et al., 2012; Lu et al., 2014), this result is quite striking. However, this timing is almost identical to the divergence time of the E and W clusters in IM (3.74 Ma; HPD: 1.31–11.80 Ma), an approach that has a different philosophy from ABC. In ABC analysis, a calibrated mutation rate for sequences is not needed for parameter estimation (such as migration rate, divergence time) (Beaumont, 2010); in contrast, the mutation rate is required to convert the rate-scaled parameter estimates to more interpretable demographic units (years or generations) in the IM model (Hey, 2010a, b). Although there are wide confidence intervals around the time estimates, the recent time estimates by two different approaches suggests that: (1) Pseudotaxus might have existed in south-east China and other locations initially – however, the lineage of P. chienii branched recently in south-east China and other members and populations of Pseudotaxus elsewhere went extinct; and (2) P. chienii might have broader ranges (perhaps including south-east China) but south-east China could have been colonized (never appeared in south-east China) or re-colonized (appeared in south-east China initially but disappeared later) recently, while populations in other locations went extinct. Whichever scenario is more plausible, the results of this study suggest that the extant P. chienii lineage is relatively young and the populations of P. chienii started to differentiate in south-east China recently (possibly started in the late Pliocene). Such a short duration of population isolation (~3.68–3.74 Ma) may be insufficient to result in species-level divergence within a slow-growing tree species with long generation time, a conclusion consistent with the idea of macroevolutionary stasis in tree lineages since the Pliocene (5.30–2.60 Ma; Huntley, 1991).
With the recent interest in phylogeographical patterns and evolutionary history of Chinese plants, Pliocene to Pleistocene origins or establishment of relict gymnosperms in central to south-east/south China have been increasingly documented. Taiwania cryptomerioides, for example, is also a relictual monotypic conifer in south China (occasionally extending to northern Vietnam). Molecular clock estimation demonstrated that the mean divergence time between two major cpDNA lineages (insular and mainland) of Taiwania occurred at ~3.23–3.41 Ma (Chou et al., 2011), a value very close to the population divergence time of P. chienii. Recently, an analysis of whole chloroplast genomic sequences of wild extant Ginkgo populations revealed that the deepest temporal footprint unexpectedly dated back to ~0.39 Ma (Hohmann et al., 2018). Although a single locus (cpDNA) may suffer from genealogical stochasticity and overestimate population divergence time (Maddison, 1997; Carstens and Knowles, 2007), previous timings are consistent with the population divergence time of P. chienii inferred from multiple nuclear sequence data, enhancing the possibility that the duration of population/lineage differentiation of relict gymnosperm taxa in central to south-east/south China may be much shorter than the ages of lineages seemingly indicate (Mao and Liu, 2012).
Fossils provide the best evidence for the presence of a species within a past window of space and time (Gavin et al., 2014). Unfortunately, the fossil record of P. chienii is very limited (Wu et al., 2007), providing little information about its evolutionary history. The situation for P. chienii is very similar to Amborella trichopoda that is sister to all other extant flowering plants (Soltis et al., 2008), on the island of New Caledonia, but has no fossil record. Amborella trichopoda has long been viewed as an autochthonous remnant of the Gondwanan biota (Heads, 2009), but geological evidence precludes the long-term existence of A. trichopoda in New Caledonia (Grandcolas et al., 2014). A palaeogeographical study of the flora of New Caledonia showed that 'Amborella' was probably represented in many places by many lineages initially, but only the sole representative (i.e. A. trichopoda) remained in New Caledonia. However, any initial New Caledonia lineage could have been lost during the Eocene, regained during the Oligocene and subsequently extinguished in other places (Lee et al., 2001). Similarly, the monsoon climate to which P. chienii is adapted did not exist before the early Miocene (~23 Ma) because south-east China was controlled by a broad, roughly W–E dry belt caused by the planetary circulation system during the early Cenozoic (Guo et al., 2008), precluding the possibility of long-term existence of Pseudotaxus in south-east China after the stem lineage initially branched out (54–65 Ma). Pseudotaxus was able to colonize south-east China only after the early Miocene (Kou et al., 2016) and the most likely colonization of P. chienii into southeast China could have occurred during the late Miocene to Pliocene when the climate deteriorated globally (Zachos et al., 2001) and the Asian monsoon was intensified (An et al., 2001), a period that is consistent with the crown ages of several relict lineages (Wang et al., 2009; Qi et al., 2012; Tian et al., 2015; Zhang et al., 2015). This conclusion implies that P. chienii could have had a broader distribution in history, the lack of fossil records might be due to the morphological similarity to Taxus (personal communication with Dr Y. Yang, Institute of Botany, The Chinese Academy of Sciences) or incomplete investigations.
In addition, the abundant fossil records of other gymnosperms, such as Metasequoia glyptostroboides, may provide clues for the evolution of P. chienii. Metasequoia glyptostroboides was once widely distributed through high latitudes of the northern hemisphere. With the climate deteriorating, its range shrank sharply and the latest fossils were found in Japan in the Pliocene or early Pleistocene. However, living M. glyptostroboides was surprisingly rediscovered in south-eastern China (LePage et al., 2005), suggesting that it might have become established in China during the early Pleistocene, possibly immigrating from Japan (LePage et al., 2005). The bulk of Metasequoia fossils, coupled with molecular dating by Chou et al. (2011), Hohmann et al. (2018) and this study, clearly demonstrates that the relict gymnosperm flora in south-east China and even the Sino-Japanese Flora are much younger than previously thought (Chen et al., 2018). Formation of the relict gymnosperm flora may have resulted from colonizations or recolonizations from other parts of the world (Thorne, 1999; Manchester et al., 2009) driven by global aridity and cooling since the Miocene (Zachos et al., 2001) and the formation of the Asian monsoon since the Miocene (Guo et al., 2008).
Strong asymmetrical historical gene flow
We hypothesized that gene flow among populations of P. chienii might be substantial enough to prevent allopatric speciation. In this study, ABC analysis found that two historical genetic admixture events occurred at 0.68 Ma (between E and CP; 95% CI: 0.16–1.58 Ma) and 0.40 Ma (between derived populations of E-CP and W; 95% CI: 0.06–0.86 Ma). IM analysis further indicated that genetic exchange between clusters was asymmetrical, with moderate to strong gene flow from east to west, whereas gene flow approached zero in the reverse direction (Fig. 4), supporting that gene flow has indeed counteracted population divergence in P. chienii.
The timing of the first admixture event is largely consistent with the population expansion time of cluster E (0.62 Ma; 95% CI: 0.08–0.98 Ma), a period corresponding to the Cromerian Glaciation immediately following the middle Pleistocene transition (0.80–1.20 Ma) (Ehlers and Gibbard, 2007). This is considered to be the largest glaciation during the Pleistocene; for example, the ice sheet during the Wangkun glaciation (700–500 ka) was 18 times larger than the present ice sheet on the Qinghai–Tibetan Plateau (Shi, 1998). This glaciation may be associated with several major intraspecific evolutionary events in other plant species of south-east China (e.g. Tian et al., 2015). It is likely that cold-tolerant P. chienii expanded its range (especially in the east populations) during favourable glacial conditions and this expansion resulted in genetic admixture. This possibility is further supported by the LAMARC analysis, which also detected an expansion event in cluster E.
The second admixture event occurred at ~0.40 Ma, a period corresponding to the Elsterian Glaciation (Marine Isotope Stage 12; Ehlers and Gibbard, 2007). However, ABC analyses showed that clusters W and CP experienced population declines starting at about 0.48 and 0.47 Ma, respectively. The population declines are also supported by the g values of LAMARC analyses and the mismatch analysis of cpDNA variation (Supplementary Data Fig. S3). These events would have reduced the possibility of genetic exchange between west and central populations. However, the central and west populations are geographically contiguous, and pollen-mediated gene flow was highly likely (Bai et al., 2014) given that the pollen of P. chienii is transmitted by wind (Su et al., 2009). This interpretation is further supported by the widely distributed chloroplast haplotype (H1) because chloroplast genomes in Taxaceae are paternally inherited (Collins et al., 2003) and transmitted by pollen, although the retention of ancestral polymorphism and insufficient marker resolution may also contribute to the sharing of haplotypes. In addition, DIYABC is usually sensitive to the strongest population change during a given time period (Cornuet et al., 2014), so we cannot exclude the possibility that clusters W and CP would have experienced slight range expansion during the Elsterian Glaciation (~0.40 Ma) after the population decline events (~0.48 and 0.47 Ma).
The finding of strong historical gene flow within P. chienii has profound implications for understanding the evolutionary history of temperate tree species in China during the late Neogene. In contrast to the expansion–contraction model for temperate tree species in response to the Quaternary climate oscillations in Europe and North America, early plant phylogeographical studies that were preferentially based on maternally inherited cpDNA sequences identified a multi-refugium model in China (particularly in central to southeast China) (Qiu et al., 2011; Liu et al., 2012). This model describes a general pattern of multiple localized refugia and little admixture among refugial populations throughout the Quaternary glacial–interglacial cycles. This model is consistent with the allopatric speciation hypothesis of Harrison et al. (2001) that population fragmentation of temperate forest species occurred during both the Quaternary glacial and interglacial periods, which may have resulted in allopatric radiations. However, to our knowledge, no clear examples of a late Pliocene–Pleistocene tree speciation event have been documented in south-east China thus far, possibly due to range shift-induced genetic exchange and pollen-mediated gene flow as demonstrated in P. chienii. This finding has two implications. First, temperate tree species in south-east China could have lower speciation rates than previously thought, and the high speciation rate in woody taxa might be ascribed to some speciose taxa (e.g. Castnopsis, Ilex, Eurya or Camellia) in evergreen broadleaved forests (Lu et al., 2018). Second, while the role of pollen-mediated gene flow has been recognized (e.g. Bai et al., 2014), the contribution of range shift to species cohesion has not been fully evaluated and needs to be further investigated in south-east China using coalescent-based methods relying on multiple nuclear loci or genomic data.
Weak climate-induced divergent selection in south-east China
Understanding the mechanisms generating differences among populations and species is a major objective in evolutionary biology. Geography and environment represent two key landscape components that can potentially influence gene flow and population connectivity (Wang and Bradburd, 2014). In this study, MMRR analysis showed that the contribution of IBD to genetic divergence of P. chienii is much higher than that of IBE. These results suggest that neutral processes associated with geography, such as mutation and genetic drift, are stronger driving factors for population divergence of P. chienii than processes associated with environments, echoing the hypothesis of limited environmental gradients within narrow geographical ranges in south-east China. In addition, MMRR analysis found that none of 19 bioclimatic variables except mean temperature of the wettest quarter (BIO 8) contributed to genetic divergence significantly, indicating that environmental gradients (using 19 climatic variables as proxies) across the range of P. chienii may not be strong enough to facilitate genetic differentiation, possibly due to the prevalence of the East Asian monsoon. PCA and multivariate statistical analysis of climatic variables did not find significant differentiation of climatic envelopes among the three genetic clusters (P = 0.207; Fig. 5B), further supporting that IBE has limited contribution to genetic divergence in P. chienii.
Divergent selection and adaptation associated with different habitats have been viewed as a major cause of allopatric speciation in plants (Rieseberg and Burke, 2001; Funk et al., 2006; Harmon et al., 2008). Because many mechanisms can generate IBE, the detection of IBE alone is not evidence of local adaptation (Wang and Bradburd, 2014). However, the effects of climatic variables such as precipitation and temperature have been implicated in cases of incipient or completed speciation events (Nosil et al., 2005; Lowry et al., 2008). It is reasonable to assume that IBE can be simplified and considered as a proxy of divergent selection. Therefore, in this case, divergent selection exerted by environmental gradients (i.e. climatic gradients) in different populations of P. chienii could be too weak to generate enough of a driving force (i.e. divergent selection or local adaptation) for speciation or incipient speciation to occur. This weak divergent selection may be partially responsible for the lack of recent tree speciation in south-east China as well.
By contrast, in south-west China and particularly the Himalaya–Hengduan Mountains (HHM) region, where the East Asian Monsoon and Indian Monsoon converge and extreme physiographical changes occur within relatively short distances, geologically recent speciation or incipient speciation events have been frequently reported (e.g. Xu et al., 2010; Jia et al., 2011; Liu et al., 2013; Li et al., 2013; Luo et al., 2016). For example, Liu et al. (2013) found that Taxus wallichiana, a species within the sister genus to Pseudotaxus, initiated incipient speciation at ~4.20 Ma, an estimate slight older than the divergence time of the three Pseudotaxus clusters. The underlying factor may be that population isolation was initiated by the rise of the Qinghai–Tibetan Plateau and then reinforced by subsequent differential ecological (climatic) adaptations (Liu et al., 2013). However, such conditions (strong physical barriers and environmental gradient) do not exist in south-east China and that is why south-west China became a cradle of plant diversity, but south-east China a preservation centre for relict lineages (Thorne, 1999).
Conclusions
The results of our study on P. chienii, taken together with those recently reported for other monotypic gymnosperms that occur in central and south-east China, clearly demonstrate that the relict gymnosperms may have become established very recently despite the fact that the stem lineages of those taxa may have had a much longer evolutionary history (Mao and Liu, 2012). The current relicts are dynamic and do not imply conservation of ancestral characters in terms of morphology, ecology and spatial distribution (Grandcolas et al., 2014). The reason that these monotypic gymnosperms have a relict status in China is that the prerequisites (enough time for strict isolation and divergent selection) for allopatric speciation in south-east China have not been fulfilled even though the opportunities for allopatric speciation prevail. This study may provide a new perspective for understanding the evolution of relict gymnosperm flora in south-east China as well as the evolution of the Sino-Japanese Flora.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: Sampling information, the distribution of chloroplast haplotypes and chloroplast genetic variation of Pseudotaxus chienii. Table S2: Primer sequences, annealing temperatures, PCR product sizes and gene ID of 14 nuclear loci used in this study. Table S3: Transcriptome sequences corresponding to the loci in Supplementary Data Table S2. Table S4: Functional categories of 14 nulcear loci against four protein databases. Table S5: Description of the prior distribution of parameters from five scenarios in Supplementary Data Fig. S2 used in ABC. Table S6: Description of the prior distribution of parameters from five scenarios in Fig. 2 used in ABC. Table S7: Description of the previous distribution of parameters from six scenarios in Fig. 3 used in ABC. Table S8: Variable sites in seven haplotypes from two cpDNA fragments. Table S9: Nucleotide variation, haplotype diversity and neutrality tests across all 14 nuclear loci. Table S10: Posterior probabilities and credibility intervals of five scenarios in Supplementary Data Fig. S2 used in ABC modelling. Table S11: Posterior probabilities and credibility intervals of five scenarios in Fig. 2 used in ABC modelling. Table S12: Posterior probabilities and credibility intervals of six scenarios in Fig. 3 used in ABC modelling. Table S13: Maximum likelihood estimates and the 95 % highest posterior density intervals of demographic parameters between each two clusters estimated in IMa2. Figure S1: The geographical distribution and haplotype network of chloroplast haplotypes in Pseudotaxus chienii. Figure S2: Five assumed scenarios of population divergence for clusters W, CP and E tested in DIYABC. Figure S3: Mismatch distribution for cpDNA sequences. Figure S4: The most likely number of clusters inferred by STRUCTURE using LnP(D) and ΔK statistics. Figure S5: STRUCTURE analysis applied to clusters W, C and E when K = 2–4 was assumed. Figure S6: Posterior probabilities for five scenarios used in Supplementary Data Fig. S2 estimated using logistic regression of 1 % of the closest datasets in DIYABC. Figure S7: Posterior probabilities for five scenarios used in Fig. 2 estimated using logistic regression of 1 % of the closest datasets in DIYABC. Figure S8: Prior and posterior distributions of demographic parameters under scenario 4 in Fig. 2 estimated using DIYABC. Figure S9: Posterior probabilities of six scenarios used in Fig. 3 for W, CP and E estimated using logistic regression of 1 % of the closest datasets in DIYABC. Figure S10: Prior and posterior distributions of demographic parameters for W (scenario 4), CP (scenario 1) and E (scenario 1) in Fig. 3 estimated using DIYABC. Figure S11: Posterior probability distributions of effective population size (θ) for each cluster and divergence time between each pair of clusters estimated by the IM model.
ACKNOWLEDGEMENTS
We are extremely grateful to a large number of individuals, too numerous to list, who provided help with fieldwork. Special thanks go to Wenxiu Zhang and Weidong Zeng for their participation in sample collection.
Funding
This work was supported by the National Natural Science Foundation of China (41461008, 31560064), a grant from Yunnan Provincial Government (2017HA014), and a grant for key discipline (ecology) of Jiangxi Agricultural University.
LITERATURE CITED
- An ZS, Kutzbach JE, Prell WL, Porter SC. 2001. Evolution of Asian monsoons and phased uplift of Himalaya-Tibetan plateau since Late Miocene times. Nature 411: 62–66. [DOI] [PubMed] [Google Scholar]
- Anacker BL, Strauss SY. 2014. The geography and ecology of plant speciation: range overlap and niche divergence in sister species. Proceedings of the Royal Society B 281: 20132980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai WN, Wang WT, Zhang DY. 2014. Contrasts between the phylogeographic patterns of chloroplast and nuclear DNA highlight a role for pollen-mediated gene flow in preventing population divergence in an East Asian temperate tree. Molecular Phylogenetics and Evolution 81: 37–48. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Beaumont MA. 2010. Approximate Bayesian computation in evolution and ecology. Annual Review in Ecology, Evolution and Systematics 41: 379–406. [Google Scholar]
- Bouillé M, Bousquet J. 2005. Trans-species shared polymorphisms at orthologous nuclear gene loci among distant species in the conifer Picea (Pinaceae): implications for the long-term maintenance of genetic diversity in trees. American Journal of Botany 92: 63–73. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Knowles LL. 2007. Estimating species phylogeny from gene-tree probabilities despite incomplete lineage sorting: an example from Melanoplus grasshoppers. Systematic Biology 56: 400–411. [DOI] [PubMed] [Google Scholar]
- Chen YS, Deng T, Zhou Z, Sun H. 2018. Is the East Asian flora ancient or not? National Science Review 5: 920–932. [Google Scholar]
- Cheng WC. 1947. New Chinese trees and shrubs. Research Notes, Forestry Institute, National Central University, Nanking, Dendrological Series 1: 1–4. (in Chinese with English abstract) [Google Scholar]
- Chou YW, Thomas PI, Ge XJ, LePage BA, Wang CN. 2011. Refugia and phylogeography of Taiwania in East Asia. Journal of Biogeography 38: 1992–2005. [Google Scholar]
- Collins D, Mill RR, Möller M. 2003. Species separation of Taxus baccata, T. canadensis, and T. cuspidata (Taxaceae) and origins of their reputed hybrids inferred from RAPD and cpDNA data. American Journal of Botany 90: 175–182. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Pudlo P, Veyssier J, et al. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30: 1187–1189. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Ehlers J, Gibbard PL. 2007. The extent and chronology of Cenozoic Global Glaciation. Quaternary International 164–165: 6–20. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre A, Päckert M, Pauls SU, et al. 2014. The role of the uplift of the Qinghai-Tibetan plateau for the evolution of Tibetan biotas. Biological Reviews 90: 236–253. [DOI] [PubMed] [Google Scholar]
- Fu LG, Jin JM. 1992. Red List of Endangered Plants in China, volume 1. Beijing: Science Press. [Google Scholar]
- Fu LG, Li N, Mill RR. 1999. Taxaceae. In: Wu ZY, Raven PH, eds. Flora of China. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press, 89–98. [Google Scholar]
- Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking, and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk DJ, Nosil P, Etges WJ. 2006. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proceedings of the National Academy of Sciences USA 103: 3209–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DG, Fitzpatrick MC, Gugger PF, et al. 2014. Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytologist 204: 37–54. [DOI] [PubMed] [Google Scholar]
- Grandcolas P, Nattier R, Trewick S. 2014. Relict species: a relict concept? Trends in Ecology and Evolution 29: 655–663. [DOI] [PubMed] [Google Scholar]
- Guo ZT, Sun B, Zhang ZS, Peng SZ, Xiao GQ, Ge JY, Hao QZ, Qiao YS, Liang MY, Liu JF, Yin QZ, Wei JJ. 2008. A major reorganization of Asian climate by the early Miocene. Climate of the Past 4: 153–174. [Google Scholar]
- Halffter G. 1987. Biogeography of the montane entomofauna of Mexico and Central America. Annual Review of Entomology 32: 95–114. [Google Scholar]
- Harmon LJ, Melville J, Larson A, Losos JB. 2008. The role of geography and ecological opportunity in the diversification of day geckos (Phelsuma). Systematic Biology 57: 562–573. [DOI] [PubMed] [Google Scholar]
- Harrison RG. 2012. The language of speciation. Evolution 66: 3643–3657. [DOI] [PubMed] [Google Scholar]
- Harrison SP, Yu G, Takahara H, Prentice IC. 2001. Diversity of temperate plants in east Asia. Nature 413: 129–130. [DOI] [PubMed] [Google Scholar]
- He Z, Li X, Yang M, et al. 2019. Speciation with gene flow via cycles of isolation and migration: insights from multiple mangrove taxa. National Science Review 6: 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heads M. 2009. Globally basal centres of endemism: the Tasman–Coral Sea region (south-west Pacific), Latin America and Madagascar/ South Africa. Biological Journal of the Linnean Society 96: 222–245. [Google Scholar]
- Hey J. 2010a The divergence of Chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Molecular Biology and Evolution 27: 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. 2010b Isolation with migration models for more than two populations. Molecular Biology and Evolution 27: 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proceedings of the National Academy of Sciences, USA 104: 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann N, Wolf EM, Rigault P, et al. 2018. Ginkgo biloba's footprint of dynamic Pleistocene history dates back only 390,000 years ago. BMC Genomics 19: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. 2009. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley B. 1991. How plants respond to climate change: migration rates, individualism and the consequences for plant communities. Annals of Botany 67: 15–22. [Google Scholar]
- Jia DR, Liu TL, Wang LY, Zhou DW, Liu JQ. 2011. Evolutionary history of an alpine shrub Hippophae tibetana (Elaeagnaceae): allopatric divergence and regional expansion. Biological Journal of the Linnean Society 102: 37–50. [Google Scholar]
- Kou Y, Cheng S, Tian S, et al. 2016. The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. Journal of Biogeography 43: 351–360. [Google Scholar]
- Kou YX, Xiao K, Lai XR, Wang YJ, Zhang ZY. 2017. Natural hybridization between Torreya jackii and T. grandis (Taxaceae) in southeast China. Journal of Systematics and Evolution 55: 25–33. [Google Scholar]
- Kuhner MK. 2006. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22: 768–770. [DOI] [PubMed] [Google Scholar]
- Lee DE, Lee WG, Mortimer N. 2001. Where and why have all the flowers gone? Depletion and turnover in the New Zealand Cenozoic angiosperm flora in relation to palaeogeography and climate. Australian Journal of Botany 49: 341–356. [Google Scholar]
- LePage BA, Williams CJ, Yang H. 2005. The Geobiology and Ecology of Metasequoia. Dordrecht: Springer Press. [Google Scholar]
- Leslie AB, Beaulieu JM, Rai HS, Crane PR, Donoghue MJ, Mathews S. 2012. Hemisphere-scale differences in conifer evolutionary dynamics. Proceedings of the National Academy of Sciences, USA 109: 16217–16221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Abbot RJ, Liu B, et al. 2013. Pliocene intraspecific divergence and Plio-Pleistocene range expansions within Picea likiangensis (Lijiang spruce), a dominant forest tree of the Qinghai-Tibet Plateau. Molecular Ecology 22: 5237–5255. [DOI] [PubMed] [Google Scholar]
- Li Z, Zou J, Mao K, et al. 2012. Population genetic evidence for complex evolutionary histories of four high altitude juniper species in the Qinghai-Tibetan Plateau. Evolution 66: 831–845. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ. 2013. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytologist 199: 1093–1108. [DOI] [PubMed] [Google Scholar]
- Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu YX. 2012. Phylogeographic studies of plants in China: advances in the past and directions in the future. Journal of Systematics and Evolution 50: 267–275. [Google Scholar]
- Lowry DB, Rockwood RC, Willis JH. 2008. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution 62: 2196–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ran JH, Guo DM, Yang ZY, Wang XQ. 2014. Phylogeny and divergence times of gymnosperms inferred from single-copy nuclear genes. PLoS ONE 9: e107679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L-M, Mao L-F, Yang T, et al. 2018. Evolutionary history of the angiosperm flora of China. Nature 554: 234–238. [DOI] [PubMed] [Google Scholar]
- Luo D, Yue JP, Sun WG, et al. 2016. Evolutionary history of the subnival flora of the Himalaya-Hengduan Mountains: first insights from comparative phylogeography of four perennial herbs. Journal of Biogeography 43: 31–43. [Google Scholar]
- Maddison WP. 1997. Gene trees in species trees. Systematic Biology 46: 523–536. [Google Scholar]
- Manchester SR, Chen ZD, Lu AM, Uemura K. 2009. Eastern Asian endemic seed plant genera and their paleogeographic history throughout the Northern Hemisphere. Journal of Systematics and Evolution 47: 1–42. [Google Scholar]
- Mao K, Liu J. 2012. Current ‘relicts' more dynamic in history than previously thought. New Phytologist 196: 329–331. [DOI] [PubMed] [Google Scholar]
- Mittermeier RA, Robles-Gil P, Mittermeier CG. 1997. Megadiversity: Earth's Biologically Wealthiest Nations. Mexico City: CEMEX/Agrupaciaon Sierra Madre, Conservation International. [Google Scholar]
- Nagalingum NS, Marshall CR, Quental TB, Rai HS, Little DP, Mathews S. 2011. Recent synchronous radiation of a living fossil. Science 334: 796–799. [DOI] [PubMed] [Google Scholar]
- Nei M. 1987. Molecular Evolutionary Genetics. New York: Columbia University Press. [Google Scholar]
- Nosil P. 2008. Speciation with gene flow could be common. Molecular Ecology 17: 2103–2106. [DOI] [PubMed] [Google Scholar]
- Nosil P, Vines TH, Funk DJ. 2005. Perspective: reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59: 705–719. [PubMed] [Google Scholar]
- Pons O, Petit RJ. 1996. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XS, Chen C, Comes HP, et al. 2012. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytologist 196: 617–630. [DOI] [PubMed] [Google Scholar]
- Qian H, Ricklefs RE. 2000. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407: 180–182. [DOI] [PubMed] [Google Scholar]
- Qiu YX, Fu CX, Comes HP. 2011. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world's most diverse temperate flora. Molecular Phylogenetics and Evolution 59: 225–244. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Burke JM. 2001. The biological reality of species: gene flow, selection, and collective evolution. Taxon 50: 47–67. [Google Scholar]
- Rosenberg NA. 2004. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes 4: 137–138. [Google Scholar]
- Sakai A. 1971. Freezing resistance of relicts from the Arcto-Tertiary flora. New Phytologist 70: 1199–1205. [Google Scholar]
- Shi Y. 1998. Evolution of the cryosphere in the Tibetan Plateau, China, and its relationship with the global change in the mid-Quaternary. Journal of Glaciology and Geocryology 20: 197–208. (in Chinese with English abstract) [Google Scholar]
- Silba J. 1996. A new species of Pseudotaxus Cheng (Taxaceae) from China. Phytologia 81: 322–328. [Google Scholar]
- Soltis DE, Bell CD, Kim S, Soltis PS. 2008. Origin and early evolution of angiosperms. Annals of the New York Academy of Sciences 1133: 3–25. [DOI] [PubMed] [Google Scholar]
- Stankowski S, Sobel JM, Streisfeld MA. 2015. The geography of divergence with gene flow facilitates multitrait adaptation and the evolution of pollinator isolation in Mimulus aurantiacus. Evolution 69: 3054–3068. [DOI] [PubMed] [Google Scholar]
- Steinbauer MJ, Field R, Grytnes JA, et al. 2016. Topography-driven isolation, speciation and a global increase of endemism with elevation. Global Ecology and Biogeography 25: 1097–1107. [Google Scholar]
- Su Y, Wang T, Ouyang P. 2009. High genetic differentiation and variation as revealed by ISSR marker in Pseudotaxus chienii (Taxaceae), an old rare conifer endemic to China. Biochemical Systematics and Ecology 37: 579–588. [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne RF. 1999. Eastern Asia as a living museum for archaic angiosperms and other seed plants. Taiwania 44: 413–422. [Google Scholar]
- Tian S, Lei SQ, Hu W, et al. 2015. Repeated range expansions and inter-/postglacial recolonization routes of Sargentodoxa cuneata (Oliv.) Rehd. et Wils. (Lardizabalaceae) in subtropical China revealed by chloroplast phylogeography. Molecular Phylogenetics and Evolution 85: 238–246. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, et al. 2012. Primer3–new capabilities and interfaces. Nucleic Acids Research 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IJ. 2013. Examining the full effects of landscape heterogeneity on spatial genetic variation: a multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution 67: 3403–3411. [DOI] [PubMed] [Google Scholar]
- Wang IJ, Bradburd GS. 2014. Isolation by environment. Molecular Ecology 23: 5649–5662. [DOI] [PubMed] [Google Scholar]
- Wang K, Yang Y. 2007. Taxonomic study on Pseudotaxus (Taxaceae). Acta Phytotaxonomica Sinica 45: 862–869. (in Chinese with English abstract) [Google Scholar]
- Wang L, Abbott RJ, Zheng W, Chen P, Wang Y, Liu J. 2009. History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Molecular Ecology 18: 709–721. [DOI] [PubMed] [Google Scholar]
- Wang T, Su YJ, Ouyang PY, et al. 2006. Using RAPD markers to detect the population genetic structure of Pseudotaxus chienii (Taxaceae), an endangered and endemic conifer in China. Acta Ecologica Sinica 26: 2313–2321. (in Chinese with English abstract) [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Whittaker RJ, Fernández-Palacios JM. 2007. Island Biogeography: Ecology, Evolution, and Conservation (second edition). New York: Oxford University Press. [Google Scholar]
- Willyard A, Syring J, Gernandt DS, Liston A, Cronn R. 2007. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for Pinus. Molecular Biology and Evolution 24: 90–101. [DOI] [PubMed] [Google Scholar]
- Woerner AE, Cox MP, Hammer MF. 2007. Recombination-filtered genomic datasets by information maximization. Bioinformatics 23: 1851–1853. [DOI] [PubMed] [Google Scholar]
- Won YJ, Hey J. 2005. Divergence population genetics of chimpanzees. Molecular Biology and Evolution 22: 297–307. [DOI] [PubMed] [Google Scholar]
- Wright S. 1949. The genetical structure of populations. Annals of Human Genetics 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wu Z, Sun H, Zhou Z, Peng H, Li D. 2007. Origin and differentiation of endemism in the flora of China. Frontiers of Biology in China 2: 125–143. [Google Scholar]
- Xu T, Abbott RJ, Milne RI, et al. 2010. Phylogeography and allopatric divergence of cypress species (Cupressus L.) in the Qinghai-Tibetan Plateau and adjacent regions. BMC Evolutionary Biology 10: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XT, Yang Y, Wang LS. 2008. Geographic distribution and potential distribution estimation of Pseudotaxus chienii. Journal of Plant Ecology 32: 1134–1145. [Google Scholar]
- Ying TS, Zhang YL, Boufford DE. 1993. The Endemic Genera of Seed Plants of China. Beijing: Science Press. [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- Zhang YH, Wang IJ, Comes HP, Peng H, Qiu YX. 2015. Contributions of historical and contemporary geographic and environmental factors to phylogeographic structure in a Tertiary relict species, Emmenopterys henryi (Rubiaceae). Scientific Reports 6: 24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YF, Abbott RJ, Jiang ZY, Du FK, Milne RI, Liu JQ. 2010. Gene flow and species delimitation: a case study of two pine species with overlapping distributions in southeast china. Evolution 64: 2342–2352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.