Abstract
Understanding the mechanisms underpinning cellular responses to microenvironmental cues requires tight control not only of the complex milieu of soluble signaling factors, extracellular matrix (ECM) connections and cell-cell contacts within cell culture, but also of the biophysics of human cells. Advances in biomaterial fabrication technologies have recently facilitated detailed examination of cellular biophysics and revealed that constraints on cell geometry arising from the cellular microenvironment influence a wide variety of human cell behaviors. Here, we create an in vitro platform capable of precise and independent control of biochemical and biophysical microenvironmental cues by adapting microcontact printing technology into the format of standard 6- to 96-well plates to create MicroContact Printed Well Plates (μCP Well Plates). Automated high-content imaging of human cells seeded on μCP Well Plates revealed tight, highly consistent control of single-cell geometry, cytoskeletal organization, and nuclear elongation. Detailed subcellular imaging of the actin cytoskeleton and chromatin within live human fibroblasts on μCP Well Plates was then used to describe a new relationship between cellular geometry and chromatin dynamics. In summary, the μCP Well Plate platform is an enabling high-content screening technology for human cell biology and cellular engineering efforts that seek to identify key biochemical and biophysical cues in the cellular microenvironment.
Keywords: High-content imaging, Microcontact printing, Actin cytoskeleton, Cell geometry, Chromatin dynamics, Microenvironment High-throughput screening
1. Introduction
Controlling human cell behavior remains an important goal for many in the regenerative medicine, cancer, and stem cell biology fields [1]. Key biophysical cues can be transmitted from the cell exterior to interior in a variety of ways involving integrins, cadherins, focal adhesion kinase, SUN and KASH nucleoskeleton proteins, YAP/TAZ transcription factors and many other proteins [2–10]. In particular, cellular geometry has been shown to influence many cell behaviors, including proliferation [11, 12], apoptosis [11, 13], differentiation [14–16], and even epigenetic reprogramming [17, 18]. Non-invasive tools have been developed to probe the biophysical properties of living cells during these processes [1,19–22]. Together, these studies point to the importance of controlling of cell shape and area with high precision during cell culture. For example, inconsistent differentiation of mesenchymal stem cells has been traced back to variations in cell shape and cytoskeletal tension inherent in standard cell culture methods [23, 24].
Microcontact printing offers a versatile and widely used technique for precise control of cell shape and area at the micron scale [25–27], and is compatible with many complementary mechanical assays [28]. Standard microcontact printing techniques employ small glass coverslips that can be difficult to manipulate by hand. Further, these techniques require individual stamps and substrates to be meticulously cleaned and carefully substrate-coated for each replicate created [11, 29, 30]. As such, it can be difficult to quickly fabricate the replicates necessary to test a wide experimental space, which is an important capability when seeking to understand and engineer the highly multivariate mechanisms underpinning human behavior.
Here we describe the creation of MicroContact-Printed Well Plates (μCP Well Plates) that adapt the spatial control and imaging capabilities of microcontact printed coverslips to the scale and universal compatibility of the standard well plate. Using this platform, we create up to 96 highly-defined, micropatterned arrays in a single step and spatially separate them by the wells of a tissue culture plate. In this format, each stage of a given experiment – including sample preparation, sterilization, cell seeding, cell treatment, and imaging – can be performed in a single, potentially automated step with conventional well plate techniques. This capability not only decreases experimental time and substrate handling, but also lowers the likelihood for inadvertent experimental errors rising from inconsistent stamping techniques or sample mislabeling. A key advantage over prior methods to pattern well plates [31] is the capability of highly consistent patterning at the single-cell level. In addition, the glass bottoms of μCP Well Plates enable flexibility of substrate patterning and functionalization by a wide variety of chemistries [32], as well as confocal and other high-resolution imaging without further handling or mounting of coverslips. We show that μCP Well Plates are suitable for both high-throughput image analysis of thousands of single-cells and high-resolution subcellular assays on living cells. Analyses at both levels are performed on μCP Well Plates with distinct geometric features of equivalent cell attachment areas to explore how the organization of the cytoskeleton mediates the relationship between cell shape, nuclear shape, and dynamics of the nuclear interior.
2. Materials and methods
Fabrication of Multi-Well Alignment Device
A multi-well alignment device for μCP Well Plate fabrication was designed with 3D modeling software (Solidworks 2014, Dassault Systemes) and fabricated from 6061 aluminum with a computer numerical control (CNC) vertical machining center (MiniMill 2, Haas) programmed with computer-aided design/manufacturing (CAD/CAM) software (MasterCAM X7, CNC Software, Inc.). Standard precision leader pins and bushings were acquired from McMaster Carr and used as received to ensure proper alignment. Three cavities were machined into the base at 127.76 mm × 85.48 mm, 116 mm × 77 mm, and 6” diameter dimensions to hold multi-well plates, micropatterned glass, and silicon master molds, respectively.
μCP Well Plate Fabrication
Double-sided adhesive (ARcare 90106) was generously donated by AR Global, Inc. and cut to 116 mm × 77 mm dimensions using a laser cutter (Universal Lasers Systems). Adhesive was placed on well plates using the alignment device and smoothed to remove air pockets. Well bottoms were then cut out using the laser cutter.
116 mm × 77 mm × 0.2 mm glass sheets were purchased at size from Coresix Precision Glass, Inc. A CHA-600 metal evaporator was used to deposit a 3.5 nm layer of titanium (to increase gold adhesion) followed by an 18 nm layer of gold onto one side of each glass sheet. Custom 6” silicon molds were ordered from FlowJEM, Inc. In 24-well plate designs, each well spot contained twelve 10×10 arrays of single-cell features (Supp. Table 1). Plates were designed to contain four well-replicates for each of five feature geometries. Molds were placed into the 6” cavity of the alignment device for casting. Sylgard 184 elastomer (Dow Corning) was used as the stamp material and cast into molds after degassing. Before setting, a transparency sheet and a flat weight were placed on top of the elastomer, spanning the width of the alignment device cavity. This insured consistent height of the stamps after setting, which was necessary to achieve consistent contact with glass sheets during patterning. Stamps were allowed to set overnight at 37°C before being carefully removed from the mold and cut by scalpel around the frame of an appropriate multi-well plate with well-bottoms already removed to ensure proper alignment.
As described previously [27], a thin layer of 2mM alkanethiol ethanol solution (Prochimia Surfaces) was spread evenly over the stamp and allowed to air dry. A gold-coated glass sheet was then placed face down into the alignment device frame, and lifted up by vacuum tooling attached to a lift arm. The stamp was placed face up in the well plate sized frame, and the glass was gently lowered onto it for transfer of micropatterned alkanethiol self-assembled monolayers (SAMs).
Chemical growth of PEG brushes was performed as described previously [27], with minor changes to reflect the larger size of glass sheets as compared to coverslips. Reactions were carried out in airtight 300 mL “Snapware” Pyrex containers (Boston Store). Sheets were placed face up in the containers and just enough PEG reaction solution (20 mL) to cover the surface was added. A solution of 164 mg of L-Ascorbic acid in 1.82 mL of deionized water was added to the container to initiate the reaction. Ambient air was flushed from the container with a nitrogen stream and the solution was well mixed before incubating at room temperature for 16 hours. Next, patterned sheets were removed from the solution with a tweezers and rinsed with MilliQ water and ethanol, followed by a two hour incubation in 70% ethanol on a room temperature shaker to remove residual reaction solution. Sheets were then dried and placed onto the multi-well plate frame using the alignment device in the same manner as during stamping. After sealing the glass sheets to the well plate frame by applying pressure by hand, μCP-Well Plates were briefly immersed in a 70% ethanol bath and allowed to air dry in a sterile tissue culture hood.
Fluorescent modification of PEG Brushes
Conjugation of fluorophores to PEG brushes was performed as descried previously [27] with volumes scaled just enough to cover glass sheets inside 300 mL Pyrex containers (~20mL).
Generation and Culture of H2B-LA Cell Line
The H2B-LA human fibroblast line was derived from the previously reported C1.2 line [18]. Gene-editing and insertion occurred in an induced pluripotent stem cell (iPSC) state to allow for clonal isolation and growth. H2B-mCherry was integrated into the C1.2-iPSC genome via CRISPR/Cas9 gene editing. The H2B-mCherry plasmid was generated by cloning the H2B-mCherry sequence (Addgene # 20972) into the EGFP sequence of the AAV-CAGGS-EGFP plasmid (Addgene #22212). This plasmid was electroporated with Cas9 and gRNA plasmids as described previously [18]. A LifeAct-GFP plasmid (Addgene #51010) was packaged in lentiviral particles by HEK-293T cells via Lipofectamine 2000 (Life Technologies) transfection with third-generation packaging vectors and harvested after 72 hours. Harvested lentiviral solutions were passed through a 0.45 μm filter and exposed to C1.2 iPSCs overnight before rinsing. iPSC lines harboring both labels were purified through puromycin selection and clonal isolation followed by fluorescent imaging and flow cytometry using a C6 Accuri (BD) to confirm expression.
H2B-LA iPSCs were then differentiated into fibroblasts using the embryoid body (EB) method [33]. EBs were created using the Aggrewell (Stem Cell Technologies) system per the manufacturer’s protocol. EBs were then cultured in non-adherent cell culture plates (Corning Inc.) suspended in 4 mL per well of “EB Medium” consisting of 80% Iscove’s Modified Dulbecco’s Medium (IMDM, Life Technologies) with 20% Fetal Bovine Serum (Life Technologies). After 4 days in suspension, aggregates were transferred onto Matrigel-coated 24-well plates and maintained in “Fibroblast Medium” consisting of DMEM-high glucose (Life Technologies) supplemented with 10% Fetal Bovine Serum (Life Technologies), 1 mM L-glutamine (Life Technologies), and 1% Penicillin/Streptomycin (Life Technologies). After approximately 9 days in culture, cells had proliferated enough to permit expansion. Cells were passaged by washing cells with PBS (Life Technologies) followed by a 5-minute incubation in trypsin (Life Technologies) at 37°C. Cells were then centrifuged, resuspended in fresh Fibroblast Media, and seeded onto tissue culture polystyrene plates (Fisher Scientific) coated with a 0.1% Gelatin A (Sigma) solution. Most cells had taken on a fibroblast-like morphology after three to five passages.
Once derived, fibroblasts were maintained in Fibroblast Medium in 5% CO2 at 37°C on gelatin-coated polystyrene plates. Media was changed once every two days, and cells were passaged approximately every four days (at 90% confluency) with trypsin as described above.
Immunofluorescence, Proliferation, and Apoptosis Assays
F-Actin stains (used in Figure 1 only) were performed at room temperature using 10 nM Phalloidin-TRITC and 0.5% Triton-X 100 (Sigma) in PBS for 30 minutes at room temperature after fixing for 15 minutes with 4% Paraformaldahyde in PBS (Sigma).
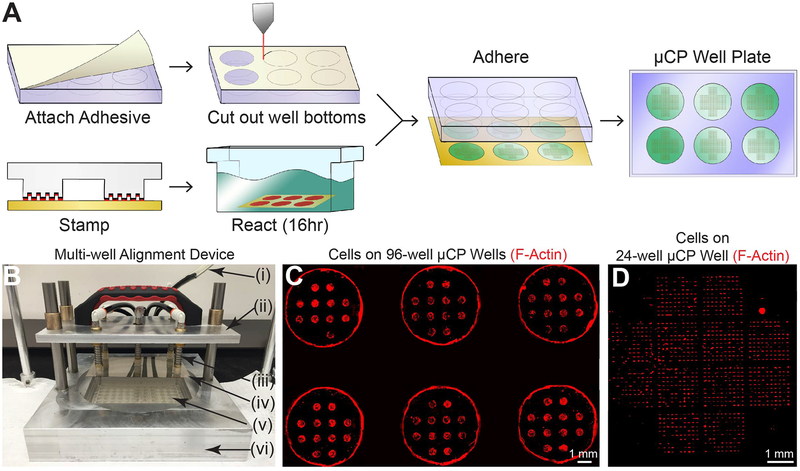
Figure 1. μCP Well Plates.
A) Workflow for creation of μCP Well Plates. B) Multi-well alignment device for μCP Well Plate assembly. Pictured: vacuum line (i), lift arm for vacuum tooling (ii), spring-leveled vacuum cup tooling (iii), micropatterned glass sheet (iv), 96-well plate with attached adhesive and cut-out well bottoms (v), and aluminum base with machined cavities designed to fit various μCP Well Plate components (vi). C) Cells on a 96-well μCP Well Plate. 6 wells of a fully patterned 96-well plate containing twelve circular features (300 μm radius) per well are shown. Human fibroblasts were seeded 72 hours prior to TRITC-phalloidin staining and imaging. Fluorescent signal at edge of wells is due to auto-fluorescence of tissue culture plastic at well edges, not cells at these locations. Inter-well areas are masked for clarity. D) Cells on 24-well μCP Well Plate. One well of a 24-well plate containing twelve 10×10 single-cell feature arrays per well is shown. Human fibroblasts were seeded 16 hours prior to TRITC-phalloidin staining and imaging.
Click-iT EdU and TUNEL assays (Life Technologies) for measuring proliferation and apoptosis, respectively, were performed per manufacturer instructions. Cells were exposed to EdU for a 12 hour period before fixing, beginning 4 hours after cell seeding.
Automated Imaging
Automated imaging was performed using a Nikon Eclipse Ti epifluorescence microscope and NIS Elements software. One full-well, multichannel stitched image per well was recorded at 10x magnification using the Nikon Perfect Focus capability to eliminate drift in the z-direction.
Image Analysis
To enable accurate measurement of fluorescent intensity across full-well images, all 10x stitched images (Figures 1–3) were corrected for non-uniform illumination across individual frames using CellProfiler software, using a 50 pixel block size and a 200 pixel smoothing filter. Illumination correction leads to increased accuracy in fluorescence intensity measurements by removing uneven illumination across the image, potentially due to imperfect optics. This standard procedure for stitched images removes the artificial “grid” appearance of many stitched images. A custom CellProfiler pipeline was then used to filter and analyze appropriately patterned cells and nuclei. The pipeline is available upon request. Image analysis of fluorescently-conjugated PEG surfaces was performed using Fiji software [34]. Feature area was calculated by thresholding into binary images and analyzing all features with fluorescent intensities below the threshold.
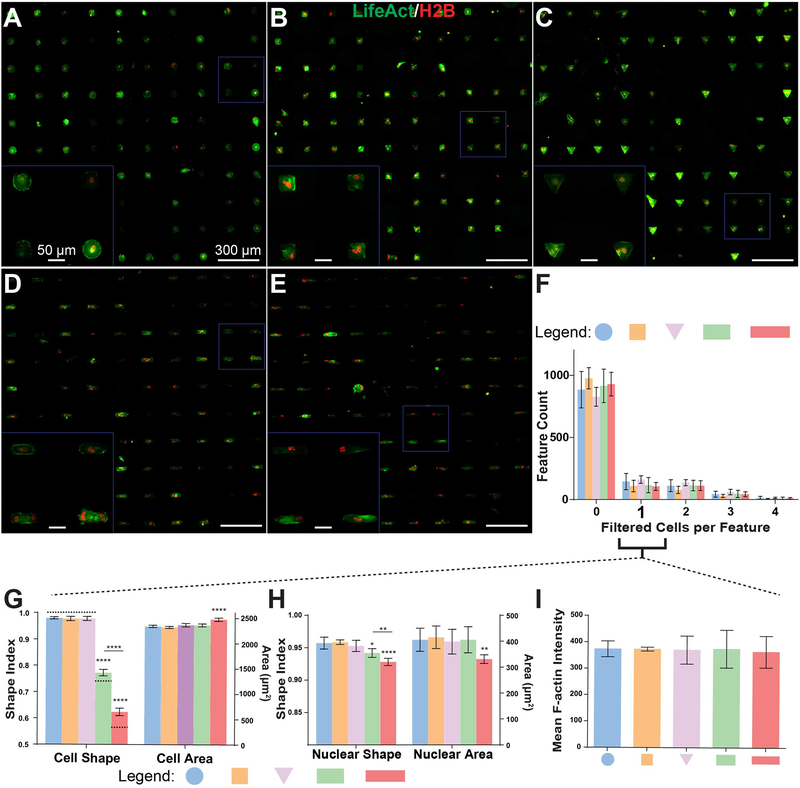
Figure 3. Cell attachment and patterning on μCP Well Plates.
A-E) 20x stitched images of H2B-mCherry- and LifeAct-GFP-labeled human fibroblast lines (H2B-LA) attached to circular (A), square (B), triangular (C), compact rectangular (D), and elongated rectangular (E) single-cell features. Cells were seeded 16 hours prior to imaging and analysis. F) Distribution of cells contained by each feature after automated removal of cells that do not meet user-specified area and shape criteria. “0” filtered cells columns correspond to the sum of both empty features and features containing cells eliminated by filtering. G-H) SI (left Y-axis) and area (right Y-axis) measurements for cell bodies (H) and nuclei (I) of features containing a single cell after filtering. Dashed bars in (H) indicate theoretical SI values for each shape. I) Mean LifeAct fluorescent intensity (averaged per-pixel across filtered LifeAct features) of patterned single-cells. All error bars denote 95% confidence intervals. Unless otherwise denoted, asterisks indicate p values from unpaired Student’s t-test compared to data from circular features (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Each bar represents data aggregated from 5 to 8 wells of a 24-well plate across two biological replicate experiments.
Order Parameter
Maximum-intensity projections of 1.5 μm or 1 μm z-stacks (1 μm for Blebbistatin trials, 1.5 μm for all other trials) obtained using a Nikon A1R confocal microscope were fed into custom Matlab code [35] to calculate a metric that quantifies the extent of F-actin alignment, what is termed an “order parameter” value for each cell. Briefly, a series of elongated Laplace of Gaussian filters was used to create a maximum response image. The image was then broken down into a grid of small overlapping windows (~30×30 pixels, spaced 5 pixels apart), which were convolved with a Gaussian filter before calculating the two-dimensional fast Fourier transform of each window. A series of line scans then determined the direction of skew in the transform and correlations in alignment were determined by comparing the direction of alignment of a given window to its neighbors via averaging the second Legendre polynomial. This value was then averaged across the entire image.
Fluorescence Recovery After Photobleaching (FRAP)
FRAP experiments were performed using a Nikon A1R confocal microscope. ~1 μm diameter spots were photobleached by 1 second exposure to a 405 nm wavelength laser at 100% power and 16 frames per second. Fluorescent images were then collected at 5% laser power every five seconds for the first 4 minutes, and every 15 seconds for an additional four minutes. Stacks were aligned using the StackReg ImageJ plugin [36] and intensity measurements of the photobleached area and whole nucleus were measured at each time point. Mobile fraction was calculated by normalizing the intensity of the photobleached region to the mean intensity of the entire nucleus as well as the intensities of the photobleached region immediately before and after bleaching to obtain a 0–1 scale. Data was then fit to a one-phase association curve of the form
by nonlinear least-squares regression where P represents the plateau value, t represents time, and k is a constant.
Statistics
All regressions, tests for significance, and confidence intervals were calculated by GraphPad software. Student’s t-tests were unpaired and two-tailed (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). p values for linear regression best-fit lines were calculated in GraphPad from F-tests and indicates the probability that the slope of the regression line is equal to zero.
3. Results
Construction and Assembly of μCP Well Plates
The workflow for creation of μCP Well Plates is described in Figure 1A. To generate the bottom surface of these plates, glass sheets were first cut to the dimensions of the bottom of standard multi-well plates and coated with a thin layer of gold (18 nm) on one side in preparation for alkanethiol-based microcontact printing [37]. Next, patterned polydimethylsiloxane (PDMS) stamps were cast from custom 6” diameter silicon molds designed to create regular arrays of single-cell features, similar to those described in a recent report [30]. Arrays were spaced with dimensions equivalent to standard multi-well tissue culture plates. These stamps are customizable to any well plate format, and we focused on 6-,12-, 24- and 96-well plates (Supp. Table 1). PDMS was subsequently allowed to set overnight and cut by scalpel to well plate dimensions.
Microcontact printing was performed on the gold-coated glass sheets as described previously [27, 38]. Briefly, self-assembled alkanethiol monolayers were transferred to gold-coated glass sheets via a PDMS stamp, after which the sheets were immersed in a polyethylene glycol (PEG) reaction solution overnight to allow for growth of hydrophilic PEG chains surrounding the designed single-cell features (Figure 1A). To prepare the well plate, medical grade double-sided adhesive sheets were cut to size and attached to the bottom of standard tissue culture plates. A laser cutter was then used to remove well bottoms by cutting through both tissue culture plastic and adhesive. Finally, patterned glass substrates were attached to the multi-well plates via the double-sided adhesive. To ensure precise control of alignment during this process, a custom multi-well alignment device (Figure 1B) was constructed and utilized to align all components of the μCP Well Plate during stamping and assembly. Plates were sterilized by full immersion in 70% ethanol to remove any potential contaminants prior to cell culture. These techniques were applied to standard multi-well plates from 6- to 96-wells with various substrate patterns (Figure 1C). For 24-well μCP plates, 1,200 single-cell features were generated per well (Figure 1D), totaling 28,800 features per plate (Supp. Table 1), or on the order of 32x more features and 24x more conditions available compared to recent reports of similar feature arrays [30, 39].
High Fidelity Patterning of Single-Cell Microarrays on μCP Well Plates
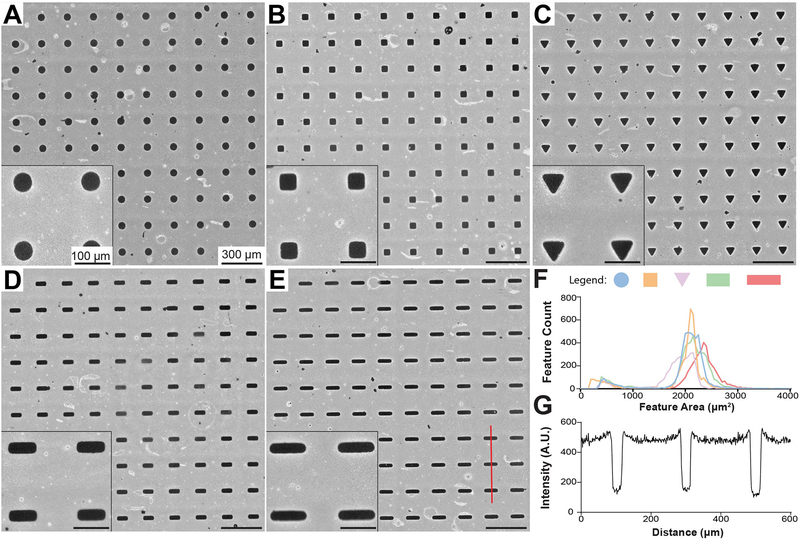
To investigate cellular responses to cell shape patterning, all features in our μCP Well Plates were designed to cover equivalent surface areas. All features in a given well were of the same shape, but separate wells contained features of one of five distinct geometries – hereafter denoted as circular, square, triangular, compact rectangular (1:2.5 aspect ratio), or elongated rectangular (1:4 aspect ratio) features (Figure 2A–E). Patterns were visible by naked eye and under phase microscopy after microcontact printing. However, to increase contrast and allow for more detailed analysis of PEG concentration and distribution, PEG brushes were functionalized with biotin and fluorescently labeled by an avidin-conjugated fluorescent probe [27]. All features were distributed near the intended surface area of 2000 μm2 (Figure 2F). Elongated rectangular features covered a slightly larger (~10% on average) surface area than the other geometries, which is a result of the desired minor axes of these geometries (15 μm) reaching the practical resolution of the photomask printer used to create photoresist for silicon master molds. Visible defects comprised <15% of all features after thresholding and were typically much smaller than the designed features (Figure 2F, minor peak on left) allowing for straightforward filtering of poorly-patterned cells by image analysis (see next section). Clear, sharp transitions were obtained at feature borders and regions between features had consistently high levels of PEG attachment (Figure 2G).
Figure 2. Analysis of Micropattern Fidelity in μCP Well Plates.
A-E) 10x stitched images of circular (A), square (B), triangular (C), compact rectangular (D), and elongated rectangular (E) single-cell features. Surface-conjugated PEG brushes were biotinylated and fluorescently labeled by an avidin-conjugated fluorescent probe to provide contrast. White areas indicate high fluorescence from conjugated PEG brushes, whereas dark areas indicate no fluorescence of the underlying substrate (gold-coated glass). F) Area distribution of single-cell features. Images were thresholded and shape properties were automatically calculated by ImageJ software. G) Intensity profile of line scan (red) across three features in (E).
μCP Well Plates Enable Precise Single-Cell Patterning and Automated High-Content Imaging
To enhance cell attachment to the exposed gold portions of the surface, a protein solution was added to each well. Single-cell features are the only portions of assembled μCP wells not occupied by anti-fouling PEG brushes [40], and we adsorbed a Matrigel coating to the gold portions for all subsequent cell culture experiments. Matrigel is a mixture of ECM proteins and associated molecules such as proteoglycans and growth factors produced by mouse sarcoma cells [41]. Although use of this complex substrate could lead to increased variability with regards to cellular organization or behavior, we reasoned that it would also engage a wide range of integrins, resulting in more stable cell attachment. Matrigel can easily be replaced by any protein able to adsorb to gold surfaces for applications requiring completely defined culture conditions. We have successfully used a recombinant vitronectin solution for this purpose (data not shown).
To allow for real-time imaging of actin and nuclear dynamics in live cells, a human fibroblast line harboring stable genomic inserts encoding Histone 2B (H2B)-mCherry fusion protein and LifeAct-GFP [42] (H2B-LA) were derived via CRISPR/Cas9 gene editing and lentiviral transduction. LifeAct is a 17 amino acid peptide derived from the yeast protein Abp 140p that, when linked to a fluorescent protein such as GFP, enables visualization of the actin cytoskeleton. The LifeAct peptide is not covalently bound to globular actin monomers (G-actin), and LifeAct binding has no discernible effect on actin dynamics [42, 43]. Gene-modified cells were added in suspension and allowed to attach to patterns for 14–18 hours before automated fluorescence imaging (Figure 3A–E).
An automated image analysis workflow was created using CellProfiler software [44] in order to assess attachment and patterning of cells (Supp. Figure 1A). Briefly, full-well stitched images were fed into CellProfiler, which filtered actin regions (cells or cell clusters) based on their spread area and other shape characteristics and removed poorly patterned or incompletely spread cells from the data set (Supp. Figure 1B). At this point, nuclei within remaining LifeAct regions were identified and analyzed for area, shape, and intensity characteristics. The number of nuclei contained within a filtered region of high LifeAct intensity was then used to determine the number of cells occupying a given feature (Supp. Figure 1C). At a seeding density of 15,000 cells per well for a 24-well μCP Plate, 100–200 correctly patterned single cells were routinely identified per well (Figure 3F), corresponding to approximately 10% of all available patterned features. Image analysis revealed that this strict filtering protocol resulted in high quality cell populations with highly consistent shape and intensity properties between replicates (Fig. 3G–I, Supp. Figure 2A–E). Minor differences in nuclear area and shape were detected based on the number of cells contained in each feature (Supp. Figure 2F–G) in addition to obvious changes in cell shape, so all subsequent analysis was performed exclusively on filtered single cells. We rejected ~90% of features identified via CellProfiler, which reflects mostly autofluorescent or apoptotic cell debris with a minority of cell clusters, poorly patterned cells, and regions of a particular well with artificially high fluorescence intensities (Supp. Figure 1B, very small purple features). In the 14–18 hour span between cell seeding and imaging, proliferation and apoptosis rates were below 5% for all geometries (Supp. Figure 2H–I) and therefore were not taken into account during subsequent analyses.
Shape characteristics of cells seeded on geometric arrays were characterized using the shape index (SI) formula [45]
| 1) |
where A represents spread area of the cell or nucleus and P represents the shape perimeter. Because perimeter measurements of shape borders can be affected greatly by image pixelation or irregular protrusions such as blebs and filopedia, we directly measured major and minor axes of each respective cell body and nucleus and estimated SI from the area and perimeter values of the resulting ellipse (Supp. Figure 3A). Shape index assessed in this manner is a measure of cell elongation, with a value of one corresponding to a perfect circle and values approaching zero with elongation. SI of cell bodies very nearly matched their theoretical predictions for each respective shape (Figure 3G, left, dashed lines). Nuclei were in turn elongated in a similar manner to their respective cell bodies, although to a lesser degree (Figure 3H), in agreement with previous results [17, 46, 47].
Spread area of patterned single-cells was constant with the exception of a <10% increase for elongated rectangular features (Figure 3G, right), mirroring results from feature characterization prior to cell attachment. Nuclei within cells patterned on elongated rectangular features did not proportionately increase in area, but had significantly lower projected areas than those of other features (Figure 3H, right), a phenomenon which has been previously reported for elongated endothelial cells [47].
Significant changes in integrated actin intensity were observed between compact and elongated geometries when analyzing 60x confocal stacks (Supp. Figure 3B). A non-significant decrease in LifeAct intensity was detected in all measured shapes after treatment with blebbistatin, a Myosin II ATPase activity inhibitor that relieves actomyosin tension and can disrupt F-actin networks [48, 49] (Supp. Figure 3C).
High-Content Imaging of Subcellular Organization
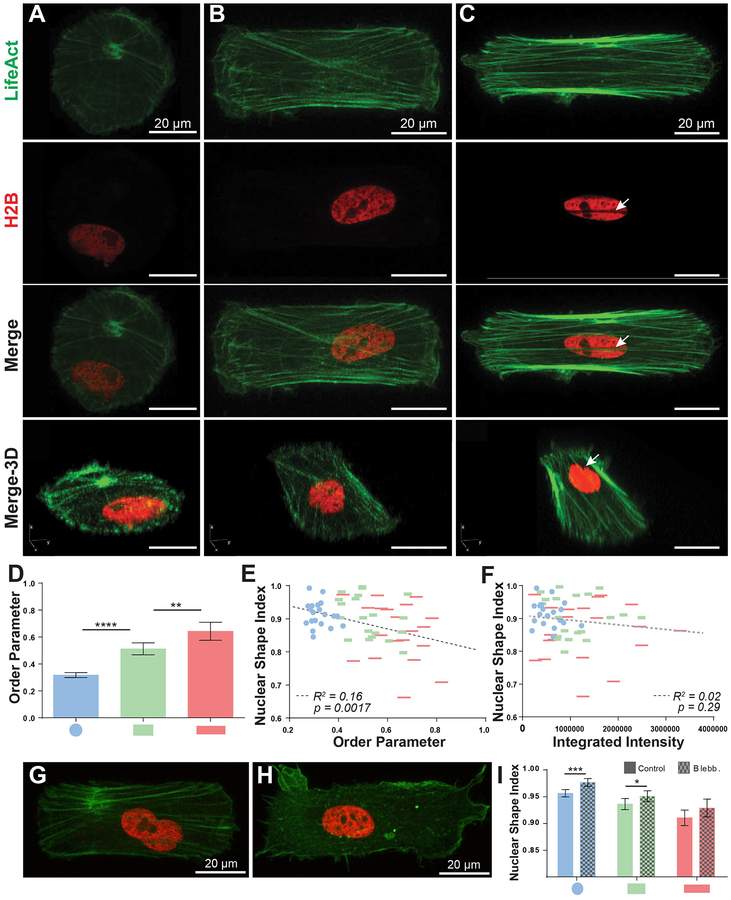
We hypothesized that actin may be involved in the observed nuclear shape dynamics in our system, as actin fibers have been reported to exert forces on the nucleus in a variety of manners, especially through actin structures directly apical to the nucleus [7, 45, 46, 47–51]. Using stacks of confocal images of single cells at this higher 60x magnification we were able to discern actin fiber alignment, which is known to correlate with cell elongation [14, 46, 49] (Figure 4A–C). We used the previously reported Order Parameter metric [35] to quantify the degree of actin alignment on a scale from 0–1, with 1 being the most aligned. We focused on circular, compact rectangular, and elongated rectangular geometries and found that order parameter increased with elongation of cell shape (Figure 4D). These results are consistent with previously reported relationships between cellular elongation and actin fiber alignment [14, 46, 49]. Furthermore, actin alignment significantly correlated to nuclear shape (Figure 4E). The p value displayed here is calculated from an F test and corresponds to the probability that the slope of the regression line is equal to zero, (i.e., the two variables are unrelated). However, regressions within each measured geometry did not reach statistical significance (Supp. Figure 4A). Nuclear shape did not significantly correlate to integrated actin intensity (Figure 4F, Supp. Figure 4B). We then measured nuclear SI of patterned single-cells after disrupting F-actin networks with blebbistatin (Figure 4G–I) [48, 49]. We detected a significant increase in nuclear SI for circular and compact geometries upon blebbistatin treatment compared to untreated controls (Figure 4I). While a less significant increase occurred in the elongated case (p=0.12), these results together suggest a complex connection between the actin cytoskeleton and nuclear shape. Notably, we observed the presence of nuclear indentations along the length of F-actin stress fibers in elongated cells (Figure 4C, arrowheads). These indentations were recently shown to be enriched for LINC complexes [50] that connect the nuclear lamina to the cytoskeleton [51–53] and highlight the influence of actin stress fibers on the nucleus.
Figure 4. Actin and nuclear organization of patterned single-cells.
A-C) 60x confocal maximum-intensity projection images of H2B-LA single-cells patterned into circular (A), compact rectangular (B), and elongated rectangular (C) geometries. Note that geometries are displayed at slightly different zoom levels. Arrowheads in (C) denote a nuclear indentation site formed by actin stress fibers. D) Order parameter of patterned single-cells (n=20). Measurements were taken from maximum-intensity projections of 10–15 1 μm z-stacks (as pictured in A-C). E-F) Single-cell scatter plots of order parameter (E) or LifeAct integrated intensity (F) vs. nuclear shape index. Dashed lines represents linear regression best-fit lines. p values for best-fit lines were calculated from F-tests and indicate the probability that the slopes of the regression lines are equal to zero. G-H) Representative images of untreated control (G) and blebbistatin-treated (H) (50 μM, 60 min exposure) cells in compact rectangular geometries. I) Nuclear shape analysis of control or blebbistatin-treated patterned cells (n>100). All error bars denote 95% confidence intervals. Asterisks indicate p values from unpaired Student’s t-test (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
Chromatin Dynamics and Cell Geometry
We next investigated whether the observed changes in cell shape, actin organization, and/or nuclear shape could lead to changes in chromatin dynamics. Histone 2B is a member of the core nucleosome [54], and fluorescently tagged H2B has been well-characterized and remains bound to DNA throughout the cell cycle, although small fractions are continually exchanged [55, 56]. Therefore, the mobility of H2B within the nucleus is a measure of (1) the mobility of the core nucleosome itself; (2) the portion of the overall H2B pool bound to DNA (its affinity); or, (3) a combination of both [57]. Changes in either of these properties reflect changes in the epigenome, which can govern gene expression and cell behavior. Fluorescence Recovery After Photobleaching (FRAP) experiments were used to assess the mobility of fluorescently tagged molecules by measuring exchange rates of fluorescent molecules in a given area. As the minutes timescale used for these experiments is far below that needed to synthesize and localize significant amounts of new fluorescent protein to the photobleached region, any measured recovery is a result of the fluorescent molecule moving into the photobleached region from the surrounding area [58]. The plateau of the fluorescent recovery curve over a given time scale is known as the “Mobile Fraction” of the molecule of interest [59, 60].
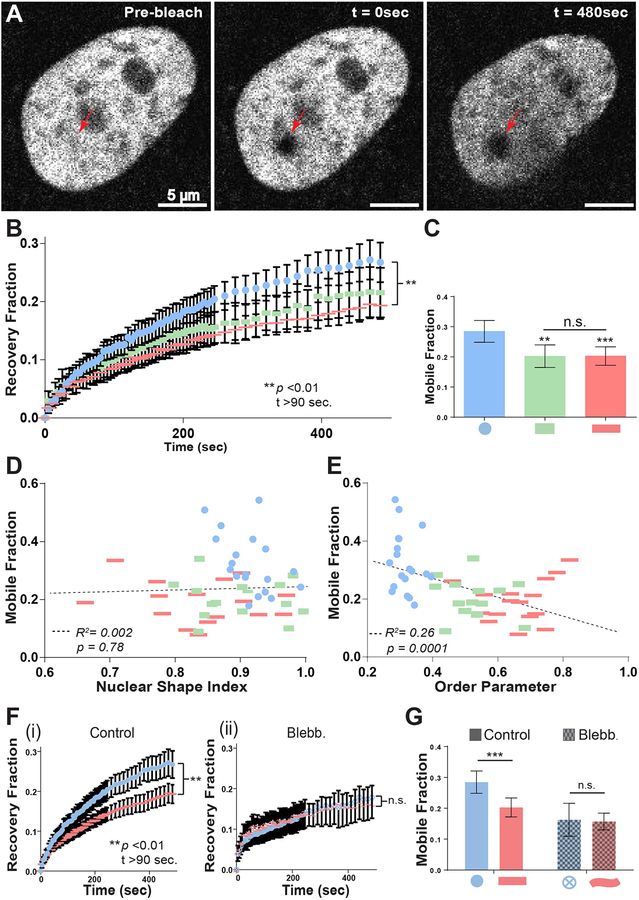
To investigate chromatin dynamics in our system, ~1 μm diameter regions in the nuclei of patterned single-cells were photobleached by exposure to a high intensity laser beam (100% power) for 1 second followed by normalized intensity measurements over an 8 minute time course (5% power) to track recovery of fluorescence (Figure 5A–B). H2B is present in at least two pools in human cells, one that recovers on the minute timescale and one that recovers over a period of hours [55]. The timescale chosen here focuses on the first pool, and is comparable to that used in similar studies measuring H2B mobility [55, 61]. H2B recovery curves of circular and elongated rectangular single-cells diverged significantly (p<0.01) at time points >90 sec., with the recovery curve for compact rectangular cells lying in between (Figure 5B). Mobile fraction was significantly reduced in both compact (p<0.01) and elongated rectangular geometries (p<0.001) compared to circular geometries (Figure 5C).
Figure 5. FRAP analysis of patterned single-cells.
A) Representative images of H2B-mCherry-labeled nuclei immediately before and after photobleaching, and at the end of the 8-minute recovery period. Arrowheads denote ~1 μm diameter photobleached area. Intensity measurements were collected every five seconds for four minutes following photobleaching, and every 15 seconds for four more minutes thereafter, and normalized to the intensity of the entire nucleus. B) Quantification of fluorescent recovery fraction in nuclei of patterned single-cells. (n=20 for compact rectangles, n=40 for circles and elongated rectangles) C) Mobile fraction of patterned single-cells. Mobile fraction was calculated as the plateau of one-phase association curve fit to the recovery fraction time course. D-E) Single-cell scatter plots of nuclear shape index (D) or order parameter (E) vs. mobile fraction. Dashed lines represent linear regression best-fit lines. p values for best-fit lines were calculated from F-tests and indicates the probability that the slopes of the regression lines are equal to zero. F) Quantification of fluorescent recovery fraction in nuclei of control (i), (n=40) and blebbistatin-treated (ii), (n=20) patterned single-cells. (50 μM, 60–120 min exposure) G) Mobile fraction of control and blebbistatin-treated patterned single-cells. All error bars denote 95% confidence intervals. Asterisks indicate p values from unpaired Student’s t-test (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
We next examined whether actin alignment and/or increased nuclear SI might be responsible for the observed decrease in H2B mobility. Accordingly, single-cell H2B mobile fraction measurements were regressed against either order parameter or nuclear SI measurements. When analyzed in this manner, H2B mobile fraction was not correlated to nuclear shape across different geometries (Figure 5D), although a significant negative correlation was detected when analyzing circular geometries alone (Supp. Figure 4C). In contrast, we detected a highly significant correlation when regressing mobile fraction against order parameter across all geometries (Figure 5E). Regressions within each geometry were not significant (Supp. Figure 4D). Disruption of the actin cytoskeleton using a blebbistatin treatment eliminated the differences in H2B mobility imposed by cell geometry (Figure 5F–G). Surprisingly, blebbistatin treatment resulted in a decrease in H2B mobility (Figure 5F–G), rather than the increase that would be expected by H2B mobility trends with respect to actin alignment and cell shape (decreased elongation ~ decreased actin alignment ~ increased H2B mobility). These results are discussed further below.
4. Discussion
We have presented here the creation of the μCP Well Plate, a novel cell culture system for quantitative imaging and mechanobiological analysis combining the single-cell geometric control, substrate flexibility, and imaging capabilities of microcontact-printed glass coverslips with the multiplexing and compatibility features of standard multi-well plates. μCP Well Plates enable rapid fabrication of many experimental replicates and conditions (>25x more than with comparable coverslip methods) in a single patterning step and high-content analysis of large, uniform single-cell populations. Unused in the current study were the capabilities to independently alter substrate and media components over dozen of conditions in a single plate, which could be of utility in drug screening, functional genomics, and a myriad of similar biotechnology applications.
μCP Well Plates for High-Content Imaging of Live Human Cells
We first demonstrated that μCP Well Plates can be patterned consistently with high precision and geometric control (Figures 1–2), and are well tolerated by human cells (Figure 3A–E). Merged with high content imaging and analysis software, μCP Well Plates were used to identify high quality single-cell populations suitable for rapid, consistent analysis of shape and intensity characteristics (Figure 3G–I). By combining multiple replicate arrays into a single plate, minor differences between populations – such as a reduction in nuclear area in cells on elongated rectangular features (Figure 3H) – can be identified in a single experiment with high confidence.
We then performed high-resolution, sub-cellular analyses of live cells using the μCP Well Plate platform to probe how cytoskeletal organization mediates the relationship between cell shape, nuclear shape, and chromatin dynamics at a single-cell level. Actin became more aligned with increasing cell elongation (Figure 4D). Nuclei within cells with high actin alignment were elongated (Figure 4E), and nuclear elongation was reversed by actomyosin inhibition (Figure 4I). Finally, increasing cell elongation gave rise to a graded decrease in Histone H2B mobility inside the nucleus (Figure 5B). While the relationship between actin and nucleosome mobility is complex and requires further study, the two most highly correlated cellular properties observed in this study across all geometries were actin alignment and H2B mobile fraction (Figure 5E), and differences in H2B mobility based on cell geometry were abolished after actin disruption (Figure 5F–G). To our knowledge, this is the first report of the mobility of a core nucleosome component being altered solely by control of cell shape.
Influence of Actin Cytoskeleton on Chromatin Dynamics
Reduction in nuclear volume in elongated endothelial cells has been shown to cause chromatin condensation [47], and chromatin hyper-condensation induced by osmotic stress or apoptosis stabilizes binding of nuclear proteins, including H2B, to DNA, resulting in decreased fluorescent recovery rates [57, 62]. Thus, aligned actin fibers in elongated cells may impart physical pressure on the nucleus, resulting in reduced nuclear volume, condensed chromatin, and lower mobility of H2B. Importantly, nuclear indentations imposed by F-actin stress fibers in elongated cells can segregate the nucleus and cause chromatin condensation even without gross changes to nuclear shape (Figure 4C, Supp. Figure 5) [50]. This interpretation is supported by recent findings that disrupting F-actin fibers (via small molecules or RNA interference) in randomly oriented cells increases H2B mobility as well as nuclear height [61].
The reduction of H2B mobile fraction after blebbistatin treatment is a surprising result, as blebbistatin relieves nuclear elongation and increases overall nuclear volume [47] – and would therefore be expected to increase H2B recovery rates. Recovery of blebbistatin-treated cells is indeed faster than all circular and rectangular control cells over the first ~60 seconds, but at longer time scales recovery is decreased (Figure 5F). Applied external force has been shown to facilitate movement of small particles throughout the nucleus [63], and reduced chromatin mobility upon blebbistatin treatment has been previously reported [39], in contrast to other inhibitors of actin organization [61]. Chemical interactions between actin fibers, the nuclear lamina, and the nucleoskeleton – such as increased LINC complex concentration at nuclear indentation sites [49] – may also be important drivers of H2B mobility, complicating simple physical interpretations.
Summary and Conclusions
Overall, more work is needed to mechanistically describe the influence of cell geometry and cytoskeletal organization on chromatin dynamics, as within the same geometry we did not observe significant relationships between actin organization and nuclear shape, nor between actin organization and chromatin dynamics. With future work, the tools described here will enable further characterization of the relationships among actin organization, cell shape, chromatin dynamics and cytoskeletal tension by providing a framework to independently control microenvironmental cues and measure subcellular dynamics in live cells at high resolution. Integration of new and emerging molecular tools, such as CRISPR labeling [64, 65], into this platform may permit visualization of these mechanobiological processes at higher resolution.
In summary, μCP Well Plates offer a flexible cell culture platform that enables mechanistic, high-throughput study of subcellular components. This platform expands our ability to precisely and independently control microenvironmental cues such as substrate composition, cell geometry, and media composition, and the platform is importantly compatible with advances in high-content imaging and single-cell analysis. These capabilities open up new frontiers in screening for molecules that could influence a multitude of human cell behaviors.
Supplementary Material
5. Acknowledgements
We acknowledge generous financial support from the National Science Foundation (CBET-1350178), National Institute for Health (NHGRI 5T32HG002760), Wisconsin Institute for Discovery (K.S.), and the Society in Science Foundation (K.S.). R.S.A. holds an Innovation in Regulatory Science Award for the Burroughs Wellcome Fund. We also would like to thank all members of the Saha lab and WID BIONATES theme for advice and support throughout this project. We finally thank Dr. Patrick Oakes for sharing his code and expertise in assessing actin alignment using image analysis in Matlab and Gavin Knight for his assistance in microcontact printing.
Abbreviations:
- μCP Well Plate
microcontact printed multiwell plate
- ECM
extracellular matrix
- FRAP
fluorescence recovery after photobleaching
- H2B
Histone 2B
- H2B-LA
H2B-mCherry and LifeAct-GFP transgenic human fibroblast line
- PDMS
polydimethylsiloxane
- PEG
polyethylene glycol
- SI
shape index
Footnotes
Conflict of interest
The authors declare no financial or commercial conflicts of interest.
7 References
- 1.Iskratsch T, Wolfenson H, and Sheetz MP, Appreciating force and shape - the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol, 2014. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giancotti FG and Ruoslahti E, Integrin signaling. Science, 1999, 285,1028–1033. [DOI] [PubMed] [Google Scholar]
- 3.Lee SE, Kamm RD, and Mofrad MR, Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech, 2007, 40, 2096–2106. [DOI] [PubMed] [Google Scholar]
- 4.Vogel V and Sheetz M, Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol, 2006, 7, 265–275. [DOI] [PubMed] [Google Scholar]
- 5.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, et al. , Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell, 2006, 127, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra SK, Hanson DA, and Schlaepfer DD, Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol, 2005, 6, 56–68. [DOI] [PubMed] [Google Scholar]
- 7.Maniotis AJ, Chen CS, and Ingber DE, Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. U S A, 1997, 94, 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starr DA and Fridolfsson HN, Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell. Dev. Biol, 2010, 26, 421–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low BC, Pan CQ, Shivashankar GV, Bershadsky A et al. , YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett, 2014, 588, 2663–2670. [DOI] [PubMed] [Google Scholar]
- 10.Aragona M, Panciera T, Manfrin A, Giulitti S et al. , A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell, 2013, 154, 1047–1059. [DOI] [PubMed] [Google Scholar]
- 11.Chen CS, Mrksich M, Huang S, Whitesides GM et al. , Geometric control of cell life and death. Science, 1997, 276, 1425–1428. [DOI] [PubMed] [Google Scholar]
- 12.Qutub AA and Popel AS, Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst. Biol, 2009, 3. doi: 10.1186/1752-0509-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Zheng W, Xie Y, Gong P et al. , Tissue-specific mechanical and geometrical control of cell viability and actin cytoskeleton alignment. Sci. Rep, 2014, 4. doi: 10.1038/srep06160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalby MJ, Gadegaard N, and Oreffo RO, Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat. Mat, 2014. 13, 558–569. [DOI] [PubMed] [Google Scholar]
- 15.McBeath R, Pirone DM, Nelson CM, Bhadriraju K et al. , Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell, 2004, 6, 483–495. [DOI] [PubMed] [Google Scholar]
- 16.Kilian KA, Bugarija B, Lahn BT, and Mrksich M Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U S A, 2010. 107, 4872–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downing TL, Soto J, Morez C, Houssin T et al. , Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mat, 2013, 12, 1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordie T, Harkness T, Jing X, Carlson-Stevermer J et al. , Nanofibrous electrospun polymers for reprogramming human cells. Cell Mol. Bioeng, 2014, 7, 379–393. [Google Scholar]
- 19.Afrin R, Yamada T, and Ikai A, Analysis of force curves obtained on the live cell membrane using chemically modified AFM probes. Ultramicroscopy, 2004, 100, 187–195. [DOI] [PubMed] [Google Scholar]
- 20.Ghibaudo M, Saez A, Trichet L, Xayaphoummine A et al. , Traction forces and rigidity sensing regulate cell functions. Soft Matt, 2008, 4, 1836–1843. [Google Scholar]
- 21.Chiou PY, Ohta AT, and Wu MC, Massively parallel manipulation of single cells and microparticles using optical images. Nature, 2005, 436, 370–372. [DOI] [PubMed] [Google Scholar]
- 22.Han SJ, Bielawski KS, Ting LH, Rodriguez ML et al. , Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys. J, 2012, 103, 640–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBeath R, Pirone DM, Nelson CM, Bhadriraju K et al. , Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell, 2004, 6, 483–495. [DOI] [PubMed] [Google Scholar]
- 24.Kilian KA, Bugarija B, Lahn BT, and Mrksich M, Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U S A, 2010, 107, 4872–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernard A, Renault JP, Michel B, Bosshard HR et al. , Microcontact printing of proteins. Adv. Mat, 2000, 12, 1067–1070. [Google Scholar]
- 26.McNulty JD, Klann T, Sha J, Salick M et al. , High-precision robotic microcontact printing (R-μCP) utilizing a vision guided selectively compliant articulated robotic arm. Lab Chip, 2014, 14, 1923–1930. [DOI] [PubMed] [Google Scholar]
- 27.Sha J, Lippmann ES, McNulty J, Ma Y et al. , Sequential Nucleophilic Substitutions Permit Orthogonal Click Functionalization of Multicomponent PEG Brushes. Biomacromolecules, 2013, 14,. 3294–3303. [DOI] [PubMed] [Google Scholar]
- 28.Geisse NA, Sheehy SP, and Parker KK, Control of myocyte remodeling in vitro with engineered substrates. In Vitro Cell. Dev. Biol. Anim, 2009, 45, 343–350. [DOI] [PubMed] [Google Scholar]
- 29.Théry M and Piel M, Adhesive micropatterns for cells: a microcontact printing protocol. Cold Spring Harb. Protoc, 2009, 2009. doi: 10.1101/pdb.prot5255. [DOI] [PubMed] [Google Scholar]
- 30.Jain N, Iyer KV, Kumar A, Shivashankar GV, Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. U S A, 2013, 110, 11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazareth EJ, Ostblom JE, Lücker PB, Shukla S et al. , High-throughput fingerprinting of human pluripotent stem cell fate responses and lineage bias. Nat. Methods, 2013, 10, 1225–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight GT, Sha J, and Ashton RS, Micropatterned, clickable culture substrates enable in situ sptiotemporal control of human PSC-derived neural tissue morphology. Chem. Commun, 2015, 51, 5238–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hockemeyer D, Soldner F, Cook EG, Gao Q et al. , A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell, 2008, 3, 346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V et al. , Fiji: an open-source platform for biological-image analysis. Nat. Methods, 2012, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aratyn-Schaus Y, Oakes PW, and Gardel ML, Dynamic and structural signatures of lamellar actomyosin force generation. Mol. Biol. Cell, 2011, 22, 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thévenaz P, Ruttimann UE, and Unser M, A Pyramid Approach to Subpixel Registration Based on Intensity. IEEE Trans. Image Process, 1998, 7, 27–41. [DOI] [PubMed] [Google Scholar]
- 37.Libioulle L, Bietsch A, Schmid H, Michel B et al. , Contact-inking stamps for microcontact printing of alkanethiols on gold. Langmuir, 1999, 15, 300–304. [Google Scholar]
- 38.Salick MR, Napiwocki BN, Sha J, Knight GT et al. , Micropattern width dependent sarcomere development in human ESC-derived cardiomyocytes. Biomaterials, 2014, 35, 4454–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talwar S, Jain N, and Shivashankar G, The regulation of gene expression during onset of differentiation by nuclear mechanical heterogeneity. Biomaterials, 2014, 35, 2411–2419. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Chen S, and Jiang S, Protein interactions with oligo (ethylene glycol)(OEG) self-assembled monolayers: OEG stability, surface packing density and protein adsorption. J. Biomater. Sci. Polym. Ed, 2007, 18, 1415–1427. [DOI] [PubMed] [Google Scholar]
- 41.Hughes CS, Postovit LM, and Lajoie GA, Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics, 2010, 10, 1886–1890. [DOI] [PubMed] [Google Scholar]
- 42.Riedl J, Crevenna AH, Kessenbrock K, Yu JH et al. , Lifeact: a versatile marker to visualize F-actin. Nat. Methods, 2008, 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riedl J, Flynn KC, Raducanu A, Gärtner F et al. , Lifeact mice for studying F-actin dynamics. Nat. Methods, 2010,. 7, 168–169. [DOI] [PubMed] [Google Scholar]
- 44.Carpenter AE, Jones TR, Lamprecht MR, Clarke C et al. , CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol, 2006, 7. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malek AM and Izumo S, Mechanism of endothelial cell shape change and cytoskeletal remodeling in response to fluid shear stress. J. Cell Sci, 1996, 109, 713–726. [DOI] [PubMed] [Google Scholar]
- 46.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS et al. , A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. U S A, 2009, 106, 19017–19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Versaevel M, Grevesse T, and Gabriele S, Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Comm, 2012, 3, 671. [DOI] [PubMed] [Google Scholar]
- 48.Goffin JM, Pittet P, Csucs G, Lussi JW et al. , Focal adhesion size controls tension-dependent recruitment of α-smooth muscle actin to stress fibers. J. Cell Biol, 2006, 172, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chrzanowska-Wodnicka M and Burridge K, Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol, 1996, 133, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Versaevel M, Braquenier JB, Riaz M, Grevesse T et al. , Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci. Rep, 2014, 4. doi: 10.1038/srep07362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crisp M, Liu Q, Roux K, Rattner JB et al. , Coupling of the nucleus and cytoplasm role of the LINC complex. J. Cell Biol, 2006, 172, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim DH, Khatau SB, Feng Y, Walcott S et al. , Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci. Rep, 2012, 2, doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chambliss AB, Khatau SB, Erdenberger N, Robinson DK et al. , The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci Rep, 2013, 3, doi: 10.1038/srep01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luger K, Mäder AW, Richmond RK, Sargent DF et al. , Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 1997, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 55.Kimura H and Cook PR, Kinetics of core histones in living human cells little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol, 2001, 153, 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuipers MA, Stasevich TJ, Sasaki T, Wilson KA et al. , Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J. Cell Biol, 2011, 192, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharya D, Mazumder A, Miriam SA, and Shivashankar GV, EGFP-tagged core and linker histones diffuse via distinct mechanisms within living cells. Biophys. J, 2006, 91, 2326–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Axelrod D, Koppel DE, Schlessinger J, Elson E et al. , Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J, 1976, 16, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellenberg J and Lippincott-Schwartz J, Dynamics and mobility of nuclear envelope proteins in interphase and mitotic cells revealed by green fluorescent protein chimeras. Methods, 1999, 19, 362–372. [DOI] [PubMed] [Google Scholar]
- 60.White J and Stelzer E, Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol, 1999, 9,. 61–65. [DOI] [PubMed] [Google Scholar]
- 61.Ramdas NM and Shivashankar G, Cytoskeletal control of nuclear morphology and chromatin organization. J. Mol. Biol, 2014, 427, 695–706. [DOI] [PubMed] [Google Scholar]
- 62.Martin RM and Cardoso MC, Chromatin condensation modulates access and binding of nuclear proteins. FASEB J, 2010, 24, 1066–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hameed FM, Rao M, and Shivashankar G, Dynamics of passive and active particles in the cell nucleus. PLoS One, 2012, 7. doi: 10.1371/journal.pone.0045843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J et al. , Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 2013, 155, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA et al. , Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl. Acad. Sci. U S A, 2015, 112, 3002–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.