Abstract
Background:
Individuals with peanut allergy range in clinical sensitivity: some can consume grams of peanut before experiencing any symptoms, while others suffer systemic reactions to 10 mg or less. Current diagnostic testing only partially predicts this clinical heterogeneity.
Objective:
We sought to identify characteristics of the peanut-specific CD4+ T cell response in peanut-allergic patients that correlate with high clinical sensitivity.
Methods:
We studied the T cell receptor β-chain (TCRβ) usage and phenotypes of peanut-activated, CD154+ CD4+ memory T cells using fluorescence-activated cell sorting, TCRβ sequencing, and RNASeq, in reactive and hyporeactive patients who were stratified by clinical sensitivity.
Results:
TCRβ analysis of the CD154+ and CD154− fractions revealed >6,000 complementarity determining region 3 (CDR3) sequences and CDR3 motifs that were significantly enriched in the activated cells and 17% were shared between peanut-allergic individuals, suggesting strong convergent selection of peanut-specific clones. These clones were more numerous among the reactive patients and this expansion was identified within effector, but not regulatory T cell populations. The transcriptional profile of CD154+ T cells in the reactive group skewed towards a polarized Th2 effector phenotype and expression of Th2 cytokines strongly correlated with peanut-specific IgE levels. There were, however, also non-Th2 related differences in phenotype. Furthermore, the ratio of peanut-specific clones in the effector versus regulatory T cell compartment, which distinguished the clinical groups, was independent of specific IgE concentration.
Conclusion:
Expansion of the peanut-specific effector T cell repertoire is correlated with clinical sensitivity, and this observation may be useful to inform our assessment of disease phenotype and to monitor disease longitudinally.
Keywords: Peanut allergy, food allergy, clinical sensitivity, CD4+ T cell, effector T cell, regulatory T cell, Th2, CD154, TCRβ sequencing, RNASeq
Graphical Abstract
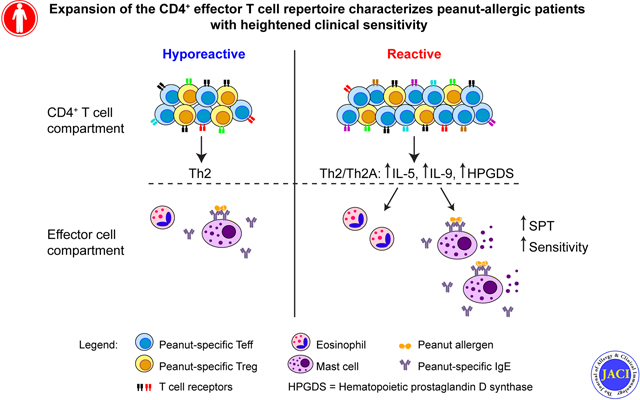
Capsule summary:
Heightened clinical sensitivity in peanut-allergic patients correlates with an expanded, more diverse, and more responsive antigen-specific effector T cell compartment, rather than a lack of regulatory T cells.
Introduction
Peanut allergy has steadily grown in prevalence, and currently affects >1% of the US population. Compared to other food allergies, peanut allergy is less frequently outgrown and more often presents with severe symptoms1. Although overall mortality is low, peanut allergy is the leading cause of death related to food-induced anaphylaxis in the US2, and the disease is associated with a high medical burden1. Reactions to allergens are mediated by activation of mast cells and basophils through the high-affinity IgE receptor, occurring when receptor-bound specific IgE is cross-linked by binding to peanut allergens - a specific immunogenic subset of those peanut proteins that elicit a high-affinity IgE response. Production of this allergen-specific IgE is T cell dependent, and a peanut-specific Th2-skewed CD4+ T cell profile characterized by increased expression of IL-4, IL-5, and IL-13 has been observed in subjects with peanut allergy3, 4.
Among individuals who are sensitized, meaning they have produced peanut-specific IgE, there is a remarkable spectrum of clinical reactivity that ranges from complete clinical tolerance to exquisite sensitivity down to low milligram amounts of ingestion5. This variation in threshold sensitivity is only weakly correlated to the serum concentration of allergen-specific IgE, and it presents one of the most significant unmet needs in clinical practice: how to identify those most at risk for allergic reactions upon (accidental) ingestion of a low amount of peanut. Threshold sensitivity is a sufficiently reproducible clinical phenotype, such that it is the FDA-endorsed primary outcome for therapies for peanut allergy currently in clinical trials6, 7 The pathophysiology of food allergy is hypothesized to depend on CD4+ Th subsets that act on B cells to maintain allergen-specific IgE8, on IgE-dependent effector cells such as mast cells and basophils, and on the epithelial barrier9 The frequency of peanut-specific effector Th cells in peripheral blood is higher in allergic patients, even during active avoidance of peanut, than in atopic or non-atopic controls who are tolerant to peanut4 Moreover, CD4+ T cell-derived transcription of IL9, which has recently been associated with a pathogenic subset of effector Th2 cells10, 11, is higher in peanut-allergic patients than in clinically tolerant individuals who produce peanut-specific IgE12.
Here, we stratified pediatric and adult peanut-allergic patients by their clinical sensitivity using the accepted standard method of double-blind placebo-controlled graded dose ingestion13. Most of the patients were reactive and experienced an objective clinical reaction at a cumulative dose of ≤443 mg peanut protein (equivalent protein content to approximately two peanut kernels). However, consistent with other studies5, fully one third of the patients were tolerant to this significant amount and we refer to them as hyporeactive patients. We compared the TCRβ usage and phenotypes of peanut-activated, CD154+ CD4+ memory T cells between these patient groups. Our data indicate that high clinical reactivity results from an expanded, more diverse and more responsive peanut-specific effector T cell population rather than a lack of regulatory T cells (at least as assessed from the peripheral blood), and that this imbalance may be a useful predictor of clinical sensitivity.
Methods
Participants
The subjects described in this study were all screened for participation in a peanut oral immunotherapy (OIT) trial (), and some were included in a high threshold peanut challenge study (), at the Food Allergy Center at Massachusetts General Hospital. All subjects were recruited with informed consent, and the study was approved by the Institutional Review Board of Partners Healthcare (protocol no. 2012P002153). Study participants with a previous diagnosis of peanut allergy, a history of peanut-induced reactions consistent with immediate hypersensitivity and confirmatory peanut- and Ara h 2-specific serum IgE concentrations (peanut-specific IgE > 5 kU/l, Ara h 2-specific IgE > 0.35 kU/l; ImmunoCAP; Thermo Fisher), underwent a double-blind placebo-controlled food challenge (DBPCFC). Increasing peanut protein doses were administered every 20 minutes to a maximum dose of 300 mg according to the following schedule: 3, 10, 30, 100, and 300 mg. Forty-one patients (66%) had an allergic reaction during this DBPCFC and were therefore labeled reactive and randomized to treatment in the peanut OIT trial, whereas 21 patients (34%) tolerated the highest dose without significant objective symptoms and were labeled hyporeactive. Six of the 21 hyporeactive patients agreed to a subsequent high-dose DBPCFC (cumulative total 7440 mg), and all six of them had an objective allergic reaction to a cumulative dose higher than 443 mg (median 3440 mg).
Cell culture, FACS for peanut-activated T cells, and FACS for Teff and Treg
Peripheral blood mononuclear cells (PBMC) were isolated from patient blood samples by means of density gradient centrifugation (Ficoll-Paque Plus; GE Healthcare). Fresh PBMC were cultured in AIM V medium (Gibco) for 20h at a density of 5×106 in 1 ml medium per well in 24-well plates, and were left unstimulated or cultured with 100 μg/ml peanut protein extract (15×106 PBMC per variable). The peanut extract was prepared by agitating defatted peanut flour (Golden Peanut and Tree Nuts) with PBS, centrifugation, and sterile-filtering. PE-conjugated anti-CD154 (clone TRAP1; BD Biosciences) was added to the cultures (20 μl/well) for the last 3h. After harvesting, the cells were labeled with AF700-conjugated anti-CD3 (clone UCHT1), APC-Cy7-conjugated anti-CD4 (RPA-T4), FITC-conjugated anti-CD45RA (HI100), PE-conjugated anti-CD154 (all from BD Biosciences), AF647-conjugated anti-CD69 (FN50; BioLegend), and Live/Dead Fixable Violet stain (L34955; Thermo Fisher). Live CD3+CD4+CD45RA− activated CD154+ and resting CD154−CD69− T cells were sorted with a FACSAria II instrument (BD Biosciences). In separate experiments, cryopreserved PBMC from the same patients were thawed and labeled with BV650-conjugated anti-CD3 (UCHT1), PE-Cy7-conjugated anti-CD4 (RPA-T4), APC-H7-conjugated anti-CD45RA (HI100) (all from BD Biosciences), BV605-conjugated anti-CD25 (BC96), BV785-conjugated anti-CD127 (A019D5) (both from BioLegend), and Live/Dead Fixable Blue stain (L23105; Thermo Fisher). Live CD3+CD4+CD45RA−CD25+CD127+ Teff and CD25++CD127− Treg were sorted with a FACSAria Fusion instrument (BD Biosciences). Sorted T cells were lysed in Buffer RLT Plus (Qiagen) + 1% β-mercaptoethanol (Sigma), and stored at −80°C, before total RNA and genomic DNA were isolated using the AllPrep DNA/RNA Micro Kit (Qiagen).
TCRβ sequencing
Genomic DNA was used to amplify and sequence the CDR3 regions (immunoSEQ assay; Adaptive Biotechnologies). The immunoSEQ approach generates an 87 base-pair fragment capable of identifying the VDJ region spanning each unique CDR3. Amplicons were sequenced using the Illumina NextSeq platform. Using a baseline developed from a suite of synthetic templates, primer concentrations and computational corrections were used to correct for the primer bias common to multiplex PCR reactions. Raw sequence data were filtered on the basis of TCRβ V, D, and J gene definitions provided by the IMGT database (www.imgt.org) and binned using a modified nearest-neighbor algorithm to merge closely related sequences and remove both PCR and sequencing errors.
Selection of enriched TCRβ CDR3 sequences in peanut-activated T cells
To select significantly enriched CdR3s, CDR3 read counts in the CD154+ and CD154−CD69− populations of each individual patient were analyzed with a G-test of independence and the resulting p-values underwent FDR correction with q < 0.0514 In addition to meeting this cutoff, selected CDR3s were further filtered by excluding those with a read count of less than 2 in the CD154+ population, and those for which the ratio of the count in the CD154+ to that in the CD154− population was less than 1. These filtering steps were used to ensure that our analysis focused on those CDR3 sequences most likely to be peanut-specific, removing CDR3s from T cells that may have responded to bystander activation.
Hamming distance and motif analysis
To determine global levels of similarity, minimum hamming distance (number of amino acid differences among CDR3s of same length) of each ps-CDR3 against all other ps-CDR3s was determined programmatically in R, taking advantage of functionality provided by the package “stringdist”15. The percentage of CDR3s at each minimum hamming distance was calculated. As a means of comparison, CDR3s from the total CD154+ and CD154− populations were sampled randomly 100 times, in the same number as the ps-CDR3s. At every sampling, minimum hamming distance was determined and median values of the percentage of CDR3s at each minimum hamming distance were calculated. For motif analysis, ps-CDR3s were trimmed to IMGT positions 107–116, which are the residues with the highest probability of antigen contact16 Subsequently, sequences were broken into motifs with a length of 4 amino acids. The proportions of those motifs among the ps-CDR3s and CDR3s from CD154− T cells were determined.
Quantitative PCR for Treg-associated genes
Total RNA from Teff and Treg was used to synthesize cDNA (iScript cDNA synthesis kit; Bio-Rad). Expression of FOXP3, CTLA4, CD25, and B2M was analyzed using the cDNA, specific primers (PrimePCR SYBR Green Assay primers; Bio-Rad), SYBR green (iTaq Universal SYBR Green Supermix; Bio-Rad), and a StepOnePlus Real-Time PCR instrument (Applied Biosystems). Data were analyzed using the 2−(ΔCt) method, which calculated expression of the target genes relative to the housekeeping gene B2M.
T cell suppression assay with Teff and Treg
FACS-sorted CD25+CD127+ Teff and CD25++CD127− Treg were cocultured with autologous bulk CD4+ responder T cells, which were isolated from PBMC with the EasySep CD4+ T cell enrichment kit (Stemcell Technologies) and labeled with CFSE cell proliferation dye (Thermo Fisher). Responder T cells were added to a 96-well U-bottom plate in 5×104/well in complete medium (RPMI + 10% FBS + Pen/Strep, all from Thermo Fisher), and Teff or Treg were added in a ratio of 1:2, 1:4, or 1:8 of Teff or Treg to responder T cells. Treg Suppression Inspector beads (Miltenyi Biotec) were added in a 1:2 ratio of beads to total T cells to induce proliferation, and cells were cultured for 5 days. After harvesting, the cells were labeled with AF700-conjugated anti-CD3, APC-Cy7-conjugated anti-CD4, and Live/Dead Fixable Violet stain. Live CD3+CD4+ responder T cell proliferation was analyzed with an LSR II instrument (BD Biosciences) and FlowJo software, and quantified as the percentage of divided (CFSElow) cells.
Gene expression analysis by RNA-Seq
Total RNA from CD154+ and CD154−CD69− T cells was used for cDNA synthesis and amplification (SMARTer ultra low input RNA kit for sequencing - v3; Clontech Laboratories). Libraries were prepared and sequenced on the Illumina HiSeq platform, at a read depth of approximately 30 million reads per sample. Paired-end RNA-Seq reads were aligned to the hg19 human reference genome with the ensemble version 75 annotation using STAR version 2.5.3a17 and gene expression was summarized using RSEM version 1.3.018. Differential expression analysis was performed using DESeq2 version 1.16.119, running under R version 3.4. Unmoderated fold changes were calculated and used in visualizations, which show genes with a minimum median expression level of Fragments Per Kilobase of transcript per Million mapped reads (FPKM) ≥ 2 in CD154+ T cells. Differentially expressed genes between the clinical groups as shown in Data file E2 had a log2 fold change ≥ 2 between CD154+ and CD154− T cells in at least one of the clinical groups, and an unadjusted P < 0.05 in the comparison between CD154+ T cells from reactive and hyporeactive patients.
Cell culture for secreted cytokine analysis
Cryopreserved PBMC were thawed and monocytes and memory CD4+ T cells were isolated using the EasySep CD14 positive selection kit and memory CD4+ T cell enrichment kit, respectively (Stemcell Technologies). Autologous monocytes and T cells were cocultured in a 1: 1 ratio (6×105 monocytes and 6×105 memory CD4+ T cells) in 0.5 ml AIM V medium per well in a 48-well plate for 3 days, and were left unstimulated or cultured with 50 μg/ml peanut extract or T-Activator CD3/CD28 beads (Thermo Fisher) in a 1:10 ratio of beads to T cells. After culture, supernatants were harvested and cytokine concentrations were measured using Cytometric Bead Array (IL-5, IL-9, IL-13, IFN-γ; BD Biosciences), Luminex (IL-4, IL-10, IL-17A; Bio-Plex, Bio-rad), and ELISA (IL-22; R&D Systems, IL-26; Millipore-Sigma).
Statistical analysis
Prism 7 (GraphPad) and R (version 3.4) were used for statistical analysis. We used the D’Agostino-Pearson omnibus normality test to assess for normal distribution. The specific parametric and non-parametric statistical tests are indicated in the figure legends.
Data Availability
The RNA-Seq and TCRb sequencing datasets generated in the course of this project have been deposited at the National Center for Biotechnology Information database of Genotypes and Phenotypes (dbGaP) and Sequence Read Archive under accession phs001897.v1.p1.
Results
Reactive patients are more strongly sensitized to peanut than hyporeactive patients
Patients with a diagnosis of peanut allergy, a history of peanut-induced reactions consistent with immediate hypersensitivity and confirmatory peanut- and Ara h 2-specific serum IgE levels (n=62), underwent a DBPCFC up to a maximum dose of 300 mg (cumulative total 443 mg) of peanut protein20, as part of their screening for a peanut OIT clinical trial (). We chose this maximum dose to target participants who were at the highest risk for persistent peanut allergy, and anticipated that this group would benefit the most from desensitization and/or clinical tolerance induction. Forty-one patients (66%) were reactive, while 21 patients (34%) tolerated the highest dose without significant objective symptoms. Six of these 21 hyporeactive patients consented to a subsequent high-dose DBPCFC (maximum dose 4000 mg, cumulative total 7440 mg), and all six patients had an objective allergic reaction to a cumulative dose higher than 443 mg (median 3440 mg).
Reactive patients had higher serum levels of whole peanut protein-specific IgE, as well as Ara h 2-specific IgE, and higher skin test reactivity to peanut than hyporeactive patients (Table E1). In contrast, hyporeactive patients tended to have a higher prevalence of co-existing atopic dermatitis than reactive patients, along with slightly higher total IgE levels. As a result, the ratio of peanut-specific IgE to total IgE, a better predictor of clinical allergy than peanut-specific IgE alone21, was higher in reactive than in hyporeactive patients (P < 0.01). Reactive patients also had a higher ratio of peanut-specific IgE to IgG4 (P < 0. 01).
Enrichment and selection of putatively peanut-specific TCRβ CDR3 sequences
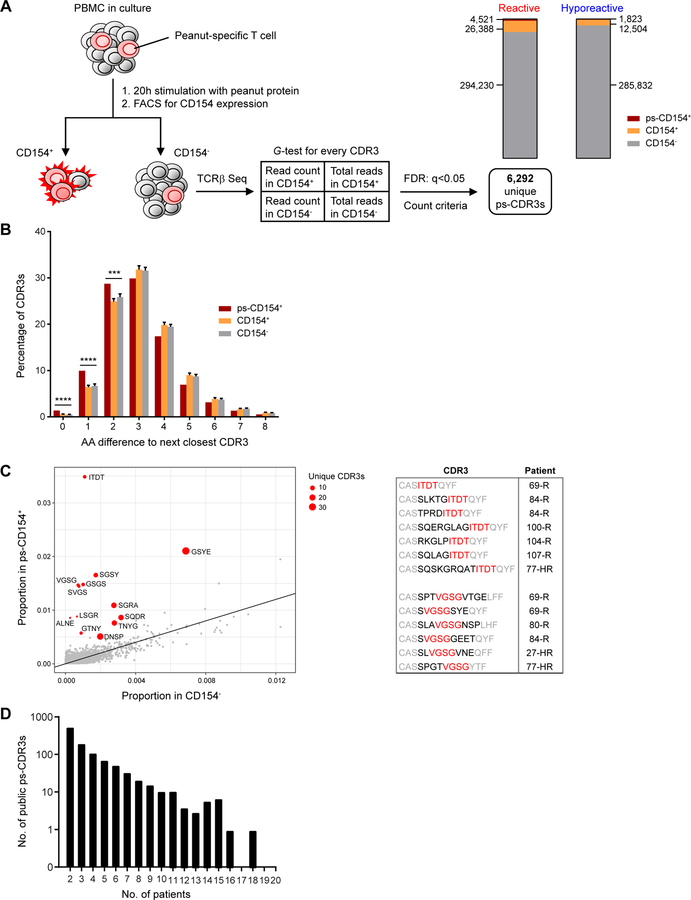
In order to compare the TCRβ usage of peanut-activated CD4+ T cells from reactive and hyporeactive patients, PBMC were isolated from blood samples and cultured for 20h with peanut protein extract. A subset of the reactive and hyporeactive patients (n=10 per group) was used for this analysis; demographic and clinical data of the individual patients included in each set of experiments in this study are shown in Table E2A–B. The selection of patients was based on the inclusion of comparable groups of reactive and hyporeactive patients with regard to age and gender, and a wide range of peanut-specific IgE levels in both groups, as well as on sample and cell availability. Activated CD154+ and resting CD154−CD69− memory CD4+ T cells were sorted by FACS (Fig. E1) and TCRβ repertoire was determined by sequencing from genomic DNA. To enrich for the subset of TCRβ sequences most likely to be peanut-specific, rather than sequences from common clones responding to bystander activation, we analyzed the counts of each unique CDR3 sequence in each sample of CD154+ cells and in the corresponding CD154− cells from the same patient. We also examined the total CDR3 counts in each sample and applied a G-test of independence14, selecting those clones with a FDR q-value < 0.05. A total of 6,292 unique CDR3 amino acid sequences were selected, corresponding to 14% of all unique CDR3s from CD154+ T cells (Fig. 1A). These putatively peanut-specific CDR3 sequences (ps-CDR3s) were significantly more similar than those selected randomly from all CD154+ T cells or all CD154− T cells, as determined by the distribution of Hamming distance between these populations (P < 0.001; Fig. 1B). In addition, we observed that specific motifs were significantly enriched in ps-CDR3s compared to CD154− T cells (Fig. 1C). These findings confirm the enrichment and convergent selection of antigen-specific T cell clones16. Most of the ps-CDR3s were private (i.e., present in only one patient), but 1,041 ps-CDR3s (17%) were public and detected in multiple individuals, ranging from two to 18 out of the 20 patients (Fig. 1D).
Fig. 1: Enrichment and selection of putatively peanut-specific TCRβ CDR3 sequences.
(A) Procedure for selection of significantly enriched, putatively peanut-specific CDR3 sequences (see Methods and Results). Stacked bar graphs show the proportions and numbers of CDR3s in the CD154−, CD154+, and putatively peanut-specific CD154+ (ps-CD154+) compartments in reactive and hyporeactive patients. (B) Minimum Hamming distance of ps-CDR3s (ps-CD154+), compared with equal-sized randomly sampled control pools of CDR3s from all CD154+ T cells or all CD154− T cells. s.d. of 100 repeat random samples of control CDR3s is shown on bars (**** P < 0.0001, *** P < 0.001, Fisher’s exact test). (C) ps-CDR3s were enriched for a subset of 4-mer amino acid motifs, as compared to CDR3s from CD154− T cells. Shown in red are motifs that were found in at least three unique ps-CDR3s, derived from at least three patients, and met a G-test and FDR cutoff of q < 0.05. The table shows the position of two of the 4-mers (in red) within the ps-CDR3s, and the patients from whom the ps-CDR3s were derived (R = reactive, HR = hyporeactive). Residues with high probability of contact with antigenic peptide are in red and black, those with low probability are in grey. (D) Distribution of the public ps-CDR3s over the patients. Shown is the number of public ps-CDR3s present in a given number of patients.
The 19 most prominent public ps-CDR3s were significantly enriched in CD154+ T cells from at least two patients and detected in at least four of the 20 patients, in any of the tested T cell subsets. To further confirm that our method for enrichment and identification of ps-CDR3s was driven by antigen-specific responses, we determined the presence of the top public ps-CDR3s in CD154+ and CD154− T cells from cow’s milk protein-stimulated PBMC cultures of 13 patients with eosinophilic esophagitis from a separate study (Fig. E2A–B). Most of the clones were present in the CD154− compartment of at least one patient, but ps-CDR3s were rare in the CD154+ compartment, indicating that enrichment of these clones in peanut-stimulated cultures resulted from antigen-specific activation.
Reactive patients have a larger and more diverse putatively peanut-specific T cell repertoire
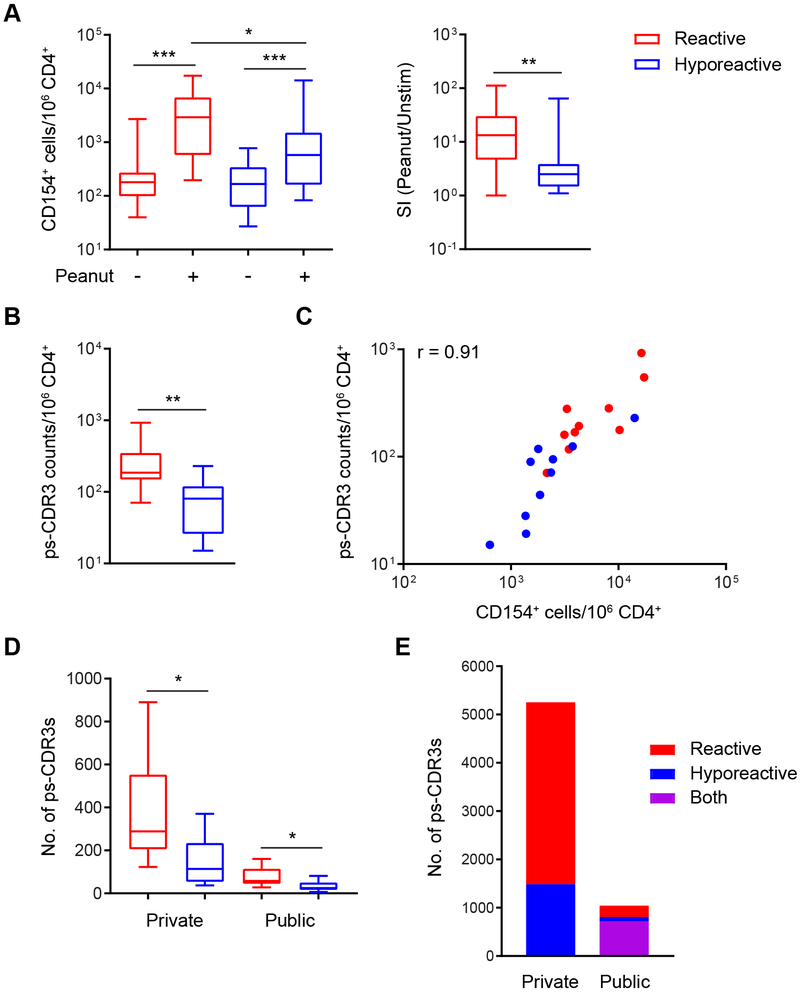
Stimulation with peanut protein induced CD154 expression on CD4+ memory T cells in both patient groups. The magnitude of that response, however, was greater in reactive patients than in hyporeactive individuals (median 2924 vs. 576 CD154+ T cells per million CD4+ T cells; P < 0.05), and the stimulation index was higher in reactive patients (median 13.3-fold vs. 2.5-fold increase in CD154+ T cells in peanut-stimulated versus unstimulated cultures; P < 0. 01), consistent with a higher frequency of peanut-specific CD4+ T cells in peripheral blood from reactive patients (Fig. 2A). This was confirmed by the observation that the counts of enriched ps-CDR3s per million CD4+ T cells were also higher in reactive patients (median 185 vs. 80; P < 0.01) (Fig. 2B). The frequency of CD154+ T cells in peanut-stimulated cultures correlated strongly with that of ps-CDR3 counts in corresponding patients (Fig. 2C). As each ps-CDR3 count represents one putatively peanut-specific CD4+ T cell, the median frequency of these cells in our entire group of allergic patients was approximately 120 per million CD4+ T cells, which corresponds with published estimates of whole allergen-specific T cell frequencies in allergic subjects10, 22, 23. The number of unique ps-CDR3s per patient was also higher in the reactive than the hyporeactive group, for private clones (median 289 vs. 114; P < 0.05) as well as public clones (median 58 vs. 25; P < 0.05) (Fig. 2D). Most of the public ps-CDR3s (716) were present in at least one patient in both clinical groups; 233 were unique to the reactive group and 92 to the hyporeactive group (Fig. 2E).
Fig. 2: Reactive patients have a larger and more diverse putatively peanut-specific CD4+ T cell repertoire.
(A) The frequency of activated, CD154+ T cells was increased in peanut-stimulated PBMC cultures as compared to unstimulated cultures in reactive as well as hyporeactive patients (n=20 per group; *** P < 0.001, Wilcoxon matched-pairs signed rank test). However, the frequency of CD154+ T cells in peanut-stimulated cultures (* P < 0.05, Mann Whitney test), as well as the stimulation index (** P < 0.01, Mann Whitney test), was higher in reactive patients. (B) The frequency of ps-CDR3 counts was higher in reactive than hyporeactive individuals (n=10 per group; ** P < 0.01, Mann Whitney test). (C) The frequency of CD154+ T cells in peanut-stimulated cultures was strongly correlated with that of ps-CDR3 counts in corresponding patients (P < 0.001, Spearman’s p). (D) The numbers of unique private and public ps-CDR3s per patient were higher in the reactive group (* P < 0.05, unpaired t-test with Welch’s correction). (E) Numbers of private and public ps-CDR3s in the clinical groups.
Of the top 19 public ps-CDR3s, the three most common public clones were highly prevalent in activated CD154+ T cells, and undetected in resting CD154− T cells from any patient (Fig. E3A–B). Eleven clones were detected in CD154+ T cells among patients of both clinical groups, while eight were only present in CD154+ T cells from reactive patients. In general, the top public clones were more frequently detected in CD154+ T cells among reactive patients than hyporeactive ones (mean 5.4 vs. 2.5 clones per patient; P < 0.01). There were no differences in the presence of these clones in CD154− T cells or ex vivo sorted regulatory T cells (Treg) between the clinical groups (Fig. E3B–C). Nevertheless, these clones were more frequently detected in effector T cells (Teff) from reactive versus hyporeactive patients (mean 3.6 vs. 1.9 clones per patient; P < 0.01) (Fig. E3D).
The putatively peanut-specific T cell repertoire of reactive patients is enriched in effector T cells
To evaluate the distribution of ps-CDR3s within effector or regulatory memory CD4+ T cell compartments, we sorted bulk CD25+CD127+ Teff and CD25++CD127− Treg from the corresponding patients in each clinical group (Fig. E4A)24 The T cell phenotype was confirmed by high expression of FOXP3, CD25, and CTLA4 (Fig. E4B), and by functional suppression (Fig. E4C) in Treg relative to the Teff. TCRβ loci of the Teff and Treg subsets were sequenced to examine the presence and frequency of ps-CDR3s (Fig. E4D). Using this method, we determined the distribution of ps-CDR3s between Teff and Treg by clinical phenotype.
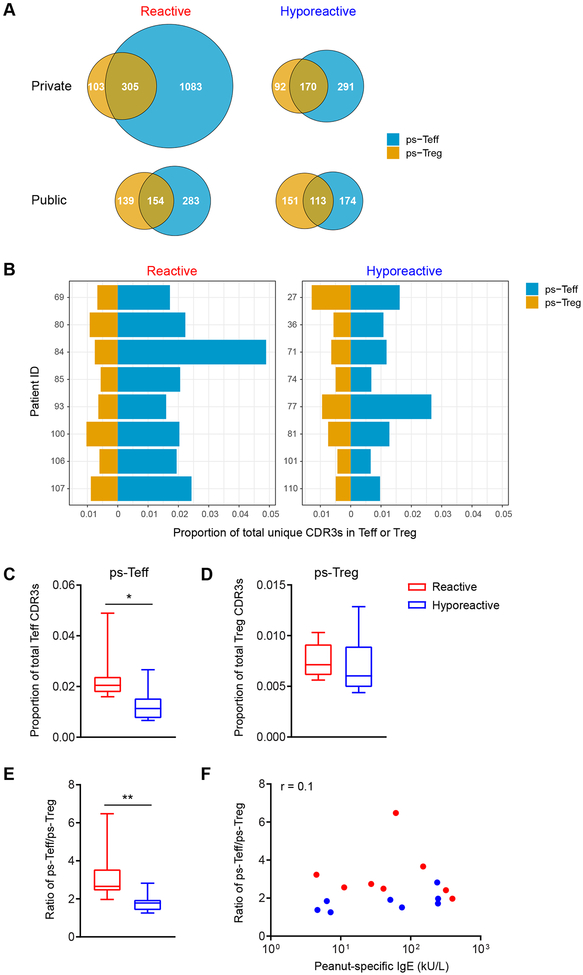
A substantial number of unique ps-CDR3s present in these subsets were found in both Teff and Treg, but the majority were only detected in one of the subsets (Fig. 3A and Fig. E5A). In general, we observed a higher degree of overlap in ps-CDR3s between Teff and Treg within each clinical group, than in Teff or Treg between the clinical groups (Fig. E5A–B). The distribution of private ps-CDR3s was highly skewed towards the Teff compartment, especially in reactive patients, whereas public ps-CDR3s were more evenly spread over Teff and Treg. The number of ps-CDR3s uniquely present in Teff was higher in reactive versus hyporeactive patients (3.7-fold higher for private clones and 1.6-fold higher for public clones), whereas the number of both private and public ps-CDR3s present only in Treg was similar (Fig. 3A). Furthermore, the proportion of ps-CDR3s in Teff was higher in reactive patients (median 0.020 vs. 0.011; P < 0.05), whereas the proportion in Treg was not different (Fig. 3B–D). As a result, the ratio between the proportions of ps-Teff and ps-Treg was higher in reactive patients (median 2.66 vs. 1.78; P < 0.01) (Fig. 3E). This ratio was not correlated with peanut-specific IgE levels in reactive or hyporeactive patients (Fig. 3F). The same outcome was observed when using absolute numbers of ps-Teff and ps-Treg instead of proportions (Fig. E6A–E). In contrast, the proportion of CDR3s derived from non-peanut-specific, CD154− T cells was not higher in Teff from reactive patients (Fig. E7). These findings indicate that the peanut-specific T cell repertoire of reactive patients is imbalanced and skewed toward the Teff compartment, and together with the data above, suggest that reactive individuals have a more expanded and diversified repertoire of peanut-specific effector T cells.
Fig. 3: The putatively peanut-specific CD4+ T cell repertoire of reactive patients is enriched in effector T cells.
(A) Numbers of unique private and public ps-CDR3s in Teff, Treg, and both, from reactive and hyporeactive patients (n=8 per group). (B-D) The proportion of ps-CDR3s in Teff, but not in Treg, was higher in reactive than hyporeactive patients (* P < 0.05, Mann Whitney test). (E) The ratio of the proportion of ps-Teff to that of ps-Treg was higher in reactive patients (** P < 0.01, Mann Whitney test). (F) The ratio of the proportion of ps-Teff to that of ps-Treg was not correlated with peanut-specific IgE concentrations in serum from the corresponding patients (Spearman’s ρ).
Putatively peanut-specific effector T cells are more responsive in reactive than in hyporeactive patients
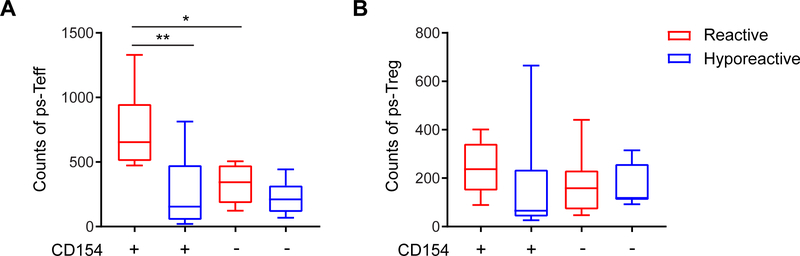
The stronger induction of CD154+ T cells in response to peanut protein in reactive versus hyporeactive patients (Fig. 2A) is consistent with an expanded ps-Teff population, as supported by the above data. However, suppression, anergy or exhaustion of peanut-specific T cells in hyporeactive patients could also contribute to this difference. To evaluate for this, we selected the ps-CDR3s present in the Teff and Treg compartments of the individual patients and analyzed the counts of these ps-Teff and ps-Treg in the CD154+ and CD154− populations from the corresponding patients following in vitro stimulation. We found that the counts of ps-Teff present in the CD154+ population were higher in reactive than hyporeactive patients (median 655 vs. 155; P < 0.01) (Fig. 4A). In contrast, within the non-responding, CD154− population, the counts of ps-Teff were not significantly different between the groups. These data are consistent with the presence of ps-Teff in hyporeactive patients that fail to respond to stimulation in vitro. Besides, the counts of ps-Treg in CD154+ and CD154− populations were not different between the clinical groups (Fig. 4B).
Fig. 4: Putatively peanut-specific effector T cells are more responsive in reactive than in hyporeactive patients.
(A) The counts of ps-Teff in CD154+ T cells from reactive patients were higher than in CD154+ T cells from hyporeactive patients (n=8 per group; ** P < 0.01, unpaired t-test), and higher than in CD154− T cells from reactive patients (* P < 0.05, paired t-test). (B) The counts of ps-Treg in CD154+ and CD154− T cells were not different between reactive and hyporeactive patients.
The transcriptional phenotype of peanut-activated CD4+ T cells differs between reactive and hyporeactive individuals
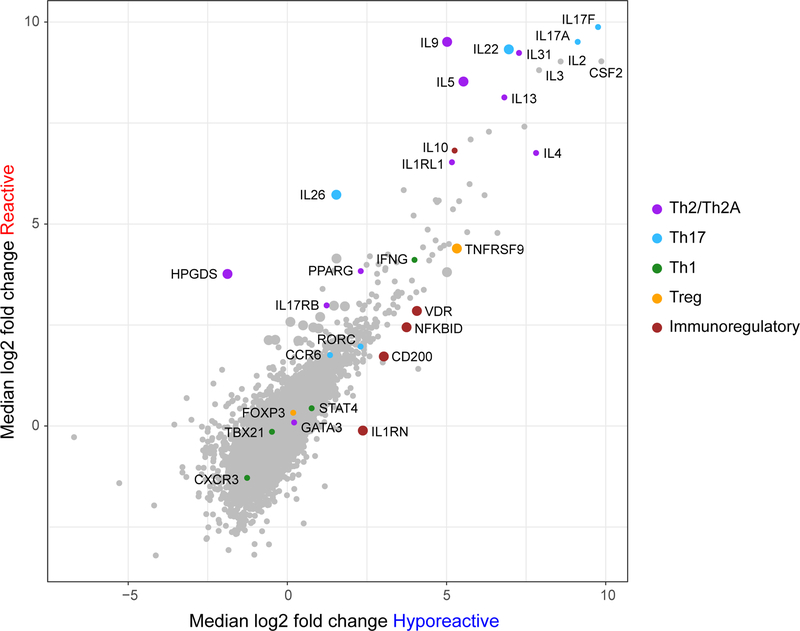
To explore differences in the transcriptional phenotype of peanut-activated CD4+ T cells between the clinical groups, activated and resting T cells from 20 additional patients (10 reactive and 10 hyporeactive) were analyzed by RNA-Seq. 1,585 genes were highly differentially expressed in peanut-activated (CD154+) T cells compared to resting (CD154−) T cells from reactive patients (P < 10−5; 1,103 up, 482 down), and 608 genes in T cells from hyporeactive patients (p < 10−5; 489 up, 119 down) (Fig. E8A, and Data file E1). Consistent with their subtly distinct clinical phenotype, there was substantial concordance of gene expression between the clinical groups, with 435 genes differentially expressed in activated T cells from reactive as well as hyporeactive patients (Fig. E8B). Th2-associated genes IL4, IL13, and IL31, as well as Th17-related genes IL17A and IL17F, were strongly induced in peanut-activated T cells from both clinical groups. Nevertheless, there were also some notable differences between the groups: we identified 31 genes that were differentially expressed between reactive and hyporeactive patients (P < 0.05; 25 up, 6 down) (Fig. 5 and Data file E2). Expression of several genes previously associated with pathogenic Th2 cells (IL5, IL9, HPGDS), and others associated with Th17 cells (IL22 and IL26), was higher in reactive than in hyporeactive patients. In contrast, a subset of genes associated with Treg (TNFRSF9/CD137) and immune regulation (NFKBID, IL1RN, VDR, CD200) was increased in hyporeactive patients.
Fig. 5: The transcriptional phenotype of peanut-activated CD4+ T cells differs between reactive and hyporeactive individuals.
Gene expression is shown as the median log2 fold change between activated CD154+ and resting CD154− T cells in reactive and hyporeactive patients (n=10 per group). The large dots indicate genes that were significantly different between reactive and hyporeactive patients (P < 0.05). For the complete list of significantly different genes between the clinical groups, see Data file E2.
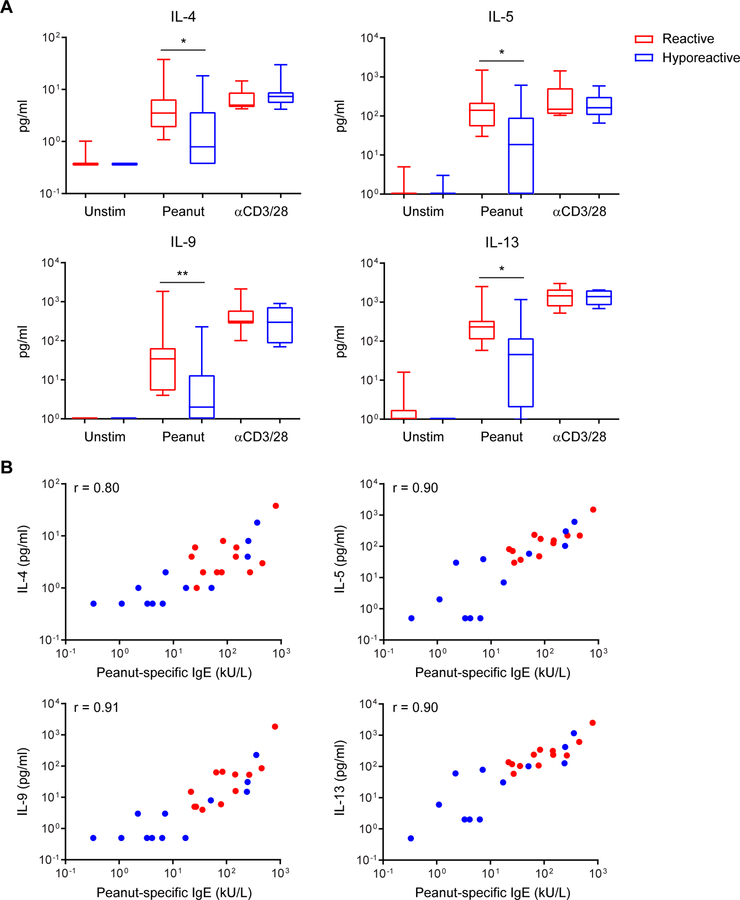
Peanut-activated CD4+ T cells from reactive patients produce higher amounts of Th2 cytokines
We confirmed the differences in gene expression for a selection of cytokines on the protein level. Co-cultures of autologous monocytes and memory CD4+ T cells from 12 reactive and 12 hyporeactive patients were stimulated with peanut protein extract or anti-CD3/CD28. In peanut-stimulated cultures, but not in polyclonally stimulated cultures, production of IL-4, IL-5, IL-9, and IL-13 was significantly higher in reactive than in hyporeactive patients (Fig. 6A). Moreover, the levels of Th2 cytokines in peanut-stimulated cultures correlated strongly with peanut-specific IgE concentrations in serum from the corresponding patients (Fig. 6B). In contrast, levels of non-Th2 cytokines including IL-17A, IL-22, IL-10, and IFN-γ (readily detected with polyclonal stimulation), were very low in peanut-stimulated cultures from most patients, and not different between the clinical groups (Fig. E9).
Fig. 6: Peanut-activated CD4+ T cells from reactive patients produce higher amounts of Th2 cytokines.
Cytokine concentrations were measured in supernatants from co-cultures of autologous monocytes and memory CD4+ T cells from reactive and hyporeactive patients (n=12 per group). (A) Production of the Th2 cytokines IL-4, IL-5, IL-9, and IL-13 was higher in peanut-activated T cells from reactive patients, but not different in polyclonally (anti-CD3/CD28) stimulated T cells (* P < 0.05, ** P < 0.01, Mann Whitney test). (B) Th2 cytokine responses in peanut-activated T cells were strongly correlated with peanut-specific IgE concentrations in serum from the corresponding patients (P < 0.001, Spearman’s ρ).
Discussion
The degree of heterogeneity in clinical status of peanut-allergic patients is emerging as increasing numbers of patients with known allergy are subjected to defined amounts of peanut protein under controlled settings in the hospital. A wide spectrum of clinical sensitivity has been observed, from the generation of peanut-specific IgE without any clinical allergy, to exquisite sensitivity. Using a functional response to in vitro stimulation with peanut protein complemented by TCRβ repertoire analysis, we have found that among patients stratified by sensitivity, the more reactive individuals exhibit a robust expansion of their antigen-specific effector T cell compartment, and that these Teff are more responsive to in vitro stimulation than those in hyporeactive patients. The expansion of the antigen-specific Teff population relative to the Treg compartment is independent of differences in specific IgE concentration, suggesting additional mechanisms of action upon clinical sensitivity and the potential utility of this measure as a biomarker for high clinical reactivity.
In addition to being more expanded and more responsive, antigen-specific Teff in reactive patients showed increased expression of the Th2-related genes IL5, IL9, and HPGDS, as well as higher production of Th2-associated cytokines, upon activation with peanut protein. These findings are consistent with previous studies, which reported increased expression of IL5, IL9, and IL13 in peanut-activated T cells from allergic patients as compared to peanut-sensitized but tolerant subjects12, 25. A recently described pathogenic effector subset of Th2 cells, also termed Th2A cells, is highly enriched in patients with allergic or eosinophilic disease, and characterized by increased expression of IL5, IL9, and HPGDS, among other markers10, 11. Our results suggest a higher frequency and/or response of peanut-specific Th2A cells in reactive patients. Furthermore, the increased production of Th2-associated cytokines with functions beyond IgE class-switching, such as IL-5 and IL-9, indicates that differences in clinical phenotype between reactive and hyporeactive patients may be driven in part by non-IgE-mediated pathology. It is possible that Th17 responses play a role in this pathology as well, since we observed strikingly high gene expression of IL17A and IL17F in peanut-activated CD4+ T cells from both groups, and higher expression of the Th17-associated cytokines IL22 and IL26 in reactive patients. Interestingly, measured secretion of IL-17A, IL-22 and IL-26 protein was lower than expected based on gene expression levels. Only modest production of IL-17A by peanut-stimulated T cells was evident in both patient groups, consistent with observations from others in peanut-allergic and tolerant subjects25, 26. Moreover, secretion of IL-22 and IL-26 protein by these cells was below the detection limit of our assays (data not shown for IL-26). Nevertheless, it would be relevant to further study the role of Th17 cytokines in peanut allergy, particularly of IL-22, as it has been implicated in both pathogenic and protective responses in allergic disease27, 28.
The T cell activation markers CD154 and CD137 have been used to distinguish antigen-activated Teff and Treg, respectively29. After a 7h incubation of PBMC with antigen, expression of these markers was mutually exclusive, as very few double-positive T cells were detected. A similar strategy was used in a recent study, which reported that differential upregulation of CD154 and CD137 efficiently distinguished peanut protein-activated Teff and Treg26 The kinetics of CD154 expression, however, have been shown to be slower in Treg than Teff30 Indeed, a recent paper reported a mixed population of CD154+ Teff and Treg after stimulating PBMC from peanut-allergic patients with peanut protein extract for 18h25, which corresponds to the incubation time used here. Although it is still likely that our CD154-based methodology favored selection of peanut-activated Teff over Treg, we did identify a substantial number of ps-CDR3s as being derived from Treg by comparing these sequences with CDR3s in ex vivo sorted Treg from corresponding patients. In addition, we observed that gene expression of CD137 (TNFRSF9) was increased in peanut-activated CD154+ T cells from reactive as well as hyporeactive patients. Importantly, we did not detect differences in the numbers or proportions of ps-Treg between the groups, suggesting that increased clinical reactivity to peanut is not due to a lack of antigen-specific Treg. This finding is consistent with the studies mentioned above, which found no evidence of a deficit in Treg specific for aeroantigens29 or peanut antigens25, 26 in allergic patients. We did, however, observe lower responsiveness of antigen-specific Teff in hyporeactive patients, along with increased expression of genes associated with Treg and immune regulation (TNFRSF9, NFKBID, IL1RN, VDR, CD200)29, 31–36 in peanut-activated T cells. It remains to be investigated whether the lower Teff responses in these patients are the result of a more balanced ratio between Teff and Treg, enhanced Treg function, anergy of Teff, or a combination of these factors.
CD154 has been shown to be effective for detection of antigen-specific CD4+ T cells by Roederer et al.37, and we have used an adapted version of their protocol for this work38. To the best of our knowledge, ours is the first study to perform TCRβ sequencing in CD154+ and CD154− T cells from antigen-stimulated PBMC cultures, and apply a statistical method to focus on the subset of activated CD154+ T cell clones that is most enriched and therefore most likely to be truly antigen-specific. The sensitivity of this method is such that we could detect striking differences in the number, proportion, and responsiveness of ps-Teff between two groups of peanut-allergic patients with a relatively subtle difference in clinical phenotype. We analyzed T cell populations in bulk rather than at the single-cell level, which prevented us from obtaining combined TCRβ and gene expression data for each individual cell. However, by using the bulk approach we could screen >100-fold higher numbers of T cells, and identify low-frequency sequences such as public ps-CDR3s and motifs, which are difficult to detect by single-cell RNASeq. In addition, an important advantage of our functional assay with peanut protein extract, as compared to selection of antigen-specific CD4+ T cells by MHCII-peptide tetramers, is that it minimizes bias in terms of epitope specificity, and can be applied in all patients, regardless of their HLA genotypes. Selection by affinity using MHCII-peptide tetramers in an individual patient is limited to a small number of known T cell epitopes in major allergens4, whereas there are many more potentially relevant epitopes present in known allergens as well as other peanut antigens39.
Our approach holds promise for application in immune monitoring over the course of tolerance-inducing therapies such as OIT, for peanut allergy as well as other allergies. Differences in clinical outcome may be correlated with variations in TCRβ usage and phenotypes of peanut-specific CD4+ T cells over time, and these factors may help in predicting the level of clinical success and informing new therapeutic strategies. To assess the clonotype stability of ps-CDR3s, we performed a preliminary analysis of TCRβ sequencing data from four placebo-treated peanut-allergic patients in peanut OIT trial , and found that of the ps-CDR3s identified at baseline, a median of 20% were detected again at the 20-week timepoint, and 16% at the 64-week timepoint. Moreover, 7% were detected at all three timepoints. These data suggest that at least a subset of ps-CDR3s is stable over a prolonged period of time. It is worth noting that we stimulated 15×106 PBMC with peanut protein extract for each patient and time point we analyzed. This is a relatively small fraction of the total number of PBMC present in peripheral blood, and it introduces a substantial sampling limitation. The percentage of overlap in ps-CDR3s between these timepoints is expected to be higher if more PBMC are used. So far, one group has published a TCRβ sequencing-based approach to monitor peanut-specific T cell responses over the course of OIT, by utilizing CFSE dilution in peanut extract-stimulated PBMC cultures and analyzing proliferating, CFSElow T cells40. The authors found an extremely diverse TCRβ repertoire in these T cells, likely due at least in part to substantial bystander activation, and noted a change in frequency of some persistent peanut-activated T cell clones during OIT. One group of clones steadily decreased in frequency during OIT, whereas another group transiently increased after 9 months and then declined after 18 months of therapy. A different study used MHCII dextramers loaded with peptides derived from the major peanut allergen Ara h 2 to select antigen-specific CD4+ T cells and perform single-cell RNASeq with samples from peanut-allergic patients undergoing OIT41. This group reported that successful OIT induced allergen-specific T cells to expand and shift toward an anergic phenotype, characterized by low expression of cytokines and the costimulatory molecule CD28. The key observations in both of these studies, however, were based on data from only two to three patients. Hence, studying substantial numbers of patients with well-defined clinical outcomes using the methodology described here could further elucidate the mechanisms behind allergy immunotherapy and help to refine this type of treatment.
In sum, we have observed that high clinical reactivity in peanut allergy correlates with an expanded, broader and more responsive antigen-specific effector T cell population, rather than a lack of regulatory T cell responses. The skewed ratio between peanut-specific Teff and Treg may be a useful predictor of clinical sensitivity, and help identify those patients who will benefit most from tolerance-inducing treatments such as OIT.
Supplementary Material
Key messages:
Reactive peanut-allergic patients have a larger, more diverse, and more Th2-skewed peanut-specific CD4+ T cell compartment in peripheral blood than hyporeactive patients.
Reactive patients show an expansion in peanut-specific effector T cells and an imbalance between effector and regulatory T cells, especially in their private repertoire.
This imbalance may be one of the causes of high clinical sensitivity and a potential biomarker, and may be altered over the course of immunotherapy with peanut.
Acknowledgements
We would like to thank our patients and their families who generously gave their time and participation, as well as Lauren Tracy, Colby Rondeau, Christine Elliot and Leah Hayden, the clinical coordinators of this study. The clinical work was performed in the Harvard Clinical and Translational Science Center supported by grants 1UL1TR001102 and 8UL1TR000170 from the National Center for Advancing Translational Science, and 1UL1RR025758 from the National Center for Research Resources. In addition, we would like to thank our colleagues at the MGH Department of Pathology Flow and Image Cytometry Research Core for their help with cell sorting, and at the MGH Next Generation Sequencing Core for their support with RNASeq. The Flow Core obtained funding from the NIH Shared Instrumentation program with grants 1S10OD012027-01A1, 1S10OD016372-01, 1S10RR020936-01, and 1S10RR023440-01A1. We would also like to acknowledge the MIT Koch Bioinformatics and Computing Core for their help with RNASeq data analysis. Their work was supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute.
Funding:
This work was supported by the NIH/NIAID (grant U19AI095261 to W.G.S. and Andrew D. Luster, K23AI130408 to Y.V.V., and K23AI121491 to S.U.P.), Sanofi U.S., the Food Allergy Science Initiative (FASI), and institutional funds from Massachusetts General Hospital.
Abbreviations:
- CDR3
Complementarity-determining region 3
- CFSE
Carboxyfluorescein succinimidyl ester
- CTLA4
Cytotoxic T-lymphocyte associated protein 4
- DBPCFC
Double-blind placebo-controlled food challenge
- DEG
Differentially expressed genes
- FACS
Fluorescence-activated cell sorting
- FDR
False discovery rate
- FOXP3
Forkhead box protein P3
- HPGDS
Hematopoietic prostaglandin D synthase
- IL1RN
Interleukin-1 receptor antagonist
- NFKBID
NFκB inhibitor delta
- OIT
Oral immunotherapy
- PBMC
Peripheral blood mononuclear cells
- TCRβ
T cell receptor β-chain
- Teff
CD4+ effector T cells
- Treg
CD4+ regulatory T cells
- VDR
Vitamin D receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflict of interest: All authors declare that they have no competing interests.
References
- 1.Sicherer SH, Sampson HA. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J Allergy Clin Immunol 2018; 141:41–58. [DOI] [PubMed] [Google Scholar]
- 2.Bock SA, Munoz-Furlong A, Sampson HA. Further fatalities caused by anaphylactic reactions to food, 2001–2006. J Allergy Clin Immunol 2007; 119:1016–8. [DOI] [PubMed] [Google Scholar]
- 3.Turcanu V, Maleki SJ, Lack G. Characterization of lymphocyte responses to peanuts in normal children, peanut-allergic children, and allergic children who acquired tolerance to peanuts. J Clin Invest 2003; 111:1065–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLong JH, Simpson KH, Wambre E, James EA, Robinson D, Kwok WW. Ara h 1-reactive T cells in individuals with peanut allergy. J Allergy Clin Immunol 2011; 127:1211–8 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hourihane JO, Allen KJ, Shreffler WG, Dunngalvin G, Nordlee JA, Zurzolo GA, et al. Peanut Allergen Threshold Study (PATS): Novel single-dose oral food challenge study to validate eliciting doses in children with peanut allergy. J Allergy Clin Immunol 2017; 139:1583–90. [DOI] [PubMed] [Google Scholar]
- 6.Investigators PGoC, Vickery BP, Vereda A, Casale TB, Beyer K, du Toit G, et al. AR101 Oral Immunotherapy for Peanut Allergy. N Engl J Med 2018; 379:1991–2001. [DOI] [PubMed] [Google Scholar]
- 7.Sampson HA, Shreffler WG, Yang WH, Sussman GL, Brown-Whitehorn TF, Nadeau KC, et al. Effect of Varying Doses of Epicutaneous Immunotherapy vs Placebo on Reaction to Peanut Protein Exposure Among Patients With Peanut Sensitivity: A Randomized Clinical Trial. JAMA 2017; 318:1798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimenez-Saiz R, Chu DK, Mandur TS, Walker TD, Gordon ME, Chaudhary R, et al. Lifelong memory responses perpetuate humoral TH2 immunity and anaphylaxis in food allergy. J Allergy Clin Immunol 2017; 140:1604–15 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W, Freeland DMH, Nadeau KC. Food allergy: immune mechanisms, diagnosis and immunotherapy. Nat Rev Immunol 2016; 16:751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wambre E, Bajzik V, DeLong JH, O’Brien K, Nguyen QA, Speake C, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitson-Salazar A, Yin Y, Wansley DL, Young M, Bolan H, Arceo S, et al. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human T(H)2 cell subpopulation with enhanced function. J Allergy Clin Immunol 2016; 137:907–18 e9. [DOI] [PubMed] [Google Scholar]
- 12.Brough HA, Cousins DJ, Munteanu A, Wong YF, Sudra A, Makinson K, et al. IL-9 is a key component of memory TH cell peanut-specific responses from children with peanut allergy. J Allergy Clin Immunol 2014; 134:1329–38 e10. [DOI] [PubMed] [Google Scholar]
- 13.Sampson HA, Gerth van Wijk R, Bindslev-Jensen C, Sicherer S, Teuber SS, Burks AW, et al. Standardizing double-blind, placebo-controlled oral food challenges: American Academy of Allergy, Asthma & Immunology-European Academy of Allergy and Clinical Immunology PRACTALL consensus report. J Allergy Clin Immunol 2012; 130:1260–74. [DOI] [PubMed] [Google Scholar]
- 14.McDonald JH. G–test of independence. In: Handbook of Biological Statistics (3rd ed.). Baltimore, Maryland: Sparky House Publishing; 2014. p. 68–76. [Google Scholar]
- 15.Van der Loo MPJ. The stringdist package for approximate string matching The R Journal 2014; 6:111–22. [Google Scholar]
- 16.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, et al. Identifying specificity groups in the T cell receptor repertoire. Nature 2017; 547:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleischer DM, Greenhawt M, Sussman G, Begin P, Nowak-Wegrzyn A, Petroni D, et al. Effect of Epicutaneous Immunotherapy vs Placebo on Reaction to Peanut Protein Ingestion Among Children With Peanut Allergy: The PEPITES Randomized Clinical Trial. JAMA 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta RS, Lau CH, Hamilton RG, Donnell A, Newhall KK. Predicting outcomes of oral food challenges by using the allergen-specific IgE-total IgE ratio. J Allergy Clin Immunol Pract 2014; 2:300–5. [DOI] [PubMed] [Google Scholar]
- 22.Akdis M, Verhagen J, Taylor A, Karamloo F, Karagiannidis C, Crameri R, et al. Immune responses in healthy and allergic individuals are characterized by a fine balance between allergen-specific T regulatory 1 and T helper 2 cells. J Exp Med 2004; 199:1567–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renand A, Farrington M, Whalen E, Wambre E, Bajzik V, Chinthrajah S, et al. Heterogeneity of Ara h Component-Specific CD4 T Cell Responses in Peanut-Allergic Subjects. Front Immunol 2018; 9:1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seddiki N, Santner-Nanan B, Martinson J, Zaunders J, Sasson S, Landay A, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med 2006; 203:1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang D, Chen X, Jones SM, Wood RA, Sicherer SH, Burks AW, et al. Single-cell profiling of peanut-responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. J Allergy Clin Immunol 2018; 141:2107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissler KA, Rasooly M, DiMaggio T, Bolan H, Cantave D, Martino D, et al. Identification and analysis of peanut-specific effector T and regulatory T cells in children allergic and tolerant to peanut. J Allergy Clin Immunol 2018; 141:1699–710 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito T, Hirose K, Nakajima H. Bidirectional roles of IL-22 in the pathogenesis of allergic airway inflammation. Allergol Int 2018. [DOI] [PubMed] [Google Scholar]
- 28.Guttman-Yassky E, Krueger JG. Atopic dermatitis and psoriasis: two different immune diseases or one spectrum? Curr Opin Immunol 2017; 48:68–73. [DOI] [PubMed] [Google Scholar]
- 29.Bacher P, Heinrich F, Stervbo U, Nienen M, Vahldieck M, Iwert C, et al. Regulatory T Cell Specificity Directs Tolerance versus Allergy against Aeroantigens in Humans. Cell 2016; 167:1067–78 e16. [DOI] [PubMed] [Google Scholar]
- 30.Litjens NH, Boer K, Betjes MG. Identification of circulating human antigen-reactive CD4+ FOXP3+ natural regulatory T cells. J Immunol 2012; 188:1083–90. [DOI] [PubMed] [Google Scholar]
- 31.Schuster M, Glauben R, Plaza-Sirvent C, Schreiber L, Annemann M, Floess S, et al. IkappaB(NS) protein mediates regulatory T cell development via induction of the Foxp3 transcription factor. Immunity 2012; 37:998–1008. [DOI] [PubMed] [Google Scholar]
- 32.Mercer F, Kozhaya L, Unutmaz D. Expression and function of TNF and IL-1 receptors on human regulatory T cells. PLoS One 2010; 5:e8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeitelhofer M, Adzemovic MZ, Gomez-Cabrero D, Bergman P, Hochmeister S, N’Diaye M, et al. Functional genomics analysis of vitamin D effects on CD4+ T cells in vivo in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 2017; 114:E1678–E87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu D, Lan B, Din Z, Chen H, Chen G. A vitamin D receptor agonist converts CD4+ T cells to Foxp3+ regulatory T cells in patients with ulcerative colitis. Oncotarget 2017; 8:53552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaine CA, Soberman RJ. The CD200-CD200R1 inhibitory signaling pathway: immune regulation and host-pathogen interactions. Adv Immunol 2014; 121:191–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherwinski HM, Murphy CA, Joyce BL, Bigler ME, Song YS, Zurawski SM, et al. The CD200 receptor is a novel and potent regulator of murine and human mast cell function. J Immunol 2005; 174:1348–56. [DOI] [PubMed] [Google Scholar]
- 37.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med 2005; 11:1113–7. [DOI] [PubMed] [Google Scholar]
- 38.Chattopadhyay PK, Yu J, Roederer M. Live-cell assay to detect antigen-specific CD4+ T-cell responses by CD154 expression. Nat Protoc 2006; 1:1–6. [DOI] [PubMed] [Google Scholar]
- 39.Birrueta G, Tripple V, Pham J, Manohar M, James EA, Kwok WW, et al. Peanut-specific T cell responses in patients with different clinical reactivity. PLoS One 2018; 13:e0204620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Begin P, Nadeau KC. Changes in peanut-specific T-cell clonotype with oral immunotherapy. J Allergy Clin Immunol 2015; 135:1636–8. [DOI] [PubMed] [Google Scholar]
- 41.Ryan JF, Hovde R, Glanville J, Lyu SC, Ji X, Gupta S, et al. Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets. Proc Natl Acad Sci U S A 2016; 113:E1286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq and TCRb sequencing datasets generated in the course of this project have been deposited at the National Center for Biotechnology Information database of Genotypes and Phenotypes (dbGaP) and Sequence Read Archive under accession phs001897.v1.p1.