Abstract
Snail family zinc finger 2 (SLUG) is related to epithelial-mesenchymal transition (EMT). Quaking (QKI) is an RNA binding protein and has been indicated to have a relationship with EMT by recent studies. The prognostic value of SLUG and QKI in breast cancer patients still needs exploration. We conducted Immunohistochemistry (IHC) to evaluate the protein expression of SLUG and QKI and the prognostic value in 108 breast cancer (BC) patients. The Bc-GenExMiner database was used to compare the mRNA levels of two genes in different subgroups of BC patients. Kaplan-Meier plotter were used for survival data of SLUG and QKI gene. We also mined the cBioPortal database for co-expression analysis of QKI and EMT markers. Our results suggested that patients with higher expression of SLUG and QKI showed shorter overall survival time. The mRNA level of SLUG and QKI were higher in ER negative, PR negative, ≤ 51 y, and TNBC patients. SLUG mRNA showed no survival significance, while higher QKI mRNA expression level was correlated with worse clinical outcome in Kaplan-Meier Plotter database. The cBioPortal database showed that QKI was correlated to SLUG as well as other EMT markers like TWIST2, VIM, and ZEB2. QKI was indicated to be a potential prognostic marker for BC patients, and combined expression of SLUG and QKI showed the best prognostic value. Co-expression analysis indicated that QKI was likely to have a correlation with SLUG and EMT.
Keywords: EMT, QKI, breast cancer, prognosis, SLUG
Introduction
The incidence of breast cancer (BC) is quite high of various malignancies in women [1]. Systemic treatment based on molecular subtypes and clinical trial results has largely improved the prognosis of patients around the world. Researchers and oncologists still work hard on finding novel prognostic markers to discover more effective therapeutic targets for patients.
The epithelial-mesenchymal transition (EMT) is a series of preserved embryonic development related processes. During EMT, cells experience dissolution of cell-cell junctions, loss of polarity and gain the features of mesenchymal cells to get migratory and invasive properties, which facilitates the ability of metastasis of tumor. To date, studies have revealed the involvement of EMT in sophisticated molecular mechanisms and multiple signal pathways [2]. EMT markers, like TWIST1, TWIST2, ZEB1, ZEB2, SNAIL, and SLUG, were reported to have the ability to initiate EMT [3,4].
SLUG (SNAI2), a member of SNAIL zinc-finger family, functions as a translation factor (TF) [5]. SLUG was reported to induce the mammary stem cell state [6] and drug resistance [7]. As one of the key factors in the EMT process, SLUG had been detected in many malignant tumors, and high expression of SLUG was certified to be correlated with poor clinical outcome in malignant neoplasms such as lung cancer [8], colorectal cancer [9], gastric cancer [10], and esophageal cancer [11]. The prognostic value of SLUG in BC patients had been indicated in several studies [12], while other studies also showed an uncertain prognostic value of SLUG [13].
QKI (quaking), member of the STAR family, contains the KH domain and works as an RNA-binding protein (RBP) [14]. The major isoforms of QKI are QKI-5, QKI-6, and QKI-7 [15]. Recent studies suggested that the loss or overexpression of QKI was related to the development of various diseases or disorders, including schizophrenia [16], myelination [17], muscle-differentiation [18] and apoptosis [19], and certainly, cancer [20]. One study showed that QKI was confirmed as one of the most important RBPs through the TGF-β induced EMT of human mammary epithelial cells [21]. Therefore, chances are that QKI may be related to the EMT process, as well as SLUG in breast cancer. There are few studies focusing on the relationship between QKI expression and clinical prognosis in breast cancer patients. Thus, research is required for prognostic value of QKI, and the correlation between QKI and EMT in breast cancer patients.
In this study, we applied immunohistochemistry (IHC) to 108 BC patients’ tumor tissues to evaluate the protein expression level of SLUG and QKI. The clinicopathologic features and survival data of SLUG and QKI expression were also analyzed. We used breast cancer gene-expression miner (BC-GenExMiner) database to assess correlations between SLUG and QKI mRNA expression level and BC patients’ clinical parameters. Then, we explored the prognostic value of SLUG and QKI mRNA expression on the Kaplan-Meier Plotter database. Last, survival analysis of genetic alterations of SLUG and QKI and co-expression analysis of QKI and EMT markers mRNA expression levels were analyzed through the cBioPortal online database.
Materials and methods
Patients
Tumor sample sections for IHC were obtained from 108 patients with primary breast cancer who were treated at Huashan Hospital affiliated to Fudan University in 2010-2011. The paraffin embedded tumor tissue sections were kindly made by the pathology department of Huashan Hospital. The patients in this study received mastectomy or breast conservative surgery followed by axillary lymph node dissection. None of the patients had received irradiation or chemotherapy before the surgery. The mean follow up time was 88.5 months (range 37-95). The study was approved by the ethics committee of Huashan Hospital, Fudan University. All patients had signed informed consent. Related clinical data are presented in Table 1.
Table 1.
Clinicopathologic variables and the protein expression levels of SLUG and QKI in BC patients according to immunohistochemistry (n = 108)
| Characteristic | n | SLUG expression | P valuea | QKI expression | P valuea | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Low | High | Low | High | ||||
| Age (years) | |||||||
| ≤ 51 | 48 | 25 | 23 | 0.281 | 17 | 31 | 0.028* |
| > 51 | 60 | 25 | 35 | 34 | 26 | ||
| Tumor size (cm) | |||||||
| ≤ 2 | 72 | 32 | 40 | 0.585 | 37 | 35 | 0.220 |
| > 2 | 36 | 18 | 18 | 14 | 22 | ||
| Lymph node status | |||||||
| Negative | 67 | 30 | 37 | 0.685 | 31 | 36 | 0.800 |
| Positive | 41 | 20 | 21 | 20 | 21 | ||
| ER status | |||||||
| Negative | 21 | 7 | 14 | 0.184 | 9 | 12 | 0.655 |
| Positive | 87 | 43 | 44 | 42 | 45 | ||
| PR status | |||||||
| Negative | 44 | 15 | 29 | 0.035* | 22 | 22 | 0.632 |
| Positive | 64 | 35 | 29 | 29 | 35 | ||
| HER-2 statusb | |||||||
| Negative | 63 | 23 | 40 | 0.066 | 36 | 27 | 0.006* |
| Positive | 38 | 21 | 17 | 11 | 27 | ||
| Ki67 index | |||||||
| ≤ 14 | 63 | 32 | 31 | 0.267 | 24 | 21 | 0.282 |
| > 14 | 45 | 18 | 27 | 27 | 36 | ||
| Clinical TNM stage | |||||||
| I~II | 90 | 43 | 47 | 0.490 | 44 | 46 | 0.438 |
| III~IV | 18 | 7 | 11 | 7 | 11 | ||
| Histologic grade | |||||||
| I~II | 85 | 41 | 44 | 0.437 | 43 | 42 | 0.178 |
| III | 3 | 9 | 14 | 8 | 15 | ||
| Histologic type | |||||||
| IDC | 84 | 32 | 52 | 0.001* | 37 | 47 | 0.216 |
| Other | 24 | 18 | 6 | 14 | 10 | ||
| E-cadherin | |||||||
| Negative | 11 | 8 | 3 | 0.064 | 5 | 6 | 0.901 |
| Positive | 97 | 42 | 55 | 46 | 51 | ||
| Distant metastasis | |||||||
| Negative | 99 | 48 | 51 | 0.172 | 49 | 50 | 0.177 |
| Positive | 9 | 2 | 7 | 2 | 7 | ||
By Chi-square test.
There were 7 patients who had no results of HER2 FISH test and the IHC showed 2+ in history.
Statistically significant.
Immunohistochemistry
The specimens were fixed in 10% formalin and embedded in paraffin wax. The paraffin-embedded cancer tissues were cut at 4 μm and then dried at 65°C for 2-4 h. Then the sections were deparaffinized in xylene and rehydrated using ethanol. Endogenous peroxidase activity was blocked by 3% hydrogen peroxidase (H2O2) at 37% for 25 min. The sections were treated with citrate buffer (pH 6.0) for antigen repair. Then the slides were rinsed in phosphate buffer solution (PBS) and blocked with serum diluted in PBS. Then the slides were incubated overnight at 4°C with primary antibody (SLUG, 1:600; Abcam, Cambridge; QKI, 1:500; Bethyl). After washing in PBS three times and incubated with the secondary antibody (Dako) for 45 min at 37°C. Staining was displayed with the DAKO DBA solution. Harris hematoxylin was used to re-stain the nucleus.
Evaluation of SLUG and QKI protein expression by IHC
Immunohistochemistry was quantified and evaluated by two pathologists using a weighted histo-score. Cytoplasmic staining for SLUG and nuclear staining for QKI were considered positive. Ten fields of vision under ×400 magnification was randomly selected to evaluate the expression status.
The positive cells were graded as follows: 0: 0-4%; 1: 5-24%; 2: 25-50%; and 3: 51-100% positive tumor cells. The staining intensity was graded as: 0: negative; 1: weak; 2 moderate; and 3: strong. The histoscore of the sections was encoded from the multiplication of positive cells percentage score and the staining intensity score. The median of all the histoscores was 4, after counting the staining of SLUG and QKI. Based on this, patients could be divided into two groups: high expression (score of 4-9), and low expression (score of 0-3).
Statistical analysis
Statistical analysis was performed using SPSS 23.0 (SPSS Inc., Chicago, IL, USA). The chi-square test was used to assess the correlation between SLUG or QKI expression level with clinical parameters. Survival analysis was performed using the Kaplan Meier method and tested by the log rank method. In the tests above, P value < 0.05 was considered significant. All tests were two-sided.
Breast cancer gene-expression miner v4.1
Breast Cancer Gene-Expression Miner v4.1 (BC-GenExMiner v4.1) is an online data-analyzing the application of breast cancer, which has consisted 36 annotated genomic datasets and developed three statistical mining functions from three basic modules: expression module, prognostic module, and correlation module. The 36 datasets were from publicly available databases (Gene Expression Omnibus, ArrayExpress, Stanford microarray database, and on the author’s individual web pages). The latest update of BCthe-GenExMiner was on 2017/12/14, with 4823 updated data of gene annotation. The expression module can help viewers to compare the expression of a target gene to clinical variables of BC patients like hormonal receptors, and nodal status [22].
The Kaplan-Meier Plotter
The Kaplan-Meier Plotter (http://kmplot.com/) is an online database developed to analyze prognostic data, including relapse free survival (RFS), overall survival (OS), distant metastasis free survival (DMFS), and post progression survival (PPS) of gene mRNA expression levels in several kinds of cancer. So far, the database contains 54,675 genes on survival of breast cancer [23], ovarian cancer [24], lung cancer [25], and gastric cancer [26]. 1809 BC patients were identified from GEO (Affymetrix microarrays only). The patient samples were split into two groups according their median mRNA levels to verify the prognostic value of an aimed gene. The high and low expression groups were compared by the hazard ratio (HR), 95% confidence intervals (CI), log-rank P value, and the K-M survival plot. The Affymetrix ID is: 213139_at (SLUG) and 212265_at (QKI).
cBioPortal database
The cBioportal database (www.cbioportal.org) [27] provides various cancer genomics dataset resources. In breast cancer datasets, the METABRIC Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016) dataset [27] was chosen in this study, which contained genomic data of 2509 patient samples. OS plot of SLUG and QKI gene alteration were derived according to the cBioPortal’s online instruction. The cBioportal was also used to perform co-expression analysis of mRNA expression from of QKI and SLUG, and other EMT markers in patient samples from METABRIC dataset.
Results
Expression level of SLUG and QKI protein in breast cancer patients
Through immunohistochemistry (IHC), we assessed the expression level of SLUG and QKI in samples of breast carcinoma patients from Huashan Hospital. SLUG was mainly expressed in the cytoplasm of tumor cells and was partially found in the nucleus of tumor cells, while QKI was mainly expressed in the nucleus of tumor cells. In some cases, QKI was also expressed in the cytoplasm of tumor cells, and could be found in tumor stroma in some cases. Of the 108 cases analyzed, 58 had a high SLUG expression (53.7%) and 57 had a high QKI expression (52.8%) (Figure 1).
Figure 1.
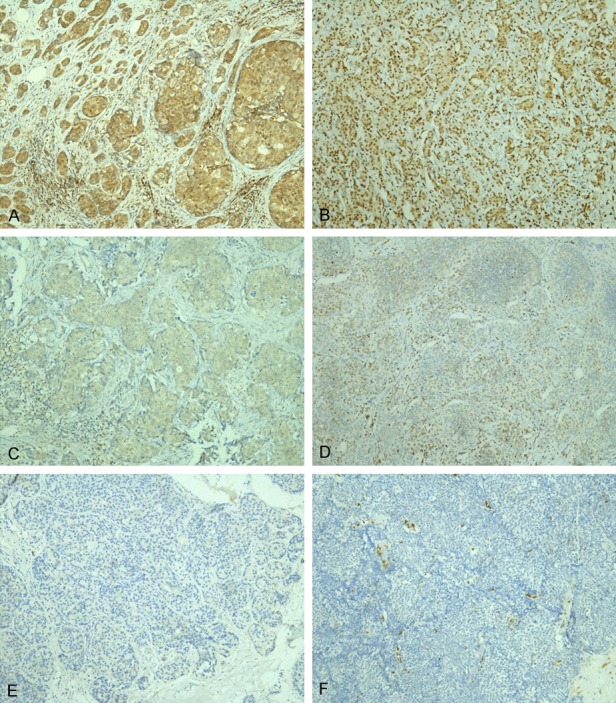
Expression of SLUG and QKI protein in invasive breast cancer (by IHC). Microscopic image of tissues sections stained for SLUG and QKI in invasive breast cancer (200×). A, B. High expression. C, D. Low expression. E, F. Negative. The left column of figures shows expression of SLUG and the right column shows expression of QKI.
Association of SLUG and QKI expression with pathologic characteristics in BC patients
To explore the relevance between SLUG or QKI expression level and breast cancer pathologic parameters, patients were divided into groups based on the different pathologic characteristics and expression levels of SLUG or QKI evaluated by IHC (Table 1). As shown, high level SLUG expression was observed to be negatively associated with progesterone receptor (PR) status (P = 0.035). The expression level of SLUG had correlation with histologic type of breast carcinoma (P = 0.001). There was no significant association between SLUG expression and parameters including age, tumor size, lymph node metastasis, TNM stage, distant metastasis, ki67 index, histological grade, estrogen receptor (ER) status, HER-2 status, and E-cadherin expression (P = 0.281, P = 0.585, P = 0.685, P = 0.490, P = 0.172, P = 0.267, P = 0.437, P = 0.184, P = 0.066, P = 0.064, respectively).
BC patients ≤ 51 y showed higher QKI expression compared to the > 51 y group (P = 0.028). The QKI expression level was observed to be positively correlated with HER2 status (P = 0.006). However, other clinicopathologic factors, including tumor size, lymph node metastasis, histological grade, histological type, TNM stage, distant metastasis, ki67 index, ER status, PR status, and E-cadherin expression showed no significant association with QKI expression (P = 0.220, P = 0.800, P = 0.178, P = 0.216, P = 0.438, P = 0.177, P = 0.282, P = 0.655, P = 0.632, P = 0.901, respectively).
Survival analysis
The Kaplan-Meier survival analysis revealed that SLUG high expression patients showed a worse prognosis for overall survival (OS) (P = 0.040), but not disease-free survival (DFS) (P = 0.139), compared to SLUG low expression patients (Figure 2A, 2B). QKI high expression BC patients showed worse prognosis for both OS (P = 0.047), and DFS (P = 0.024) (Figure 2C, 2D).
Figure 2.
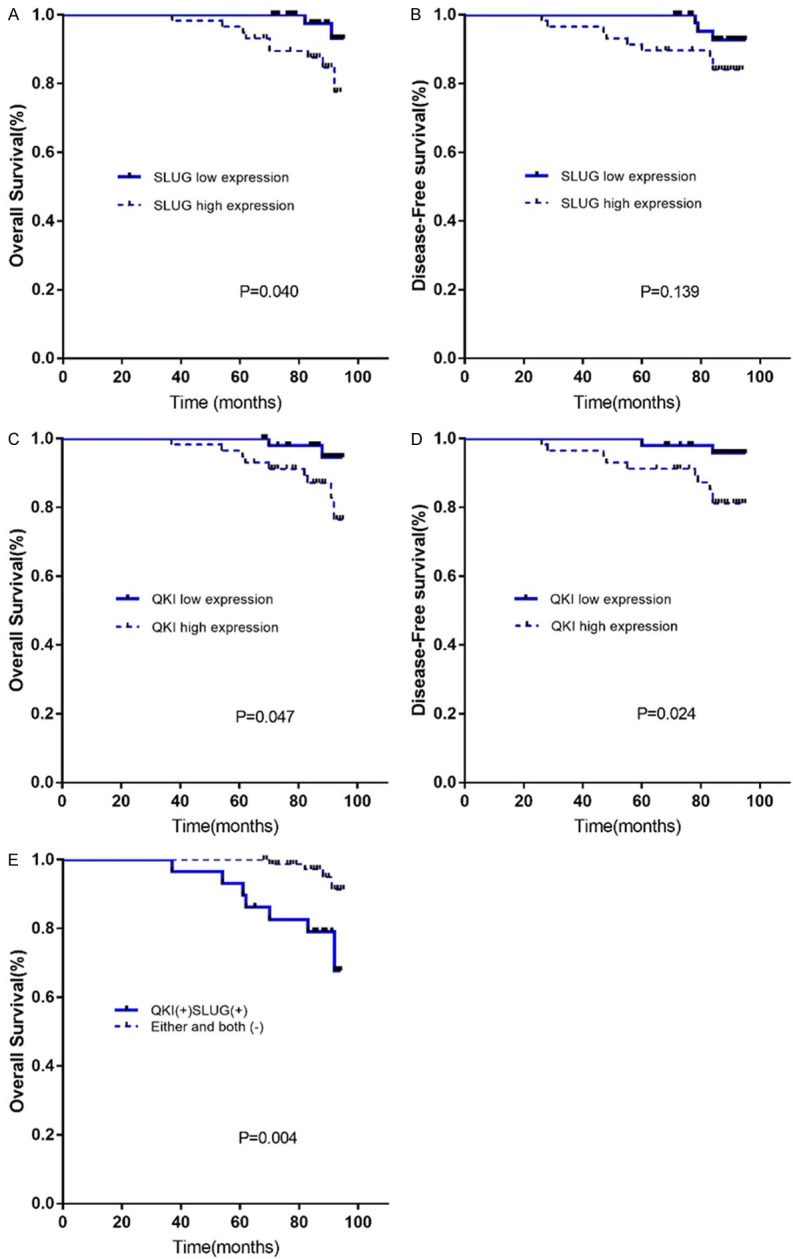
Kaplan-Meier analysis of (A) OS and (B) DFS of breast cancer patients. High expression of SLUG predicted worse overall prognosis but not disease-free survival. Kaplan-Meier analysis result of (C) OS and (D) DFS analysis of breast cancer patients showed that high expression of QKI protein had worse OS and DFS. (E) Patients with high expression of both SLUG and QKI protein had worse OS.
Then we combined the expression status of SLUG and QKI into two groups: both SLUG and QKI high expression, and patients with SLUG or QKI or both low expression. Considering that SLUG expression level did not show difference in DFS, we chose to analyze the OS of both SLUG and QKI high expression group and the other group. AS shown in Figure 2E, Kaplan-Meier analysis suggested that patients with both SLUG and QKI high expression had worse prognosis for OS (P=0.004), and the difference was significant.
The relationship between mRNA levels of SLUG and QKI and clinicopathological parameters of breast cancer patients
The BC-GenExMiner was carried out to compare the mRNA expression level of SLUG and QKI among groups of patients based on various clinicopathologic parameters. ER and PR status were found to be negatively correlated with both SLUG and QKI mRNA expression. The ≤ 51 y group was showed to have higher SLUG and QKI mRNA expression level than the > 51 y group. There was no significant difference between HER2(+) and HER2(-) group in SLUG or QKI mRNA expression. The expression levels of SLUG and QKI were both remarkably upregulated in TNBC patients. As regards nodal status, SLUG mRNA level showed no significant difference between lymph node positive and lymph node negative patients. The expression level of QKI was unexpectedly higher in the nodal negative breast cancer patients compared to nodal positive ones (Table 2).
Table 2.
The relationship between mRNA expression of SLUG and QKI and clinicopathologic parameters of breast cancer (from the breast cancer gene miner v4.1)
| Parameters | Cases | SLUG mRNA | P value | Cases | QKI mRNA | P value |
|---|---|---|---|---|---|---|
| Age | ||||||
| ≤ 51 | 1392 | ↑ | 0.0066* | 1411 | ↑ | 0.00472* |
| > 51 | 2209 | - | 2128 | - | ||
| Nodal status | ||||||
| Negative | 2493 | - | 0.1090 | 2322 | ↑ | < 0.0001* |
| Positive | 1561 | - | 1670 | - | ||
| ER (IHC) | ||||||
| Negative | 1559 | ↑ | < 0.0001* | 1501 | ↑ | < 0.0001* |
| Positive | 3987 | - | 3970 | - | ||
| PR (IHC) | ||||||
| Negative | 946 | ↑ | 0.0319* | 1076 | ↑ | < 0.0001* |
| Positive | 1439 | - | 1545 | - | ||
| HER2 (IHC) | ||||||
| Negative | 1409 | - | 0.9976 | 1596 | - | 0.7742 |
| Positive | 201 | - | 217 | - | ||
| TNBC | ||||||
| Not | 4099 | - | 0.0001* | 417 | - | < 0.0001* |
| TNBC | 374 | ↑ | 4152 | ↑ |
Statistically significant.
The prognostic value of SLUG and QKI mRNA levels in breast cancer
In addition to the survival analysis from the clinical cases in this research, the prognostic value of SLUG and QKI mRNA expression levels was also studied by the online Kaplan-Meier Plotter database. According to Kaplan-Meier curve and log-rank test, the expression level of SLUG mRNA showed no correlation with the relapse free survival (RFS) (P = 0.3), overall survival (OS) (P = 0.35), distant metastasis free survival (DMFS) (P = 0.22), and post-progression survival (PPS) (P = 0.27) (Supplementary Table 1). Increased QKI mRNA expression level was correlated with worse OS (HR = 1.26; 95% CI: 1.02-1.56, P = 0.035), RFS (Hazard ratio, (HR) = 1.14; 95% CI: 1.02-1.27, P = 0.019), and PPS (HR = 1.52; 95% CI: 1.19-1.94, P = 0.00082), but not DMFS (HR = 1.2; 95% CI: 0.99-1.46, P = 0.061), which did not show statistical significance (Figure 3).
Figure 3.
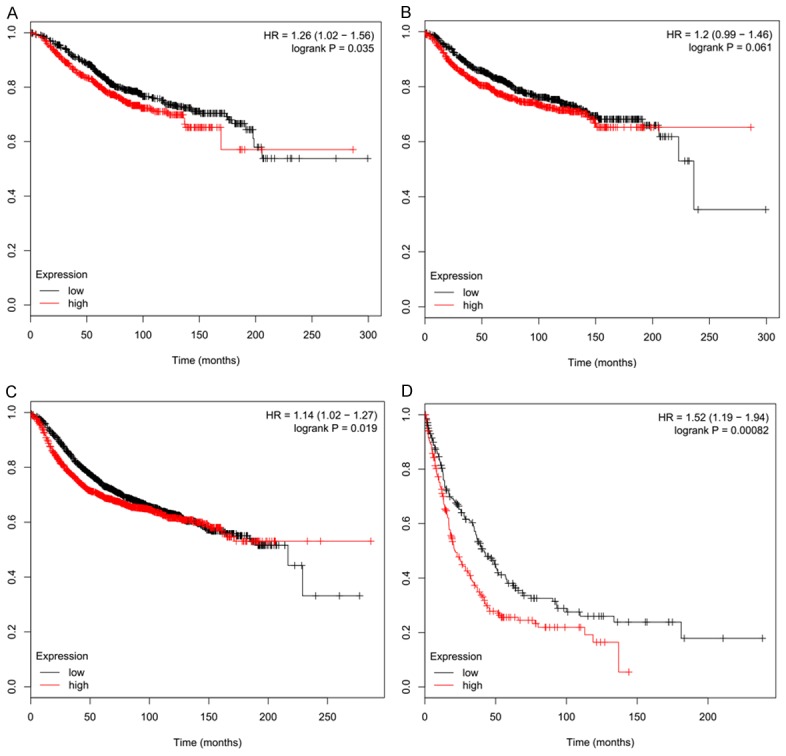
Higher QKI mRNA expression of showed worse (A) OS, (C) RFS, and (D) PPS from Kaplan-Meier Plotter dataset. Expression of QKI did not show significant difference in (B) DFS in patients.
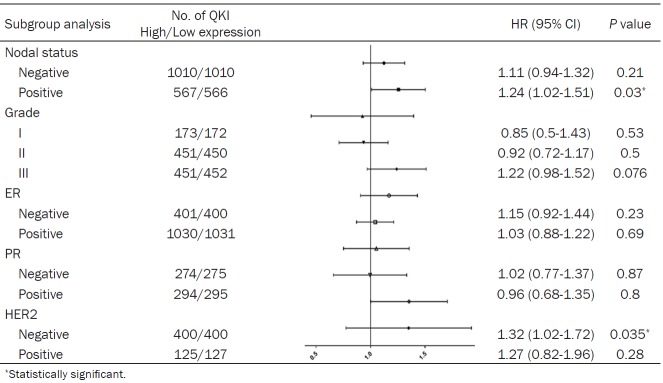
Then the correlation of QKI mRNA expression and clinical outcome in BC patients of different subgroups of ER, PR, HER2, and other factors were studied further. Unfortunately, higher QKI mRNA expression level did not show worse OS in all kinds of subgroups (Supplementary Table 2). Then RFS was chosen for analysis, for its more significant difference between high and low QKI levels and the larger sample of population. Higher QKI mRNA expression level was correlated to worse RFS in lymph node positive patients (HR = 1.24; 95% CI: 1.02-1.51, P = 0.03) and HER2-negative breast cancer patients (HR = 1.32; 95% CI: 1.02-1.72, P = 0.035) (Table 3).
Table 3.
Correlation of QKI mRNA expression and clinical survival of breast cancer patients with different clinicopathologic factors (from Kaplan-Meier Plotter) (RFS)
Survival and co-expression analysis of cBioPortal
By analyzing the dataset of Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016) using the cBioPortal online database, survival plot showed that breast cancer patients with QKI and SLUG gene alterations had worse overall survival compared to those without QKI or SLUG gene alterations (Figure 4).
Figure 4.
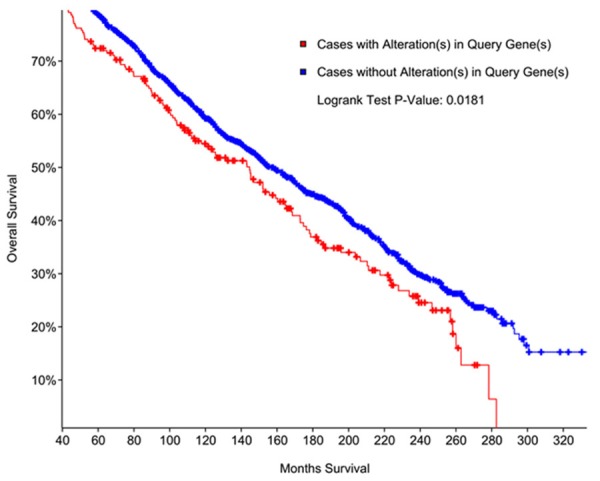
Kaplan-Meier overall survival analysis of patients with or without gene alterations of SLUG and QKI.
In the co-expression analysis, the mRNA expression levels of SLUG and QKI in METABRIC were positively correlated (Figure 5A, P < 0.05). The co-expression correlations of other EMT-related markers were also analyzed by cBioPortal. The mRNA expression levels of VIM, TWIST2, and ZEB2 in breast cancer patients were found to be positively correlated with the expression of QKI, as shown in Figure 5B-D.
Figure 5.
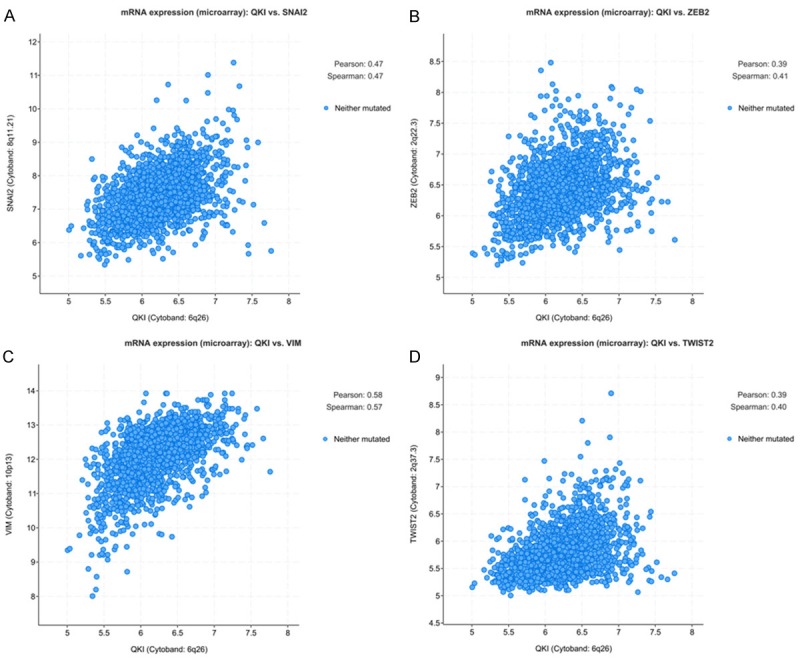
(A) Correlation of SLUG and QKI mRNA expression from METABIC dataset. Correlation of QKI mRNA expression and (B) ZEB2 (C) VIM (D) TWIST2 from METABIC dataset.
Discussion
In our study, IHC expression level of SLUG had correlation with histologic type (P = 0.0001) and PR status (P = 0.035), and the QKI expression level was positively related to HER2 status (P = 0.006), and negatively related to patients’ age (P = 0.028). The Kaplan-Meier survival analysis based on IHC evaluation showed that the overall survival of patients with low SLUG expression was better than those with higher SLUG expression (P = 0.040). The overall survival and disease-free survival of patients with low QKI expression were better than patients with high QKI expression (P = 0.047, P = 0.024). Patients with both higher level expression of SLUG and QKI showed worse prognosis. These results indicated that high protein expression level of QKI may predict a worse outcome of BC patients, while the predictive value of SLUG overexpression was not as good as QKI. The limited number of patients in this study should also be considered when assessing the prognostic value of these two genes.
And then the BC GenExMiner database showed that higher expression of SLUG and QKI was related to ER and PR down regulation, lower patients age, and TNBC patients, which were worse outcome suggesting prognostic factors of BC patients. And parameter subgroups, like SLUG expression in different PR status, QKI expression in age subgroups showed similar results between IHC analysis and BC GenExMiner analysis. Therefore, the result of IHC analysis and BC GenExMiner showed that patients with higher expression levels of SLUG and QKI tended to have a worse clinical prognosis.
The data from KM Plotter showed a correlation between SLUG or QKI mRNA expression and patients’ outcome. Different SLUG mRNA expression level showed no significant difference on their prognosis of OS, RFS, DMFS, and PPS. High expression of QKI mRNA level was related to the worse OS, RFS, and PPS, but not DMFS.
Dozens of researchers had verified the oncogenic function and prognosis value of SLUG. Some studies demonstrated that SLUG can promote metastasis of various cancers, and the mechanisms included promoting EMT by down regulation of E-cadherin [28], which was commonly observed in the initiation of the EMT process [12], activating the expression of MMP1, which can participate in the breakdown of extracellular matrix to initiate the metastasis of tumor [29], and inducing the hormone therapy resistance through repressing ER α expression [30]. Meanwhile, some studies showed indeterminate results for prognostic value of SLUG [13,31]. The survival analysis based on the IHC score in this study and the KM Plotter online data did not show strong evidence that higher expression level of SLUG was a risk factor for prognosis of breast carcinoma patients. One meta-analysis about the prognostic value of SLUG in different kinds of tumor indicated that the protein expression, rather than mRNA transcription of SLUG should be considered as a potential prognosis factor of tumors [32], which was corresponding to the result of our study. Besides, the publication bias of SLUG studies should also be considered when evaluating its prognostic value. Another meta-analysis showed that SLUG expression was significantly associated with poor prognosis of metastatic breast cancer patients [33]. So, the failure of the IHC SLUG expression level of this study to predict the DFS of BC patients may also due to that most of the population were early-stage BC patients.
The IHC results and KM Plotter survival analysis of patients with different QKI expression levels were compatible. However, it should be noticed that other probes for QKI showed a reverse result, which indicated that patients with higher expression of QKI mRNA had better prognosis of RFS (Supplementary Table 3). However, high expression of these QKI probes did not show better OS, DMFS, and PPS. Therefore, we chose the probe that showed significant difference in OS, RFS, and PPS at the same time, which suggested a consistent result with this study. Previous papers studied QKI showed both oncogenic and tumor suppressing effect of it. QKI was observed to be down regulated in lung cancer [34], gastric cancer [35], and prostate cancer [36]. On the other hand, He et al. reported the upregulation of QKI and its ability to promote oncogenesis in esophageal carcinoma [37]. QKI-5 could repress the expression of FOXO1, a tumor suppress gene in breast cancer [38]. Hence, the function and mechanism of QKI in tumorigenesis remain to be elucidated more deeply.
The METABRIC dataset showed that patients with both gene alteration of SLUG and QKI had worse overall survival. Yet, independent SLUG or QKI gene alteration did not show prognostic value (Supplementary Figure 1). Our IHC survival analysis result showed that patients with both SLUG and QKI high expression had worse OS, compared to the other group of patients, which resembled the METABRIC result. These two results might indicate a possibility that SLUG and QKI can cooperatively promote the progression of tumor. More studies are warranted for this assumption in breast cancer.
Moreover, the co-expression analysis of QKI and SLUG, and QKI and other EMT markers from METABRIC dataset revealed an interrelationship between QKI and EMT. This result was in line with Simon et al.’s research which demonstrated that QKI may be the key factor promoting the generation of several circRNAs during the process from epithelial to mesenchymal [21]. Yang et al. also determined that QKI had a potential role in promoting mesenchymal splicing patterns through motif analysis [39]. One recently published study found that QKI could regulate the splicing and function of the FLNB, which could play a causal role in the regulation of EMT [40].
In summary, this research explored the potential prognostic value of SLUG and QKI in breast carcinoma patients and had several implications. The IHC evaluation and survival analysis showed that the elevated expression of QKI was related to worse clinical outcome in breast cancer patients. High level of SLUG expression can only predict worse OS, but not DFS. Then, overexpression of SLUG and QKI mRNA was more frequently observed in ER-negative, PR-negative, and TNBC breast cancer patients. Third, the data from KM plotter showed that high QKI, but not SLUG mRNA expression level could indicate a worse outcome for BC patients. Lastly, the analysis of METABRIC dataset showed the positive correlation between QKI and SLUG, and other EMT markers as well. Patients with gene alteration of both SLUG and QKI tended to have worse overall survival. Therefore, QKI is expected to be further studied for its prognostic value and molecular mechanism of occurrence and progression of breast carcinoma.
Conclusion
Based on IHC and survival analysis in our experiments, and the survival data from KM plotter, the prognostic value of SLUG was not very notable, while QKI showed a better prognostic value in BC patients. The combined SLUG and QKI expression suggested a worse prognosis in BC patients. The correlation of QKI and EMT was verified in a co-expression analysis of METABRIC dataset, indicating a relationship of QKI and EMT, and the possibility that SLUG and QKI may cooperatively promote tumorigenesis in breast cancer.
Acknowledgements
This work was kindly supported by Science and Technology Commission of Shanghai Municipality, China (No. 15411952503 to Dr Q.Z.); And the National Nature Science Foundation of China (No. 81602326 to Dr Y.T.J), National Natural Science Foundation of China (No. 81772796 to Dr X.P.L).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Pan R, Zhu M, Yu C, Lv J, Guo Y, Bian Z, Yang L, Chen Y, Hu Z, Chen Z, Li L, Shen H China Kadoorie Biobank Collaborative Group. Cancer incidence and mortality: a cohort study in China, 2008-2013. Int J Cancer. 2017;141:1315–1323. doi: 10.1002/ijc.30825. [DOI] [PubMed] [Google Scholar]
- 2.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallerand H, Robert G, Pasticier G, Ravaud A, Ballanger P, Reiter RE, Ferrière JM. The epithelial-mesenchymal transition-inducing factor TWIST is an attractive target in advanced and/or metastatic bladder and prostate cancers. Urol Oncol. 2010;28:473–9. doi: 10.1016/j.urolonc.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype. Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 5.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–66. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 6.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–28. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih JY, Tsai MF, Chang TH, Chang YL, Yuan A, Yu CJ, Lin SB, Liou GY, Lee ML, Chen JJ, Hong TM, Yang SC, Su JL, Lee YC, Yang PC. Transcription repressor Slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res. 2005;11:8070–8. doi: 10.1158/1078-0432.CCR-05-0687. [DOI] [PubMed] [Google Scholar]
- 9.Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–22. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro Alves C, Rosivatz E, Schott C, Hollweck R, Becker I, Sarbia M, Carneiro F, Becker KF. Slug is overexpressed in gastric carcinomas andmay act synergistically with SIP1 and Snail in the down-regulation of E-cadherin. J Pathol. 2007;211:507–15. doi: 10.1002/path.2138. [DOI] [PubMed] [Google Scholar]
- 11.Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S, Aikou T. Slug expression in the e-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1174–80. [PubMed] [Google Scholar]
- 12.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 13.van Nes JG, de Kruijf EM, Putter H, Faratian D, Munro A, Campbell F, Smit VT, Liefers GJ, Kuppen PJ, van de Velde CJ, Bartlett JM. Co-expression of SNAIL and TWIST determines prognosis in estrogen receptor-positive early breast cancer patients. Breast Cancer Res Treat. 2012;133:49–59. doi: 10.1007/s10549-011-1684-y. [DOI] [PubMed] [Google Scholar]
- 14.Hall MP, Nagel RJ, Fagg WS, Shiue L, Cline MS, Perriman RJ, Donohue JP, Ares M Jr. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA. 2013;19:627–38. doi: 10.1261/rna.038422.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teplova M, Hafner M, Teplov D, Essig K, Tuschl T, Patel DJ. Structure-function studies of STAR family quaking proteins bound to their in vivo RNA target sites. Genes Dev. 2013;27:928–940. doi: 10.1101/gad.216531.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci U S A. 2006;103:7482–7. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu HY, Dawson MR, Reynolds R, Hardy RJ. Expression of QKI proteins and MAP1B identifies actively myelinating oligodendrocytes in adult rat brain. Mol Cell Neurosci. 2001;17:292–302. doi: 10.1006/mcne.2000.0941. [DOI] [PubMed] [Google Scholar]
- 18.van der Veer EP, de Bruin RG, Kraaijeveld AO, de Vries MR, Bot I, Pera T, Segers FM, Trompet S, van Gils JM, Roeten MK, Beckers CM, van Santbrink PJ, Janssen A, van Solingen C, Swildens J, de Boer HC, Peters EA, Bijkerk R, Rousch M, Doop M, Kuiper J, Schalij MJ, van der Wal AC, Richard S, van Berkel TJ, Pickering JG, Hiemstra PS, Goumans MJ, Rabelink TJ, de Vries AA, Quax PH, Jukema JW, Biessen EA, van Zonneveld AJ. Quaking, an RNA-binding protein, is a critical regulator of vascular smooth muscle cell phenotype. Circ Res. 2013;113:1065–75. doi: 10.1161/CIRCRESAHA.113.301302. [DOI] [PubMed] [Google Scholar]
- 19.Guo W, Jiang T, Lian C, Wang H, Zheng Q, Ma H. QKI deficiency promotes FoxO1 mediated nitrosative stress and endoplasmic reticulum stress contributing to increased vulnerability to ischemic injury in diabetic heart. J Mol Cell Cardiol. 2014;75:131–40. doi: 10.1016/j.yjmcc.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Li ZZ, Kondo T, Murata T, Ebersole TA, Nishi T, Tada K, Ushio Y, Yamamura K, Abe K. Expression of Hqk encoding a KH RNA binding protein is altered in human glioma. Jpn J Cancer Res. 2002;93:167–77. doi: 10.1111/j.1349-7006.2002.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Jézéquel P, Frénel JS, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 24.Győrffy B, Bottai G, Lehmann-Che J, Kéri G, Orfi L, Iwamoto T, Desmedt C, Bianchini G, Turner NC, de Thè H, André F, Sotiriou C, Hortobagyi GN, Di Leo A, Pusztai L, Santarpia L. TP53 mutation-correlated genes predict the risk of tumor relapse and identify MPS1 as a potential therapeutic kinase in TP53-mutated breast cancers. Mol Oncol. 2014;8:508–19. doi: 10.1016/j.molonc.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolós V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 29.Shen CJ, Kuo YL, Chen CC, Chen MJ, Cheng YM. MMP1 expression is activated by Slug and enhances multi-drug resistance (MDR) in breast cancer. PLoS One. 2017;12:e0174487. doi: 10.1371/journal.pone.0174487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Wu Y, Abbatiello TC, Wu WL, Kim JR, Sarkissyan M, Sarkissyan S, Chung SS, Elshimali Y, Vadgama JV. Slug contributes to cancer progression by direct regulation of ERα signaling pathway. Int J Oncol. 2015;46:1461–72. doi: 10.3892/ijo.2015.2878. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Lemma S, Karihtala P, Haapasaari KM, Jantunen E, Soini Y, Bloigu R, Pasanen AK, Turpeenniemi-Hujanen T, Kuittinen O. Biological roles and prognostic values of the epithelial-mesenchymal transition-mediating transcription factors Twist, ZEB1 and Slug in diffuse large B-cell lymphoma. Histopathology. 2013;62:326–33. doi: 10.1111/his.12000. [DOI] [PubMed] [Google Scholar]
- 32.Huang C, Zhang P, Zhang D, Weng X. The prognostic implication of slug in all tumour patients - a systematic meta-analysis. Eur J Clin Invest. 2016;46:398–407. doi: 10.1111/eci.12608. [DOI] [PubMed] [Google Scholar]
- 33.Imani S, Hosseinifard H, Cheng J, Wei C, Fu J. Prognostic value of EMT-inducing transcription factors (EMT-TFs) in metastatic breast cancer: a systematic review and meta-analysis. Sci Rep. 2016;6:28587. doi: 10.1038/srep28587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Li X, Sun C, Shi C, Hua D, Yu L, Wen Y, Hua F, Wang Q, Zhou Q, Yu S. Quaking-5 suppresses aggressiveness of lung cancer cells through inhibiting β-cateninsignaling pathway. Oncotarget. 2017;8:82174–82184. doi: 10.18632/oncotarget.19066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bian Y, Wang L, Lu H, Yang G, Zhang Z, Fu H, Lu X, Wei M, Sun J, Zhao Q, Dong G, Lu Z. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem Biophys Res Commun. 2012;422:187–93. doi: 10.1016/j.bbrc.2012.04.138. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Zhang G, Wei M, Lu X, Fu H, Feng F, Wang S, Lu W, Wu N, Lu Z, Yuan J. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic and prognostic protein. Cancer Biol Ther. 2014;15:108–18. doi: 10.4161/cbt.26722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Z, Yi J, Liu X, Chen J, Han S, Jin L, Chen L, Song H. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15:51. doi: 10.1186/s12943-016-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Yu F, Jin L, Yang G, Ji L, Wang F, Lu Z. Post-transcriptional repression of FOXO1 by QKI results in low levels of FOXO1 expression in breast cancer cells. Oncol Rep. 2014;31:1459–65. doi: 10.3892/or.2013.2957. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, Xing Y, Carstens RP. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol Cell Biol. 2016;36:1704–19. doi: 10.1128/MCB.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Choi PS, Chaffer CL, Labella K, Hwang JH, Giacomelli AO, Kim JW, Ilic N, Doench JG, Ly SH, Dai C, Hagel K, Hong AL, Gjoerup O, Goel S, Ge JY, Root DE, Zhao JJ, Brooks AN, Weinberg RA, Hahn WC. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. Elife. 2018:7. doi: 10.7554/eLife.37184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.