Abstract
Background
Hepatocellular carcinoma (HCC) is one of the most common cancers, whereas the molecular mechanism remains largely unknown. PRAS40 (encoded by AKT1S1) phosphorylation was increased in human melanoma, prostate cancer and lung cancer specimens, which was considered as the results of Akt activation. However the mechanism in detail and its role in HCC stay elusive.
Methods
PRAS40 expression and phosphorylation were analyzed in HCC specimens, and the survival rates of patients were investigated. Functional analyses of PRAS40 in HCC were performed in vivo and in vitro. The miR-124-3p binding sites in PRAS40 were investigated using luciferase assay. MiR-124-3p expression in HCC specimens was examined by In Situ hybridization, and the correlation to PRAS40 level was evaluated.
Findings
The phosphorylation, protein and mRNA levels of PRAS40 were increased significantly in HCC specimens from our cohorts and TCGA database, which was positively correlated to the poor prognosis of HCC patients. Compared to Akt1s1+/+ mice, hepatocarcinogenesis was suppressed in Akt1s1−/− mice, and the activation of Akt was impaired. PRAS40 depletion resulted in the inhibition of HCC cellular proliferation. Tumor suppressor miR-124-3p was found to downregulate PRAS40 expression by targeting its 3′UTR. MiR-124-3p levels were inversely correlated to PRAS40 protein and phosphorylation levels in HCC specimens. The proliferation inhibition by miR-124-3p mimics was partially reversed by exogenous PRAS40 introduction in HCC cells.
Interpretation
PRAS40 hyperexpression induced by loss of miR-124-3p contributes to PRAS40 hyperphosphorylation and hepatocarcinogenesis. These results could be expected to offer novel clues for understanding hepatocarcinogenesis and developing approaches.
Keywords: Hepatocellular carcinoma, Carcinogenesis, Cell signaling, Microrna, Molecular oncology
Research in context.
Evidence before this study:
We investigated all the manuscripts on PRAS40 in tumor included in PubMed, and found that PRAS40 phosphorylation was reported to be increased in human melanoma, prostate cancer and non-small cell lung cancer specimens. However, it remains elusive whether the hyperphosphorylation of PRAS40 is caused only by the activation of the upstream Akt pathway, or is the result of its own hyperexpression. Further, the regulation of PRAS40 phosphorylation was reported to be related to several pathways, whereas the regulation of PRAS40 expression remains largely unknown except for the regulation by EWS in Ewing's sarcoma.
Added value of this study
Here, we report the significant increase of the protein and mRNA levels of PRAS40 in HCC tissue samples from both our cohorts and TCGA database, similar as the increase of p-PRAS40, suggesting that the increase of PRAS40 phosphorylation in HCC was mainly caused by the augmentation of PRAS40 expression. Both the expression and phosphorylation of PRAS40 is positively correlated to the poor prognosis of HCC patients, and hepatogenesis was suppressed in Akt1s1−/− mice, showing the critical role of PRAS40 in HCC. Further, miR-124-3p levels are inversely correlated to PRAS40 protein levels in both human and mouse HCC specimens, and miR-124-3p targets PRAS40 expression.
Implications of all the available evidence
PRAS40 may be a potential therapeutic target for HCC. MiR-124-3p could be considered as an approach targeting PRAS40 in HCC.
Alt-text: Unlabelled box
1. Introduction
Liver cancer is one of the most common cancers, and hepatocellular carcinoma (HCC) is the most frequent primary liver cancer. The incidence of HCC is increasing, whereas the treatment options are limited and often inefficient [1]. The poor understanding of the molecular mechanism of HCC carcinogenesis and progression is considered as the main obstruction.
PRAS40 (encoded by AKT1S1) was primarily found as a 14-3-3 binding protein [2], characterized as a substrate for Akt [3] and a component of mTOR complex 1 [3], [4], [5], [6], [7]. PRAS40 plays an essential role in cell survival, proliferation, metastasis, apoptosis, senescence, immunoregulation and protein degradation in different species [3,[8], [9], [10]]. Hyperphosphorylation of PRAS40 has been reported in human melanoma, prostate cancer and non-small cell lung cancer [10], [11], [12]. However, it remains elusive whether the hyperphosphorylation of PRAS40 is caused only by the activation of the upstream Akt pathway, or is the result of its own hyperexpression. PRAS40 knockdown suppresses the cellular proliferation by deregulating apoptosis [9,10], which might be associated with p53 upregulation in a Ribosomal protein 11-dependent manner [13]. In addition, PRAS40 is phosphorylated by Rab11-family interacting protein 4, mediating the migration and invasion of HCC cells [14]. However, it needs to be clarified whether the expression level of PRAS40 associates with HCC progression.
Previously, we reported that as an mRNA target of EWS, PRAS40 protein expression is inhibited by EWS [9]. PRAS40 protein level is increased due to the haploinsufficiency of EWS in Ewing sarcoma family tumor cell lines [9], whereas the regulation system for PRAS40 expression in the tumors without EWS aberrant expression remains largely unknown. MicroRNAs (miRNAs) control the expression of the target mRNAs by pairing to them to alter transcription, mRNA stability or translation [15], [16], [17], [18]. Either the loss or gain of miRNA function could contribute to tumorigenesis by regulating the expression of oncogenes or tumor suppressor genes, respectively. Aberrant expression of miRNAs, such as miR-21, miR-29 and miR-221, has been reported in HCC [19], [20], [21]. MiR-124-3p is highly conserved and encoded by three genes miR-124-1, miR-124-2, and miR-124-3 located on 8p23.1, 8q12.3 and 20q13.33, respectively. MiR-124-3p is one of the most expressed miRNAs in brain, and plays important roles in the development and function of nervous system [22], [23], [24]. Recently, miR-124 has been found to be able to control tumorigenesis [25], [26], [27], [28], immunology [29], [30], [31], metabolism [32], reproduction [33] and so on. MiR-124 is downregulated in various tumors, resulting in the hyperexpression of multiple targets, such as cyclin-dependent kinase 4 [34], CBL [35], C—C motif ligand 2 and Interleukin-8 [36], polypyrimidine tract-binding protein 1 [37], Interleukin-6, Signal transducer activator of transcription 3 (STAT3) [29] and Nuclear factor kappa B [28,38], which contributes to tumorigenesis and metastasis. In HCC, miR-124 is hypoexpressed due to the hypermethylation of the promoters [39,40] or the hypoexpression of the Hepatocyte nuclear factor 4α (HNF4α) [25,28]. MiR-124 inhibits the metastasis of HCC by targeting Rho Associated Protein Kinase 2 and Enhancer of zeste homolog 2 [26], or targeting transcription factor Specificity protein 1 to suppress the expression of integrin αV indirectly [41]. Therefore, miR-124 works as a tumor suppressive miRNA, while the mechanism in detail needs to be determined further.
Herein we reported the hyperexpression and hyperphosphorylation of PRAS40 in human HCC, and a positive correlation between increased expression and phosphorylation levels of PRAS40 and the overall survival of HCC patients. PRAS40 depletion resulted in a suppression of hepatocarcinogenesis and cellular proliferation. Further tumor suppressor miR-124-3p could bind the 3′UTR of PRAS40 and downregulate the expression of PRAS40. In addition, miR-124-3p level presents an inverse correlation to PRAS40 level in HCC specimens. Therefore the hypoexpression of miR-124-3p led to an abnormally increased expression of PRAS40, which contributes to PRAS40 phosphorylation, HCC carcinogenesis and progression. These results could be expected to offer novel clues for developing HCC treatment.
2. Materials and methods
2.1. Patient samples
The matched primary cancer tissue and their corresponding adjacent peri‑cancer tissue of HCC in cohort 1 (n = 22) were obtained from the Second Affiliated Hospital of Dalian Medical University. Tissue arrays HLiv-HCC180Bch-01 (n = 44, Cohort 2) and OD-CT-DgLiv01-012 (n = 50 for PRAS40 and p-PRAS40 staining; n = 49 for miR-124-3p staining, Cohort 3) were purchased from Shanghai Outdo Biotech Co. All the patients had not received preoperative anticancer treatment. This study was approved by the Medical Ethics Committees of Dalian Medical University (ME201904), and all the materials were sufficiently anonymised.
2.2. Akt1s1 knockout mice construction
This study was approved by the animal care and use committee of Dalian Medical University (AEE17054). Methods were carried out in accordance with the approved guidelines of Dalian Medical University. Mice were maintained in a temperature- and humidity-controlled room on a 12-h light/dark cycle at Specific Pathogen Free Experimental Animal Center of Dalian Medical University.
Akt1s1 knockout mouse was created based on CRISPR/Cas9 system by Beijing Biocytogen (Beijing, China). The exons 2–5 at the EGE-HYT-006 locus in the mouse genome were deleted, which was positively confirmed by PCR product sequencing. None of the top ten possible off-targets were confirmed. The wild type and mutant alleles for Akt1s1 mutant mice were detected in a multiplex PCR reaction using a pair of primers (Forward: AACTGGATGCATGAGAATCGGGACT; Reverse: GGGGAACCGGGATACAATTGTCAGG).
3. Results
3.1. PRAS40 expression and phosphorylation are upregulated in HCC
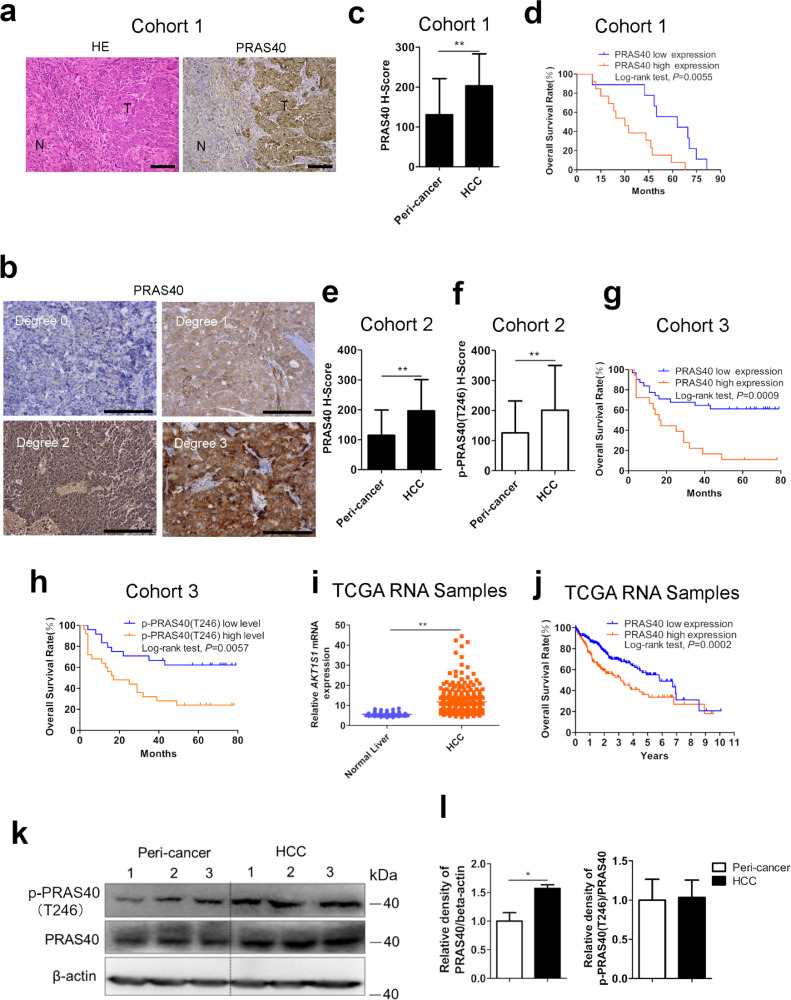
To determine the possible role of PRAS40 in HCC carcinogenesis and progression in vivo, we first analyzed the protein levels by IHC in 3 independent patient cohorts (Fig. 1a–h, Supplementary Table 1–5, and Supplementary Fig. 1). A significant increase of PRAS40 staining in HCC tissue was observed compared to the peri‑cancer region (Fig. 1a). Based on staining intensity (Fig. 1b) and extent, we calculated the H-Scores of PRAS40 staining [42]. The IHC results of 22 (Cohort 1, Supplementary Table 1) and 44 pairs (Cohort 2, Supplementary Table 3) of HCC and peri‑cancer clinical tissue samples showed significantly higher scores of PRAS40 staining in HCC tissue than the paired peri‑cancer tissue, respectively (P<0.01 from Student's t-test, Fig. 1c, e and Supplementary Fig. 1). P-PRAS40 staining showed a similar tendency (P<0.01 from Student's t-test, Fig. 1f, Supplementary Fig. 1). Next we studied the RNA-seq results in TCGA LIHC (Liver hepatocellular carcinoma) database (The Cancer Genome Atlas) [43]. The expression of AKT1S1 mRNA in 371 HCC specimens (median FPKM value=10.55) showed a significantly higher level than that in 50 normal liver samples (median FPKM value=5.42, P<0.01, Fig. 1i). Among these samples, AKT1S1 DNA copy number was investigated in 97 HCC specimens and 59 normal liver samples, whereas no significant change was clarified (Supplementary Fig. 2) (https://www.oncomine.com). To confirm the significance of the augmentation of PRAS40 protein and phosphorylation levels in HCC, we next constructed a DEN-induced HCC model in mice, and the results suggested that PRAS40 protein and phosphorylation levels were increased in HCC tissue significantly (Fig. 1k and l). The ratio of p-PRAS40/PRAS40 was similar in both HCC and peri‑cancer tissue, suggesting that the increase of p-PRAS40 level in HCC tissue was mainly caused by the augmentation of PRAS40 expression (Fig. 1l). Further we compared the protein levels of PRAS40 in 7 HCC cell lines and normal hepatocyte cell line THLE-3. PRAS40 protein levels were higher in all of the HCC cells than that in normal hepatocytes (Supplementary Fig. 3).
Fig. 1.
The protein levels of PRAS40 in HCC tissue and its correlation to the survival rate of HCC patients. a–d. Analyses of 22 pairs of primary HCC and peri‑cancer tissue samples in Cohort 1. HE and IHC staining of PRAS40 in HCC and peri‑cancer tissue (a). Degrees indicating the intensity of PRAS40 staining in representative HCC tissue (b). H-scores multiplied by the intensity and extent of PRAS40 staining in HCC and peri‑cancer tissue (c). The correlation of PRAS40 protein level to the survival rate of HCC patients (d). e-f. H-scores of PRAS40 staining (e) and p-PRAS40 staining (f) in 44 pairs of primary HCC and peri‑cancer tissue samples in Cohort 2. g-h. The correlation of PRAS40 protein level (g) and phosphorylation level (h) to the survival rate of 50 HCC patients in Cohort 3. i-j. RNA-seq results of AKT1S1 mRNA in HCC and normal liver tissue samples in public TCGA dataset. The relative AKT1S1 mRNA levels were compared in 371 cases of HCC and 50 cases of normal liver tissue (i). The correlation of AKT1S1 mRNA level to the survival rate of 365 HCC patients (j). k-l. PRAS40 protein levels in the livers of DEN-injected Akt1s1+/+ mice were evaluated by Western blotting (k). The quantitative results were shown in l. Scale bars, 100μm. N, non-tumor; T, tumor. Bars, SD. **, P<0.01 from Student's t-test.
3.2. High expression and phosphorylation of PRAS40 are positively correlated to the poor prognosis of HCC patients
To understand the clinical relevance of PRAS40 hyperexpression or hyperphosphorylation in HCC further, we examined the possible correlation between PRAS40 or p-PRAS40 IHC staining in HCC specimens and patient survival rate after surgery. According to the staining H-Scores, we grouped the HCC specimens as PRAS40 or p-PRAS40 low level (H-Score<150) and PRAS40 or p-PRAS40 high level (H-Score≥150). High expression of PRAS40 protein (13 and 18 cases in Cohort 1 and 3, respectively) was positively associated with a lower overall survival rate of HCC patients compared to low expression PRAS40 protein (9 and 32 cases in Cohort 1 and 3, respectively)(P = 0.0055 from log-rank test, Fig. 1d; P = 0.0009 from log-rank test, Fig. 1g). High level of p-PRAS40 (24 cases in Cohort 3) was positively associated with a lower overall survival rate of HCC patients compared to low level of p-PRAS40 (26 cases in Cohort 3)(P = 0.0057 from log-rank test, Fig. 1h). Further multivariate Cox regression analyses indicated that high level of either PRAS40 or p-PRAS40 is an independent prognostic factor for poor survival of patients with HCC (P = 0.016, Cohort 1, Supplementary Table 2; P = 0.003 for PRAS40, P = 0.028 for p-PRAS40, Cohort 3, Supplementary Table 5).
Analyses of the 365 HCC patients’ survival information included in TCGA LIHC database suggested that high level of AKT1S1 mRNA (FPKM value ≥11.99, 141 cases) was positively associated with a lower overall survival rate of HCC patients compared to low AKT1S1 mRNA level (FPKM value <11.99, 224 cases) (P = 0.0002 from log-rank test, Fig. 1j) (The Human Protein Atlas) [44]. Collectively, these results suggest that high level of PRAS40 is associated with the poor prognosis of HCC patients.
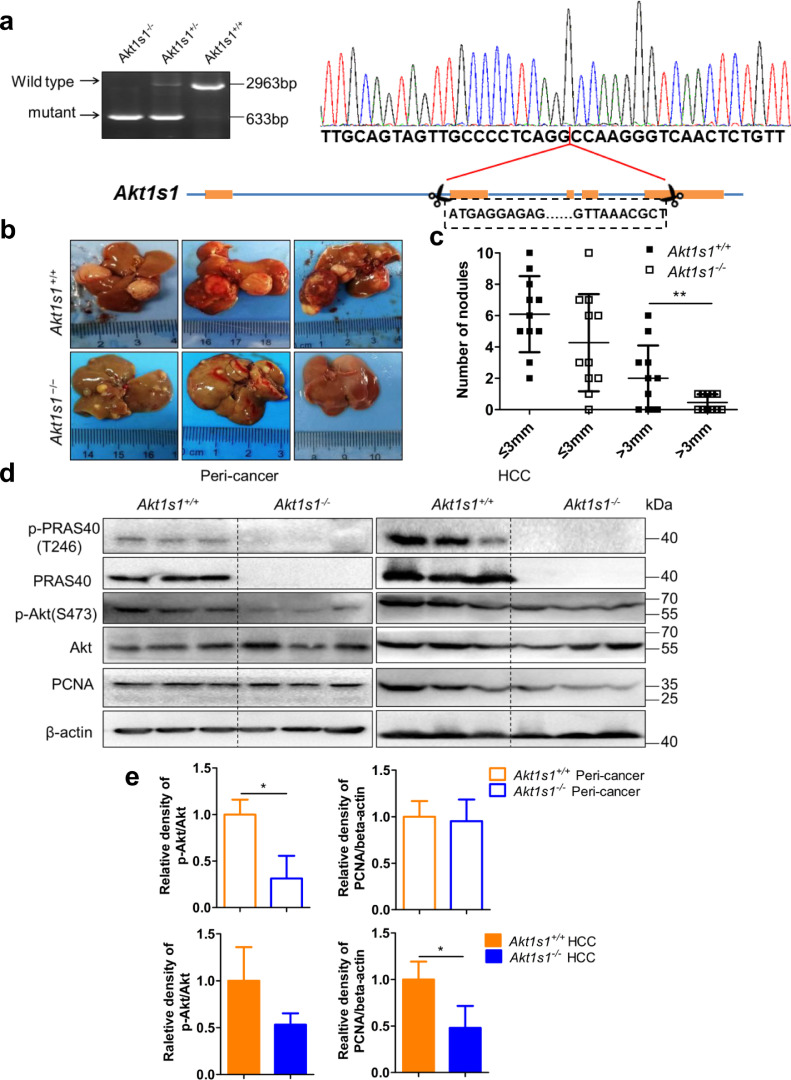
3.3. PRAS40 depletion suppresses DEN-induced hepatocarcinogenesis
To study the significance of PRAS40 in hepatocarcinogenesis, we constructed Akt1s1+/− mice, which were used to form Akt1s1−/− mice after backcrossed six generations to C57BL/6 N genetic background (Fig. 2a). Fourteen-day-old Akt1s1−/− or Akt1s1+/+ male mice were applied a single intaperitoneal injection of DEN (25 mg/kg, n = 11). At 42 weeks old, all DEN-treated Akt1s1+/+ mice developed HCC (11/11), whereas 10 out of 11 Akt1s1−/− mice developed HCC. The number of the tumors with larger size (>3 mm) formed in Akt1s1−/− livers was greatly less than those in Akt1s1+/+ livers (P<0.01 from Student's t-test, Fig. 2b and c). To investigate the mechanism of tumor suppression induced by PRAS40 depletion, we next examined the alteration of Akt phosphorylation. We found that the phosphorylation of Akt was obviously decreased in both the peri‑cancer and HCC tissue of Akt1s1−/− mice, compared to those in Akt1s1+/+ mice. In contrast, the level of PCNA, a proliferation marker, was decreased only in HCC but not peri‑cancer tissue of Akt1s1−/− mice (Fig. 2d and e).
Fig. 2.
HCC formation in Akt1s1+/+ and Akt1s1−/− mice. a. Genotyping results of the mice and the schematic diagram of the design of Akt1s1−/− mice. b. The representative livers of DEN-injected Akt1s1+/+ and Akt1s1−/− mice. c. Quantitative results of the tumors formed in Akt1s1+/+ and Akt1s1−/− mice, n = 11. d. Western blotting detected by the antibodies indicated in the peri‑cancer and HCC samples of Akt1s1+/+ and Akt1s1−/− mice. e. The quantitative results of Western blotting. Bars, SD. **, P<0.01 from Student's t-test.
3.4. PRAS40 depletion represses the cellular proliferation of HCC cells in vitro
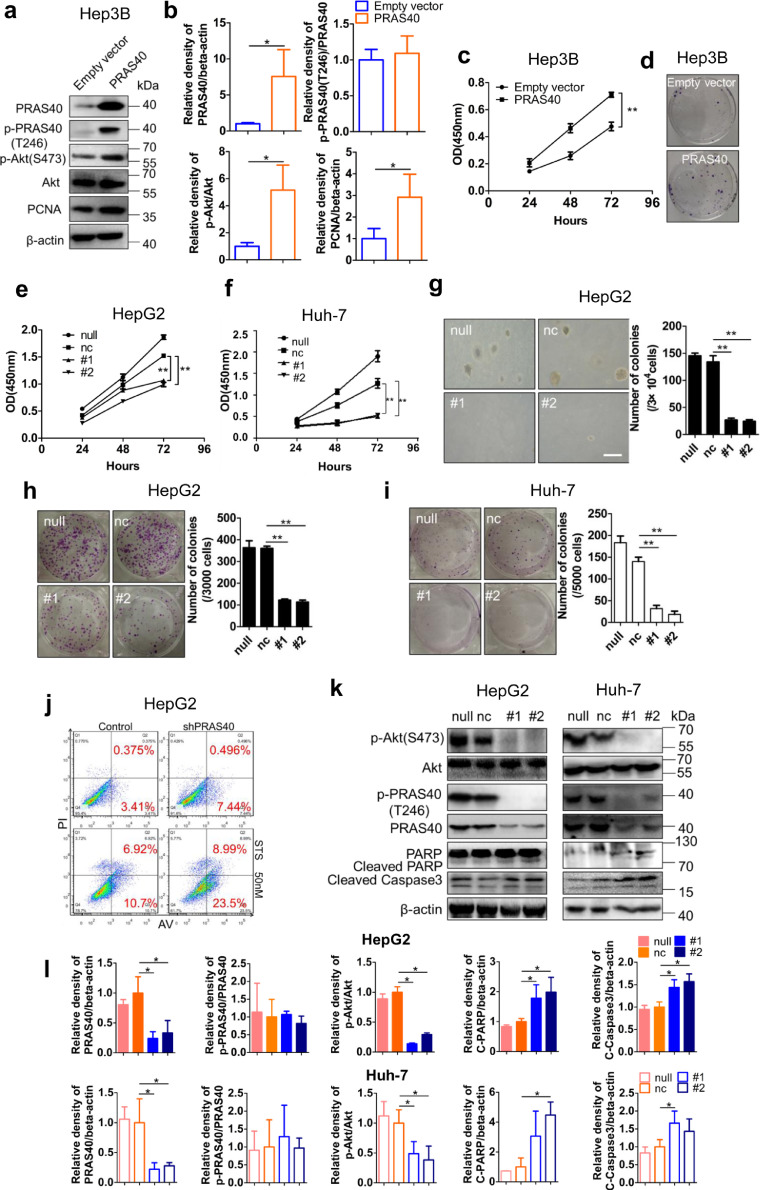
The above observations prompted us to explore the potential biological function of PRAS40 in HCC cell proliferation in vitro. Initially, we overexpressed PRAS40 in Hep3B cells which showed a comparatively low level of PRAS40 protein (Supplementary Fig. 3). The phosphorylation of Akt and the expression of PCNA were both upregulated (Fig. 3a and b). Further the PRAS40-overexpressing cells presented a remarkable enhancement of cell growth in cell viability assay (P<0.01 from Student's t-test, Fig. 3c) and more colonies (Fig. 3d), compared with the empty vector-transfected cells.
Fig. 3.
The effects of PRAS40 depletion on HCC cell growth in vitro. a-d. Hep3B cells were introduced with the virus containing empty vector or PRAS40 expression vector. Cell lysates were analyzed by Western blotting with the antibodies indicated (a). Quantitative results of 2 independent experiments (b). Cell viability assays were performed in triplicate at the indicated times (c). Colony formation assay results were shown (d). HepG2 or Huh-7 cells were transfected without (null) or with negative control (nc), or PRAS40 (#1, #2) siRNAs (e-l). Cell viability assays were performed in triplicate at the indicated times in HepG2 (e) and Huh-7 (f) cells. Soft agar assays were performed in HepG2 cells, and the quantitative results of 2 independent experiments were presented in right panels, Scale bar, 50 μm (g). Colony formation assays were performed in HepG2 (h) and Huh-7 (i) cells. Quantitative results of 2 independent experiments were presented in right panels. HepG2 cells were stained with annexin V (AV) and PI and analyzed by flow cytometry (j). HepG2 and Huh-7 cell lysates were analyzed by Western blotting with the antibodies indicated (k-l). C-PARP, cleaved PARP; C-—Caspase 3, cleaved Caspase 3. Bars, SD. **, P<0.01 from Student's t-test.
Next, we knocked down the expression of PRAS40 with siRNAs in HepG2 and Huh-7 cells (Fig. 3e–l). Both HepG2 and Huh-7 cells transfected with PRAS40 siRNAs exhibited remarkable decline in cell viability to 27–57% of those by the cells without siRNA transfection (P<0.01 from Student's t-test), whereas those transfected with non-specific siRNA did not show significant change (Fig. 3e and f). The number of the colonies formed in soft agar by PRAS40 knockdown cells was as little as about 17% (P<0.01 from Student's t-test) of those formed by the cells without transfection, whereas the cells transfected with non-specific siRNA did not show any significant change (Fig. 3g). The capacity of anchorage-dependent colony formation was also evaluated. PRAS40 depleted cells showed much fewer and smaller colonies than control cells, and the colony numbers were only 10–33% (P<0.01 from Student's t-test) of those formed by the cells without transfection, whereas those transfected with non-specific siRNA showed more than 77% of the colony number formed by those without transfection (Fig. 3h and i). These results suggest that PRAS40 is involved in the promotion of cellular proliferation.
PRAS40 deregulates apoptosis in melanoma and Ewing's sarcoma cells [9,10,45]. Here we examined the effects of PRAS40 depletion on apoptosis by flow cytometry. Early apoptotic cells, represented by the ratio of the PI−/Annexin V+ cells, were increased to 7.44% in PRAS40 shRNA transfected cells from 3.41% in the control shRNA transfected cells. When the cells were treated with an apoptosis inducer, staurosporine (STS), early apoptotic cells were increased to 23.5% in PRAS40 shRNA transfected cells from 10.7% in the control shRNA transfected cells (Fig. 3j). The levels of cleaved PARP, cleaved caspase 3, apoptosis markers, were found to be increased in PRAS40-depleted HepG2 and Huh-7 cells (Fig. 3k and l). Furthermore, the levels of phosphorylated Akt were downregulated remarkably in PRAS40-knockdown cells (#1 and #2). These data suggest that PRAS40 may deregulate apoptosis through upregulating Akt activation in HCC.
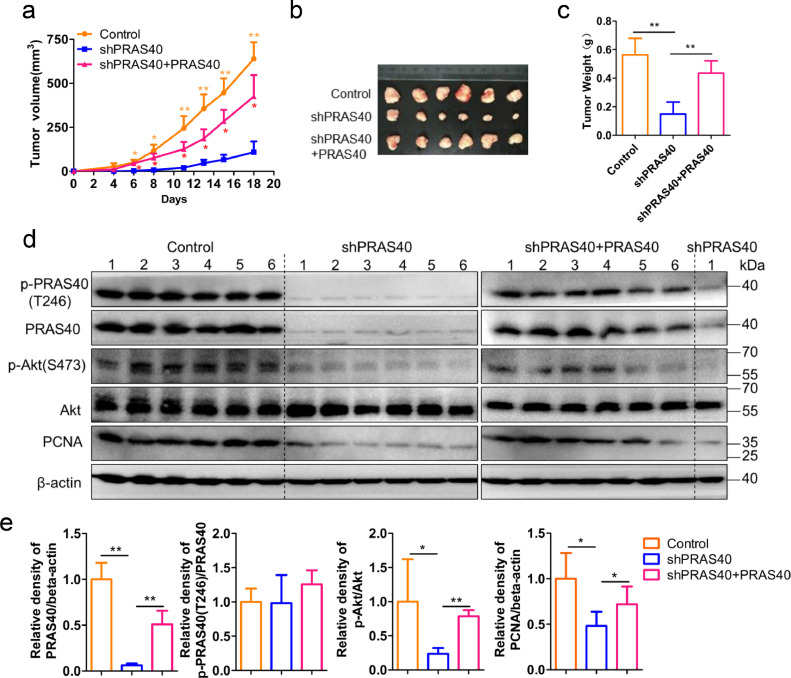
3.5. PRAS40 depletion represses the cellular proliferation of HCC cells in vivo
According to the in vitro results, we further explored the possibility that PRAS40 depletion suppresses the growth of HCC xenografts in mice. From 6 days after tumor cell injection, tumor growth was notably inhibited in the tumor xenografts formed by the PRAS40-knockdown HepG2 cells compared to the control cells (Fig. 4a). The tumor volume and weight formed in the PRAS40-knockdown groups were significantly reduced (P<0.01–0.05 from Student's t-test) compared to the control groups (Fig. 4a–c). While when shRNA resistant PRAS40 expression vector was introduced together with PRAS40 shRNA into HepG2 cells, the tumor volume and weight formed were comparable to the control group (Fig. 4a–c). In addition, Akt phosphorylation and PCNA expression were downregulated remarkably in PRAS40-knockdown cells, which were restored to the levels in control cells (Fig. 4d and e). These data suggest that PRAS40 knockdown significantly suppressed Akt phosphorylation and the tumor growth of HCC xenografts in vivo.
Fig. 4.
The effects of PRAS40 depletion on HCC xenograft formation in nude mice. HepG2 cells were introduced with control, PRAS40 shRNA (shPRAS40) or PRAS40 shRNA together with shRNA-resistant PRAS40 expression vector (shPRAS40+PRAS40). a-d. The cells of each group were inoculated into the two posterior flanks of the mice, n = 6. Tumor volumes were recorded every 2–3 days (a). On day 19, tumors were dissected and obtained (b-c). Pictures were taken at the sacrificed time (b). Tumor weights were measured (c). Bars, SD. *, P<0.05; **, P<0.01 (Student's t-test). d. Cell lysates were applied to Western blotting. e. Quantitative results of the six samples from each group. Bars, SD. *, P<0.05; **, P<0.01 (Student's t-test).
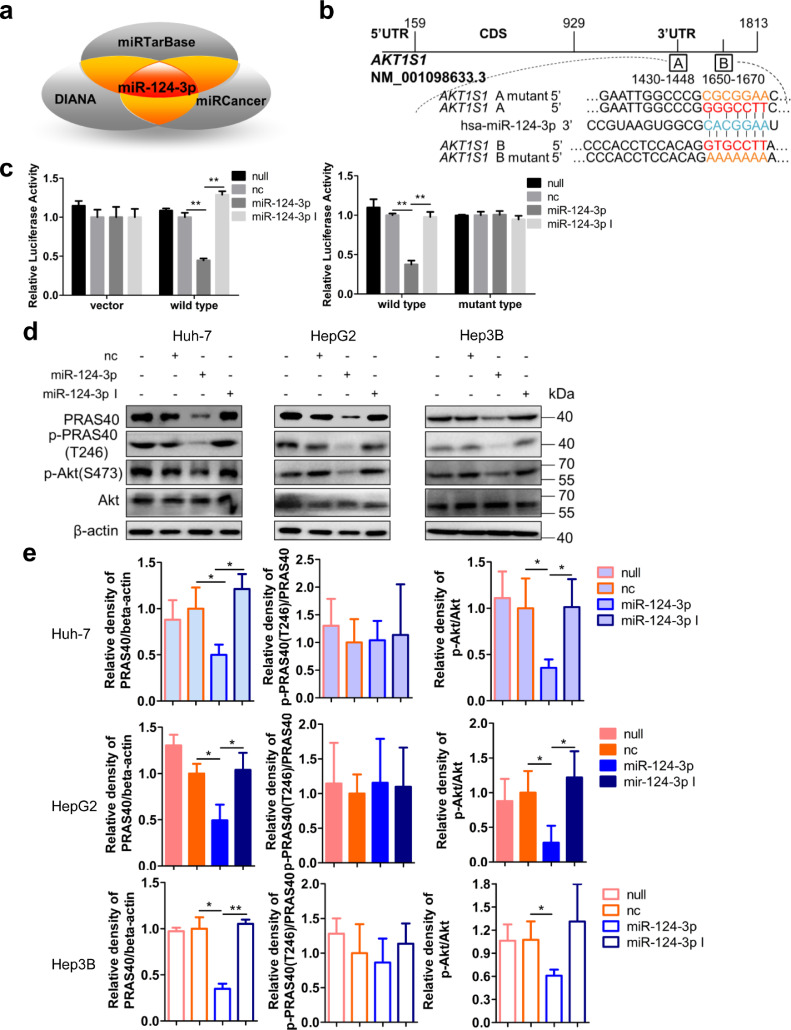
3.6. MiR-124-3p targets PRAS40 3′UTR
For the mechanism of PRAS40 hyperexpression in HCC, since we did not find AKT1S1 DNA amplification in HCC samples in public TCGA database, and miRNAs play important roles in the regulation of mRNA and protein expression, we considered the possibilities of the regulation by miRNAs. We aimed to identify those miRNAs that target AKT1S1 3′UTR serving as tumor suppressors in the pathogenesis of HCC. We predicted the miRNAs which bind AKT1S1 3′UTR using both miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and DIANA-TarBase (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index), and selected the miRNAs downregulated in HCC (miRCancer) as candidates (Fig. 5a). MiR-124-3p binding sites within the AKT1S1 3′UTR are conserved between humans and mice. To clarify whether miR-124-3p directly recognizes the predicted binding sites within AKT1S1 3′UTR, we cloned wild type or binding sites-mutant type AKT1S1 3′UTR into a luciferase reporter vector (Fig. 5b). As expected, cotransfection with miR-124-3p mimic reduced the luciferase activity of the reporter including AKT1S1 3′UTR (wild type) to 38% of that produced by cotransfection with negative control miRNA in HeLa cells (P<0.01 from Student's t-test). On the contrary, cotransfection with miR-124-3p inhibitor did not reduce the luciferase activity of the reporter including AKT1S1 3′UTR. While neither miR-124-3p nor negative control miRNA altered the luciferase activity of the control reporter without AKT1S1 3′UTR (vector) (Fig. 5c). In contrast, the luciferase activity of the reporter including AKT1S1 3′UTR with two mutations of miR-124-3p binding sites (mutant type) was not altered by either miR-124-3p mimic or inhibitor, as same as the negative control miRNA (Fig. 5c). Together, these results indicate that PRAS40 is a direct target of miR-124-3p.
Fig. 5.
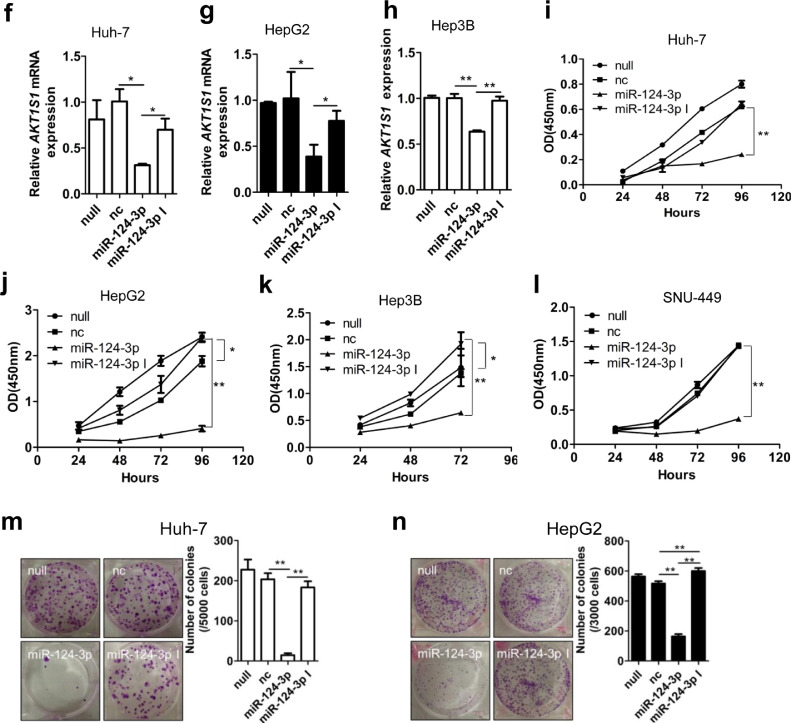
PRAS40 is the target of miR-124-3p and the effects of miR-124-3p on cell growth. a. The prediction of the miRNAs binding AKT1S1 3′UTR. b. Schematic illustration of the predicted miR-124-3p binding sites in AKT1S1 3′UTR and the design of mutant type of AKT1S1 3′UTR. c. HeLa cells were cotransfected with pCIneo-Luc (vector), pCIneo-Luc-AKT1S1 (wild type) or pCIneo-Luc-AKT1S1 mutant (mutant type) reporter vector and nonspecific control miRNA (nc), miR-124-3p or miR-124-3p inhibitor (miR-124-3p I) together with Renilla luciferase expression vector phRL-SV40. Firefly luciferase activities were measured and normalized to Renilla luciferase activities. The experiments were performed in triplicate. d-n. HCC cells were transfected without ( null) or with negative control (nc), miR-124-3p mimic or miR-124-3p inhibitor (miR-124-3p I). The PRAS40 protein levels were analyzed by Western blotting in Huh-7, HepG2 and Hep3B cells (d). Quantitative results of 2 independent experiments (e). The AKT1S1 mRNA levels were analyzed by real time PCR in Huh-7 (f), HepG2 (g) and Hep3B cells (h cells in triplicate. Cell viability assays were performed in triplicate at the indicated time points in Huh-7 (i), HepG2 (j), Hep3B (k), and SNU-449 (l) cells. Colony formation assays were performed in Huh-7 (m) and HepG2 (n) cells. Quantitative results of 2 independent experiments were shown in right panels (m-n). Bars, SD. *, P<0.05; **, P<0.01 (Student's t-test).
3.7. Forced miR-124-3p expression downregulates PRAS40 expression and cellular proliferation
To verify whether miR-124-3p represses the expression of PRAS40 in HCC cells, we next transfected miR-124-3p mimic into Huh-7, HepG2 or Hep3B cells. The western blot results showed that compared to negative control miRNA transfection, miR-124-3p mimic but not inhibitor introduction significantly decreased PRAS40 protein levels. Simultaneously, a significant decrease of Akt phosphorylation in miR-124-3p mimic- but not inhibitor- expressing cells was confirmed (Fig. 5d and e). The real time PCR results showed that miR-124-3p mimic introduction significantly decreased AKT1S1 mRNA levels to 36–60% of that produced by negative control miRNA transfection (P<0.05 from Student's t-test), whereas miR-124-3p inhibitor did not show remarkable changes (Fig. 5f–h). With subsequent functional assays, all the cell viability assays and the colony formation assays revealed impaired cell growth in the cells transfected with miR-124-3p mimic (Fig. 5i–n). The cells transfected with miR-124-3p mimic presented a remarkable decline in cell viability to 21–45% of that in the cells transfected with negative control miRNA (P<0.01 from Student's t-test). Similarly, the capacity of colony formation was also greatly decreased by miR-124-3p mimic transfection to 7–32% of that by the cells transfected with negative control miRNA (P<0.01 from Student's t-test). However, transfection with miR-124-3p inhibitor moderately augmented the cell viability to 1.3 folds (P<0.05 from Student's t-test) and the colony number to 1.2 folds (P<0.01 from Student's t-test) of that observed in negative control miRNA transfected HepG2 cells, while upregulated the cell viability to 1.6 folds (P<0.05 from Student's t-test) of that observed in negative control miRNA transfected Hep3B cells.
3.8. MiR-124-3p levels are inversely correlated to PRAS40 protein levels in HCC tissue
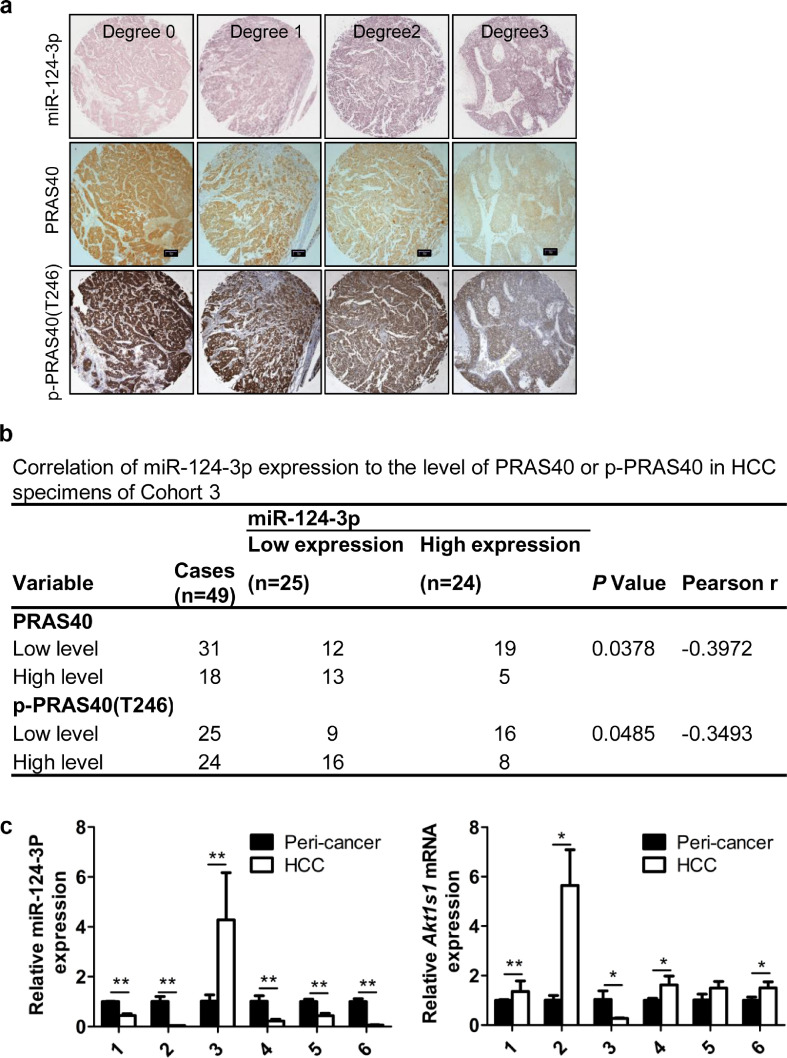
To clarify the correlation between the levels of PRAS40 protein and miR-124-3p in vivo, we further examined the expression levels of miR-124-3p in 49 cases of HCC tissue (Cohort 3). According to the staining scores, we grouped the HCC specimens as miR-124-3p low expression (score<150) and miR-124-3p high expression (score≥150). Low levels of miR-124-3p were more likely to be seen in HCC with high level of PRAS40 protein (P = 0.0378 from Fisher's exact test, Pearson r=−0.3972) or p-PRAS40 (P = 0.0485 from Fisher's exact test, Pearson r=−0.3493) (Fig. 6a and b). Further, we examined the miR-124-3p levels in mouse tissue. The real time PCR results showed that compared with the peri‑cancer tissue, miR-124-3p level was remarkably reduced in the HCC tissue of 5/6 of Akt1s1+/+ mice, while PRAS40 levels were notably augmented in the same HCC tissue. In addition, the levels of miR-124-3p and PRAS40 were opposite in all 6 mice (Fig. 6c). Therefore the level of miR-124-3p inversely correlates to PRAS40 protein level significantly in vivo.
Fig. 6.
The correlation between the levels of miR-124-3p and PRAS40 protein. a. ISH staining of miR-124-3p and IHC staining of PRAS40 and p-PRAS40 were performed in 49 clinical HCC specimens in Cohort 3 (same samples used in Fig. 1G-H). Scale bars, 100μm. b. Correlation analyses between the staining scores of miR-124-3p and PRAS40 or p-PRAS40, P value is calculated by Fisher's exact test and a Pearson correlation coefficient (r) is shown. c. Real time PCR in the peri‑cancer and HCC samples of Akt1s1+/+ mice, n = 6. The experiments were performed in triplicate. Bars, SD. *, P<0.05; **, P<0.01 (Student's t-test).
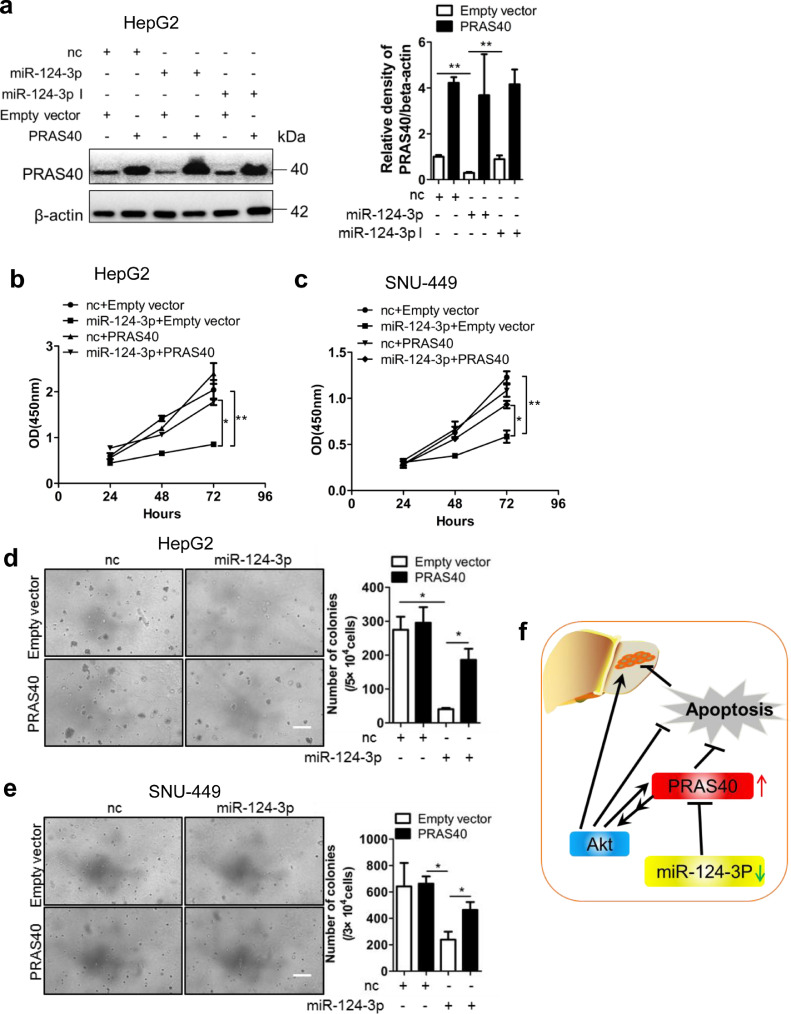
3.9. The repression of cellular proliferation by miR-124-3p is partially restored by PRAS40 overexpression
To confirm the relationship between miR-124-3p and PRAS40 further, we next introduced both miR-124-3p mimic and full length PRAS40 into HepG2 cells. The western blot results showed that endogenous PRAS40 protein level was notably lower in the cells introduced with miR-124-3p mimic than those introduced with negative control miRNA. However, exogenous PRAS40 level expressed by the vector lacking the 3′UTR region of PRAS40 was not changed at all (Fig. 7a and b). Both HepG2 and SNU-449 cells introduced with miR-124-3p mimic exhibited remarkable decline in cell viability to 42–48% of those by the cells with negative control miRNA introduction (P<0.01 from Student's t-test), while those transfected with miR-124-3p mimic together with PRAS40 expression vector showed significant increase in cell viability to 1.6–2 folds of those miR-124-3p mimic only introduced cells (P<0.05 from Student's t-test, Fig. 3c and d). In soft agar assays, transfection of miR-124-3p mimic resulted in a significantly smaller number and size of colonies compared to the negative control miRNA transfection (15–37%, P<0.05 from Student's t-test). In addition, the cells cotransfected with PRAS40 and miR-124-3p showed 1.9–4.6 folds of colony numbers of those formed in the cells with miR-124-3p mimic transfection (P<0.05 from Student's t-test, Fig. 7e and f). These data suggested that miR-124-3p downregulated PRAS40 expression to suppress cell growth, which could be partially reversed by PRAS40. Thus PRAS40 should function at the downstream of miR-124-3p in HCC cellular proliferation.
Fig. 7.
The effects of exogenous PRAS40 on the growth repression induced by miR-124-3p. a-f. HCC cells were cotransfected with negative control (nc) or miR-124-3p and empty vector or PRAS40 expression vector. PRAS40 protein levels were analyzed by Western blotting (a), and quantitative results of 2 independent Western blottings were shown in right panel. Cell viability assays (b-c) and soft agar assays (d-e) were performed in HepG2 and SNU-449 cells. Quantitative results of 2 independent soft agar assays were shown in right panels. Scale bars, 100μm. Bars, SD. *, P<0.05; **, P<0.01 (Student's t-test). f. Schematic graph representing the working model of PRAS40 in HCC.
4. Discussion
HCC is the fifth most common malignancy and the third most frequent reason of cancer death [46]. Effective treatments are anticipated, especially for the advanced HCC. Understanding the carcinogenesis is believed to be helpful to develop novel approaches to HCC. Herein, we focus on PRAS40 which is a substrate of Akt and a component of mTORC1. We report the upregulation of PRAS40 expression and phosphorylation in HCC and the positive correlation between the high phosphorylation, protein or mRNA level of PRAS40 and the poor prognosis of HCC patients (Fig. 1, Supplementary Table 2, 5, Supplementary Fig. 1 and 3). More importantly, we found that PRAS40 depletion dramatically inhibited HCC carcinogenesis and progression by the suppression of Akt signaling both in vivo and in vitro (Fig. 2, Fig. 3, Fig. 4). Thus the essential role of PRAS40 in HCC carcinogenesis has been confirmed in human specimens, mice models and cell lines. In the further study, a miR-124-3p/PRAS40 axis was identified (Fig. 5, Fig. 6, Fig. 7), which provides an explanation for the mechanism of PRAS40 hyperexpression and the tumor suppressive role of miR-124-3p in HCC.
The level of p-PRAS40 was reported to be upregulated in human melanoma, prostate cancer and non-small cell lung cancer samples, which is considered as the results of upstream Akt pathway activation [10,11]. Here we found that besides phosphorylation level, the protein and mRNA levels of PRAS40 were increased significantly in HCC clinical samples compared to the peri‑cancer samples in our cohorts and public TCGA database (Fig. 1). High levels of PRAS40 protein and mRNA were both found to be positively correlated to the poor prognosis of HCC patients in our results and TCGA data (Fig. 1d, g, j, Supplementary Table 2 and 5). In addition, in the DEN-induced HCC tissue, PRAS40 and p-PRAS40 levels were also upregulated remarkably, and the ratio of p-PRAS40/PRAS40 was similar in both HCC and peri‑cancer samples which suggests that PRAS40 hyperphosphorylation is mainly caused by PRAS40 hyperexpression (Fig. 1k and l). Thus we raised a novel possibility that PRAS40 hyperexpression could contribute to hepatocarcinogenesis independently of the upstream Akt pathway activation, which may provide a reference for other cancers. Furthermore, the data that HCC repression in Akt1s1−/− mice (Fig. 2), cell growth decrease and HCC xenograft impairment in PRAS40 knockdown cells (Figs. 3 and 4) solidly proved the significantly essential role of PRAS40 in HCC carcinogenesis and promotion.
For the mechanism that high level of PRAS40 contributes to hepatocarcinogenesis, the consistence of the downregulation of Akt phosphorylation in both Akt1s1−/− mice (Fig. 2d and e) and PRAS40 knockdown cells (Fig. 3k,l and 4d,e), and the upregulation of Akt phosphorylation in PRAS40-overexpressing cells (Fig. 3a and b) suggests that PRAS40 controls a positive feedback to Akt activation, leading to cell proliferation. These results are identical to the data of ischemic brains in Akt1s1−/− mice reported by another group [47]. Further studies are necessary to clarify this point.
The next question is how PRAS40 expression is increased in HCC. Since the DNA amplification is not found in HCC clinical samples in TCGA database (Supplementary Fig. 2), we considered the mRNA regulation. Recent studies present plentiful evidences on miRNAs controlling the expression of the target mRNAs [15], [16], [17], [18]. We next focused on miR-124-3p which is predicted to target AKT1S1 3′UTR (Fig. 5a). Being a tumor suppressor, miR-124-3p is downregulated in a couple of tumors including HCC, and shows an inverse correlation to PRAS40 or p-PRAS40 level (Fig. 6). Our results suggest that miR-124-3p downregulates the protein and mRNA levels of PRAS40 through binding AKT1S1 3′UTR (Fig. 5c–h), and inhibits Akt phosphorylation and cell growth (Fig. 5d,e and i–n). On the contrary, the miR-124-3p inhibitor transfection did not show notable effects on the protein and mRNA levels of PRAS40, Akt phosphorylation and cell growth in HCC cells (Fig. 5c–n), which is considered as the results of the miR-124-3p hypoexpression in HCC cells [25,26,41].
Further we tried to confirm the role of the miR-124-3p/PRAS40 axis in HCC progression. The data suggested that exogenous PRAS40 restored but not fully reversed the repression of cell growth by miR-124-3p (Fig. 7a–f). Since there are multiple target genes of miR-124-3p in HCC, other target genes such as Specificity protein 1, Rho Associated Protein Kinase 2 and Enhancer of zeste homolog 2, could be still repressed by miR-124-3p when only PRAS40 was introduced. The complexity of this network needs to be verified further.
The essential factor for liver development and hepatocyte function, HNF4α, promotes the transcription of miR-124. In HCC, HNF4α downregulation results in a low expression of miR-124 [25,28]. On the other hand, CpG islands are present in all of the promoters of miR-124-1, miR-124-2 and miR-124-3, leading to the hypermethylation of miR-124 promoter and its hypoexpression [39]. Accordingly downregulated miR-124-3p level results in the upregulation of PRAS40 expression, sequentially inducing Akt activation to initiate hepatocarcinogenesis (Fig. 7g). Collectively, our work has provided evidences to support that miR-124-3p/PRAS40 axis plays an important role in hepatocarcinogenesis, which offered a novel reference to target HCC.
Author contributions
ZQ and LH designed research; ZQ, TZ, LS, HF, HL, JW, SZ, TZ, LG, LJ, HZ, GH, TM, YW and LH performed the experiments and analyzed the data; LH wrote the manuscript. All authors read and approved the manuscript.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgements
This work was supported by the Climbing Scholars Supporting Program of Liaoning Province, National Natural Science Foundation of China (81772971), and Liaoning Provincial Program for Top Discipline of Basic Medical Sciences. All of the funders had not any role in study design, data collection, data analysis, interpretation, and writing of the report. We thank all the Huang lab members for the kind support.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.102604.
Appendix. Supplementary materials
References
- 1.Siegel R., Ward E., Brawley O., Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Kovacina K.S., Park G.Y., Bae S.S., Guzzetta A.W., Schaefer E., Birnbaum M.J. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 3.Oshiro N., Takahashi R., Yoshino K., Tanimura K., Nakashima A., Eguchi S. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca B.D., Smith E.M., Lee V.H., MacKintosh C., Proud C.G. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 5.Sancak Y., Thoreen C.C., Peterson T.R., Lindquist R.A., Kang S.A., Spooner E. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Vander Haar E., Lee S.I., Bandhakavi S., Griffin T.J., Kim D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Harris T.E., Roth R.A., Lawrence J.C., Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 8.Yu F., Narasimhan P., Saito A., Liu J., Chan P.H. Increased expression of a proline-rich Akt substrate (PRAS40) in human copper/zinc-superoxide dismutase transgenic rats protects motor neurons from death after spinal cord injury. J Cerebral Blood Flow and Metabolism. 2008;28:44–52. doi: 10.1038/sj.jcbfm.9600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L., Nakai Y., Kuwahara I., Matsumoto K. PRAS40 is a functionally critical target for EWS repression in Ewing sarcoma. Cancer Res. 2012;72:1260–1269. doi: 10.1158/0008-5472.CAN-11-2254. [DOI] [PubMed] [Google Scholar]
- 10.Madhunapantula S.V., Sharma A., Robertson G.P. PRAS40 deregulates apoptosis in malignant melanoma. Cancer Res. 2007;67:3626–3636. doi: 10.1158/0008-5472.CAN-06-4234. [DOI] [PubMed] [Google Scholar]
- 11.Shipitsin M., Small C., Giladi E., Siddiqui S., Choudhury S., Hussain S. Automated quantitative multiplex immunofluorescence in situ imaging identifies phospho-S6 and phospho-PRAS40 as predictive protein biomarkers for prostate cancer lethality. Proteome Sci. 2014;12:40. doi: 10.1186/1477-5956-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent E.E., Elder D.J., Thomas E.C., Phillips L., Morgan C., Pawade J. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. Br J Cancer. 2011;104:1755–1761. doi: 10.1038/bjc.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havel J.J., Li Z., Cheng D., Peng J., Fu H. Nuclear PRAS40 couples the Akt/mTORC1 signaling axis to the RPL11-HDM2-p53 nucleolar stress response pathway. Oncogene. 2015;34:1487–1498. doi: 10.1038/onc.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu F., Deng X., Yang X., Jin H., Gu D., Lv X. Hypoxia upregulates Rab11-family interacting protein 4 through HIF-1alpha to promote the metastasis of hepatocellular carcinoma. Oncogene. 2015;34:6007–6017. doi: 10.1038/onc.2015.49. [DOI] [PubMed] [Google Scholar]
- 15.Telonis A.G., Magee R., Loher P., Chervoneva I., Londin E., Rigoutsos I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017;45:2973–2985. doi: 10.1093/nar/gkx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahadi A., Sablok G., Hutvagner G. miRTar2GO: a novel rule-based model learning method for cell line specific microRNA target prediction that integrates Ago2 CLIP-Seq and validated microRNA-target interaction data. Nucleic Acids Res. 2017;45:e42. doi: 10.1093/nar/gkw1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dooley J., Lagou V., Garcia-Perez J.E., Himmelreich U., Liston A. miR-29a-deficiency does not modify the course of murine pancreatic acinar carcinoma. Oncotarget. 2017;8:26911–26917. doi: 10.18632/oncotarget.15850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engreitz J.M., Sirokman K., McDonel P., Shishkin A.A., Surka C., Russell P. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Y., Fang J.H., Yun J.P., Yang J., Zhang Y., Jia W.H. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 21.Pineau P., Volinia S., McJunkin K., Marchio A., Battiston C., Terris B. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makeyev E.V., Zhang J., Carrasco M.A., Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visvanathan J., Lee S., Lee B., Lee J.W., Lee S.K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo A.S., Staahl B.T., Chen L., Crabtree G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatziapostolou M., Polytarchou C., Aggelidou E., Drakaki A., Poultsides G.A., Jaeger S.A. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng F., Liao Y.J., Cai M.Y., Liu Y.H., Liu T.H., Chen S.P. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 27.Bhaskaran V., Nowicki M.O., Idriss M., Jimenez M.A., Lugli G., Hayes J.L. The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma. Nat Commun. 2019;10:442. doi: 10.1038/s41467-019-08390-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning B.F., Ding J., Liu J., Yin C., Xu W.P., Cong W.M. Hepatocyte nuclear factor 4alpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology. 2014;60:1607–1619. doi: 10.1002/hep.27177. [DOI] [PubMed] [Google Scholar]
- 29.Koukos G., Polytarchou C., Kaplan J.L., Morley-Fletcher A., Gras-Miralles B., Kokkotou E. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–852. doi: 10.1053/j.gastro.2013.07.001. e842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponomarev E.D., Veremeyko T., Barteneva N., Krichevsky A.M., Weiner H.L. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin F., Hu H., Xu M., Zhan S., Wang Y., Zhang H. Serum microRNA profiles serve as novel biomarkers for autoimmune diseases. Front Immunol. 2018;9:2381. doi: 10.3389/fimmu.2018.02381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw T.A., Singaravelu R., Powdrill M.H., Nhan J., Ahmed N., Ozcelik D. MicroRNA-124 regulates fatty acid and triglyceride homeostasis. iScience. 2018;10:149–157. doi: 10.1016/j.isci.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng R., Chin J.S., Yew J.Y., Bushati N., Cohen S.M. miR-124 controls male reproductive success in Drosophila. eLife. 2013;2:e00640. doi: 10.7554/eLife.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R., Zhang S., Chen X., Li N., Li J., Jia R. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166. doi: 10.1186/s12943-018-0911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Chen L., Wu Z., Wang M., Jin F., Wang N. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer. 2016;16:826. doi: 10.1186/s12885-016-2862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Fan Q., Li J., Song J., Gu Y. MiR-124 down-regulation is critical for cancer associated fibroblasts-enhanced tumor growth of oral carcinoma. Exp Cell Res. 2017;351:100–108. doi: 10.1016/j.yexcr.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Ferrarese R., GRt Harsh, Yadav A.K., Bug E., Maticzka D., Reichardt W. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest. 2014;124:2861–2876. doi: 10.1172/JCI68836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao Y., Wang J., Yan W., Zhou Y., Chen Y., Zhou K. Dysregulated miR-124 and miR-200 expression contribute to cholangiocyte proliferation in the cholestatic liver by targeting IL-6/STAT3 signalling. J Hepatol. 2015;62:889–896. doi: 10.1016/j.jhep.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 39.Furuta M., Kozaki K.I., Tanaka S., Arii S., Imoto I., Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 40.Anwar S.L., Albat C., Krech T., Hasemeier B., Schipper E., Schweitzer N. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer. 2013;133:660–670. doi: 10.1002/ijc.28068. [DOI] [PubMed] [Google Scholar]
- 41.Cai Q.Q., Dong Y.W., Wang R., Qi B., Guo J.X., Pan J. MiR-124 inhibits the migration and invasion of human hepatocellular carcinoma cells by suppressing integrin alphaV expression. Sci Rep. 2017;7:40733. doi: 10.1038/srep40733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bollag G., Hirth P., Tsai J., Zhang J., Ibrahim P.N., Cho H. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research Network Electronic address wbe, cancer genome atlas research N. comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046. e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 45.Lv D., Liu J., Guo L., Wu D., Matsumoto K., Huang L. PRAS40 deregulates apoptosis in Ewing sarcoma family tumors by enhancing the insulin receptor/Akt and mTOR signaling pathways. Am J Cancer Res. 2016;6:486–497. [PMC free article] [PubMed] [Google Scholar]
- 46.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 47.Xiong X., Xie R., Zhang H., Gu L., Xie W., Cheng M. PRAS40 plays a pivotal role in protecting against stroke by linking the Akt and mTOR pathways. Neurobiol Dis. 2014;66:43–52. doi: 10.1016/j.nbd.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.