Abstract
Acute myocardial infarction (AMI) results from long-term diminished blood supply diminishment (ischemia) to the heart, and the main reason for ischemia is hypoxia. BCL2 interaction protein 3 (BNIP3) can be upregulated by hypoxia and participates in the mediation of hypoxia-activated apoptosis in cardiac myocyte death. The purpose of this study was to interrogate the mechanism of BNIP3 in hypoxia-activated cardiac myocyte injury. Cell viability and apoptosis were evaluated by Cell counting kit 8 (CCK-8), 5-ethynyl-2’-deoxyuridine (EdU), TdT-mediated dUTP Nick-End Labeling (TUNEL), and caspase-3 activity assays. Molecular interactions were assessed by RNA immunoprecipitation (RIP) and pull-down assays. Gene levels were assessed via quantitative real-time PCR and western blot. BNIP3 expression was upregulated by hypoxia in H9c2 cells. We found that circ-BNIP3 (hsa_circ_0005972), whose annotated gene was BNIP3, was induced by hypoxia and positively regulated BNIP3 expression. Knockdown of BNIP3 or circ-BNIP3 reversed the effect of hypoxia in attenuating H9c2 cell viability and inducing apoptosis. circ-BNIP3 sponged miRNA-27a-3p (miR-27a-3p) to upregulate BNIP3 expression. Moreover, eukaryotic translation initiation factor 4A3 (EIF4A3) bound with the upstream region of the circ-BNIP3 mRNA transcript and induced circ-BNIP3 expression in H9c2 cells. EIF4A3-induced circ-BNIP3 aggravated hypoxia-caused injury of H9c2 cells through targeting miR-27a-3p/BNIP3 pathway, indicating circ-BNIP3 as a new target for relieving hypoxia-induced injury of cardiac myocytes.
Keywords: hypoxia-activated injury, H9c2 cells, circ-BNIP3, BNIP3, miR-27a-3p
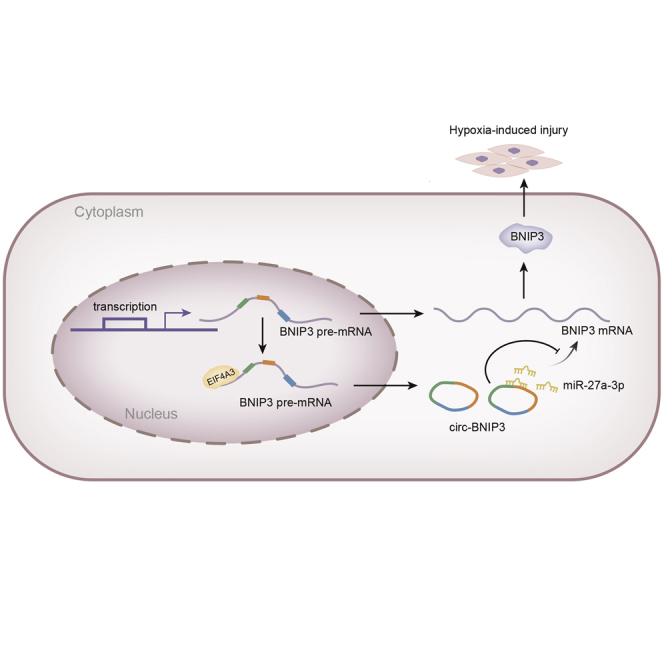
Graphical Abstract
Introduction
Myocardial infarction (MI), resulting from hypoxia or acute, persistent ischemia, is recognized as a severe manifestation of the coronary artery diseases that can lead to irreversible heart muscle damage and the progression of heart failure.1, 2, 3 The major clinical symptoms of MI include severe dyspnea, persistent chest pain, syncope, and fever.4,5 Although tools for preventing and treating MI have been improved to some extent, MI still contributes to large numbers of deaths globally.6, 7, 8, 9
Circular RNAs (circRNAs), a newly-identified class among noncoding RNAs (ncRNAs), are characterized by a closed and continuous loop structure lacking in the 5′ end cap or 3′ terminal poly(A) tail.10,11 circRNAs are more stable than linear RNA and are resistant to the effects of RNA exonucleases, cap removal, or adenylation.12 Over the past years, studies have revealed the role of circRNAs in regulating cellular biological processes, including apoptosis and proliferation.13,14 Recently, the influences of circRNAs in cardiomyocyte injury have been increasingly revealed. For example, circ-NCX1 mediated ischemic myocardial injury by microRNA-133a-3p (miR-133a-3p).15 circRNA autophagy-related circular RNA (ACR) attenuated myocardial ischemia/reperfusion injury through inhibiting autophagy via Pink1/FAM65B pathway.16 Hsa_circ_0005972, whose annotated gene was BCL2 interacting protein 3 (BNIP3), is a newly identified circRNA by our study, and it has never been related to MI before.
BNIP3 belongs to the BH3-only subfamily of Bcl-2 family proteins, which can heterodimerize and antagonize the activities of prosurvival proteins, including Bcl-XL and Bcl-2, as well as induce cell apoptosis.17,18 Although BNIP3 expression is unable to be detected in the majority of organs normally, including in heart, it can be activated under hypoxia.17,19 Also, it has been reported that BNIP3 protein overexpression in some cultured cell lines can lead to membrane insertion, as well as the initiation of the cell death pathway with necrosis-like features.17 Moreover, a study has pointed out that hypoxia and acidosis can induce BNIP3 to activate cardiac myocyte death.20 However, the mechanism of BNIP3 in hypoxia-induced injury of cardiomyocytes remains elusive. Also, the relation between BNIP3 and circ-BNIP3 has never been explored before.
The purpose of the present study was to uncover the mediation mechanism of BNIP3 in hypoxia-caused injury in H9c2 cells. We confirmed that BNIP3 was increased by hypoxia, and BNIP3 silence ameliorated the hypoxia-activated injury of H9c2 cells. We searched on circBase and first identified that circ-BNIP3 was associated with BNIP3 and had positive regulation on BNIP3 expression. We validated that circ-BNIP3 was upregulated under hypoxia and its knockdown reversed hypoxia-caused injury of H9c2 cells. Mechanistically, we validated that circ-BNIP3 sponged miRNA-27a-3p to upregulate BNIP3. Moreover, we explored the mechanism of circ-BNIP3 upregulation and found that EIF4A3 bound to BNIP3 mRNA at the upstream region of circ-BNIP3 mRNA transcripts and facilitated mRNA splicing and induced circ-BNIP3 expression.
Results
Hypoxia Led to Injury of H9c2 Cells and Upregulated BNIP3 Expression
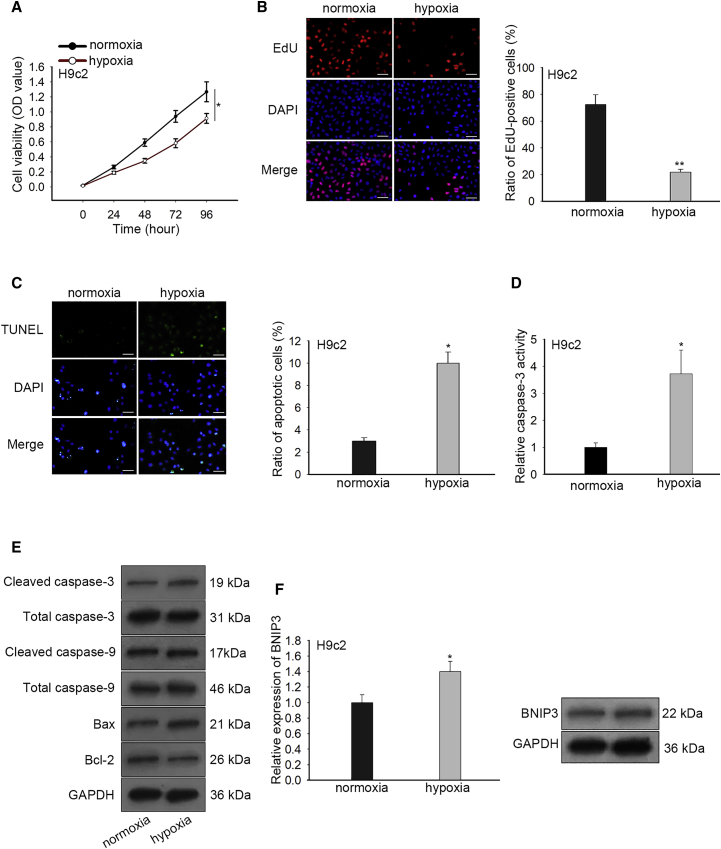
At first, we incubated H9c2 cells in the hypoxic incubator to construct hypoxic H9c2 cells. Thereafter, we tested viability and apoptosis of H9c2 cells under hypoxia versus normoxia treatment. Consequently, we observed through CCK-8 that the viability of H9c2 cells was attenuated by hypoxia compared with normoxia (Figure 1A). Also, EdU assay validated that H9c2 cell viability was decreased in response to hypoxia (Figure 1B). These results showed that hypoxia impaired viability of H9c2 cells. Besides, TUNEL assay demonstrated that the apoptotic H9c2 cells increased in presence with hypoxia (Figure 1C). To further evaluate the apoptosis level of H9c2 cells under hypoxia, we determined caspase-3 activity. As a result, hypoxia treatment induced caspase-3 activity in H9c2 cells (Figure 1D). In addition, the protein levels of cleaved-caspase-3/9 and Bax were improved by hypoxia, with the levels of total caspase-3/9 unchanged, whereas the protein level of anti-apoptosis Bcl-2 diminished upon hypoxia (Figure 1E). These data indicated that hypoxia led to H9c2 cell injury by abrogating viability and facilitating apoptosis. Then, we examined expression of BNIP3 under hypoxia in H9c2 cells. Results showed increased BNIP3 mRNA and protein levels under hypoxia in H9c2 cells (Figure 1F), implying the participation of BNIP3 in hypoxia-caused injury of H9c2 cells.
Figure 1.
Hypoxia Led to Injury of H9c2 Cells and Upregulated BNIP3 Expression
(A and B) CCK-8 and EdU assays (scale bar = 100 μm) detected that -cell viability of H9c2 cells in hypoxia group was lower than that in normoxia control. (C and D) TUNEL (scale bar = 100 μm) and caspase-3 activity analyses were used to assess the increased apoptosis rate of H9c2 cells under hypoxia versus normoxia control. (E) Western blot analysis determined the higher levels of cleaved caspase-3/9, Bax and lower level of Bcl-2 in H9c2 cells under hypoxia versus normoxia control. (F) Quantitative real-time PCR and western blot analyses were utilized to evaluate the increased level of BNIP3 in H9c2 cells under hypoxia versus normoxia control. Bar graphs were shown as mean ± SD. *p < 0.05, **p < 0.01.
BNIP3 Silence Reversed Hypoxia-Activated Injury of H9c2 Cells
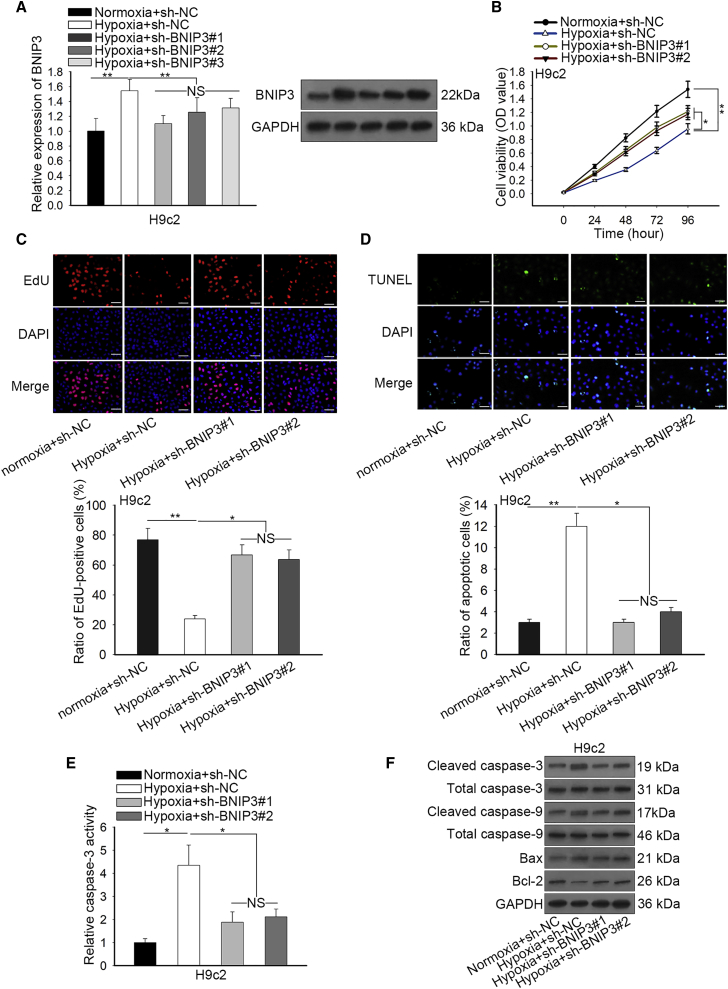
Next, we probed the function of BNIP3 in regulating hypoxia-stimulated injury in H9c2 cells. Quantitative real-time PCR and western blot analyses revealed that transfection of short hairpin RNA targeting BNIP3 (sh-BNIP3#1), sh-BNIP3#2, or sh-BNIP3#3 decreased the expression of BNIP3 in hypoxia-treated H9c2 cells (Figure 2A). Because sh-BNIP3#1/2 presented better effect in reducing BNIP3 expression, we used them for subsequent loss-of-function assays. We observed by CCK-8 and EdU assays that hypoxia hampered the viability of H9c2 cells, and such an effect could be counteracted by silencing BNIP3 (Figures 2B and 2C). Additionally, apoptosis of H9c2 cells facilitated by hypoxia treatment was abrogated by BNIP3 knockdown (Figures 2D and 2E). Concordantly, the induced levels of pro-apoptotic cleaved-caspase-3/9 and Bax in hypoxic H9c2 cells were reduced under knockdown of BNIP3, whereas the reduced level of anti-apoptotic Bcl-2 in hypoxic H9c2 cells was recovered by knockdown of BNIP3 (Figure 2F). In sum, the abovementioned data indicated that BNIP3 silence reversed hypoxia-stimulated injury of H9c2 cells.
Figure 2.
BNIP3 Silence Reversed Hypoxia-Caused Injury of H9c2 Cells
(A) H9c2 cells underwent hypoxia treatment, with normoxia as control, and were transfected with sh-NC and sh-BNIP3#1/2/3, respectively, under hypoxia. Quantitative real-time PCR and western blot detections of the decreased expression level of BNIP3 in hypoxia-treated H9c2 cells transfected with sh-BNIP3#1/#2/#3 compared with sh-NC group. (B and C) CCK-8 and EdU (scale bar = 100 μm) assays detected the increased cell viability of hypoxia-treated H9c2 cells after silencing of BNIP3. (D and E) TUNEL (scale bar = 100 μm) and caspase-3 activity analyses were applied to estimate the decreased apoptosis rate of hypoxia-treated H9c2 cells transfected with sh-BNIP3#1/#2. (F) Western blotting determined the negative effect of silenced BNIP3 on the levels of cleaved caspase-3/9, . Bax as well as the positive effect on the level of Bcl-2 in hypoxia-treated H9c2 cells. Bar graphs were shown as mean ± SD.*p < 0.05, **p < 0.01. NS: no significance.
circ-BNIP3 Was Upregulated by Hypoxia and Positively Regulated BNIP3 in H9c2 Cells
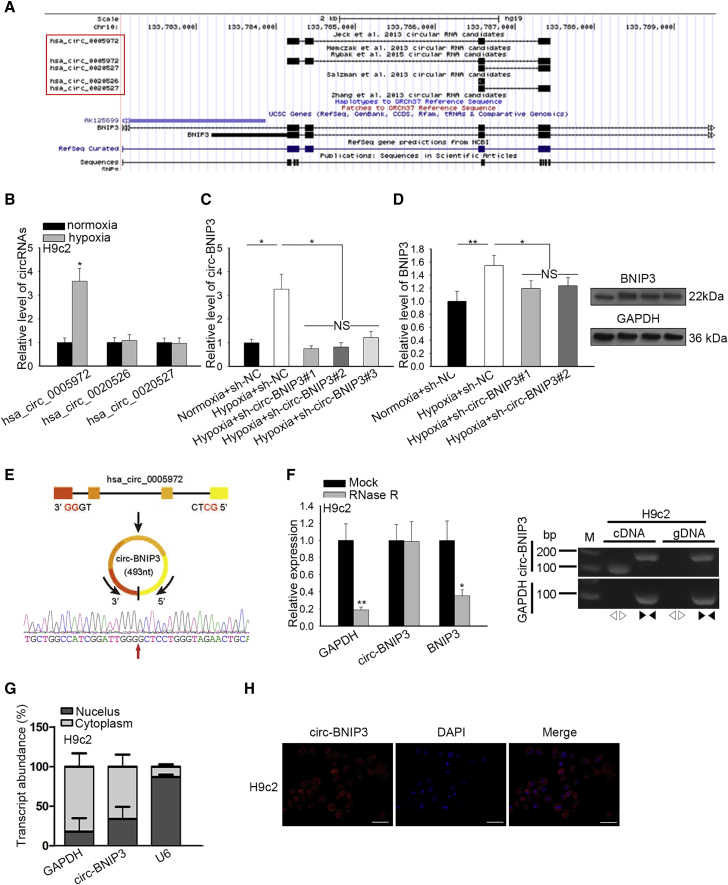
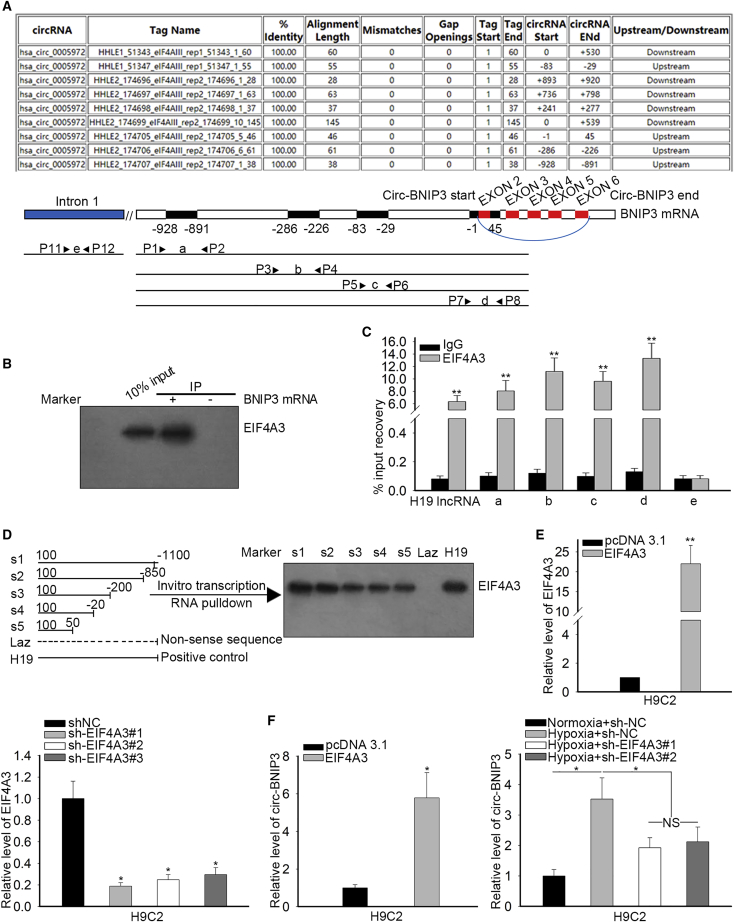
Then, we tried to explore the mechanism underlying the upregulation of BNIP3. It has been reported that circRNA can regulate the expression of its host gene21 and participate in hypoxia-induced cell injury.15 Therefore, we tried to find the circRNAs related to BNIP3 through browsing circBase (http://www.circbase.org/). Results showed that 3 circRNAs were transcribed from BNIP3 gene, which were hsa_circ_0005972, hsa_circ_0020526, and hsa_circ_0020527 (Figure 3A). To figure out the association of the 3 circRNAs with hypoxia-induced damage in H9c2 cells, we examined their expressions under hypoxia. Results showed that only hsa_circ_0005972 was upregulated responding to hypoxia in H9c2 cells (Figure 3B), indicating that hsa_circ_0005972 might participate in the regulation of hypoxia-induced injury of H9c2 cells. Therefore, we termed it as circ-BNIP3 and further investigated its effect on BNIP3 expression. As presented in Figure S1A, quantitative real-time PCR indicated that circ-BNIP3 expression was overtly increased in response to hypoxia in neonatal rat ventricular cardiomyocyte (NRVM). We confirmed that introducing sh-circ-BNIP3#1/2/3 could impair the inductive effect of hypoxia on circ-BNIP3 expression in H9c2 cells, and sh-circ-BNIP3#1 and 2 were more efficient than sh-circ-BNIP3#3 (Figure 3C), so sh-circ-BNIP3#1/2 were used for later experiments. We found that the hypoxia-induced expression of BNIP3 at mRNA and protein levels was abrogated by circ-BNIP3 silence in H9c2 cells (Figure 3D), which meant that circ-BNIP3 positively regulated BNIP3 expression.
Figure 3.
circ-BNIP3 Was Upregulated by Hypoxia and Positively Regulated BNIP3 in H9c2 Cells
(A) Three circRNAs (hsa_circ_0005972, hsa_circ_0020526, and hsa_circ_0020527) transcribed from BNIP3 were obtained from circBase. The hsa_circ_0005972 was re-termed as circ-BNIP3. (B) Expression of the abovementioned miRNAs was evaluated in hypoxic and normoxic H9c2 cells by quantitative real-time PCR. (C) H9c2 cells underwent hypoxia treatment, with normoxia as control, and were transfected with sh-NC and sh-circ-BNIP3#1/2/3, respectively, under hypoxia. Quantitative real-time PCR analysis for circ-BNIP3 expression in H9c2 cells of each group. (D) Quantitative real-time PCR and western blot determined BNIP3 expression in H9c2 cells of each group. (E) The schematic diagram of formation of circ-BNIP3 and the circRNA sequencing results. (F) Left: relative RNA levels were examined by quantitative real-time PCR and normalized to the value determined in the mock group. Right: circ-BNIP3 amplified by divergent primers were in cRNA rather than in gDNA. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was linear control. (G and H) Circ-BNIP3 was found to be enriched in the cytoplasm of H9c2 cells according to the data of subcellular fractionation assay and FISH (scale bar = 100 μm). Bar graphs were shown as mean ± SD. *p < 0.05, **p < 0.01. NS: no significance.
Moreover, we tried to confirm the circular feature of circ-BNIP3. We validated through circRNA sequencing that circ-BNIP3 was located at chromosome 10 (chr10): 133784141–133787447 with a spliced length of 493 bp (Figure 3E). Also, circ-BNIP3, rather than BNIP3, cannot be digested by RNase R (Figure 3F, left), confirming that circ-BNIP3 had a structure of covalently closed continuous loop.22, 23, 24 Moreover, circ-BNIP3 amplified by divergent primers was detectable in cDNA rather than gDNA (Figure 3F, right). We further identified that circ-BNIP3 was located in cytoplasm of H9c2 cells by subcellular fractionation and fluorescence in situ hybridization (FISH) staining analysis (Figures 3G and 3H). These results suggested that circ-BNIP3 was a bona fide circRNA.
circ-BNIP3 Silence Reversed Hypoxia-Induced Damage of H9c2 Cells
Then, the biological role of circ-BNIP3 in hypoxia-induced damage of H9c2 cells was evaluated. The inhibitive effect of hypoxia on H9c2 cell viability was abrogated by the depletion of circ-BNIP3 (Figures 4A and 4B). The hypoxia-induced apoptosis of H9c2 cells was reversed by the knockdown of circ-BNIP3 (Figures 4C and 4D). The induced levels of cleaved-caspase-3/9 and Bax and reduced levels of Bcl-2 under hypoxia could be reversed by silencing circ-BNIP3 (Figure 4E). Taken together, circ-BNIP3 silence reversed hypoxia-induced injury of H9c2 cells.
Figure 4.
circ-BNIP3 silence reversed hypoxia-produced injury of H9c2 cells
H9c2 cells underwent hypoxia treatment, with normoxia as control and were transfected with sh-NC and sh-circ-BNIP3#1/2, respectively, under hypoxia. (A and B) CCK-8 and EdU (scale bar = 100 μm) assays tested the decreased cell viability of hypoxia-treated H9c2 cells transfected with sh-circ-BNIP3#1/#2 compared with that transfected with sh-NC (C and D) TUNEL (scale bar = 100 μm) and caspase-3 activity analyses for detecting the decreased apoptosis of hypoxia-treated H9c2 cells transfected with sh-circ-BNIP3#1/#2 compared with that transfected with sh-NC. (E) Western blot was utilized to determine the expressions of apoptosis-associated proteins in H9c2 cells in each group. Bar graphs were shown as mean ± SD. *p < 0.05, **p < 0.01. NS: no significance.
circ-BNIP3 Sponged miR-27a-3p to Upregulate BNIP3 in H9c2 Cells
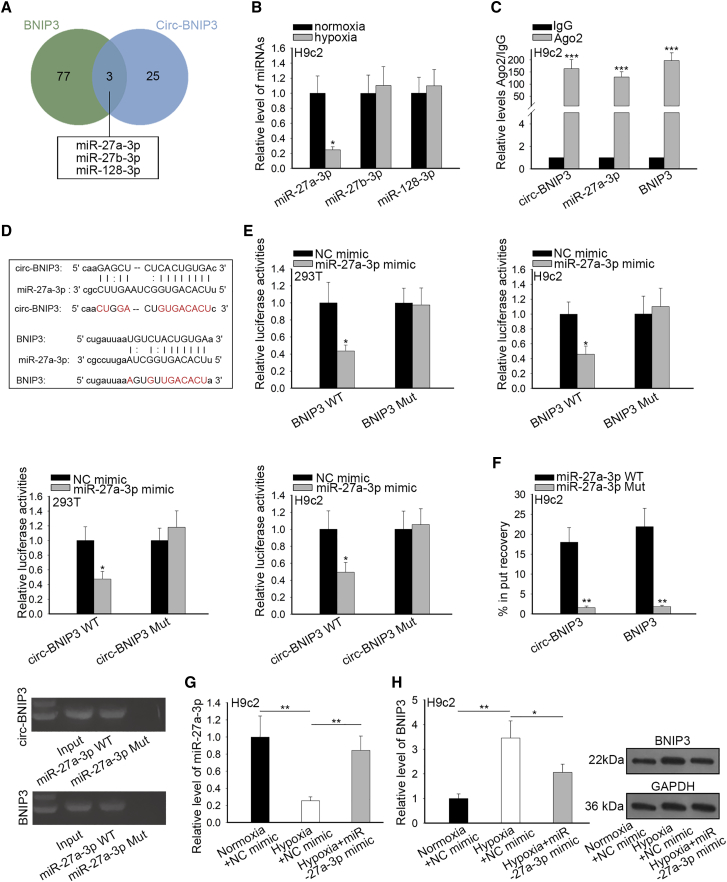
In subsequence, we investigated how circ-BNIP3 regulated BNIP3 expression. Mounting studies have revealed that circRNAs could regulate gene expressions through performing as miRNA sponge, including in regulating cardiomyocyte injury.15,25 Hence, we speculated that circ-BNIP3 regulated BNIP3 expression through sponging miRNA in H9c2 cells. We searched Starbase (http://starbase.sysu.edu.cn/) to identify the shared miRNAs interacting with both circ-BNIP3 and BNIP3 mRNA. As presented in Figure 5A, the intersection of the Venn pattern showed 3 miRNAs shared by circ-BNIP3 and BNIP3 mRNA, which were miR-27a-3p, miR-27b-3p, and miR-128-3p (Figure 5A). Further, we determined the involvement of 3 miRNAs in hypoxia-induced injury in H9c2 cells. Results of quantitative real-time PCR analysis showed that only miR-27a-3p was downregulated upon hypoxia versus normoxia control in H9c2 cells (Figure 5B), indicating the association of miR-27a-3p in hypoxia-induced injury in H9c2 cells. Also, previous studies have reported that miR-27a-3p could aggravate proliferation and hamper apoptosis in cancer cells.26 Therefore, we selected miR-27a-3p for further investigation. RNA immunoprecipitation (RIP) assay confirmed the enrichment of circ-BNIP3, miR-27a-3p, and BNIP3 mRNA in the precipitates of Ago2 (Figure 5C). Moreover, we obtained the binding sequences on circ-BNIP3 and BNIP3 mRNA for miR-27a-3p from Starbase (Figure 5D). Luciferase reporter assays were conducted in both H9c2 and 293T cells to further investigate the interaction of miR-27a-3p with circ-BNIP3 and BNIP3. Results showed that overexpression of miR-27a-3p resulted in decreased luciferase activity of wild-type (WT)-circ-BNIP3 and WT-BNIP3, instead of mutant (mut)-circ-BNIP3 and mut-BNIP3 (Figure 5E). Additionally, pull-down assay confirmed the abundant expression of circ-BNIP3 and BNIP3 mRNA in miR-27a-3p WT pull-down (Figure 5F). Then, we validated that overexpression of miR-27a-3p enhanced miR-27a-3p expression reduced in H9c2 cells treated with hypoxia (Figure 5G) and that overexpression of miR-27a-3p reversed the inductive effect of hypoxia on BNIP3 expression in H9c2 cells (Figure 5H). These results hinted that circ-BNIP3 sponged miR-27a-3p to upregulate BNIP3 in H9c2 cells.
Figure 5.
circ-BNIP3 Sponged miR-27a-3p to Upregulate BNIP3 in H9c2 Cells
(A) Three miRNAs (miR-27a-3p, miR-27b-3p, and miR-128-3p) that can simultaneously bind with circ-BNIP3 and BNIP3 were obtained by Starbase and the intersection was shown as a Venn plot (B) Quantitative real-time PCR examination of the expressions of 3 miRNAs under hypoxia and normoxia control in H9c2 cells.miR-27a-3p was significantly downregulated under hypoxia compared to miR-27b-3p and miR-128-3p. (C) RIP assay was used for detection of the enrichment of miR-27a-3p, circ-BNIP3 and BNIP3 in response to anti-Ago2 compare to the negative control IgG, (D) The binding sites on circ-BNIP3 and BNIP3 mRNA for miR-27a-3p were obtained from Starbase, and the mutant sites (red characters) were designed for luciferase reporter assays. (E) Luciferase reporter assays were conducted in both H9c2 and 293T cells to detect the binding of miR-27a-3p on circ-BNIP3 and BNIP3 mRNA. (F) Pull-down assay confirmed the enrichment of circ-BNIP3 and BNIP3 mRNA in miR-27a-3p-Wt pull-downcompared with miR-27a-3p-WT (G) Increasing of miR-27a-3p in hypoxia-treated H9c2 cells was identified by RT-qPCR after transfected with relative miRNA mimics. (H) RT-qPCR and western blot detected that the levels of BNIP3 mRNA and protein increased under treatments of hypoxia+NC mimic were decreased after upregulation of miR-27a-3p in H9c2 cells. Bar graphs were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
EIF4A3 Induced circ-BNIP3 Expression in H9c2 Cells
Further, we probed the mechanism of circ-BNIP3 upregulation in H9c2 cells. Previous studies demonstrated that EIF4A3, a key regulator of RNA splicing,27 induced circ-matrix metallopeptidase 9 (MMP9) expression via binding to the upstream region of MMP9 mRNA and induce circular RNA formation.28 Herein, we found through Circular RNA Interactome (https://circinteractome.nia.nih.gov/) that EIF4A3 had 4 binding sites on the upstream region of BNIP3 mRNA transcript (Figure 6A). The higher level of EIF4A3 was examined in H9c2 and NRVM cells after treated with hypoxia (Figure S1B). We conduced pull-down assay using BNIP3 mRNA and confirmed the enrichment of EIF4A3 protein in the pull-down of BNIP3 mRNA rather than control (Figure 6B). To detect whether EIF4A3 bound at the predicted regions on the BNIP3 mRNA transcript, we carried out RIP assay using EIF4A3 antibody, followed by quantitative real-time PCR detection using the primers designed according to four binding regions (a, b, c, and d). Results confirmed that fragment a, b, c, and d were all enriched in EIF4A3 precipitates (Figure 6C). Also, the RNA constructs containing 1, 2, 4, and 4 EIF4A3 binding sites (s1, s2, s3, and s4) were used for pull-down assay. Western blot analysis after pull-down assay illustrated that the depletion of EIF4A3 sites on circ-BNIP3 mRNA transcript resulted in lessened enrichment of EIF4A3 protein (Figure 6D), further confirmed the requirement of the predicted sites for the binding of EIF4A3. Later, we detected the effect of EIF4A3 on circ-BNIP3 expression. We confirmed the overexpression and knockdown of EIF4A3 in H9c2 cells by quantitative real-time PCR analysis (Figure 6E). We verified that overexpression of EIF4A3 induced circ-BNIP3 expression and that knockdown of EIF4A3 reversed the inductive effect of hypoxia on circ-BNIP3 expression in H9c2 cells (Figure 6F). To conclude, the data above suggested that EIF4A3 induced circ-BNIP3 expression in H9c2 cells.
Figure 6.
EIF4A3 Induced circ-BNIP3 Expression in H9c2 Cells
(A) The binding sites for EIF4A3 on the BNIP3 mRNA transcript at the upstream region of circ-BNIP3 were obtained from Circular RNA Interactome. (B) Pull-down assay confirmed the enrichment of EIF4A3 protein in BNIP3 mRNA pull-down. (C) Quantitative real-time PCR following the EIF4A3-RIP assay confirmed the binding of EIF4A3 on the predicted binding region (a, b, c, and d) of BNIP3 mRNA. H19, a known interacting long noncoding RNA (lncRNA) with EIF4A3, was the positive control. The intron 1 of BNIP3 mRNA was used as negative control. (D) The schematic diagram of 5 RNA constructs with EIF4A3 binding sites, which were truncated to different degrees (s1–s5). Laz and H19 were the negative control and positive control. The pull-down assay was conducted to analyze the interplay between EIF4A3 and BNIP3 mRNA (s1–s5). (E) The overexpression and knockdown of EIF4A3 in H9c2 cells were confirmed by quantitative real-time PCR analysis. (F) The expression of circ-BNIP3 under EIF4A3 overexpression in H9c2 cells treated with or without hypoxia was analyzed upon quantitative real-time PCR analysis. Bar graphs were shown as mean ± SD. *p < 0.05, **p < 0.01. NS: no significance.
circ-BNIP3 Regulated Hypoxia-Induced Injury of H9c2 Cells in a BNIP3-Dependent Manner
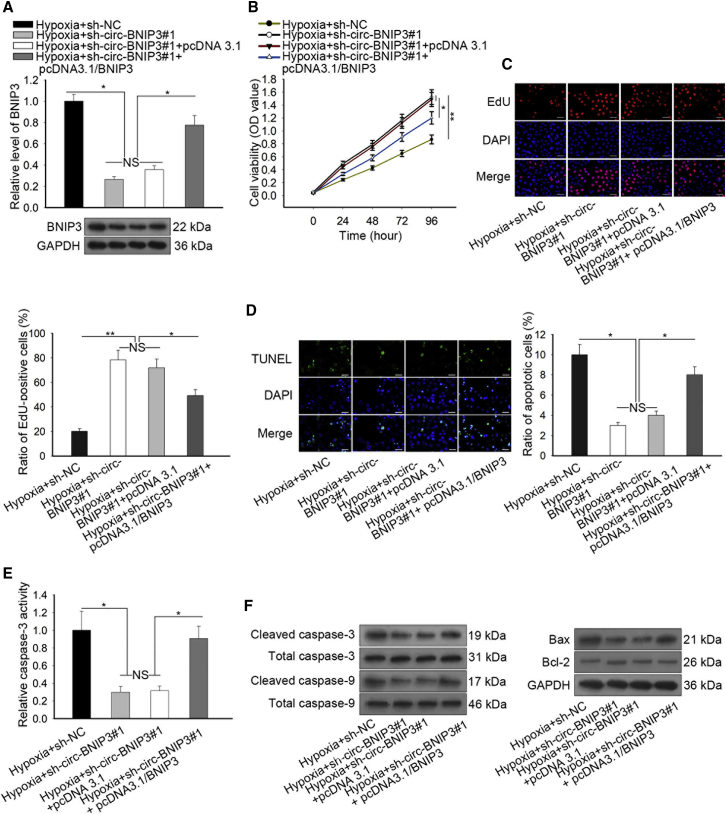
Finally, we carried out rescue assays to probe whether BNIP3 participated in circ-BNIP3-mediated hypoxia-induced injury of H9c2 cells. First, we confirmed that transfection of pcDNA3.1/BNIP3 rescued the silence of BNIP3 caused by circ-BNIP3 knockdown under hypoxia (Figure 7A). CCK-8 and EdU analyses demonstrated that silencing circ-BNIP3 facilitated the viability of hypoxia-treated H9c2 cells, and such an effect was counteracted by overexpressing BNIP3 (Figures 7B and 7C). The apoptosis of H9c2 cells under hypoxia was impeded by circ-BNIP3 depletion, and such an effect was impaired by overexpression of BNIP3 (Figures 7D and 7E). Also, the decreased levels of cleaved-caspase-3/9 and Bax and the increased level of Bcl-2 caused by circ-BNIP3 inhibition in hypoxia were reversed by overexpression of BNIP3 in H9c2 cells (Figure 7F). Therefore, it was suggested that circ-BNIP3 regulated hypoxia-induced injury of H9c2 cells in a BNIP3-dependent manner.
Figure 7.
circ-BNIP3 Regulated Hypoxia-Induced Injury of H9c2 Cells in a BNIP3-Dependent Manner
(A) The level of BNIP3 decreased in circ-BNIP3-downregulated cells was recovered by the overexpression of BNIP3. (B and C) CCK-8 and EdU (scale bar = 100 μm) assays were operated to detect that the increased cell viability of H9c2 cells was recovered again after co-transfected with pcDNA3.1/BNIP3. (D and E) TUNEL (scale bar = 100 μm) and caspase-3 activity analyses showed that the decreased apoptosis rate of circ-BNIP3-downregulated H9c2 cells was increased after overexpression of BNIP3. (F) Western blot analysis was used to determine the expressions of apoptosis-related proteins in H9c2 cells of each group. Bar graphs were shown as mean ± SD. *p < 0.05, **p < 0.01. NS: no significance.
Discussion
MI is known to be a result of hypoxia or acute, persistent ischemia, and can contribute to irreversible heart muscle damage and heart failure.1, 2, 3 Statistics revealed that MI caused large numbers of deaths globally.6,9 Therefore, exploring the molecular mechanism underlying the hypoxia-induced injury of cardiomyocytes can potentially benefit the improvement of MI treatment.
BNIP3 is reputed as a crucial regulator of apoptosis belonging to the BH3-only subfamily of Bcl-2 family proteins.17,18 Although in many organs, including heart, BNIP3 expression is reported to be undetectable under normal condition, hypoxia is discovered to be able to induce BNIP3 expression.17,19 Mounting studies have revealed that BNIP3 positively regulated hypoxia-induced cell injury. For example, miR-210 targeted BNIP3 to protect rat adrenal gland pheochromocytoma (PC-12) cells from hypoxia-induced injury.29 Tetramethylpyrazine alleviated the hypoxia-induced apoptosis of myocardial cells via inhibiting Hypoxia inducible factor (HIF)-1α/c-jun N-terminal kinase (JNK)/p38 signaling and insulin like growth factor binding protein 3 (IGFBP3)/BNIP3 axis, and upregulating PI3K/Akt survival signaling.30 Furthermore, it has been reported that hypoxia and acidosis can induce BNIP3 to activate cardiac myocyte death.20 These findings suggested that BNIP3 is an important regulator of hypoxia-resulted in injury for cardiomyocytes. In concordance, this study confirmed that BNIP3 could be upregulated in H9c2 cells under hypoxia, and depletion of BNIP3 reversed the effect of hypoxia in retarding viability and facilitating apoptosis in H9c2 cells.
Furthermore, we explored the mechanism of BNIP3 overexpression in H9c2 cells under hypoxia. Previously, it has been revealed that circRNAs are a novel class of ncRNAs able to regulate the expression of its host gene.21 Also, circRNAs are increasingly reported to play vital roles in hypoxia-caused injury of cardiomyocytes.15,16 Therefore, we speculated that BNIP3 might be regulated by circRNA in H9c2 cells. We identified 3 circRNAs associated with BNIP3, and only circ-BNIP3 (hsa_circ_0005972) was upregulated under hypoxia in H9c2 cells, indicating the involvement of circ-BNIP3 in hypoxia-produced injury of H9c2 cells. Loss-of-function assays indicated that targeting circ-BNIP3 can reverse hypoxia-induced injury of H9c2 cells.
Mechanistically, circRNAs have been demonstrated to modulate gene expression through competitive RNA (ceRNA) network, whereby circRNAs sponged miRNAs to prevent downstream mRNAs from miRNA-mediated degradation,31 including in cardiomyocyte injury.15 In our study, we first identified through Starbase that miR-27a-3p was among the 3 miRNAs shared by circ-BNIP3 and BNIP3. We chose miR-27a-3p because we found that it could only be downregulated by hypoxia in H9c2 cells, indicating that miR-27a-3p might be a negative regulator of hypoxia-produced injury in H9c2 cells. Also, a former study reported that miR-27a-3p can aggravate proliferation and hamper apoptosis in cancer cells,26 but the role of miR-27a-3p in hypoxia-induced injury of cardiomyocytes remained elusive until this study. We subsequently validated the interaction of miR-27a-3p with circ-BNIP3 and BNIP3 and that miR-27a-3p negatively regulated BNIP3 expression in H9c2 cells, indicating that circ-BNIP3 upregulated BNIP3 through targeting miR-27a-3p. Rescue assays suggested that circ-BNIP3 regulated hypoxia-generated injury of H9c2 cells through BNIP3.
Furthermore, we interrogated the upstream mechanism of circ-BNIP3 upregulation in H9c2 cells. A former study demonstrated that EIF4A3 bound to the upstream region of circ-MMP9 mRNA transcripts to facilitate circular RNA formation and induce circ-MMP9 expression.28 Herein, we identified through online bioinformatics tool Circular RNA Interactome that EIF4A3 potentially bound to 4 sites on BNIP3 mRNA at the upstream region of the circ-BNIP3 mRNA transcripts. We confirmed through RIP and pull-down assays that EIF4A3 interacted with BNIP3 mRNA at the upstream binding sites. Upregulation of EIF4A3 induced circ-BNIP3 expression in H9c2 cells, whereas downregulation of EIF4A3 reversed the hypoxia-induced circ-BNIP3 expression, indicating that targeting EIF4A3 can regulate the upregulation of circ-BNIP3 in hypoxia-injured H9c2 cells.
In conclusion, our paper first demonstrated that EIF4A3-induced circ-BNIP3 aggravated hypoxia-injured H9c2 cells by targeting miR-27a-3p/BNIP3, representing the first discovery of circ-BNIP3 in hypoxia-caused H9c2 cell injury and that circ-BNIP3 might be a novel target for MI treatment. However, more experiments based on in vivo models are needed to determine the therapeutic significance of targeting circ-BNIP3.
Materials and Methods
Cell Culture and Treatment
The cell line H9c2 was derived from rat embryonic ventricular cardiomyocytes, which were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and subsequently kept in DMEM with 10% (v/v) FBS, 100 U/mL penicillin and 100 lg/mL streptomycin in 5% CO2 at the temperature of 37°C. H9c2 cells were grown in an incubator containing 94% N2, 5% CO2, and 1% O2 to simulate hypoxia injury.
CCK-8 Assay
To evaluate viability, we seeded H9c2 cells in 96-well plates and tested via CCK-8 assay (Dojindo, Kumamoto, Japan). After addition of CCK-8 solution in each well, cells were then cultured. The absorbance of 450 nm was monitored via ThermoMax microplate reader (Olympus).
EdU Assay
The treated H9c2 cell line was put on sterile coverslips in 24-pore plates. EdU kit (RiboBio) was utilized for assessing cell proliferative ability based on the instruction provided by supplier. Images were captured by Laser confocal microscopy (FV300, Olympus, Japan). Nuclei was double dyed with EdU and 4’,6-diamidino-2-phenylindole (DAPI; Beyotime, Shanghai, China) to assess cell viability.
TUNEL Staining Assay
For TUNEL staining assay, H9c2 cells were first immobilized in 1% formaldehyde and mixed with 0.2% Triton X-100. Thereafter, cells were treated with fluorescein 2'-Deoxyuridine 5'-Triphosphate (dUTP)-end labeling (ApoAlert DNA Fragmentation Assay Kit; Clontech, Mountain View, CA, USA) and DAPI. At length, cell lines were observed by use of fluorescence microscope (NIKON TE200-U, Tokyo, Japan).
Detection of Capase-3 Activity
Caspase-3 activity was measured for evaluating cell apoptosis. The Caspase-3 Activity Kit was bought from Solarbio (Beijing, China). First, total protein was isolated from H9c2 cells and put into 96-well culture dishes with 90 μL reaction buffer and caspase-3 substrate. Post 4 h incubation at 37°C, caspase-3 activity was determined with a microplate reader (Tecan, Switzerland) at 405 nm.
Western Blotting
The proteins were reaped from radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Shanghai, China) with addition of protease inhibitors (Roche, Guangzhou, China), followed by quantitation with the BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Waltham, MA, USA). Following the user guide, western blot system was constructed with a Bio-Rad Bis-Tris Gel system. Primary antibodies (dilution: 1:1,000) were put in 5% blocking buffer and co-cultured all night at 4°C. Secondary antibodies labeled by horseradish peroxidase were then used to culture the polyvinylidene difluoride (PVDF) membrane at indoor temperature (approximately 25°C) for just 1 h. The protein bands were examined through Image Lab Software (Bio-Rad, Shanghai, China).
RNA Extraction and Quantitative Real-Time PCR
Total RNA of H9c2 cells using the TRIzol reagent (Invitrogen, CA, USA) for separation was reverse-transcribed to generate cDNA. This reaction was implemented at 95°C for 3 min for 35 cycles, 95°C for 12 s, and 58°C for 30 s. Quantitative real-time PCR analysis was conducted with One Step SYBR PrimeScript PLUS RT-RNA PCR Kit (TaKaRa Biotechnology, Tokyo, Japan). Relative expression of genes was normalized to that of GAPDH or U6 (the internal controls) using 2-ΔΔCt method.
Cell Transfection and Plasmids
H9c2 cells and hypoxic H9c2 cells were planted in 6-well plates until reached 70% cell confluence. Cell transfection was conducted by Lipofectamine 2000 (Invitrogen, CA, USA). Short hairpin RNAs (shRNAs) against BNIP3, circ-BNIP3, or EIF4A3 were constructed by RiboBio (Guangzhou, China) and named as sh-BNIP3#1/2/3, sh-circ-BNIP3#1/2/3, or sh-EIF4A3#1/2/3. miR-27a-3p mimics and negative control (miR-NC) were designed and synthesized by Genechem Company (Shanghai, China). To overexpress EIF4A3, we transfected cells with pcDNA3.1/EIF4A3 and the empty pcDNA3.1 vector. 48 h later, collected cells were further analyzed.
Nuclear-Cytoplasmic RNA Isolation Assay
Cytoplasmic and nuclear RNA isolation was carried out following the protocol of PARIS Kit (Invitrogen, USA). H9c2 cells were lysed in cell fractionation buffer and subsequently centrifuged. The supernatant was collected, and the remaining lysates were rinsed in cell fractionation buffer and later centrifuged. The nuclei were lysed by cell disruption buffer and the isolated RNAs were assessed through quantitative real-time PCR.
FISH Assay
FISH assay was operated using PARIS kit (Life Technologies, Waltham, MA, USA). The needed RNA FISH probes were designed and synthesized via RiboBio (Guangzhou, China). Cells cultured with 4% formaldehyde for a quarter were rinsed in PBS. Thereafter, air-dried cells were dealt with FISH probe (40 nM) in hybridization buffer. After washing and dehydrating, cell nuclei were subjected to DAPI staining. An Olympus fluorescence microscope (IX73; Olympus, Tokyo, Japan) was used to capture the images.
RIP Assay
RIP was experimented with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA). Cells (1 × 107) were collected and cultivated in RIP lysis buffer, followed by immunoprecipitated with Ago2 antibody (Cell Signaling Technology, MA, USA). The final retrieved RNA was subjected to quantitative real-time PCR analysis. Normal mouse immunoglobulin G (IgG) controls were assayed simultaneously.
Luciferase Reporter Assay
The reporter plasmids containing psiCHECK-circ-BNIP3 WT/Mut and psiCHECK-BNIP3 WT/Mut were designed and constructed by Promega Corporation (Fitchburg, WI, USA) based on the bioinformatics-predicted results. The sequences of WT circ-BNIP3 and BNIP3 mRNA were obtained by PCR amplification. H9c2 cells were subjected to co-transfection of the aforementioned plasmids and miR-27a-3p mimic or NC mimic using Lipofectamine 2000. At 48 h after transfection, luciferase activity was assessed by a Dual Luciferase Reporter Assay System (Promega).
RNA Pull-Down Assay
Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA) was operated for RNA pull-down assay in line with the standard method. BNIP3 mRNA expression vectors were truncated to different degrees (s1 s5) and generated. The biotinylated BNIP3 DNA probe (GenePharma, Shanghai, China) was coated with beads via dissolution of the probe and cultured with Dynabeads M-280 Streptavidin (Invitrogen, CA, USA) for nearly 10 min at indoor temperature. Eventually, cell lysates were cultured with the probe-coated beads, followed by analysis by quantitative real-time PCR method.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was operated through the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, USA) as per the guidebook. Chromatin was crosslinked and sonicated into 200 to 1,000 bp fragments and immunoprecipitated with 2 μg of anti-EIF4A3 antibody (Cell Signaling Technology, USA) and 2 μg IgG with rotation for whole night at 4°C. 30 μL of magnetic beads were added to each tube for 2 h at 4°C. At last, the purified immunoprecipitated chromatin was analyzed by quantitative real-time PCR.
Statistical Analysis
Experimental results from at least three separate biological replications were presented as mean ± SD. Statistical analyses were performed on SPSS Vision 19.0 (SPSS, Chicago, IL, USA) and plotted with GraphPad PRISM 6 (GraphPad, San Diego, CA, USA). The p values were calculated using Student’s t test or one-way ANOVA. Threshold of p values was set as 0.05 to indicate a statistically significant difference.
Availability of Data and Materials
Research data are not shared.
Author Contributions
Y.L. and Y.X.: manuscript, study design, methodology, formal analysis. S.R.: data curation and investigation. J.X. and Y.W.: resources.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Thank you to all involved in this study.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.017.
Supplemental Information
References
- 1.Kannel W.B., Cupples L.A., Gagnon D.R. Incidence, precursors and prognosis of unrecognized myocardial infarction. Adv. Cardiol. 1990;37:202–214. doi: 10.1159/000418828. [DOI] [PubMed] [Google Scholar]
- 2.Roger V.L., Go A.S., Lloyd-Jones D.M., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., et al Executive Summary: Heart Disease and Stroke Statistics—2012 Update. Circulation. 2012;125:188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- 3.Boersma E., Mercado N., Poldermans D., Gardien M., Vos J., Simoons M.L. Acute myocardial infarction. Lancet. 2003;361:847–858. doi: 10.1016/S0140-6736(03)12712-2. [DOI] [PubMed] [Google Scholar]
- 4.Lippi G., Sanchis-Gomar F., Cervellin G. Chest pain, dyspnea and other symptoms in patients with type 1 and 2 myocardial infarction. A literature review. Int. J. Cardiol. 2016;215:20–22. doi: 10.1016/j.ijcard.2016.04.045. [DOI] [PubMed] [Google Scholar]
- 5.Coventry L.L., Bremner A.P., Williams T.A., Jacobs I.G., Finn J. Symptoms of myocardial infarction: concordance between paramedic and hospital records. Prehosp. Emerg. Care. 2014;18:393–401. doi: 10.3109/10903127.2014.891064. [DOI] [PubMed] [Google Scholar]
- 6.Do R., Stitziel N.O., Won H.-H., Jørgensen A.B., Duga S., Angelica Merlini P., Kiezun A., Farrall M., Goel A., Zuk O., NHLBI Exome Sequencing Project Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiedler J., Jazbutyte V., Kirchmaier B.C., Gupta S.K., Lorenzen J., Hartmann D., Galuppo P., Kneitz S., Pena J.T., Sohn-Lee C. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720–730. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 8.Mythili S., Malathi N. Diagnostic markers of acute myocardial infarction. Biomed. Rep. 2015;3:743–748. doi: 10.3892/br.2015.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acconcia M.C., Caretta Q., Romeo F., Borzi M., Perrone M.A., Sergi D., Chiarotti F., Calabrese C.M., Sili Scavalli A., Gaudio C. Meta-analyses on intra-aortic balloon pump in cardiogenic shock complicating acute myocardial infarction may provide biased results. Eur. Rev. Med. Pharmacol. Sci. 2018;22:2405–2414. doi: 10.26355/eurrev_201804_14833. [DOI] [PubMed] [Google Scholar]
- 10.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 11.Granados-Riveron J.T., Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2016;1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong Z., Huang M., Lv M., He Y., Duan C., Zhang L., Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 14.He R., Liu P., Xie X., Zhou Y., Liao Q., Xiong W., Li X., Li G., Zeng Z., Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M., Ding W., Tariq M.A., Chang W., Zhang X., Xu W., Hou L., Wang Y., Wang J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–5869. doi: 10.7150/thno.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L.Y., Zhai M., Huang Y., Xu S., An T., Wang Y.H., Zhang R.C., Liu C.Y., Dong Y.H., Wang M. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2018;26:1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vande Velde C., Cizeau J., Dubik D., Alimonti J., Brown T., Israels S., Hakem R., Greenberg A.H. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 2000;20:5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray R., Chen G., Vande Velde C., Cizeau J., Park J.H., Reed J.C., Gietz R.D., Greenberg A.H. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 2000;275:1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- 19.Bruick R.K. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubasiak L.A., Hernandez O.M., Bishopric N.H., Webster K.A. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc. Natl. Acad. Sci. USA. 2002;99:12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X., Wang J., Zhang C., Lin C., Zhang J., Zhang W., Zhang W., Lu Y., Zheng L., Li X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 2018;246:166–179. doi: 10.1002/path.5125. [DOI] [PubMed] [Google Scholar]
- 22.Qu S., Zhong Y., Shang R., Zhang X., Song W., Kjems J., Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 24.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Z., Zhou W., Duan L., Zhang J., Lu X., Jin L., Yu Y. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J. Cell. Physiol. 2019;234:3887–3896. doi: 10.1002/jcp.27162. [DOI] [PubMed] [Google Scholar]
- 26.Yuan, Kluiver Y.,J., Koerts J., de Jong D., Rutgers B.,F., Abdul Razak R., Terpstra M., Plaat B.E., Nolte I.M., Diepstra A. miR-24-3p Is Overexpressed in Hodgkin Lymphoma and Protects Hodgkin and Reed-Sternberg Cells from Apoptosis. Am. J. Pathol. 2017;187:1343–1355. doi: 10.1016/j.ajpath.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Chan C.C., Dostie J., Diem M.D., Feng W., Mann M., Rappsilber J., Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R., Zhang S., Chen X., Li N., Li J., Jia R., Pan Y., Liang H. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer. 2018;17:166. doi: 10.1186/s12943-018-0911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan Y., Zhang X., Zhang Y., Dong Y. MicroRNA-210 Protects PC-12 Cells Against Hypoxia-Induced Injury by Targeting BNIP3. Front. Cell. Neurosci. 2017;11:285. doi: 10.3389/fncel.2017.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin K.H., Kuo W.W., Jiang A.Z., Pai P., Lin J.Y., Chen W.K., Day C.H., Shen C.Y., Padma V.V., Huang C.Y. Tetramethylpyrazine Ameliorated Hypoxia-Induced Myocardial Cell Apoptosis via HIF-1α/JNK/p38 and IGFBP3/BNIP3 Inhibition to Upregulate PI3K/Akt Survival Signaling. Cell. Physiol. Biochem. 2015;36:334–344. doi: 10.1159/000374076. [DOI] [PubMed] [Google Scholar]
- 31.Zhong Y., Du Y., Yang X., Mo Y., Fan C., Xiong F., Ren D., Ye X., Li C., Wang Y. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Research data are not shared.