Abstract
The cytochrome P450 2D6 (CYP2D6) gene locus is challenging to accurately genotype due to numerous single nucleotide variants and complex structural variation. Our goal was to determine whether the CYP2D6 genotype‐phenotype correlation is improved when diplotype assignments incorporate structural variation, identified by the bioinformatics tool Stargazer, with next‐generation sequencing data. Using CYP2D6 activity measured with substrates dextromethorphan and metoprolol, activity score explained 40% and 34% of variability in metabolite formation rates, respectively, when diplotype calls incorporated structural variation, increasing from 36% and 31%, respectively, when diplotypes did not incorporate structural variation. We also investigated whether the revised Clinical Pharmacogenetics Implementation Consortium (CPIC) recommendations for translating genotype to phenotype improve CYP2D6 activity predictions over the current system. Although the revised recommendations do not improve the correlation between activity score and CYP2D6 activity, perhaps because of low frequency of the CYP2D6*10 allele, the correlation with metabolizer phenotype group was significantly improved for both substrates. We also measured the function of seven rare coding variants: one (A449D) exhibited decreased (44%) and another (R474Q) increased (127%) activity compared with reference CYP2D6.1 protein. Allele‐specific analysis found that A449D is part of a novel CYP2D6*4 suballele, CYP2D6*4.028. The novel haplotype containing R474Q was designated CYP 2D6*138 by PharmVar; another novel haplotype containing R365H was designated CYP2D6*139. Accuracy of CYP2D6 phenotype prediction is improved when the CYP2D6 gene locus is interrogated using next‐generation sequencing coupled with structural variation analysis. Additionally, revised CPIC genotype to phenotype translation recommendations provides an improvement in assigning CYP2D6 activity.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑Cytochrome P450 2D6 (CYP2D6) is a clinically relevant pharmacogene, but the predictive value of current pharmacogenetic tests is limited by the technical challenges posed by structural variation at the highly polymorphic CYP2D6 gene locus.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑How much of the variability in CYP2D6 activity is explained by single nucleotide variants alone, and does inclusion of structural variation data improve the association? How do revisions to the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommendations for translating CYP2D6 genotype to phenotype improve activity prediction?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑CYP2D6 activity score is more predictive of CYP2D6 activity when diplotype assignments are made using both single nucleotide variant and structural variation data. The new CPIC‐recommended genotype to phenotype translation method improved the relationship between phenotype and CYP2D6 activity compared with the current system.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑Targeted genotyping platforms may miss important CYP2D6 single nucleotide and structural variation, resulting in incorrect phenotype predictions. Greater precision can be obtained using targeted next‐generation sequencing in combination with a bioinformatics tool like Stargazer.
The combination of a wide range of clinically relevant substrates and a well‐studied relationship between genetic variation and enzymatic activity makes cytochrome P450 2D6 (CYP2D6) a logical candidate for clinical pharmacogenetic testing.1, 2 To guide the implementation of pharmacogenomic testing, the Clinical Pharmacogenetics Implementation Consortium (CPIC) has published six guidelines for drugs metabolized by CYP2D6 (i.e., codeine, tamoxifen, atomoxetine, selective serotonin reuptake inhibitors, tricyclic antidepressants, and antiemetics). A high degree of variability in CYP2D6 activity has been observed among individuals as well as populations, which can largely be attributed to variation in the CYP2D6 gene.3, 4 The CYP2D6 gene is notoriously difficult to interrogate,5 however, due to challenges imposed by the complexity of the CYP2D gene locus, which includes a highly homologous pseudogene, CYP2D7, and a high degree of CYP2D6 variation.3, 6, 7, 8 Due to such variation, targeted genotyping panels may miss rare, clinically relevant variants that are present in an individual but not interrogated, leading to phenotype misclassification.3, 7 This is particularly problematic in diverse populations where there are limited or no data regarding genetic variation.4, 7, 9
In addition to characterizing all single nucleotide variants and indels (referred to collectively as SNVs for brevity) when assigning CYP2D6 diplotypes, it is important to consider structural variation. Gene deletions, duplications, and multiplications, as well as gene rearrangements between CYP2D6 and CYP2D7, known as hybrid genes, have all been observed. Structural variation is not uncommon: the CYP2D6*5 gene deletion occurs at a frequency of 2–6% worldwide, whereas duplications/multiplications of functional gene units, such as CYP2D6*1xN or *2xN, occur at frequencies up to 12%.4, 7, 10, 11
Many current CYP2D6 pharmacogenetic testing panels are designed to detect common alleles as well as nonspecific copy number variations (e.g., testing for the presence of deletion and/or duplications, but not necessarily specifying the variation). This approach may result in ambiguous diplotype calls, however, leading to uncertainty in phenotype, or CYP2D6 activity score (AS) assignments. For instance, a duplication observed in an individual genotyped as CYP2D6*1/*4 could result in either a CYP2D6*1x2/*4 (AS = 2) or a CYP2D6*1/*4x2 (AS = 1). Additionally, the presence ofso‐called hybrid genes (CYP2D6‐2D7 and CYP2D7‐2D6) may lead to false positive or negative calls using traditional approaches. For instance, polymerase chain reaction (PCR)‐based CYP2D6 copy number assays may produce erroneous results in the presence of hybrid genes that lack PCR probe binding sites.12, 13, 14 Overestimation or underestimation of CYP2D6 activity is possible if undetected structural variation leads to a default allele assignment of CYP2D6*1.
Genotyping panels are amenable to automated diplotype assignment because there are a small number of variants. More in‐depth interrogation, however, requires “manual” assignment of haplotypes and diplotypes by comparing the variants found in a sample to those listed in CYP2D6 haplotype definition tables.10, 15, 16, 17 The large amount of data obtained via CYP2D6 sequencing make the manual method cumbersome and prone to error, highlighting the need to turn the mental algorithm of manual allele identification into a bioinformatics tool. One such tool, Stargazer, combines SNV calls with structural variation data from next‐generation sequencing to improve the accuracy of diplotype calls and greatly reduces the labor involved.18
In clinical settings, CYP2D6 AS is generally grouped into one of four phenotype classifications: poor metabolizer (PM), intermediate metabolizer (IM), normal metabolizer (NM), and ultrarapid metabolizer (UM). Current CPIC definitions, upon which the majority of findings of this paper are based, group each phenotype by a range of AS: 0, 0.5, 1–2, and> 2, respectively, for PMs, IMs, NMs, and UMs. Recently, CPIC has reached consensus on new standardized CYP2D6 genotype to phenotype translation criteria; major changes include downgrading the score for the CYP2D6*10 allele from 0.5 to 0.25 and redefining AS ranges. Under the new definitions, PM remains an AS = 0, whereas the IM range is expanded to 0.25–1, the NM range is 1.25–2.25, and UMs include scores > 2.25.19, 20
The first major goal of this study was to assess the importance of structural variation in assigning CYP2D6 diplotypes (e.g., the degree to which the correlation between CYP2D6 AS and in vitro CYP2D6 activity is improved when structural variation data are included relative to diplotype assignments made without structural variation data). In light of the recent consensus on CPIC's new CYP2D6 guidelines, an additional goal was to assess whether the revised phenotype groups were more predictive of CYP2D6 activity than the current groups. Due to discovery of novel CYP2D6 variants in our sample set, a final goal was to characterize the effect of rare CYP2D6 variants on activity and describe novel haplotypes. As a general approach, we assessed CYP2D6 activity in a panel of 314 human liver tissue samples using two CYP2D6 probe substrates, dextromethorphan and metoprolol, and assigned CYP2D6 diplotypes to each sample using next‐generation sequencing data from the PGRNseq platform coupled with Stargazer, a CYP2D6 allele‐calling bioinformatics tool.
Methods
Chemicals and reagents
Dextromethorphan, metoprolol tartrate, NADPH, potassium phosphate, EDTA, and acetonitrile were purchased from Sigma‐Aldrich (St. Louis, MO). Dextrorphan‐d3 and Carvedilol‐d3 were purchased from Cerilliant (Round Rock, TX), and C/D/N Isotopes (Pointe‐Claire, Quebec, Canada), respectively.
Human liver samples
Human liver tissue samples (n = 314) were obtained from the University of Washington Human Liver Bank (n = 48) and the St. Jude Liver Resource at St. Jude Children's Research Hospital (n = 266). Institutional review boards at both sites approved the collection and use of these samples for research. All links between archived tissues and donors were destroyed; further details described previously.21 Human liver microsome (HLM) preparation and total protein quantitation were previously described.21
Protein and messenger RNA quantitation
Microsomal CYP2D6 protein content was quantitated using a surrogate peptide‐based liquid chromatography‐tandem mass spectrometrymethod.21, 22 Methods for messenger RNA (mRNA) quantitation were previously described.23
DNA isolation, CYP2D6 sequencing, RNA‐seq analysis, and CYP2D6 allele and AS assignment
Genomic DNA was isolated from human livers as described.21 The next‐generation sequencing platform PGRNseqversion 1.1 was used to identify CYP2D6 genomic variation, including both SNV and structural variation data. PGRNseq captures all CYP2D6 exons as well as 2 kb of upstream and 1 kb of downstream sequence. It also captures the CYP2D7 pseudogene.24 Allele and subsequent star allele diplotype assignments were first made manually using PGRNseq SNV data only. Allele definitions are according to the Pharmacogene Variation Consortium (PharmVar).16, 17 Manual diplotype assignment was performed by cross‐referencing SNVs found in each sample to CYP2D6 allele definitions on PharmVar, taking a general approach described previously.3 The Stargazer algorithm was then used to detect CYP2D6 structural variation from the next‐generation sequencing data and the star allele diplotype assignments determined manually were corrected accordingly.18 Values for AS calculations were assigned to each allele based on CPIC criteria.25, 26, 27, 28, 29, 30
CYP2D6 metabolite formation rate in HLMs
HLM incubations were conducted using a metabolite formation approach to estimate intrinsic clearance, defined as maximum velocity (Vmax) divided by Michaelis–Menten constant (Km).31, 32, 33 Optimization experiments confirmed that substrate concentrations were below Km, incubation times and total HLM protein content were within linear conditions of metabolite formation, and< 10% substrate was depleted over the incubation time (data not shown). Incubations were performed in triplicate using 20 μg of total HLM protein diluted in 100 mM potassium phosphate, pH 7.4 and 1 mM EDTA buffer. HLMs were pre‐incubated with substrate (1.5 μM dextromethorphan or 4 μM metoprolol) for 5 minutes at 37°C. nicotinamide adenine dinucleotide phosphate(final concentration 1 mM) was added to start the reaction and samples were incubated for 20 minutes (metoprolol) and 30 minutes (dextromethorphan) at 37°C, respectively. Reactions were terminated by the addition of ice‐cold acetonitrile.
Metabolite and parent drug quantitation
Quantitation of dextromethorphan, dextrorphan, metoprolol, and α‐hydroxymetoprolol (detailed in Supplementary Methods ) was performed with modifications to published methods on an Agilent Technologies G1956B mass spectrometer coupled to an Agilent 1200 series high‐performance liquid chromatography using a Zorbax SB‐C18 2.1 mm × 150 mm × 5 μm column (Agilent Technologies, Santa Clara, CA).34, 35, 36 The column was maintained at 30°C with a flow rate of 0.3 mL/minutes for dextrorphan, and 35°C with a flow rate of 0.25 mL/minutes for α‐hydroxymetoprolol.
Functional characterization of rare CYP2D6 variants
Rare variants were characterized in vitro using a cytochrome P450 S. cerevisiae model (detailed in Supplementary Methods ).37, 38, 39, 40, 41, 42 Methods include construction of yeast expression plasmids: three controls (wild‐type, inactive, and intermediate) and seven rare coding CYP2D6 variants (P8S, S168A, D198N, P267L, V338M, A449D, and R474Q). Each construct was functionally characterized using both a click chemistry‐compatible ticlopidine 5′‐carboxypropargyl amide probe that has specificity for CYP2D6 activity and a fluorogenic CYP2D6 substrate, Vivid 7‐ethoxymethoxy‐3‐cyanocoumarin.
Confirmation of novel haplotypes
Allele‐specific long‐range PCR and Sanger sequencing were used to determine CYP2D6 haplotypes in two liver bank samples, as previously described.43
Statistical analysis
Samples were included for statistical analysis if both PGRNseq and activity data for at least one probe substrate were available (n = 314). All analyses were performed using RStudio (RStudio, Boston, MA). Multiple linear regression with robust SEs was used to assess the contribution of selected predictors to the variability observed in CYP2D6 activity (dextrorphan and α‐hydroxymetoprolol formation rates) by assessing improvements in goodness of fit (R 2). Donor age and sex were not included as covariates due to a lack of significance in a primary analysis, which is consistent with the literature.44 Although ethnicity has been described to contribute to variation in CYP2D6 activity, we did not include it as a covariate because almost all liver donors were of European descent (95.5%). We also excluded liver disease and concomitant CYP2D6 inhibitors because the data were incomplete for most donors. Liver collection site was included as a covariate due to significant inter‐site differences in mean enzyme activity.
Results
Liver donor demographics
Donor demographics (n = 314) were presented previously (Table S1 ).21, 23 Data on concomitant medications were missing or incomplete for most donors and, thus, are not reported; concomitant use of a strong CYP2D6 inhibitor (fluoxetine) was reported for only one donor.
Correlation between CYP2D6 mRNA and protein content and CYP2D6 activity
Mean CYP2D6 activity, expressed as formation rates of dextrorphan from dextromethorphan and α‐hydroxymetoprolol from metoprolol, varied significantly between liver collection sites, consistent with our collaborators' reports from these samples.21, 23 Therefore, liver collection site was included as a covariate in all analyses. Formation rates of dextrorphan and α‐hydroxymetoprolol were well correlated with one another (R 2 = 0.90). Both dextrorphan and α‐hydroxymetoprolol metabolic formation rates were significantly correlated with CYP2D6 mRNA (Figure 1 a,b; R 2 values of 0.24 and 0.21, respectively) and protein content (Figure 1 c,d; R 2 values of 0.51 and 0.46, respectively).
Figure 1.
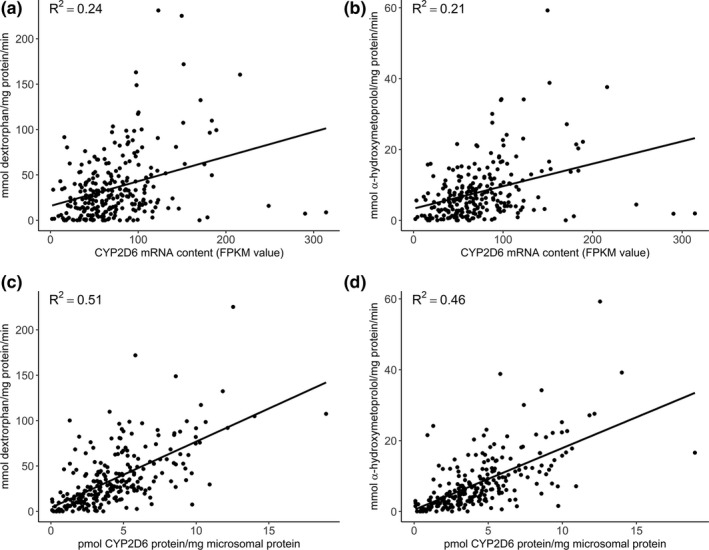
Association among CYP2D6 metabolite formation rate and CYP2D6 messenger RNA (mRNA) and protein content. CYP2D6 metabolite formation rate was correlated with CYP2D6 mRNA content quantitated by RNA‐seq: dextrorphan formation rate (a) and alpha‐hydroxymetoprolol formation rate (b); with CYP2D6 protein content quantitated by liquid‐chromatography tandem mass spectrometry: dextrorphan formation rate (c) and alpha‐hydroxymetoprolol formation rate (d). FPKM, fragments per kilobase of transcript per million mapped reads
CYP2D6 allele and diplotype frequencies and AS re‐assignment following structural variation analysis
We identified 25 different major CYP2D6 haplotypes in the liver bank samples using the Stargazer algorithm from next‐generation sequencing data (Table 1), comprising 73 unique diplotypes (Table S2 ). Nine normal function alleles, with an aggregate frequency of 54.1%, were assigned a value of 1 to calculate the AS. Seven decreased function alleles (aggregate frequency of 18.9%) received a value of 0.5. Seven no function alleles (26.1% of the total alleles) received a value of 0. We identified two increased function alleles, which received a value of 2. Of the 25 unique alleles, 8 were structural variants. Representative examples of structural variants detected by Stargazer are displayed in Figure S1 . Allele frequencies reflect the predominantly European descent of the samples; the three most common alleles were CYP2D6*1 (32.0%), *2(14.3%), and *4 (13.5%), which is consistent with published reports for this population.4
Table 1.
Distribution of CYP2D6 haplotypes (n = 628) identified in liver bank samples
| Allele | n (%) | Activity |
|---|---|---|
| SNVs and indels | ||
| *1 | 201 (32.0%) | Normal |
| *2 | 90 (14.3%) | Normal |
| *33 | 5 (0.8%) | Normal |
| *34 | 1 (0.2%) | Normal |
| *35 | 37 (5.9%) | Normal |
| *39 | 1 (0.2%) | Normal |
| *43 | 2 (0.3%) | Normal |
| *9 | 22 (3.5%) | Decreased |
| *10 | 15 (2.4%) | Decreased |
| *17 | 4 (0.6%) | Decreased |
| *29 | 1 (0.2%) | Decreased |
| *41 | 70 (11.1%) | Decreased |
| *59 | 5 (0.8%) | Decreased |
| *3 | 7 (1.1%) | None |
| *4 | 85 (13.5%) | None |
| *6 | 6 (1.0%) | None |
| *20 | 3 (0.5%) | None |
| Structural variants | ||
| *1 x 2 | 3 (0.5%) | Increased |
| *2 x 2 | 3 (0.5%) | Increased |
| *41 x 2 | 1 (0.2%) | Normal |
| *13 + *2 | 1 (0.2%) | Normal |
| *36 + *10 | 2 (0.3%) | Decreased |
| *4N + *4 | 4 (0.6%) | None |
| *68 + *4 | 35 (5.6%) | None |
| *5 | 24 (3.8%) | None |
SNV, single nucleotide variation.
AS were calculated for diplotypes identified from SNV data alone (17 alleles) and for diplotypes identified with the inclusion of structural variation data (25 alleles) using the current CPIC translational guidelines. Of the samples analyzed, 70 (22.3%) had incorrectly assigned diplotypes based on SNV data alone. Of importance, the inclusion of structural variation changed the AS for 25 of those samples, representing ~ 8% of the investigated liver tissue samples. Figure 2 describes the diplotypes of the 70 samples with structural variation alleles as well as the changes in AS assignments. Both SNV and structural variation data were derived from PGRNseq next‐generation sequencing results. The column on the left contains diplotypes assigned manually using SNV data alone, the corresponding AS, and the number of samples with that diplotype. The column on the right contains diplotypes assigned by Stargazer using the aforementioned SNV data as well as structural variation data, the corresponding AS, and the number of samples with that diplotype. For example, using SNV data alone, 39 samples were assigned a diplotype of CYP2D6*1/*1, which has an AS = 2. When diplotypes were re‐assigned using Stargazer, 8 of the samples were *1/*5, resulting in a decrease in AS to 1. The CYP2D6*5 gene deletion was identified in 25 samples, occurring at an allele frequency of 3.8%. The importance of including CYP2D6*5 is evident as the corrected diplotype calls decreased the AS of 18 samples (5.7%). The remaining seven samples with CYP2D6*5 had another no function allele; thus, AS did not change. A total of seven duplicated alleles were identified (i.e., CYP2D6*1x2, *2x2, and *41x2); these are duplications of functional and decreased function alleles, increasing the AS for samples in which they were identified. We also detected a number of hybrid tandem structures: CYP2D6*4N+*4, *68 + *4, *36 + *10, and *13 + *2 (previously known as *77 + *2). Because CYP2D6*4N, *68, *36, and *13 are nonfunctional, their inclusion did not change the AS assignment.12, 13, 14
Figure 2.
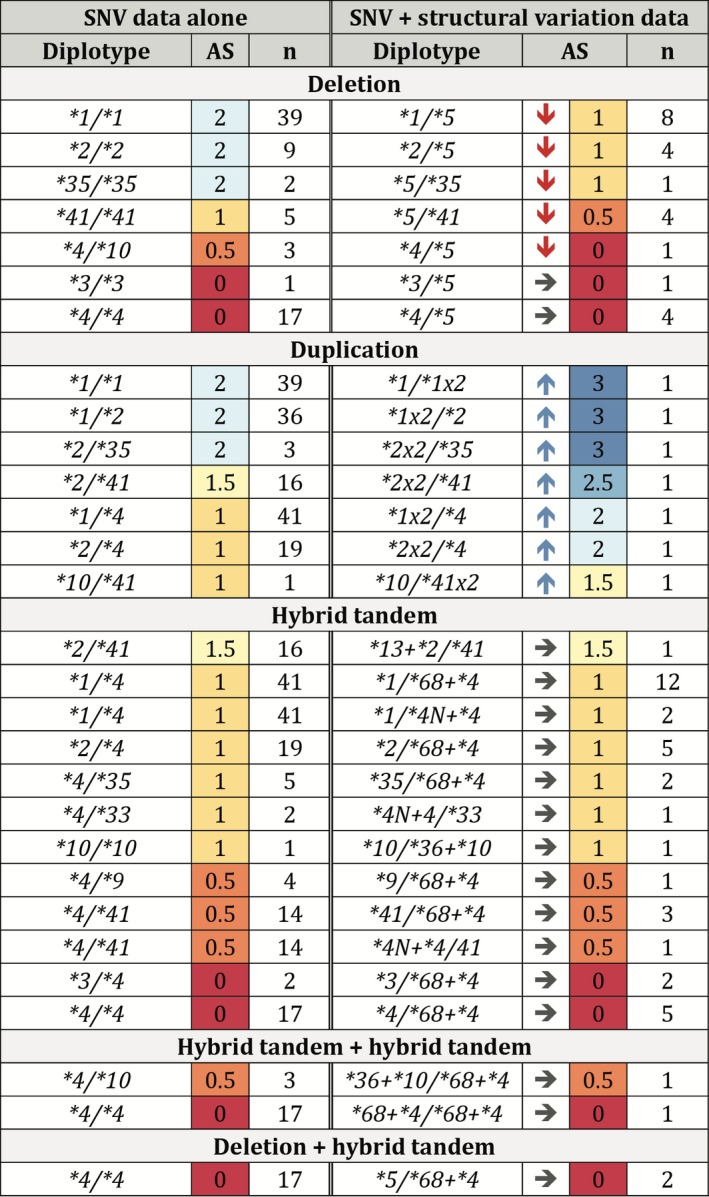
Diplotypes and activity scores assigned with single nucleotide variation (SNV) data alone and with Stargazer structural variation data. Columns on the left show diplotypes and activity scores (AS) assigned using allele calls from SNV data alone. Corrected diplotypes and AS, based on Stargazer allele assignments, are displayed in the columns on the right. AS are color‐coded as follows: 3 (dark blue); 2.5 (medium blue); 2 (light blue); 1.5 (light yellow); 1 (dark yellow); 0.5 (orange); and 0 (red). Arrows indicate the direction of the change in AS assignments with the incorporation of structural data: decrease (↓), increase (↑), and no change (→).
Correlation between CYP2D6 AS and metabolic activity
The inclusion of structural data for CYP2D6 diplotype calling improved the predictive value of the AS assignments for both probe substrates, dextromethorphan and metoprolol (Figure 3 a–d; Table 2). Using SNV data alone, AS predicted 0.36 and 0.31 of the variability in CYP2D6 activity for dextromethorphan and metoprolol, respectively. With structural variation data included, these coefficients increased to 0.40 and 0.34, respectively.
Figure 3.
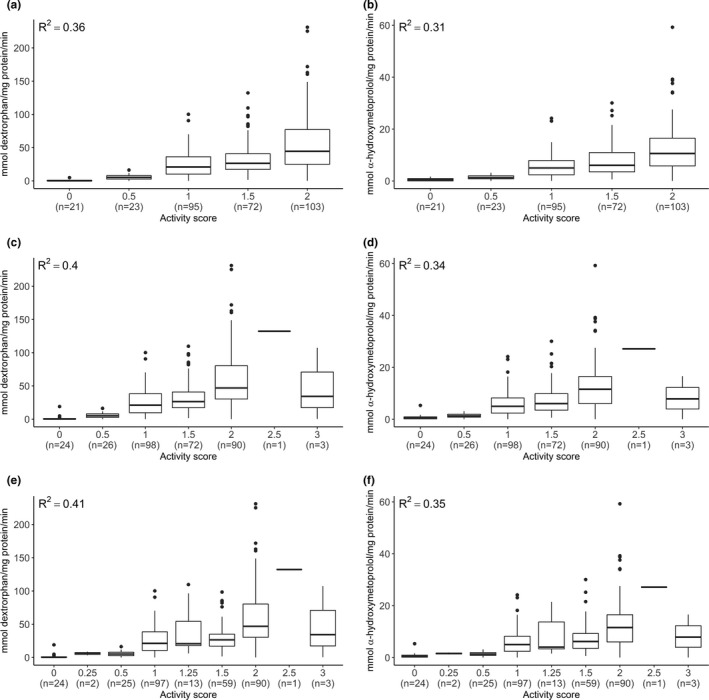
Association between CYP2D6 metabolite formation rate and CYP2D6 activity score (AS). Dextrorphan formation rate by AS assigned with single nucleotide variation (SNV) data alone (a), with Stargazer (c), and with revised Clinical Pharmacogenetics Implementation Consortium (CPIC) definitions (e). Alpha‐hydroxymetoprolol formation rate by activity score assigned with SNV data alone (b), with Stargazer (d), and with revised CPIC definitions (f). Boxes represent interquartile range with interior line representing the median. Error bars represent 1.5× the interquartile range. Number of samples in each AS category is given in parentheses.
Table 2.
Multiple linear regression: comparison of association between CYP2D6 activity and AS assigned using SNV data only or including structural variation data from Stargazer
| Dextromethorphan | Metoprolol | |||||
|---|---|---|---|---|---|---|
| β | SE | P value | β | SE | P value | |
| Prediction of CYP2D6 activity using AS assigned using SNV data only | ||||||
| (Intercept)a | −3.71 | 2.41 | 0.12 | −0.15 | 0.40 | 0.71 |
| AS = 0.5 | 6.51 | 2.90 | 0.03 | 1.13 | 0.49 | 0.02 |
| AS = 1 | 22.82 | 3.34 | 4.54 × 10 −11 | 4.61 | 0.66 | 1.38 × 10 −11 |
| AS = 1.5 | 32.21 | 3.60 | 3.61 × 10 −17 | 7.26 | 0.85 | 7.08 × 10 −16 |
| AS = 2 | 52.99 | 4.49 | 8.35 × 10 −27 | 11.59 | 0.99 | 4.36 × 10 −26 |
| Site: UW | 30.63 | 6.02 | 6.29 × 10 −7 | 5.17 | 1.31 | 1.05 × 10 −4 |
| Degrees of freedom | 308 | 289 | ||||
| R 2/adj. R 2 | 0.36/0.35 | 0.31/0.30 | ||||
| Prediction of CYP2D6 activity using activity scores assigned using Stargazer, based on both SNV and structural variation data | ||||||
| (Intercept)a | −2.08 | 2.18 | 0.34 | 0.19 | 0.40 | 0.64 |
| AS = 0.5 | 3.17 | 2.98 | 0.29 | 0.43 | 0.53 | 0.42 |
| AS = 1 | 23.23 | 3.17 | 2.24 × 10 −12 | 4.77 | 0.66 | 3.64 × 10 −12 |
| AS = 1.5 | 29.75 | 3.33 | 3.76 × 10 −17 | 6.73 | 0.83 | 1.07 × 10 −14 |
| AS = 2 | 54.98 | 4.55 | 8.86 × 10 −28 | 11.99 | 1.05 | 2.91 × 10 −25 |
| AS = 2.5 | 105.89 | 5.72 | 7.32 × 10 −52 | 22.24 | 1.20 | 5.90 × 10 −51 |
| AS = 3 | 49.46 | 26.20 | 0.06 | 8.00 | 3.98 | 0.04 |
| Site: UW | 28.52 | 5.76 | 1.24 × 10 −6 | 4.73 | 1.27 | 2.47 × 10 −4 |
| Degrees of freedom | 306 | 295 | ||||
| R 2/adj. R 2 | 0.40/0.39 | 0.34/0.33 | ||||
AS, activity score; β, β‐coefficient; UW, University of Washington; SNV, single nucleotide variation. Bold values indicate a P < 0.05.
AS = 0 is incorporated into the intercept.
Comparison of the current and revised CPIC guidelines for CYP2D6 genotype to phenotype translation
The revised CPIC system recommends assigning a lower value (0.25 instead of 0.5) to the CYP2D6*10 allele due to its severely decreased function.19 This change affected 16 liver bank samples: AS was decreased by 0.25 in 15 samples and 0.5 in 1 sample. The R 2 for the correlation between AS and CYP2D6 activity increased only slightly from 0.40 to 0.41 for dextrorphan formation rate (Figure 3 c,e) and from 0.34 to 0.35 for α‐hydroxymetoprolol (Figure 3 d,f). Using the revised method, the number of PMs and UMs did not change relative to current CPIC definitions, but 98 NMs were reclassified as IMs. The R 2 increased from 0.27 to 0.31 for dextrorphan formation rate (Figure 4 a,c) and 0.21 to 0.26 for α‐hydroxymetoprolol (Figure 4 b,d).
Figure 4.
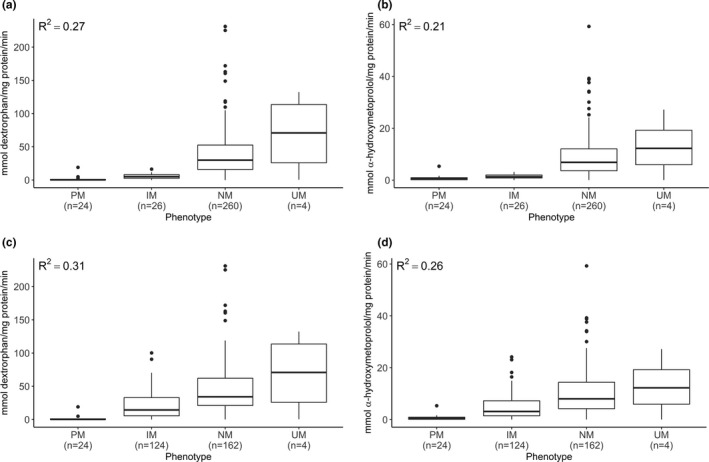
Comparison between current and revised Clinical Pharmacogenetics Implementation Consortium (CPIC) genotype to phenotype translation tables for dextromethorphan and metoprolol. Dextrorphan formation rate by phenotype (IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer) assigned with current (a) and revised (c) CPIC definitions; alpha‐hydroxymetoprolol formation rate by phenotype assigned with current (b) and revised (d) CPIC definitions. Boxes represent interquartile range with interior line representing the median. Error bars represent 1.5× the interquartile range. Number of samples in each phenotype category is given in parentheses.
Functional characterization of rare CYP2D6 variants
A total of 180 SNVs were identified by PGRNseq analysis in the 314 livers tested (Table S3 ). To explore the potential contribution of rare coding SNVs, we selected seven nonsynonymous SNVs with Combined Annotation Dependent Depletion scores of 0–32 for functional characterization. These SNVs were expressed individually in a CYP2D6*1 construct, not in the context of the haplotype in which they were identified. Five exhibited metabolic activity comparable to the reference (CYP2D6.1) enzyme (100%), whereas one (rs79392742; A449D) displayed decreased activity (44.4 ± 6.9%) comparable with the decreased activity P34S control (35.4 ± 4.7%; Figure 5). This finding was confirmed using Vivid7‐ethoxymethoxy‐3‐cyanocoumarin, a standard fluorogenic CYP2D6 substrate (Figure S2 ) and consistent with the SWISS‐MODEL,45 which predicted that A449D (Combined Annotation Dependent Depletion score of 32) could interfere with heme binding (Figure S3 ). The liver carrying A449D was genotyped as CYP2D6*1/*4. Subsequent analysis utilizing allele‐specific long‐range PCR revealed that A449D is located on the CYP2D6*4 allele, constituting a novel suballele, which was designated CYP2D6*4.028 by PharmVar.15, 43 Allele‐specific analysis identified that another nonsynonymous SNV (rs1058172; R365H) was found on a novel haplotype and designated CYP2D6*139 by PharmVar.15, 43 Another variant (rs141756339; R474Q), showed increased activity compared with CYP2D6.1 protein (127 ± 8.1%; Figure 5), which was also confirmed using Vivid7‐ethoxymethoxy‐3‐cyanocoumarin (Figure S2 ). The variant was found in a sample that was initially called CYP2D6*2/*41 by Stargazer; Sanger sequencing of an allele‐specific long‐range PCR product identified R474Q on the CYP2D6*41 haplotype.43 This allele received a novel star designation by PharmVar: CYP2D6*138.15, 43
Figure 5.
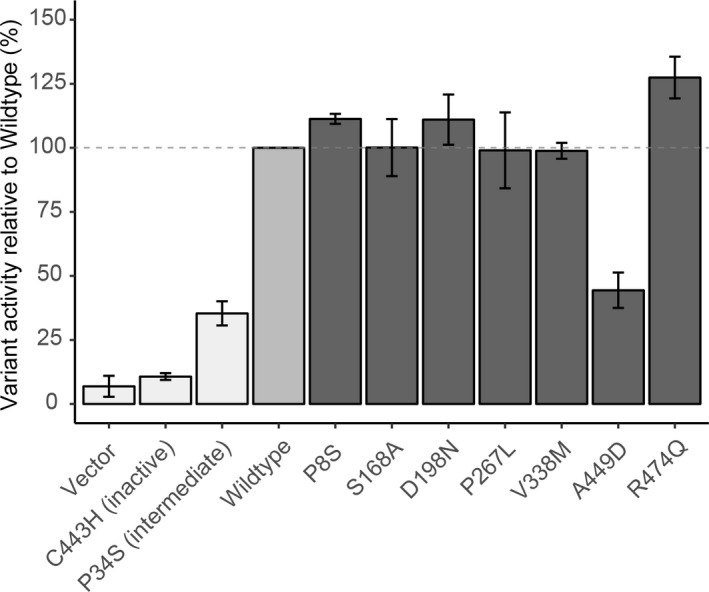
Functional characterization of rare CYP2D6 coding variants with CYP2D6 probe tic‐ABP1P. Each CYP2D6 variant was induced in an isogenic yeast strain and function was characterized with a tic‐ABP1P CYP2D6 probe. Fluorescence was normalized to that of CYP2D6 wildtype (medium grey; horizontal dashed line represents 100% activity). Control strains (light grey): empty vector, inactive variant (C443H), and decreased function variant (P34S). Rare coding variant strains (dark gray): P8S, S168A, D198N, P267L, V338M, A449D, and R474Q. Error bars indicate SEs from at least two independent replicates.
Discussion
To accurately assign CYP2D6 diplotype and subsequent translation into phenotype, it is essential to consider both SNVs and structural variation. Pharmacogenetic tests designed to detect a limited number of alleles and nonspecific copy number variation may miss rare, clinically relevant variants that are present in an individual, but not interrogated, or result in ambiguous diplotypes that yield a range of possible activity scores. In this study, the PGRNseq next‐generation sequencing platform was coupled with the Stargazer interpretive platform to include all SNVs and structural variation to predictmetabolizer status. We found that the inclusion of structural variation is essential to accurately assign CYP2D6 diplotype and AS, improving the prediction of CYP2D6 activity compared with the assignment based on SNV data alone. It follows that the identification of gene deletions, gene duplications/multiplications, and hybrid alleles maximizes the clinical utility of CYP2D6 pharmacogenetic tests.
When diplotypes were assigned based on SNV data alone, AS explained 36% and 31% of the variation in CYP2D6 activity for dextromethorphan and metoprolol, respectively. When diplotypes were reassigned based on the Stargazer algorithm that utilizes both SNV and structural variation data, these numbers increased to ~ 40% and 34%, respectively. Samples with incorrect AS assignments contained alleles that contribute to “extreme” phenotypes: the CYP2D6*5 allele harboring a gene deletion or alleles carrying two or more normal or decreased function gene copies including CYP2D6*1xN, 2xN, and 41xN. Of the six samples with gene duplications, three had diplotypes that would have been assigned the same AS regardless of which allele was duplicated (*1/*1x2, *2/*1x2, and *35/*2x2), two had diplotypes that would have been assigned an AS = 1 rather than 2 if the other allele had been duplicated (*4/*1x2 and *4/*2x2), and one would have been assigned an AS = 2 rather than 2.5 (*41/*2x2). If our samples had been tested on a panel that only reported a nonspecific duplication signal, it would have been impossible to assign a definitive diplotype to half of the samples with duplications. As a result, AS would be calculated as a range of possible values, which may not be particularly informative and confusing to prescribers as well as patients. The CYP2D6*4N, *36, *68, and *13 alleles harboring CYP2D7 sequences are nonfunctional, and, thus, did not affect AS assignments when in tandem arrangements with alleles identified via SNV‐only analysis. A CYP2D6*68 + *4 allele for example, receives a value of 0 regardless whether it is called as CYP2D6*68 + *4 or as CYP2D6*4. They cannot simply be ignored, however, as their presence can interfere with PCR‐based genotyping assays, resulting in incorrect phenotype predictions. This is exemplified by a previous study that found that 4 of 33 (12%) duplications detected in a European cohort were actually hybrid tandem alleles.14
Only one diplotype was irreconcilable between the two assignment methods. Using SNV data alone, the sample seemed to be homozygous for 100C>T and heterozygous for 1847G>A (positions according to the CYP2D6 RefSeq NG_008376.3) and was called CYP2D6*4/*10. Stargazer analysis, however, determined that this sample had a copy number of 1 and called CYP2D6*4/*5 based on the presence of 1847G>A. The heterozygous variant call, which showed a significant allele imbalance, was most likely due to a mapping error and interpreted as hemizygosity by Stargazer. The sample's CYP2D6 activity (< 0.5 mmol/mg protein/minutes for both probe drugs) supports a CYP2D6*4/*5 call.
The metabolite formation rates for samples with an AS = 3 were lower than expected and the small number of samples (n = 3) with functional gene duplications makes interpretation difficult. One sample in particular, genotyped as a CYP2D6*1/*1x2, had low CYP2D6 activity for both probe drugs. It is possible that this is due to inferior liver tissue and/or HLM quality, a hypothesis supported by the observation that CYP3A4 activity, determined using the same batch of HLMs (unpublished data), was also quite low. In general, variation not accounted for in this study may be from phenoconversion caused by donors' concomitant medications, variation in genes not considered in this investigation, or distant regulatory SNVs, such as rs5758550, which has been described to impact CYP2D6 expression.46, 47
CPIC's CYP2D6 guidelines provide recommendations based on metabolizer phenotypes (e.g., PM, IM, NM, and UM) that are defined based on AS. Due to interlaboratory discrepancies in criteria used to assign genotype to phenotype, a CPIC‐led CYP2D6 expert working group created a new consensus‐based genotype‐phenotype translation guideline.19 One notable change is downgrading CYP2D6*10 (to better reflect the level of decreased function) to a value of 0.25 instead of 0.5 for AS calculation. We found that the amount of variability explained by AS did not change after accounting for the CYP2D6*10 downgrade, which may be explained by the low frequency of this allele in our sample set. In contrast, phenotypes assigned using the new translation method explained more variability in CYP2D6 activity than the current system. The latter was driven by the reclassification of 98 samples currently defined as NMs now defined as IMs in the new system. A downside to the new translation table is that it adds uncertainty when nonspecific duplications are detected in conjunction with a diplotype, such as CYP2D6*1/*4. Under the current system, an assignment of NM would be made regardless of which allele is duplicated as NM encompasses an AS range of 1–2. If the new system is used, the patient's genotype would be either IM or NM depending on which allele is duplicated. Hence, when using the new CPIC recommended translation method, it is even more important to interrogate structural variants in order to avoid ambiguous phenotype assignments. To the best of our knowledge, this is the first study comparing the current and new CPIC translation systems and demonstrates that phenotype determined with the new system is a better predictor of CYP2D6 activity.
Within the region of the CYP2D6 locus included in PharmVar, the liver bank samples contained 44 SNVs that were not included in any defined CYP2D6 haplotype, 7 of which have not yet been assigned anrs number, and 6 that are missense SNVs that could potentially alter function. We evaluated the catalytic activity of seven rare nonsynonymous variants and showed that A449D causes decreased activity similar to P34S (100C>T), a diagnostic, or core, SNV for a number of decreased and nonfunctional alleles, including CYP2D6*10. In contrast, R474Q exhibited increased activity in comparison to the reference protein. These nonsynonymous variants were evaluated for function individually in the reference background, and not within the context of their haplotype. Subsequent sequencing of allele‐specific PCR products revealed that A449D is part of a novel nonfunctional CYP2D6*4 haplotype, CYP2D6*4.028. Unless this amino acid change is identified on a functional allele, it is not clinically relevant. The R474Q amino acid change was found on an allele harboring SNVs otherwise defining the decreased function CYP2D6*41 allele. Because R474Q increases function compared with thereference protein, this amino acid change may offset the reduction in activity observed for CYP2D6*41. The activity of the tissue sample carrying the variant, however, was consistent with its initial AS assignment of 1.5.
In summary, for a gene locus as complex as CYP2D6, analysis of SNVs alone is clearly not sufficient for making accurate diplotype calls and phenotype predictions. Many of the currently available CYP2D6 pharmacogenetic tests are designed to detect the most common alleles and interrogate whether gene duplication genes are present, but do not necessarily determine copy number or specify which allele is duplicated; furthermore, few laboratories test for hybrid genes. The combination of Stargazer and next‐generation sequencing offers a solution to many of the limitations of existing tests. First, it can detect rare alleles typically not included on genotyping panels. Second, the approach identifies the nature of the duplicated gene copy rather than detecting a nonspecific, and potentially ambiguous, duplication signal. Third, Stargazer can identify hybrid gene structures, including those in tandem arrangements with nonhybrid alleles, solving the problem of incorrect duplication calls or miscalls due to the interference of these complex structures. Our findings also highlight the value of gene resequencing when predicting phenotype. At the population level, the consideration of rare variation may enhance clinical outcomes for precision pharmacotherapy. If we are to strive for precision in the therapeutic treatment of disease, a feasible first step, as described in this study, is to consider the full complement of structural variants, SNVs, and indels for CYP2D6 diplotype assignment.
Funding
This work was supported by the Northwest Alaska‐Pharmacogenomics Research Network (NWA‐PGRN; U01GM092676 and P01GM116691); the Center for Exposures, Diseases, Genomics, and Environment (EDGE; P30ES007033); NIGMS R24GM115277 and R01GM132162; and the support of A.G. for the Pharmacogene Variation Consortium (PharmVar, NIGMS R24GM12393). M.J.D. is a Senior Fellow in the Genetic Networks program at the Canadian Institute for Advanced Research and is supported in part by a Faculty Scholars grant from the Howard Hughes Medical Institute. D.M.F. is a CIFAR Azrieli Global Scholar.
Conflict of Interest
The authors declare no competing interests for this work.
Author Contributions
R.D., S.B.L., A.G., K.E.T., and E.L.W. wrote the manuscript. R.D., S.B.L., D.D.S., D.A.N., K.E.T., and E.L.W. designed the research. R.D., S.B.L., L.H.W., M.F., and A.G. performed the research. R.D., S.B.L., A.G., T.A.T., D.A.N., K.E.T., and E.L.W. analyzed the data. K.C., B.P., B.R.P., M.G.M., M.J.D., D.M.F., A.E.R., and E.S. contributed new reagents/analytical tools.
Supporting information
Table S1.
Table S2.
Table S3.
Figure S1.
Figure S2.
Figure S3.
Supplementary Methods.
Acknowledgments
We would like to acknowledge the technical support of Mrs. Wendy Wang, Children's Mercy Kansas City, for allele‐specific Sanger sequencing to characterize the novel CYP2D6 haplotypes. The authors also thank Linda Risler at the Clinical Pharmacokinetics Laboratory at the University of Washington for training in HLM incubations.
References
- 1. Gardiner, S.J. & Begg, E.J. Pharmacogenetics, drug‐metabolizing enzymes, and clinical practice. Pharmacol. Rev. 58, 521–590 (2006). [DOI] [PubMed] [Google Scholar]
- 2. Lauschke, V.M. & Ingelman‐Sundberg, M. Prediction of drug response and adverse drug reactions: from twin studies to next generation sequencing. Eur. J. Pharm. Sci. 130, 65–77 (2019). [DOI] [PubMed] [Google Scholar]
- 3. Gaedigk, A. Complexities of CYP2D6 gene analysis and interpretation. Int. Rev. Psychiatry 25, 534–553 (2013). [DOI] [PubMed] [Google Scholar]
- 4. Gaedigk, A. , Sangkuhl, K. , Whirl‐Carrillo, M. , Klein, T. & Leeder, J.S. Prediction of CYP2D6 phenotype from genotype across world populations. Genet. Med. 19, 69–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nofziger, C. & Paulmichl, M. Accurately genotyping CYP2D6: not for the faint of heart. Pharmacogenomics 19, 999–1002 (2018). [DOI] [PubMed] [Google Scholar]
- 6. Hicks, J.K. , Swen, J.J. & Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr. Drug Metab. 15, 218–232 (2014). [DOI] [PubMed] [Google Scholar]
- 7. Yang, Y. , Botton, M.R. , Scott, E.R. & Scott, S.A. Sequencing the CYP2D6 gene: from variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics 18, 673–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nofziger, C. et al PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 107, 154–170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fohner, A. et al Pharmacogenetics in American Indian populations: analysis of CYP2D6, CYP3A4, CYP3A5, and CYP2C9 in the Confederated Salish and Kootenai Tribes. Pharmacogenet. Genomics 23, 403–414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gene‐specific Information Tables for CYP2D6 . PharmGKB. <https://www.pharmgkb.org/page/cyp2d6RefMaterials>. Accessed January 29,2019).
- 11. Jarvis, J.P. , Peter, A.P. & Shaman, J.A. Consequences of CYP2D6 copy‐number variation for pharmacogenomics in psychiatry. Front. Psychiatry 10, 432 (2019). 10.3389/fpsyt.2019.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaedigk, A. et al Identification of novel CYP2D7‐2D6 hybrids: non‐functional and functional variants. Front. Pharmacol. 1, 121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Black, J.L. , Walker, D.L. , O'Kane, D.J. & Harmandayan, M. Frequency of undetected CYP2D6 hybrid genes in clinical samples: impact on phenotype prediction. Drug Metab. Dispos. Biol. Fate Chem. 40, 111–119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaedigk, A. et al CYP2D7‐2D6 hybrid tandems: identification of novel CYP2D6 duplication arrangements and implications for phenotype prediction. Pharmacogenomics 11, 43–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. PharmVar . <https://www.pharmvar.org/gene/CYP2D6>. Accessed January 16, 2019.
- 16. Gaedigk, A. et al The Pharmacogene Variation (PharmVar) consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharmacol. Ther. 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaedigk, A. et al The evolution of Pharmvar. Clin. Pharmacol. Ther. 105, 29–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee, S. et al Stargazer: a software tool for calling star alleles from next‐generation sequencing data using CYP2D6 as a model. Genet. Med. 21, 361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caudle, K. E. et al Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from CPIC and DPWG. Clin. Transl. Sci. 13, 116–124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. CYP2D6 Genotype to Phenotype Standardization Project . <https://cpicpgx.org/resources/cyp2d6-genotype-to-phenotype-standardization-project/>. Accessed July 29,2019.
- 21. Shirasaka, Y. et al Interindividual variability of CYP2C19‐catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J. 16, 375–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prasad, B. & Unadkat, J.D. Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J. 16, 634–648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanner, J.‐A. et al Predictors of variation in CYP2A6 mRNA, protein, and enzyme activity in a human liver bank: influence of genetic and nongenetic factors. J. Pharmacol. Exp. Ther. 360, 129–139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gordon, A.S. et al PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet. Genomics 26, 161–168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crews, K.R. et al Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95, 376–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hicks, J.K. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hicks, J.K. et al Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bell, G.C. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin. Pharmacol. Ther. 102, 213–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goetz, M.P. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy. Clin. Pharmacol. Ther. 103, 770–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown, J. T. et al Clinical Pharmacogenetics Implementation Consortium guideline for cytochrome P450 (CYP2D6) genotype and atomoxetine therapy. Clin. Pharmacol. Ther. 106, 94–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Houston, J.B. , Kenworthy, K.E. & Galetin, A. Typical and atypical enzyme kinetics In Drug metabolizing Enzymes: Cytochrome P450 and Other Enzymes in Drug Discovery and Development (eds. Lee J. S., Obach R. S., Fisher M. B.) 211–254 (CRC Press, Boca Raton, FL, 2003). [Google Scholar]
- 32. Wienkers, L.C. & Stevens, J. C. Cytochrome P450 reaction phenotyping In Drug Metabolizing Enzymes: Cytochrome P450 and Other Enzymes in Drug Discovery and Development (eds. Lee J.S., Obach R.S., Fisher M.B.) 255–310 (CRC Press, Boca Raton, FL, 2003). [Google Scholar]
- 33. Venkatakrishnan, K. , von Moltke, L.L. , Obach, R.S. & Greenblatt, D.J. Drug metabolism and drug interactions: application and clinical value of in vitro models. Curr. Drug Metab. 4, 423–459 (2003). [DOI] [PubMed] [Google Scholar]
- 34. Zhu, B. et al Assessment of cytochrome P450 activity by a five‐drug cocktail approach. Clin. Pharmacol. Ther. 70, 455–461 (2001). [DOI] [PubMed] [Google Scholar]
- 35. Ryu, R.J. et al Pharmacokinetics of metoprolol during pregnancy and lactation. J. Clin. Pharmacol. 56, 581–589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McConnachie, L.A. , Phillips, B. , Bajpai, M. , Shen, D.D. & Ho, R.J.Y. Only truncated, not complete cytochrome p450 2D6 RNA transcript and no detectable enzyme activity are expressed in human lymphocytes. Drug Metab. Dispos. Biol. Fate Chem. 31, 1103–1107 (2003). [DOI] [PubMed] [Google Scholar]
- 37. McDonald, M.G. et al Expression and functional characterization of breast cancer‐associated cytochrome P450 4Z1 in Saccharomyces cerevisiae. Drug Metab. Dispos. Biol. Fate Chem. 45, 1364–1371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gietz, R.D. & Schiestl, R.H. High‐efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007). [DOI] [PubMed] [Google Scholar]
- 39. Wright, A.T. & Cravatt, B.F. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem. Biol. 14, 1043–1051 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vatsis, K.P. , Peng, H.‐M. & Coon, M.J. Abolition of oxygenase function, retention of NADPH oxidase activity, and emergence of peroxidase activity upon replacement of the axial cysteine‐436 ligand by histidine in cytochrome P450 2B4. Arch. Biochem. Biophys. 434, 128–138 (2005). [DOI] [PubMed] [Google Scholar]
- 41. Yoshioka, S. , Takahashi, S. , Hori, H. , Ishimori, K. & Morishima, I. Proximal cysteine residue is essential for the enzymatic activities of cytochrome P450cam. Eur. J. Biochem. 268, 252–259 (2001). [DOI] [PubMed] [Google Scholar]
- 42. Auclair, K. , Moënne‐Loccoz, P. & Ortiz de Montellano, P.R. Roles of the proximal heme thiolate ligand in cytochrome p450(cam). J. Am. Chem. Soc. 123, 4877–4885 (2001). [DOI] [PubMed] [Google Scholar]
- 43. Gaedigk, A. , Riffel, A.K. & Leeder, J.S. CYP2D6 haplotype determination using long range allele‐specific amplification. J. Mol. Diagn. 17, 740–748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bebia, Z. et al Bioequivalence revisited: influence of age and sex on CYP enzymes. Clin. Pharmacol. Ther. 76, 618–627 (2004). [DOI] [PubMed] [Google Scholar]
- 45. Schwede, T. , Kopp, J. , Guex, N. & Peitsch, M.C. SWISS‐MODEL: an automated protein homology‐modeling server. Nucleic Acids Res. 31, 3381–3385 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shah, R.R. & Smith, R.L. Addressing phenoconversion: the Achilles' heel of personalized medicine. Br. J. Clin. Pharmacol. 79, 222–240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang, D. , Papp, A.C. & Sun, X. Functional characterization of CYP2D6 enhancer polymorphisms. Hum. Mol. Genet. 24, 1556–1562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Table S2.
Table S3.
Figure S1.
Figure S2.
Figure S3.
Supplementary Methods.


