While the GAS M protein has been the leading vaccine target for decades, the bacteria encode many other virulence factors of interest for vaccine development. In this work, we show that emm-like genes are encoded in a remarkable majority of GAS genomes and expressed at a level similar to that for the emm gene. In collaboration with the U.S. Centers for Disease Control, we developed molecular definitions of the different emm and emm-like gene families. This clarification should abrogate mistyping of strains, especially in the area of whole-genome typing. We have also updated the emm-typing collection by removing emm-like gene sequences and provided in-depth analysis of Mrp and Enn protein sequence structure and diversity.
KEYWORDS: Streptococcus pyogenes, Mga regulon, M-like proteins, global GAS diversity
ABSTRACT
The core Mga (multiple gene activator) regulon of group A Streptococcus (GAS) contains genes encoding proteins involved in adhesion and immune evasion. While all GAS genomes contain genes for Mga and C5a peptidase, the intervening genes encoding M and M-like proteins vary between strains. The genetic make-up of the Mga regulon of GAS was characterized by utilizing a collection of 1,688 GAS genomes that are representative of the global GAS population. Sequence variations were examined with multiple alignments, and the expression of all core Mga regulon genes was examined by quantitative reverse transcription-PCR in a representative strain collection. In 85.2% of the sampled genomes, the Mga locus contained genes encoding Mga, Mrp, M, Enn, and C5a peptidase proteins. These isolates account for 53% of global infections. Only 9.1% of genomes did not contain either an mrp or an enn gene. The pairwise identity within Enn (68.6%) and Mrp (83.2%) protein sequences was higher than within M proteins (44.7%). Gene expression varied between strains tested, but high expression was recorded for all genes in at least one strain. Previous nomenclature issues were clarified with molecular gene definitions. Our findings support a shift in focus in the GAS research field to further consider the role of Mrp and Enn in virulence and vaccine development.
IMPORTANCE While the GAS M protein has been the leading vaccine target for decades, the bacteria encode many other virulence factors of interest for vaccine development. In this work, we show that emm-like genes are encoded in a remarkable majority of GAS genomes and expressed at a level similar to that for the emm gene. In collaboration with the U.S. Centers for Disease Control, we developed molecular definitions of the different emm and emm-like gene families. This clarification should abrogate mistyping of strains, especially in the area of whole-genome typing. We have also updated the emm-typing collection by removing emm-like gene sequences and provided in-depth analysis of Mrp and Enn protein sequence structure and diversity.
INTRODUCTION
Group A Streptococcus (GAS) is a human-specific bacterial pathogen responsible for a range of different conditions affecting different tissue types, including pharyngeal epithelium, keratinocytes, and deeper tissues during invasive diseases (1). GAS infection also leads to autoimmune sequelae, such as rheumatic heart disease, which is responsible for around 320,000 of the more than 500,000 deaths worldwide each year attributed to GAS (2).
GAS produce many virulence factors to aid in establishing and propagating human infection. The GAS M protein is an important and well-characterized virulence factor that is crucial for adhesion to primary tissue sites, invasion of nonimmune cells, and evasion of the host immune system (3). The gene encoding M protein, emm, is found in the Mga (multiple gene activator) regulon. Each GAS strain carries a single variant of the emm gene, whose sequence of the 5′ 180 nucleotides forms the basis for the emm-typing scheme (4, 5). emm typing has differentiated more than 220 emm types to date (6). A convenient feature of emm genes is that their signal sequence encoding regions universally contain a conserved 19-bp sequence (7). This feature has greatly simplified the assignment of emm types from short-read genomic sequences since they are invariably very closely linked to the upstream 19-bp primer 1 sequence (8). Genomic sequencing has also revealed that certain historically established emm types have been assigned to sequences located in emm-like genes due to the annealing of emm-typing primers to emm-like genes (9). A more recent classification, called the emm-cluster-typing scheme, groups strains into 48 clusters based on full M protein sequence homology and functional properties (10–12).
The core Mga regulon spans from the ubiquitous genes for Mga (mga), a transcriptional regulator, to the C5a peptidase (scpA) (13). Other genes that can be present in the Mga regulon include genes encoding the M-like proteins (emm-like genes), Mrp (mrp), and Enn (enn). In some genomes the locus also encodes protein H (sph), a surface protein involved in immune evasion (14); streptococcal inhibitor of complement (SIC; sic), a secreted virulence factor also involved in immune evasion (15); and proteins closely related to SIC (CRS; crs) and distantly related to SIC (DRS; drs) (16). Depending on the emm-like gene content of this locus, strains are classified into five emm patterns (A to E) (17).
As well as regulating Mga regulon gene expression, the Mga protein directly affects transcription of genes involved in the early stages of infection and is chiefly active during the exponential growth phase. Mga indirectly affects expression of over 10% of the GAS genome (18, 19). Limited evidence suggests that mrp and enn genes are expressed between 4- and 32-fold less than the emm gene in the strains analyzed (20, 21).
M-like proteins are fibrillar coiled-coil proteins that extend from the surface of the bacteria and share structural characteristics similar to M proteins (22). The virulence potential of these proteins is relatively unclear, although they have been shown to share binding properties with M proteins (22). The vaccine potential of Mrp has been recently investigated, since antibodies against Mrp have been shown to elicit protection in animal models of infection (23) and increased the bactericidal activity of anti-M antisera (24).
In this study, we carefully describe the Mga core regulon of GAS based on a genetically diverse worldwide study of 2,083 GAS genomes (25). We applied particular emphasis to the genetic description of Mrp and Enn, since these two proteins remain poorly characterized to date. We also sought to address the mislabeling of certain emm types by molecular clarification of gene families.
(This information was presented in part at the 19th Lancefield International Symposium for Streptococci and Streptococcal Diseases in Fiji in 2017 and at the workshop of f-TALES: Big Data in Life Sciences and Biomedicine in Belgium in 2017.)
RESULTS
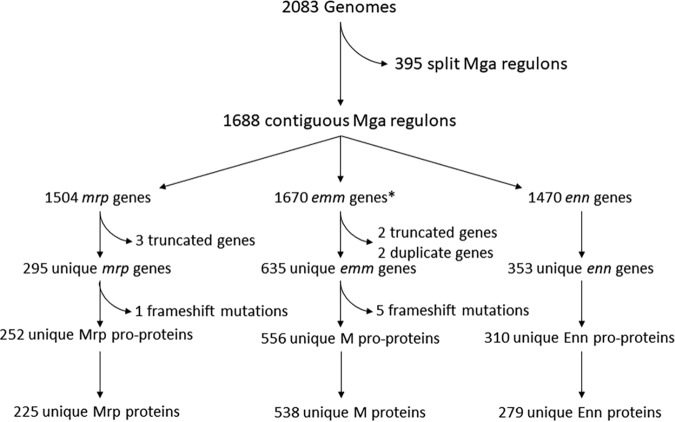
The Mga regulon was located in a single contig in 1,688 genomes belonging to 130 different emm types, 39 emm clusters, and 262 phylogroups from the 2,083 global genome database (Fig. 1) (25).
FIG 1.
Details of the genome and gene collections. Details on the process by which the genomes and genes available were selected for further analysis. The final collection of alleles provides the best representation of global diversity of the gene families available, while avoiding the possibility of confounding by sequence ambiguities. *, a total of 19 genomes had substantial sequence ambiguities in the emm gene domain but nevertheless contained emm-typing sequences. These genes were excluded from gene family analyses, but genomes were included in Mga composition analyses.
Defining mrp, enn, and emm.
mga-, mrp-, emm-, enn-, and scpA-specific oligonucleotide probes were designed to facilitate identification of gene families (Table 1). The mrp gene was defined as being an open reading frame (ORF) downstream of the mga gene, upstream of the emm gene, and containing the mrp-specific probe. The enn gene was defined as being an ORF downstream of the emm gene, upstream of the scpA gene, and containing the enn probe sequence. The emm gene was defined as containing either the emm probe 1 at the 5′ end or the emm probe 2 at the 3′ end of the gene. Chimeric emm genes were defined as containing the emm probe 1 at the 5′ end and the enn probe at the 3′ end of the gene. Sph genes were observed as large ORFs downstream of emm genes that did not contain either the emm or the enn probe and were identified by BLAST search.
TABLE 1.
Nucleotide probes for in silico identification of Mga regulon genes
| Gene | Flexibilitya (no. of nt) | Sequence | Probe in unique alleles (%) |
|---|---|---|---|
| mga | 3 | GAGATTGAAAAACAGTACGATGTTATCGTGACAGATGTTATGGT | 386/390 (99.0) |
| mrp | 1 | AACCAAGAAAAAGAAAAGTTAGAAGC | 295/295 (100) |
| emm (3′) | 1 | AACAAAGAGCTTGAAGAA | 623/635 (98.1) |
| emm (5′) | 0 | TATTSGCTTAGAAAATTAA | 624/635 (98.3)b |
| enn | 1 | TCTGAGTTAACRCAAGCRAARRYTCAACTYKY | 350/352 (99.4) |
| scpA | 1 | GAAGTAACAGTAACAGTTCACAACAAATCTGATAAACCTCAAGAGTTGTATTA | 550/553 (99.5) |
The flexibility number refers to the number of mismatched nucleotide (nt) bases allowed to provide 99 to 100% specificity and sensitivity. All emm genes contained at least one of the two emm probes; there were 32 genomes with 10 distinct emm alleles that contained the 3′ probe but not the 5′ probe and 18 genomes with 7 distinct emm alleles that contained the 5′ probe but not the 3′ probe.
Whatmore et al. (7).
In the 1,688 genomes analyzed, there were 176 emm subtypes represented in more than one genome. Of these, more than one unique mrp allele was present in 46% of cases (n = 81) and more than one unique enn allele was present in 57% of cases (n = 100). Therefore, the previous nomenclature system in which the emm-like genes were named based on the emm type they were isolated from (26, 27) was not optimal. Thus, a systematic Mrp and Enn nomenclature was established, in which each unique protein sequence has a numerical identifier (e.g., Mrp1), and any allele that produces the same protein sequence is named as a subtype (e.g., Mrp1.1). This nomenclature is hosted on the website of the GAS reference laboratory at U.S. Centers for Disease Control and Prevention (CDC; https://www2a.cdc.gov/ncidod/biotech/strepblast.asp), and the alleles in each genome are listed in Table S3 in the supplemental material. Where possible, the association between previously designated mrp and enn sequences are noted in the lists of unique alleles (Table S4). However, since the new nomenclature is unlinked from the emm type of the strain, the previous names could unfortunately not be retained.
Composition of Mga regulons.
Of the 1,688 genomes analyzed, all contain mga and scpA genes, which ranged from 6,016 to 11,641 bp apart. The length of each gene family within the core Mga regulon displayed some variability (Fig. S1), the most variable being emm and the genes encoding transposases, while the least variably sized genes were mga genes and pgs (X92371.1), a gene encoding Pgs, a 15.5-kDa protein of unknown function (CAA63115).
Length and distribution of Mga regulon genes. Scatterplot comparing the range of sizes of gene families present in the Mga regulon. The central line represents the mean size, and the bars represent the SD and the colored circles indicate the range of sizes in the collection. Sizes ranged from the large scpA gene family (mean = 3,500 bp; SD = 92.9bp) to the small pgs gene family (mean = 461 bp; SD = 27.1 bp). The emm genes and transposase genes showed the most variability in length (emm: mean = 1,218 bp; SD = 153.6 bp; transposase: mean = 853 bp; SD = 256.5 bp); however, the latter is attributable to the presence of three major size variants. Download FIG S1, PDF file, 0.3 MB (267.6KB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
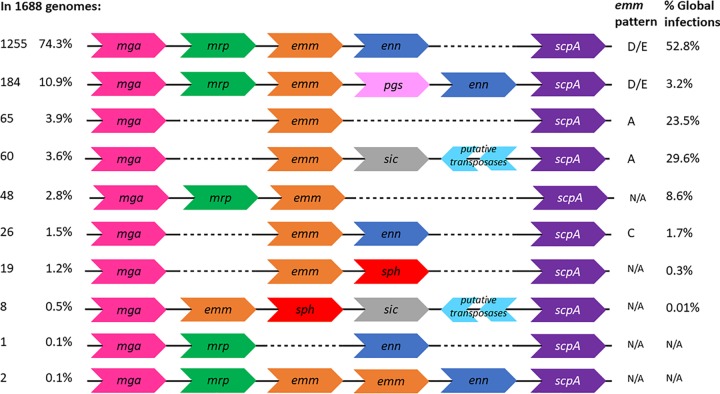
All of the genomes in the database possessed either an emm or emm-like gene, and the vast majority (85.2%) contained a gene for all three. Importantly, 74.3% of the genomes (n = 1,255) have a core Mga regulon consisting of: mga, mrp, emm, enn, and scpA, specifically in this stated order (Fig. 2). In the remaining 10.9% of isolates (n = 184), the Mga regulon also included a pgs gene between the emm and enn genes.
FIG 2.
Configurations of Mga regulons. In the large collection of contiguous Mga regulon sequences, we identified 10 possible configurations of the regulon based on presence and positions of genes. All Mga regulons began and ended with the mga and scpA genes and could contain genes for Mrp, Emm, Enn, Pgs, protein H, SIC, and transposases. The most frequent Mga regulon configuration, with genes for the trio of M and M-like proteins, was present in around 74% of genomes, from emm types that are responsible for around half of global infections.
The 85.2% of genomes which contained genes for mrp, emm and enn would be a D or E pattern under the emm-pattern typing system. Pattern C, defined as containing an emm and enn gene, was present in only 1.5% of isolates (n = 25). Among the genomes without a gene encoding M-like proteins, around half (3.9% of total, n = 65) had only an emm gene between mga and scpA (emm-pattern A), while 3.6% (n = 60) also harbored genes for SIC and for two transposases downstream of the emm gene. The genomes containing emm but neither enn nor mrp genes belonged to only 15 different emm types, mostly belonging to A to C emm clusters (Table S5). There were no pattern B isolates within the collection, i.e., encoding two emm genes with no other M-like genes. A total of 47 genomes contained an mrp and emm gene but no enn gene. In two genomes there were two copies of the same emm gene between mrp and enn genes, and six genomes did not contain an emm gene but contained an mrp and enn gene. These variants have not previously been assigned to an emm pattern. Isolates with the same emm type tended to encode the same pattern of proteins in their Mga regulon.
A gene encoding protein H (sph) was present in 1.6% of isolates (n = 28), of which eight also contained a gene for SIC and genes for two transposases downstream. In all isolates containing a sic gene there were also genes present for two transposases (n = 67). The Mga regulons encoding protein H, transposases, SIC and pgs belonged to few emm types (Table S5).
Although Mga proteins are highly conserved (93.9% pairwise identity), we observed two distinct variants, which differ by 21% at the amino acid level (0.254 substitutions per site, Fig. S2A). The minor variant was present in 10.6% of genomes, exclusively in strains that do not encode an Mrp protein, i.e., strains from A and C patterns and strains encoding SIC and protein H. The other variant was present in the remaining 89.4% of genomes. There was very little diversity within each protein variant (98% sequence identity, 0.01 substitutions per site). The amino acid diversity of Mga proteins was more evenly distributed along the length of the proteins than the other proteins present in the Mga regulon (Fig. S2B). This is in concordance with previous findings of 24.5% nucleotide diversity between two divergent mga alleles (13, 28).
Sequence identity within Mga regulon protein families. (A) Mga proteins are highly conserved (93.9% pairwise identity); however, two clear variants can be observed, MgaA and MgaB. MEGA was used to calculate genetic distances between all Mga proteins. Within both subgroups, the substitutions per site were 0.01 but between the groups was 0.25. (B) Details of the amino acid sequence identity within the different protein families present in the Mga locus. The percent identity is displayed along the length of mature protein consensus sequences. Mrp, M, Enn, and H proteins contain regions of low sequence identity in their N-terminal domains and become more conserved towards the C-terminal domains. Of the M and M-like proteins, Mrp proteins have the highest average pairwise identity (83.2%), followed by Enn proteins (68.6%), and M proteins have the lowest (44.7%). pgs sequences do not contain neither signal peptide (no YSIRK sequence) nor LPXTG motif indicative of streptococcal surface proteins. It is therefore unlikely to be a surface expressed virulence factor like many other Mga regulon-encoded proteins. Download FIG S2, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
emm.
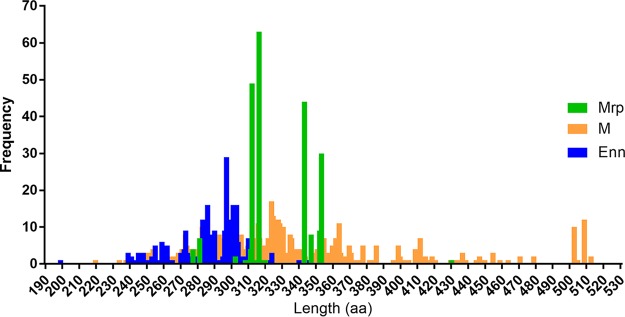
The M protein has been genetically analyzed previously and was not a major focus of this study (6). The mature M protein sequences ranged in size from 220 to 513 amino acids in length (Fig. 3) and had an average pairwise identity of 44.7%.
FIG 3.
Distribution of M and M-like protein lengths. Distribution of the lengths of all M and M-like proteins from the collection. Bars represent the number of genomes in the collection that contained a protein of the size indicated on the x axis. M proteins show the most diversity in protein lengths and can be both the smallest or the largest of the trio of proteins. Enn proteins are typically smaller and have less variable length distribution, and Mrp proteins are largely restricted to four possible lengths.
mrp.
An mrp gene was present in 88.9% of genomes described (n = 1,501) and included 295 unique alleles based on individual single nucleotide polymorphisms and 225 unique mature protein sequences (Fig. 1). Mrp protein sequences ranged from 277 to 430 amino acids in length (Fig. 3) and had an average pairwise identity of 83.2% (Fig. S2B). While Mrp proteins shared functional characteristics with M proteins (22), they were more homogenous in sequence. Unlike the wide distribution of emm gene length, genes for mrp had more restricted variability of length, since the majority of genes fit into four distinct length classes (Fig. 3).
In place of C-repeat sequences in the C-terminal region of M, Mrp proteins have A-repeat sequences (26, 29) (Fig. S3). These A-repeats were 35 amino acids in length, spanned a region of uninterrupted alpha-helix, and had no intervening sequence. In 95% of Mrp protein sequences, three distinct A-repeats could be identified and in 99% of sequences there were at least two A-repeats. The number of repeats was at least partly responsible for the observed difference in gene lengths. Numbered from the N terminus, the A1 repeats contained 95% pairwise identity, and the A2 and A3 repeats contained 99% pairwise identity. When aligned, all A repeat sequences contained 74% average pairwise identity.
Mrp A repeat and Enn C repeat consensus sequences. Sequence logos generated by Geneious based on multiple sequence alignments of the A- and C-repeat regions of Mrp (A) and Enn (B) proteins, respectively. Letter height is determined based on the likelihood of the presence of the amino acid in each position. Sequences are highly homogenous and contain the leucine residues essential for correct coil formation. Download FIG S3, PDF file, 0.3 MB (305.2KB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
enn.
An enn gene was present in 87.1% of genomes (n = 1,470), and of these, there were 352 unique alleles. The genes encoded 276 unique mature protein sequences following removal of signal sequences and cleaved regions (Fig. 1). Mature proteins ranged in length from 199 to 346 amino acids (Fig. 3) and had an average pairwise identity of 68.6% (Fig. S2B). Enn proteins therefore presented genetic diversity that is intermediate between the high diversity of M and the low diversity of Mrp and were generally the smaller of this trio of protein families (Fig. 3).
The repeat region of Enn proteins contained C-repeats (30) which are predicted to form alpha-helices disrupted by small regions of random coil and divided by linker regions of 7, 14, or 28 amino acids (Fig. S3). The repeats were less homogenous than the A repeats of Mrp: the C1 repeat was present in 100% of sequences and had 94% sequence identity, the C2 repeat was present in 94% of sequences and had 94% sequence identity, and the C3 repeat was present in 37% of sequences and had 93% sequence identity. The number of repeats present and the combination of linker regions had a large effect on the protein lengths.
Following the variable region at the N terminus, Enn proteins had either an EQ-rich central core (n = 154/278; 55% of sequences) with significant similarity to the analogous region in M proteins or, in 39% of sequences (n = 109/278), an 18-amino-acid consensus sequence (EKEKEDLKTTLAKTTKEN). There was greater sequence similarity between the N-terminal 50 first amino acids from Enn proteins with the 18-amino-acid core than EQ-rich cores, with 58 and 34% pairwise identities, respectively.
Sequence similarities across different regions of proteins.
All M and M-like protein sequences were preceded by a signal peptide, typically 41 amino acids in length. The sequences were highly homogenous within each protein family and only slightly less so between the different families (Table 2). The most C-terminal part of the proteins contained the LPXTG sortase motif, which allows attachment of the protein to the bacterial cell wall. This was also the region of the most sequence homogeneity in the mature M and M-like proteins, and all proteins became increasingly heterogeneous more distally (Fig. S2B).
TABLE 2.
Amino acid sequence identities in different regions of M and M-like proteinsa
| Protein(s) | Amino acid identity (%) |
|||
|---|---|---|---|---|
| Signal peptide | First 50 amino acids | 51st to repeat region | Repeat region to LPXTG | |
| Mrp | 98.0 | 43.2 | 91.6 | 96.1 |
| Emm | 81.8 | 15.0 | 41.4 | 87.9 |
| Enn | 96.2 | 27.3 | 56.2 | 93.7 |
| Mrp + Emm + Enn | 78.4 | 59.2 | ||
The variability between protein families was too great in the regions from the first amino acids of the mature protein to the repeat regions to perform a meaningful multiple alignment across the three protein families.
Expression analysis.
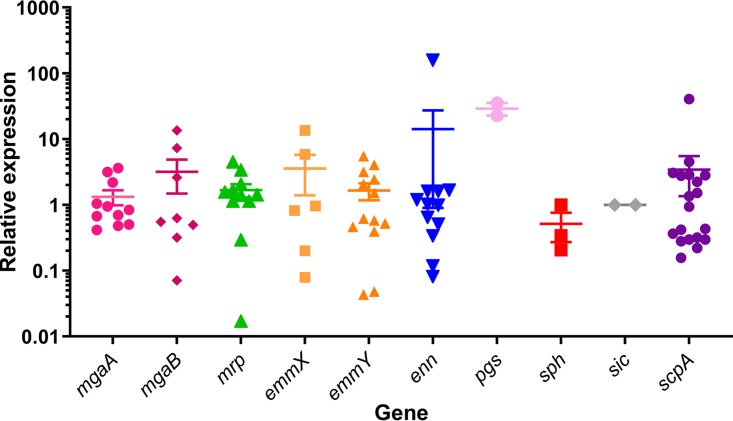
To better characterize expression of the Mga regulon components, gene expression was analyzed during exponential growth of 19 representative isolates grown in rich broth, conditions known to maximize emm gene expression (20) (Table S1). The expression of all genes was observed at levels comparable to emm, with similar variability (Fig. 4).
FIG 4.
Expression analysis of Mga regulon genes. cDNA from 19 isolates grown to mid-log phase in rich medium were analyzed for the expression of Mga regulon genes. The isolates were selected to be representative of all possible Mga regulon configurations and emm cluster diversity where possible. Primers were designed to amplify all members of the gene family where possible (mrp, enn, pgs, sph, and scpA) and to amplify a subset where sequence diversity necessitates. The dot plot symbols represent the mean value of the four qPCR analyses for each isolate, and the error bars represent the standard errors for all isolates for each gene.
Details of isolates used for expression analyses. Isolates used for expression analyses were selected first to be representative of the different Mga regulon configurations identified in this study and of different emm clusters. All protein-coding genes present in the different Mga regulon configurations were examined for the ability to be expressed, in more than one isolate when possible. The isolates included in the large genomic study by Davies et al. are indicated by asterisk (*). Download Table S1, DOCX file, 0.01 MB (15.5KB, docx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In silico emm-typing ambiguities.
Using the current CDC emm-typing database, there were 2,192 emm-typing sequences present in the analyzed Mga regulons. After analyses of positional information and the presence of gene-defining oligonucleotide probes (Table 1), as well as BLAST searches, it was determined that 529 of the emm-typing sequences were present in genes other than emm . The presence of two emm-typing sequences in two genes in the same locus (the emm gene and a non-emm gene) has high potential for mistyping. emm sequencing regions for emm types emm134.1 and emm226.0 were present in 20 sph genes encoding three distinct protein H alleles. Sequencing regions for the emm types emm141.0 and emm156.0 were also present in 44 mrp genes encoding nine distinct Mrp proteins. The remaining 465 emm sequence typing regions in non-emm genes were in enn genes (Table S6). Utilizing whole-genome-sequencing (WGS) data to derive emm types of GAS is becoming increasingly common practice; however, there is the possibility for the detection of an emm-like gene in place of the emm gene (9). A well-curated database of emm-typing sequences is essential to reduce potential mistyping using WGS and bioinformatic pipelines. This study utilized a globally informed whole-genome-based platform to thoroughly refine the emm sequence database to differentiate PCR-derived emm types into those relating to mrp, enn, and sph genes. Of note, 18 different emm-typing sequences were not found in emm genes but only in mrp, enn, or sph genes. Athey et al. previously identified 12 of these emm types (9), and in this work we identified emm141 in mrp63, emm134.1, and emm226 in three sph genes and emm203, emm134, and emm166 in eight, seven, and four enn genes, respectively. Such sequences have been removed from the emm sequence typing database and correctly renamed in the appropriate M-like database on the website of the GAS reference laboratory at the CDC (https://www2a.cdc.gov/ncidod/biotech/strepblast.asp).
Chimeric proteins.
Chimeric emm genes containing the N-terminal portion of the emm gene and C-terminal portion of the enn gene were present in 17 genomes belonging to six different emm types (emm types 4 [n = 12], 9, 44, 58, 73, and 82). These genes contained the 5′ emm probe and the enn probe at their 3′ ends (Fig. S4). This phenomenon was recently described in M4 isolates (31). Of note, all belonged to a specific Mga configuration with mrp and emm genes and were exceptions in their emm type.
Production of chimeric M/Enn proteins. Schematic of possible mechanisms of action resulting in the chimeric emm/enn genes found in 13 genomes. Recombination Detection Program (RDP) v4.97 was used to identify potential parent and recombinant sequences using a collection, including predicted chimeric genes and the emm and enn genes from the same emm types that contained both genes separately and the five novel chimeric genes were detected as well as likely enn parent genes (A). Software was able to predict the parent alleles of the chimeric gene, in some instances with high statistical probability (B) and identify the breakpoint in the alignment based on pairwise identity between the sequences (C). Triangles indicate the location of gene-specific probes, notably the absence of the 3′ emm (yellow) probe in the recombinant gene but presence of both the 5′ emm (red) and enn probes (dark blue). Download FIG S4, PDF file, 0.05 MB (49KB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
This study provides for the first time a comprehensive, genome-based, genetic description of the GAS Mga regulon and, in particular, the M-like proteins Mrp and Enn. We also provide a molecular definition using conserved oligonucleotide probes for emm and emm-like genes that allows for proper identification and will improve strain typing.
With the increasing adoption by public health laboratories of WGS for GAS typing in place of PCR sequence typing of the emm gene, it is critical to differentiate between emm and emm-like genes. Although the current system utilizing the 5′ emm probe is efficient for detection of emm genes (7), this could be further improved by incorporating the 3′ emm probe and the other gene family specific probes described in this study into a pipeline for emm gene typing. Updating the emm-typing collection to reflect the emm or emm-like genes further decreases the risk of identifying the incorrect gene or detecting two “emm” genes in a single genome.
Approximately 85% of genetically diverse GAS genomes were found to encode M, Mrp, and Enn proteins; this is striking because, compared to M, Mrp and Enn have been relatively poorly characterized to date. The retention of these genes suggests an important survival advantage, since pathogenic bacteria are under strong selection pressures. Isolates encoding Mrp and Enn are epidemiologically relevant, causing more than half of global GAS infections (Fig. 2), particularly in developing nations and in the indigenous populations of Australia and New Zealand. Indeed, in the latter populations D4 strains, which all encode the trio of M, Mrp, and Enn proteins, are considered endemic and have been linked to the development of rheumatic fever following skin infection (32, 33). M proteins from D4 strains are relatively small and have been shown to not induce a high M-type-specific antibody response (34). In these cases, it is conceivable that the M-like proteins perform roles otherwise performed by M proteins. The emm type is predictive of the composition of the Mga in many instances. This suggests the regulon has evolved as a whole in order to fill a functional niche. No D4 cluster M protein has been shown to bind fibrinogen (35); however, all Mrp proteins have one or two fibrinogen-binding motifs (36), and all D4 emm-types contain an mrp gene. In high-income settings where specific emm types, such as M1 and M12, are responsible for the majority of infections, the proportion of M-like protein-producing strains would be lower (8, 37).
In contrast to previous studies which found very low expression of mrp and enn genes compared to emm genes (20), we observed, under the growth conditions described here, high expression of all emm and emm-like genes in at least one isolate. Importantly, all genes present in core Mga regulons were capable of being transcribed.
The emergence of a range of emm types deriving from possible gene fusion between emm and enn genes suggests this may be a significant mechanism for the bacteria to alter function or evade immune recognition. This phenomenon, recently identified in M4 isolates (31), appears to have occurred in diverse emm types in the United States, a high-income-nation setting where there is low diversity of circulating emm types (38, 39).
The prevalence of emm-like genes, in addition to their genetic similarities and comparable expression with emm genes, indicates the importance of their encoded proteins to GAS virulence. The results presented here will thus aid further genetic and biological characterization of the Mga regulon in order to better understand its role in virulence and vaccine development.
MATERIALS AND METHODS
Genome collection and global epidemiological data.
We analyzed Mga regulon genes in a 2,083-GAS genome database representative of worldwide geographic and clinical diversity (25). A previously published global database of GAS infection (38) was used to assess the frequencies of each Mga configuration among global infections.
Bioinformatics.
The Mga regulon was derived from 2,083 GAS genome assemblies (25) based on the coordinates of the mga and scpA genes. Mga regulons that were not contained within a single contig (395/2,083) were excluded from the analysis. Annotation was performed using Geneious 10.1.2 based on gene orientation and BLAST searches to initially define emm, enn, and mrp gene families. To facilitate gene identification and solve nomenclature ambiguities, nucleotide probes to identify emm, enn, and mrp were developed based on regions of high sequence identity, with high sensitivity and specificity for each gene family. We also used the previously described emm-typing sequence as an emm probe (7). The specificity of probes was determined by Geneious 10.1.12 motif search within whole GAS genomes. Alleles with sequence ambiguities and frameshift mutations resulting in truncations were excluded from unique gene and protein sequence analyses (Fig. 1). Unique genes were differentiated by single nucleotide variations, and genes that produced proteins with the same amino acid sequences were annotated as subtypes (e.g., mga13.0 and mga13.1). The emm-typing database available from the CDC website (www.cdc.gov/streplab) was used to annotate emm genes and to search for emm-typing sequences within emm-like genes.
Mature protein sequences of M and M-like proteins were derived by removing the 41 to 42 amino acid signal peptide based on the EMBOSS 6.5.7 tool sigcleave at the amino terminus and the cleaved region following the threonine of the sortase LPXTG signal at the carboxy terminus. Repeat regions were identified by comparison to published sequences for M and M-like proteins (3, 30). Domains were defined as the N-terminal 50 amino acids, the 51st amino acid to the beginning of A- or C-repeat sequences and the repeat sequences to the mature protein’s last residue (22).
Multiple alignments were performed with the MAFFT program using the global pairwise iterative refinement (G-INS-i) method which uses the Needleman-Wunsch algorithm and BLOSUM62 scoring matrix (40). The percent identities at each position along amino acid sequences were graphed using Geneious 10.1.2, and genetic distances between groups of mga alleles were calculated using MEGA version X (41). RDP v4.97 (42) was used to detect recombination events and map gene breakpoints.
Expression analysis.
A representative collection of 19 GAS strains were selected for expression analyses, to include a diverse array of Mga regulon configurations and emm clusters (Table S1). Reverse transcriptase quantitative PCR was performed on the 19 representative GAS strains. Bacteria were grown at 37°C with 5% CO2 in Todd-Hewitt broth (Carl Roth, Karlsruhe, Germany) with 0.5% yeast extract (Carl Roth) until exponential phase (optical density at 600 nm, 0.4 to 0.6), and 1 ml of culture was harvested at 5,000 × g for RNA extraction. Bacteria were washed once with distilled water, and pellets were lysed with enzymatic lysis buffer consisting of 9.5 mg/ml lysozyme, 20 mM Tris-HCl, 3 mM EDTA, and 1% Triton X-100. Further lysis and separation of aqueous phase was achieved using PureZOL RNA isolation reagent (Bio-Rad; 732-6880), and RNA extraction was performed according to manufacturer’s instructions (Aurum Bio-Rad; 732-6870). After extraction, genomic DNA contamination was removed using Turbo DNase treatment (Invitrogen; AM1907), and the RNA yield and purity was estimated using a QuickDrop spectrophotometer (Molecular Devices). Reverse transcriptase (NEB; E6560) was performed using approximately 300 ng of RNA and a 6 μM concentration of the random primer mix provided for 1 h at 42°C after an initial incubation for 5 min at 25°C. Target genes from 5 μl of cDNA were amplified in 20-μl reactions with Luna Universal SYBR qPCR master mix (NEB; M3003) in a Bio-Rad CX96 real-time PCR detection system with conditions as follows: 95°C for 60 s, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Dissociation curves were calculated for each reaction to confirm product specificity. No-reverse-transcriptase and no-template controls were performed for each extraction and each pair of primers. Results were analyzed with qBase+ (Biogazelle) software, and the relative expression compared to recA was calculated for each sample. Specific primers were used (see Table S2), and amplification efficiencies were calculated. cDNA was produced twice from each strain, each sample was analyzed twice by qPCR, and the four relative expression values were averaged.
Primers used in expression analyses by RT-qPCR. Primer efficiencies were calculated using known concentration of genomic DNA of a subset of strains for each gene (shown in boldface). Based on the composition of the Mga regulon of the strain, expression was examined using universal primer sets where possible. The mga gene from strains that encoded the trio of Mrp, M, and Enn proteins was amplified by mgaB primers, and the mga gene from the other strains was amplified by mgaA primers. Three pairs of primers were used for the different emm genes: emmX for strains from clade X emm clusters (10), emmY-1 for A to C emm pattern strains from clade Y emm clusters, and emmY-2 for clade Y emm clusters that encoded emm-like genes, plus the M6 isolate. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins and alleles present in the Mga regulons of all genomes analyzed in this study. Download Table S3, XLSX file, 0.2 MB (156.7KB, xlsx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
mrp and enn alleles in this collection. Download Table S4, XLSX file, 0.3 MB (338.9KB, xlsx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of emm patterns, clusters, and types in different Mga regulon configurations. The vast majority of genomes in this collection encoded the trio of M and M-like proteins and included emm types from many multiprotein clusters, some single protein clusters from both clades X and Y and four emm types that have yet to be assigned a cluster (N/C). The remaining configurations were more restricted to both emm type and cluster. Strikingly, all genomes that encoded pgs belonged to the E3 cluster, and all genomes that did not encode mrp or enn belonged to only 15 different emm types. Global infection data are from Steer et al. (LID, 2009). Download Table S5, XLSX file, 0.02 MB (15.2KB, xlsx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
M-like genes with emm-sequence typing. emm sequence typing regions were detected in many emm-like genes, with 100% sequence identity, predominantly in enn genes. The 19 emm types (plus nine subtypes) largely belong to either the E5 emm cluster or were not assigned to an emm cluster and aside from emm137.0 were not otherwise present in emm genes. Download Table S6, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
H.R.F. is supported by a Ph.D. fellowship granted by the Royal Academy of Medicine from Belgium and sponsored by GSK Vaccines. The work is supported by the Belgian Fonds National de la Recherche Scientifique research grants (PDR T.0255.16 and CDR J.0019.17). The funders had no role in study design and analysis.
REFERENCES
- 1.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. 2014. Disease manifestations and pathogenic mechanisms of group a streptococcus. Clin Microbiol Rev 27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watkins DA, Johnson CO, Colquhoun SM, Karthikeyan G, Beaton A, Bukhman G, Forouzanfar MH, Longenecker CT, Mayosi BM, Mensah GA, Nascimento BR, Ribeiro ALP, Sable CA, Steer AC, Naghavi M, Mokdad AH, Murray CJL, Vos T, Carapetis JR, Roth GA. 2017. Global, regional, and national burden of rheumatic heart disease, 1990–2015. N Engl J Med 377:713–722. doi: 10.1056/NEJMoa1603693. [DOI] [PubMed] [Google Scholar]
- 3.Smeesters PR, McMillan DJ, Sriprakash KS. 2010. The streptococcal M protein: a highly versatile molecule. Trends Microbiol 18:275–282. doi: 10.1016/j.tim.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Beall B, Facklam R, Thompson T. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol 34:953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Active Bacterial Core Surveillance/Emerging Infections Program Network, Sakota V, Jackson D, Franklin AR, Beall B. 2003. Array of M protein gene subtypes in 1064 recent invasive group A streptococcus isolates recovered from the active bacterial core surveillance. J Infect Dis 188:1587–1592. doi: 10.1086/379050. [DOI] [PubMed] [Google Scholar]
- 6.McMillan DJ, Dreze PA, Vu T, Bessen DE, Guglielmini J, Steer AC, Carapetis JR, Van Melderen L, Sriprakash KS, Smeesters PR. 2013. Updated model of group A streptococcus M proteins based on a comprehensive worldwide study. Clin Microbiol Infect 19:E222–E229. doi: 10.1111/1469-0691.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whatmore AM, Kapur V, Sullivan DJ, Musser JM, Kehoe MA. 1994. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol 14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 8.Chochua S, Metcalf BJ, Li Z, Rivers J, Mathis S, Jackson D, Gertz RE Jr, Srinivasan V, Lynfield R, Van Beneden C, McGee L, Beall B. 2017. Population and whole genome sequence based characterization of invasive group A streptococci recovered in the United States during 2015. mBio 8:e01422-17. doi: 10.1128/mBio.01422-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Athey TB, Teatero S, Li A, Marchand-Austin A, Beall BW, Fittipaldi N. 2014. Deriving group A Streptococcus typing information from short-read whole-genome sequencing data. J Clin Microbiol 52:1871–1876. doi: 10.1128/JCM.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanderson-Smith M, De Oliveira DM, Guglielmini J, McMillan DJ, Vu T, Holien JK, Henningham A, Steer AC, Bessen DE, Dale JB, Curtis N, Beall BW, Walker MJ, Parker MW, Carapetis JR, Van Melderen L, Sriprakash KS, Smeesters PR, M Protein Study Group. 2014. A Systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis 210:1325–1338. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shulman ST, Tanz RR, Dale JB, Steer AC, Smeesters PR. 2014. Added value of the emm cluster typing system to analyze group A streptococcus epidemiology in high-income settings. Clin Infect Dis 59:1651–1652. doi: 10.1093/cid/ciu649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smeesters PR, Laho D, Beall B, Steer AC, Van Beneden CA. 2017. Seasonal, geographic, and temporal trends of emm clusters associated with invasive group A streptococcal infections in US multistate surveillance. Clin Infect Dis 64:694–695. doi: 10.1093/cid/ciw807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollingshead SK, Readdy TL, Yung DL, Bessen DE. 1993. Structural heterogeneity of the emm gene cluster in group A streptococci. Mol Microbiol 8:707–717. doi: 10.1111/j.1365-2958.1993.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 14.Akesson P, Cooney J, Kishimoto F, Bjorck L. 1990. Protein H: a novel IgG binding bacterial protein. Mol Immunol 27:523–531. doi: 10.1016/0161-5890(90)90071-7. [DOI] [PubMed] [Google Scholar]
- 15.Akesson P, Sjoholm AG, Bjorck L. 1996. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem 271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 16.Hartas J, Sriprakash KS. 1999. Streptococcus pyogenes strains containing emm12 and emm55 possess a novel gene coding for distantly related SIC protein. Microb Pathog 26:25–33. doi: 10.1006/mpat.1998.0244. [DOI] [PubMed] [Google Scholar]
- 17.Bessen DE, Smeesters PR, Beall BW. 2018. Molecular epidemiology, ecology, and evolution of group A streptococci. Microbiol Spectr 6. doi: 10.1128/microbiolspec.CPP3-0009-2018. [DOI] [PubMed] [Google Scholar]
- 18.Hondorp ER, McIver KS. 2007. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol 66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- 19.McIver KS, Scott JR. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol 179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bessen DE, Fischetti VA. 1992. Nucleotide sequences of two adjacent M or M-like protein genes of group A streptococci: different RNA transcript levels and identification of a unique immunoglobulin A-binding protein. Infect Immun 60:124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podbielski A, Flosdorff A, Weber-Heynemann J. 1995. Molecular characterization of the M type 49 group A streptococcal (GAS) virR gene. Dev Biol Stand 85:153–157. [PubMed] [Google Scholar]
- 22.Frost HR, Sanderson-Smith M, Walker M, Botteaux A, Smeesters PR. 2018. Group A streptococcal M-like proteins: from pathogenesis to vaccine potential. FEMS Microbiol Rev 42:193–204. doi: 10.1093/femsre/fux057. [DOI] [PubMed] [Google Scholar]
- 23.Dale JB, Niedermeyer SE, Agbaosi T, Hysmith ND, Penfound TA, Hohn CM, Pullen M, Bright MI, Murrell DS, Shenep LE, Courtney HS. 2015. Protective immunogenicity of group A streptococcal M-related proteins. Clin Vaccine Immunol 22:344–350. doi: 10.1128/CVI.00795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courtney HS, Niedermeyer SE, Penfound TA, Hohn CM, Greeley A, Dale JB. 2017. Trivalent M-related protein as a component of next generation group A streptococcal vaccines. Clin Exp Vaccine Res 6:45–49. doi: 10.7774/cevr.2017.6.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies MR, McIntyre L, Mutreja A, Lacey JA, Lees JA, Towers RJ, Duchene S, Smeesters PR, Frost HR, Price DJ, Holden MTG, David S, Giffard PM, Worthing KA, Seale AC, Berkley JA, Harris SR, Rivera-Hernandez T, Berking O, Cork AJ, Torres R, Lithgow T, Strugnell RA, Bergmann R, Nitsche-Schmitz P, Chhatwal GS, Bentley SD, Fraser JD, Moreland NJ, Carapetis JR, Steer AC, Parkhill J, Saul A, Williamson DA, Currie BJ, Tong SYC, Dougan G, Walker MJ. 2019. Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat Genet doi: 10.1038/s41588-019-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heath DG, Boyle MD, Cleary PP. 1990. Isolated DNA repeat region from fcrA76, the Fc-binding protein gene from an M-type 76 strain of group A streptococci, encodes a protein with Fc-binding activity. Mol Microbiol 4:2071–2079. doi: 10.1111/j.1365-2958.1990.tb00567.x. [DOI] [PubMed] [Google Scholar]
- 27.Stenberg L, O’Toole P, Lindahl G. 1992. Many group A streptococcal strains express two different immunoglobulin-binding proteins, encoded by closely linked genes: characterization of the proteins expressed by four strains of different M-type. Mol Microbiol 6:1185–1194. doi: 10.1111/j.1365-2958.1992.tb01557.x. [DOI] [PubMed] [Google Scholar]
- 28.Haanes EJ, Cleary PP. 1989. Identification of a divergent M protein gene and an M protein-related gene family in Streptococcus pyogenes serotype 49. J Bacteriol 171:6397–6408. doi: 10.1128/jb.171.12.6397-6408.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Toole PW, O’Toole P, Stenberg L, Rissler M, Lindahl G. 1992. Two major classes in the M protein family in group A streptococci. Proc Natl Acad Sci U S A 89:8661–8665. doi: 10.1073/pnas.89.18.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podbielski A, Weber-Heynemann J, Cleary PP. 1994. Immunoglobulin-binding FcrA and Enn proteins and M proteins of group A streptococci evolved independently from a common ancestral protein. Med Microbiol Immunol 183:33–42. doi: 10.1007/bf00193629. [DOI] [PubMed] [Google Scholar]
- 31.DebRoy S, Li X, Kalia A, Galloway-Pena J, Shah BJ, Fowler VG, Flores AR, Shelburne SA. 2018. Identification of a chimeric emm gene and novel emm pattern in currently circulating strains of emm4 group A streptococcus. Microb Genom 4. doi: 10.1099/mgen.0.000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson DA, Smeesters PR, Steer AC, Morgan J, Davies M, Carter P, Upton A, Tong SY, Fraser J, Moreland NJ. 2016. Comparative M-protein analysis of Streptococcus pyogenes from pharyngitis and skin infections in New Zealand: implications for vaccine development. BMC Infect Dis 16:561. doi: 10.1186/s12879-016-1891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks T, Smeesters PR, Steer AC. 2012. Streptococcal skin infection and rheumatic heart disease. Curr Opin Infect Dis 25:145–153. doi: 10.1097/QCO.0b013e3283511d27. [DOI] [PubMed] [Google Scholar]
- 34.Frost HR, Laho D, Sanderson-Smith ML, Licciardi P, Donath S, Curtis N, Kado J, Dale JB, Steer AC, Smeesters PR. 2017. Immune cross-opsonization within emm clusters following group A streptococcus skin infection: broadening the scope of type-specific immunity. Clin Infect Dis 65:1523–1531. doi: 10.1093/cid/cix599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanderson-Smith ML, Dinkla K, Cole JN, Cork AJ, Maamary PG, McArthur JD, Chhatwal GS, Walker MJ. 2008. M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J 22:2715–2722. doi: 10.1096/fj.07-105643. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Courtney HS. 2011. Promotion of phagocytosis of Streptococcus pyogenes in human blood by a fibrinogen-binding peptide. Microbes Infect 13:413–418. doi: 10.1016/j.micinf.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Nelson GE, Pondo T, Toews KA, Farley MM, Lindegren ML, Lynfield R, Aragon D, Zansky SM, Watt JP, Cieslak PR, Angeles K, Harrison LH, Petit S, Beall B, Van Beneden CA. 2016. Epidemiology of invasive group A streptococcal infections in the United States, 2005–2012. Clin Infect Dis 63:478–486. doi: 10.1093/cid/ciw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. 2009. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 39.Smeesters PR, McMillan DJ, Sriprakash KS, Georgousakis MM. 2009. Differences among group A streptococcus epidemiological landscapes: consequences for M protein-based vaccines? Expert Rev Vaccines 8:1705–1720. doi: 10.1586/erv.09.133. [DOI] [PubMed] [Google Scholar]
- 40.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Length and distribution of Mga regulon genes. Scatterplot comparing the range of sizes of gene families present in the Mga regulon. The central line represents the mean size, and the bars represent the SD and the colored circles indicate the range of sizes in the collection. Sizes ranged from the large scpA gene family (mean = 3,500 bp; SD = 92.9bp) to the small pgs gene family (mean = 461 bp; SD = 27.1 bp). The emm genes and transposase genes showed the most variability in length (emm: mean = 1,218 bp; SD = 153.6 bp; transposase: mean = 853 bp; SD = 256.5 bp); however, the latter is attributable to the presence of three major size variants. Download FIG S1, PDF file, 0.3 MB (267.6KB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence identity within Mga regulon protein families. (A) Mga proteins are highly conserved (93.9% pairwise identity); however, two clear variants can be observed, MgaA and MgaB. MEGA was used to calculate genetic distances between all Mga proteins. Within both subgroups, the substitutions per site were 0.01 but between the groups was 0.25. (B) Details of the amino acid sequence identity within the different protein families present in the Mga locus. The percent identity is displayed along the length of mature protein consensus sequences. Mrp, M, Enn, and H proteins contain regions of low sequence identity in their N-terminal domains and become more conserved towards the C-terminal domains. Of the M and M-like proteins, Mrp proteins have the highest average pairwise identity (83.2%), followed by Enn proteins (68.6%), and M proteins have the lowest (44.7%). pgs sequences do not contain neither signal peptide (no YSIRK sequence) nor LPXTG motif indicative of streptococcal surface proteins. It is therefore unlikely to be a surface expressed virulence factor like many other Mga regulon-encoded proteins. Download FIG S2, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mrp A repeat and Enn C repeat consensus sequences. Sequence logos generated by Geneious based on multiple sequence alignments of the A- and C-repeat regions of Mrp (A) and Enn (B) proteins, respectively. Letter height is determined based on the likelihood of the presence of the amino acid in each position. Sequences are highly homogenous and contain the leucine residues essential for correct coil formation. Download FIG S3, PDF file, 0.3 MB (305.2KB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Details of isolates used for expression analyses. Isolates used for expression analyses were selected first to be representative of the different Mga regulon configurations identified in this study and of different emm clusters. All protein-coding genes present in the different Mga regulon configurations were examined for the ability to be expressed, in more than one isolate when possible. The isolates included in the large genomic study by Davies et al. are indicated by asterisk (*). Download Table S1, DOCX file, 0.01 MB (15.5KB, docx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Production of chimeric M/Enn proteins. Schematic of possible mechanisms of action resulting in the chimeric emm/enn genes found in 13 genomes. Recombination Detection Program (RDP) v4.97 was used to identify potential parent and recombinant sequences using a collection, including predicted chimeric genes and the emm and enn genes from the same emm types that contained both genes separately and the five novel chimeric genes were detected as well as likely enn parent genes (A). Software was able to predict the parent alleles of the chimeric gene, in some instances with high statistical probability (B) and identify the breakpoint in the alignment based on pairwise identity between the sequences (C). Triangles indicate the location of gene-specific probes, notably the absence of the 3′ emm (yellow) probe in the recombinant gene but presence of both the 5′ emm (red) and enn probes (dark blue). Download FIG S4, PDF file, 0.05 MB (49KB, pdf) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in expression analyses by RT-qPCR. Primer efficiencies were calculated using known concentration of genomic DNA of a subset of strains for each gene (shown in boldface). Based on the composition of the Mga regulon of the strain, expression was examined using universal primer sets where possible. The mga gene from strains that encoded the trio of Mrp, M, and Enn proteins was amplified by mgaB primers, and the mga gene from the other strains was amplified by mgaA primers. Three pairs of primers were used for the different emm genes: emmX for strains from clade X emm clusters (10), emmY-1 for A to C emm pattern strains from clade Y emm clusters, and emmY-2 for clade Y emm clusters that encoded emm-like genes, plus the M6 isolate. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins and alleles present in the Mga regulons of all genomes analyzed in this study. Download Table S3, XLSX file, 0.2 MB (156.7KB, xlsx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
mrp and enn alleles in this collection. Download Table S4, XLSX file, 0.3 MB (338.9KB, xlsx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of emm patterns, clusters, and types in different Mga regulon configurations. The vast majority of genomes in this collection encoded the trio of M and M-like proteins and included emm types from many multiprotein clusters, some single protein clusters from both clades X and Y and four emm types that have yet to be assigned a cluster (N/C). The remaining configurations were more restricted to both emm type and cluster. Strikingly, all genomes that encoded pgs belonged to the E3 cluster, and all genomes that did not encode mrp or enn belonged to only 15 different emm types. Global infection data are from Steer et al. (LID, 2009). Download Table S5, XLSX file, 0.02 MB (15.2KB, xlsx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
M-like genes with emm-sequence typing. emm sequence typing regions were detected in many emm-like genes, with 100% sequence identity, predominantly in enn genes. The 19 emm types (plus nine subtypes) largely belong to either the E5 emm cluster or were not assigned to an emm cluster and aside from emm137.0 were not otherwise present in emm genes. Download Table S6, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2020 Frost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.