Abstract
The rapid increase in multidrug resistant pathogens is a major health concern that could bring mankind back to the pre-antibiotic era. Streptococcus pneumoniae is a highly recombinogenic opportunistic pathogen that causes a variety of deadly diseases and rapidly develops resistance to current antibiotic treatments. S. pneumoniae pathogenicity is dependent on a cell-density communication mechanism, or quorum sensing (QS), termed the competence regulon. In this work, we set out to design signal-based QS modulators capable of affecting the two specificity groups found in S. pneumoniae. Through systematic analysis and rational design, we were able to construct peptide-based pan group QS activators and inhibitors with activities in the nanomolar range. These novel analogs are privileged scaffolds for the development of anti-virulence therapeutics against S. pneumoniae infections.
Graphical Abstract
The best of both worlds: The development of a pneumococcal CSP1-CSP2 hybrid peptide allows the identification of a pan group quorum sensing inhibitor with activities in the nanomolar range.
Introduction
Similar to multicellular organisms, bacteria can coordinate their physiological behaviors to acquire group advantages by working collectively to effectively exhibit phenotypes such as motility, competence, biofilm formation and sporulation, which were otherwise unachievable.[1–3] To this end, bacteria sense the population cell density in the surrounding medium. This concentration-dependent phenomenon is known as quorum sensing (QS).[4] QS involves producing, secreting and detecting a class of signaling molecules known as auto-inducers.[4–5] Many gram-negative bacteria produce small signaling molecules called acyl homoserine lactones (AHLs) as their auto-inducer, whereas most gram-positive bacteria depend on peptide-based signaling molecules termed auto-inducing peptides (AIPs).[4,6–9]
Streptococcus pneumoniae is an opportunistic pathogen that inhabits the nasopharynx of humans and can be deadly if it spreads to other parts of the body.[10] S. pneumoniae has been found to cause many different diseases, including bacteremia, meningitis, and pneumonia.[11–12] It is estimated that there are 900,000 cases of pneumococcal pneumonia infections annually in the United States. Of these, 5–7% are fatal and 400,000 result in hospitalization.[13] In addition, due to its highly recombinogenic nature, S. pneumoniae is able to rapidly develop resistance against antimicrobial therapeutics. [14–17] Therefore, there is a dire need to develop novel strategies to treat this prevalent pathogen. Previous studies have shown that controlling QS could be a valid alternative strategy for attenuating S. pneumoniae infections.[18–20] Specifically, Lau and co-workers have shown that inhibition of the competence regulon lead to attenuation of the acquisition of antibiotic resistance genes in vivo.[18]
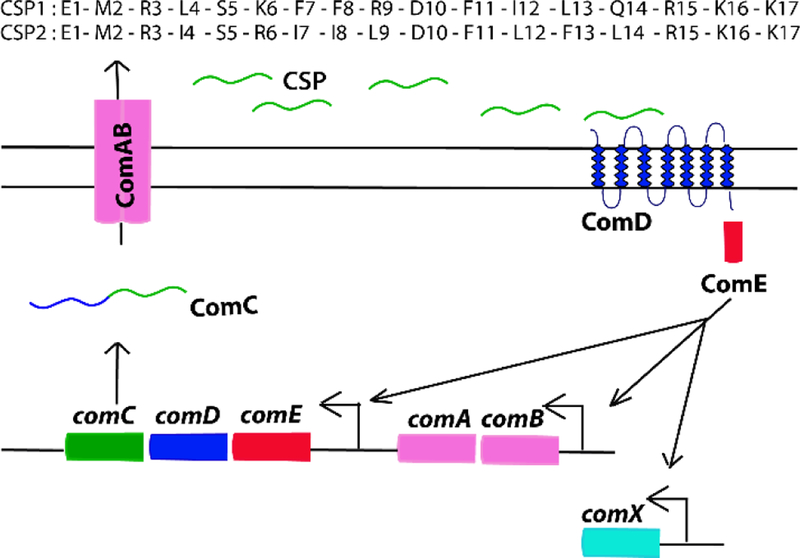
In S. pneumoniae, the major QS circuitry is termed the competence regulon.[21] This circuitry is centered on a 17-amino acid peptide pheromone termed the competence stimulating peptide (CSP), and its activation of a two-component regulatory system, once an extracellular threshold concentration is reached.[22] At this concentration, CSP effectively binds to its cognate transmembrane histidine kinase receptor, ComD, to initiate the QS signaling cascade. The activated ComD receptor phosphorylates the response regulator, ComE, which leads to expression of a variety of early and late genes associated with competence, virulence factor production and biofilm formation. The comABCDE genes code the QS circuitry components, including the pro-CSP, ComC, which is processed and exported outside the cell through the ComAB transporter as the mature CSP. The comX gene, an alternative sigma factor, is responsible for inducing different physiological behaviors and is the master regulator of pneumococcus infections (Figure 1).[22–25] Different strains of S. pneumoniae can be classified into two pherotypes or specificity groups (Group 1 and Group 2) based on the signaling molecule they produce (CSP1 or CSP2) with their corresponding receptors, ComD1 or ComD2.[26] The two CSP signals share eight identical residues (50% homology).[27]
Figure 1. The competence regulon QS circuitry in S. pneumoniae.
The prepeptide ComC is being processed and secreted by the ComAB transporter as the mature CSP signal. At high concentration, CSP activates the transmembrane histidine kinase receptor ComD. Activated ComD phosphorylates ComE, which then autoactivates the competence QS circuit and upregulates the expression of genes involved in attaining genetic competence, virulence factor production and biofilm formation through ComX. The sequences of CSP1 and CSP2 are shown at the top.
Previously, we reported the rational design of a double modified CSP2 analog, CSP2-E1Ad10, which was a potent antagonist against group 2 S. pneumoniae and was able to attenuate pneumonia in a mouse model of infection.[20] In the present study, we continued our efforts to develop a pan group QS inhibitor against both S. pneumoniae specificity groups. To this end, we utilized the CSP2 scaffold and systematically modified its residues to resemble more CSP1 with the goal of achieving analogs with pan group activities. Through this analysis, we identified both a pan group activator and a pan group inhibitor.
Results and Discussion
Our main goal in this study was to design a pan group QS inhibitor. In a previous work, we identified a ComD2 inhibitor, CSP2-E1Ad10, which also exhibited weak inhibition of ComD1, thereby being the closest thing to a pan-group QS inhibitor.[28] We therefore chose CSP2 as a lead scaffold to begin the design of pan group inhibitors. Our approach was to first develop CSP2-based pan group QS activators and then convert them into pan group inhibitors by replacing the glutamic acid at position 1 (E1) with alanine, as this modification was found to be crucial and sufficient to turn S. pneumoniae CSP-based QS activators into competitive inhibitors [18–19,28] Moreover, since the last two residues in both CSP1 and CSP2 (K16 and K17) were found to be dispensable,[28] we chose a truncated version of CSP2, CSP2-des(K16-K17) (termed here CSP2(15)), as our starting scaffold.
All the CSP2 analogs were constructed by using standard solid phase peptide synthesis protocols on Wang resin using a microwave-assisted automated synthesizer.[29] The crude peptides were purified to homogeneity through reverse phase high performance liquid chromatography (RP-HPLC) to >95% purity and their identity confirmed by mass spectrometry (for full details see SI). To evaluate the ability of the analogs to modulate QS in both specificity groups, we utilized the beta galactosidase cell-based gene reporter assay with the two previously reported group 1 and 2 reporter strains, D39pcomX::lacZ and TIGR4pcomX::lacZ, which are wild type strains producing their native CSP and also carrying the lacZ gene under the control of the comX promoter.[18]
Single and Double modifications
CSP1 and CSP2 differ at positions 4, 6, 7, 8, 9, 12, 13, and 14. We have previously systematically modified CSP2(15) at these positions with the corresponding residues present in CSP1 and found that CSP2(15)-R6K and CSP2(15)-I8F exhibit significant loss in potency.[20] Therefore, we excluded these two modifications when conducting a systematic double modification evaluation, to afford 15 double modified analogs. Analysis of the double modification library revealed four residues, 4, 9, 13, and 14, that their combinations result in analogs, 2, 4, and 12, capable of activating ComD1 while maintaining the activity against ComD2 (Table 1). We therefore selected these four residues for further analysis.
Table 1.
EC50 values of the double modified CSP2 analogs against the ComD1 and ComD2 receptorsa
| ComD1 | ComD2 | |||||
|---|---|---|---|---|---|---|
| Peptide Number | Peptide Name | Sequence | EC50 (nM)b | 95% CIc | EC50 (nM)b | 95% CIc |
| CSP1e | EMRLSKFFRDFILQRKK | 10.3 | 6.27–16.8 | 526 | 498–556 | |
| CSP2e | EMRISRIILDFLFLRKK | 1650 | 1190–2300 | 50.7 | 40.6–63.2 | |
| CSP2(15)f | EMRISRIILDFLFLR | --d | -- | 21.9 | 11.5–41.6 | |
| 1 | CSP2(15)-I4LI7F | EMRLSRFILDFLFLR | --d | -- | >1000 | -- |
| 2 | CSP2(15)-I4LL9R | EMRLSRIIRDFLFLR | 685 | 469–999 | 58.2 | 42.3–80.1 |
| 3 | CSP2(15)-I4LL12I | EMRLSRIILDFIFLR | --d | -- | 19.6 | 8.60–44.8 |
| 4 | CSP2(15)-I4LF13L | EMRLSRIILDFLLLR | 610 | 414–897 | 9.16 | 7.49–11.2 |
| 5 | CSP2(15)-I4LL14Q | EMRLSRIILDFLFQR | --d | -- | 5.51 | 2.31–13.1 |
| 6 | CSP2(15)-I7FL9R | EMRISRFIRDFLFLR | 661 | 304–1437 | 149 | 73.7–304 |
| 7 | CSP2(15)-I7FL12I | EMRISRFILDFIFLR | --d | -- | 26.3 | 20.5–33.8 |
| 8 | CSP2(15)-I7FF13L | EMRISRFILDFLLLR | --d | -- | 67.2 | 30.6–147 |
| 9 | CSP2(15)-I7FL14Q | EMRISRFILDFLFQR | --d | -- | 82.6 | 53.0–128 |
| 10 | CSP2(15)-L9RL12I | EMRISRIIRDFIFLR | --d | -- | 77.6 | 45.4–132 |
| 11 | CSP2(15)-L9RF13L | EMRISRIIRDFLLLR | --d | -- | 78.1 | 46.6–130 |
| 12 | CSP2(15)-L9RL14Q | EMRISRIIRDFLFQR | 408 | 361–462 | 15.2 | 6.14–37.5 |
| 13 | CSP2(15)-L12IF13L | EMRISRIILDFILLR | --d | -- | 44.5 | 22.5–88.0 |
| 14 | CSP2(15)-L12IL14Q | EMRISRIILDFIFQR | --d | -- | 72.2 | 49.6–104 |
| 15 | CSP2(15)-F13LL14Q | EMRISRIILDFLLQR | --d | -- | 22.5 | 19.2–26.2 |
See experimental section for detail of reporter strains. See supporting information for methods and plots of agonism dose response curves. All assays were performed in triplicates.
EC50 values were determined by testing peptides over a range of concentrations.
95% confidence interval.
EC50 not determined due to the analog’s low induction in primary agonism screening assay.
Data from ref. 28.
Data from ref. 20.
Triple and Quadruple modifications
We utilized the results from the double modification library to design and construct a set of triple and quadruple modified analogs. To this end, we synthesized four triple-modified analogs (all combinations of triple modifications in positions 4, 9, 13, and 14), as well as a quadruple-modified analog bearing modification in all four positions. Out of the five analogs, 17 and 20 exhibited improved activity against the ComD1 receptor, while retaining the activity against the ComD2 receptor (Table 2).
Table 2.
EC50 values of the triple & quadruple modified CSP2 analogs against the ComD1 and ComD2 receptorsa
| ComD1 | ComD2 | |||||
|---|---|---|---|---|---|---|
| Peptide Number | Peptide Name | Sequence | EC50 (nM)b | 95% CIc | EC50 (nM)b | 95% CIc |
| 16 | CSP2(15)-I4LL9RF13L | EMRLSRIIRDFLLLR | >1000 | -- | 79.4 | 53.2–118 |
| 17 | CSP2(15)-I4LL9RL14Q | EMRLSRIIRDFLFQR | 166 | 110–249 | 19.3 | 11.2–33.1 |
| 18 | CSP2(15)-I4LF13LL14Q | EMRLSRIILDFLLQR | 874 | 663–1150 | 11.4 | 7.20–18.0 |
| 19 | CSP2(15)-L9RF13LL14Q | EMRISRIIRDFLLQR | >1000 | -- | 18.6 | 9.28–35.3 |
| 20 | CSP2(15)-I4LL9RF13LL14Q | EMRLSRIIRDFLLQR | 193 | 160–233 | 17.3 | 15.7–19.2 |
See experimental section for detail of reporter strains. See supporting information for methods and plots of agonism dose response curves. All assays were performed in triplicates.
EC50 values were determined by testing peptides over a range of concentrations.
95% confidence interval.
Quintuple modification
To validate that we have identified the optimized structures for pan group activation, we selected the highly potent quadruple-modified peptide analog, 20, and added an additional modification at positions 6, 7, 8 or 12. Analysis of the four analogs validated that modifying CSP2(15) in either one of these positions (6, 7, 8 or 12) lead to reduction in activity against the ComD2 receptor (Table 3). Out of the four analogs, 23 was found to be the most active analog, with approximately 2.5-fold increase in activity against ComD1 and approximately 2.5-fold decrease in activity against ComD2, compared to the quadruple-modified analog, 20, suggesting that further improvement in activity against the ComD1 receptor would be at the expense of reduced activity against the ComD2 receptor.
Table 3.
EC50 values of the quintuple modified CSP2 analogs against the ComD1 and ComD2 receptorsa
| ComD1 | ComD2 | |||||
|---|---|---|---|---|---|---|
| Peptide Number | Peptide Name | Sequence | EC50 (nM)b | 95% CIc | EC50 (nM)b | 95% CIc |
| 21 | CSP2(15)-I4LR6KL9RF13LL14Q | EMRLSKIIRDFLLQR | >1000 | -- | 141 | 85.5–235 |
| 22 | CSP2(15)-I4LI7FL9RF13LL14Q | EMRLSRFIRDFLLQR | 266 | 144–492 | >1000 | -- |
| 23 | CSP2(15)-I4LI8FL9RF13LL14Q | EMRLSRIFRDFLLQR | 75.3 | 59.4–95.4 | 41.0 | 21.2–79.0 |
| 24 | CSP2(15)-I4LL9RL12IF13LL14Q | EMRLSRIIRDFILQR | >1000 | -- | 158 | 64.5–387 |
See experimental section for detail of reporter strains. See supporting information for methods and plots of agonism dose response curves. All assays were performed in triplicates.
EC50 values were determined by testing peptides over a range of concentrations.
95% confidence interval.
E1A and d10 modifications
Having several pan group activator scaffolds in hand, we set out to develop pan group QS inhibitors. Previous work by the Lau group and our group revealed that modification of Glu1 to alanine is sufficient to convert CSP-based QS activators into competitive inhibitors.[18–19, 28] Moreover, our structure-function analysis of CSP2 revealed that modifying Asp 10 with its enantiomer (D-Asp, CSP2-d10) increases CSP2 activity against the ComD2 receptor by 20-fold.[28] We therefore hypothesized that incorporation of the E1A modification in the optimized pan group activator scaffolds, with or without the additional d10 modification, would result in potent pan group QS inhibitors. For this analysis, we chose to work on the full length CSP2 scaffold, since in a previous work we found that the truncated CSP2(15) scaffold bearing the E1A and d10 modifications lost its weak inhibitory activity against the ComD1 receptor, compared to the full length CSP2-E1Ad10, that exhibited weak pan group inhibitory propensity.[20]
We selected the four most potent pan group activators identified in the double to quintuple modification libraries: 12,17,20 and 23, and added the E1A, with or without the d10 modification, to their full length CSP2 scaffolds, expecting to convert these analogs to pan group inhibitors. Unfortunately, none of the analogs exhibited pan group inhibitory activity, two analogs were found to be potent ComD2 inhibitors, one analog was found to be a potent ComD1 inhibitor, while the rest of the analogs were found to be relatively inactive against both groups (Table 4). These results indicate that direct conversion of a pan-group activator into a pan-group inhibitor is not feasible. This trend further validates our previous observation where CSP2(15)-I4Ld10, which was found to be a pan group activator (EC50 values of 234 nM and 3.28 nM against ComD1 and ComD2, respectively), could not be converted into a pan group inhibitor by adding the E1A modification.[20]
Table 4.
IC50 values of the E1A & d10 modifications of CSP2 analogs against the ComD1 and ComD2 receptorsa
| ComD1 | ComD2 | |||||
|---|---|---|---|---|---|---|
| Peptide Number | Peptide Name | Sequence | IC50 (nM)b | 95% CIc | IC50 (nM)b | 95% CIc |
| CSP2-E1Ad10e | AMRISRIILdFLFLRKK | >1000 | -- | 56.5 | 53.5–59.6 | |
| 25 | CSP2-E1AL9RL14Q | AMRISRIIRDFLFQRKK | >1000 | -- | --d | -- |
| 26 | CSP2-E1AL9Rd10L14Q | AMRISRIIRdFLFQRKK | --d | -- | --d | -- |
| 27 | CSP2-E1AI4LL9RL14Q | AMRLSRIIRDFLFQRKK | >1000 | -- | >1000 | -- |
| 28 | CSP2-E1AI4LL9Rd10L14Q | AMRLSRIIRdFLFQRKK | >1000 | -- | --d | -- |
| 29 | CSP2-E1AI4LL9RF13LL14Q | AMRLSRIIRDFLLQRKK | >1000 | -- | --d | -- |
| 30 | CSP2-E1AI4LL9Rd10F13LL14Q | AMRLSRIIRdFLLQRKK | >1000 | -- | 168 | 119–238 |
| 31 | CSP2-E1AI4LI8FL9RF13LL14Q | AMRLSRIFRDFLLQRKK | 455 | 270–767 | --d | -- |
| 32 | CSP2-E1AI4LI8FL9Rd10F13LL14Q | AMRLSRIFRdFLLQRKK | >1000 | -- | 161 | 135–193 |
See experimental section for detail of reporter strains. See supporting information for methods and plots of antagonism dose response curves. All assays were performed in triplicates.
IC50 values were determined by testing peptides over a range of concentrations.
95% confidence interval.
IC50 not determined due to the analog’s low activity in primary antagonism screening assay.
Data from ref. 28.
Non-proteogenic conservative modifications
Our inability to construct a pan group inhibitor led us to hypothesize that the fourth position in the CSPs sequences (Leu for CSP1, Ile for CSP2) plays a key role in receptor specificity. Therefore, we decided to further examine the fourth position by incorporating conservative point modifications at this position to modulate receptor specificity. We started this analysis by using the lead ComD2 inhibitor, which also exhibited weak inhibitory activity against ComD1, CSP2-E1Ad10, as the starting scaffold and replaced isoleucine at the fourth position with homoleucine, norleucine, and norvaline. Biological evaluation of the three resulting analogs revealed that the norvaline modification is the most accommodated one, leading to the most potent ComD2 inhibitor, while exhibiting weak inhibitory activity against the ComD1 receptor (Table 5). In a final attempt to directly convert pan group activators into pan group inhibitors, we took the quintuple modified analog, 23 and inserted norvaline in the fourth position, along with the E1A modification (with or without the d10 modification). Expectedly, the resulting two analogs did not exhibit potent pan group inhibitory activity (Table 5).
Table 5.
IC50 values of the non-proteogenic modifications of CSP2 analogs against the ComD1 and ComD2 receptorsa
| ComD1 | ComD2 | |||||
|---|---|---|---|---|---|---|
| Peptide Number | Peptide Name | Sequence | IC50 (nM)b | 95% CIc | IC50 (nM)b | 95% CIc |
| 33 | CSP2-E1AI4Nvad10 | AMRNvaSRIILdFLFLRKK | >1000 | -- | 42.2 | 34.1–52.0 |
| 34 | CSP2-E1AI4Nled10 | AMRNleSRIILdFLFLRKK | >1000 | -- | 79.1 | 74.7–84.0 |
| 35 | CSP2-E1AI4HLeud10 | AMRhLeuSRIILdFLFLRKK | >1000 | -- | 97.6 | 62.3–151 |
| 36 | CSP2-E1AI4NvaI8FL9RF13LL14Q | AMRNvaSRIFRDFLLQRKK | >1000 | -- | >1000 | — |
| 37 | CSP2-E1AI4NvaI8FL9Rd10F13LL14Q | AMRNvaSRIFRdFLLQRKK | --d | -- | 229 | 89.7–585 |
See experimental section for detail of reporter strains. See supporting information for methods and plots of antagonism dose response curves. All assays were performed in triplicates.
IC50 values were determined by testing peptides over a range of concentrations.
95% confidence interval.
IC50 not determined due to the analog’s low activity in primary antagonism screening assay.
Systematic alterations of CSP2-E1AI4Nvad10
Due to our failure to directly convert pan group activators into inhibitors we decided to change strategies and start the design of pan group inhibitors from our most promising analog in hand, 33, as the lead inhibitor scaffold. Based on our initial results, the key positions that could be modified while maintaining biological activity against both groups are 4, 8, 9, 13 and 14. As the fourth position was already modified to norvaline in the lead inhibitor scaffold, we decided to systematically modify the other four positions, starting with all the single point modification possibilities and continuing to all the double and triple modification possibilities. To our satisfaction, of the 12 analogs, one peptide, 41, was found to effectively inhibit both ComD receptors, having IC50 values of 510 nM and 18.2 nM against the ComD1 and ComD2 receptors, respectively, making it the first S. pneumoniae nanomolar-range pan group inhibitor (Table 6).
Table 6.
IC50 values of the E1A, I4Nva & d10 modifications of CSP2 analogs against the ComD1 and ComD2 receptorsa
| ComD1 | ComD2 | |||||
|---|---|---|---|---|---|---|
| Peptide Number | Peptide Name | Sequence | IC50 (nM)b | 95% CIc | IC50 (nM)b | 95% CIc |
| 33 | CSP2-E1AI4Nvad10 | AMRNvaSRIILdFLFLRKK | >1000 | -- | 42.2 | 34.1–52.0 |
| 38 | CSP2-E1AI4NvaI8Fd10 | AMRNvaSRIFLdFLFLRKK | >1000 | -- | 42.9 | 16.7–110 |
| 39 | CSP2-E1AI4NvaL9Rd10 | AMRNvaSRIIRdFLFLRKK | >1000 | -- | 225 | 115–439 |
| 40 | CSP2-E1AI4Nvad10F13L | AMRNvaSRIILdFLLLRKK | >1000 | -- | 43.9 | 34.0–57.5 |
| 41 | CSP2-E1AI4Nvad10L14Q | AMRNvaSRIILdFLFQRKK | 510 | 416–625 | 18.2 | 10.8–30.5 |
| 42 | CSP2-E1AI4NvaI8FL9Rd10 | AMRNvaSRIFRdFLFLRKK | >1000 | -- | 41.6 | 22.4–77.0 |
| 43 | CSP2-E1AI4NvaI8Fd10F13L | AMRNvaSRIFLdFLLLRKK | >1000 | -- | 32.1 | 19.6–52.3 |
| 44 | CSP2-E1AI4NvaI8Fd10L14Q | AMRNvaSRIFLdFLFQRKK | >1000 | -- | 33.1 | 15.8–69.0 |
| 45 | CSP2-E1AI4NvaL9Rd10F13 | AMRNvaSRIIRdFLLLRKK | >1000 | -- | 407 | 176–940 |
| 46 | CSP2-E1AI4NvaL9Rd10L14Q | AMRNvaSRIIRdFLFQRKK | >1000 | -- | --d | -- |
| 47 | CSP2-E1AI4Nvad10F13LL14Q | AMRNvaSRIILdFLLQRKK | >1000 | -- | 53.5 | 39.4–73.0 |
| 48 | CSP2-E1AI4NvaI8FL9Rd10F13L | AMRNvaSRIFRdFLLLRKK | >1000 | -- | 183 | 77.2–436 |
| 49 | CSP2-E1AI4NvaI8FL9Rd10L14Q | AMRNvaSRIFRdFLFQRKK | >1000 | -- | 51.6 | 40.5–65.5 |
See experimental section for detail of reporter strains. See supporting information for methods and plots of antagonism dose response curves. All assays were performed in triplicates.
IC50 values were determined by testing peptides over a range of concentrations.
95% confidence interval. dIC50 not determined due to the analog’s low activity in primary antagonism screening assay.
The L14Q modification is the key
To gain additional insight and determine the role of both the fourth and fourteenth positions in pan group inhibition, we designed two final analogs, having the original side chain residue of the two native CSPs at position four (Ile and Leu). Interestingly, both resulting analogs exhibited pan group inhibition (Table 7). Expectedly, the analog bearing Leu, the original residue in CSP1, was more active towards ComD1 while the analog bearing lie, the original residue in CSP2, was more active towards ComD2. However, both analogs were less active than the analog bearing the norvaline residue, suggesting that the alkyl branching in Leu and lie is detrimental to binding ComD1 and ComD2, likely due to steric clashes with the receptor binding pocket. Overall, these new results indicate that the L14Q modification is the key for constructing CSP2-based pan group inhibitors.
Table 7.
IC50 values of the L14Q modification CSP2 analogs against the ComD1 and ComD2 receptorsa
| ComD1 | ComD2 | |||||
|---|---|---|---|---|---|---|
| Peptide Number | Peptide Name | Sequence | IC50 (nM)b | 95% CIc | IC50 (nM)b | 95% CIc |
| 50 | CSP2-E1AdlOL14Q | AMRISRIILdFLFQRKK | 766 | 463–1269 | 49.5 | 21.2–116 |
| 51 | CSP2-E1AI4Ld10L14Q | AMRLSRIILdFLFQRKK | 503 | 307–825 | 87.2 | 36.3–210 |
See experimental section for detail of reporter strains. See supporting information for methods and plots of antagonism dose response curves. All assays were performed in triplicates.
IC50 values were determined by testing peptides over a range of concentrations.
95% confidence interval.
Conclusion
In this study, we sought to identify pan group modulators of the S. pneumoniae competence QS circuitry by systematically altering the CSP2 scaffold with the residues present in CSP1. The approach was to merge these two CSPs into a hybrid sequence capable of effectively binding both the ComD1 and ComD2 receptors. Our analysis revealed that the residue requirements for pan group activators and inhibitors are different, as opposed to the simple E1 A modification that was found to be sufficient in converting ComD1 or ComD2 activators into competitive inhibitors, resulting in the inability to directly convert pan group activators into pan group inhibitors. Instead, to our satisfaction, a different strategy that utilized a potent ComD2 inhibitor as the starting scaffold yielded the first nanomolar-range pan group inhibitor, 41. This analog is a privileged scaffold for the design of future S. pneumoniae anti-infective therapeutics.
Experimental Details
Chemical Reagents and Instrumentation
All chemical reagents and solvents were purchased from Sigma-Aldrich and used without further purification. Water (18 MΩ) was purified using a Millipore Analyzer Feed System. Solid-phase resins were purchased from Advanced ChemTech or Chem-Impex International.
Reversed-phase high-performance liquid chromatography (RP-HPLC) was performed using a Shimadzu system equipped with a CBM-20A communications bus module, two LC-20AT pumps, an SIL-20A auto sampler, an SPD-20A UV/VIS detector, a CTO-20A column oven, and an FRC-10A fraction collector. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) data were obtained on a Bruker Microflex spectrometer equipped with a 60 Hz nitrogen laser and a reflectron. In positive ion mode, the acceleration voltage on Ion Source 1 was 19.01 kV. Exact mass (EM) data were obtained on an Agilent Technologies 6230 TOF LC/MS spectrometer. The samples were sprayed with a capillary voltage of 3500 V and the electrospray ionization (ESI) source parameters were as follows: gas temperature of 325 °C at a drying gas flow rate of 3 L/min at a pressure of 25 psi.
Solid Phase Peptide Synthesis
All the CSP2 analogs were synthesized in a microwave synthesizer using standard Fmoc-based solid phase peptide synthesis (SPPS) procedures on 4-Benzyloxybenzyl alcohol (Wang) resin using commercially available Fmoc-protected amino acids. Pre-loaded Fmoc-L-Arg(pbf) Wang resin (0.308 mmol/g) or Fmoc-L-Lys(Boc) Wang resin (0.332 mmol/g) were used for peptides that required an arginine or lysine at the C-terminus, respectively (for the full SPPS protocols, see the Supporting Information).[29]
Peptide Purification
Crude peptides were purified with RP-HPLC. The crude peptide was dissolved in 20 mL ACN:H2O (1:3) and purified in 5 mL portions on a Phenomenex Kinetex (5 μm, 10 mm, 250 mm, 110 Å) C18 column with a 5 mL/min flow rate; mobile phase A= 18 MΩ water + 0.1 % TFA and mobile phase B = ACN + 0.1 % TFA. The collected fraction was lyophilized overnight and dissolved again in 5 mL ACN:H2O (1:3) for a second prep run. Preparative HPLC methods were used to separate the crude peptide mixture to different chemical components using a linear gradient (first prep 5% B → 45% B over 40 min, and second prep 27% B → 37% B over 30 min). Then, an analytical Phenomenex Kinetex C18 column (5 μm, 4.6 mm, 250 mm, 110 Å) was used to quantify the purity of the desired fractions using a linear gradient (5% B → 95% B over 27 min). Purities were determined by integration of peaks with UV detection at 220 nm. Only peptide fractions that were purified to homogeneity (>95%) were used for the biological assays. EM was used to validate the identity of synthesized peptides. The observed mass-to-charge (m/z) ratio of the peptide was compared to the expected m/z ratio for each peptide (see Tables S-1 and S-2).
Biological Assays
Biological Reagents and Strain Information
All standard biological reagents were purchased from Sigma-Aldrich. Horse serum (defibrinated) was stored at 4 °C until use in bacterial growth conditions. To examine the ability of the synthesized CSP analogs to modulate the ComD receptors, and thus, the QS circuit in S. pneumoniae, beta-galactosidase assays were performed using D39pcomX::lacZ (group I) and TIGR4pcomX::lacZ (group II) reporter strains.[18]
Bacterial Growth Conditions
Frozen stocks of individual pneumococcal strains, D39pcomX::lacZ and TIGR4pcomX::lacZ were streaked onto a THY agar plate supplemented with 5% horse serum with chloramphenicol at a final concentration of 4 μg/mL. The plate was incubated for 8 h in a CO2 incubator (37 °C with 5% CO2). Fresh colonies were picked into sterilized cultural tubes containing 5 mL of THY broth supplemented with chloramphenicol at a final concentration of 4 μg/mL and the cultures were incubated in a CO2 incubator overnight (15 h). Overnight cultures were then diluted (1:50 for D39pcomX::lacZ; 1:10 for TIGR4pcomX::lacZ) with THY and the resulting solution was incubated in a CO2 incubator for 3–4 hours, until the bacteria reached the desired optical density (OD600 of 0.30–0.35 for D39pcomX::lacZ and 0.20–0.25 for TIGR4pcomX::lacZ) as determined by using a plate reader.
β-Galactosidase assay:
The β-gal assay was performed as we previously described (for full details of the assay protocols see the Supporting Information).[28,30–31]
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health (R35GM128651) and the National Science Foundation (CHE-1808370). S. pneumoniae D39pcomX::lacZ and TIGR4pcomX::lacZ reporter strains were generous gifts from G. W. Lau (University of Illinois at Urbana-Champaign).
Footnotes
Supporting Information
Full details of peptide synthesis and characterization, the beta-galactosidase bioassay, initial screening results, and dose response curves for CSP2 analogs. This information is available online.
The authors declare no competing financial interest.
References
- [1].Bassler BL, Losick R, Cell 2006,125, 237–246. [DOI] [PubMed] [Google Scholar]
- [2].Camilli A, Bassler BL, Science 2006, 311, 1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Parsek MR, Greenberg EP, Trends Microbiol 2005,13, 27–33. [DOI] [PubMed] [Google Scholar]
- [4].Waters CM, Bassler BL, Annu Rev Cell Dev Biol 2005, 21, 319–346. [DOI] [PubMed] [Google Scholar]
- [5].Rutherford ST, Bassler BL, Cold Spring Harb Perspect Med 2012, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ng W-L, Bassler BL, Annu Rev Genet 2009, 43, 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP, FEMS Microbiol Rev 2001, 25, 365–404. [DOI] [PubMed] [Google Scholar]
- [8].Galloway WR, Hodgkinson JT, Bowden SD, Welch M, Spring DR, Chem Rev 2011, 111, 28–67. [DOI] [PubMed] [Google Scholar]
- [9].Miller MB, Bassler BL, Annu Rev Microbiol 2001, 55, 165–199. [DOI] [PubMed] [Google Scholar]
- [10].Bogaert D, Hermans P, Adrian P, Rümke H, de Groot R, Vaccine 2004, 22,2209–2220. [DOI] [PubMed] [Google Scholar]
- [11].O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Lancet 2009, 374, 893–902. [DOI] [PubMed] [Google Scholar]
- [12].Mehr S, Wood N, Paediatr Respir Rev 2012, 13, 258–264. [DOI] [PubMed] [Google Scholar]
- [13].Huang SS, Johnson KM, Ray GT, Wroe P, Lieu TA, Moore MR, Zell ER, Linder JA, Grijalva CG, Metlay JP, Vaccine 2011, 29, 3398–3412. [DOI] [PubMed] [Google Scholar]
- [14].Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, science 2011, 331, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hansman D, Devitt L, Miles H, Riley I, Med J Aust 1974, 353–356. [DOI] [PubMed] [Google Scholar]
- [16].Klugman KP, Clin Microbiol Rev 1990, 3,171–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tankovic J, Perichon B, Duval J, Courvalin P, Antimicrob Agents Chemother 1996, 40, 2505–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhu L, Lau GW, PLoS Pathog 2011, 7, e1002241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Duan C, Zhu L, Xu Y, Lau GW, PloS one 2012, 7, e44710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Koirala B, Lin J, Lau GW, Tal-Gan Y, Chembiochem 2018,19, 2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pestova EV, Håvarstein LS, Morrison DA, Mol Microbiol 1996, 21, 853–862. [DOI] [PubMed] [Google Scholar]
- [22].Hâvarstein LS, Coomaraswamy G, Morrison DA, Proc Natl Acad Sei USA 1995, 92, 11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Galante J, Ho ACY, Tingey S, Charalambous BM, Curr Pharm Des 2015, 21, 25–30. [DOI] [PubMed] [Google Scholar]
- [24].Lau GW, Haataja S, Lonetto M, Kensit SE, Marra A, Bryant AP, McDevitt D, Morrison DA, Holden DW, Mol Microbiol 2001, 40, 555–571. [DOI] [PubMed] [Google Scholar]
- [25].Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, Andrew PW, Pozzi G, Mol Microbiol 2006, 61, 1196–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, Simon D, Morrison DA, J Bacteriol 1996,178, 6087–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Johnsborg O, Kristiansen PE, Blomqvist L, Håvarstein LS, J Bacteriol 2006, 188, 1744–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang Y, Koirala B, Sanchez LA, Phillips NR, Hamry SR, Tal-Gan Y, Acs Chem Biol 2017,12, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chan WC, White PD, Fmoc solid phase peptide synthesis: a practical approach, Vol. 222, Oxford University Press; Oxford, 2000. [Google Scholar]
- [30].Koirala B, Hillman RA, Tiwold EK, Bertucci MA, Tal-Gan Y, Beilstein J Org Chem 2018,14, 1769–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Koirala B, Phillips NR, Tal-Gan Y, ACS Med Chem Lett 2019, DOI: 10.1021/acsmedchemlett.1029b00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.