Abstract
Endoplasmic reticulum (ER) stress activates the unfolded protein response (UPR), which reduces levels of misfolded proteins. However, if ER homeostasis is not restored and the UPR remains chronically activated, cells undergo apoptosis. The UPR regulator, PKR-like endoplasmic reticulum kinase (PERK), plays an important role in promoting cell death when persistently activated; however, the underlying mechanisms are poorly understood. Here, we profiled the microRNA (miRNA) transcriptome in human cells exposed to ER stress and identified miRNAs that are selectively induced by PERK signaling. We found that expression of a PERK-induced miRNA, miR-483, promotes apoptosis in human cells. miR-483 induction was mediated by a transcription factor downstream of PERK, activating transcription factor 4 (ATF4), but not by the CHOP transcription factor. We identified the creatine kinase brain-type (CKB) gene, encoding an enzyme that maintains cellular ATP reserves through phosphocreatine production, as being repressed during the UPR and targeted by miR-483. We found that ER stress, selective PERK activation, and CKB knockdown all decrease cellular ATP levels, leading to increased vulnerability to ER stress–induced cell death. Our findings identify miR-483 as a downstream target of the PERK branch of the UPR. We propose that disruption of cellular ATP homeostasis through miR-483–mediated CKB silencing promotes ER stress–induced apoptosis.
Keywords: unfolded protein response (UPR), endoplasmic reticulum stress (ER stress), stress response, microRNA (miRNA), endoplasmic reticulum (ER), translation, translation control, activating transcription factor-4 (ATF-4), PKR-like endoplasmic reticulum kinase (PERK), ATP, F1F0-ATPase, fluorescence resonance energy transfer (FRET), creatine kinase B (CKB)
Introduction
Eukaryotic cells employ the endoplasmic reticulum (ER)4 organelle to fold secreted and membrane proteins, synthesize hydrophobic lipids and sterols, and store free calcium (1). Physiologic, pathologic, or environmental processes that interfere with ER functions lead to a condition known as ER stress (2), which is detected by a conserved intracellular signal transduction mechanism, the unfolded protein response (UPR) (3). The UPR initiates transcriptional and translational programs that seek to restore ER homeostasis.
In mammals, the UPR is controlled by three ER-resident transmembrane sensors: inositol-requiring enzyme 1 (IRE1), PKR-like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6) (3). Each of these UPR regulators has ER stress–sensing luminal domains coupled across the ER membrane to cytosolic effector domains that initiate distinct signal transduction cascades during the UPR (3). In particular, PERK responds to ER stress by oligomerizing to activate its cytosolic kinase domain, which specifically phosphorylates a serine at position 51 of the α subunit of eukaryotic initiation factor 2 (eIF2α) (4). eIF2α partners with eIF2β and eIF2γ to form the heterotrimeric eIF2 that is essential for canonical translational initiation at AUG start codons, but phosphorylation of eIF2α at serine 51 abrogates eIF2-dependent protein translation, leading to global slowdown of protein synthesis and diminished protein folding demands on the ER (5, 6). However, translation of many UPR genes, including activating transcription factor 4 (Atf4), C/EBP homologous protein (Chop), Gadd34, and BiP/Grp78, persists or even increases despite phosphorylation of eIF2α because the 5′-UTRs of these transcripts contain upstream open reading frames or internal ribosome entry sites that enable them to bypass p-eIF2α–mediated translational slowdown or employ noncanonical modes of translational initiation during the UPR (7–13). Thus, the PERK arm of the UPR dampens protein translation and also triggers transcriptional programs by production of transcription factors such as activating transcription factor 4 (ATF4) and C/EBP-homologous protein (CHOP).
The transcriptional and translational programs governed by IRE1, PERK, and ATF6 help cells to cope with and adapt to ER stress by improving the fidelity of protein folding, enhancing the elimination of irreparably damaged ER proteins, and alleviating the protein-folding demands upon the ER through translational attenuation. The requirement of the UPR in maintaining ER homeostasis is exemplified by the exquisite sensitivity of cells bearing knockouts of UPR genes to ER stress-induced damage and cell death and, conversely, the enhanced resistance to ER stress when UPR-associated proteins are artificially activated (14–19). However, if the transcriptional and translational programs of the UPR fail to restore ER homeostasis and remove ER stress, persistent UPR activation creates a maladaptive cellular environment that can culminate in cell death. The mechanisms by which UPR signaling shifts from the restoration of ER homeostasis to the promotion of cell death are under active investigation, as this has important implications for cell survival and function in many disease conditions.
Metazoan cells contain an abundance of small noncoding microRNAs (miRs) that function in post-transcriptional gene silencing. miR genes are present throughout the genome and can be found within introns or exons of other genes. miRs are initially transcribed as a much longer primary miRNA precursor in the nucleus (20). Progressive processing by Drosha/DGCR8 and Dicer enzymes ultimately yields the mature 18–23-nucleotide miR in the cytosol (20). miRs typically regulate target genes by binding to cognate sequences within the 3′-UTR of target mRNAs, leading to translational inhibition and mRNA degradation (20). By this mechanism, miRs play an important role in regulating a wide variety of cellular signaling events through modulating the protein levels of their target genes (21). A growing number of studies have implicated a role for miRs in the UPR (22). Numerous miRs have been identified that target and down-regulate expression of UPR regulators (23–29). Conversely, UPR-induced transcription factors such as CHOP can directly up-regulate specific miR genes (30, 31). These findings suggest that miRs sculpt the UPR by regulating expression of UPR genes, and the UPR also induces miRs as part of its transcriptional programs to influence cellular responses to ER stress.
In this present study, we hypothesized that the UPR may induce miRs that influence the cell life/death choice in response to ER stress. We screened a commercially assembled human microRNA gene panel for miRs up-regulated in HEK293 cells exposed to chemical forms of ER stress. We identified a small group specifically induced by the PERK arm of the UPR under experimental conditions when unabated PERK signaling causes cell death. We found that one of these, miR-483, promoted cell death when expressed in human cells. We provide evidence that miR-483 decreased cell viability by targeting expression of creatine kinase brain-type, leading to loss of cellular ATP stores.
Results
Comprehensive analysis of ER stress-regulated human miRNAs
Cells respond to ER stress by activating potent transcriptional programs that up-regulate many mRNAs. We hypothesized that ER stress also broadly reshapes the microRNA transcriptome, similar to its effects on the mRNA transcriptome. To investigate this potential role, we profiled the expression of 754 human miRNAs tiled on printed array cards (TaqMan array human MicroRNA A+B cards, Thermo Fisher Scientific). We probed these cards with total RNA samples collected from HEK293 cells treated for 24 h with solvent control or tunicamycin, an agent that blocks N-linked glycosylation to create potent ER stress, and performed quantitative RT-PCR to determine the expression levels of all 754 miRNAs in our experimental samples. Using this strategy, we identified numerous miRNAs whose levels were profoundly regulated positively and negatively by ER stress (Fig. 1A). We focused our subsequent analysis on miR-215 and miR-483-5p, two of the most highly ER stress–induced miRNAs, because they were previously found to influence cell growth and differentiation in experimental cancer models. In glioma and colorectal cell lines, miR-215 promoted tumor differentiation and survival (32, 33). In animal tumor models, miR-483-5p prevented metastases (34). Another highly ER stress–induced microRNA was miR-616*. Interestingly, we found that the gene encoding miR-616 lay within intron 2 of DDIT3/Chop, a proapoptotic transcription factor strongly up-regulated by ER stress. This genomic pairing of miR-616* within DDIT3/Chop was unique to primates and not seen in mice or lower organisms. Based on these functional and genomic observations, we selected miR-215, miR-483, and miR-616* for additional investigation to see whether they influenced cellular survival or death in response to ER stress.
Figure 1.
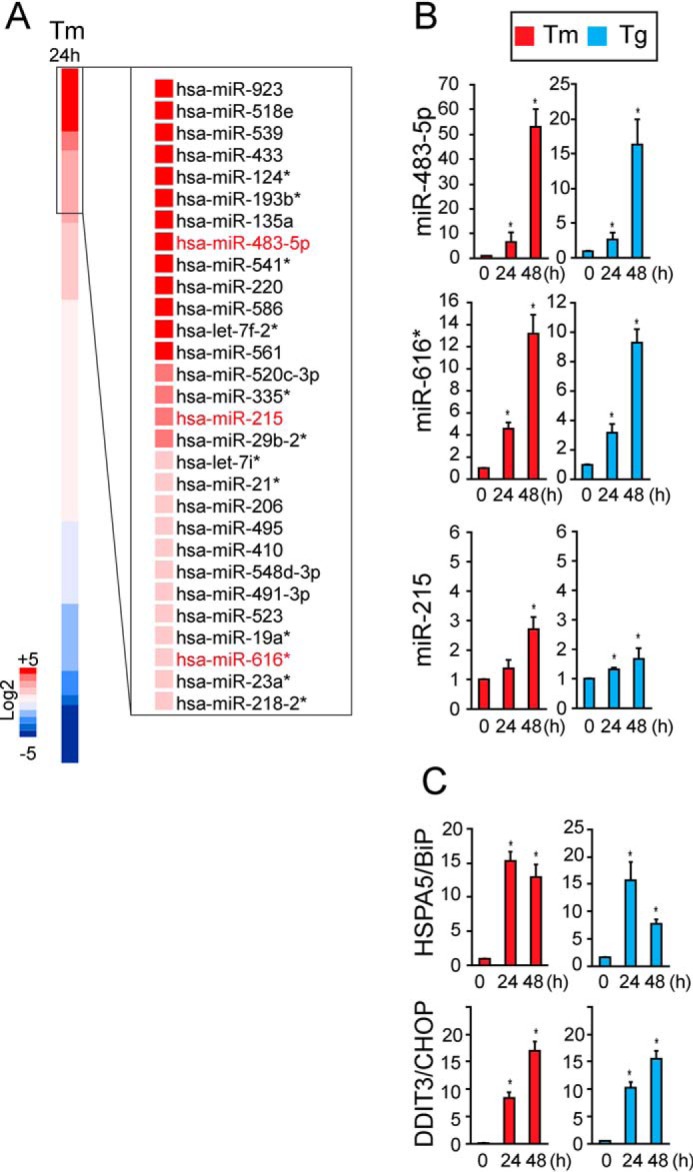
Identification of human miRNAs induced by ER stress. A, HEK293 cells were treated with tunicamycin (Tm) at 5 μg/ml for 24 h, and total RNA was isolated. The expression profiles of 754 human miRNAs from Tm-treated samples (n = 3) were determined using Taqman miRNA array A+B cards and are shown relative to untreated samples in the heat map. Three miRNAs induced by Tm treatment, miR-483-5p, miR-215, and miR-616*, were selected for further analysis. B and C, HeLa cells were treated with Tm (5 μg/ml) or 500 nm thapsigargin (Tg) for 0, 24, or 48 h. Total RNA was collected, and miR-483-5p, miR-215, and miR-616* levels were measured by qPCR and shown relative to levels at 0 h. In C, HSAP5/BiP and DDIT3/Chop mRNA levels were measured by qRT-PCR and shown relative to 0 h. Values are expressed as mean ± S.D. (error bars) of at least three independent experiments. *, p < 0.05, Student's t test.
Next, we examined whether miR-215, miR-483, and miR-616* induction was evident in other human cell types and in response to other sources of ER stress. To do this, we cultured HeLa cells and treated them with tunicamycin or thapsigargin, agents that induce potent ER stress by inhibiting protein glycosylation or disrupting ER calcium stores, respectively. We collected total RNA after 24 or 48 h of drug treatment and performed qRT-PCR to measure miR-215, miR-483, and miR-616* levels. We found progressive induction of all three microRNAs after chemical ER stress induction (Fig. 1B), similar to the transcriptional up-regulation of genes induced by ER stress, such as BiP and Chop (Fig. 1C) (18). These results supported our qRT-PCR array findings that miR-215, miR-483, and miR-616* are induced by ER stress.
miRs-215, 483-5p, and 616* are induced by the PERK arm of the unfolded protein response
In human cells, ER stress-induced transcriptional programs are activated by the UPR signal transduction cascades controlled by IRE1, PERK, and ATF6. To determine whether the UPR regulated the expression of miR-215, miR-483, and miR-616*, we selectively activated the IRE1, PERK, or ATF6 pathways in HeLa cells and then measured miRNA transcript levels by qRT-PCR. To activate the transcriptional program initiated by IRE1, we expressed the spliced X-box–binding protein 1 (XBP1s) transcription factor specifically produced by IRE1's endoribonuclease function (35, 36). We confirmed increased XBP1s transcriptional activity after transfection through marked increase in mRNA levels of ERdj4, a target gene of XBP1s (Fig. 2A) (37). By contrast, mRNA levels of Chop and BiP, target genes of the PERK and ATF6 pathways (7, 37), were not statistically significantly increased with XBP1s expression (Fig. 2A). To activate the PERK pathway, we treated HeLa cells expressing Fv2E-PERK, a chimeric fusion of the PERK kinase domain with two FK506-binding domains, with AP20187, a small-molecule ligand previously demonstrated to drive the homodimerization, trans-phosphorylation, and activation of Fv2E-PERK (10, 19, 38). Prior studies demonstrated that AP20187 dimerization of Fv2E-PERK selectively activated the translational and transcriptional programs of the PERK branch of the UPR without activating IRE1 or ATF6 in cells (10, 19, 38–40). Consistent with our prior studies, the addition of AP20187 to cells bearing Fv2E-PERK caused marked induction of Chop, a target gene robustly up-regulated by PERK activation (Fig. 2B). By contrast, BiP, a marker for ATF6 signaling, and spliced Xbp1 mRNA, a marker of IRE1 activation, were not induced with AP20187 addition (Fig. 2, B and D). To activate the ATF6 pathway, we used HeLa cells expressing the 373-amino acid cytosolic transcriptional activator fragment of ATF6 (ATF6(373)) under the control of a doxycycline-regulated promoter (40–42). Prior studies demonstrated that the addition of doxycycline lead to expression of the ATF6 transcriptional activator fragment and potent induction of the BiP gene, without triggering Xbp1 mRNA splicing or activating the PERK pathway (39, 40, 42). Consistent with our prior studies, the addition of doxycycline to cells bearing the TetOn-ATF6(373) construct led to marked induction of BiP, with lesser effects on Chop and no splicing of Xbp-1 mRNA (Fig. 2, C and D).
Figure 2.
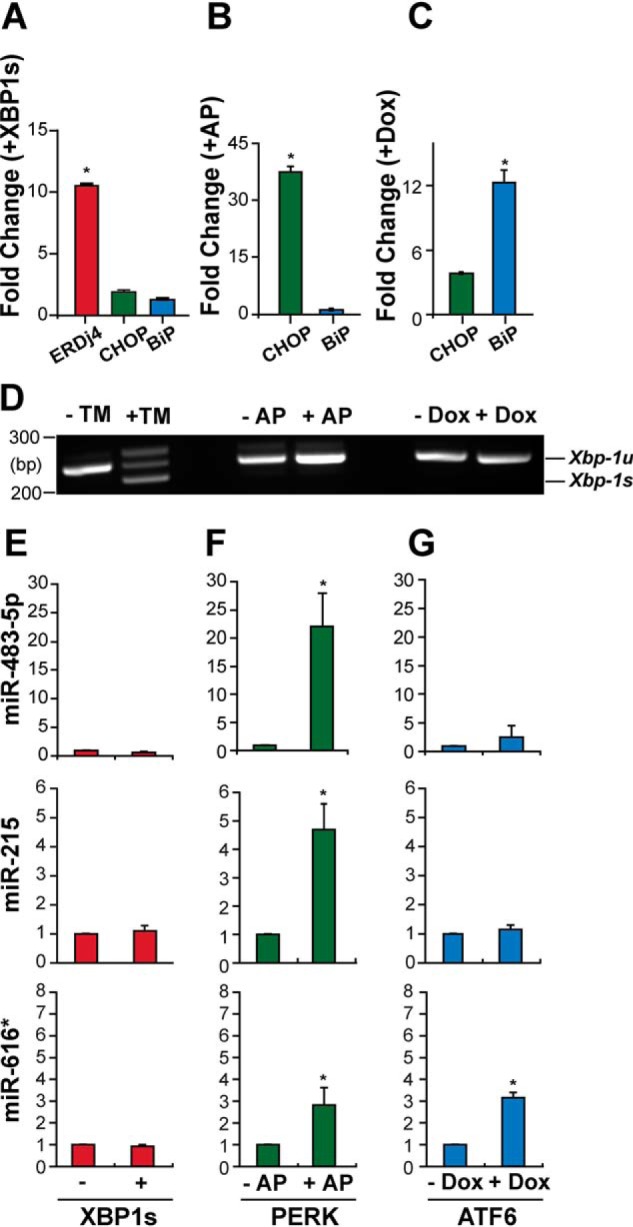
The PERK branch of the UPR selectively induces miR-483-5p, miR-215, and miR-616*. A, HeLa cells were transduced with XBP1s for 72 h. Total RNA was collected, and ERdj4, Chop, and BiP mRNA levels were measured by qRT-PCR and shown relative to GFP-transduced cells. Values are expressed as mean ± S.D. (error bars) of at least three independent experiments. *, p < 0.05, analysis of variance. B, C, and D, HeLa cells expressing Fv2E-PERK or Tet-regulated ATF6(373) were treated with AP20187 (1 nm) or doxycycline (100 μg/ml) for 48 h as indicated. BiP and Chop levels were measured by qRT-PCR, and Xbp-1 mRNA splicing was determined by RT-PCR. As a positive control, in D, Xbp-1 mRNA splicing was determined in cells treated with tunicamycin (5 μg/ml). E, HeLa cells were transduced with XBP1s (+) or GFP (−). After 3 days, miR-483-5p, miR-215, and miR-616* levels were quantified by qPCR and shown relative to GFP-transduced levels. F, HeLa cells stably expressing a chemical dimerizer–inducible Fv2E-PERK were treated with AP20187 (1 nm). After 48 h, miRNA expression levels were quantified by qPCR from AP20187-treated cells (+AP) and shown relative to N,N-dimethylformamide-treated samples (−AP). G, HeLa cells stably expressing a Dox-regulated transcriptional activator domain of ATF6 were treated with 100 μg/ml Dox. After 48 h, miRNA expression levels were quantified by qPCR from untreated cells (−Dox) and shown relative to Dox-treated cells (+Dox). Values are expressed as mean ± S.D. of at least three independent experiments. *, p < 0.05, Student's t test.
Next, we applied these validated XBP1s, Fv2E-PERK, and doxycycline (Dox)-regulated ATF6(373) systems to determine how UPR signaling regulated the three ER stress-induced miRs selected for analysis. We found that XBP1s expression did not induce miR-215, miR-483, and miR-616* (Fig. 2E). By contrast, selective PERK activation using the Fv2E-PERK system up-regulated all three miRs (Fig. 2F). Selective activation of the ATF6 pathway using the Dox-inducible ATF6(373) system up-regulated miR-616* but had no significant effects on the levels of miR-215 or miR-483 (Fig. 2G). These results showed that the PERK signaling pathway was the most potent of the three signaling arms of the UPR for induction of miR-215, miR-483, and miR-616*.
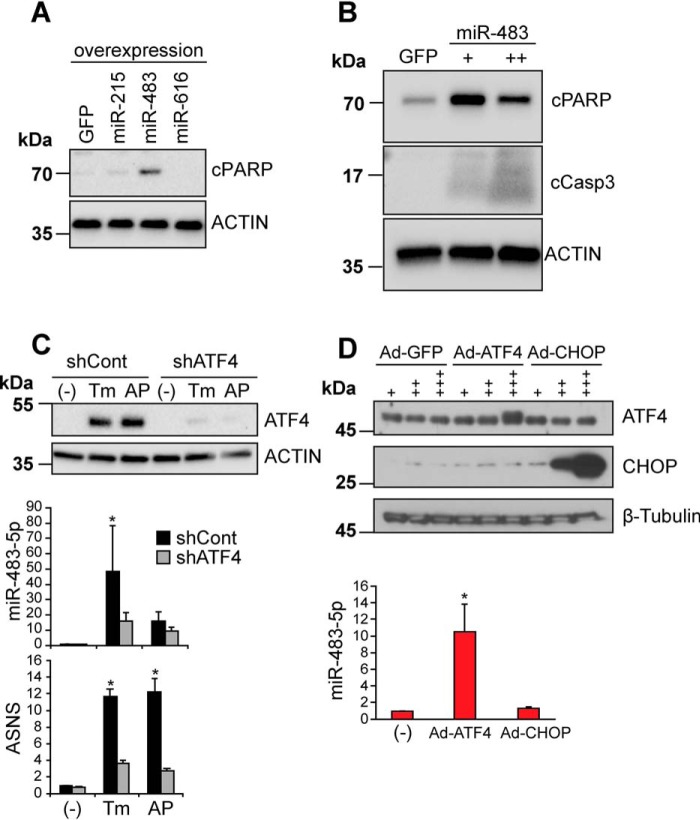
Overexpression of miR-483 promotes cell death
Unabated ER stress or sustained signaling by the PERK arm of the UPR promotes cell death by mechanisms that are poorly understood. We investigated whether the ER stress and PERK-induced expression of miR-215, miR-483, and miR-616* influenced cell survival by overexpressing them in HeLa cells by lentiviral transduction. We used the psiCHECK-2 microRNA Biosensor luciferase reporter assay (Promega) to empirically confirm that these overexpressed miRs were functional by showing silencing of Renilla luciferase and a reduced ratio of Renilla/firefly luciferase luminescence using this assay (Fig. S1). When we examined protein lysates collected after 48 h of miR expression, we found strong production of cleaved PARP and cleaved caspase-3, markers of apoptosis, in cells expressing miR-483 (Fig. 3, A and B). By contrast, we observed little cleaved PARP production with miR-215 or miR-616* expression (Fig. 3A). These findings implicated miR-483 as a mediator of PERK-induced cell death. Because ATF4 and CHOP are also induced by PERK and promote cell death by inducing downstream target genes during the UPR (39, 43, 44), we next examined whether the up-regulation of miR-483 by PERK signaling operated through ATF4 or CHOP. We used adenoviruses to overexpress ATF4 or CHOP, which were previously shown to potently increase levels of functional ATF4 or CHOP proteins (39, 43). We confirmed selective production of ATF4 or CHOP protein in transduced cells, compared with GFP-transduced cells (Fig. 3D). When we transduced cells with ATF4 for 48 h, we found strong induction of miR-483 (Fig. 3D). By contrast, CHOP expression failed to induce miR-483 (Fig. 3D).
Figure 3.
miR-483 expression induces apoptosis and is up-regulated by ATF4. A, HeLa cells were transduced with lentivirus expressing GFP, miR-483-5p, miR-215, or miR-616* for 48 h. Cell lysates were immunoblotted for apoptosis marker cleaved PARP (cPARP) or actin (loading control). B, HeLa cells were transduced with lentivirus expressing GFP, single miR-483 transduction, or double miR-483 transduction for 48 h. Cell lysates were immunoblotted for cPARP, cleaved caspase-3, or actin (loading control). C, HeLa[Fv2E-PERK] cells stably expressing a short hairpin sequence against Atf4 (shATF4) or a scrambled control (shCont) were treated with Tm (5 μg/ml) or AP20187 (1 nm) for 48 h. Cell lysates were immunoblotted for ATF4 or actin (loading control). Total RNA was collected, and miR-483 levels were analyzed by qRT-PCR and shown relative to levels in cells with no drug treatment (−). ASNS is a transcriptional target of ATF4 and was also evaluated by qRT-PCR to confirm reduced ATF4 transcriptional function in shATF4-treated cells. Representative immunoblots are shown, and values are expressed as mean ± S.D. (error bars) of at least three independent experiments. *, p < 0.05 Student's t test. D, top, cells were transduced with adenovirus expressing GFP, ATF4, or Chop at 109 (+), 1010 (++), or 1011 (+++) pfu, and protein lysates were immunoblotted for ATF4 or CHOP. β-Tubulin was used as a loading control. Bottom, HeLa cells were transduced with adenovirus expressing ATF4 or CHOP at 1011 pfu. Total RNA was collected after 48 h, and miR-483 levels were analyzed by qRT-PCR and are shown relative to untransduced controls (−). Values are expressed as mean ± S.D. of at least three independent experiments. *, p < 0.05, Student's t test.
To further test the link between ATF4 and miR-483, we introduced short hairpin RNAs against Atf4 mRNA sequences (shATF4) into the HeLa[Fv2E-PERK] cells. This shATF4 construct was previously demonstrated to robustly knock down ATF4 in HT1080 and DLD1 human cell lines (45). We confirmed that ATF4 production was substantially diminished in our HeLa cells expressing shATF4 compared with control scrambled mRNA hairpin sequences after tunicamycin or AP20187 exposure (Fig. 3C, top). We confirmed that ASNS, a downstream target gene of ATF4, was not normally induced when ATF4 was knocked down (Fig. 3C, bottom) (17, 43). When we measured miR-483 levels under these conditions, we found significantly reduced miR-483 production when ATF4 was knocked down compared with controls. These findings placed ATF4 upstream of miR-483 induction and suggested that ATF4 may directly up-regulate miR-483 transcription.
miR-483 silences creatine kinase brain-type during the UPR
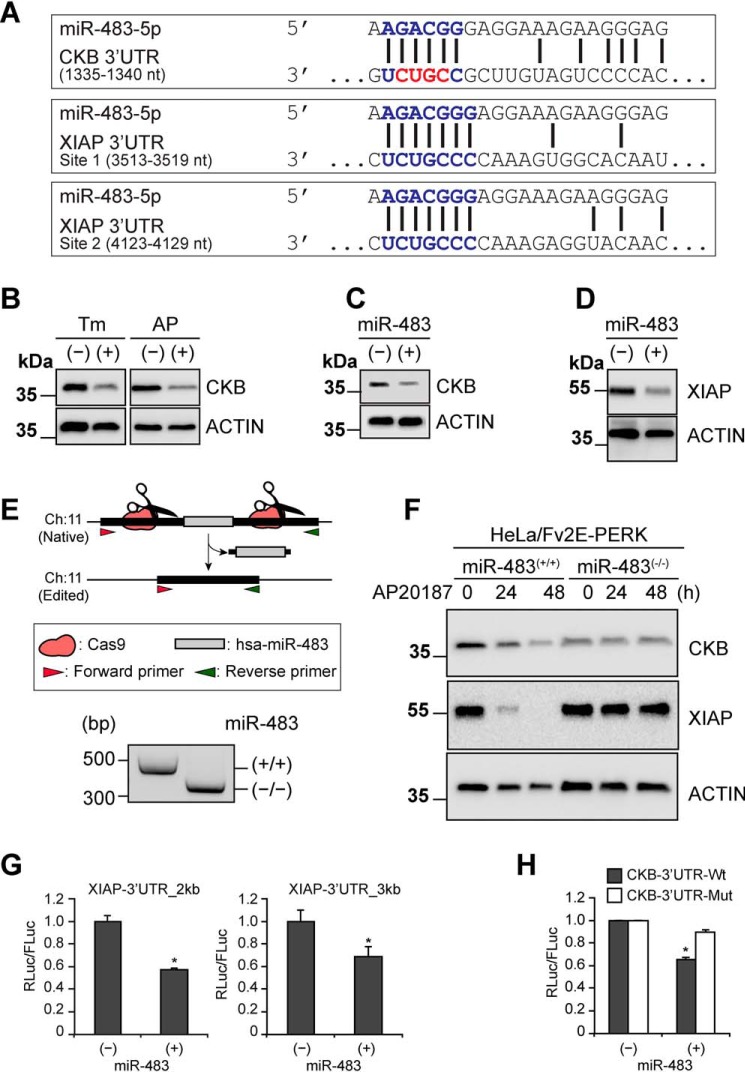
To explore the mechanism by which miR-483 caused cell death during the UPR, we identified potential mRNA targets based upon the presence of bioinformatically predicted miR-483–binding sites within the 3′-UTR of mRNAs using the TargetScan algorithm (http://www.targetscan.org/vert_72/) (81).5 We focused on two predicted miR-483 target mRNAs that encoded for proteins with functions required for cell homeostasis and whose loss could increase cell death upon miRNA-483–mediated gene silencing. The creatine kinase brain-type (CKB) mRNA contained a miR-483 binding site in its 3′-UTR that was conserved across mammalian CKB mRNAs (Fig. 4A). CKB catalyzes the production of phosphocreatine to ensure that cells have sufficient ATP for cellular energy demands. Recent experiments demonstrated that miR-483 overexpression suppressed tumor liver metastasis by down-regulating CKB in the hepatic microenvironment of experimental animal models (34). Another mRNA encoding X-linked inhibitor of apoptosis (XIAP) also contained multiple putative miR-483–binding sites in its 3′-UTR (Fig. 4A), although these were not conserved in Xiap mRNAs from other species. XIAP prevents cell death by directly inhibiting and promoting the degradation of caspase enzymes (46). We previously demonstrated that XIAP protein levels fall in response to ER stress and PERK activation (39). These observations suggested that miR-483 impaired cell viability via two different mechanisms: 1) suppression of XIAP to release the brake against caspases and 2) suppression of CKB to disrupt cellular ATP stores.
Figure 4.
miR-483 targets creatine kinase brain-type (CKB) mRNA and down-regulates XIAP. A, predicted miR-483–binding sites in the 3′-UTR of CKB and XIAP. The nucleotides mutated in the CKB luciferase reporter are highlighted in red. B, HeLa[Fv2E-PERK] cells were treated with Tm (5 μg/ml) or AP20187 (1 nm) for 48 h), and CKB and actin (loading control) were detected by immunoblotting. C and D, cells were transduced with miR-483 or scrambled control, and CKB, XIAP, or actin (loading control) was detected by immunoblotting. E, CRISPR-Cas9 deletion of miR-483 in HeLa[Fv2E-PERK] cells. Two guide RNAs were designed from chromosome 11 sequences flanking miR-483 and inserted into the PX330 CRISPR vector. HeLa[Fv2E-PERK] cells were transfected, and puromycin-resistant colonies were subcloned. Individual clones were genotyped for deletion of the miR-483 region by PCR using forward and reverse primers flanking the deleted region. Correctly edited clones generated a ∼330-bp PCR product compared with a ∼480-bp PCR amplicon in clones retaining the miR-483 region. A representative gel is shown. F, HeLa[Fv2E-PERK] parental cells or cells edited for miR-483 were treated with AP20187 (1 nm) for the indicated durations. CKB, XIAP, and actin (loading control) levels were detected by immunoblotting. G, miR-483 was transfected into cells expressing firefly luciferase (FLuc) or Renilla luciferase (RLuc) fused to Xiap mRNA 3′-UTR lacking all predicted miR-483–binding sites (Xiap-3′UTR-2kb) or bearing intact miR-483–binding sites (Xiap-3′UTR-3kb). RLuc/FLuc activity was measured by luminometer and shown relative to untransfected samples. H, miR-483 was transfected into cells expressing FLuc or RLuc fused to WT CKB mRNA 3′-UTR (CKB-3′UTR-Wt) or mutated at the miR-483–binding site (CKB-3′UTR-Mut). RLuc/FLuc activity was measured by luminometer and shown relative to untransduced samples. For G and H, values are expressed as mean ± S.D. (error bars) of at least three independent experiments. *, p < 0.05, Student's t test.
In support of this model, we saw clear drops in XIAP and CKB protein levels after miR-483 expression (Fig. 4, C and D) comparable with the drops in XIAP and CKB levels seen after tunicamycin treatment or selective PERK activation (Fig. 4B) (39). To further test miR-483's role in suppressing XIAP and CKB, we used CRISPR-Cas9 gene editing to delete miR-483 from HeLa[Fv2E-PERK] cells (Fig. 4E). Then we selectively activated the PERK pathway using AP20187. In HeLa[Fv2E-PERK] cells retaining miR-483, we found a progressive reduction in XIAP and CKB levels upon AP20187 treatment, but this was abolished in cells deleted for miR-483 (Fig. 4F). These findings provided experimental evidence linking miR-483 expression to down-regulation of XIAP and CKB protein levels.
To determine whether miR-483 directly silenced the Xiap or CKB mRNAs through putative binding sites in the 3′-UTR, we used Renilla luciferase mRNA reporters fused to the 3′-UTRs of Xiap or CKB lacking or mutated at the miR-483 binding sites (34). Next, we expressed miR-483 in cells bearing these luciferase 3′-UTR reporters. We found that Renilla luciferase fused to the Xiap 3′-UTR lacking both putative miR-483–binding sites was still suppressed by miR-483 (Fig. 4G). By contrast, mutation of the miR-483–binding site in the 3′-UTR of CKB rendered the Renilla luciferase-CKB-3′-UTR reporter construct insensitive to miR-483 suppression (Fig. 4H). These findings provided evidence that miR-483 directly targeted the CKB mRNA to silence its expression, whereas it indirectly down-regulated XIAP. Our findings indicate that miR-483 directly silences CKB and indirectly gene-silences XIAP after induction by PERK signaling.
ER stress suppresses CKB to deplete cellular ATP stores
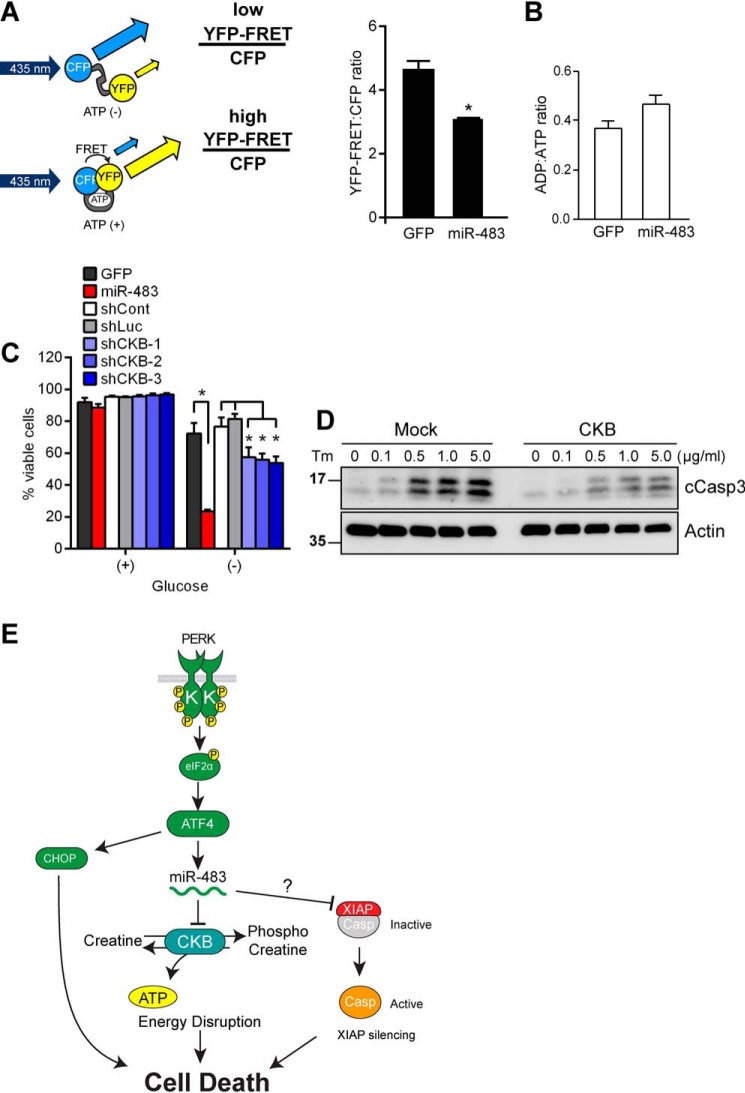
To explore how cellular ATP levels changed during the UPR, we created HeLa cells bearing a fluorescence-based intracellular reporter of ATP concentrations, ATeam (adenosine 5′-triphosphate indicator based on ϵ subunit for analytical measurements) (47). In this reporter, a cyan fluorescent protein (CFP) and a yellow fluorescent protein (YFP) are linked by the small ATP-binding ϵ subunit of a bacterial F0F1-ATP synthase (Fig. 5A). When ATP is absent, the ϵ subunit linker is distended, allowing for distinct CFP fluorescence (Fig. 5A). Upon ATP binding to the ϵ subunit, the linker contracts to bring the CFP and YFP in close proximity, thereby enabling excitation of acceptor YFP by donor CFP (YFP-FRET signal) (Fig. 5A). Prior studies had demonstrated that the ATeam construct enabled sensitive real-time identification of HeLa cells with depleted ATP levels by quantification of the YFP-FRET/CFP ratio in live cells (47).
Figure 5.
CKB suppression during the UPR depletes cellular ATP stores. A, the YFP-FRET/CFP ratio was measured by flow cytometry in HeLa cells stably expressing the ATeam reporter and transduced with lentivirus expressing GFP or miR-483 for 24 h. Values are expressed as mean ± S.D. (error bars) of at least three independent experiments. *, p < 0.05, Student's t test. B, HeLa cells were transduced with lentivirus expressing GFP or miR-483 for 24 h. Cell lysates were collected, and the ADP/ATP ratio was measured using the ADP/ATP ratio assay kit (MAK135, Sigma), Values are expressed as mean ± S.D. of at least three independent replicates. p > 0.05, Student's t test. C, HeLa cells transduced with lentivirus expressing GFP, miR-483, control shRNAs (scrambled or anti-luciferase sequences), or shRNAs against CKB were incubated in standard or glucose-free medium for 8 h. Total and viable cell counts were performed using trypan blue exclusion. Values are expressed as mean ± S.D. of at least three independent experiments. *, p < 0.05, two-way analysis of variance and Tukey's test for multiple comparisons. D, HeLa cells were transfected with CKB and treated with Tm for 72 h at the indicated concentrations. Protein lysates were immunoblotted for cleaved caspase-3 (cCasp3) or actin (loading control). E, model of disruption of intracellular ATP homeostasis through PERK-mediated induction of miR-483 in response to ER stress.
We transduced HeLa cells stably expressing the ATeam reporter with miR-483 or GFP. We found a substantial decrease in the YFP/CFP ratio in cells expressing miR-483 (Fig. 5A). Consistent with our live-cell measurements, we also saw a trending decrease in ATP levels using an enzymatic assay of lysates prepared from cells expressing miR-483 (Fig. 5B). Additionally, following glucose starvation, miR-483–transduced cells exhibited decreased cell viability compared with GFP-transduced control cells, whereas CKB shRNA-transduced cells were also less viable than cells expressing control shRNA sequences (Fig. 5C). These findings suggested that miR-483–dependent silencing of CKB leads to cellular ATP depletion with deleterious consequences for cell viability.
To further determine how CKB protein levels impacted cell survival during ER stress, we tested whether restoration of CKB could prevent ER stress-induced cell death. We transfected cells with CKB and exposed them to increasing doses of tunicamycin. We confirmed increased CKB protein levels by immunoblotting in transfected cells compared with mock-treated cells (Fig. S2). After 72 h, we saw increased cell death morphologically and biochemically with increased production of cleaved caspase-3 on immunoblotting, and this was partially attenuated in cells overexpressing CKB (Fig. 5D). These results provide evidence that CKB played a cytoprotective role during the UPR, and its down-regulation by PERK's induction of miR-483 increased cellular vulnerability to ER stress–induced cell death.
Discussion
Unchecked ER stress leads to cell death, and dysregulated PERK activity can drive this process (18, 19, 43, 48). In this study, we identified novel signal transduction steps by which PERK signaling impairs cell viability (Fig. 5E). First, we found that the PERK arm of the UPR selectively up-regulated miR-483 through the actions of the ATF4 transcription factor. Then we demonstrated that miR-483 silenced two genes regulating vital cellular processes: XIAP and CKB. miR-483 directly targeted the 3′-UTR of CKB, leading to its silencing, consistent with prior report of this interaction in metastatic cancer cells (34). Next, we provided evidence that miR-483 overexpression depleted cellular ATP stores. Based on these findings, we propose that dysregulated PERK signaling disrupts ATP homeostasis and metabolically weakens the cell. Coupled with other maladaptive changes induced by extended UPR signaling, the cell readily succumbs to cell death in the face of continued ER stress.
Our identification of miR-483 as part of the PERK signaling cascade adds to the growing number of genes induced by the PERK arm of the UPR that elicit maladaptive cellular effects. The best-characterized example is CHOP. CHOP's proapoptotic properties were initially noted because of the enhanced ability of Chop−/− cells to survive chemically induced ER stress in vitro (44) and were later reaffirmed, in vivo, by the partial amelioration of pathology and improved tissue function when Chop was deleted in some mouse models of diseases associated with ER stress (49–53). CHOP's transcriptional program imposes protein-folding demands on the ER and disrupts the luminal ER redox environment during the UPR (43, 54), and in some cell types, CHOP directly induces cell death executioners (48, 55, 56). However, CHOP expression does not trigger cell death, and Chop deletion does not change outcomes in many other ER stress disease models (39, 43, 57–60). By contrast, ATF4 can mediate ER stress-induced cell death both in vitro and in vivo (61–65). ATF4 can biochemically partner with CHOP via their bZIP domains and shares many transcriptional targets with CHOP (7, 43, 66). ATF4 also engages apoptotic pathways by stimulating TRAIL death receptors and suppressing anti-apoptotic IAP molecules (39, 67). These findings implicate both ATF4 and CHOP as PERK-induced transcription factors that drive the maladaptive phase of the UPR leading to cell death. Indeed, ATF4 and CHOP co-expression caused increased protein expression, increased oxidative stress, reduced ATP/ADP ratios, and more cell death (43).
This reduction of ATP during the UPR was previously proposed to be a consequence of increased energy consumption arising from ATF4-CHOP–driven protein expression. Our current study provides a direct mechanism into how ATP is depleted during the UPR, through our identification of an ER stress– and PERK–induced miR-483 that directly silences the CKB enzyme required for maintaining cellular ATP stores. ATF4 promotes the loss of ATP via miR-483 induction and suppression of phosphocreatine generation, but ATF4 also concomitantly promotes new protein synthesis by activation of the ATF4-CHOP transcriptional program (43). Clearly, increased energy demand in the face of falling energy supplies is unsustainable for the cell. Reducing energy demands and restoring ATP levels could be accomplished if ATF4, CHOP, and miR-483 levels returned to baseline. However, in situations with persistent ER stress that lead to dysregulated PERK signaling, sustained induction of ATF4, CHOP, and miR-483 could inexorably worsen imbalances in cellular energetics and ultimately culminate in an inviable metabolic environment where apoptosis is triggered. This imbalance in cellular energetics created by dysregulated PERK signaling is an important determinant in the decision to undergo cell death in response to chronic ER stress.
miR-483 joins the growing list of small RNAs regulated by PERK signaling (24–26, 30, 31, 68). Interestingly, many of these PERK-modulated miRs also influenced cell survival in response to ER stress. For instance, miR-211 is induced rapidly after ER stress by the PERK arm of the UPR, but miR-211 levels decline with extended ER stress (24). miR-211's targets include Chop and circadian cycle regulators, and silencing of these genes promotes cell survival (24, 68). Induction of miR-211 could contribute to the protective effects of PERK activation in response to acute ER stress. By contrast, the PERK-regulated miR-106b-25 cluster targets the Bim Bcl-2 family pro-apoptosis factor, and extended experimental PERK signaling (>24 h) down-regulated miR-106b-25, leading to increased Bim protein (26). Loss of miR-106b-25 and ensuing increase in BIM is another maladaptive consequence of extended PERK signaling. In our studies, we identified miR-483 induction in cells exposed to extended experimental ER stress or PERK activation (>24 h). miR-483–mediated silencing of CKB operates at similar time points as the loss of miR-106b-25 silencing of Bim. Together, these distinct maladaptive effects of extended PERK signaling—loss of CKB and rise of BIM—influence the final cellular decision to undergo cell death. These studies suggest that miRs are dynamically regulated in response to acute and extended PERK signaling and help shape the protective or maladaptive effects on the cell.
miR-483 has been readily detected in blood, ascites, and pleural fluid samples from patients with a wide variety of cancers. miR-483-5p was significantly overexpressed in plasma from both hepatocellular carcinoma and chronic lymphocytic leukemia/lymphoma patients (69, 70). miR-483-5p was also up-regulated in circulating blood samples from patients with adrenocortical carcinoma, and miR-483-5p overexpression correlated with significantly worse survival and recurrence in this cancer (71–73). Multiple myeloma patients with increased circulating miR-483-5p had significantly worse progression-free survival compared with patients with low plasma levels of miR-483-5p (74). Based on these cancer epidemiologic associations, clinical tests for miR-483-5p are under development for potential use as a minimally invasive biomarker for cancer diagnosis and prognosis (75). A role for ER stress and PERK signaling in up-regulating circulating miR-483-5p in these different cancers has not been examined. However, cancers thrive under many adverse environmental conditions linked to ER stress and UPR activation, including hypoxia, starvation, and oxidative stress (76). Based on our findings, the increased miR-483-5p levels found in these patients could plausibly arise from increased ER stress and PERK activation in these cancers. If future studies of these cancers validate this conjecture, plasma testing of miR-483-5p could be used as a biomarker to quantify the amount of ER stress or the strength of UPR activation in tumors. In turn, these miR-483-5p–positive cancers could be particularly responsive to the growing number of small molecules that modulate the UPR (77–80). In particular, our studies predict that pharmacologic inhibition of the PERK branch of the UPR or the integrated stress response will be potent in driving down ATF4-CHOP-miR-483 levels, whereas agents that enhance phosphorylation of eIF2α will have the opposite effect.
Experimental procedures
Cell culture
HEK293 cells, HeLa cells, Huh7 cells, Raji cells, and their transfectants were maintained in DMEM (Corning Cellgro, Manassas, VA), 4.5 g/liter glucose, supplemented with 10% fetal bovine serum (Corning Cellgro), 100 units of penicillin, 100 μg/ml streptomycin (Invitrogen), and nonessential amino acids (Invitrogen) at 37 °C under 5% CO2. For glucose starvation experiments, cells were cultured in DMEM with 10% fetal bovine serum, 100 units of penicillin, 100 μg/ml streptomycin, and no glucose, sodium pyruvate, or l-glutamine.
TaqMan array miRNA profiling
Total RNA from HEK293 cells treated with and without 5 μg/ml tunicamycin for 24 h was extracted using the mirVana miRNA isolation kit (Ambion, Foster City, CA) according to the manufacturer's instructions. 1 μg of RNA was reverse-transcribed using Megaplex RT Primers (human pool A and B version 2.1, Applied Biosystems) and the TaqMan miRNA reverse transcription kit (Applied Biosystems). Real-time qRT-PCR with human TaqMan array cards (A and B) was performed on Applied Biosystems 7900HT system. Data analysis was performed using DataAssist Software version 2.0 (Applied Biosystems).
qRT-PCR, RT-PCR, and qPCR analyses
For qRT-PCR analysis, total RNA was collected by the RNeasy mini kit (Qiagen, Hilden, Germany), and 1 μg of RNA was used for cDNA synthesis by iScript (Bio-Rad). Diluted cDNA samples were used for real-time PCR in SYBR Green qPCR supermix (Bio-Rad) on a CFX96 thermal cycler (Bio-Rad). For quantitative measurement of BiP transcripts, the following primer pair was used: 5′-CGGGCAAAGATGTCAGGAAAG-3′ (forward) and 3′-TTCTGGACGGGCTTCATAGTAGAC-5′ (reverse). For quantitative measurement of CHOP transcripts, the following primer pair was used: 5′-ACCAAGGGAGAACCAGGAAACG-3′ (forward) and 3′-TCACCATTCGGTCAATCAGAGC-5′ (reverse). For quantitative measurement of ERdj4 transcripts, the following primer pair was used: 5′-CATCAGAGCGCCAAATCAAG-3′ (forward) and 5′-CATCAGAGCGCCAAATCAAG-3′ (reverse). For quantitative measurement of ASNS transcripts, the following primer pair was used: 5′-ATCAGATGAACTTACGCAGGG-3′ and 3′-AGTTCAAGACCATGGGCAG-5′ (reverse). For RT-PCR detection of Xbp-1 mRNA splicing, the following primer pair was used: 5′-TTACGAGAGAAAACTCATGGC-3′ (forward) and 3′-GGGTCCAAGTTGTCCAGAATGC-5′ (reverse).
For qPCR analysis, total RNA, including the miRNA fraction, was prepared using the miRNeasy mini kit (Qiagen), and 1 μg of RNA was subjected to an RT reaction with the miScript II RT kit (Qiagen) and proprietary miR-483-5p, miR-616*, and miR-215 primers (Qiagen miScript Primer Assay). Real-time qPCR for mature miRNAs was carried out by an miScript SYBR Green PCR kit using specific miRNA assay primers obtained from Qiagen on a CFX96 thermal cycler. Expression of the U6 small RNA was used as an internal control (Qiagen miScript primer assay).
Conditional activation of individual UPR pathways
Stable transfectants of HeLa cells with chemical-genetic constructs to regulate the PERK pathway (HeLa/Fv2E-PERK) or ATF6 (HeLa/TO-ATF6(373)) were established previously (39). PERK signaling in HeLa/Fv2E-PERK cells and ATF6 signaling in HeLa/TO-ATF6(373) were specifically activated by AP20187 (ARIAD) or by withdrawing Dox from the culture medium, respectively. To selectively activate the IRE1-XBP1 pathway, enforced expression of spliced XBP1 was introduced by lentiviral transduction of human XBP1s. Lentivirus packaging was performed with the integration-defective lentiviral system to avoid random integration of the transgene (LENTI-Smart NIL, Invivogen, San Diego, CA).
ATP measurements
The ATeam1.03 cDNA (containing the ATeam reporter), a kind gift from Dr. Imamura (47), was cloned into pLVX-AcGFP-Blast vector (Clontech/Takara BioScience, San Diego), where AcGFP cDNA was removed and linearized with XhoI and NotI digestion, and pLVX-ATeam-Blast was constructed. For measurement of intracellular ATP concentration, pATeam-Blast was lentivirally transduced into HeLa/Fv2E-PERK cells, and HeLa/ATeam cells were established. Flow cytometry analysis was performed on a BD FACSCanto at the Flow Cytometry Core at the San Diego Center for AIDS Research; compensation was performed using UltraComp eBeads (Fisher, 501129038) bound to antibodies of the corresponding wavelengths. Dead cells were excluded using LIVE/DEAD Fixable Far Red Stain (Thermo Fisher Scientific, L34973), and singlet discrimination was performed using FSC-H versus FSC-W gating. The CFP, YFP, and YFP acceptor (excited by 435 nm) fluorescent signals from the ATeam reporter were quantified from 10,000 events. Data analysis was done using Flowing Software.
ADP/ATP ratio values in cell lysates were determined using an ADP/ATP ratio assay kit (Sigma-Aldrich, MAK135) as directed.
CRISPR/Cas9 gene editing
To create miR-483–deleted cells, two sgRNAs were designed. Two sets of synthesized DNA oligonucleotides (sense-left, caccgGCATTGCTGTGGGGGAGAGG; antisense-left, aaacCCTCTCCCCCACAGCAATGCc; sense-right, caccgGGCACCACCTAGGAGGCTGG; antisense-right, aaacCCAGCCTCCTAGGTGGTGCCc) were obtained from Valuegene (San Diego, CA). Each set of oligonucleotides was phosphorylated with T4 PNK (New England Biolabs) by incubation for 30 min at 37 °C, annealed in ligation buffer (New England Biolabs) by incubation for 5 min at 95 °C, and declined in temperature to 25 °C at 0.1 °C/s on a thermal cycler. The oligonucleotide reaction mixture was diluted 100× in water and inserted into linearized PX330 (a gift from Feng Zhang) by BbsI with a quick DNA ligation kit (New England Biolabs), and PX330-miR483-Left and PX330-miR483-Right plasmids were constructed. 1 μg of each plasmid was mixed with 0.2 μg of pBabe-puro-empty vector. The plasmid mixture was transfected into HeLa-Fv2E-PERK cells. At 48 h after transfection, culture medium was replaced with fresh DMEM containing 2.5 μg/ml puromycin to select for cells expressing transgene. After puromycin selection, single cells were isolated in 96-well plates. Clones were expanded, and deletion of miR-483 was confirmed by PCR amplification of the genomic DNA flanking the miR-483 gene with the following primer set (forward, GGTGCCAGCCAGTCCTTG; reverse, CCACAACCAGAGGGACACC), and PCR products were resolved on 2.5% agarose gel in 1× TBE buffer.
Viral transduction
Adenoviral transduction for GFP, ATF4, or CHOP was described previously (39).
pLVX-AcGFP-miR-483, -miR-215, and -miR-616 were constructed using Gibson assembly. Briefly, genomic regions harboring each miRNA with ∼200-bp flanking regions were amplified from HEK293's genomic DNA, and purified products were subsequently cloned into pLVX-AcGFP-N1 digested with NotI and XbaI using Gibson assembly. Lentivirus plasmid was premixed with lentivirus-packaging plasmids, psPAX2, and pMD2G, at a 3:2:1 ratio, and co-transfected into 293FT cells with PEI-Max (Polysciences Inc., Fisher). Supernatant containing virus particles was collected at 48 and 72 h after transfection. The LMP-shRNA nonsilencing (shCont) and LMP-shRNA human ATF4 (shATF4) were created previously (39), and retrovirus particles were created in Phoenix-ampho cells as described previously (39). HeLa/Fv2E-PERK cells were retrovirally transduced with shCont or shATF4 and established as HeLa/shCont and HeLa/shATF4 by puromycin selection. pBabe-puro-CKB for enforced expression of human CKB was provided by Dr. Tavazoie (34) and was used for establishment of HeLa/CKB cells.
Luciferase 3′-UTR reporters
The psiCheck2 luciferase reporter assays (Promega, Madison, WI) were performed as described previously (34). The WT and mutant CKB luciferase constructs were provided by Dr. Tavazoie (34). The Xiap 3′-UTR containing putative miR-483–binding sites or truncated to omit both miR-483–binding sites was cloned into the psiCheck2 Dual-Luciferase reporter vector (34). Cells were co-transfected with miRNA expression constructs and luciferase reporter constructs, and Renilla and firefly luciferase activities were determined using a dual-luciferase assay kit per the psiCheck2 manufacturer's instructions (Promega) (34). The psiCheck2 luciferase reporter was used to functionally validate expression of transduced miRs, by cloning synthetic miR-binding sites into the 3′-UTR of Renilla luciferase, and measuring the ratio of Renilla and firefly luciferase activities per the manufacturer's instructions.
Western blot analysis
For Western blot analysis, 10–20 μg of protein was prepared in SDS lysis buffer (62.5 mm Tris-HCl, pH 6.8, 2% (w/v) SDS, 10% glycerol), mixed with 100 mm DTT in 1× LDS buffer (Invitrogen), and incubated at 70 °C for 5 min prior to loading onto a 4–15% gradient SDS-polyacrylamide gel (Bio-Rad). After SDS-PAGE, samples were transferred to polyvinylidene difluoride membranes (Bio-Rad) in 1× Tris/glycine buffer containing 20% methanol and transferred at 250 mA for 1 h. The membrane was blocked in 5% skim milk in TBS buffer containing 0.1% Tween 20 (TBST) for 1 h at room temperature and was subsequently incubated with primary antibody in 5% BSA in TBST buffer at 4 °C overnight with gentle shaking. After incubation with primary antibody, the membrane was washed in TBST buffer three times at room temperature and then incubated with secondary antibody conjugated with horseradish peroxidase (HRP) for 1 h at room temperature. HRP-catalyzed chemiluminescent signal using SuperSignal West Pico or Femto solution (Pierce) was detected on ChemiDoc (Bio-Rad). Primary antibodies used were as follows: cleaved PARP (Cell Signaling: 9541, 1:1000), ATF4 (Santa Cruz Biotechnology: (C-20) sc-200, 1:1000), ATF4 (Cell Signaling: 11815S, 1:2000), CHOP (Santa Cruz Biotechnology, Inc.: sc-575, 1:1000), CKB (Santa Cruz Biotechnology: sc-15157, 1:1000), cleaved caspase-3 (Cell Signaling: 9664, 1:1000), actin (Millipore: 1:10,000), XIAP (BD Biosciences: 610717, 1:1000), and tubulin (Sigma: T9026, 1:2000). HRP-conjugated anti-mouse and anti-rabbit secondary antibodies were from Cell Signaling (1:3000) or Jackson Laboratory (1:2000).
Author contributions
N. H., M. Z., and J. H. L. conceptualization; N. H., K. C., J. H., L. C., M. Z., E. H. K., and J. H. L. data curation; N. H., K. C., M. Z., and J. H. L. formal analysis; N. H., L. C., M. Z., E. H. K., and J. H. L. funding acquisition; N. H., K. C., J. H., and E. H. K. validation; N. H., K. C., J. J. R., J.-M. L., M. Z., and J. H. L. investigation; N. H. visualization; N. H., K. C., J. J. R., J.-M. L., J. H., and E. H. K. methodology; N. H., K. C., M. Z., and J. H. L. writing-original draft; N. H., M. Z., and J. H. L. project administration; C. A., J. H., M. Z., and J. H. L. writing-review and editing; J. H., M. Z., E. H. K., and J. H. L. resources; L. C. and J. H. L. software; M. Z. and J. H. L. supervision.
Supplementary Material
Acknowledgments
We thank A. Pasquinelli, R. L. Wiseman, and members of the Lin laboratory for helpful suggestions. We thank S. Tavazioe for the CKB plasmids, C. Koumenis for the shATF4 vectors, F. Zhang for the PX330 plasmid, and H. Imamura for the ATeam reporter.
This work was supported by the Genomics Core at the University of California San Diego Center for AIDS Research (Grant AI36214) and the Flow Cytometry Core at the San Diego Center for AIDS Research (Grant P30 AI036214); the Veterans Affairs San Diego Health Care System; the San Diego Veterans Medical Research Foundation; National Institutes of Health Grants R01EY027335, R01NS088485, and P50AG005131; and Veterans Affairs Merit awards I01BX002284 and I01RX002340. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- IRE1
- inositol-requiring enzyme 1
- PERK
- PKR-like endoplasmic reticulum kinase
- ATF4 and ATF6
- activating transcription factor 4 and 6, respectively
- eIF2
- eukaryotic initiation factor 2
- miRNA
- microRNA
- qPCR and qRT-PCR
- quantitative PCR and RT-PCR, respectively
- CHOP
- C/EBP-homologous protein
- CKB
- creatine kinase brain-type
- XIAP
- X-linked inhibitor of apoptosis
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- DMEM
- Dulbecco's modified Eagle's medium
- HRP
- horseradish peroxidase
- Tm
- tunicamycin
- Dox
- doxycycline
- FLuc
- firefly luciferase
- RLuc
- Renilla luciferase
- PARP
- poly(ADP-ribose) polymerase.
References
- 1. Alberts B. (2015) Molecular Biology of the Cell, 6th Ed., Garland Science, Taylor and Francis Group, New York [Google Scholar]
- 2. Hiramatsu N., Chiang W. C., Kurt T. D., Sigurdson C. J., and Lin J. H. (2015) Multiple mechanisms of unfolded protein response-induced cell death. Am. J. Pathol. 185, 1800–1808 10.1016/j.ajpath.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- 4. Harding H. P., Zhang Y., and Ron D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- 5. Wek R. C., Jiang H. Y., and Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 10.1042/BST0340007 [DOI] [PubMed] [Google Scholar]
- 6. Harding H. P., Calfon M., Urano F., Novoa I., and Ron D. (2002) Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18, 575–599 10.1146/annurev.cellbio.18.011402.160624 [DOI] [PubMed] [Google Scholar]
- 7. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., and Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- 8. Starck S. R., Tsai J. C., Chen K., Shodiya M., Wang L., Yahiro K., Martins-Green M., Shastri N., and Walter P. (2016) Translation from the 5′ untranslated region shapes the integrated stress response. Science 351, aad3867 10.1126/science.aad3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vattem K. M., and Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 10.1073/pnas.0400541101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu P. D., Harding H. P., and Ron D. (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 10.1083/jcb.200408003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Young S. K., Palam L. R., Wu C., Sachs M. S., and Wek R. C. (2016) Ribosome elongation stall directs gene-specific translation in the integrated stress response. J. Biol. Chem. 291, 6546–6558 10.1074/jbc.M115.705640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Palam L. R., Baird T. D., and Wek R. C. (2011) Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 10.1074/jbc.M110.216093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hellen C. U., and Sarnow P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612 10.1101/gad.891101 [DOI] [PubMed] [Google Scholar]
- 14. Harding H. P., Zhang Y., Bertolotti A., Zeng H., and Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- 15. Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., and Kaufman R. J. (2007) ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 13, 351–364 10.1016/j.devcel.2007.07.005 [DOI] [PubMed] [Google Scholar]
- 16. Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., and Mori K. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 13, 365–376 10.1016/j.devcel.2007.07.018 [DOI] [PubMed] [Google Scholar]
- 17. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., and Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- 18. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., and Walter P. (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science 318, 944–949 10.1126/science.1146361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin J. H., Li H., Zhang Y., Ron D., and Walter P. (2009) Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One 4, e4170 10.1371/journal.pone.0004170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krol J., Loedige I., and Filipowicz W. (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 21. Mendell J. T., and Olson E. N. (2012) MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maurel M., and Chevet E. (2013) Endoplasmic reticulum stress signaling: the microRNA connection. Am. J. Physiol. Cell Physiol. 304, C1117–C1126 10.1152/ajpcell.00061.2013 [DOI] [PubMed] [Google Scholar]
- 23. Byrd A. E., Aragon I. V., and Brewer J. W. (2012) MicroRNA-30c-2* limits expression of proadaptive factor XBP1 in the unfolded protein response. J. Cell Biol. 196, 689–698 10.1083/jcb.201201077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chitnis N. S., Pytel D., Bobrovnikova-Marjon E., Pant D., Zheng H., Maas N. L., Frederick B., Kushner J. A., Chodosh L. A., Koumenis C., Fuchs S. Y., and Diehl J. A. (2012) miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol. Cell 48, 353–364 10.1016/j.molcel.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gupta A., Hossain M. M., Read D. E., Hetz C., Samali A., and Gupta S. (2015) PERK regulated miR-424(322)-503 cluster fine-tunes activation of IRE1 and ATF6 during unfolded protein response. Sci. Rep. 5, 18304 10.1038/srep18304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gupta S., Read D. E., Deepti A., Cawley K., Gupta A., Oommen D., Verfaillie T., Matus S., Smith M. A., Mott J. L., Agostinis P., Hetz C., and Samali A. (2012) Perk-dependent repression of miR-106b-25 cluster is required for ER stress-induced apoptosis. Cell Death Dis. 3, e333 10.1038/cddis.2012.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahmadi A., Khansarinejad B., Hosseinkhani S., Ghanei M., and Mowla S. J. (2017) miR-199a-5p and miR-495 target GRP78 within UPR pathway of lung cancer. Gene 620, 15–22 10.1016/j.gene.2017.03.032 [DOI] [PubMed] [Google Scholar]
- 28. Jiang L., Zang D., Yi S., Li X., Yang C., Dong X., Zhao C., Lan X., Chen X., Liu S., Liu N., Huang H., Shi X., Wang X., and Liu J. (2016) A microRNA-mediated decrease in eukaryotic initiation factor 2α promotes cell survival during PS-341 treatment. Sci. Rep. 6, 21565 10.1038/srep21565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A., Li D., Hou Z., Lv K., Kan G., Cao H., Wu H., Song J., Pan X., Sun Q., et al. (2013) miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 19, 93–100 10.1038/nm.3026 [DOI] [PubMed] [Google Scholar]
- 30. Behrman S., Acosta-Alvear D., and Walter P. (2011) A CHOP-regulated microRNA controls rhodopsin expression. J. Cell Biol. 192, 919–927 10.1083/jcb.201010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu Z., Bu Y., Chitnis N., Koumenis C., Fuchs S. Y., and Diehl J. A. (2016) miR-216b regulation of c-Jun mediates GADD153/CHOP-dependent apoptosis. Nat. Commun. 7, 11422 10.1038/ncomms11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu J., Sun T., Wang H., Chen Z., Wang S., Yuan L., Liu T., Li H. R., Wang P., Feng Y., Wang Q., McLendon R. E., Friedman A. H., Keir S. T., Bigner D. D., et al. (2016) MiR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell 29, 49–60 10.1016/j.ccell.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones M. F., Hara T., Francis P., Li X. L., Bilke S., Zhu Y., Pineda M., Subramanian M., Bodmer W. F., and Lal A. (2015) The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 112, E1550–E1558 10.1073/pnas.1503370112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loo J. M., Scherl A., Nguyen A., Man F. Y., Weinberg E., Zeng Z., Saltz L., Paty P. B., and Tavazoie S. F. (2015) Extracellular metabolic energetics can promote cancer progression. Cell 160, 393–406 10.1016/j.cell.2014.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., and Ron D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- 36. Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- 37. Shoulders M. D., Ryno L. M., Genereux J. C., Moresco J. J., Tu P. G., Wu C., Yates J. R. 3rd, Su A. I., Kelly J. W., and Wiseman R. L. (2013) Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3, 1279–1292 10.1016/j.celrep.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu P. D., Jousse C., Marciniak S. J., Zhang Y., Novoa I., Scheuner D., Kaufman R. J., Ron D., and Harding H. P. (2004) Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23, 169–179 10.1038/sj.emboj.7600030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hiramatsu N., Messah C., Han J., LaVail M. M., Kaufman R. J., and Lin J. H. (2014) Translational and posttranslational regulation of XIAP by eIF2α and ATF4 promotes ER stress-induced cell death during the unfolded protein response. Mol. Biol. Cell 25, 1411–1420 10.1091/mbc.e13-11-0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chiang W. C., Hiramatsu N., Messah C., Kroeger H., and Lin J. H. (2012) Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest. Ophthalmol. Vis. Sci. 53, 7159–7166 10.1167/iovs.12-10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hiramatsu N., Joseph V. T., and Lin J. H. (2011) Monitoring and manipulating mammalian unfolded protein response. Methods Enzymol. 491, 183–198 10.1016/B978-0-12-385928-0.00011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okada T., Yoshida H., Akazawa R., Negishi M., and Mori K. (2002) Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366, 585–594 10.1042/bj20020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., and Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., and Ron D. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ye J., Kumanova M., Hart L. S., Sloane K., Zhang H., De Panis D. N., Bobrovnikova-Marjon E., Diehl J. A., Ron D., and Koumenis C. (2010) The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29, 2082–2096 10.1038/emboj.2010.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salvesen G. S., and Duckett C. S. (2002) IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3, 401–410 10.1038/nrm830 [DOI] [PubMed] [Google Scholar]
- 47. Imamura H., Nhat K. P., Togawa H., Saito K., Iino R., Kato-Yamada Y., Nagai T., and Noji H. (2009) Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U.S.A. 106, 15651–15656 10.1073/pnas.0904764106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu M., Lawrence D. A., Marsters S., Acosta-Alvear D., Kimmig P., Mendez A. S., Paton A. W., Paton J. C., Walter P., and Ashkenazi A. (2014) Cell death: opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 345, 98–101 10.1126/science.1254312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oyadomari S., Koizumi A., Takeda K., Gotoh T., Akira S., Araki E., and Mori M. (2002) Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Invest. 109, 525–532 10.1172/JCI0214550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Silva R. M., Ries V., Oo T. F., Yarygina O., Jackson-Lewis V., Ryu E. J., Lu P. D., Marciniak S. J., Ron D., Przedborski S., Kholodilov N., Greene L. A., and Burke R. E. (2005) CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. J. Neurochem. 95, 974–986 10.1111/j.1471-4159.2005.03428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pennuto M., Tinelli E., Malaguti M., Del Carro U., D'Antonio M., Ron D., Quattrini A., Feltri M. L., and Wrabetz L. (2008) Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot-Marie-Tooth 1B mice. Neuron 57, 393–405 10.1016/j.neuron.2007.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu J., Ruas J. L., Estall J. L., Rasbach K. A., Choi J. H., Ye L., Boström P., Tyra H. M., Crawford R. W., Campbell K. P., Rutkowski D. T., Kaufman R. J., and Spiegelman B. M. (2011) The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1α/ATF6α complex. Cell Metab. 13, 160–169 10.1016/j.cmet.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu Y., Park K. K., Yang L., Wei X., Yang Q., Cho K. S., Thielen P., Lee A. H., Cartoni R., Glimcher L. H., Chen D. F., and He Z. (2012) Differential effects of unfolded protein response pathways on axon injury-induced death of retinal ganglion cells. Neuron 73, 445–452 10.1016/j.neuron.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., and Ron D. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Puthalakath H., O'Reilly L. A., Gunn P., Lee L., Kelly P. N., Huntington N. D., Hughes P. D., Michalak E. M., McKimm-Breschkin J., Motoyama N., Gotoh T., Akira S., Bouillet P., and Strasser A. (2007) ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129, 1337–1349 10.1016/j.cell.2007.04.027 [DOI] [PubMed] [Google Scholar]
- 56. Teske B. F., Fusakio M. E., Zhou D., Shan J., McClintick J. N., Kilberg M. S., and Wek R. C. (2013) CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell 24, 2477–2490 10.1091/mbc.e13-01-0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nashine S., Bhootada Y., Lewin A. S., and Gorbatyuk M. (2013) Ablation of C/EBP homologous protein does not protect T17M RHO mice from retinal degeneration. PLoS One 8, e63205 10.1371/journal.pone.0063205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adekeye A., Haeri M., Solessio E., and Knox B. E. (2014) Ablation of the proapoptotic genes chop or ask1 does not prevent or delay loss of visual function in a P23H transgenic mouse model of retinitis pigmentosa. PLoS One 9, e83871 10.1371/journal.pone.0083871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chiang W. C., Kroeger H., Sakami S., Messah C., Yasumura D., Matthes M. T., Coppinger J. A., Palczewski K., LaVail M. M., and Lin J. H. (2015) Robust endoplasmic reticulum-associated degradation of rhodopsin precedes retinal degeneration. Mol. Neurobiol. 52, 679–695 10.1007/s12035-014-8881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chiang W. C., Joseph V., Yasumura D., Matthes M. T., Lewin A. S., Gorbatyuk M. S., Ahern K., LaVail M. M., and Lin J. H. (2016) Ablation of Chop transiently enhances photoreceptor survival but does not prevent retinal degeneration in transgenic mice expressing human P23H rhodopsin. Adv. Exp. Med. Biol. 854, 185–191 10.1007/978-3-319-17121-0_25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lange P. S., Chavez J. C., Pinto J. T., Coppola G., Sun C. W., Townes T. M., Geschwind D. H., and Ratan R. R. (2008) ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J. Exp. Med. 205, 1227–1242 10.1084/jem.20071460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baleriola J., Walker C. A., Jean Y. Y., Crary J. F., Troy C. M., Nagy P. L., and Hengst U. (2014) Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell 158, 1159–1172 10.1016/j.cell.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bhootada Y., Kotla P., Zolotukhin S., Gorbatyuk O., Bebok Z., Athar M., and Gorbatyuk M. (2016) Limited ATF4 expression in degenerating retinas with ongoing ER stress promotes photoreceptor survival in a mouse model of autosomal dominant retinitis pigmentosa. PLoS One 11, e0154779 10.1371/journal.pone.0154779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kline C. L., Van den Heuvel A. P., Allen J. E., Prabhu V. V., Dicker D. T., and El-Deiry W. S. (2016) ONC201 kills solid tumor cells by triggering an integrated stress response dependent on ATF4 activation by specific eIF2α kinases. Sci. Signal. 9, ra18 10.1126/scisignal.aac4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ishizawa J., Kojima K., Chachad D., Ruvolo P., Ruvolo V., Jacamo R. O., Borthakur G., Mu H., Zeng Z., Tabe Y., Allen J. E., Wang Z., Ma W., Lee H. C., Orlowski R., et al. (2016) ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci. Signal. 9, ra17 10.1126/scisignal.aac4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Newman J. R., and Keating A. E. (2003) Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300, 2097–2101 10.1126/science.1084648 [DOI] [PubMed] [Google Scholar]
- 67. Iurlaro R., Püschel F., León-Annicchiarico C. L., O'Connor H., Martin S. J., Palou-Gramón D., Lucendo E., and Muñoz-Pinedo C. (2017) Glucose deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol. Cell Biol. 37, e00479–16 10.1128/MCB.00479-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bu Y., Yoshida A., Chitnis N., Altman B. J., Tameire F., Oran A., Gennaro V., Armeson K. E., McMahon S. B., Wertheim G. B., Dang C. V., Ruggero D., Koumenis C., Fuchs S. Y., and Diehl J. A. (2018) A PERK-miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat. Cell Biol. 20, 104–115 10.1038/s41556-017-0006-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moussay E., Wang K., Cho J. H., van Moer K., Pierson S., Paggetti J., Nazarov P. V., Palissot V., Hood L. E., Berchem G., and Galas D. J. (2011) MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 108, 6573–6578 10.1073/pnas.1019557108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shen J., Wang A., Wang Q., Gurvich I., Siegel A. B., Remotti H., and Santella R. M. (2013) Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: miR-483-5p as a potential biomarker. Cancer Epidemiol. Biomarkers Prev. 22, 2364–2373 10.1158/1055-9965.EPI-13-0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Soon P. S., Tacon L. J., Gill A. J., Bambach C. P., Sywak M. S., Campbell P. R., Yeh M. W., Wong S. G., Clifton-Bligh R. J., Robinson B. G., and Sidhu S. B. (2009) miR-195 and miR-483-5p identified as predictors of poor prognosis in adrenocortical cancer. Clin. Cancer Res. 15, 7684–7692 10.1158/1078-0432.CCR-09-1587 [DOI] [PubMed] [Google Scholar]
- 72. Patterson E. E., Holloway A. K., Weng J., Fojo T., and Kebebew E. (2011) MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer 117, 1630–1639 10.1002/cncr.25724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chabre O., Libé R., Assie G., Barreau O., Bertherat J., Bertagna X., Feige J. J., and Cherradi N. (2013) Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr. Relat. Cancer 20, 579–594 10.1530/ERC-13-0051 [DOI] [PubMed] [Google Scholar]
- 74. Qu X., Zhao M., Wu S., Yu W., Xu J., Xu J., Li J., and Chen L. (2014) Circulating microRNA 483-5p as a novel biomarker for diagnosis survival prediction in multiple myeloma. Med. Oncol. 31, 219 10.1007/s12032-014-0219-x [DOI] [PubMed] [Google Scholar]
- 75. Perge P., Butz H., Pezzani R., Bancos I., Nagy Z., Pálóczi K., Nyírőo G., Decmann Á., Pap E., Luconi M., Mannelli M., Buzás E. I., Tóth M., Boscaro M., Patócs A., and Igaz P. (2017) Evaluation and diagnostic potential of circulating extracellular vesicle-associated microRNAs in adrenocortical tumors. Sci. Rep. 7, 5474 10.1038/s41598-017-05777-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cubillos-Ruiz J. R., Bettigole S. E., and Glimcher L. H. (2017) Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell 168, 692–706 10.1016/j.cell.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sidrauski C., Acosta-Alvear D., Khoutorsky A., Vedantham P., Hearn B. R., Li H., Gamache K., Gallagher C. M., Ang K. K., Wilson C., Okreglak V., Ashkenazi A., Hann B., Nader K., Arkin M. R., et al. (2013) Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498 10.7554/eLife.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Axten J. M., Medina J. R., Feng Y., Shu A., Romeril S. P., Grant S. W., Li W. H., Heerding D. A., Minthorn E., Mencken T., Atkins C., Liu Q., Rabindran S., Kumar R., Hong X., et al. (2012) Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J. Med. Chem. 55, 7193–7207 10.1021/jm300713s [DOI] [PubMed] [Google Scholar]
- 79. Das I., Krzyzosiak A., Schneider K., Wrabetz L., D'Antonio M., Barry N., Sigurdardottir A., and Bertolotti A. (2015) Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348, 239–242 10.1126/science.aaa4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tsaytler P., Harding H. P., Ron D., and Bertolotti A. (2011) Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332, 91–94 10.1126/science.1201396 [DOI] [PubMed] [Google Scholar]
- 81. Agarwal V., Bell G. W., Nam J. W., and Bartel D. P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.