Abstract
Small RNAs (smRNAs) are important regulators of many biologic processes and are now most frequently characterized using Illumina sequencing. However, although standard RNA sequencing library preparation has become routine in most sequencing facilities, smRNA sequencing library preparation has historically been challenging because of high input requirements, laborious protocols involving gel purifications, inability to automate, and a lack of benchmarking standards. Additionally, studies have suggested that many of these methods are nonlinear and do not accurately reflect the amounts of smRNAs in vivo. Recently, a number of new kits have become available that permit lower input amounts and less laborious, gel-free protocol options. Several of these new kits claim to reduce RNA ligase-dependent sequence bias through novel adapter modifications and to lessen adapter-dimer contamination in the resulting libraries. With the increasing number of smRNA kits available, understanding the relative strengths of each method is crucial for appropriate experimental design. In this study, we systematically compared 9 commercially available smRNA library preparation kits as well as NanoString probe hybridization across multiple study sites. Although several of the new methodologies do reduce the amount of artificially over- and underrepresented microRNAs (miRNAs), we observed that none of the methods was able to remove all of the bias in the library preparation. Identical samples prepared with different methods show highly varied levels of different miRNAs. Even so, many methods excelled in ease of use, lower input requirement, fraction of usable reads, and reproducibility across sites. These differences may help users select the most appropriate methods for their specific question of interest.
Keywords: small RNA sequencing, miRNA sequencing, RNA sequencing, Illumina Library Prep
INTRODUCTION
Small RNAs (smRNAs) have been identified as important regulators of many cellular processes.1–3 They can be divided into many different subtypes with different functions including microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), enhancer RNAs, etc. Many smRNAs were initially characterized by Northern blot, a method that has been largely replaced by quantitative PCR (qPCR) and Illumina (San Diego, CA, USA) sequencing. These newer methods have been a significant advance because they allow for increased resolution of nearly identical smRNA species and for the discovery of novel smRNAs. These methods can also be used to identify synthetic molecules such as small interfering RNAs and clustered regularly interspaced short palindromic repeats (CRISPR) guide RNAs, which has increased the importance of these methods.
Because of their small size, smRNAs are poorly characterized by classic RNA sequencing methodologies that use random priming; instead, they require specialized protocols for library preparation. These specialized methods originally were based on sequential ligation of defined adapter oligos to the 3′ and 5′ end of the molecules using RNA ligases followed by amplification with the defined sequences. These methods were extremely labor-intensive, involving several gel purification steps. Furthermore, several studies have suggested that both sequencing and qPCR based methodologies are biased; some smRNAs are substantially underrepresented, and others are highly overrepresented—a phenomenom called “jackpotting,” which can result from adapter ligation bias or PCR amplification bias.2, 4–7 Although the results are highly reproducible within a single method, their absolute quantification can be off by orders of magnitude, limiting their utility to relative quantification. New methods using circularization or polyadenylation followed by template switching have recently become commercially available, and hybridization methods have improved in their sensitivity and accuracy. In addition, innovative chemistries allow lower input amount as well as less laborious gel-free protocol options. In addition, novel adapter modifications have been introduced in an attempt to reduce RNA ligase-dependent sequence bias and to lessen adapter-dimer contamination in the resulting sequencing libraries.8, 9
In this study, we systematically compared 9 commercial smRNA library preparation kits across multiple study sites. In addition, NanoString (Seattle, WA, USA), a PCR free probe hybridization technology, was included as an orthogonal approach for smRNA detection. Kits were evaluated on the diversity of library composition, linearity of detection, and ease of use. We used synthetic equimolar miRNAs both alone and in the context of RNA from a Dicer knockout (Dicer−/−) cell line,10 as well as human brain reference (HBR) RNA. We observed that, although several methodologies were able to eliminate jackpotting of specific miRNAs (that is, no miRNAs >10× over median), no methods completely addressed the issue of bias: all kits continue to show that at least 50% of miRNAs are observed at over 2-fold from the median. Several of the newer methods did reduce the input amount required and streamlined the protocol. In addition, the kits differed significantly in their ability to detect different types of smRNAs and in the frequency with which they observed nonbiological reads (such as primer dimers). These results, coupled with other recent comparisons of smRNA library preparation kits,6, 7 can help researchers determine the most appropriate methodologies for their own studies.
MATERIALS AND METHODS
Study design
Sixteen genomics core facilities were selected from the Association of Biomedical Research Facilities membership. All of the participating cores routinely perform smRNA or miRNA library preparation for laboratories at their institutes. Each site prepared between 1 and 3 library types. Kits were assigned to each site to minimize the overlap of methods, and sites were not selected based on prior experience with specific chemistries. Each smRNA kit was tested independently at 4 separate sites using a standardized protocol discussed and approved by the manufacturer on a phone call or web conference prior to execution as described below. For each kit at each site, samples were prepared in technical duplicate to assess the technical variability of the method. Indices were assigned by the group to prevent overlapping among libraries. All vendor-supplied reagents were shipped directly to the test site to minimize upstream handling and prevent unnecessary freeze-thaw cycles. The total RNA samples used in this study were prepared at a centralized location, divided into aliquots, and shipped overnight on dry ice to each test site. Upon completion, each site quantified each library by fluorometry and Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) or a similar method, and libraries were sent to a single location for normalizing, pooling, and sequencing. Library pools were multiplexed and sequenced over 2 single-end, 75-bp runs on an Illumina NextSeq500 to achieve a target read depth of 5–10 million reads for analysis. Data were distributed to designated sites for downstream processing.
Sample preparation
miRXplore Universal Reference
miRXplore Universal Reference (MUR) was obtained from Miltenyi Biotec (130-093-521; Bergisch Gladbach, Germany) and was prepared following the manufacturer’s instructions to create a stock solution containing 5 fmol of each miRNA. This stock solution was used as input to the library preparation methods described below.
MUR + Dicer −/− RNA
RNA was isolated from Dicer−/− mesenchymal stem cells (CRL-3221; American Type Culture Collection, Manassas, VA, USA),10 a generous donation from Dr. Phil Sharp’s laboratory at Massachusetts Institute of Technology and the Koch Institute for Integrative Cancer Research. Cells were stored at −80°C until further processing. MUR plus Dicer−/− (MUR-D) samples were prepared by adding 1 μl of a 1:200 dilution of MUR RNA per 1 μg of Dicer−/− RNA. With this ratio, miRNAs represent about 2% of the total RNA pool, well within the range typically observed in cells.
HBR
This was obtained from Thermo Fisher Scientific (AM7962; Waltham, MA, USA), prepared according to the manufacturer’s instructions, and divided into aliquots for shipment to customer site prior to storage at −80°C.
Library preparation
Nine library preparation kits were evaluated. Based on library construction biochemistry, there were 3 technology groups: sequential ligation, template switching, and circularization. In addition, ligation plus probe hybridization was also evaluated.
Sequential ligation
Illumina TruSeq Small RNA Library Prep Kit (protocol 15004197 v.02; Illumina)
The amount of input RNA was 1 μg for HBR, 1 μg for MUR-D, and 100 pg for MUR. User-supplied reagents including T4 ligase (Epicentre, Madison, WI, USA) and Superscript III reverse transcriptase (Thermo Fisher Scientific) were purchased separately by each site. Libraries were prepared according to the manufacturer’s protocol with a modification in size selection. Instead of agarose gel purification, Pippin Prep instrument (Sage Science, Beverly, MA, USA) and 3% agarose dye-free cassettes with internal standards (CP3010; Sage Science) were used under the following conditions: base pair start = 110 bp, base pair end = 160 bp, range = broad, target peak size = 147 bp. Libraries were amplified using 11 cycles of PCR for both MUR and MUR-D samples. The procedure was performed end-to-end in 1 d without the use of stopping points.
Lexogen Small RNA-Seq Library Prep Kit (kit 058.08, protocol 052UG128V0100; Lexogen, Vienna, Austria)
The amount of input RNA was 100 ng for HBR, 100 ng for MUR-D, and 100 pg for MUR. Libraries were prepared in a single day without the use of stopping points, with the following modifications to the protocol. Step 5.1.1: A3 diluted 0.5× for MUR-D, 0.3× for MUR. Step 5.1.5: 50 μl ethanol used for all samples. Step 5.1.11: A5 diluted 0.5× for MUR-D, 0.3× for MUR. Step 5.1.15: Reverse transcription primer diluted 0.5× for MUR-D and 0.3× for MUR samples. Step 9: eluted in 12 μl EB Buffer (Qiagen, Germantown, MD, USA), with entire volume carried forward to Step 10. Library PCR was performed using 16 cycles for MUR-D and 22 cycles for MUR samples.
New England Biolabs NEBNext Small RNA Library Prep Set (kit E3700, protocol E3700S; New England Biolabs, Ipswich, MA, USA)
The amount of input RNA was 100 ng for HBR, 100 ng for MUR-D, and 35 pg for MUR. Library preparation was performed either in 1 d or allowed to incubate at 4°C overnight following the PCR amplification step, using 15 cycles for all sample types. The reverse transcription reaction was not heat-inactivated. The size selection of libraries was performed using a 3% dye-free gel cassette on a Pippin Prep instrument, as for Illumina libraries (described above).
Qiagen QIAseq miRNA Library Kit (kit 331502, protocol 11/2016; Qiagen, Germantown, MD, USA)
The amount of input RNA was 10 ng for HBR, 10 ng for MUR-D, and 35 pg for MUR. Libraries were prepared in a single day or used the optional stopping point following the cDNA cleanup step. Library amplification PCR was performed using 19 cycles for MUR-D and 22 cycles for MUR samples based on Table 12 in the QIAseq protocol and consultation with Qiagen applications scientists.
PerkinElmer NextFlex Small RNA-Seq Kit v.3 (kit 5132-05, protocol V16.06; PerkinElmer, Waltham, MA, USA)
The amount of input RNA was 250 ng for HBR and MUR-D and 100 pg for MUR. The protocol was performed with the following modifications. Step A4: adapters were used undiluted for MUR-D and were diluted 1:4 for MUR samples. Step A6: adapter ligation reactions were incubated overnight at 20°C. Library PCR was performed using 18 cycles for both MUR-D and MUR samples.
Trilink Biotechnologies CleanTag Small RNA Library Prep Kit (kit L-3206, protocol L-3206v7; Trilink Biotechnologies, San Diego, CA, USA)
The amount of input RNA was 10 ng for HBR and MUR-D and 35 pg for MUR. Library preparation was performed in a single day following the protocol, with the following modifications. CleanTag 3′ and CleanTag 5′ adapters were diluted 1:4 for MUR-D, HBR, and MUR samples. In the reverse transcription reaction, DTT was diluted 1:10 (100 mM) for MUR samples only. Reverse transcription reactions were set up as follows. The sample and 2 μl of reverse transcriptase were combined and heated to 70°C for 2 min, after which the tubes were spun down and the remainder of reaction components were added. Library PCR was conducted using 18 cycles for all samples. Amplified libraries were size-selected to retain only fragments between 100 and 200 bp, using Ampure XP beads (Beckman Coulter, La Brea, CA, USA).
Template switching
Takara Bio SMARTer smRNA Kit (kit 635029, protocol 040816; Takara Bio, Kusatsu, Japan)
The amount of input RNA was 1 ng for MUR-D and HBR and 35 pg for MUR. Libraries were prepared according to the manufacturer’s protocol with the following modifications. Step A6: Polyadenylation master mix was prepared without the addition of ATP. Step V.E22: 20 μl of supernatant was transferred for the next steps. Library amplification was performed using 16 and 11 cycles for MUR-D and MUR samples, respectively. Libraries fragments from 148 to 185 bp were size-selected on a Sage Science Pippin Prep instrument using a 3% dye-free agarose gel, as recommended in the protocol. Library preparation was performed in 1 d without the use of stopping points.
Diagenode CATS Small RNA-Seq Kit (kit C05010044, protocol v.2|09.17; Diagenode, Liège, Belgium)
The amount of input RNA for HBR and MUR-D was 10 ng and 35 pg for MUR. Libraries were prepared with the following modifications. Step 1.1: Dephosphorylation reagent was diluted 5 times for MUR samples. Step 2.8: Reverse transcription primer M was used for MUR-D and HBR, and reverse transcription primer L was used for MUR. Step 3.15: Twelve cycles were used for MUR-D and HBR and 18 cycles for MUR in PCR preamplification. Step 3.16: AMPure XP bead cleanup was performed in full prior to size selection following Optional Enrichment instructions in Step I. No size selection, only cleanup, was performed for MUR.
Circularization using Somagenics RealSeq-AC miRNA Library Kit (kit 500-00012, protocol 20170811_RealSeq-AC; Somagenics, Santa Cruz, CA, USA)
An early-release beta version of the Somagenics protocol was used in this study, and users may wish to test with the latest version of their chemistry. The amount of input RNA for HBR and MUR-D was 250 ng and 35 pg for MUR. Libraries were prepared according to protocols with the following modifications. Step 1: Adapter ligation was carried out using 0.5 μl undiluted stock for MUR-D and HBR samples and 3 μl of undiluted adapter stock for MUR samples. Step 2: Adaptor blocking was conducted with a temperature gradient from 65 to 37°C in 5 min, after which libraries were stored overnight prior to circularization. Step 6: Library PCR was performed using 13 cycles for MUR-D and HBR and 7 cycles for MUR samples. Amplified libraries were size-selected to remove small fragments using Ampure XP beads.
Ligation + array hybridization using NanoString nCounter miRNA Expression Assay (kit CSO-MIR3-12, protocol MAN-0009-05; NanoString)
The amount of input RNA was 100 ng for MUR-D and 10 pg for MUR. Steps 1–14 were performed at individual test sites, frozen at −20°C, and shipped overnight on dry ice to a single location. Hybridization, cartridge loading, and scanning for all samples were performed in parallel by a single technician. Data were processed using nSolver software prior to further downstream analysis.
Adapter removal of smRNA sequencing reads
The adapter removal and quality control were performed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), with specific parameters for the kits and specific adapters or random bases around the target sequences. In Trim Galore, the parameter “–length” was set to 18 to discard reads that became shorter than length 18.
miRNA quantification using miRDeep2
miRDeep210 was used in this study. First, mapper.pl was used to get collapsed reads in fasta format. The script quantifier.pl was used to quantify the expression levels of the miRNAs in different samples. For MUR and MUR-D samples, mature miRNA sequences and their corresponding precursors sequences were extracted from the file downloaded from the miRBase database (ftp://mirbase.org/pub/mirbase/22/miRNA.xls.zip). Eight hundred ninety-five of the miRNAs in the standard are included in miRBase v.22. Then the expression levels of the miRNAs were quantified using quantifier.pl in mirDeep2. For HBR samples, miRNAs and their corresponding precursors were downloaded from the mirBase database. In house scripts were used to extract the raw read counts and normalized read counts from the output files of quatifier.pl.
The heat maps were generated using R package pheatmap (https://github.com/raivokolde/pheatmap). In the figures, log2 transformation was applied to the read counts.
RESULTS
To assess the strengths of the various smRNA kits, we used 3 different standardized RNA reference samples (Fig. 1A). First, to determine the biases of each chemistry we used the MUR, an equimolar pool of over 950 synthetic, unmodified, HPLC-purified RNA oligonucleotides corresponding to mature miRNAs from human, mouse, rat, and related viruses. Although this standard is widely used to benchmark miRNA detection methods, its utility is limited because the synthetic miRNAs lack the dynamic range and other background RNAs that are characteristic of a real biological sample. RNA from the HBR (AM7962; Thermo Fisher Scientific), a commercially available sample from the 2006 Microarray Quality Control study,11 was used as a second standard because it is commonly used to characterize miRNA chemistries by other studies and provides a highly complex normal sample. However, because the actual characterization of the miRNAs in this sample is not known, it has limited use for quantifying bias. To address these concerns, we created a third standard, MUR-D. This standard is derived by using total RNA from Dicer-deleted mesenchymal stem cells, which lack almost all mature miRNA,12 and supplementing it with the MUR, which gives both rigorous known quantification as well as a more biologically representative context for the study.
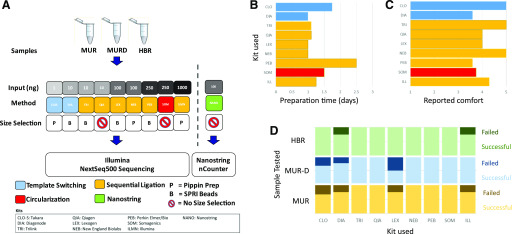
FIGURE 1.
Study design and workflow metrics. A) Schematic of study design is shown. MUR, MUR-D, and HBR were processed using 9 different smRNA profiling methods at 4 sites each. The general methodologies included sequential ligation (Illumina, TriLink, Qiagen, NEB, PerkinElmer, Lexogen), template switching (Takara, Diagenode), circularization (Somagenics), and NanoString probe-based hybridization. B) Total start-to-finish preparation time for each kit as reported by sites. C) Mean “ease of use” reported by each site (scale: 1 = uncomfortable, 5 = comfortable). D) Library preparation success rates. Light color blocks are successfully produced libraries. Dark colors are failed libraries. CLO and CLO-S, Takara Bio (Clontech) SMARTer smRNA-Seq Kit; DIA, Diagenode CATS Small RNA-Seq Kit; ILL and ILMN, Illumina TruSeq Small RNA Library Prep Kit; LEX, Lexogen Small RNA-Seq Library Prep Kit; NANO, NanoString nCounter miRNA Expression Assay; NEB, New England Biolabs NEBNext Small RNA Library Prep Set; PEB, PerkinElmer NextFlex Small RNA-Seq Kit v.3; QIA, Qiagen QIAseq miRNA Library Kit; SOM, Somagenics RealSeq-AC miRNA Library Kit; TRI, Trilink CleanTag Small RNA Library Prep Kit.
smRNA chemistries were derived from a broad variety of sources, reflecting several different methods for library preparation. Typical smRNA library preparation for Illumina sequencing is based on sequential ligation of oligonucleotides to the 5′ and 3′ ends of the smRNAs. Most of the kits tested, including Illumina TruSeq Small RNA Library Prep Kit, Lexogen Small RNA-Seq Library Prep Kit, New England Biolabs NEBNext Small RNA Library Prep Set, PerkinElmer (formerly Bioo Scientific) NextFlex Small RNA-Seq Kit v.3, Qiagen QIAseq miRNA Library Kit, and Trilink CleanTag Small RNA Library Prep Kit all use variants of this methodology. Two additional strategies have recently been made commercially available. The Takara SMARTer smRNA Kit and the Diagenode CATS Small RNA-Seq Kit both begin by polyadenylation of the 3′ nucleotide of the smRNA followed by reverse transcription and template switching. The Somagenics RealSeq-AC beta kit uses a circularization strategy with only a single intramolecular ligation to minimize bias. Finally, NanoString small RNA hybridization was used as a nonsequencing methodology for comparison. All kits were tested at 4 sites following consultation with technical experts from the manufacturers, and the libraries were sequenced at a single location.
Initial quality of the libraries was determined by fragment analysis and qPCR quantification. Technical staffs were asked to judge the ease of use of the preparation and to measure the actual time of library preparation (Fig. 1B–D). Template switching protocols appeared to be the most challenging, with significant numbers of failed samples (Fig. 1D). Despite a lower level of comfort with the Somagenics circularization methodology and the PerkinElmer kit, all sites were able to successfully create libraries with these kits. Significant time differences were observed between the different protocols. Most sites reported that that the protocols took slightly over 1 d of lab time to complete. The PerkinElmer kit was a notable exception, taking 2.5 d to complete, a full day longer than other methods. Libraries that failed quality control were excluded from further analysis.
Data from the Miltenyi Biotec miRXplore standards were used to determine bias of the different chemistries. RNA reads from the spike-in only (MUR) and Dicer−/− (MUR-D) libraries were aligned to the miRNAs included in the control, and uniquely mapping reads were reported. Although a very similar number of the input miRNAs were detectable in each sample (Supplemental Fig. S1) a significant number of miRNAs appear to be highly underrepresented [<5 counts per million (CPM)] in most of the ligation-based kits for both MUR and MUR-D, with the exception of the PerkinElmer kit, which has randomized ligation nucleotides (Fig. 2A, B).
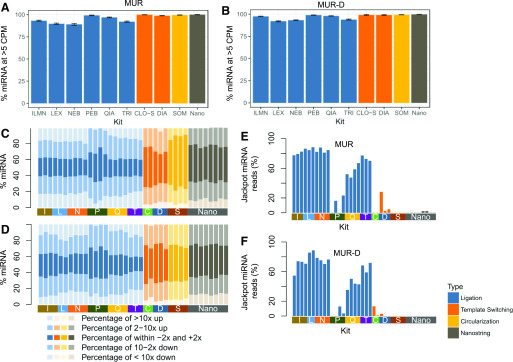
FIGURE 2.
miRNA detection and bias. Percentages of Miltenyi Biotec miRXplore miRNA detected above 5 CPM in MUR (A) and in MUR-D (B) are shown. Percentages and amplitude of Miltenyi Biotec miRXplore miRNA detected that deviated from the median are shown in MUR (C) or MUR-D (D). The darkest shade is within 2-fold of the median; the medium shade is 2–10-fold either up or down vs. the expected value; the lightest shade is >10× either increased or decreased. Percentages of reads increased >10× from median in MUR (E) or MUR-D (F). CLO-S and C, Takara Bio (Clontech) SMARTer smRNA-Seq Kit; DIA and D, Diagenode CATS Small RNA-Seq Kit; ILMN and I, Illumina TruSeq Small RNA Library Prep Kit; LEX and L, Lexogen Small RNA-Seq Library Prep Kit; Nano, NanoString nCounter miRNA Expression Assay; NEB and N, New England Biolabs NEBNext Small RNA Library Prep Set; PEB and P, PerkinElmer NextFlex Small RNA-Seq Kit v.3; QIA and Q, Qiagen QIAseq miRNA Library Kit; SOM and S, Somagenics RealSeq-AC miRNA Library Kit; TRI and T, Trilink CleanTag Small RNA Library Prep Kit.
To better quantify the bias, we identified the miRNAs with the median CPM for each sample and measured the fraction of miRNAs close to that median (±2×), somewhat amplified or lost (±2–10×), and either jackpotted (>10× median) or dropped out (<0.1× median, Fig. 2C, D). Critically, the fraction of miRNAs close to the median varied widely, ranging from about 20% in many of the sequential ligation methods to about 60% in the Somogenics samples working with only the Miltenyi Biotec controls. Although the low fraction of reads within 2-fold of the median for most sequential ligation protocols was striking, the results are consistent with previous findings.1, 5, 7, 13, 14 Importantly, in the context of total RNA, no kit, including NanoString, preserved even 50% of the miRNAs within 2-fold of median. However, the circularization and template switching–based methods had significantly larger fractions of material within the 2-fold range. A crucial aspect to this increase is the absence of overrepresented or jackpotted miRNAs in these preparations as well as the PerkinElmer chemistry. The jackpotted miRNAs not only reduced the number of reads close to the median, but they also could take up a significant fraction of all the miRNA reads (up to 80%; Fig. 2E, F), requiring increased read depth to detect other miRNAs.
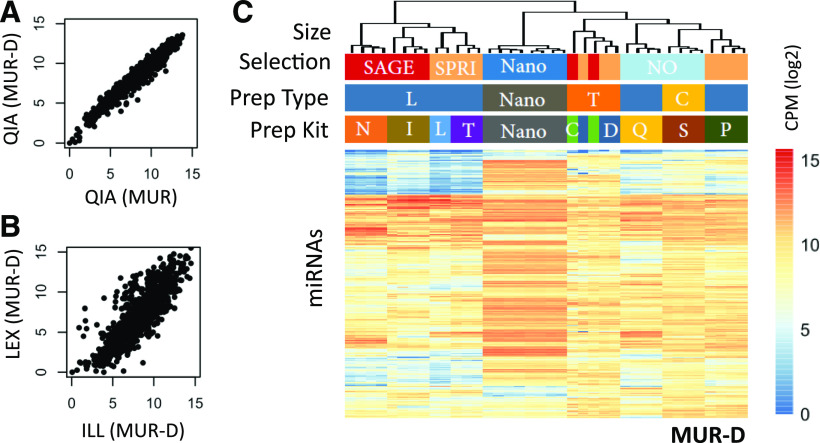
Although the absolute quantification of miRNAs has been a known challenge, most smRNA studies have focused on relative quantification, which has been shown to be preserved within kits.4, 6, 7We observed similar results, with a high degree of correlation within kits even when comparing the miRXplore samples alone with the same miRNAs in the context of total RNA from the Dicer−/− cells (Fig. 3A and Supplemental Fig. S2). When comparing between kits, we found significant deviation, even among jackpotted and dropout miRNAs (Fig. 3B). Global comparison of the different methodologies showed very little correlation, with the notable exception of the 2 template switching methods that clustered tightly together (Fig. 3C and Supplemental Fig. S3).
FIGURE 3.
Relative expression of miRNAs. A) Correlation scatter plots of miRNAs detected in MUR and MUR-D for the QIA method, with miRNAs scored in log2 mapped reads per million. B) Correlation scatter plot of miRNAs detected in MUR-D using LEX vs. ILL kits, with miRNAs scored in log2 mapped reads per million. C) Unsupervised hierarchical clustering heatmap of log2 transformed CPM across all methods. C (prep kit), Takara Bio (Clontech) SMARTer smRNA-Seq Kit; C (prep type), circularization; D, Diagenode CATS Small RNA-Seq Kit; ILL and I, Illumina TruSeq Small RNA Library Prep Kit; L (prep type), sequential ligation; LEX and L (prep kit), Lexogen Small RNA-Seq Library Prep Kit; N, New England Biolabs NEBNext Small RNA Library Prep Set; Nano, NanoString nCounter miRNA Expression Assay; NO, No size selection; P, PerkinElmer NextFlex Small RNA-Seq Kit v.3; QIA and Q, Qiagen QIAseq miRNA Library Kit; S, Somagenics RealSeq-AC miRNA Library Kit; SAGE, pippin prep; SPRI, solid phase reversible immobilization; T (prep kit), Trilink CleanTag Small Library Prep Kit; T (prep type), template switching.
Efficient analysis of smRNAs not only requires the identification and quantification of the miRNAs but is also dependent on the fraction of usable reads. Reads can be excluded because of short inserts (e.g., primer dimer) or poor-quality sequences. Additionally, degraded fragments of other RNAs, such as rRNAs and tRNAs, may or may not be of interest to the researcher. Many of the different strategies for library preparation are based on increasing the fraction of usable reads over the original Illumina protocol.
To address the fraction of usable reads, we focused on the MUR-D and HBR samples, because the spike-in alone sample poorly represents typical experiments. Overall, the ligation-based chemistries performed better than the newer methods, with fewer reads discarded as too short (<17 nt) or fewer clear experimental artifacts, though there was some significant site-to-site variability with many kits. Generally, the ligation-based protocols had fewer discarded reads in the MUR-D sample (20–30%; Fig. 4A, B) than did the brain reference (10–60%; Fig. 4C, D). Among the ligation chemistries, the Illumina kit and PerkinElmer kit had more short reads in HBR than comparable kits, whereas the Lexogen and Trilink kits showed more poor-quality reads (Fig. 4D). With the template switching and circularization methods, we found many more short inserts and a much lower fraction of reads in the 18–25-nt range. Overall, the fraction of reads in each size range varied dramatically, even within a kit, suggesting size optimization is one of the most challenging aspects of all of the protocols. The Qiagen and Illumina kits had some of the most consistent large fraction of reads in the 18–25-nt range for miRNAs, whereas the Lexogen and Trilink kits were biased toward slightly longer reads, particularly in the HBR sample.
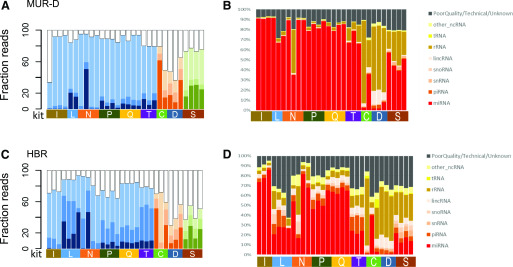
FIGURE 4.
Detection of smRNAs in complex samples. Insert sizes in MUR-D (A) and HBR (B) are shown with inserts <18 nt as white, 18–23 nt as the lightest shade, 24–35 nt as the medium shade, and >35 nt as the darkest shade. Data from the different method types are colored (ligation, blue; polyA tailing, orange; circularization, green). The right panels show the smRNA target distribution for MUR-D (C) and HBR (D). Mapped targets are noted in the key. Unmapped targets are shown in black. C, Takara Bio (Clontech) SMARTer smRNA-Seq Kit; D, Diagenode CATS Small RNA-Seq Kit; I, Illumina TruSeq Small RNA Library Prep Kit; L, Lexogen Small RNA-Seq Library Prep Kit; lincRNA, long intervening noncoding RNA; N, New England Biolabs NEBNext Small RNA Library Prep Set; ncRNA, noncoding RNA; P, PerkinElmer NextFlex Small RNA-Seq Kit v.3; Q, Qiagen QIAseq miRNA Library Kit; S, Somagenics RealSeq-AC miRNA Library Kit; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; T, Trilink CleanTag Small Library Prep Kit.
smRNAs can come from a broad variety of sources, including piRNAs, tRNAs, rRNAs, enhancer RNAs, etc. Categorizing the reads within the HBR and MUR-D samples, we observed that the libraries from the ligation-based chemistries showed a higher fraction of miRNA-derived reads, whereas fragments of rRNA, tRNA, and long noncoding RNAs were more common in the other 2 methods, particularly the Diagenode and Takara kits. Results were more variable from the HBR samples run on the Lexogen and Trilink kits, showing much lower amounts of miRNAs and higher amounts of piRNAs than the similar methods. Notably, the difference was also seen between PerkinElmer (the 1 ligation-based method that had low levels of jackpotting) and the nonligation methods, suggesting the effect is not strictly due to ligation bias.
DISCUSSION
Investigators now have an abundance of library preparation methods to select from when designing experiments aimed at exploring smRNA biology. This presents a great opportunity to match specific research aims with optimal methodological parameters such as input amount and detection of miRNAs or other smRNA classes. Recent studies have compared many of the available methods,6, 7 but each study was limited, either in terms of the number of methods evaluated or the number of sites. Furthermore, they paid little attention to soft metrics such as time and ease of prep, which are important practical considerations for use in research labs, core facilities, and service providers. This study comprehensively evaluated 9 different kits as well as NanoString probe hybridization, spanning 4 different methodological approaches, using commercially available samples and synthetic spike-in standards. Samples were prepared at 4 laboratory sites for each method to assess smRNA detection, quantification, and inter- and intrasite reproducibility in order to inform best-practice and method selection decisions in a core facility setting (Table 1).
TABLE 1.
Summary of kits tested
| Kit | Method | miRNA detection | Reproduci-bility | miRNA discovery | Size selection | Prep time, d | Reduced jackpotting | Low input | UMI correction |
|---|---|---|---|---|---|---|---|---|---|
| Takara | Template switching | X | X | X | Pippin Prep | 1.75 | X | X | |
| Diagenode | Template switching | X | X | X | SPRI | 1 | X | X | |
| Trilink | Sequenital ligation | X | X | X | SPRI | 1.2 | |||
| Qiagen | Sequenital ligation | X | X | X | None | 1.2 | X | ||
| Lexogen | Sequenital ligation | X | X | X | SPRI | 1 | |||
| New England Biolabs | Sequenital ligation | X | X | X | Pippin Prep | 1 | |||
| PerkinElmer | Sequenital ligation | X | X | X | SPRI | 2.5 | X | ||
| Illumina | Sequenital ligation | X | X | X | Pippin Prep | 1.3 | |||
| Somagenics | Circular-ization | X | X | X | None | 1.5 | X | ||
| NanoString | Probe hybrid-ization | X | X | None | 2 | X |
SPRI, solid phase reversible immobilization; UMI, unique molecular identifier.
Our results are consistent with previous reports demonstrating a wide range of method-specific smRNA quantification bias.4–7, 9 Importantly, kit-specific biases were reproducible both across sites and between sample replicates, even when comparing miRXplore spike-in alone with the spike-in plus Dicer1−/− RNA background, suggesting that all of the methods may be appropriate options for relative differential expression analysis. However, because of strong kit-specific biases, experimental designs must be limited to a single preparation method, which is an especially important consideration if the design includes preexisting public or legacy data. Clinical biomarker applications may require further validation using an orthogonal detection method such as NanoString as an alternative to next-generation sequencing library prep methodology.
The data clearly show that there is not a single approach that is appropriate for all sample types and applications. Experimental considerations, such as availability of starting material, tolerance of jackpotted sequences, and, most importantly, other data sets being compared, must be carefully considered. There is still much room to improve existing methods, as well as an opportunity to develop novel approaches especially with regard to reducing sequence specific bias. To this end, we propose the continued use of the Dicer−/− with a miRXplore spike-in as an improved benchmarking standard, because it provides both a complex mixture of full-length and smRNAs and a known value for miRNAs to be tested.
Supplementary Material
This article includes supplemental data. Please visit http://jbt.abrf.org/ to obtain this information.
ACKNOWLEDGMENTS
The authors are grateful to David Nadziejka (Van Andel Institute, Grand Rapids, MI, USA) for helpful discussions and comments on the manuscript. S.S.L. is funded by the National Cancer Institute of the U.S. National Institutes of Health (NIH) under award P30-CA14051 and by the National Institute of Environmental Health Sciences of the NIH under award P30-ES002109. J.S. is funded by Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Support Award RP170002. The authors declare no conflicts of interest.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. [DOI] [PubMed] [Google Scholar]
- 2.Garzon R, Pichiorri F, Palumbo T, et al. . MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. Metazoan microRNAs. Cell. 2018;173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linsen SE, de Wit E, Janssens G, et al. . Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods. 2009;6:474–476. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs RT, Sun Z, Zhuang F, Robb GB. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS One. 2015;10:e0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giraldez MD, Spengler RM, Etheridge A, et al. . Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat Biotechnol. 2018;36:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barberán-Soler S, Vo JM, Hogans RE, Dallas A, Johnston BH, Kazakov SA. Decreasing miRNA sequencing bias using a single adapter and circularization approach. Genome Biol. 2018;19:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shore S, Henderson JM, Lebedev A, et al. . Small RNA library preparation method for next-generation sequencing using chemical modifications to prevent adapter dimer formation. PLoS One. 2016;11:e0167009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baran-Gale J, Kurtz CL, Erdos MR, et al. . Addressing bias in small RNA library preparation for sequencing: a new protocol recovers microRNAs that evade capture by current methods. Front Genet. 2015;6:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravi A, Gurtan AM, Kumar MS, et al. . Proliferation and tumorigenesis of a murine sarcoma cell line in the absence of DICER1. Cancer Cell. 2012;21:848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi L, Reid LH, Jones WD, et al. ; MAQC Consortium . The Microarray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JX, Rastetter RH, Wilhelm D. Non-coding RNAs: an introduction. Adv Exp Med Biol. 2016;886:13–32. [DOI] [PubMed] [Google Scholar]
- 14.Hafner M, Renwick N, Brown M, et al. . RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.