Abstract
Gram-negative bacterial infections are a significant public health concern, and the lack of new drug classes for these pathogens is linked to the inability of most drug leads to accumulate inside Gram-negative bacteria1-7. Here we report the development of a web application, called eNTRyway, which predicts compound accumulation (in E. coli) from its structure. eNTRyway, in conjunction with structure-activity relationship and x-ray data, was utilized to re-design Debio-1452, a Gram-positive-only antibiotic8, into versions that accumulate in E. coli and possess antibacterial activity against high-priority Gram-negative pathogens. The lead compound Debio-1452-NH3 operates as an antibiotic via the same mechanism as Debio-1452, namely potent inhibition of the enoyl-acyl carrier protein reductase FabI as validated by in vitro enzyme assays and generation of bacterial isolates with spontaneous target mutations. Debio-1452-NH3 is well tolerated in vivo, reduces bacterial burden in mice, and rescues mice from lethal infections with clinical isolates of Acinetobacter baumannii, Klebsiella pneumoniae, and Escherichia coli. This work provides tools for the facile discovery and development of high-accumulating compounds in E. coli, and a general blueprint for the conversion of Gram-positive-only compounds into broad-spectrum antibiotics.
During the last half-century seven new antibiotic classes have been FDA approved9, none of which are useful against the most problematic Gram-negative “ESKAPE”10 pathogens. These organisms, namely Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, are responsible for ~75% of infections and deaths from antibiotic-resistant bacteria;2 alarmingly, the number of patients to die from these Gram-negative infections is up almost 4-fold in just eight years2. Despite tremendous advances in genomics and high-throughput screening in the last 50 years, antibiotic classes to treat Gram-negative bacteria that are structurally distinct or act through mechanisms outside the common targets6 have remained elusive. This lack of new drug classes for the problematic Gram-negatives is consistent with the low hit rate reported from extensive antimicrobial high-throughput screening campaigns4. This discovery challenge is not ascribed to poor target activity, but rather to challenges of compound accumulation in Gram-negative pathogens, given the impermeability of their cell membranes coupled with promiscuous efflux pumps5.
In contrast to the discouraging results in screening for Gram-negative actives, there have been an abundance of compounds discovered that have promising activity against Gram-positive bacteria. Present amongst these compounds are both natural products and synthetic compounds, many of which act through mechanisms outside the common targets, that is, they are not ribosome binders, cell wall biosynthesis inhibitors, or DNA gyrase inhibitors6. Importantly, the vast majority of these compounds would kill Gram-negative bacteria if they could accumulate inside these pathogens. Unfortunately, no method for the conversion of Gram-positive-only compounds into broad-spectrum antibiotics has been widely applicable. Creative approaches to this problem such as attachment to siderophores11, cotreatment with efflux pump inhibitors12, and potentiation by non-lytic polymyxins13 are under active investigation. An alternative approach is tuning the physiochemical properties of antibiotics to favor accumulation inside the bacterial cell. If accumulation of these drugs inside Gram-negative bacteria could be optimized by employing routine medicinal chemistry strategies – analogous to the now-common practice of improving solubility or pharmacokinetics – then the attrition of antibiotics in early discovery stage might be mitigated. Such a strategy would rely on an appropriate knowledgebase detailing the physicochemical parameters allowing compound accumulation in Gram-negative bacteria.
Toward this end we recently reported the results from a prospective study examining the ability of diverse compounds to accumulate in E. coli14. This work revealed that compounds with certain physicochemical properties – the presence of an ionizable nitrogen, low three-dimensionality, and low numbers of rotatable bonds (codified as the “eNTRy rules”7) – have a significantly higher probability of accumulating in E. coli14. To assist in implementation of the eNTRy rules, here we report a convenient, open-access web-based cheminformatics tool (called eNTRyway) to provide binary classification for compound accumulation in E. coli. Using eNTRyway to predict compounds that have increased potential to accumulate in E. coli in combination with structure-activity relationship and x-ray data enables the design of compounds that are not only active against the designated bacterial target but also possess whole-cell activity.
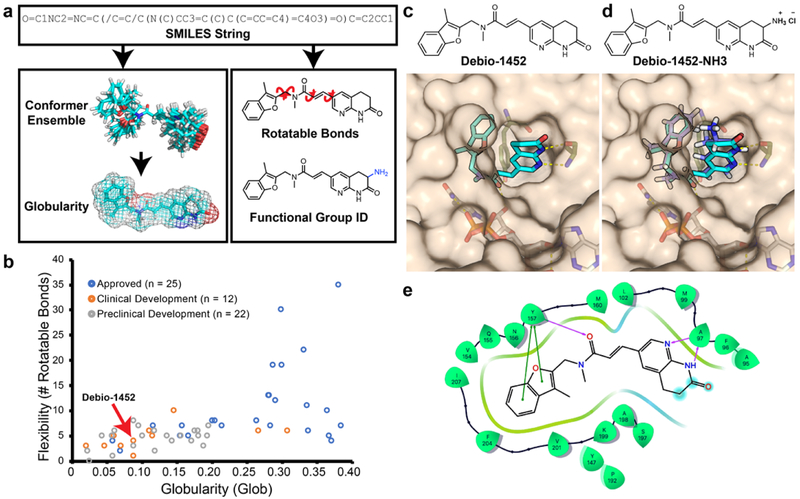
The initial methodology for calculating eNTRy rule parameters relied on proprietary algorithms to perform conformer generation and globularity calculation14. With the goal of developing a free, easy-to-use web application, the first step was to implement these calculations with open-source libraries. While several conformer generation methods were evaluated, ultimately the software Open Babel’s genetic algorithm was found to rapidly and accurately generate an ensemble of conformers15. Globularity calculation was implemented with linear algebra modules from NumPy16. Upon establishing accurate and accessible calculation methods for the eNTRy rules, a web application (www.entry-way.org) was built in which users submit a SMILES string of a molecule of interest. RDkit parses the SMILES string and makes an initial estimate of the 3D coordinates, Open Babel’s genetic algorithm generates a library of conformers, and a custom Python program calculates globularity (Fig. 1a). The rotatable bond counting and functional group detection is performed separately using the 2D structure (Fig. 1a). Results from these calculations are displayed to the user and are compared to breakpoints provided by the eNTRy rules; for specific examples of visual outputs from entry-way.org please see Extended Data Fig. 1.
Fig. 1. Overview of eNTRyway molecule processing.
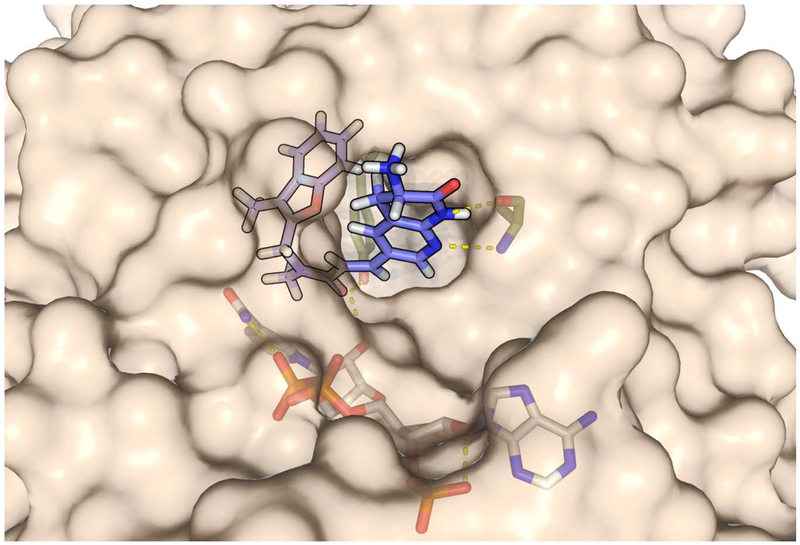
a. Molecules are submitted as SMILES strings, and after an initial estimate of the 3D structure, a conformer space is systematically explored from which an average globularity is calculated. Particular functional groups and number of rotatable bonds are determined directly from 2D structure. b. Physiochemical properties of existing Gram-positive-only antibiotics, Debio-1452 is indicated with red arrow (for specific compounds see Supplementary Table 1). c. Solvent exposure of the naphthyridinone of Debio-1452 (hydrogen atoms are not shown, carbon atoms are shown in cyan, nitrogen atoms are shown in blue, oxygen atoms are shown in red) when bound to S. aureus FabI (PDB: 4FS3) suggests sites for modification. d. Molecular docking of amine-containing derivative, Debio-1452-NH3 (hydrogen atoms are shown in white, carbon atoms are shown in lavender, nitrogen atoms are shown in blue, and oxygen atoms are shown in red). overlaid with the crystal structure of Debio-1452 (hydrogen atoms are not shown, carbon atoms are shown in cyan, nitrogen atoms are shown in blue, oxygen atoms are shown in red). Additional derivative docking scores are shown in Supplementary Table 3, and a non-overlapping pose of Debio-1452-NH3 is shown in Extended Data Fig. 2. e. Ligand interaction diagram of interaction shown in panel c, cyan spheres indicate positions of solvent exposure.
Applying eNTRyway to almost 70 Gram-positive only FDA-approved antibiotics, drugs currently in clinical development, and lead compounds in pre-clinical stages indicates that there are numerous candidates for conversion at all stages of development (Fig. 1b, structures and properties listed in Supplementary Table 1). Through this analysis Debio-1452 emerged as an attractive candidate for transformation to a Gram-negative active version. With a low globularity (Glob = 0.093) and few rotatable bonds (RB = 4), Debio-1452 (Fig. 1c) conforms to two of the three eNTRy rules, lacking only a primary amine. Debio-1452 (also called AFN-1252) originated in the early 2000s from lead optimization efforts of a hit discovered in a target-based (biochemical) high-throughput screen at GlaxoSmithKline for inhibitors of the enoyl-acyl carrier reductase enzyme FabI17,18. FabI plays a pivotal role in the elongation cycle of the bacterial fatty acid biosynthesis pathway (FAS-II); for Gram-negative pathogens the FAS-II pathway is essential to survival and inhibition of the FAS-II pathway cannot be bypassed during host infections through uptake of exogenous nutrients19. Notably, components of the bacterial FAS-II pathway are distinct from those found in the analogous mammalian FAS-I pathway20. This essentiality and orthogonality to the mammalian counterpart makes components of the FAS-II pathway attractive biological targets for antimicrobial drug discovery, and the potential for targeting FAS-II is exemplified by the success of triclosan and isoniazid, both of which inhibit FabI or a closely related enzyme21,22. Although redundant isoforms of FabI are present in certain bacterial species, such as FabV of P. aeruginosa23, which can compensate for FabI inhibition, FabI is the sole enoyl-acyl carrier reductase for the other Gram-negative ESKAPE pathogens including Enterobacter spp., A. baumannii, and K. pneumoniae24.
While Debio-1452 displays significant antibiotic activity against Staphylococcus aureus (MIC = 0.008 μg/mL), activity against Gram-negative ESKAPE pathogens and E. coli is lacking, with no activity up to its aqueous solubility limit (MIC > 32 μg/mL, Table 1); however, Debio-1452 is very potent in E. coli with compromised efflux or permeability defects (ΔtolC or ΔrfaC, Table 1)8, suggesting that whole cell activity against Gram-negative bacteria is limited by low cellular accumulation and not by on-target activity. Debio-1452 has marked efficacy in animal models of S. aureus infections25 and in patients in an open-label clinical trial26. Debio-1452 thus is a constituent of a new antimicrobial drug class, hits a compelling antibiotic target and possesses structural features making it appropriate for conversion to a Gram-negative active via the eNTRy rules. While over 100 derivatives of Debio-1452 have been prepared27-31, none are reported to have robust activity against the Gram-negative ESKAPE pathogens.
Table 1. Antimicrobial activity of Debio-1452 and derivatives.
The aqueous solubility limit of Debio-1452 prevents determining actual MIC values that are above 32 μg/mL. MIC values were determined using the micro-dilution broth method as outlined by the Clinical and Laboratory Standards Institute (http://clsi.org/) (CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 11th ed. CLSI standard M07. Wayne, PA: Clinical and Laboratory Standards Institute; 2018). All experiments were performed in biological triplicate.
| Bacterial Strain | MIC (μg/mL) | ||
|---|---|---|---|
| Debio-1452 | Debio-1452-NH3 | 8 | |
| WT Gram-positive | |||
| S. aureus ATCC29213 | 0.008 | 0.03 | 0.016 |
| Gram-negative permeability mutant | |||
| E. coli ΔtolC JW5503 | 0.031 | 0.062 | 0.125 |
| E. coli ΔrfaC JW3596 | 0.5 | 0.25 | 0.25 |
| WT Gram-negative | |||
| E. coli MG1655 | >32 | 4 | >32 |
| E. coli BAA-2340 | >32 | 4 | >32 |
| E. coli AR-0493 | >32 | 4 | >32 |
| E. cloacae ATCC 29893 | >32 | 8 | >32 |
| K. pneumoniae BAA-1705 | >32 | 8 | >32 |
| K. pneumoniae S47889 | >32 | 8 | >32 |
| A. baumannii W41979 | >32 | 4 | >32 |
| A. baumannii F19521 | >32 | 4 | >32 |
| P. aeruginosa PA01 | >32 | >64 | >32 |
As Debio-1452 already possessed favorable physicochemical properties in terms of globularity and flexibility, conversion into an analogue with increased potential for E. coli accumulation and Gram-negative antibacterial activity required the strategic introduction of a primary amine. Examination of the co-crystal structure of Debio-1452 bound to S. aureus FabI (Fig. 1c, e) showed the 3-position of the naphthyridinone ring (adjacent to the lactam carbonyl) to be the most solvent-exposed region of the molecule32, consistent with antibacterial data suggesting that substituents were tolerated at this position28,29. eNTRyway predicted that key compounds with amines at this location would accumulate in E. coli, highlighting the 3-position of the naphthyridinone ring as a promising point for incorporation of an amine (Supplementary Table 2). Additionally, computational docking of these compounds, including Debio-1452-NH3 (Fig. 1d, Supplementary Table 3, & Extended Data Fig. 2), suggested amine modification would not be disruptive to binding the FabI target.
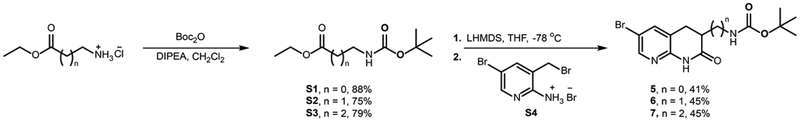
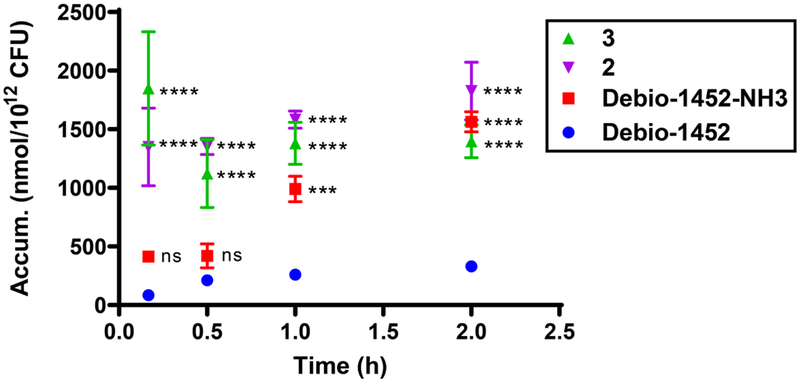
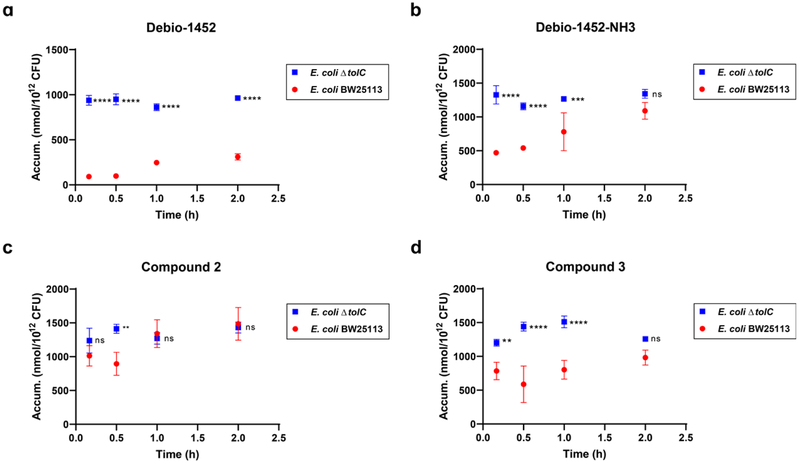
The synthesis of the amine functionalized acryloaminopyridines was performed in a modular fashion with a late-stage Heck coupling of acrylamide 4 with functionalized naphthyridinones 5-7 (prepared as described in Extended Data Fig. 3) followed by deprotection of 8-10 to yield target amine-containing compounds 1-3 (Extended Data Fig. 4). Compounds 1-3 were assessed for their ability to accumulate in E. coli MG1655 using the standard LC-MS/MS assay14. While Debio-1452 showed no statistically significant accumulation over negative controls, the derivatives with ionizable nitrogens accumulated in E. coli; the accumulation in E. coli MG1655 over the course of 2 h is shown in Extended Data Fig. 5, and accumulation in an efflux-deficient strain of E. coli (ΔtolC) is shown in Extended Data Fig. 6.
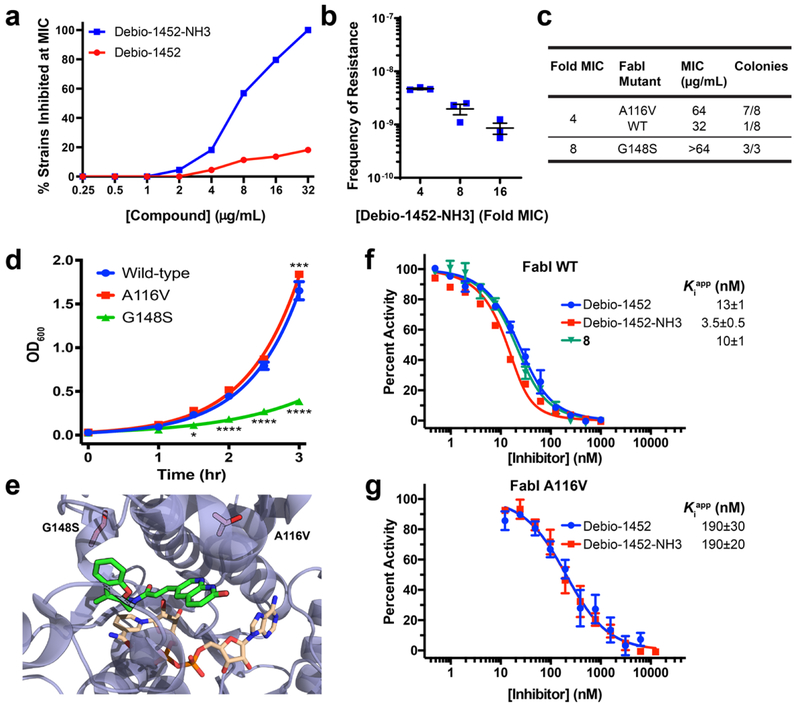
Debio-1452-NH3 retained potent antibacterial activity similar to parent Debio-1452 when assessed against Gram-positive S. aureus, and against E. coli strains with defective efflux (ΔtolC) or modified outer membranes (ΔrfaC) which render them susceptible to Gram-positive-only antibiotics (Table 1). Importantly, Debio-1452-NH3 also possesses antibacterial activity (MIC = 4 μg/mL) in E. coli strains with intact outer membranes, whereas Debio-1452 has no activity in these strains (MIC > 32 μg/mL, Table 1); while compounds 2 and 3 are also active, Debio-1452-NH3 is the most active of the modified compounds (Supplementary Table 4). Debio-1452-NH3 is active against strains of Gram-negative pathogens E. cloacae, K. pneumoniae, and A. baumannii, with MIC values from 4-8 μg/mL, while Debio-1452 and compound 8 (possessing a non-ionizable nitrogen at the 3-position of the naphthryidinone ring) are very potent against S. aureus and E. coli ΔtolC and ΔrfaC strains, but have no activity against Gram-negatives with intact outer membranes (Table 1). As anticipated, despite reaching statistically significant levels of accumulation (Extended Data Fig. 7), these amine analogues were not active against P. aeruginosa (Table 1, Supplementary Table 4), as these bacteria possess an additional FabI isoform, FabV, which enables rescue of the FAS-II pathway from inhibition of FabI23. Debio-1452-NH3 was further evaluated for its antibiotic activity against a panel of clinical isolates. Multidrug-resistant Enterobacteriaceae clinical isolates were notably more susceptible to Debio-1452-NH3 than Debio-1452 (Fig. 2a).
Fig. 2. Debio-1452-NH3 Mode of Action Studies.
a. Antimicrobial activity of Debio-1452 and Debio-1452-NH3 against a panel of Enterobacteriaceae clinical isolates. All experiments were performed in biological triplicate. b. Spontaneous frequency of E. coli MG1655 resistance to Debio-1452-NH3 as a function of concentration. Data shown represent the average of three independent experiments. Error bars represent standard error of the mean. c. Relative prevalence of FabI variants in E. coli resistant to Debio-1452-NH3. Resistant colonies of E. coli MG1655 were isolated following selection at 4- and 8-fold the MIC of Debio-1452-NH3. d. Fitness of E. coli isolates that harbor the FabI variant conferring resistance to Debio-1452-NH3 (either A116V or G148S) were evaluated relative to parental strain. Data points represent average of four independent experiments. Error bars represent standard error of the mean. Measurements were compared by repeated measure two-way ANOVA. Tukey’s multiple comparisons test was used to compare strains at each timepoint. Statistical significance from the wild-type strain (E. coli MG1655) is indicated with asterisks (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). e. Location of E. coli FabI amino acid substitutions conferring resistance to Debio-1452-NH3 (red residues, A116V and G148S). PDB: 4JQC. f. In vitro inhibition of E. coli FabI and (g) the A116V FabI variant by Debio-1452 and derivatives. Initial reaction rates were used to calculate percent activity relative to inhibitor-free reaction. Dose-response curves were fit to Morrison’s quadratic to determine apparent Ki. Data is represented as average of three independent experiments. Error bars represent standard error of the mean.
To assess the antibacterial mode-of-action of Debio-1452-NH3, E. coli MG1655 colonies resistant to this compound were generated via the large-inoculum method. The frequency of resistance (FoR) for Debio-1452-NH3 was 10−8–10−9 (Fig. 2b), similar to the FoR observed for Debio-1452 in S. aureus33. Sequencing of fabI in E. coli MG1655 resistant to Debio-1452-NH3 revealed amino acid substitutions near the active site of FabI, at positions A116 and G148 (Fig. 2c, 2e, and Extended Data Fig. 8). E. coli harboring FabI A116V, which possessed MIC values of 64 μg/mL with Debio-1452-NH3, showed little impact on fitness as assessed by bacterial growth rates, while strains harboring FabI G148S, which possessed MIC values of >64 μg/mL, had significantly reduced fitness (Fig. 2d). In addition, the FabI inhibitor triclosan showed minimal cross-resistance to G148S mutants, and no cross-resistance to A116V mutants (Supplementary Table 5).
Key compounds were also evaluated for their ability to inhibit E. coli FabI in vitro utilizing a standard biochemical assay34. The results of these experiments revealed Debio-1452, Debio-1452-NH3, and compound 8 to all be potent FabI inhibitors (Fig. 2f). Site directed mutagenesis of fabI allowed for production and in vitro enzymatic evaluation of the altered enzymes, and the inhibitors showed reduced potency against FabI A116V (Fig. 2g). Taken together these results point to Debio-1452-NH3 killing Gram-negative bacteria through inhibition of FabI.
Debio-1452-NH3 showed little toxicity to cultured mammalian cells at 72 hours (Supplementary Table 4). In addition, the activity of Debio-1452-NH3 was only marginally altered (2-fold higher MIC in S. aureus) in the presence of serum, while Debio-1452 showed an 8-fold increase in MIC in the presence of serum (Supplementary Table 4); in E. coli, the MIC of Debio-1452-NH3 increased 4-fold in the presence of human serum albumin (Supplementary Table 4). The lack of mammalian cell toxicity and the small MIC shift in the presence of serum encouraged the exploration of Debio-1452-NH3 in vivo. Debio-1452-NH3 was found to be well tolerated in mice at doses of 50 mg/kg daily (IP injection, once-a-day for 5 days), and pharmacokinetic analysis (Extended Data Fig. 9) suggested the potential for efficacy in mouse models of infection.
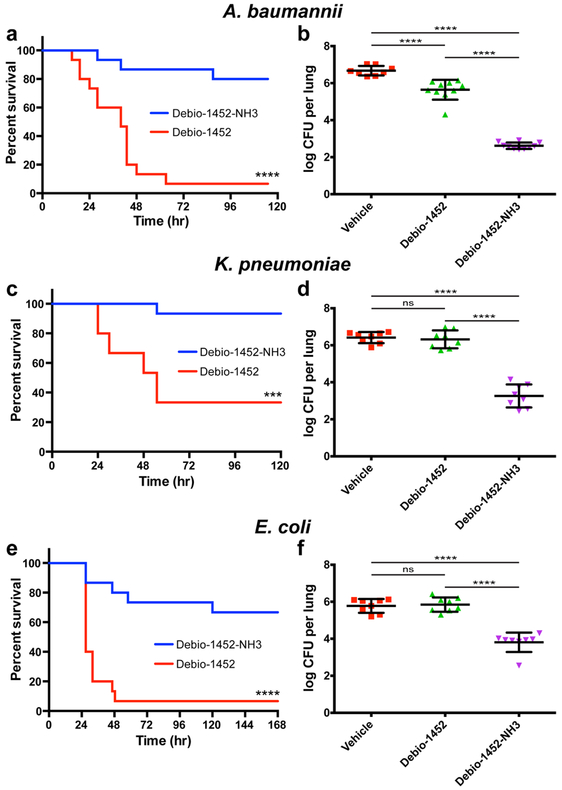
For the efficacy models Debio-1452 and Debio-1452-NH3 were assessed side-by-side in mouse models of infection with A. baumannii, K. pneumoniae, and E. coli, evaluating both overall survival and bacterial burden for each type of infection, using strains against which Debio-1452-NH3 has MIC values of 4-8 μg/mL. Mice were infected with A. baumannii W41979, a clinical isolate resistant to carbapenems and cephalosporins, to induce sepsis. Four doses of Debio-1452 or Debio-1452-NH3 were administered once-per-day (50 mg/kg, IV) to separate groups of mice, resulting in significantly increased survival of the Debio-1452-NH3 treated mice (Fig. 3a), and other experiments with A. baumannii showed Debio-1452-NH3 reduced bacterial burden in a mouse model of acute pneumonia (Fig. 3b). Analogous experiments demonstrated the efficacy of Debio-1452-NH3 against mice infected with carbapenem-resistant K. pneumoniae and colistin-resistant E. coli (Fig. 3c-f). Additionally, the efficacy of Debio-1452-NH3 was assessed in four additional mouse models of Gram-negative infection, utilizing strains that had higher MIC values (three strains of K. pneumoniae and one strain of A. baumannii, MIC = 16 μg/mL for all four strains). Debio-1452-NH3 was effective in reducing bacterial burden in mouse lungs during acute pneumonia in all four of these models (Extended Data Fig. 10).
Fig. 3. In vivo efficacy of Debio-1452-NH3.
a. Kaplan-Meier survival curve of mouse efficacy model of A. baumannii sepsis. Seven-week old CD-1 mice were infected with A. baumannii W41979 (MIC = 4 μg/mL with Debio-1452-NH3) (2.6x108 CFU/mouse, 15 mice per group) via IV injection. Mice were treated once-a-day for 4 days with FabI inhibitor (IV, 50 mg/kg). Log-rank test, p < 0.0001. b. Acute pneumonia infections initiated in CD-1 mice with A. baumannii W41979 (2.1x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (10 mice per group) 8, 30, and 48 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 72 h post-infection. c. Kaplan-Meier survival curve of in vivo efficacy model of K. pneumoniae sepsis. Seven-week old CD-1 mice were infected with K. pneumoniae BAA-1705 (MIC = 8 μg/mL with Debio-1452-NH3) (1.08x108 CFU/mouse, 15 mice per group) via IV injection. Mice were treated once-a-day for 4 days with FabI inhibitor (IV, 50 mg/kg). Log-rank test, p < 0.0001. d. K. pneumoniae bacterial burden study. Acute pneumonia infections initiated in CD-1 mice with K. pneumoniae BAA-1705 (4.4x108 CFU/mouse, intranasal) were treated with vehicle or FabI inhibitor (8 mice per group) 6, 23, and 45 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 72 h post-infection. e. Kaplan-Meier survival curve of in vivo efficacy model of E. coli sepsis. Seven-week old CD-1 mice were infected with E. coli AR-0493 (MIC = 4 μg/mL with Debio-1452-NH3) (1.6x108 CFU/mouse, 15 mice per group) via IV injection. Mice were treated once-a-day for 4 days with FabI inhibitor (IV, 50 mg/kg). Log-rank test, p < 0.001. f. E. coli bacterial burden study. Acute pneumonia infections initiated in CD-1 mice with E. coli AR-0493 (1.73x108 CFU/mouse, intranasal) were treated with vehicle or FabI inhibitor (8 mice per group) 6 and 28 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. Drugs were formulated in 20% sulfobutyl ether(7) β-cyclodextrin from solid immediately before treatment. For the Kaplan-Meier survival curves (a, c, e), the two-tailed log-rank (Mantel-Cox) test was used to compare survival curves. For the bacterial burden studies (b,d,f), data are shown as mean with error bars representing standard deviation of the mean. For the bacterial burden studies, significance was determined by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons. Statistical significance is indicated with asterisks (ns, not significant when p > 0.05 (d, p = 0.9732; f, p = 0.9843), *** p < 0.001, **** p < 0.0001).
The extreme challenge of identifying new classes of antibiotics for Gram-negative pathogens is apparent from the well-documented lack of success of large (millions of compounds) high-throughput screening campaigns4, and this dearth of leads has directly led to the current situation where no new antibacterial classes for Gram-negative infections have been FDA approved in over 50 years1. Although most all Gram-positive-only compounds would kill Gram-negatives if they were not accumulation limited, in many cases even years of effort and the synthesis of hundreds of compounds has not provided Gram-negative active versions of high-value Gram-positive-only antibiotics. Thus, while optimization of bioavailability, solubility, and other important parameters has been a triumph of modern drug discovery, building in Gram-negative activity remains out of reach7.
The description herein of eNTRyway should streamline Gram-negative antibiotic discovery efforts through its ability to predict compound accumulation in E. coli in silico, therefore reducing the number of compounds that need to be synthesized and evaluated. Debio-1452 is an interesting case study, as the original lead was discovered in a high-throughput biochemical screen17,18. The repeated inability to build-in whole cell accumulation into lead compounds arising from biochemical screens4, coupled with the reality that all classes of approved antibiotics were discovered in whole-cell screening9, has led to the sensible suggestion that whole-cell screens be prioritized over target-based biochemical HTS in antibacterial drug discovery35. However, the demonstration herein that eNTRyway can be used to assist identification of FabI inhibitors that exhibit whole-cell accumulation in E. coli and consequently whole-cell activity in Gram-negative bacteria including E. coli, E. cloacae, K. pneumoniae, and A. baumannii suggests the intriguing possibility that Gram-negative activity might also be readily built-in for other Gram-positive-only compounds, and also for some of the numerous existing hits from biochemical HTS. Indeed, as shown in Fig. 1b there are dozens of attractive starting points for these efforts, including many compounds that are active against targets not currently represented by approved antibiotics and would thus be less likely to encounter pre-existing resistance3.
While appending of an amine onto an otherwise hydrophobic organic compound has the potential to be disruptive to target engagement, known structure-activity relationships and X-ray data can be leveraged to select the proper position for modification. The majority of compounds identified by eNTRyway as good conversion targets have well-described SAR, published crystal structures, and target engagement validation (Fig. 1b, Supplementary Table 1). This abundance of characterization data engenders optimism that many of these compounds can be converted to versions with activity against Gram-negative pathogens. While the eNTRy rules were developed from studies on E. coli, the successful conversions of deoxynybomycin14 and now Debio-1452 to compounds that have activity against multiple Gram-negative ESKAPE pathogens suggests overlap in the chemical features required for compound accumulation in problematic Gram-negative organisms. The dozens of antibiotics that are safe to humans that possess a primary amine7 demonstrate that there is no general problem with this functional group, but of course toxicity of any new compound will need to be evaluated. As demonstrated herein with Debio-1452-NH3, addition of a primary amine will likely provide a general in vivo pharmacokinetic advantage for an antibiotic (compared to the parent compound lacking an amine) in terms of reducing protein binding and enhancing activity in serum. The availability of eNTRyway allows rapid in silico prediction of compound accumulation in E. coli and subsequent prioritization of compounds to synthesize and evaluate, and Debio-1452-NH3 is a promising lead for further development. eNTRyway is one of a growing number of web tools designed to facilitate antibacterial research36, and we anticipate this web application will assist in the development of many antibiotics with activity against Gram-negative pathogens.
Methods
Bacterial strains.
S. aureus ATCC 29213, E. coli MG1655, E. coli BAA-2340, E. coli BAA-2469, E. coli BAA-2471, E. cloacae BAA-2341, E. cloacae BAA-2468, K. pneumoniae BAA-1705, K. pneumoniae BAA-2342, K. pneumoniae BAA-2470, K. pneumoniae BAA-2472, and K. pneumoniae BAA-2473 were obtained from ATCC. E. coli BW25113, E. coli JW5503, and E. coli JW3596 were obtained from the KEIO Collection. E. cloacae ATCC 29893 was provided by W. van der Donk (UIUC). E. coli, K. pneumoniae, and A. baumannii AR strains were obtained from the CDC and FDA AR Isolate Bank. E. coli F20987, E. coli M66623, E. cloacae S28901.1, K. pneumoniae M14723, K. pneumoniae M67198, K. pneumoniae M67297, K. pneumoniae S20595, K. pneumoniae S47889, A. baumannii W41979, A. baumannii F19521, A. baumannii M13100, and A. baumannii WO22 were obtained from the University of Illinois Chicago Medical School. A. baumannii KB304, A. baumannii KB343, and A. baumannii KB357 were provided by J. Quale.
Antimicrobial susceptibility tests.
Susceptibility testing was performed in biological triplicate, using the micro-dilution broth method as outlined by the Clinical and Laboratory Standards Institute. Bacteria were cultured with cation-adjusted Muller-Hinton broth (Sigma-Aldrich, Cat# 90922) in round-bottom 96-well plates (Corning, Cat# 3788). Human serum (pooled gender, 0.2 μm filtered) was purchased from BioIVT (Hicksville, NY). Human serum albumin was purchased from Sigma-Aldrich (Cat# A1653).
Accumulation assay.
The accumulation assay was performed in triplicate in batches of twelve samples (4 compounds total, 3-time points, 1 sample/time point), with each batch containing tetracycline as a positive control. E. coli MG1655, E. coli BW25113, E. coli ΔtolC, or P. aeruginosa PAO1was used for these experiments. For each replicate, 2.5 mL of an overnight culture of E. coli was diluted into 250 mL of fresh Luria Bertani (LB) broth (Lennox), or 5 mL of an overnight culture of P. aeruginosa was diluted in 250 mL of fresh Tryptic Soy Broth (TSB), and grown at 37 °C with shaking to an optical density (OD600) of 0.70. The bacteria were pelleted at 3,220 r.c.f. for 10 min at 4 °C and the supernatant was discarded. The pellets were re-suspended in 40 ml of phosphate buffered saline (PBS) and pelleted as before, and the supernatant was discarded. The pellets were resuspended in 10.8 mL of fresh PBS and aliquoted into twelve 1.7 mL Eppendorf tubes (890 μL each). The number of colony-forming units (CFUs) was determined by a calibration curve. The samples were equilibrated at 37°C with shaking for 5 min, compound was added (final concentration = 50 μM), and then samples were incubated at 37 °C with shaking for either 10 min, 30 min, 1 hour, or 2 hours. These time points are short enough to minimize metabolic and growth changes (no changes in OD600 or CFU count observed). After incubation, 800 μL of the cultures were carefully layered on 700 μL of silicone oil [9:1 AR20 (Acros, Cat#174665000)/Sigma High Temperature (Sigma-Aldrich, Cat#175633), cooled to −78°C]. Bacteria were pelleted through the oil by centrifuging at 13,000 r.c.f. for 2 min at room temperature (supernatant remains above the oil); the supernatant and oil were then removed by pipetting. To lyse the samples, each pellet was resuspended in 200 μL of water, and then they were subjected to three freeze-thaw cycle of three minutes in liquid nitrogen followed by three minutes in a water bath at 65°C. The lysates were pelleted at 13,000 r.c.f. for 2 min at room temperature and the supernatant was collected (180 μL). The debris was re-suspended in 100 μL of methanol and pelleted as before. The supernatants were removed and combined with the previous supernatants collected. Finally, remaining debris was removed by centrifuging at 20,000 r.c.f. for 10 min at room temperature.
Supernatants were analyzed with the 5500 QTRAP LC/MS/MS system (Sciex, Framingham, MA) in the Metabolomics Lab of Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign. Software Analyst 1.6.2 was used for data acquisition and analysis. The 1200 series HPLC system (Agilent Technologies, Santa Clara, CA) includes a degasser, an autosampler, and a binary pump. The LC separation was performed on an Agilent Zorbax SB-Aq column (4.6 x 50mm, 5μm) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetontrile). The flow rate was 0.3 mL/min. The linear gradient was as follows: 0-3 min, 100% A; 10-15 min, 2% A; 16-20.5 min, 100% A. The autosampler was set at 15 °C. The injection volume was 1 μL. Mass spectra were acquired under positive electrospray ionization (ESI) with the voltage of 5,500 V. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 33, 65, and 60 psi, respectively. Multiple reaction monitoring (MRM) was used for quantitation with external calibration: Debio-1452 m/z 376.1 --> m/z 244.1; Debio-1452-NH3 m/z 391.2 --> m/z 171.0; Compound 3 m/z 419.3 --> m/z 258.0; Compound 2 m/z 405.2 --> m/z 201.1. The limit of quantitation of each compound (S/N = 10) is 20 nM, 5 nM, 10 nM, and 10 nM, respectively. All compounds reached nM concentrations above the limit of quantitation.
Cell culture.
IMR90 cells were obtained from ATCC. IMR90 cells were grown in EMEM with 10% fetal bovine serum (Gemini Benchmark, Cat# 100-106), 100 U/mL penicillin, and 100 μg/mL streptomycin. All cells were cultured at 37°C in a 5% CO2 environment. Cell lines were authenticated externally by commercial vendor and were inspected visually in-house. Cell lines were not tested for mycoplasma contamination. Media was prepared by the University of Illinois School of Chemical Sciences Cell Media Facility. Sex and age of cell lines: IMR90 (Female, 16 weeks gestation).
Cell viability.
Cells seeded (IMR90: 5,000 cells/well) a 96-well plate (Greiner Bio-One, Cat# 655180) and were allowed to attach overnight. Cells were treated with investigational compounds in DMSO (for compound screening compounds were tested at 30 μM, 1% DMSO final, 100 μL/well; for IC50 determination: concentrations of compounds tested were 10 nM to 300 μM, 1% DMSO final, 100 μL/well)37. Raptinal (100 μM) was used as a dead control.38 On each plate at least 3 technical replicates per compound were performed. After 72 h post-treatment, cell viability was assessed using the Alamar Blue method39. Stock Alamar Blue solution [10 μL, 440 μM resazurin (Sigma-Aldrich, Cat# R7017) in sterile 1X PBS] was added to each well, and plate was incubated for 3-4. Conversion of Alamar Blue was measured with plate reader (SpectraMax M3, Molecular Devices) by fluorescence (ex 555 nm, em 585 nm, cutoff 570 nm, autogain). Percent death was determined by normalizing to DMSO-treated cells and Raptinal-treated cells. For IC50 determination, the data were plotted as compound concentration versus percent dead cells and fitted to a logistic-dose response curve using OriginPro 2015 (OriginLab, Northampton, MA). The data were generated in triplicate, and IC50 values are reported as the average of three separate experiments along with standard error of the mean.
Molecular docking.
Docking of Debio-1452 derivatives into FabI crystal structures was performed with the Small-Molecule Drug Discovery Suite 2018-4 (Schrodinger, New York, NY). Co-crystal structures of Debio-1452 bound to E. coli FabI (PDB: 4JQC) and S. aureus FabI (PDB: 4FS3) were prepared using the Protein Prep Wizard with default settings and used to build receptor grids. For S. aureus FabI allowed rotation for Tyr157 and NADP hydroxyls. For E. coli FabI allowed rotation for Tyr156 and NAD hydroxyls. Positional constraints were applied on the benzofuran ring (center of mass must be within 2 Å of initial position). H-bonding constraints were applied to Tyr156 and Ala95 (E. coli numbering). Ligands were prepared with LigPrep and amines were protonated. Both enantiomers of each ligands were docked with Glide XP. The poses of higher scoring enantiomers were refined and ΔΔGbind was calculated using Prime MM-GBSA. Protein residues within 5 Å of the ligand were sampled using the hierarchical sampling procedure in Prime MM-GBSA.
Selection of resistant mutants.
Resistant mutants were selected via the large inoculum method. Briefly, E. coli MG1655 (1.8x109 CFU) were plated on 100 mm plates of LB agar containing 64, 32, and 16 μg/mL Debio-1452-NH3. Colonies were visible after incubating at 37°C for 48 h. Resistant colonies were confirmed by streaking on selective media with the same concentration of Debio-1452-NH3.
Sequencing of fabI.
FabI was amplified by colony PCR. Colonies were picked and diluted in 100 μL sterile H2O. PCR reactions were setup by combining 25 μL MiFi Mix (Bioline, London, UK), 1 μL 20μM primer mix (EcFabI-PCR-FOR and EcFabI-PCR-REV), 10 μL template, and 14 μL H2O. Reaction was performed on C1000 Thermal Cycler (Bio-Rad, Hercules, CA) with the following conditions: initial denature 95°C, 3 min; denature 95°C, 15 s; anneal 57°C, 15 s; extend 72°C, 30 s; final extend 3 min; 35 cycles. 5 μL portion of PCR reaction mixture was analyzed by agarose gel to confirm single 1.4 kbp product. PCR products were purified using GeneJET PCR Purification Kit (Thermo Scientific). PCR amplicons were submitted to the Core DNA Sequencing Facility at the University of Illinois at Urbana-Champaign for Sanger sequencing with overlapping internal primers (EcFabI-Seq-FOR and EcFabI-Seq-REV). All primers were obtained from Integrated DNA Technologies (Coralville, IA).
| Primer | Sequence |
|---|---|
| EcFabI-PCR-FOR | 5’- GGGGCCAGCGTTTCTTTTTC -3’ |
| EcFabI-PCR-REV | 5’- AAACATGGAGACGGTGCTGG -3’ |
| EcFabI-Seq-FOR | 5’- ATAGCTACTCACAGCCAGGT -3’ |
| EcFabI-Seq-REV | 5’- GAAGGGGAGAAAGACGGATC -3’ |
Plasmids.
Expression vectors for FabI were a generous gift from Peter Tonge (Stonybrook University, NY). Site directed mutagenesis was performed with NEB Q5 Site Directed Mutagenesis Kit according to kit instructions with the primers and annealing temperatures listed below. Mutations were confirmed by Sanger sequencing using T7 promoter and T7 terminator primers.
| Species | Variant | Forward Primer | Reverse Primer | Ta |
|---|---|---|---|---|
| E. coli | A116V | 5′- TTCAAAATTGTCCACGACATCAGCTC -3′ | 5′- GCCTTCACGGGTAACGGC -3′ | 67 |
| E. coli | G148S | 5′- TTCCTACCTTAGCGCTGAGCG -3′ | 5′- AGGGTCAGCAGGGCAGAA -3′ | 67 |
Expression and purification of E. coli FabI.
E. coli BL21 (DE3) pLysS (Novagen) were transformed with pET15-ecFabI. Overnight culture (10 mL) in LB supplemented with 50 μg/mL ampicillin from single colony was diluted into 1 L LB + 50 μg/mL ampicillin. Bacteria were grown at 37°C, with shaking at 250 rpm until OD600 reached 0.8. Culture was cooled to 18°C and induced with 0.5 mM IPTG for 18 h at 18°C. Bacteria were harvested by centrifugation (5,000xg, 10 min, 4°C). Cell pellets were flash frozen and store at −20°C pending purification. Frozen cell pellets were thawed on ice and resuspended (5 mL per gram wet pellet, typically 20-30 mL) with 0.5% CHAPS, 1 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL pepstatin A, and 2 μg/mL aprotinin in binding buffer [20 mM Tris (pH 7.9), 500 mM NaCl, 5 mM imidazole]. Bacteria were lysed by sonication on ice (30%, 10 s pulse, 20 s rest, 5 min total). Lysate was clarified by centrifugation (35,000xg, 1 h, 4°C) and filtration through 0.2 μm syringe filter. Lysate was incubated with 5 mL Co-NTA agarose (HisPur cobalt resin, Thermo-Fisher Scientific Cat#89965, pre-equilibrated with binding buffer) for 30 min at 4°C with gentle rocking. Agarose-containing lysate was transferred to column and flow through discarded. Column was washed with 2 column volumes of binding buffer followed by 10 column volumes of wash buffer [20 mM Tris (pH 7.9), 500 mM NaCl, 60 mM imidazole]. Protein was eluted with 15 mL elution buffer [20 mM Tris (pH 7.9), 500 mM NaCl, 300 mM imidazole]. Fractions containing protein were identified by SDS-PAGE, subjected to dialysis against FabI storage buffer [60 mM PIPES (pH 8.0), 150 mM NaCl, 1 mM EDTA], and concentrated with Amicon spin filter. Protein solution was aliquoted, flash frozen in liquid nitrogen, and stored at −80°C. Protein concentration was determined by BCA assay (ThermoFisher Cat#23227).
In vitro FabI inhibition assay.
NADH (Sigma-Aldrich, Cat# N8129) and crotonoyl-CoA (Sigma-Aldrich, Cat# 28007) were both diluted into activity buffer [100 mM potassium glutamate (pH 7.8)], and working solution was dispensed into two columns of round-bottom 96-well plate (330 μL per well). Inhibitors, dissolved in DMSO at 200X final concentration, were added to each well with a multichannel pipette (2.48 μL per well). After thorough mixing, this mixture was transferred to UV-transparent plate (UV-STAR half-area 96-well plate, Greiner Bio-One, Cat#675801, 100 μL per well, three technical replicates per inhibitor concentration). Plate was placed in plate-reader (SpectraMax 190, Molecular Devices) under temperature control (25°C), and temperature was allowed to equilibrate. Fresh aliquot of enzyme from −80°C was thawed on ice and diluted in activity buffer. Enzyme working solution was added to plate (50 μL per well, 20 nM enzyme final). After shaking for 5 s, reaction progress was monitored by absorbance at 340 nm every 15 s for 90 min. Linear portion of reaction progress curve was fit using plate-reader software (SoftMax Pro 7.0). Percent activity was calculated relative to DMSO-only and no-enzyme controls in each plate. Percent activity curves were fit to Morrison’s quadratic using Graphpad Prism 8.2.1.441 to obtain apparent Ki.
Mouse MTD of Debio-1452-NH3.
The protocol was approved by the IACUC at the University of Illinois at Urbana-Champaign (Protocol Number:16144). In these studies, 10- to 12-week-old female C57BL/6 mice purchased from Charles River were used. Mice were randomly chosen and divided into subsequent groups. No additional randomization was used to allocate experimental groups; blinding was not performed for subsequent quantitation. The maximum tolerated dose (MTD) of single compound was determined first. Debio-1452 amine analogues were formulated in 20% sulfobutyl ether(7) β-cyclodextrin in sterile water. Debio-1452-NH3 (1), and compounds 2 and 3 were given by IP injection and mice were monitored for signs of toxicity for 2 weeks (single dose). For multiple doses, the compound was given by daily IP for 5 consecutive days and mice were monitored for signs of toxicity for 1 month. MTD was the highest dosage with acceptable toxicity (e.g. < 20% weight loss). Single dose MTDs were initially determined. Debio-1452-NH3 (1) was well tolerated at a single dose of 50 mg kg−1 and compound 3 was well tolerated at a single dose of 100 mg kg−1. Further analysis showed that Debio-1452-NH3 (1) was well tolerated with daily dosing of 50 mg kg−1 for 5 consecutive days. The MTD of Debio-1452-NH3 (1) was used to inform the dosing schedule used in subsequent efficacy studies.
Pharmacokinetic assessment.
The protocol was approved by the IACUC at the University of Illinois at Urbana-Champaign (Protocol Number: 16144). 10- to 12-week-old female C57BL/6 mice were purchased from Charles River Laboratories and acclimated for 4-7 days. All animals were housed in a pathogen-free environment and received sterile food and water. Mice were randomly chosen and divided into subsequent groups. No additional randomization was used to allocate experimental groups; blinding was not performed for subsequent quantitation. Debio-1452-NH3 was formulated in 20% sulfobutyl ether(7) β-cyclodextrin in sterile water. mice were treated with Debio-1452-NH3 (50 mg kg−1) via i.p. injection, with three mice per time point (0, 15, 30, 45, 60, 120, 240 480, 960, and 1440 min). At specified time points, mice were killed and blood was collected, centrifuged and the serum was frozen at −80 °C until analysis. The proteins in a 10-μl aliquot of serum were precipitated by the addition of 50 μl of methanol with the addition of 10 μl of internal standard. The sample was then vortexed and centrifuged to remove the proteins. Supernatants were analyzed with the 5500 QTRAP LC/MS/MS system (Sciex, Framingham, MA) in the Metabolomics Lab of Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign. Software Analyst 1.6.2 was used for data acquisition and analysis. The 1200 series HPLC system (Agilent Technologies, Santa Clara, CA) includes a degasser, an autosampler, and a binary pump. The LC separation was performed on an Agilent Zorbax SB-Aq column (4.6 x 50mm, 5μm) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetontrile). The flow rate was 0.35 mL/min. The linear gradient was as follows: 0-2 min, 100% A; 8-12 min, 2% A; 12.1-17 min, 100% A. The autosampler was set at 10 °C. The injection volume was 5 μL. Mass spectra were acquired under positive electrospray ionization (ESI) with the voltage of 5,500 V. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 33, 65, and 60 psi, respectively. Multiple reaction monitoring (MRM) was used for quantitation with external calibration: Debio-1452-NH3 m/z 391.2 --> m/z 171.0. The limit of quantitation of (S/N = 10) is 1 nM. Pharmacokinetic parameters were calculated with a one-compartment model using a nonlinear regression program (Phoenix WinNonlin Version 8.1, Certara USA Inc., Princeton, NJ 08540 USA).
Bacterial sepsis survival model.
The protocol was approved by the IACUC at the University of Illinois at Urbana-Champaign (Protocol Number: 17271). Six- to eight week -old male CD-1 mice were purchased from Charles River Laboratories and acclimated for 4-7 days. All animals were housed in a pathogen-free environment and received sterile food and water. Mice were randomly chosen and divided into subsequent groups. No additional randomization was used to allocate experimental groups; blinding was not performed for subsequent quantitation. For the preparation of each inoculum, overnight cultures of clinical isolates were diluted into LB broth and grown to log-phase growth at 37 °C. Infection was established via 100 μL retro-orbital injection of bacteria: A. baumannii (W41979) 2.6x108 CFU/mouse, K. pneumoniae (BAA-1705) 1.08x108 CFU/mouse, or E. coli (AR-0493) 1.6x108 CFU/mouse. Mice were treated once-a-day for four days with either Debio-1452 Tosylate or Debio-1452-NH3 (retro-orbital, 50 mg/kg). Drugs were formulated in 20% sulfobutyl ether(7) β-cyclodextrin from solid immediately before treatment. For survival analyses, a Kaplan-Meier Log Rank Survival Test was performed using GraphPad Prism 8.2.1.441.
Acute pneumonia bacterial burden model.
The protocol was approved by the IACUC at the University of Illinois at Urbana-Champaign (Protocol Number: 17271). Acclimated CD-1 mice (see above) were infected via intranasal inoculation of bacteria: A. baumannii (W41979) 2.1x108 CFU/mouse, K. pneumoniae (BAA-1705) 4.4x108 CFU/mouse, or E. coli (AR-0493) 1.73x108 CFU/mouse, A. baumannii (M13100) 2.1x108 CFU/mouse, intranasal, K. pneumoniae (S20595) 1.9x108 CFU/mouse, intranasal, K. pneumoniae (BAA1473) 3.4x108 CFU/mouse, intranasal, K. pneumoniae (AR0347) 2.5x108 CFU/mouse, intranasal. Infected mice were then treated once daily for three days with either vehicle, Debio-1452 Tosylate, or Debio-1452-NH3 (retro-orbital, 50 mg/kg). At 48 h post-infection (E. coli (AR-0493), A. baumannii (M13100), K. pneumoniae (S20595, BAA1473, AR0347)) or 72 h post-infection (A. baumannii (W41979) and K. pneumoniae (BAA-1705)), CFU were determined in the lungs through serial dilutions. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison tests.
eNTRyway Development
The eNTRyway web application accepts SMILES strings provided by users. After validating the SMILES string, an initial guess of 3D coordinates was performed using the ETKDG method followed by brief minimization with the Merck Molecular Force Field (MMFF94). An ensemble of conformers is generated using Open Babel’s genetic algorithm with the following options: “--conformer --nconf 200 --mutability 5 --children 5 --convergence 5 --score energy -writeconformers” The globularity and PBF of each conformer are calculated, and the mean values are stored for each molecule. Rotatable bonds are counted according to the following rules: (1) must be a single or triple bond, (2) must include two heavy atoms, (3) cannot be terminal, (4) cannot be in a ring, (5) if a single bond that includes one sp hybridized atom, not rotatable. Functional groups are detected with Open Babel using the following SMARTS filters:
Primary Amine: '[\$([N;H2;X3][CX4]),\$([N;H3;X4+][CX4])]'
Secondary Amine: '[\$([CX4][N;H1;X3][CX4]),\$([CX4][N;H2;X4+][CX4])]'
Tertiary Amine: '[NX3]([CX4])([CX4])[CX4]'
A Rails web interface was developed to handle job submission and scheduling. Deployed on AWS Elastic Beanstalk, the web interface is accessible at entryway.org. Alternatively, the analysis workflow can be run on command-line using the entry-cli tool (https://github.com/HergenrotherLab/entry-cli).
Statistical analyses.
GraphPad Prism 8.2.1.441 was used for data analysis and figure generation. Data are shown as the mean ± s.e.m. Statistical significance was determined by one-way ANOVA (with Tukey’s multiple comparisons tests) for two groups at a single time point, two-way ANOVA (with Sidak’s multiple comparison’s test) for two groups at multiple time points, or two-way ANOVA (with Tukey’s multiple comparisons tests) for three or more groups at multiple time points. Kaplan-Meier survival curves were compared using the two-tailed log-rank (Mantel-Cox) test. P < 0.05 was considered statistically significant. In this study, no statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during the experiments and outcome assessments.
Data availability.
The main data supporting the findings of this study are available within the article and the Supplementary Information. Source data underlying Figs. 2, 3 and Extended Data Figs. 5, 6, 7, 9, and are provided in as Source Data files. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Code availability.
Source code for eNTRyway for local use is available on Github (https://github.com/HergenrotherLab/entry-cli).
Extended Data
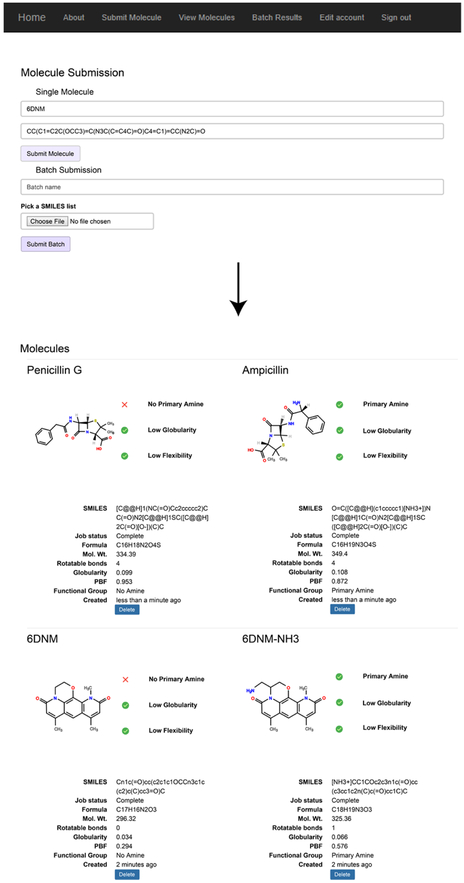
Extended Data Fig. 1. Overview of eNTRyway functionality and examples of visual outputs.
eNTRyway classifies molecules that are likely capable of accumulating in E. coli. eNTRyway calculates physiochemical properties of molecules and compares to a training set of compounds. SMILES strings for this analysis are generated in Open Babel using canonicalized atom order. Compounds containing an ionizable nitrogen (primary amines are most favorable), low three-dimensionality (globularity ≤ 0.25), and low flexibility (rotatable bonds ≤ 5) have a significantly higher probability of accumulating in E. coli. It is important to note that localized polarity and sterics about the amine are important considerations when implementing the eNTRy rules as these factors are not explicitly included in the eNTRYway results. Previously, it was found that increased amphiphilic moment (vsurf_A) correlated with increased accumulation while sterically encumbered primary amines displayed decreased accumulation.14 Using eNTRyway to reanalyze the diverse compound collection published in Richter et al14 (Tables S2-S4; 188 compounds), we found that the accuracy is 84% (Supplementary Table 6).
Extended Data Fig. 2. Docking of Debio-1452-NH3 in S. aureus FabI.
Extended Data Fig. 3. Synthesis of modified naphthyridinones.
di-tert-butyl dicarbonate; DIPEA, diisopropylamine; LHMDS, Lithium bis(trimethylsilyl)amide; THF, tetrahydrofuran; Cy, cyclohexyl; DMA, dimethylacetamide.
Extended Data Fig. 4. Synthesis of amine-containing derivatives of Debio-1452.
Cy, cyclohexyl; DIPEA, diisopropylamine; DMA, dimethylacetamide.
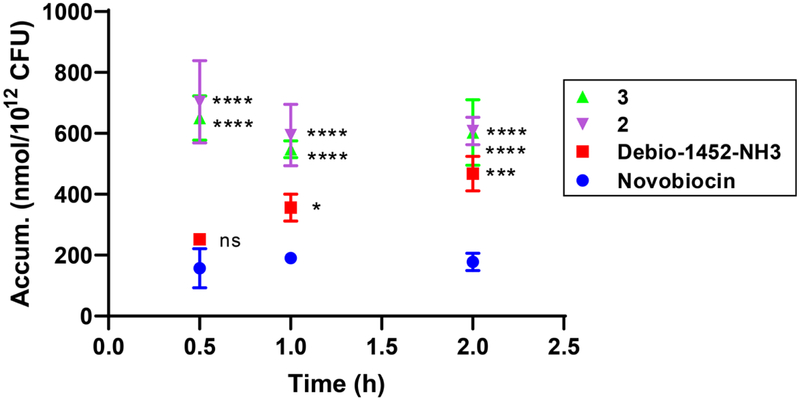
Extended Data Fig. 5. Time-course accumulation of Debio-1452 and derivatives in E. coli MG1655.
Bacterial cells were washed with and suspended in PBS. Cells were exposed to compound (50 μM final) and incubated at 37°C with shaking for 10 min, 30 min, 1 h, and 2 h. All cells were viable at these time points. Extra-cellular compound was removed, cells were lysed, and amount of compound in lysate was quantified by LC-MS/MS. Accumulation reported in nmol per 1012 colony-forming units (CFUs). Data shown represents the average of three independent experiments. Error bars represent the standard deviation of the mean. Measurements were compared by ordinary two-way ANOVA. Tukey’s multiple comparisons test was used to compare compounds at each timepoint. Statistical significance from Debio-1452 is indicated with asterisks (ns, not significant when p > 0.05 (Debio-1452-NH3, 10 min, p = 0.1719; Debio-1452-NH3, 0.5 h, p = 0.5522), *** p < 0.001, **** p < 0.0001).
Extended Data Fig. 6. Time-course accumulation of Debio-1452 and derivatives in E. coli BW25113 and E. coli ΔtolC JW5503.
a. Debio-1452 accumulation over time. b. Debio-1452-NH3 accumulation over time. c. Compound 2 accumulation over time. d. Compound 3 accumulation over time. Bacterial cells were washed with and suspended in PBS. Cells were exposed to compound (50 μM final) and incubated at 37°C with shaking for 10 min, 30 min, 1 h, and 2 h. All cells were viable at these time points. Extra-cellular compound was removed, cells were lysed, and amount of compound in lysate was quantified by LC-MS/MS. Accumulation reported in nmol per 1012 colony-forming units (CFUs). Data shown represents the average of three independent experiments. Error bars represent standard deviation of the mean. Measurements were compared by ordinary two-way ANOVA. Sidak’s multiple comparisons test was used to compare compounds at each timepoint. Statistical significance of E. coli ΔtolC data points relative to E. coli BW25113 data points is indicated with asterisks (ns, not significant when p > 0.05 (b, 2.0 h, p = 0.0897; c, 10 min, p = 0.3571; c, 1.0 h, p = 0.9728; c, 2.0 h, p = 0.9894; d, 2.0 h, p = 0.0791), ** p < 0.01, *** p < 0.001, **** p < 0.0001).
Extended Data Fig. 7. Time-course accumulation of Debio-1452 derivatives in P. aeruginosa PAO1.
Bacterial cells were washed with and suspended in PBS. Cells were exposed to compound (50 μM final) and incubated at 37°C with shaking for 30 min, 1 h, and 2 h. All cells were viable at these time points. Extra-cellular compound was removed, cells were lysed, and amount of compound in lysate was quantified by LC-MS/MS. Accumulation reported in nmol per 1012 colony-forming units (CFUs). Data shown represents the average of three independent experiments. Error bars represent the standard deviation. Measurements were compared by ordinary two-way ANOVA. Tukey’s multiple comparisons test was used to compare compounds at each timepoint. Statistical significance from novobiocin, a negative antibiotic control, is indicated with asterisks (ns not significant when p > 0.05 (Debio-1452-NH3, 0.5 h, p = 0.3609, * p < 0.05, *** p < 0.001, **** p < 0.0001).
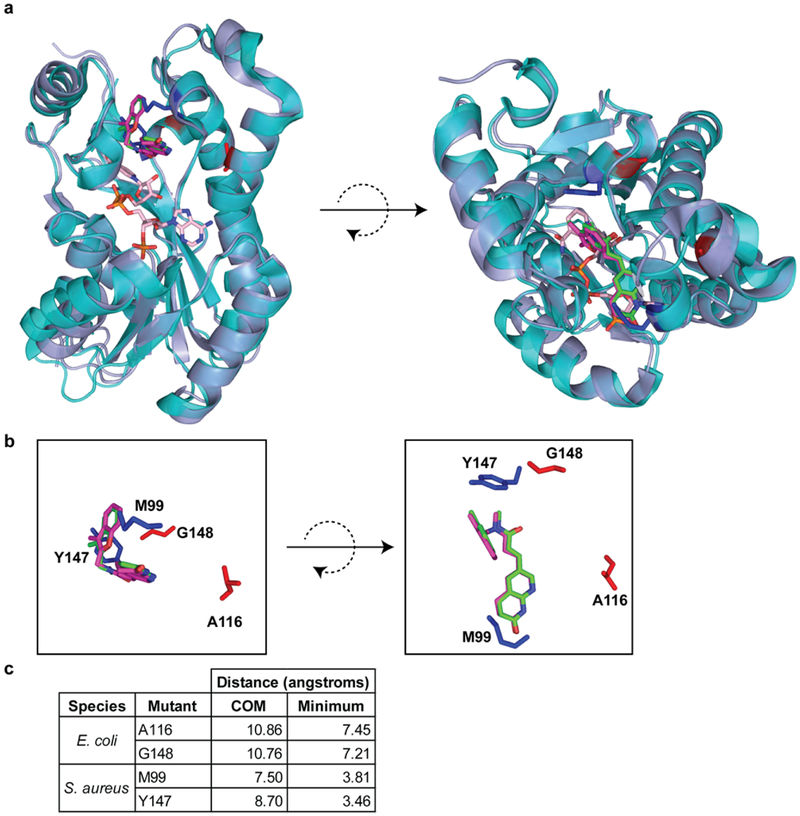
Extended Data Fig. 8. Location of mutated residues that confer resistance to Debio-1452 and Debio-1452-NH3.
a. Structural alignment of E. coli FabI (periwinkle, PDB 4JQC) and S. aureus FabI (turquoise, PDB 4FS3). Residues mutated in E. coli are highlighted in red. Residues mutated in S. aureus are highlighted in blue. Debio-1452 from E. coli structure is green and from S. aureus is magenta. NADPH from S. aureus structure is pink. b. Same poses as a but without cartoon representation of protein backbone and NADPH. c. Distances from mutation sites and Debio-1452, either between centers of mass (COM) or the minimum pairwise distance between atoms within mutated residues and Debio-1452.
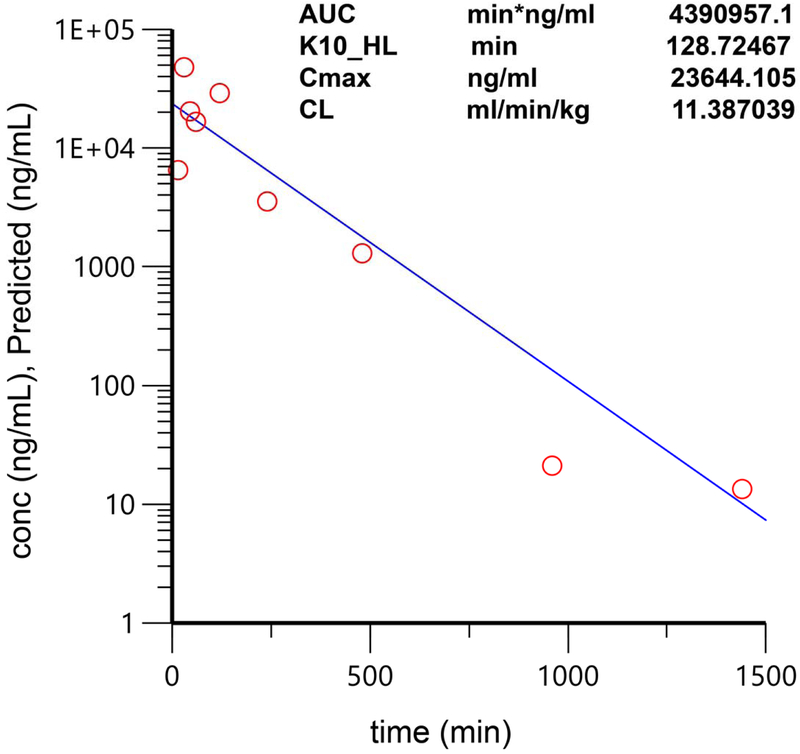
Extended Data Fig. 9. Pharmacokinetic analysis of Debio-1452-NH3.
C57/BL6 mice were treated with 50 mg/kg Debio-1452-NH3 via intraperitoneal injection, with three mice per time point (0, 15, 30, 45, 60, 120, 240 480, 960, and 1440 min). After the indicated time points, mice were sacrificed and the serum concentrations of Debio-1452-NH3 were determined by LC-MS/MS. Data are shown mean and represent the average of three independent experiments. The pharmacokinetic parameters shown in the figure above were calculated with a one-compartment model using a nonlinear regression program (Phoenix WinNonlin Version 8.1, Certara USA Inc., Princeton, NJ 08540 USA).
Extended Data Fig. 10. In vivo efficacy of Debio-1452-NH3 with strains that are less susceptible in cell culture.
a. Acute pneumonia infections initiated in CD-1 mice with A. baumannii M13100 (2.1x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6.5 and 23 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. b. Acute pneumonia infections initiated in CD-1 mice with K. pneumoniae S20595 (1.9x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6.5 and 22.5 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. c. Acute pneumonia infections initiated in CD-1 mice with K. pneumoniae BAA2473 (3.4x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6 and 22.5 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. d. Acute pneumonia infections initiated in CD-1 mice with K. pneumoniae AR0347 (2.5x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6 and 23 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. Drugs were formulated in 20% sulfobutyl ether(7) β-cyclodextrin from solid immediately before treatment. For the bacterial burden studies (a-d), data are shown as mean and error bars represent standard deviation. Significance was determined by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons. Statistical significance is indicated with asterisks (ns, not significant when p > 0.05 (a, p = 0.8582; b, p = 0.1019; c, p = 0.7600; d, p = 0.7536), **** p < 0.0001). Debio-1452-NH3 has an MIC of 16 μg/mL for all four strains used in these mouse models.
Supplementary Material
Acknowledgments
We are grateful to the NIH (AI136773, GM118575) and U. of Illinois for funding this work. We thank Prof. Peter Tonge (SUNY Stonybrook) for the FabI expression vector, Prof. Levent Dirikolu (School of Veterinary Medicine, Louisiana State University) for pharmacokinetic data analysis and Prof. John Cronan for helpful and insightful comments regarding this work.
Footnotes
Competing interest. The University of Illinois has filed patents on some compounds described in this manuscript.
References
- 1.Lewis K Platforms for antibiotic discovery. Nat Rev Drug Discov 12, 371–387, doi:nrd3975 [pii] 10.1038/nrd3975 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Cassini A et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19, 56–66, doi: 10.1016/S1473-3099(18)30605-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theuretzbacher U et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect Dis 19, e40–e50, doi: 10.1016/S1473-3099(18)30513-9 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Tommasi R, Brown DG, Walkup GK, Manchester JI & Miller AA ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 14, 529–542, doi: 10.1038/nrd4572nrd4572 [pii] (2015). [DOI] [PubMed] [Google Scholar]
- 5.Tommasi R, Iyer R & Miller AA Antibacterial Drug Discovery: Some Assembly Required. ACS Infect Dis 4, 686–695, doi: 10.1021/acsinfecdis.8b00027 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Silver LL in Topics in Medicinal Chemistry Vol. 25 Ch. 2, 31–67 (2017). [Google Scholar]
- 7.Richter MF & Hergenrother PJ The challenge of converting Gram-positive-only compounds into broad-spectrum antibiotics. Ann N Y Acad Sci 1435, 18–38, doi: 10.1111/nyas.13598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlowsky JA, Kaplan N, Hafkin B, Hoban DJ & Zhanel GG AFN-1252, a FabI inhibitor, demonstrates a Staphylococcus-specific spectrum of activity. Antimicrob Agents Chemother 53, 3544–3548, doi: 10.1128/AAC.00400-09AAC.00400-09 [pii] (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silver LL Challenges of antibacterial discovery. Clin Microbiol Rev 24, 71–109, doi:24/1/71[pii] 10.1128/CMR.00030-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice LB Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197, 1079–1081, doi: 10.1086/533452 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Ghosh M et al. Siderophore Conjugates of Daptomycin are Potent Inhibitors of Carbapenem Resistant Strains of Acinetobacter baumannii. ACS Infect. Dis 4, 1529–1535, doi: 10.1021/acsinfecdis.8b00150 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Li X-Z, Plésiat P & Nikaido H The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev 28, 337–418, doi: 10.1128/CMR.00117-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett D et al. Potentiation of Antibiotic Activity by a Novel Cationic Peptide: Potency and Spectrum of Activity of SPR741. Antimicrob. Agents Chemother 61, e00200–00217, doi: 10.1128/AAC.00200-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter MF et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545, 299–304, doi: 10.1038/nature22308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Boyle NM et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33, doi: 10.1186/1758-2946-3-33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliphant TE Guide to NumPy. (Trelgol Publishing, 2006). [Google Scholar]
- 17.Seefeld MA et al. Inhibitors of bacterial enoyl acyl carrier protein reductase (FabI): 2,9-disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents. Bioorg. Med. Chem. Lett 11, 2241–2244 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Payne DJ et al. Discovery of a novel and potent class of FabI-directed antibacterial agents. Antimicrob Agents Chemother 46, 3118–3124 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao J & Rock CO Exogenous fatty acid metabolism in bacteria. Biochimie 141, 30–39, doi: 10.1016/j.biochi.2017.06.015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asturias FJ et al. Structure and molecular organization of mammalian fatty acid synthase. Nat. Struct. Mol. Biol 12, 225–232, doi: 10.1038/nsmb899 (2005). [DOI] [PubMed] [Google Scholar]
- 21.McMurry LM, Oethinger M & Levy SB Triclosan targets lipid synthesis. Nature 394, 531–532, doi: 10.1038/28970 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Rawat R, Whitty A & Tonge PJ The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc. Natl. Acad. Sci. U.S.A 100, 13881–13886, doi: 10.1073/pnas.2235848100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu L, Lin J, Ma J, Cronan JE & Wang H Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother 54, 689–698, doi: 10.1128/AAC.01152-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsons JB & Rock CO Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opin. Microbiol 14, 544–549, doi: 10.1016/j.mib.2011.07.029 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banevicius MA, Kaplan N, Hafkin B & Nicolau DP Pharmacokinetics, pharmacodynamics and efficacy of novel FabI inhibitor AFN-1252 against MSSA and MRSA in the murine thigh infection model. J. Chemother 25, 26–31, doi: 10.1179/1973947812Y.0000000061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafkin B, Kaplan N & Murphy B Efficacy and Safety of AFN-1252, the First Staphylococcus-Specific Antibacterial Agent, in the Treatment of Acute Bacterial Skin and Skin Structure Infections, Including Those in Patients with Significant Comorbidities. Antimicrob. Agents Chemother 60, 1695–1701, doi: 10.1128/AAC.01741-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takhi M et al. Discovery of azetidine based ene-amides as potent bacterial enoyl ACP reductase (FabI) inhibitors. Eur. J. Med. Chem 84, 382–394, doi: 10.1016/j.ejmech.2014.07.036 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Ramnauth J et al. 2,3,4,5-Tetrahydro-1H-pyrido[2,3-b and e][1,4]diazepines as inhibitors of the bacterial enoyl ACP reductase, FabI. Bioorg. Med. Chem. Lett 19, 5359–5362, doi: 10.1016/j.bmcl.2009.07.094 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Sampson PB et al. Spiro-naphthyridinone piperidines as inhibitors of S. aureus and E. coli enoyl-ACP reductase (FabI). Bioorg. Med. Chem. Lett 19, 5355–5358, doi: 10.1016/j.bmcl.2009.07.129 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Christie SMR, Jinhong; Johnson Michael E. Enoyl reductase inhibitors with antibacterial activity. US patent 2,0180,072,666 A1 (2018).
- 31.Gerusz VE, Sonia; Oxoby Mayalen; Denis Alexis. Novel heterocyclic acrylamides and their use as pharmaceuticals. US patent 8,846,711 B2 (2012).
- 32.Kaplan N et al. Mode of action, in vitro activity, and in vivo efficacy of AFN-1252, a selective antistaphylococcal FabI inhibitor. Antimicrob. Agents Chemother 56, 5865–5874, doi: 10.1128/AAC.01411-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao J, Maxwell JB & Rock CO Resistance to AFN-1252 arises from missense mutations in Staphylococcus aureus enoyl-acyl carrier protein reductase (FabI). J. Biol. Chem 288, 36261–36271, doi: 10.1074/jbc.M113.512905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivaraman S, Zwahlen J, Bell AF, Hedstrom L & Tonge PJ Structure-activity studies of the inhibition of FabI, the enoyl reductase from Escherichia coli, by triclosan: kinetic analysis of mutant FabIs. Biochemistry 42, 4406–4413, doi: 10.1021/bi0300229 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Jackson N, Czaplewski L & Piddock LJV Discovery and development of new antibacterial drugs: learning from experience? J. Antimicrob. Chemother 73, 1452–1459, doi: 10.1093/jac/dky019 (2018). [DOI] [PubMed] [Google Scholar]
- 36.https://revive.gardp.org/, http://www.antibioticdb.com/, https://www.pewtrusts.org/en/research-and-analysis/data-visualizations/2014/antibiotics-currently-in-clinical-development
- 37.Lee HY et al. Reactive Oxygen Species Synergize To Potently and Selectively Induce Cancer Cell Death. ACS Chem Biol 12, 1416–1424, doi: 10.1021/acschembio.7b00015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palchaudhuri R et al. A Small Molecule that Induces Intrinsic Pathway Apoptosis with Unparalleled Speed. Cell Reports 13, 2027–2036, doi: 10.1016/j.celrep.2015.10.042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llabani E et al. Diverse compounds from pleuromutilin lead to a thioredoxin inhibitor and inducer of ferroptosis. Nat Chem 11, 521–532, doi: 10.1038/s41557-019-0261-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the findings of this study are available within the article and the Supplementary Information. Source data underlying Figs. 2, 3 and Extended Data Figs. 5, 6, 7, 9, and are provided in as Source Data files. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.