Abstract
Here, we present a potent RNA vaccine approach based on a novel bipartite vector system using trans-amplifying RNA (taRNA). The vector cassette encoding the vaccine antigen originates from an alphaviral self-amplifying RNA (saRNA), from which the replicase was deleted to form a transreplicon. Replicase activity is provided in trans by a second molecule, either by a standard saRNA or an optimized non-replicating mRNA (nrRNA). The latter delivered 10- to 100-fold higher transreplicon expression than the former. Moreover, expression driven by the nrRNA-encoded replicase in the taRNA system was as efficient as in a conventional monopartite saRNA system. We show that the superiority of nrRNA- over saRNA-encoded replicase to drive expression of the transreplicon is most likely attributable to its higher translational efficiency and lack of interference with cellular translation. Testing the novel taRNA system in mice, we observed that doses of influenza hemagglutinin antigen-encoding RNA as low as 50 ng were sufficient to induce neutralizing antibodies and mount a protective immune response against live virus challenge. These findings, together with a favorable safety profile, a simpler production process, and the universal applicability associated with this bipartite vector system, warrant further exploration of taRNA.
Keywords: vaccine, RNA vaccine, saRNA, taRNA, alphavirus, influenza vaccine, trans-replication, replicon
This study describes the development of trans-amplifying RNA, a bipartite vaccine vector derived from the alphaviral genome. This vector induced protective immune response in mice to influenza HA with only 50 ng of antigen encoding RNA, a 400× lower dose compared to mRNA and 25× lower compared to self-amplifying RNA.
Introduction
RNA-based immunization for the prevention of infectious diseases is considered an attractive alternative to conventional vaccine approaches. RNA elicits potent, protective immune responses against various pathogens1, 2 and offers advantages over the use of live and inactivated virus, subunit vaccines, and other nucleic-acid-based formats. In particular, RNA is non-infectious, non-integrating and, by virtue of rapid degradation by normal cellular processes, is only transiently active. RNA can be administered repeatedly to both prime and boost immune responses and is not limited by anti-vector immunity. Moreover, the RNA backbone engages pattern recognition receptors in the host cell, thereby naturally adjuvanting the response to the encoded immunogen.3 Importantly, RNA enables rapid, cost-efficient, cell- and animal-material-free, scalable production without the use of egg- or cell-based culture. Thus, RNA may facilitate how vaccines are made and has the potential to enable a rapid response to emerging infections.
Two major types of RNAs are being pursued as vaccines for the prevention of diseases such as influenza: one is self-amplifying RNA (saRNA), the other non-replicating mRNA (nrRNA).4 While nrRNA is a synthetic analog of natural mature mRNA, with a synthetic cap analog and engineered UTRs that flank the coding region, saRNA vaccines are mostly derived from the bicistronic genomes of plus-stranded RNA viruses such as alphaviruses. For vaccine engineering, genes in the alphavirus genomic RNA encoding non-structural proteins are maintained, whereas the structural protein genes under the control of a subgenomic promotor are replaced with the vaccine antigen of interest. The non-structural proteins autoproteolytically maturate and assemble to the multi-enzyme replicase complex, which is capable of replicating its own template RNA. The replicase acts as an RNA-dependent RNA polymerase and amplifies saRNA in cis, resulting in high vector copy numbers in the cells of vaccinated hosts and consequently high vaccine antigen levels.5, 6 Accordingly, lower dose levels of saRNA than nrRNA may suffice to induce comparable immune responses.7, 8
Replicase is also capable of amplifying RNAs in trans, as exemplified by so-called transreplicons (TRs).9 TRs are engineered from saRNA by deleting the replicase. The replicase activity is encoded on a co-transfected RNA and interacts in the cytoplasm in a diffusion-controlled manner with TRs. The specificity of replicase:TR interaction is ensured by conserved sequence elements at the very 5′- and 3′-terminal regions that the TRs inherit from the parental saRNA.9 The replicase can be provided in different ways. One is uncapped nrRNA preceded by an internal ribosomal entry site (IRES), which is either in vitro transcribed or generated from transfected plasmids in cells overexpressing T7 RNA polymerase.9, 10, 11 Alternatively, the replicase is provided by stably transfected expression plasmids12 or encoded by an unrelated saRNA.11
Such trans-amplifying RNA (taRNA) systems, which split the TR and the replicase activity onto two vectors, are typically used for studying mechanisms and structural requirements of alphaviral RNA replication9, 11, 12, 13, 14 or for the production of recombinant propagation-defective alphaviral particles for which TRs encoding alphaviral structural genes act as helper RNA.15, 16 As a platform for infectious disease vaccines, these systems have not been systematically explored. From a vaccine development standpoint, taRNA-based split-vector systems may be advantageous over saRNA with regard to safety, versatility, and manufacturing.
In this paper, we present a novel RNA vaccine platform based on taRNA. It consists of a TR encoding the vaccine antigen and a second molecule coding for an in trans-acting alphaviral replicase. We compared saRNA and nrRNA as alternative vectors for delivery of the replicase activity. We used a nrRNA design specifically optimized for long RNA half-life and high translational efficiency.17, 18, 19, 20 We report high transgene expression of our taRNA system. Nanogram doses of antigen-encoding TR were sufficient to induce robust immunity protective against live virus challenge. We show that in trans replication mediated by nrRNA-encoded replicase activity is far superior to that provided by saRNA-encoded replicase activity.
Our data motivate further exploration of taRNA-based vaccines, which bear the promise of enabling fast and cost-efficient production of large numbers of vaccine doses as required for rapidly evolving or emerging viral pathogens.
Results
Expression from taRNA in Conjunction with nrRNA-Delivered Replicase Activity Is as Efficient as Expression from saRNA
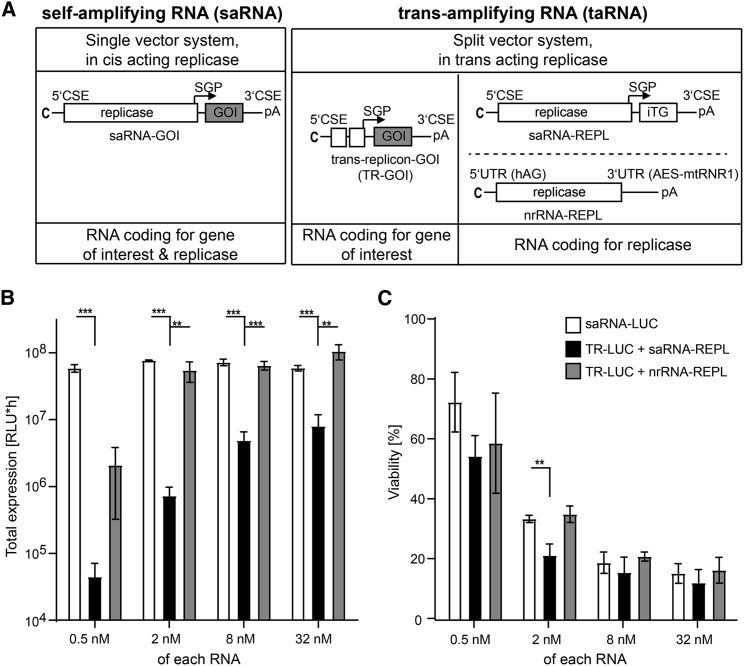
We engineered taRNA as a split-vector system with an in trans-acting replicase (Figure 1A). The starting material was saRNA from Semliki Forest virus (SFV), a single-vector system with an in cis-acting replicase. We generated the TR component by deleting the replicase gene from the saRNA and retaining those parts of the coding sequence that overlap with the SFV 5′ conserved sequence element (CSE) and the subgenomic promoter (SGP), as described recently.9 Sequences downstream of the SGP, including the gene of interest (GOI) and the 3′ CSE, were maintained from the originating saRNA. For the replicase component of the split-vector system, we designed two alternative formats. One was a saRNA identical to the one encoding the GOI, except that it encoded an irrelevant transgene (iTG). Since the purpose of this RNA was to deliver the replicase activity but not a GOI, we refer to this format as saRNA-REPL. The other format encoding the replicase gene was a synthetic nrRNA (nrRNA-REPL). The nrRNA design was pharmacologically optimized for stability and translational efficiency by a beta-s-ARCA(D2) cap,20 the human alpha-globin 5′ UTR,17 a 3′ UTR representing a fusion of motifs derived from amino-terminal enhancer of split (AES) mRNA and mitochondrially encoded 12S rRNA (mtRNR1),19 and an unmasked poly(A) tail.21
Figure 1.
Expression from Trans-amplifying RNA in Conjunction with nrRNA-Delivered Replicase Activity Is as Efficient as Expression from Self-Amplifying RNA
(A) RNA vaccine platforms in this study. Self-amplifying RNA (saRNA, left) is a single-vector system encoding replicase that acts in cis to amplify the complete saRNA, and a second open reading frame with a gene of interest (GOI). Trans-amplifying RNA system (taRNA, right) is a split-vector system consisting of a transreplicon (TR), which encodes the GOI only, and a second RNA delivering alphaviral replicase. TR-GOI is amplified in trans by a replicase either encoded on saRNA (saRNA-REPL) bearing an irrelevant transgene (iTG) or on non-replicative mRNA (nrRNA-REPL). RNA structural elements for replication of saRNA or TRs are located in conserved sequence elements (CSE) at the 5′ and 3′ ends and in the subgenomic promotor (SGP) upstream of the GOI. The UTRs (5′-human alpha globin UTR [hAG] and 3′-AES/mtRNR1-UTR) in the nrRNA-REPL lack viral CSE function and therefore do not promote replication by replicase. All RNAs are capped (C) and bear poly(A) tails (pA). (B) Expression capacity and (C) cytotoxicity of saRNA and taRNA in BHK-21 cells. Vectors bearing firefly luciferase (LUC) as GOI were electroporated into BHK-21 cells (3 × 106 cells per sample) in equimolar amounts. (B) Total luciferase expression was measured for 72 h and approximated by calculating the area under the curve (AUC) for each sample. (C) Relative viability of cells was assessed by measuring viability after 48 h and normalization to the mock control. Mean and SD of three independent experiments are shown. Unpaired Student’s t test was performed (*p < 0.05; **p < 0.01; ***p < 0.001; not annotated p > 0.05). Raw datasets used for calculating the AUC of the expression time course, as well as the full viability time course, are provided in Figure S1.
To compare the two alternative taRNA split-vector approaches, we electroporated BH-K21 cells with increasing equimolar amounts of these vector systems encoding luciferase as reporter; a conventional saRNA single-vector-encoding luciferase was used as a control. Expression of luciferase from saRNA was very high and largely dose independent. Expression levels achieved by taRNA driven by nrRNA-REPL were comparable to those of the saRNA single vector system (except at the lowest dose). In contrast, expression levels achieved by taRNA in conjunction with saRNA-REPL did not reach this benchmark (Figure 1B; Figure S1A). As expected for SFV-based vectors at higher doses, all three systems resulted in reduction of cell viability starting at 24 h after electroporation (Figure 1C; Figure S1B).
Higher Translational Efficiency and Lack of Interference with Cellular Translation Contribute to Superiority of nrRNA- over saRNA-Encoded Replicase to Drive TR Expression
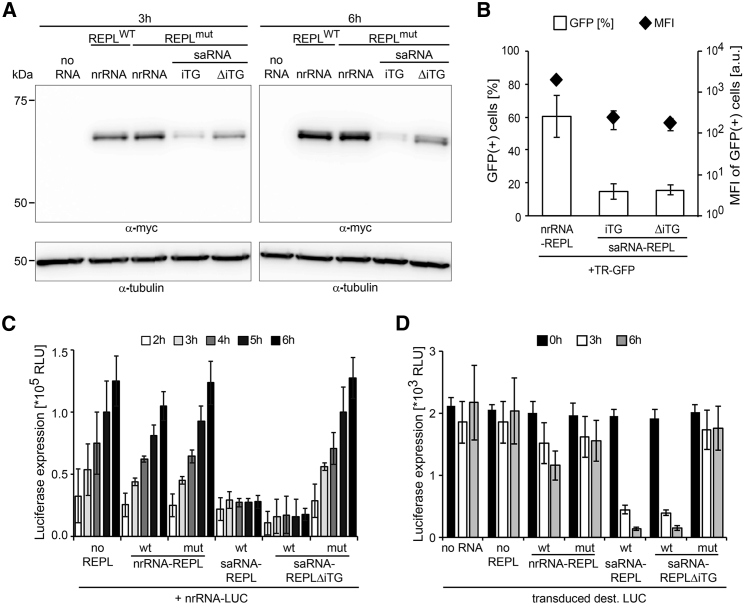
Next, we wanted to understand why nrRNA-encoded replicase was superior to saRNA-encoded replicase in complementing the taRNA split-vector system. To investigate whether the translation efficiency of the replicase open reading frame (ORF) depends on the vector backbone, we introduced several essential controls. One control entailed quantifying replicase expression in transfected cells in a model without RNA replication; we used a replicase mutant (mut-REPL),9 which is deficient in polymerase activity. This enabled the analysis of replicase translation from exclusively in vitro transcribed and transfected RNA molecules and neutralized de novo saRNA synthesis as a confounding factor. Second, we included a saRNA variant with a mutant SGP and full deletion of the transgene ORF (saRNA-REPL-ΔiTG) to control for the possibility that the large “unused” second ORF (iTG) downstream of the SGP in saRNA-REPL may impair expression from this construct (e.g., by inducing nonsense-mediated mRNA decay22). The amount of replicase protein generated in cells transfected with nrRNA-REPL was the same for wild-type (WT)- and mut-REPL (Figure 2A), indicating that the mutation did not affect protein stability. The expression of the mutant replicase was higher from nrRNA compared to saRNA encoding an iTG (Figure 2A; Figure S2A). Furthermore, expression of mutant replicase was higher with saRNA lacking the iTG as compared to saRNA encoding an iTG, irrespective of absence (Figure 2A) or presence of TRs (Figure S2A), confirming our assumption that nonsense-mediated mRNA decay (NMD) would affect replicase levels. Expression of active replicase was only higher with nrRNA-REPL as compared with saRNA-REPL early after electroporation (3 h) (Figure S2A). Notably, replicase accumulated faster and to higher levels in cells transfected with saRNA-REPL-ΔiTG compared to saRNA-REPL, indicating higher replication rates or maintenance of superior replicase translation of the saRNA lacking the SGP and second ORF.
Figure 2.
Higher Translational Efficiency and Lack of Interference with Cellular Translation Contribute to Superiority of nrRNA- over saRNA-Encoded Replicase to Drive TR Expression
(A) Replicase expression levels encoded on either nrRNA or saRNA. 4 × 106 BHK-21 cells per sample were electroporated with 15 μg of nrRNA encoding either WT or mut replicase or 15 μg of saRNA encoding mut replicase with an iTG (here firefly luciferase) or without the iTG. Cells were lysed 3 h and 6 h after electroporation, and replicase protein levels were assessed by western blot (representative experiment). (B) Transfection rate of TR vectors replicated by saRNA. 1 × 106 BHK-21 cells per sample were co-electroporated with 0.1 μg TR-GFP and 2.5 μg of nrRNA-REPL, a saRNA-REPL with an iTG, or a saRNA-REPL lacking the iTG. The next day, transfection rate and expression level per cell were assessed by flow cytometric quantification of the frequency of GFP-positive cells (white bars) and the MFI of GFP (black diamonds). (C) nrRNA translation in cells co-transfected with taRNA or saRNA. 0.5 × 106 BHK-21 cells per sample were electroporated with nrRNA encoding firefly luciferase (nrRNA-LUC) (3.5 nM) and TR encoding SecNLuc as iTG (3.2 nM). 32 nM of further RNAs were co-electroporated as indicated: either nrRNA-REPL, saRNA-REPL without iTG, or saRNA-REPL encoding GFP as iTG. Replicase was either active (WT) or an inactive mutant (mut). From 2 h to 6 h after transfer, nrRNA-LUC translation was measured as a surrogate for cellular translation. (D) Expression of a cell-made reporter transcript. BHK-21 cells were generated by lentiviral transduction to express destabilized luciferase (Luc2CP). 0.5 × 106 transduced BHK-21 cells per sample were co-electroporated with a TR encoding SecNLuc as iTG (3.2 nM) and 32 nM of either nrRNA-REPL, saRNA-REPL without iTG, or saRNA-REPL encoding GFP as iTG. Luc2CP expression was measured at the indicated time points; cells electroporated without RNA (no RNA) served as reference. (B–D) Data are shown as mean and SD of three independent experiments. Further data are provided in Figure S2.
When any of the three replicase-encoding molecules was co-transfected with a TR coding for GFP (Figure 2B) or for secreted nano-luciferase (Figure S2B), nrRNA-REPL outperformed the saRNA-encoded replicases with regard to the fraction of transfected cells as well as the intensity of expression in transfected cells. The saRNA-REPL-ΔiTG performed comparably to saRNA-REPL, indicating that the efficiency of translation from the TR is not determined by the amount of active replicase protein alone that is provided by these saRNA variants (Figure S2A). Unexpectedly, the replication of the TR in nrRNA-REPL- and saRNA-REPL-transfected cells, as assessed by qPCR (Figure S2C), did not differ, indicating that the superiority of nrRNA in providing replicase activity occurs at the translational level. We investigated this hypothesis with two independent approaches. First, we assessed the expression of a co-transfected nrRNA coding for luciferase (nrRNA-LUC) in the presence of either the saRNA or the taRNA split-vector systems. Second, we generated a stably transduced BHK-21 cell line expressing destabilized luciferase (Luc2CP) and measured Luc2CP levels in response to saRNA or taRNA transfection. We studied the first 6 h after saRNA transfection, when cellular viability is not yet compromised by the intrinsically cytotoxic SFV-replicase (Figure S1B). The translation of co-transfected nrRNA-LUC was unaffected by taRNA in conjunction with nrRNA-REPL but strongly inhibited when co-transfected with saRNA-REPL or saRNA-REPL-ΔiTG (Figure 2C). Translational inhibition did not occur in the presence of replication-incompetent saRNA-mut-REPL-ΔiTG. Similarly, the use of both saRNA versions with WT replicase reduced promoter-driven expression of Luc2CP within 3 h and at a much greater extent than taRNA replication driven by nrRNA-REPL (Figure 2D), while the transcript level of Luc2CP was not affected (Figure S2D). These data suggest that saRNA replication rather than TR-replication impairs cellular translation.
These data indicate that a higher translational activity as well as a less-pronounced inhibitory effect on cellular translation of nrRNA contribute to the superiority of nrRNA-REPL over saRNA-REPL in driving expression of high amounts of protein from taRNA.
Immunization with Influenza Hemagglutinin (HA)-Encoding taRNA Profoundly Reduces the Doses Required for Inducing Protective Immune Responses in Mice
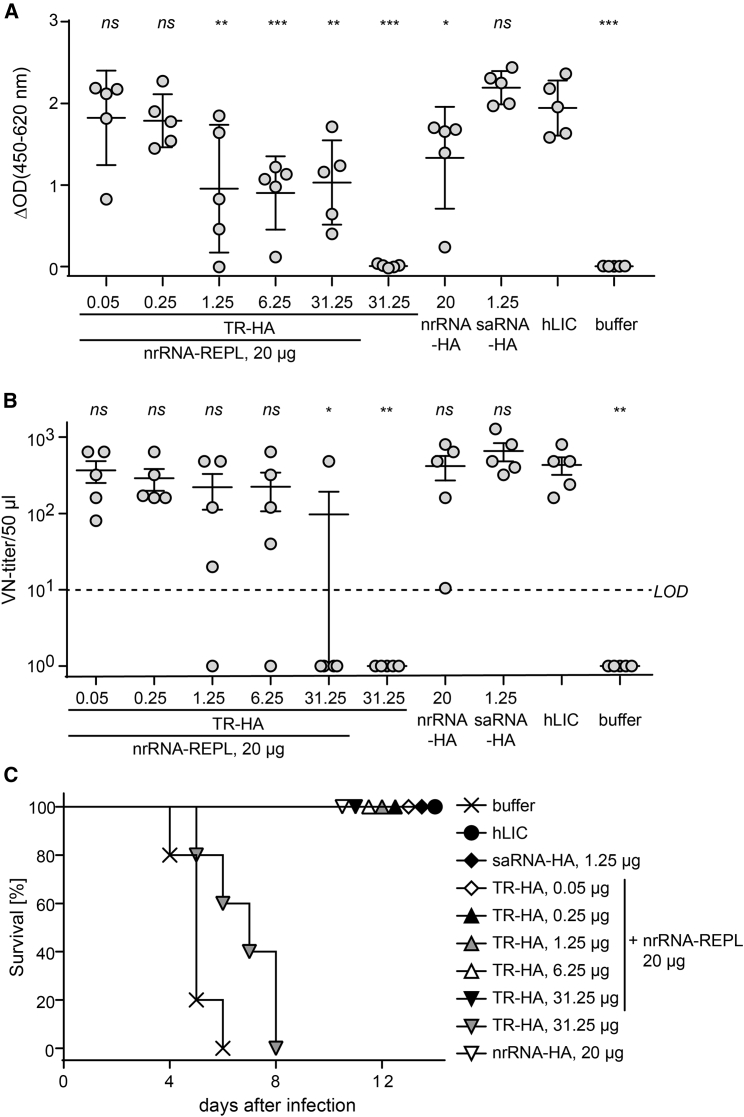
Recently, we showed that 20 μg nrRNA and 1.25 μg saRNA encoding the influenza HA antigen are required to robustly achieve protective immunity against influenza in mice; these doses are comparable to 2.4 HA units from human licensed vaccine (hLIC) injected intramuscularly.8 We investigated the potency of taRNA in inducing protective immune responses by benchmarking against our former data. To this aim, we immunized mice intradermally two times with the taRNA split-vector system, titering TR-HA over a dose range of 0.05–31.25 μg combined with 20 μg nrRNA-REPL. All groups immunized with taRNA developed HA-specific antibody responses. The two lowest doses of TR-HA (50 and 250 ng) were most effective and did not significantly differ from intramuscularly administered hLIC (Figure 3A). TR RNA without nrRNA-REPL did not yield an antibody response. Next, we analyzed the functional virus-neutralizing (VN) antibodies, which only develop if the HA antigen adopts the correct conformation on the immunized host’s cell surface. In all taRNA-immunized groups, VN antibodies were detected (Figure 3B) and mice survived influenza virus challenge (Figure 3C) with minimal loss of body weight and no signs of illness (Figure S2). Again, the most robust neutralizing antibody response resulted from the lowest TR RNA dose (Figure 3B).
Figure 3.
Immunization with Influenza HA-Encoding taRNA Profoundly Reduces the Doses Required for Inducing Protective Immune Responses in Mice
(A–C) Immune response to influenza HA assessed by (A) ELISA measuring total anti-HA IgG antibody amount, (B) virus neutralization test (VNT), and (C) survival of animals after challenge infections. BALB/c mice were immunized by intradermal injection of varying amounts of TR encoding the hemagglutinin antigen of influenza virus A/California/07/2009(H1N1) (TR-HA) in conjunction with 20 μg nrRNA-REPL. Reference groups received either 20 μg nrRNA-HA or 1.25 μg saRNA-HA. Negative controls received 20 μL buffer without RNA or the highest amount of TR-HA without nrRNA-REPL. Intramuscular injection of 2.4 HA units from human licensed vaccine (hLIC) served as a positive control. Animals were immunized twice at days 0 and 21. Serum for VNT and ELISA was sampled on study day 55 to perform serological analysis. Challenge infection with 10-fold MLD50 of viable influenza A/California/4/2009 (H1N1) was done on study day 56. (A and B) Data are shown as mean and SD of the groups (n = 5). One-way ANOVA was used to calculate statistical significance of difference to the hLIC vaccination (*p < 0.05; **p < 0.01; ***p < 0.001; ns p > 0.05). Dashed line in (B) indicates the limit of detection [LOD] of the VNT.) Further data are provided in Figure S3.
In summary, we found that the taRNA split-vector system is highly dose efficient in conferring protective immunity.
Production of TR Can Be Simplified without Compromising the Immunogenicity of the taRNA Split-Vector Vaccine
The taRNA split-vector system as compared to the conventional saRNA-based approach has the critical advantage that the invariant nrRNA-REPL component could be produced in advance in large scale, ready to be combined with seasonally or on-demand-produced TRs encoding interchangeable vaccine antigens. The real-world utility of such a vaccine system would critically depend on the degree of simplification in the production of the TR component.
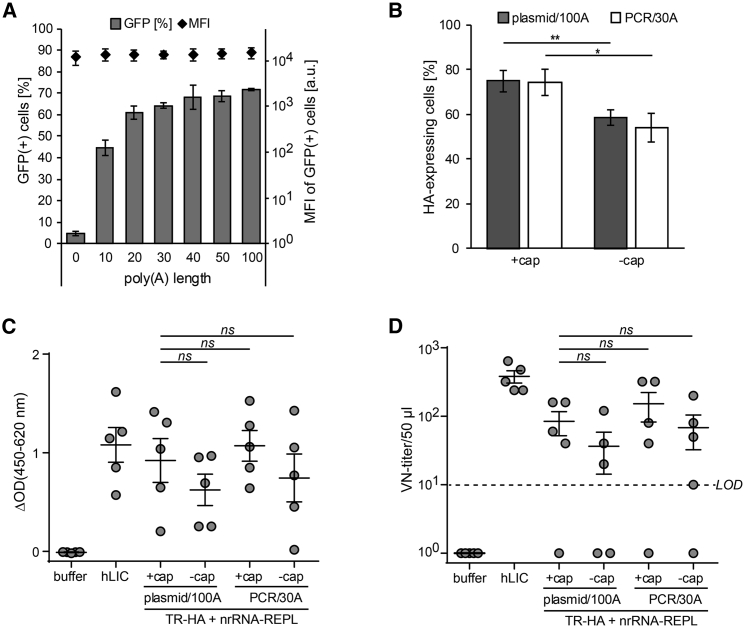
The alphaviral replicase not only replicates RNA, it also caps and poly-adenylates novel RNA copies. We wondered whether this feature could be leveraged to omit the capping reaction and to tolerate a shorter poly(A) tail for initiation of replication.23 A shorter poly(A) tail would also facilitate the use of PCR-amplicons instead of linearized plasmids as DNA templates for in vitro transcription of RNA, resulting in further simplification and cost reduction in replicase production. In fact, we found that in conjunction with nrRNA-REPL for an uncapped reporter TR, a poly(A)-tail length of only 30 adenosine residues (30A) sufficed to reach the maximal transfection rate and expression levels per cell achieved with the 100A-long tail we had in our standard construct (Figure 4A). Next, we generated influenza virus HA-coding TRs in four different versions: transcribed either from a linearized plasmid with a 100A-long poly(A) tail or from a PCR-amplicon with 30A poly(A) tail, each with or without capping. Analysis of protein levels produced by cells transfected with these constructs revealed that the type of DNA template and of associated poly(A) tail length was irrelevant for HA-expression levels, whereas capping had a slight benefit but did not substantially impact expression (Figure 4B).
Figure 4.
The Production Process for TR Can Be Simplified without Compromising the Strong Immunogenicity of the taRNA Split-Vector Vaccine
(A) Effect of shortening poly(A) tail on transfection and expression. TR encoding GFP (TR-GFP) with varying poly(A) tail lengths were produced from PCR amplicons (0 to 50A) or linearized plasmid (100A) and left uncapped. 1 μg TR-GFP and 2 μg nrRNA-encoding replicase (nrRNA-REPL) were then co-electroporated into 0.5 × 106 BHK-21 cells. The next day, the frequency of GFP-positive cells (bars) and the MFI of GFP in these cells (black diamonds) was assessed by flow cytometry. Data show mean and SD of three independent experiments. (B) Expression of capped and uncapped TR-HA. 1 μg capped or uncapped TR-HA transcribed from linearized plasmids or PCR products were co-eletroporated with 2 μg nrRNA-REPL into 0.5 × 106 BHK-21 cells. HA expression was assessed by flow cytometry upon labeling with HA-specific antibodies. Data shown as mean and SD of three independent experiments; significance of differences between capped and uncapped TR were calculated by unpaired t tests; *p < 0.05; not annotated p > 0.05. (C and D) Humoral immune response in vaccinated mice assessed as (C) ELISA for total anti-HA IgG antibody and (D) virus neutralization test (VNT). BALB/c mice were immunized twice intradermally (study day 0 and 21) with 50 ng capped or uncapped TR-HA used in (B) and 20 μg capped nrRNA-REPL. Controls received only buffer or an intramuscular injection of 2.4 HA units from human licensed vaccine (hLIC). 55 days after the first immunization, serum was sampled for serological analysis. Mean and SD of the groups (n = 5) are indicated. One-way ANOVA was used to calculate statistical significance of difference to the vaccination using simplified TRs compared to capped plasmid-produced TR (*p < 0.05; **p < 0.01; ***p < 0.001; ns p > 0.05). Dashed line in (D) indicates the limit of detection (LOD) of the VNT.
50 ng of each of these four TR-HA variants, each combined with 20 μg nrRNA-REPL, were used to immunize mice intradermally. We found that total antibody and VN antibody titers did not significantly differ between vaccines derived from plasmid and from PCR-based DNA templates. Uncapped TR variants trended toward inducing lower immune responses, but in conjunction with the PCR-based DNA-template process resulted in robust titers of neutralizing antibodies (Figures 4C and 4D).
Overall, our findings suggest that TR production can be simplified without an unreasonable loss of protective potency.
Discussion
In this study, we present a novel taRNA split-vector system derived from alphaviral saRNA and show that it qualifies as a highly potent prophylactic vaccine. One of our key findings is that antigen expression by the taRNA system depends on the nature of the co-transfected RNA encoding the replicase activity in trans.
Robust vaccine antigen expression and immune response is achieved when a pharmacologically optimized nrRNA molecule is used to complement the system with replicase activity. With nrRNA-REPL, the production of replicase protein relies completely on the RNA copies transfected into the cell, and the replicase amplifies exclusively the antigen-encoding TR. saRNA-REPL, in contrast, produces replicase both for self-amplification and amplification of the TR. Our data suggest that this dual activity brings disadvantages for the saRNA-REPL variant of the taRNA system. The 5′ and 3′ UTRs of saRNA-REPL contain stem-loop motifs that act as promoters for RNA replication, and it is known that RNAs with secondary structures within the 5′ UTR are intrinsically poorly translatable.24, 25 The structural design of the nrRNA19 we used for encoding replicase was specifically optimized in this regard. This could explain why we found higher amounts of replicase protein produced from nrRNA-REPL compared to saRNA, in particular so swiftly after electroporation or in the absence of de novo RNA synthesis. We also showed that in saRNA-REPL, the second ORF downstream to the replicase contributes to impaired translation of the replicase itself, most likely by triggering nonsense-mediated RNA decay, which has been reported to restrict alphavirus replication.22 Despite higher replicase levels achieved with saRNA deleted for this downstream ORF, the expression of the TR was not augmented. Overall, our data suggest that the advantage of using nrRNA for expressing replicase is governed at the level of translation within the first few hours after RNA transfer, and further studies are underway to further explore the exact mechanism.
Furthermore, it is conceivable that replicase molecules translated from nrRNA-REPL can more easily dissociate and relocalize to the TR, whereas replicase translated from saRNA-REPL is prone to bind to sequence elements in the vicinity, and thus is expected to preferentially promote cis replication as observed during genome replication in a number of RNA viruses.26, 27 Our observation that translation of an unrelated nrRNA was strongly inhibited in cells transfected with saRNA-REPL, but not with nrRNA-REPL, indicates a broader interference from saRNA-REPL with the cellular translation machinery. Further investigations are needed for a deeper understanding of how nrRNA-REPL outperforms saRNA-REPL as a complementing element of a taRNA split-vector system.
As a result of superior vaccine antigen delivery, the mice we immunized with nanogram doses of TR RNA encoding influenza HA antigen in conjunction with nrRNA-encoded replicase mounted robust protective immune responses against influenza. To our knowledge, this is the first study using taRNA as a vaccine, whereas previously described RNA-based influenza vaccines were based on either self-amplifying or non-replicating RNA (reviewed in Scorza and Pardi1).
Our group has recently reported the dose levels required in this mouse influenza challenge model with other naked RNA-based single-vector systems to achieve an outcome equivalent to that generated with 2.4 HA units of intramuscularly (i.m.)-injected hLIC. These were determined to be in the range of 20 μg for nrRNA (encoding the vaccine antigen only without any replicase component) and of 1.25 μg of conventional Semliki Forest-virus-based saRNA construct administered intramuscularly.8 Other studies reported potent immune responses against influenza HA by leveraging lipid nanoparticle (LNP)-formulated RNA. To our knowledge, 100 ng was the lowest reported dose of RNA shown to be immunogenic in mice, when applied as LNP-saRNA and injected twice intramuscularly.28 Moreover, a single intradermal dose of 400 ng of nucleoside-modified LNP-nrRNA protected mice from a homologous lethal virus challenge.29 Our taRNA system induced the best immune response, with as little as 50 ng of naked, uncomplexed TR RNA encoding the vaccine antigen; higher vaccine doses showed a negative impact on immunogenicity. We expect further improvement of dose efficiency by leveraging LNP formulations.
Splitting the vaccine antigen and the replicase components by encoding them on independent vectors is favorable for various reasons. First, the split-vector system comes with a safety advantage. Pseudotyped alphaviruses have unknown replicative competence and pathogenicity. For instance, the glycoproteins of vesicular stomatitis virus and rabies virus are in principle capable of packaging saRNA.30, 31 Without strong proof that any given glycoprotein lacks the intrinsic capacity to form membranous particles and package saRNA, safety concerns for the use of such systems to translate and develop prophylactic antiviral vaccines remain and have to be addressed. Consequently, e.g., German health authorities classify cells treated with a saRNA construct as biosafety level 2, unless the specific glycoprotein of interest is explicitly proven to be incapable of packaging and transferring saRNA.
Second, uncoupling the antigen delivery and the replicase activity is favorable from a versatility and efficiency point of view, since each of both components can be independently optimized. For instance, nrRNA-REPL can be optimized for replicase translation irrespective of RNA functional regions such as the 51-nt CSE. Similarly, the 5′ end of the TR, comprising all structural elements for replication efficiency, can be modified without the need to conserve the amino acid sequence of the replicase N terminus. While saRNA replication does not tolerate nucleoside modifications, nrRNA translation does; moreover, nucleoside-modified nrRNA achieves superior immune responses compared with conventional nrRNA.29, 32, 33 Within the taRNA system, the replicase-coding nrRNA could be nucleoside-modified to reduce activation of cellular innate immune receptors and to maintain translation.34 Further, the taRNA split-vector system is not limited to the SFV backbone but generally applicable to alphaviral vectors. taRNA made of non-cytotoxic replicases (e.g., mutant SFV35, 36) could be especially interesting.
Third, the taRNA system comes with the promise of easy, time- and cost-efficient manufacturing. The improvement of dose efficiency of TRs contributes to this effect, as the same batch of the TR vaccine serves a larger number of individuals. Large-scale production of long RNA at high yield and good integrity is more challenging than production of short RNA constructs. Whereas the length of an antigen-coding saRNA is 7.6 kb plus the antigen, with 7.3 kb of the vector coding for the replicase, the taRNA system is based on the immunologically relevant TR with a length of 1.5 kb plus the antigen and a 7.8-kb-long nrRNA-REPL. Another way to simplify the production process without losing the immunogenicity outcome in vivo is by omitting in vitro capping and shortening poly(A) tails of the TR. For a seasonal vaccine such as influenza, nrRNA-REPL as the invariable component could be pre-produced at large scale and stored, and new process development and production on demand would only be required for the variable, highly dose-efficient antigen-encoding RNA.
In conclusion, we consider this taRNA-based vaccine platform well suited and universally applicable for rapidly evolving pathogens as well as newly emerging infectious diseases.
Materials and Methods
Cell Culture
All growth media, fetal calf serum (FCS), and supplements were supplied by Life Technologies/Gibco unless stated otherwise. BHK-21 cells (ATCC; CCL10) and MDCK-II cells (ATCC; CRL-2936) were grown in Eagle’s minimal essential medium supplemented with 10% FCS. Cells were grown at 37°C in humidified atmosphere equilibrated to 5% CO2.
Animals
Female BALB/c_Rj mice, 6–8 weeks of age, were purchased from Janvier LABS (Le Genest-Saint-Isle, France). All animal experiments were conducted according to German laws and guidelines for animal welfare and approved by the local authorities (Landesuntersuchungsamt Koblenz/Rhineland-Palatinate, Germany).
RNA and Lentiviral Vectors
For in vitro synthesis of mRNA encoding SFV replicase (accession number KP699763) or HA of A/California/7/2009 (H1N1; accession number ACQ55359) the respective coding sequences were inserted into SpeI and XhoI sites of the latest generation of immuno-pharmacologically improved vectors.17, 19, 20, 21 In brief, the human alpha-globin 5′ UTR ensures high translational activity, while the AES-mtRNR1 sequences in the 3′ UTR increase RNA stability. Furthermore, a segmented 100-nt poly(A) tail interrupted by a short linker (A30LA70, where L = GCAUAUGACU) improves plasmid stability in E. coli, similar to what was described recently by others.18 Template plasmids for in vitro synthesis of TR and saRNA were derived from the pST1 plasmid backbone by inserting sequences flanked by viral 5′ and 3′ UTRs between the T7 promoter and the segmented poly(A) using PCR-based cloning techniques. TR and saRNA UTRs were not altered. saRNA sequences were derived from SFV isolate L10, clone SFV4 (accession number KP699763) as described.37 To generate saRNA lacking the downstream ORF (saRNA-ΔiTG) the subgenomic 5′ UTR and coding region of the iTG were deleted by PCR-based cloning. Furthermore, a high number of synonymous mutations was introduced to the 3′-terminal sequence of nsP4 overlapping with the SGP (5′-ACCTCTAtGGaGGaCCccGtcTaGTaaGgTgA-3′; mutations in lowercase letters). The TR was generated similarly to what was previously described,9 by deleting large parts of the replicase ORF from saRNA, keeping only the 5′-terminal 221 nt of nsP1 and the 3′-terminal 984 nucleotides of nsP4. To generate the lentiviral-vector-expressing destabilized Luc2CP, Luc2CP coding sequence was taken from pGL4.12[luc2CP] (Promega, Madison, WI, USA) and inserted into the lentiviral backbone pLenti6.4/R4R2/V5-DEST multisite Gateway destination vector under the control of the human EF1a promoter using the LR-clonase II enzyme mix according the manufacturer’s instructions (Life Technologies, Darmstadt, Germany). Packaging vectors pCMVΔR8.91 (encoding HIV gag-pol) and M620 (encoding the GALV envelop) were kindly provided from Manuel Grez (Georg Speyer Haus, Frankfurt, Germany).
In Vitro RNA Transcription
For in vitro runoff transcription, all template plasmids were linearized using type IIS restriction enzyme SapI, which generates an unmasked poly(A) tail.21 Some templates were generated by PCR. To this aim, TR-GFP or TR-HA were amplified from the respective plasmid using a forward primer covering the T7-polymerase promoter and a reverse primer to add poly(A) tails of various length as indicated. RNA was transcribed from these PCR products with or without cap-analog. Synthesis and purification of RNA were previously described.21, 38 Concentration and purity of synthetic RNA was assessed by spectrophotometry (NanoDrop 2000c, PeqLab) and RNA integrity by on-chip electrophoresis on the 2100 BioAnalyzer (Agilent).
Production of Lentiviral Supernatants and Cell Transduction
Lentiviral particles were generated by co-transfection of HEK293T cells with the lentiviral vector together with pCMVΔR8.91 and M620 using TransitLT1 (Mirus Bio, Madison, WI, USA). Crude lentiviral supernatants were harvested and loaded onto plates coated with 20 μg/mL Retronectin (Takahara, Clonetech Laboratries, USA) by centrifugation (1.500 × g, 15 min, 15°C). After washing with PBS to remove unbound virus particles, BHK-21 cells were plated and incubated overnight at 37°C for transduction. Infected bulk populations expressing Luc2CP were used for the experiments.
Influenza Virus
Throughout the studies, the A/California/4/2009 (Cf4/H1N1), with a 99% identity in segment 4 HA coding sequence to A/California/7/2009 (Cf7/H1N1), but with better viral replication on cells, was used. The original virus stock was a kind gift from the Institute of Immunology, Friedrich-Loeffler-Institut, Isle of Riems, Germany. Cf4/H1N1 was grown on MDCK-II cells. For virus stock production, supernatants of infected cells were cleared by low-speed centrifugation and stored at −80°C; virus titer was determined as described.8
Antibody Analysis
For antibody analysis, blood samples were taken under isoflurane anesthesia via the retroorbital venous plexus on days 20, 35, and 55 after the first immunization. Virus neutralization test (VNT) to determine VN antibodies in the serum was performed based on the WHO’s Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza (WHO Global Influenza Surveillance Network) and as described before.8 In brief, serum levels of HA-neutralizing antibodies were quantified by incubating serial dilutions of the mouse sera for 2 h with 100 tissue culture infection dose 50 (TCID50) of active Cf4/H1N1 influenza virus and then applied onto a confluent MDCK cell monolayer in 96-well plates. Upon incubation for 3 days with MDCK cell culture supernatant was harvested and mixed 1:2 with 0.5% chicken red blood cells (RBCs; Lohmann Tierzucht, Cuxhaven, Germany). RBC agglutination was visually observed in round-bottom 96-well-plates, and the VN serum titer was recorded as the inverse of the lowest dilution that inhibited agglutination (VNT titer/50 μL serum).
For ELISA, recombinant Cf4/H1N1-HA protein (Life Sciences, Idstein, Germany) was biotinylated utilizing the EZ-Link Sulfor-NHS-LC-biotinylation kit according to supplier’s protocol (Thermo Fisher Scientific, Germany). 96-well streptavidin plates (VWR, Darmstadt, Germany) were coated with the biotinylated HA protein at 4°C overnight. Upon washing and blocking, serum samples were screened for HA-specific antibodies by incubation on plates for 1 h at 37°C. After incubation and including assay controls, plates were incubated with horseradish peroxidase (HRP)-labeled secondary anti-mouse immunoglobulin G (IgG) antibody for another 45 min at 37°C before 3, 3', 5, 5'-tetramethylbenzidine (TMB) one substrate (BIOTREND, Cologne, Germany) was applied. Colorimetric detection was monitored and optical density read at 450 nm calculated to a wavelength reference of 620 nm (Δ450–620nm)
Immunizations and Viral Challenge Infections
For RNA immunization, mice were put under isoflurane anesthesia and the dorsal area was shaved for single intradermal injections on day 0 and 21. RNAs were mixed and resolved in a total volume of 20 μL RNase-free 0.15 mM NaCl. hLIC served as a positive control and was given as an intramuscular injection with 2.4 μg HA units in 20 μL of 0.15 mM NaCl likewise on days 0 and 21 of the study.
To evaluate the vaccine protection against Influenza virus infection, on day 55 after study start, immunized mice were exposed to a 10-fold MLD50 of Cf4/H1N1. While anesthetized with an intraperitoneal administered mixture of ketamine/xylazine (120 mg/kg ketamine, Hameln Pharma, Hameln, Germany; 16 mg/kg xylazine, Rompun, Bayer, Leverkusen, Germany), the virus in 30 μL of 0.15 mM NaCl was dispensed via the intranasal route. Mouse-lethal-dose-50 (MLD50) was identified by an endpoint titration described elsewhere.8 To evaluate the vaccine protection against influenza virus infection, immunized mice were challenged intranasally with a 10-fold MLD50 of Cf4/H1N1 in 30 μL while anesthetized with ketamine/xylazine mixture intraperitoneally (120 mg/kg ketamine, Hameln Pharma, Hameln, Germany; 16 mg/kg xylazine, Rompun, Bayer, Leverkusen, Germany). Mice were examined daily. Euthanization followed latest 14 days after challenge infection or when study endpoint criteria (e.g., > 25% weight loss compared to day of infection) were fulfilled.
RNA Transfection
RNA (nrRNA, saRNA, and taRNA) was electroporated into BHK-21 cells (0.5–5 million cells/electroporation; in dependency on required total cell number) at room temperature by applying defined pulses (750 V/cm; one pulse of 16 ms) with a square-wave electroporator (BTX ECM 830, Harvard Apparatus, Holliston, MA, USA). For strictly equimolar transfers, splitting of the saRNA to generate taRNA was taken into account, meaning that, for instance, 0.5 nM saRNA coding for a GOI was compared to either 0.5 nM nrRNA coding for replicase plus 0.5 nM TR coding for the GOI, or to 0.5 nM saRNA coding for replicase and an unrelated transgene plus 0.5 nM TR coding for the GOI. For all other experiments, molarities or amounts of RNAs were used as indicated in the figure legends. After transfection, cells were incubated without refreshing medium until analysis.
Luciferase and Viability Assay
Luciferase assays with transfected cells were performed with the Bright-Glo luciferase assay system or NanoGlo assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions as described recently.39 Viability of transfected cells was assessed using a luminescence-based method assaying ATP concentration after 48 h (CellTiter-Glo; Promega, Madison, WI, USA) according to the instructions of the manufacturer. Relative viability was calculated by normalizing the signal of each sample to the signal of the mock control. Bioluminescence (photons per second [p/s]) of both assays was measured using a microplate luminescence reader Infinite M200 (Tecan Group, Männedorf, Switzerland).
Flow Cytometric Analysis
For flow cytometric analysis of fluorescent protein expression, the cells were harvested, washed with PBS, and fixed with PBS containing 2% formaldehyde. Expression of fluorescent proteins was assessed using FACS Canto II flow cytometer (BD Bioscience, Heidelberg, Germany) and the companion Diva software.
Western Blots
Cells were lysed using RIPA buffer. Lysates were supplemented with 4× Laemmli buffer,40 separated with self-made 7.5% SDS-PAGE gels and wet-blotted onto nitrocellulose membranes (GE Healthcare, Marlborough, MA, USA) using the Trans-Blot Cell (Bio-Rad, Berkeley, CA, USA). Afterward, nitrocellulose membranes were blocked with 5% non-fat dry milk in 1× PBS supplemented with Tween 20 (PBS-T) and incubated with appropriate dilutions of primary antibodies (anti-myc clone 9E10 [Sigma-Aldrich, #M4439]; anti-tubulin clone B-5-1-2 [Sigma-Aldrich, #T5168]) and secondary antibodies. Replicase expression was detected using a myc-tag that was fused in frame into the variable domain of the nsP3 subunit. To ensure equal loading protein concentration in cell lysates was measured by Pierce BCA protein assay (Thermo Fisher Scientific, Waltham, MA, USA) and controlled by the detection of α-tubulin. Chemiluminescent signals were developed with Lumi-Light western blotting substrate (Roche) and were recorded using the LAS 4000 system (GE Healthcare LS). For densitometric analysis, we used the software Image Quant TL v7.0 (GE Healthcare LS).
Statistics
The data of independent experiments were summarized and displayed as mean ± SD. All statistical analysis was performed with GraphPad Prism 8. Tests applied to the experiments are detailed in the respective figure legends (unpaired Student’s t test, one-way ANOVA). No statistical methods were applied to pre-determine sample size for animal experiments.
Author Contributions
Conceptualization, T.B., M.P.; Methodology, T.B., M.P., A.V., S.E., K.C.W., E.H.; Investigation, A.V., S.E., T.H., S.B., R.B.; Writing – Original Draft, T.B., M.P.; Writing – Review & Editing, A.V., K.C.W., R.B., Ö.T., U.S.; Visualization, T.B., M.P., Ö.T., U.S.; Supervision, U.S.
Conflicts of Interest
U.S., T.B., M.P., and Ö.T. are inventors on patents and patent applications that cover parts of this article. S.E., U.S., Ö.T., A.V., and K.C.W. are employees at BioNTech Corporation (Mainz, Germany), a privately owned company developing therapeutic RNA. All others have no potential conflict of interest.
Acknowledgments
We gratefully acknowledge Karen Chu for proofreading the manuscript and Katalin Kariko for scientific discussions. The authors also thank TRON’s cloning unit for creating some of the vectors and Bernadette Jesionek for excellent animal facility care.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.09.009.
Supplemental Information
References
- 1.Scorza F.B., Pardi N. New Kids on the Block: RNA-Based Influenza Virus Vaccines. Vaccines (Basel) 2018;6:6E20. doi: 10.3390/vaccines6020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundstrom K. RNA-based drugs and vaccines. Expert Rev. Vaccines. 2015;14:253–263. doi: 10.1586/14760584.2015.959932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iavarone C., O’hagan D.T., Yu D., Delahaye N.F., Ulmer J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines. 2017;16:871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 4.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 5.Lundstrom K. Biology and application of alphaviruses in gene therapy. Gene Ther. 2005;12(Suppl 1):S92–S97. doi: 10.1038/sj.gt.3302620. [DOI] [PubMed] [Google Scholar]
- 6.Tuomi K., Kädäridäinen L., Söderlund H. Quantitation of Semlike Forest virus RNAs in infected cells using 32-P equilibrium labelling. Nucleic Acids Res. 1975;2:555–565. doi: 10.1093/nar/2.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spuul P., Balistreri G., Hellström K., Golubtsov A.V., Jokitalo E., Ahola T. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J. Virol. 2011;85:4739–4751. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanz M.A., Garcia-Moreno M., Carrasco L. Inhibition of host protein synthesis by Sindbis virus: correlation with viral RNA replication and release of nuclear proteins to the cytoplasm. Cell. Microbiol. 2015;17:520–541. doi: 10.1111/cmi.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallio K., Hellström K., Balistreri G., Spuul P., Jokitalo E., Ahola T. Template RNA length determines the size of replication complex spherules for Semliki Forest virus. J. Virol. 2013;87:9125–9134. doi: 10.1128/JVI.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Utt A., Quirin T., Saul S., Hellström K., Ahola T., Merits A. Versatile Trans-Replication Systems for Chikungunya Virus Allow Functional Analysis and Tagging of Every Replicase Protein. PLoS ONE. 2016;11:e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blakney A.K., McKay P.F., Shattock R.J. Structural Components for Amplification of Positive and Negative Strand VEEV Splitzicons. Front. Mol. Biosci. 2018;5:71. doi: 10.3389/fmolb.2018.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartholomeeusen K., Utt A., Coppens S., Rausalu K., Vereecken K., Ariën K.K., Merits A. A Chikungunya Virus trans-Replicase System Reveals the Importance of Delayed Nonstructural Polyprotein Processing for Efficient Replication Complex Formation in Mosquito Cells. J. Virol. 2018;92:e00152-18. doi: 10.1128/JVI.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smerdou C., Liljeström P. Two-helper RNA system for production of recombinant Semliki forest virus particles. J. Virol. 1999;73:1092–1098. doi: 10.1128/jvi.73.2.1092-1098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark C., Kennedy S.I. The generation and propagation of defective-interfering particles of Semliki Forest virus in different cell types. Virology. 1978;89:285–299. doi: 10.1016/0042-6822(78)90060-0. [DOI] [PubMed] [Google Scholar]
- 17.Stadler C.R., Bähr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P., Karikó K., Türeci Ö., Sahin U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 18.Trepotec Z., Geiger J., Plank C., Aneja M.K., Rudolph C. Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life. RNA. 2019;25:507–518. doi: 10.1261/rna.069286.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlandini von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., Diken M., Löwer M., Vallazza B., Beissert T. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2018;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., Darzynkiewicz E., Huber C., Türeci O., Sahin U. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17:961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 21.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci O., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 22.Balistreri G., Horvath P., Schweingruber C., Zünd D., McInerney G., Merits A., Mühlemann O., Azzalin C., Helenius A. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe. 2014;16:403–411. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Hardy R.W., Rice C.M. Requirements at the 3′ end of the sindbis virus genome for efficient synthesis of minus-strand RNA. J. Virol. 2005;79:4630–4639. doi: 10.1128/JVI.79.8.4630-4639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 25.Babendure J.R., Babendure J.L., Ding J.-H., Tsien R.Y. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang Y., Gillam S. Rubella virus RNA replication is cis-preferential and synthesis of negative- and positive-strand RNAs is regulated by the processing of nonstructural protein. Virology. 2001;282:307–319. doi: 10.1006/viro.2001.0862. [DOI] [PubMed] [Google Scholar]
- 27.Weiland J.J., Dreher T.W. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc. Natl. Acad. Sci. USA. 1993;90:6095–6099. doi: 10.1073/pnas.90.13.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hekele A., Bertholet S., Archer J., Gibson D.G., Palladino G., Brito L.A., Otten G.R., Brazzoli M., Buccato S., Bonci A. Rapidly produced SAM(®) vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolls M.M., Webster P., Balba N.H., Rose J.K. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell. 1994;79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 31.Jia F., Miao H., Zhu X., Xu F. Pseudo-typed Semliki Forest virus delivers EGFP into neurons. J. Neurovirol. 2017;23:205–215. doi: 10.1007/s13365-016-0486-8. [DOI] [PubMed] [Google Scholar]
- 32.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;169:176. doi: 10.1016/j.cell.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pepini T., Pulichino A.-M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debastitis J.C., Maruggi G., Otten G.R., Geall A.J. Induction of an IFN-Mediated Antiviral Response by a Self-Amplifying RNA Vaccine: Implications for Vaccine Design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundstrom K., Abenavoli A., Malgaroli A., Ehrengruber M.U. Novel Semliki Forest virus vectors with reduced cytotoxicity and temperature sensitivity for long-term enhancement of transgene expression. Mol. Ther. 2003;7:202–209. doi: 10.1016/s1525-0016(02)00056-4. [DOI] [PubMed] [Google Scholar]
- 36.Tamm K., Merits A., Sarand I. Mutations in the nuclear localization signal of nsP2 influencing RNA synthesis, protein expression and cytotoxicity of Semliki Forest virus. J. Gen. Virol. 2008;89:676–686. doi: 10.1099/vir.0.83320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beissert T., Koste L., Perkovic M., Walzer K.C., Erbar S., Selmi A., Diken M., Kreiter S., Türeci Ö., Sahin U. Improvement of In Vivo Expression of Genes Delivered by Self-Amplifying RNA Using Vaccinia Virus Immune Evasion Proteins. Hum. Gene Ther. 2017;28:1138–1146. doi: 10.1089/hum.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreiter S., Konrad T., Sester M., Huber C., Türeci O., Sahin U. Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA. Cancer Immunol. Immunother. 2007;56:1577–1587. doi: 10.1007/s00262-007-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koste L., Beissert T., Hoff H., Pretsch L., Türeci Ö., Sahin U. T-cell receptor transfer into human T cells with ecotropic retroviral vectors. Gene Ther. 2014;21:533–538. doi: 10.1038/gt.2014.25. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson J.O., Ostwald K., Kåbjörn C., Andersson M. A method for protein assay in Laemmli buffer. Anal. Biochem. 1994;219:144–146. doi: 10.1006/abio.1994.1243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.