Abstract
The Hippo pathway is a key signaling pathway in the control of organ size and development. The most distal elements of this pathway, the TEAD transcription factors, are regulated by several proteins, such as YAP (Yes‐associated protein), TAZ (transcriptional co‐activator with PDZ‐binding motif) and VGLL1‐4 (Vestigial‐like members 1–4). In this article, combining structural data and motif searches in protein databases, we identify two new TEAD interactors: FAM181A and FAM181B. Our structural data show that they bind to TEAD via an Ω‐loop as YAP/TAZ do, but only FAM181B possesses the LxxLF motif (x any amino acid) found in YAP/TAZ. The affinity of different FAM181A/B fragments for TEAD is in the low micromolar range and full‐length FAM181A/B proteins interact with TEAD in cells. These findings, together with a recent report showing that FAM181A/B proteins have a role in nervous system development, suggest a potential new involvement of the TEAD transcription factors in the development of this tissue.
Keywords: FAM181A, FAM181B, hippo pathway, nervous system, omega loop, protein–protein interaction, TEAD, YAP
1. INTRODUCTION
The Hippo pathway, which is well conserved in metazoans, is linked to various biological processes, such as cell growth/proliferation and tissue homeostasis/regeneration.1 The deregulation of this pathway in cancer has attracted a lot of interest in recent years, because its study may lead to the development of new anticancer drugs.2, 3, 4, 5 Furthermore, because of its role in the control of organ growth and regeneration, modulators of the Hippo pathway could prove to be interesting new molecules in regenerative medicine.2, 6, 7 The TEAD (TEA/ATTS domain) transcription factors are the most distal elements of this pathway.8, 9 These proteins, which have no transcriptional activity on their own, need to interact with different partners to modulate gene transcription. The four human TEAD proteins are regulated by YAP (Yes‐associated protein), its paralog TAZ (transcriptional co‐activator with PDZ‐binding motif) and VGLL1‐4 (vestigial‐like).10, 11, 12 Human cells express only one or a subset of the TEAD genes, suggesting that these transcription factors have tissue‐selective functions (e.g., 13). For example, the autosomal dominant eye disease Sveinsson's chorioretinal atrophy is linked to a mutation of the TEAD1 gene.14 The YAP and TAZ genes are broadly expressed.4, 12 VGLL1‐3 shows a tissue‐specific expression pattern, while VGLL4 is more ubiquitously expressed.11 The structures of TEAD in complex with YAP,15, 16 TAZ,17 VGLL118 and VGLL419 reveal that these proteins bind to an overlapping region at the surface of TEAD. The TEAD‐binding domains of YAP and TAZ are very similar and are made of three distinct secondary structure elements: a β‐strand, an α‐helix and an Ω‐loop.15, 16, 17 The TEAD‐binding domains of VGLL1 and VGLL4 also contain a β‐strand and an α‐helix, but they lack the Ω‐loop, which is key for interaction between YAP/TAZ and TEAD.18, 19 The structures of the TAZ:TEAD17 and VGLL4:TEAD19 complexes show that TAZ and VGLL4 can bind to two TEAD molecules simultaneously, but this has not yet been reported for YAP or VGLL1‐3. The four human TEAD proteins have a similar affinity for peptides mimicking the minimal TEAD‐binding domain of YAP, TAZ and VGLL120 suggesting that these different regulators can compete with each other to gain access to TEAD. YAP/TAZ and VGLL1‐4 are currently the most studied TEAD interactors, but publications suggest that other proteins may also modulate the activity of these transcription factors. For example, the p160 coactivator proteins have been reported to potentiate transcription from a TEAD response element.21 This prompted us to look for new TEAD interactors. Instead of conducting broad interactome or ChiP studies combined with complex bioinformatics analyses, we used a strategy focused on our current knowledge of the structure of the YAP:TEAD complex. Since Ω‐loops do not often mediate protein–protein interactions22, 23, 24 and because an Ω‐loop is key to the formation of the YAP:TEAD complex, we decided to look for proteins that contain this specific recognition motif. To that end, we combined the current structural data on the YAP:TEAD complex and searches in protein databases with small degenerate sequences. This approach led to the identification of the FAM181A and FAM181B proteins that bind to TEAD via an Ω‐loop both in biochemical assays and in cells.
2. RESULTS
2.1. Identification of the FAM181A and FAM181B proteins
The X‐ray structures of the YAP:TEAD complex15, 16, 20, 25, 26 together with the data obtained from mutational studies of YAP27, 28 allowed us to define residues from the Ω‐loop of YAP that are key for the interaction with TEAD. This information was used to search different databases with small degenerated sequences mimicking the Ω‐loop region of YAP (residues 85–99, YAP85–99) (Figure S1, Material and Methods). The FAM181A and FAM181B proteins were identified using this approach. These proteins are present in species from various animal classes, such as mammals, insects and fish (Appendix S1). FAM181A and FAM181B contain a putative Ω‐loop (Figure 1), but FAM181B also possesses the LxxLF motif (x = any amino acid) found in the α‐helix of the TEAD‐binding domain of YAP (and TAZ).29 Nonetheless, the number of amino acids between this motif and the Ω‐loop is greater in FAM181B (50 residues) than in YAP (14 residues) (Figure 1). A survey of the literature shows that little is known about FAM181A/B, and only one publication has studied the function of these proteins.30 This in vivo study reveals that FAM181A/B play a role in nervous system development and function. Interestingly, the authors have noticed that a short sequence present in FAM181A/B displays a high similarity with YAP. This sequence corresponds to FAM181A190–205 and FAM181B220–235, which we identified as putative Ω‐loops. The published data also show that FAM181A/B localize to the nucleus, suggesting that they could potentially interact with TEAD, but the authors did not probe this interaction. Altogether, the presence of a putative Ω‐loop in FAM181A/B and their nuclear localization prompted us to determine whether they can bind to TEAD.
Figure 1.

Primary sequence of the human FAM181A and FAM181B proteins. (a). Primary sequence of FAM181A and FAM181B from UniProt. The amino acids in bold correspond to FAM181A127–205 and FAM181B157–237. The residues belonging to the Ω‐loop are in blue. Val190FAM181A and Val220FAM181B are in orange. The LxxLF motif present in FAM181B is colored green. (b). The primary sequences of YAP57‐99, FAM181A127–205 and FAM181B157–235 were manually aligned. The secondary structure adopted by YAP upon binding to TEAD is indicated.16 α: α‐helix; Ω: Ω‐loop. Dashes (−) indicate gaps. The colors are the same as in (a)
2.2. Biochemical characterization of the interaction between the Ω‐loop of FAM181A/B and TEAD
Since the isolated Ω‐loop region of YAP (YAP85–99) has a measurable affinity for TEAD (~70 μM),31 the corresponding regions of FAM181A/B could also interact with TEAD. We noticed that Val84YAP is conserved in FAM181A (Val190FAM181A) and in FAM181B (Val220FAM181B) (Figure 1). As this residue may have a steric shielding effect on YAP folding27, it was included in the synthetic peptides mimicking YAP and FAM181A/B. The ability of YAP84–99, YAP85–99, FAM181A190–205 and FAM181B220–235 to inhibit the YAP:TEAD interaction was measured in a TR‐FRET assay (Figure S2).32 In agreement with earlier data31, YAP85–99 inhibits the YAP:TEAD interaction with a high double‐digit micromolar IC50 (Table 1). The addition of Val84YAP dramatically increases potency—YAP84–99 is 10 times more potent than YAP85–99 (Table 1)—revealing that this residue is important for the YAP:TEAD interaction. The potency of FAM181A190–205 is similar to that of YAP84–99, while FAM181B220–235 is slightly less potent (Table 1). To demonstrate that the FAM181A/B peptides bind to TEAD, we used a Surface Plasmon Resonance (SPR) assay (Figure 2). The dissociation constants (K d) measured at equilibrium by SPR with the four peptides are in good agreement with the IC50 measured in the TR‐FRET assay (less than twofold difference) and they confirm that YAP84–99 has a higher affinity for TEAD than YAP85–99 (Table 1). FAM181A190–205 and FAM181B220–235 bind to TEAD in a dose‐dependent manner. The signal measured at equilibrium (R max eq), similar to that determined for YAP84–99, is close to the maximum feasible signal (R max th) (Figure 2) indicating that both peptides bind to TEAD with a stoichiometry of 1. The affinity of FAM181A190–205 for TEAD is similar to that of YAP84–99, while FAM181B220–235 binds less tightly (Table 1). Altogether, the results obtained by TR‐FRET and SPR reveal that FAM181A190–205 and FAM181B220–235 compete with YAP for binding to TEAD and that they bind to this transcription factor with a low micromolar affinity, FAM181A190–205 having a lower Kd than FAM181B220–235.
Table 1.
Measure of the potency of YAP, FAM181A and FAM181B derivatives
| TR‐FRET IC50 (μM) | SPR K d (μM) | |
|---|---|---|
| FAM181A190–205 | 4.8 ± 0.4 | 2.8 ± 0.2 |
| FAM181B220–235 | 27.0 ± 0.5 | 18 ± 1 |
| YAP84–99 | 6.1 ± 0.3 | 3.68 ± 0.06 |
| YAP85–99 | 84 ± 2 | 58.4 ± 0.8 |
| FAM181A127–205 | 27 ± 2 | 9.8 ± 0.3a |
| FAM181B157–237 | 6.7 ± 0.5 | 7.3 ± 0.2 |
The inhibition (IC50) of the YAP:TEAD interaction by different YAP, FAM181A and FAM181B fragments was measured in a TR‐FRET assay. The affinity (K d) of the different YAP and FAM181A/B derivatives for TEAD4 was determined at equilibrium by surface plasmon resonance. The values correspond to the averages and standard errors of n ≥ 2 experiments.
Apparent K d (see text for explanation).
Figure 2.
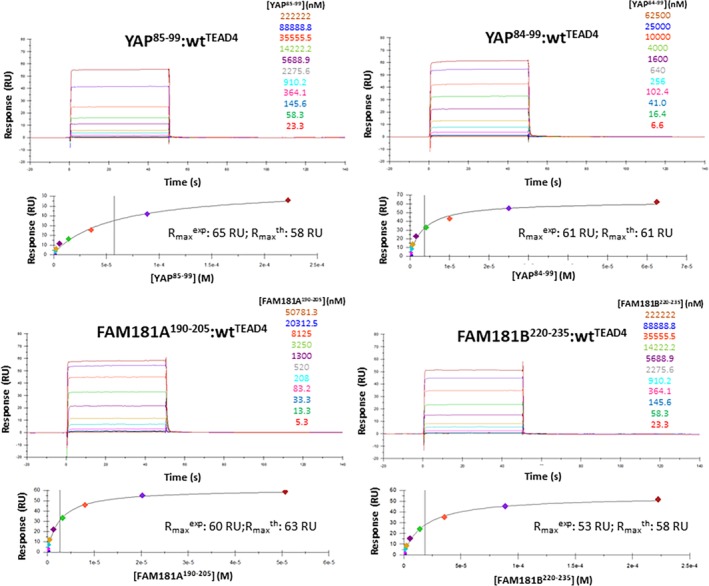
Binding of YAP85–99, YAP84–99, FAM181A190–205 and FAM181B220–335 to TEAD4. Biotinylated N‐Avitagged TEAD4217‐434 was immobilized on sensor chips, and the binding of different concentrations of YAP85–99, YAP84–99, FAM181A190–205 and FAM181B220–335 was measured at 298 K by Surface Plasmon Resonance. The upper panels show representative sensorgrams and the lower panels the corresponding binding isotherms from which Kd values (at equilibrium) were derived. The sensorgrams were globally fitted with a 1:1 interaction model using the Biacore T200 evaluation software (Biacore, Sweden). The concentrations used are indicated. The signal measured at equilibrium (Rmax eq) and the calculated maximum feasible signal (Rmax th) are given
2.3. Determination of the structure of the FAM181A/B:TEAD complexes
To establish whether FAM181A190–205 and FAM181B220–235 adopt an Ω‐loop conformation upon binding to TEAD, we determined the structures of the FAM181A190–205:TEAD4 (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6SEN) and FAM181B220–235:TEAD4 (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6SEO) complexes by X‐ray crystallography (Table S1). The superimposition of these two structures with that of YAP60–100 in complex with TEAD4 (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6GE3)26 shows that YAP84–100, FAM181A190–205 and FAM181B220–235 adopt a similar Ω‐loop conformation upon binding to TEAD4 and they interact with the same surface area (Figure 3). In the following, we shall focus our analysis on residues conserved between YAP and FAM181A/B, which have been described as important for the YAP:TEAD interaction.27, 28 Met192FAM181A/Leu222FAM181B, Leu197FAM181A/Leu227FAM181B and Phe201FAM181A/Phe231FAM181B show hydrophobic interactions with TEAD4 similar to those of Met86YAP, Leu91YAP and Phe95YAP (Figure 4a). These three residues also form a hydrophobic core that contributes to the stabilization of the bound Ω‐loop. Trp202FAM181A/Phe232FAM181B are located at the top of this hydrophobic core, helping to stabilize the bound Ω‐loop, and they form a cation‐π interaction with Arg193FAM181A/Arg223FAM181B (Figure 4b). Phe96YAP and Arg87YAP show the same interactions in the YAP:TEAD complex (Figure 4b). The salt bridge between Arg89YAP and Asp272TEAD4 and the hydrogen bonds between Ser94YAP and Glu263TEAD4:Tyr429TEAD4 are also present in the FAM181A/B:TEAD4 complexes. Arg195FAM181A/Arg225FAM181B are in the vicinity of Asp272TEAD4 (Figure 4c) and Ser200FAM181A/Ser230FAM181B are within hydrogen bond distance from Glu263TEAD4:Tyr429TEAD4 (Figure 4d). Pro198FAM181A/Pro228FAM181B hold the same position as Pro92YAP at the binding interface (Figure 4d). This proline residue, which is strictly conserved in all the sequences obtained in our database search (Figure S1), is probably important for establishment of the Ω‐loop conformation. Overall, the structural data confirm that FAM181A190–205 and FAM181B220–235 form an Ω‐loop upon binding to TEAD and that the key interactions of YAP with TEAD are also present in the FAM181A/B:TEAD complexes.
Figure 3.
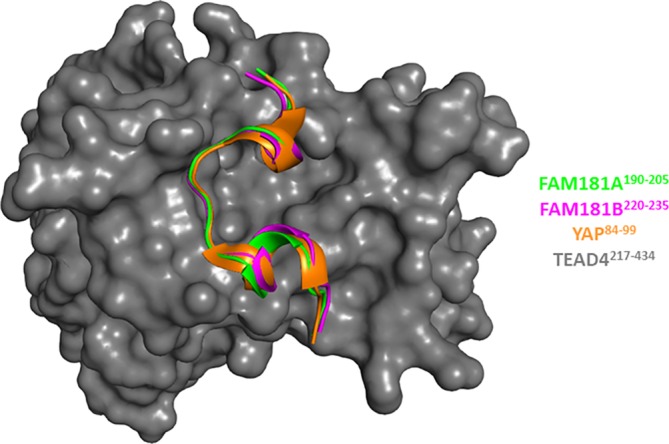
Structures of the FAM181A190–205:TEAD4 and FAM181B220–235:TEAD4 complexes. The structures of the FAM181A190–205:TEAD4 (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6SEN) and FAM181B220–235:TEAD4 complexes (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6SEO) have been superimposed on that of the YAP60–100:TEAD4 complex (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6GE3 26). FAM181A190–205, FAM181B220–235 and YAP84–100 are represented by green, magenta and orange ribbons, respectively. TEAD4 surface is colored gray. The picture was drawn with PyMOL (Schrödinger Inc., Cambridge, Massachusetts)
Figure 4.
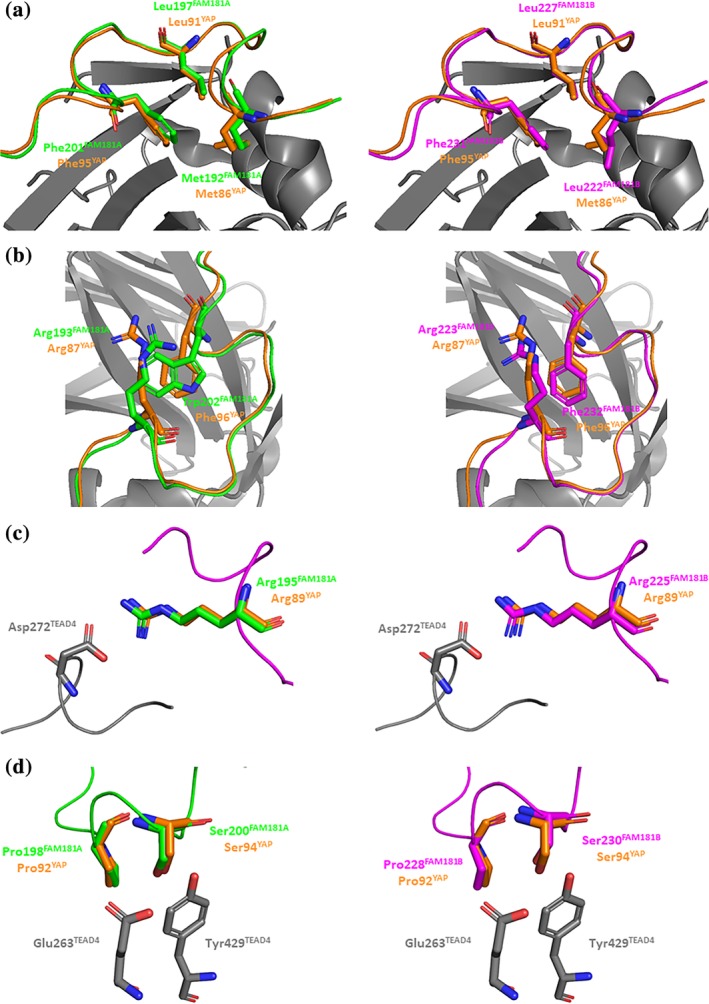
Key interactions between FAM181A190–205/FAM181B220–235 and TEAD4. Left panels. The structures of the FAM181A190–205:TEAD4 (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6SEN) and YAP60–100:TEAD4 (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6GE3 26) complexes have been superimposed. The residues of FAM181A, YAP and TEAD are represented by green, orange and gray sticks, respectively. Right panels. The structures of the FAM181B220–235:TEAD4 complexes (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6SEO) and YAP60–100:TEAD4 (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6GE3 26) complexes have been superimposed. The residues of FAM181B, YAP and TEAD are represented by magenta, orange and gray sticks, respectively. (a). Hydrophobic core of the Ω‐loop. (b). Residues involved in the stabilization of the hydrophobic core. (c). Salt bridge with Asp272TEAD4. (d). Hydrogen bonds with Glu263TEAD4:Tyr429TEAD4 and position of the conserved proline at the binding interface. The picture was drawn with PyMOL (Schrödinger Inc., Cambridge, Massachusetts)
2.4. Study of longer FAM181A/B fragments
FAM181B and YAP contain an LxxLF motif (x = any amino acid) located N‐terminally of the Ω‐loop (Figure 1b). In the YAP:TEAD complex, these three conserved residues make hydrophobic interactions in the α‐helix binding pocket of TEAD.15, 16 To determine the involvement of this motif (FAM181B165–169) in the FAM181B:TEAD interaction, we purified FAM181B157–237 (Figure S3), which contains the LxxLF motif and the Ω‐loop (Figure 1b). The TR‐FRET and SPR results show that FAM181B157–237 has a higher affinity for TEAD than FAM181B220–235 (Table 1, Figures S2 and S4). However, the potency gain is relatively small, fourfold, compared with the 84‐fold gain observed with YAP [YAP61–99, which contains the LxxLF motif (YAP65–69) has a K d = 44 nM31]. We next studied whether the LxxLF motif of FAM181B binds to TEAD at the same place as YAP. Val389TEAD4 is located in the α‐helix binding pocket of TEAD and it shows van der Waals interactions with Phe69YAP from the LxxLF motif. The Val389AlaTEAD4 mutation destabilizes the YAP:TEAD complex by 1.7 kcal/mol (ΔΔG = ΔGmut – ΔGwt).28 The K d of FAM181B157–237 for Val389AlaTEAD4 is 36 ± 2 μM (Figure S4), showing that the mutation reduces binding by 0.94 kcal/mol. We also investigated whether this mutation affects the binding of FAM181A, which lacks an LxxLF motif. The FAM181A127–205 construct, in which the number of residues at the N‐terminus of the Ω‐loop is similar to that of FAM181B157–237, was purified (Figure 1b and Figure S3). FAM181A127–205 has an apparent K d of 9.8 ± 0.3 μM for wtTEAD4 (an accurate K d could not be determined because FAM181A127–205 binds non‐specifically to sensor chips at high concentrations) and of 14.7 ± 0.1 μM for Val419AlaTEAD4 (Table 1, Figure S4). Therefore, the Val419AlaTEAD4 mutation has little effect (ΔΔG ~ 0.2 kcal/mol) on the FAM181A:TEAD interaction. This suggests that FAM181A127–205, which has no LxxLF motif, does not come into contact with Val419TEAD4. The FAM181A127–189 region may not interact with TEAD and, if it remains in solution, this long (63 amino acids) and probably flexible fragment might destabilize the bound Ω‐loop, decreasing its affinity for TEAD as observed in our experiments (FAM181A127–205 IC50/K d > FAM181A190–205 IC50/K d; Table 1). To further confirm that the LxxLF motif from FAM181B interacts with TEAD4 in a manner similar to that of the corresponding motif from YAP, we conducted molecular dynamics simulations. An initial model of TEAD4217–434 in complex with FAM181B154–236 was constructed using the FAM181B220–235:TEAD4217–434 crystal structure (pdb http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6SEO) and by homology to YAP16 the N‐terminal region of FAM181B154–236 was modeled as a β‐strand (FAM181B154–159) and an α‐helix (FAM181B160–172). In this initial model, an arbitrary conformation allowing the α‐helix and Ω‐loop sequences to be connected in 3D was given to the linker region (FAM181B173–219). Starting from this initial model, a molecular dynamics simulation of 10 ns with an explicit water solvation model was run using the Desmond module (default parameters) in the molecular modeling package Maestro (Schrodinger Inc., Cambridge, Massachusetts). The final conformation obtained after this simulation shows that Phe169FAM181B from the LxxLF motif is in the vicinity of Val389TEAD4 and that it occupies the same position at the binding interface as the corresponding residue from YAP, Phe69YAP Figure 5. This result is in agreement with the experimental data obtained with the Val389AlaTEAD4 mutant. The simulation also gives an idea of the dynamics of the linker region. The β‐strand:α‐helix and the Ω‐loop regions of FAM181B154–236 were quite stable during the simulation (Figure S5). Only small fluctuations of some residue side chains were observed, while the main chains retained their secondary structures. However, the linker region (FAM181B173–219) showed substantial flexibility with no observable regular secondary structure and a tendency to fold back on the α‐helix region towards the end of the simulation (Figure S5). This shows that, once bound to TEAD, FAM181B154–236 remains quite flexible because of the presence of a long linker between its α‐helix and its Ω‐loop.
Figure 5.
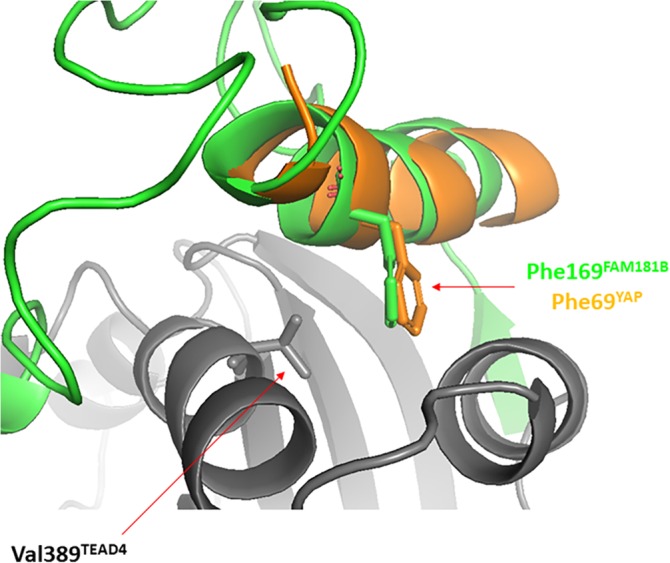
Interaction between the LxxLF motif and TEAD. A simulation of molecular dynamics was run using the FAM181B154–236:TEAD217–434 complex (see text for details). The figure represents a superimposition between the conformation obtained after 10 ns simulation and the structure of the YAP:TEAD complex (PDB http://bioinformatics.org/firstglance/fgij//fg.htm?mol=3KYS 16). FAM181B154–236 and TEAD217–434 and colored in green and gray, respectively. The YAP61–74 region from the YAP:TEAD structure is represented in orange. The picture was drawn with PyMOL (Schrödinger Inc., Cambridge, Massachusetts)
We next studied the structure of unbound FAM181A127–205 and FAM181B157–237 by circular dichroism (CD). In contrast to the CD spectrum obtained with TEAD4217–434, which is characteristic of a well‐folded protein (Figure S6a), the CD spectra of FAM181A127–205 and FAM181B157–237 show that—under our experimental conditions—these two fragments do not adopt a well‐defined structure in solution. We also analyzed the amino acid sequence of full‐length FAM181A/B with PrDOS (http://prdos.hgc.jp), a protein disorder prediction server.33 This in silico analysis suggests that the regions corresponding to FAM181A127–205 and FAM181B157–237 are probably disordered in the context of the full‐length proteins (Figure S6b).
Altogether, the data we obtained with FAM181A127–205 and FAM181B157–237 indicate that these two protein fragments are flexible both in solution and once bound to TEAD, suggesting that this high flexibility may have a negative contribution to their binding to TEAD.
2.5. Study of the FAM181A/B:TEAD interaction in cells
The findings described above show that fragments of FAM181A/B and TEAD4 are able to interact and form stable complexes in a cell‐free environment. To study this interaction in greater detail, we decided to investigate whether the full‐length FAM181A/B and TEAD proteins can interact with each other in cells. To this end, N‐terminally V5‐tagged TEAD4 and (FLAG)3‐tagged FAM181A/B were co‐transfected into HEK293FT cells. A V5‐mediated immunoprecipitation of TEAD4 was performed and V5 and FLAG antibodies were used to detect TEAD4 and FAM181A/B, respectively, by Western blot. FAM181A and FAM181B are efficiently co‐immunoprecipitated with wtTEAD4 (Figure 6). The FAM181A/B proteins were not detected in similar experiments carried out in the absence of V5‐tagged TEAD4 (empty vector control), indicating that these proteins do not bind in a non‐specific manner to the antibodies or beads (Figure 6). To establish whether the observed interaction is specific, we used a mutated form of TEAD4, Asp272AlaTEAD4. This mutation, which abolishes the formation of a salt bridge between Asp272TEAD4 and Arg195FAM181A/Arg225FAM181B (Figure 4c), reduces the affinity for TEAD4 of FAM181A127–205 (K d > 16 μM, Figure S4) and FAM181B157–237 (K d > 75 μM, Figure S4). The Asp272AlaTEAD4 mutation significantly affects the interaction between TEAD4 and FAM181A/B in co‐immunoprecipitation experiments (Figure 6), showing that the observed interactions between the wild‐type proteins are specific. Similar results were previously obtained with YAP, which interacts with Asp272TEAD4 via Arg89YAP (Figure 4c).28 We also repeatedly observed that the expression levels of FAM181A were significantly higher in co‐transfection experiments performed with wtTEAD4 but not with Asp272AlaTEAD4 (Figure 6). Therefore, we quantified by ImageJ software (https://imagej.nih.gov/ij/) the IP over input ratio, normalized to the respective empty control within each experiment. Across three independent experiments, which exhibited a similar pattern, Asp272AlaTEAD4 consistently displayed an approximately fivefold reduction of co‐IP capacity with FAM181A relative to wtTEAD4. By similar quantification for FAM181B, Asp272AlaTEAD4 displayed an approximately fourfold reduction of co‐IP capacity relative to wtTEAD4. The observed effect of wtTEAD4 on the expression levels of FAM181A suggests that the formation of a complex between these two proteins may help enhancing the stability of FAM181A (at least in HEK293FT background and in conditions where this protein is overexpressed).
Figure 6.
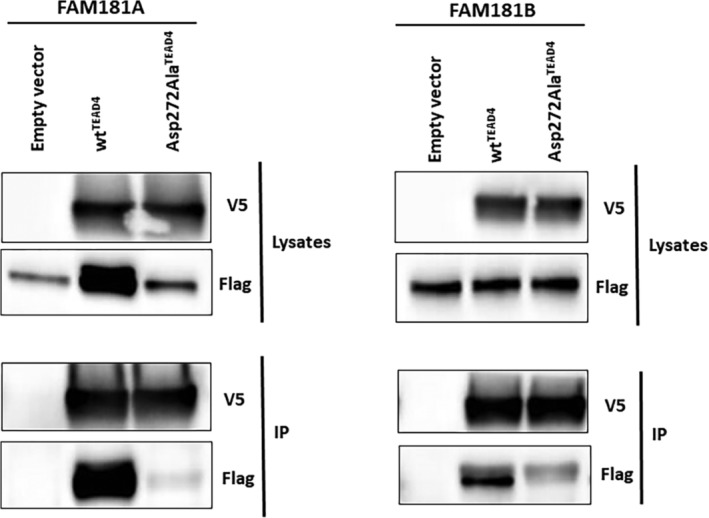
Interaction between FAM181A/B and TEAD4 in cells. N‐terminally V5‐tagged wtTEAD4 or Asp272AlaTEAD4 were co‐transfected with (FLAG)3‐FAM181A/B into HEK293FT cells. TEAD4 was immunoprecipitated with a V5 specific antibody and co‐immunoprecipitated FAM181A/B was determined by anti‐FLAG Western blot. Left panel co‐transfection FAM181A:TEAD4. Right panel co‐transfection FAM181B:TEAD4. To provide a better view without signal saturation, a shorter exposure of total FAM181B is shown (flag in the lysates sub‐panel). IP, Immunoprecipitated fraction
Overall, these data show that FAM181A/B and TEAD4 interact in a specific manner in the context of the full‐length proteins in a cellular environment. This, together with the biochemical and structural data, supports the findings that FAM181A and FAM181B are TEAD interactors.
3. DISCUSSION
The TEAD transcription factors bind to several proteins that modulate their activity. Currently, YAP, TAZ and VGLL1‐4 are the best‐characterized TEAD interactors. The TEAD‐binding domain of YAP and TAZ is formed of a β‐strand, an α‐helix and an Ω‐loop, while the current data show that the TEAD‐binding domain of VGLL1‐4 contains only a β‐strand and an α‐helix. In this article, we identify two new TEAD interactors, FAM181A and FAM181B. We show that a region of these two proteins adopts an Ω‐loop conformation upon binding to TEAD and that the residues from this secondary structure element engage with TEAD interactions similar to those of the corresponding residues from YAP. In a fashion similar to that of YAP, FAM181B also possesses an LxxLF motif (note that a VxxHF is present in VGLL1‐418). Site‐directed mutagenesis experiments and molecular modeling studies suggest that the LxxLF motif of FAM181B and YAP bind to TEAD in a similar manner. FAM181A/B fragments containing only the Ω‐loop or the Ω‐loop plus the LxxLF motif (FAMB181B) have an affinity for TEAD in the low micromolar range. In transient transfection experiments, full‐length FAM181A/B and TEAD interact with each other, and complex formation is prevented by a mutation that disrupts a key interaction at the Ω‐loop binding pocket. Altogether, this indicates that FAM181A/B and TEAD can interact with each other both in biochemical assays and in a cellular environment.
The affinity for TEAD of the different FAM181A/B fragments that we have studied is lower than that of similar YAP, TAZ and VGLL1 constructs.31, 34 This suggests that FAM181A/B may not easily gain access to TEAD in cells in the presence of these other proteins. However, the nuclear localization of TEAD regulators can be modulated by the Hippo pathway, and the phosphorylation of YAP, for example, leads to its sequestration in the cytoplasm.12, 35 This suggests that, under specific physiological conditions, FAM181A/B may not need to compete with other TEAD regulators to form a complex with this transcription factor. Therefore, despite their lower affinity for TEAD, FAM181A/B proteins might be able to form transient complexes with this transcription factor in vivo.
Marks et al. have shown that the loss of FAM181B function did not lead to an obvious phenotype in mice, and they hypothesize that this lack of effect could be due to a possible redundancy between FAM181A and FAM181B.30 This suggests that, to elucidate the biological relevance of the FAM181A/B:TEAD interaction, in vivo experiments will have to be conducted where the role of FAM181A and FAM181B is evaluated at the same time. While such studies are beyond the scope of this manuscript, our findings provide ideas on tools that could be used for such experiments. The analysis of the structure of the FAM181A/B:TEAD complexes suggest different mutations of FAM181A/B that can be designed to prevent the interaction with TEAD. For example, our data on the Asp272AlaTEAD4 mutation and earlier findings on the effect of the Arg89AlaYAP mutation in cells16 suggest that Arg195AlaFAM181A and Arg225AlaFAM181B could be utilized as probes to determine the effect of the disruption of FAM181A/B:TEAD interactions in an in vivo set up. The cellular data obtained with Ser94AlaYAP 16 also indicate that the Ser200AlaFAM181A/Ser230AlaFAM181B mutations could be used to disrupt the FAM181A/B:TEAD complex in vivo. Preliminary experiments we carried out in HEK293FT cells using a reporter gene under the control of a YAP optimized promoter28 did not show a modulation of TEAD transcriptional activity by FAM181A/B, suggesting that perhaps the conditions used to determine whether FAM181A/B exert an effect on TEAD transcriptional activity should be more physiological. For example, Honda et al. have utilized a luciferase gene under the control of a MYH7 promoter in C2C12 cells (mouse myoblasts) to study the effect of VGLL2 (selectively expressed in muscle cells) on TEAD.36 Therefore, to determine the impact of FAM181A/B on TEAD transcriptional activity, reporter gene assays may have to be conducted in cells derived from a neural lineage using a reporter gene under the control of the promoter of a gene that is specifically expressed in that cell type. Drosophila has been an instrumental model organism for studying the role of the Hippo pathway in development, and it is a very attractive system for studying the FAM181A/B genes. Unfortunately, our motif search only identified Yorkie, the drosophila homolog of YAP, but not FAM181A/B, suggesting that these proteins are not present in flies (Appendix S1).
In summary, the discovery of FAM181A/B as new TEAD interactors paves the way for future in vivo studies to elucidate the relevance of this interaction in the context of the development of the nervous system and to gain better knowledge on the physiological role of the FAM181A/B:TEAD complex.
4. MATERIAL AND METHODS
4.1. Database search
The Motif Search algorithm from GenomeNet (http://www.genome.jp) was used to interrogate the GenBank, Uniprot, RefSeq and PDBSTR databases. Three operators were utilized to create degenerate sequence motifs: x represents any amino acid; [AB] means either amino acid A or B; {A} means any amino acid except A. The amino acid sequence of human YAP corresponding to residues 85–99, which adopts an Ω‐loop conformation upon binding to TEAD, was used as template sequence for the database search. Structural data and results from structure–function studies were combined to identify the residues from YAP85–99 which do not interact with TEAD in the YAP:TEAD complex (e.g., residues with the side chain pointing to the solvent).15, 16, 20, 25, 26, 27, 28 On the basis of this initial analysis, several positions were annotated with x (85‐P‐x‐R‐x‐R‐x‐L‐P‐x‐S‐F‐F‐x‐x‐P‐99) indicating that any amino acid can occupy them. The first search was used to determine whether amino acids other than a serine could be present at position‐94 (Motif_01, Figure S1). The sequences obtained were individually analyzed using the structural data to determine whether the amino acids identified at position‐94 could engage in favorable interactions with TEAD. All the sequences identified contained a glutamate residue at position‐94. According to the structure of the YAP:TEAD complex, such residues should not be tolerated at the binding interface, because this would result in a steric clash with Tyr429TEAD4 and repulsive electrostatic interactions with Glu263TEAD4.26 On the basis of this result, a serine was maintained at position‐94 and a search with Motif_02 was conducted (Figure S1). This methodology was repeated with a total of 14 motifs to obtain the final consensus sequence: P‐x‐[RKHS]‐x‐R‐x‐[LMF]‐P‐x‐S‐F‐[FW]‐x‐x‐P. The databases were interrogated with this motif to generate the list of the sequences used in this communication (Appendix S2).
4.2. Peptides
The synthetic peptides (both N‐acetylated and C‐amidated) were purchased from Biosynthan (Germany). The purity (>90%) and the chemical integrity of the peptides was determined by liquid chromatography–mass spectrometry (LC–MS) from 10 mM stock solutions in 90:10 (v/v) dimethyl sulfoxide (DMSO):water.
4.3. Proteins
The amino acid sequences of full length FAM181A (UniProt Q8N9Y4) and FAM181B (UniProt A6NEQ2) were back‐translated into an Escherichia coli codon‐optimized DNA sequence by an in‐house tool and synthesized by GeneArt (Thermo Fisher Scientific, Germany). The DNA fragment encoding FAM181A127–205 was PCR‐amplified with primers comprising LguI restriction sites and cloned into a pET‐derived vector with an N‐terminal His6‐Gly·Ser spacer‐Lipoyl‐Gly·Ser spacer‐HRV3C affinity purification and solubilizing tag.37 The DNA fragment encoding FAM181B159–237 was PCR‐amplified with primers comprising LguI restriction sites for seamless cloning into a pET‐derived vector with an N‐terminal His6‐Gly·Ser spacer‐Rbx‐Gly·Ser spacer‐HRV3C affinity purification and solubilizing tag.38 The FAM181B fragment in the resulting expression plasmid was N‐terminally extended by adding residues 157 and 158 through site‐directed mutagenesis with the QuikChange II Lightning Site‐Directed Mutagenesis kit (Agilent, Santa Clara, California). The DNA sequence of all expression constructs was verified by Sanger sequencing. Recombinant FAM181A127–205 and FAM181B157–237 proteins were purified using identical protocols. The expression plasmid was transformed into NiCo21 (DE3) competent E. coli cells (New England Biolabs, Ipswich, Massachusetts). TB medium supplemented with 50 mM MOPS was inoculated with a bacterial pre‐culture and incubated under constant shaking at 37°C. At OD600 = 0.8, the culture was chilled to 18°C, and protein expression was induced by addition of 0.2 mM Isopropyl β‐D‐1‐thiogalactopyranoside. After overnight expression, the bacteria were harvested by centrifugation for 20 min at 6,000g, frozen on dry ice and stored at −80°C. The cell pellets were thawed and suspended in buffer A (50 mM Tris‐HCl, 300 mM NaCl, 30 mM imidazole, pH 8.0) supplemented with cOmplete protease inhibitor (Roche, Switzerland) and TurboNuclease (Merck, Germany). The cells were mechanically disrupted by three passages through an EmulsiFlex C3 homogenizer (Avestin, Canada), and insoluble cell debris was removed by centrifugation for 30 min at 40,000g. The clarified cell lysate was loaded onto two 1 ml HisTrap HP columns (GE Healthcare, UK) mounted in series on an ÄKTA Pure chromatography system (GE Healthcare). Contaminating proteins were washed away with 10 column volumes of buffer A, and the His‐tagged protein was eluted with a linear gradient over 10 column volumes to 100% buffer B (buffer A with 300 mM imidazole). The N‐terminal purification tag was cleaved off overnight at 5°C by His6‐tagged HRV 3C protease during dialysis against buffer A. The cleaved protein was passed over the re‐equilibrated HisTrap HP columns to remove the cleaved tag, HRV3C‐protease, and contaminating host cell proteins. The fractions containing the FAM181A/B proteins were pooled, concentrated with Amicon Ultra‐15 3 K centrifugal filter unit (Merck, Germany) and loaded onto a HiLoad Superdex 75 16/600 pg size exclusion column (GE Healthcare, United Kingdom) equilibrated with 50 mM Tris‐HCl, 150 mM NaCl, 10% glycerol, pH 8.0. The fractions containing pure protein were pooled and concentrated to about 2 mg/ml in an Amicon filter unit (Merck, Germany). Purity and concentration of the protein samples were determined by RP‐UHPLC, measuring the absorbance at 210 nm. The concentration was calculated using a BSA standard curve as reference. Identity and molecular weight of the FAM181A/B proteins were confirmed by LC–MS. The final yield was about 10 mg/L expression culture. The different TEAD4 variants were purified as previously described.28
4.4. Time‐resolved FRET, surface Plasmon resonance and circular dichroism
Biotinylated N‐Avitagged‐TEAD4217–434 (1 nM; wtTEAD4) and LANCE Eu‐W1024 Streptavidin (0.5 nM, PerkinElmer, Waltham, Massachusetts) were pre‐incubated for 1 hr at room temperature in 50 mM HEPES pH 7.4, 100 mM KCl, 0.05% (v/v) Tween‐20, 0.25 mM TCEP, 1 mM, and 0.05% (w/v) BSA. N‐terminally Cy5‐labelled YAP60–100 (20 nM) and serial dilutions of the peptides/protein fragments to be tested were added and incubated in white 384‐well plates (Greiner Bio‐One International, Austria) for 1 hr at room temperature. DMSO was present at 2% in the assay. The solubility of the peptides in assay buffer was measured by dynamic light scattering with a Dyna Prot device (Wyatt technology Corp., Germany). The fluorescence in the TR‐FRET assay was measured with a Genios Pro reader (Tecan, Switzerland) (50 μs delay between excitation and fluorescence, 75 μs integration time, excitation wavelength 340 nm, emission wavelengths 620 and 665 nm). Data analyses were carried out using the TR‐FRET 665/620 nm emission ratio. The IC50 values were obtained by nonlinear regression analysis with GraphPad Prism (GraphPad Software, San Diego, California). The Surface Plasmon Resonance and Circular Dichroism experiments were conducted as previously described.28
4.5. Structural biology
The untagged wtTEAD4217–434 protein used for crystallization was obtained as described previously.25 Complex crystals between myristoylated wtTEAD4217–434 and the FAM181 peptides were grown at 293 K using the sitting drop vapor diffusion method. wtTEAD4217–434 (7.4 mg/ml) and FAM181A190–205 or FAM181B220–235 (0.5 mM) were pre‐incubated in 25 mM Tris‐HCl pH 8.0, 100 mM NaCl and 1 mM TCEP (molar ratio peptide:protein = 1.7). For crystallization, the FAM181A/B:TEAD4 complexes were mixed with an equal volume of reservoir solution (0.3 + 0.3 μl). Reservoir solutions: FAM181A190–205:TEAD4 complex—200 mM (NH4)2SO4, 100 mM CH3COONa(H2O)3 pH 4.59 and 35% pentaerythritol ethoxylate (15/4 EO/OH); FAM181B220–235:TEAD4 complex—50 mM (NH4)2SO4, 50 mM Bis‐Tris pH 6.5 and 30% pentaerythritol ethoxylate (15/4 EO/OH). For data collection, the FAM181A190–205:TEAD4 crystals were directly shock‐cooled in liquid nitrogen while the FAM181B220–235:TEAD4 crystals were first soaked for a few seconds in the reservoir solution containing 30% glycerol before shock‐cooling in liquid nitrogen. X‐ray diffraction data were collected at the Swiss Light Source (SLS, beamline X10SA) using a Pilatus pixel detector. Raw diffraction data were analyzed and processed using the autoPROC39/STARANISO (Global Phasing Ltd., UK) toolbox. The structures were solved by molecular replacement with PHASER40 using as search model the coordinates of previously solved in‐house structures of TEAD4. The software COOT41 and BUSTER (Global Phasing Ltd.) were used for iterative rounds of model building and structure refinement. The refined coordinates of the complex structures have been deposited in the Protein Data Bank (http://www.wwpdb.org) with the accession numbers 6SEN (FAM181A190–205:TEAD4) and 6SEO (FAM181B220–235:TEAD4).
4.6. Cellular biology
Cloning. nV5‐tagged TEAD4 cDNA constructs (wtTEAD4 and Asp272AlaTEAD4) were described previously.28 FAM181A (RefSeq NM_138344.5) was amplified by standard PCR from a pDONR221‐FAM181A (human codon‐optimized) clone, obtained from GeneArt (Thermo Fisher Scientific, Germany), using primers containing N‐terminal (FLAG)3 epitope. The PCR product was cloned by Gateway reaction into pcDNA3.1 Hygro‐DEST (Invitrogen, Carlsbad, California), according to the manufacturer's protocol. FAM181B (RefSeq NM_17885.4) was amplified by standard PCR from a pcDNA‐DEST40‐FAM181B (human codon‐optimized) clone using primers containing N‐terminal (FLAG)3 epitope. The PCR product was cloned by Gateway reaction into pcDNA3.1 Hygro‐DEST (Invitrogen), according to the manufacturer's protocol.
Cell culture and transfections. HEK293FT cells, cell culture handling and Lipofectamine 2000‐based transient transfections were described previously.28
TEAD/FAM181 co‐immunoprecipitation (co‐IP). For FAM181A co‐IP experiments, HEK293FT cells were transfected with nV5‐TEAD4 and (FLAG)3‐FAM181B constructs (1:1 ratio), and lysed in NP40 buffer (Invitrogen) containing PhosSTOP and Protease Inhibitor Cocktail (both from Roche, Switzerland) 48 hr after transfection. For FAM181B co‐IP experiments, HEK293FT cells were transfected with nV5‐TEAD4 and (FLAG)3‐FAM181B constructs (1.5:1 ratio), and the proteins were extracted from nuclei using the NE‐PER™ Nuclear (Thermo Fisher Scientific, Germany) 48 hr after transfection. Lysates (100 μg) were then incubated with V5 antibody overnight (FAM181B co‐IP) or for 2 hr (FAM181A co‐IP) under rotation at 4°C, followed by incubation with Dynabeads Protein G (Invitrogen) for 2 hr under rotation at 4°C. Immunoprecipitates were washed three times with NP40 buffer containing protease and phosphatase inhibitors, eluted with Laemmli Sample Buffer (BioRad, Hercules, California) by incubation at 95°C for 5 min and resolved by standard SDS‐PAGE gel electrophoresis and Western blotting. Antibodies for IP: V5 (Invitrogen). Antibodies for Western blot: FLAG (Sigma‐Aldrich, St. Louis, Michigan) and V5 (Cell Signaling Technology, Danvers, Massachusetts) as primary antibodies; HRP‐anti‐rabbit (Cell Signaling Technology) as secondary antibody.
The TEAD transcription factors are regulated by different proteins. Combining structural data and motif searches in protein databases FAM181A and FAM181B were identified as new TEAD interactors. Biochemical and structural data reveal that FAM181A/B bind to TEAD via an Ω‐loop and this interaction was also demonstrated in a cellular context. The FAM181A/B:TEAD interaction might play a role in the development of the nervous system as FAM181A/B are specifically expressed in this tissue.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Appendix: S1 Table S1. Crystallographic data collection and refinement statistics for the FAM181A/B:TEAD4 complexes. Figure S1. Sequences of the motifs used to interrogate the databases. Figure S2. Inhibition curves obtained in the TR‐FRET assay. Figure S3. LC–MS analyses of FAM181A127–205 and FAM181B157–237. Figure S4. Binding of FAM181B157–237 and FAM181A127–205 to wtTEAD4, Val389AlaTEAD4 and Asp272AlaTEAD4. Figure S5. Molecular dynamics simulation of the FAM181B154–236:TEAD4217–434 complex. Figure S6. Structural study of FAM181A/B.
Appendix: S2 Amino acid sequences of the proteins obtained with the final motif search.
ACKNOWLEDGMENTS
The authors thank Aude Izaac, Suzanne Chau, Aurélie Winterhalter, Patrick Graff and René Hemmig for excellent technical assistance. X‐ray data collection was performed on the X10SA beamline at the Swiss Light Source, Paul Scherrer Institute, Villingen, Switzerland.
Bokhovchuk F, Mesrouze Y, Delaunay C, et al. Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors. Protein Science. 2020;29:509–520. 10.1002/pro.3775
Fedir Bokhovchuk and Yannick Mesrouze contributed equally to this work.
REFERENCES
- 1. Wang Y, Yu A, Yu FX. The hippo pathway in tissue homeostasis and regeneration. Protein Cell. 2017;8:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Santucci M, Vignudelli T, Ferrari S, et al. The hippo pathway and YAP/TAZ‐TEAD protein‐protein interaction as targets for regenerative medicine and cancer treatment. J Med Chem. 2015;58:4857–4873. [DOI] [PubMed] [Google Scholar]
- 3. Zhang K, Qi HX, Hu ZM, et al. YAP and TAZ take center stage in cancer. Biochemistry. 2015;54:6555–6566. [DOI] [PubMed] [Google Scholar]
- 4. Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holden JK, Cunningham CN. Targeting the hippo pathway and cancer through the TEAD family transcription factors. Cancer. 2018;10:E81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moya IM, Halder G. Hippo‐YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol. 2018;20:211–226. [DOI] [PubMed] [Google Scholar]
- 7. Wang J, Liu S, Heallen T, Martin JF. The hippo pathway in the heart: Pivotal roles in development, disease, and regeneration. Nat Rev Cardiol. 2018;15:672–684. [DOI] [PubMed] [Google Scholar]
- 8. Landin‐Malt A, Benhaddou A, Zider A, Flagiello D. An evolutionary, structural and functional overview of the mammalian TEAD1 and TEAD2 transcription factors. Gene. 2016;591:292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin KC, Park HW, Guan KL. Regulation of the hippo pathway transcription factor TEAD. Trends Biochem Sci. 2017;42:862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hilman D, Gat U. The evolutionary history of YAP and the hippo/YAP pathway. Mol Biol Evol. 2011;28:2403–2417. [DOI] [PubMed] [Google Scholar]
- 11. Simon E, Faucheux C, Zider A, Thézé N, Thiébaud P. From vestigial to vestigial‐like: The Drosophila gene that has taken wing. Dev Genes Evol. 2016;226:297–315. [DOI] [PubMed] [Google Scholar]
- 12. Callus BA, Finch‐Edmondson ML, Fletcher S, Wilton SD. YAPing about and not forgetting TAZ. FEBS Lett. 2019;593:253–276. [DOI] [PubMed] [Google Scholar]
- 13. Jacquemin P, Sapin V, Alsat E, Evain‐Brion D, Dollé P, Davidson I. Differential expresssion of the TEF family of transcription factors in the murine placenta and during differenciation of primary trophoblasts in vitro . Dev Dyn. 1998;212:423–436. [DOI] [PubMed] [Google Scholar]
- 14. Fossdal R, Jonasson F, Kristjansdottir GT, et al. A novel TEAD1 mutation is the causative allele in Sveinsson's chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration). Hum Mol Genet. 2004;13:975–981. [DOI] [PubMed] [Google Scholar]
- 15. Chen L, Chan SW, Zhang XQ, et al. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Z, Zhao B, Wang P, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaan HYK, Chan SW, Tan SKJ, et al. Crystal structure of TAZ‐TEAD complex reveals a distinct interaction mode from that of YAP‐TEAD complex. Sci Rep. 2017;7:2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pobbati AV, Chan SW, Lee I, Song H, Hong W. Structural and functional similarity between Vgll1‐TEAD and YAP‐TEAD complexes. Structure. 2012;20:1135–1140. [DOI] [PubMed] [Google Scholar]
- 19. Jiao S, Wang H, Shi Z, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. [DOI] [PubMed] [Google Scholar]
- 20. Bokhovchuk F, Mesrouze Y, Izaac A, et al. Molecular and structural characterization of a TEAD mutation at the origin of Sveinsson's chorioretinal atrophy. FEBS J. 2019;286:2381–2398. [DOI] [PubMed] [Google Scholar]
- 21. Belandia B, Parker MG. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem. 2000;275:30801–30805. [DOI] [PubMed] [Google Scholar]
- 22. Leszczynski JF, Rose GD. Loops in globular proteins: A novel category of secondary structure. Science. 1986;234:849–855. [DOI] [PubMed] [Google Scholar]
- 23. Ring CS, Kneller DG, Langridge R, Cohen FE. Taxonomy and conformational analysis of loops in proteins. J Mol Biol. 1992;224:685–699. [DOI] [PubMed] [Google Scholar]
- 24. Fetrow JS. Omega loops: Nonregular secondary structures significant in protein function and stability. FASEB J. 1995;9:708–717. [PubMed] [Google Scholar]
- 25. Mesrouze Y, Meyerhofer M, Bokhovchuk F, et al. Effect of the acylation of TEAD4 on its interaction with co‐activators YAP and TAZ. Protein Sci. 2017;26:2399–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mesrouze Y, Bokhovchuk F, Izaac A, et al. Adaptation of the bound intrinsically disordered protein YAP to mutations at the YAP:TEAD interface. Protein Sci. 2018;27:1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Z, Lin Z, Zhou Z, et al. Structure‐based design and synthesis of potent cyclic peptides inhibiting the YAP‐TEAD protein‐protein interaction. ACS Med Chem Lett. 2014;5:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mesrouze Y, Bokhovchuk F, Meyerhofer M, et al. Dissection of the interaction between the intrinsically disordered YAP protein and the transcription factor TEAD. Elife. 2017;6:e25068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen L, Loh PG, Song H. Structural and functional insights into the TEAD‐YAP complex in the hippo signaling pathway. Protein Cell. 2010;1:1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marks M, Pennimpede T, Lange L, Grote P, Herrmann BG, Wittler L. Analysis of the Fam181 gene family during mouse development reveals distinct strain‐specific expression patterns, suggesting a role in nervous system development and function. Cancer. 2016;575:438–451. [DOI] [PubMed] [Google Scholar]
- 31. Hau JC, Erdmann D, Mesrouze Y, et al. The TEAD4‐YAP/TAZ protein‐protein interaction: Expected similarities and unexpected differences. Chembiochem. 2013;14:1218–1225. [DOI] [PubMed] [Google Scholar]
- 32. Mesrouze Y, Erdmann D, Zimmermann C, Fontana P, Schmelzle T, Chène P. Different recognition of TEAD transcription factor by the conserved β‐strand:Loop:α‐helix motif of the TEAD‐binding site of YAP and VGLL1. ChemistrySelect. 2016;1:2993–2997. [Google Scholar]
- 33. Ishida T, Kinoshita K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mesrouze Y, Hau JC, Erdmann D, et al. The surprising features of the TEAD4‐Vgll1 protein‐protein interaction. Chembiochem. 2014;15:537–542. [DOI] [PubMed] [Google Scholar]
- 35. Shreberk‐Shadek M, Oren M. New insights into YAP/TAZ nucleo‐cytoplasmic shuttling: New cancer therapeutic opportunities? Mol Oncology. 2019;13:1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Honda M, Tsuchimochi H, Hitachi K, Ohno S. Transcriptional cofactor Vgll2 is required for functional adaptations of skeletal muscle induced by chronic overload. J Cell Physiol. 2019;234:15809–15824. [DOI] [PubMed] [Google Scholar]
- 37. Lebendiker M, Danieli T. Production of prone‐to‐aggregate proteins. FEBS Lett. 2014;588:236–246. [DOI] [PubMed] [Google Scholar]
- 38. Kohli BM, Ostermeier C. A Rubredoxin based system for screening of protein expression conditions and on‐line monitoring of the purification process. Protein Expr Purif. 2003;28:362–367. [DOI] [PubMed] [Google Scholar]
- 39. Vonrhein C, Flensburg C, Keller P, et al. Data processing and analysis with the autoPROC toolbox. Acta Cryst D. 2011;67:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. PHASER crystallographic software. J Appl Cryst. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P, Cowtan K. Coot: Model‐building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix: S1 Table S1. Crystallographic data collection and refinement statistics for the FAM181A/B:TEAD4 complexes. Figure S1. Sequences of the motifs used to interrogate the databases. Figure S2. Inhibition curves obtained in the TR‐FRET assay. Figure S3. LC–MS analyses of FAM181A127–205 and FAM181B157–237. Figure S4. Binding of FAM181B157–237 and FAM181A127–205 to wtTEAD4, Val389AlaTEAD4 and Asp272AlaTEAD4. Figure S5. Molecular dynamics simulation of the FAM181B154–236:TEAD4217–434 complex. Figure S6. Structural study of FAM181A/B.
Appendix: S2 Amino acid sequences of the proteins obtained with the final motif search.


