ABSTRACT
Multipotent progenitor populations are necessary for generating diverse tissue types during embryogenesis. We show the RNA polymerase-associated factor 1 complex (Paf1C) is required to maintain multipotent progenitors of the neural crest (NC) lineage in zebrafish. Mutations affecting each Paf1C component result in near-identical NC phenotypes; alyron mutant embryos carrying a null mutation in paf1 were analyzed in detail. In the absence of zygotic paf1 function, definitive premigratory NC progenitors arise but fail to maintain expression of the sox10 specification gene. The mutant NC progenitors migrate aberrantly and fail to differentiate appropriately. Blood and germ cell progenitor development is affected similarly. Development of mutant NC could be rescued by additional loss of positive transcription elongation factor b (P-TEFb) activity, a key factor in promoting transcription elongation. Consistent with the interpretation that inhibiting/delaying expression of some genes is essential for maintaining progenitors, mutant embryos lacking the CDK9 kinase component of P-TEFb exhibit a surfeit of NC progenitors and their derivatives. We propose Paf1C and P-TEFb act antagonistically to regulate the timing of the expression of genes needed for NC development.
KEY WORDS: Paf1 complex, Neural crest, Stem cells, Transcription pausing, Zebrafish mutant, P-TEFb
Highlighted Article: Polymerase-associated factor 1 complex function is required to maintain the neural crest progenitor population in zebrafish.
INTRODUCTION
RNA Pol II-mediated transcription is a highly regulated process with factors modifying initiation, productive elongation and termination (Chen et al., 2018; Nechaev and Adelman, 2011; Zhou et al., 2012). Studies of the RNA polymerase-associated factor 1 complex (Paf1C) have linked it to each stage of RNA Pol II-mediated transcription, but its precise roles in these events are still unclear. The Paf1C, which is composed of five canonical protein components, Paf1, Ctr9, Cdc73, Rtf1 and Leo1, is recruited to RNA Pol II molecules following initiation of gene transcription, where it has been proposed to function as a scaffold or ‘platform’ on which factors interact to effect regulation of transcription (Jaehning, 2010; Van Oss et al., 2017). Paf1C has been implicated in facilitating histone modifications, transcription initiation and progression, and RNA termination and processing, but it is best known for its role in promoting elongation at early stages of transcription (Chen et al., 2009; Costa and Arndt, 2000; Gerlach et al., 2017; Kim et al., 2010; Nordick et al., 2008; Penheiter et al., 2005; Rozenblatt-Rosen et al., 2009; Sheldon et al., 2005; Squazzo et al., 2002; Van Oss et al., 2016; Vos et al., 2018a).
Paf1C is not simply a general modulator of transcription, but rather contributes to specific biological processes. Paf1C has been linked to the control of pluripotency in embryo stem cells in culture (Ding et al., 2009; Ponnusamy et al., 2009). It is essential for the expression of a subset of genes in yeast, and mutations affecting the complex have discrete tissue-specific phenotypes in Drosophila and vertebrates (Akanuma et al., 2007; Bahrampour and Thor, 2016; Kim et al., 2012; Langenbacher et al., 2011; Mosimann et al., 2006, 2009; Nguyen et al., 2010; Penheiter et al., 2005). Although Paf1C has been most often associated with positive aspects of transcription, paradoxically, several studies have indicated that Paf1C can also have an inhibitory role: Paf1C has been shown to function antagonistically to positive elongation factors during the development of erythroid progenitors and in oligodendrocyte differentiation (Bai et al., 2010; Kim et al., 2012). Given the range of phenotypes associated with loss of Paf1C function and the diversity of biochemical activities associated with the Paf1C, it is likely that Paf1C has cell- or tissue-specific functions dependent on the presence of other regulatory factors (Van Oss et al., 2017).
Finally, Paf1C has been implicated in maintaining promoter-proximal paused RNA Pol II (Chen et al., 2015), although its role in this process is still controversial (Van Oss et al., 2017; Vos et al., 2018b). The release of promoter-proximal RNA Pol II is a crucial regulatory step in gene activation (Adelman and Lis, 2012; Gaertner and Zeitlinger, 2014). Pause release is promoted by recruitment of the positive transcription elongation factor b (P-TEFb) complex, the activity of which requires the CDK9 kinase. P-TEFb phosphorylates, displaces and modifies negative regulators of transcription elongation in addition to promoting phosphorylation of residues in the C-terminal domain (CTD) of RNA Pol II, thereby stimulating productive RNA Pol II elongation (Li et al., 2018; Lu et al., 2016; Peterlin and Price, 2006). Although the biological significance of promoter-proximal paused RNA Pol II is unclear, speculation has focused on its potential role in multipotent precursor cells (Min et al., 2011). Pausing may serve as a regulatory mechanism to effect a rapid transcription response to stimuli or to coordinate the synchronous induction of multiple genes in response to developmental cues (Adelman et al., 2009; Adelman and Lis, 2012; Boettiger and Levine, 2009; Gariglio et al., 1981; Gerlach et al., 2017; Henriques et al., 2013; Levine, 2011; Mayer et al., 2017; Rahl et al., 2010; Rougvie and Lis, 1988).
Here, we report that unbiased forward genetic screens reveal Paf1C is needed to maintain multipotent progenitors of the neural crest (NC) lineage. The NC is one of the earliest stem cell-like populations to arise in the embryo (Le Douarin and Kalcheim, 1999). It is a uniquely vertebrate population of multipotent cells that contributes to a dramatic range of tissues, including craniofacial cartilage and bone, pigment cells of the skin and inner ear, the enteric and peripheral nervous systems, neurons and glia, and neuroendocrine cells. The NC arises through a gene regulatory network (reviewed by Simoes-Costa and Bronner, 2015) initiated by the convergence of multiple growth factor signaling pathways that activate a transcription factor cascade leading to induction and specification of precursors, and finally establishment of a definitive premigratory NC progenitor population at the dorsal surface of the neural tube. The progenitors then migrate along defined routes responding to environmental cues that shape their terminal positions and differentiation (Bronner and Simões-Costa, 2016; Vega-Lopez et al., 2017). Knowledge of the dynamic cellular and genetic events that mark the origins of NC precursor cells and their developmental progression toward a dedicated progenitor state makes this lineage particularly useful for associating gene functions with the attainment of specific stages of NC identity.
We find the Paf1C is required in vivo to maintain multipotent progenitors of the NC and additional embryonic stem cell populations, including primitive blood progenitors and primordial germ cells. Loss of zygotic expression of any of the proteins comprising the Paf1C produces embryos that are severely deficient in NC-derived tissues. Our results reveal an antagonistic relationship between Paf1C and the pause-release factor P-TEFb in maintaining the multipotent NC precursor population. Loss of the essential CDK9 kinase component of P-TEFb suppresses the NC phenotype of paf1 mutants. Consistent with the interpretation that limiting expression of some genes helps maintain progenitors, cdk9 mutant embryos exhibit a surfeit of NC tissue. Our results indicate that the antagonistic activity of Paf1C and P-TEFb on RNA Pol II transcription activity is essential to maintain the multipotential NC progenitor population in vivo.
RESULTS
paf1 is essential for neural crest formation
alyronz12 (aln) mutant embryos have a severe loss of NC-derived tissues, with a complete lack of melanophores (Fig. 1A,B) (Cretekos and Grunwald, 1999). alnz12 is a complex mutation consisting of an insertion of multiple rearranged plasmid elements and a deletion of genomic sequences near the south telomere of chromosome 15 (Fig. S1) (Cretekos and Grunwald, 1999). We proceeded to identify the single gene responsible for the NC phenotype. We found an overlapping deficiency, c4 (Fisher et al., 1997), that failed to complement the NC functions affected by alnz12 (Fig. 1C). The region deleted in both mutations defined a ‘critical region’ containing the gene responsible for the aln NC phenotype. A new ENU-induced allele, alnz24, was isolated from a non-complementation screen. alnz12/alnz24 transheterozygotes and alnz24 homozygotes appear remarkably similar to the original alnz12 mutant, with a severe block to NC migration and differentiation, indicated by the absence of crestin expression in the trunks of 24 h post-fertilization (hpf) mutant embryos and the absence of pigmentation in 48 hpf mutant embryos (Fig. 1D-G). Map crosses indicated alnz24 segregated as a simple recessive mutation and placed it within the alnz12/c4 critical region (Fig. S1 and data not shown).
Fig. 1.
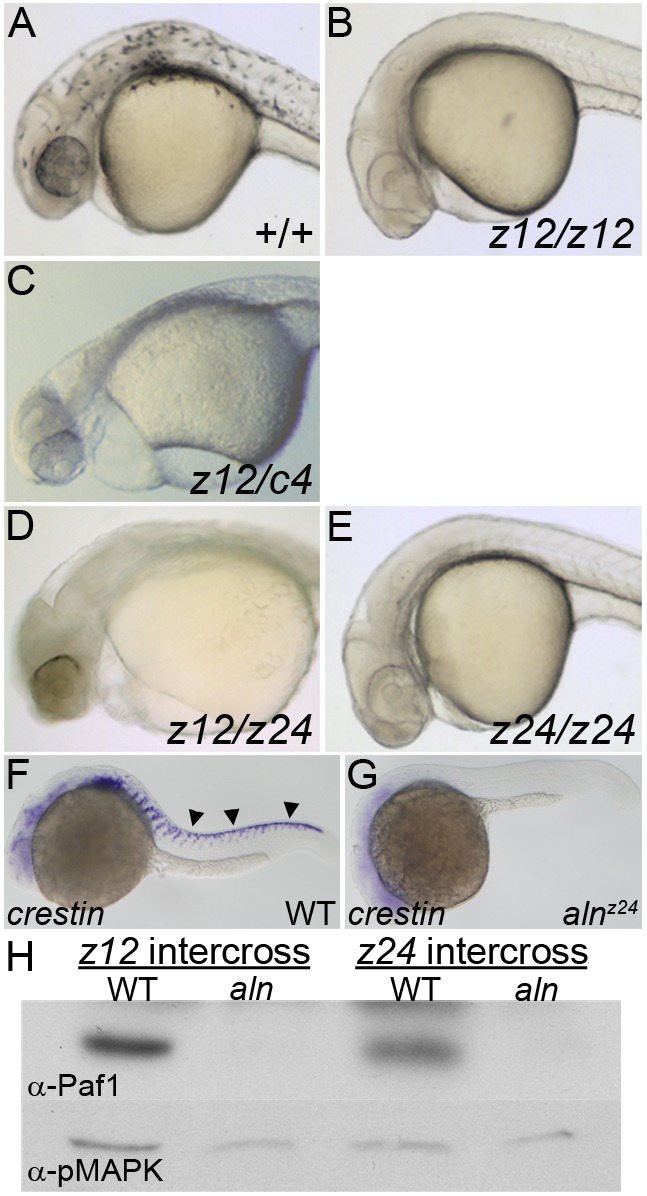
alnz24 is a null mutation that regulates NC formation and encodes RNA polymerase II-associated factor 1 (Paf1). (A-E) 36 hpf wild-type and aln mutant embryos. Wild-type embryos (A) have pigmented melanophores lacking in embryos harboring combinations of aln mutant alleles: (B) homozygous for the alnz12 deletion mutation, (C) transheterozygous for the alnz12 and c4 deletions, (D) transheterozygous for the alnz12 deletion and alnz24 ENU-induced mutations, and (E) homozygous for the alnz24 mutation. (F,G) crestin expression detected by whole-mount in situ hybridization in 24 hpf wild-type sibling (F) and alnz24 mutant (G) embryos, indicating a complete absence of migrating trunk NC in alnz24. Arrowheads indicate premigratory and migrating trunk NC. A-G are lateral views with anterior to the left. (H) Paf1 protein is not detected by immunoblot analysis in 36 hpf alnz12 or alnz24 mutant embryos, but is readily detected in wild-type control siblings. Immunoblot detection of pMAPK serves as a protein-loading control.
The aln NC phenotype is due to loss of function of a single gene, paf1, which encodes the Paf1 protein associated with the eponymous transcription factor complex. Morpholino oligonucleotide (MO) knockdown of selected candidate genes within the critical region revealed that embryos lacking paf1 function had striking resemblance to aln mutants. Sequencing of paf1 cDNA derived from wild-type and mutant embryos identified a C>A transversion (nucleotide position 843 of the cDNA, NM_001024453) that resulted in a premature stop codon at amino acid 281 in the alnz24 allele (Fig. S1). Consistent with the genetic analyses indicating the aln alleles are null mutations, 36 hpf alnz12 and alnz24 mutant embryos lacked Paf1 protein (Fig. 1H). As the paf1 transcript and its protein product are maternally supplied and broadly expressed (Fig. S1), the alnz24 phenotype reflects the earliest requirements for zygotic paf1 function.
Two additional experiments confirmed loss of paf1 gene function was responsible for the entire aln NC phenotype. First, overexpression of wild-type but not alnz24 mutant (843C>A) paf1 mRNA rescued melanophore formation in alnz12 or alnz24 mutant embryos (Fig. S2). Second, all wild-type eggs injected with a paf1 splice-blocking MO (paf1 SB MO; see Materials and Methods) developed as phenocopies of the alnz24 mutant, an effect that could be rescued completely by co-injection with wild-type paf1 mRNA (Fig. S2).
Establishment but not maintenance of NC progenitors in zygotic aln/paf1 mutants
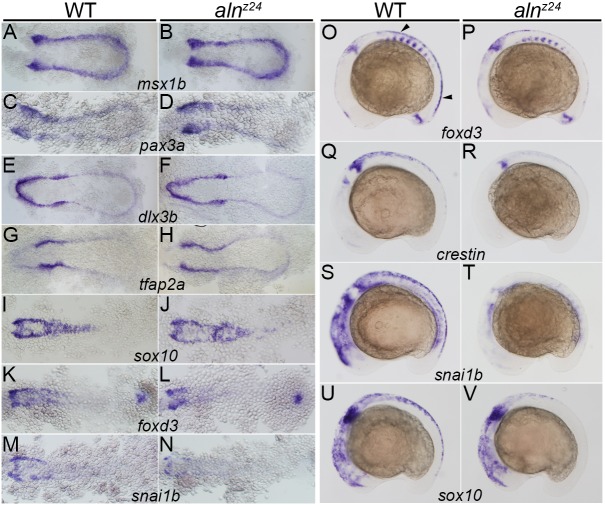
Given the complete loss of trunk NC derivatives in alnz24, we analyzed expression of key transcription factor genes that mark NC development (Simoes-Costa and Bronner, 2015) to identify the first stage of NC development that requires zygotic paf1 gene function (Fig. 2). The earliest known markers expressed by precursors of the NC are msx1b and pax3a, which mark cells at the border of the non-neural and neural ectoderm, the region in which NC is induced. At 11-11.5 hpf, msx1b and pax3a were expressed comparably in wild-type and alnz24 mutant embryos (Fig. 2A-D). Expression of genes that function in early NC specification downstream of msx1b and pax3a, including dlx3b, tfap2a and sox10, also appeared unaffected in 11.5-12 hpf alnz24 mutant embryos (Fig. 2E-J). However, shortly after this period, at 12-12.5 hpf, mutant embryos exhibited slight reductions in the expression of NC markers, such as foxd3 and snai1b (Fig. 2K-N).
Fig. 2.
Zygotically supplied Paf1 is not required for the initial induction or specification of the premigratory NC, but is required at later stages for development of the full premigratory and migrating NC population. Gene expression detected by whole-mount in situ hybridization in 11-11.5 hpf (A-D), 11.5-12 hpf (E-J), 12-12.5 hpf (K-N) and 16 hpf (O-V) wild-type sibling and alnz24 mutant embryos. Expression of msx1b (A,B), pax3a (C,D), dlx3b (E,F), tfap2a (G,H) and sox10 (I,J) is similar in alnz24 mutants and wild-type siblings. A slight decrease in foxd3 (K,L) and snai1b (M,N) expression is detected in alnz24 embryos when compared with wild-type siblings. Expression of foxd3 (O,P), crestin (Q,R), snai1b (S,T) and sox10 (U,V) are all reduced or absent in the trunk region of 16 hpf alnz24 mutant embryos. A-N are dorsal views with anterior towards the left. O-V are lateral views with anterior towards the left. Arrowheads indicate premigratory NC.
As premigratory NC emerges at the dorsal aspect of the neural keel, expression of NC markers dissipates in alnz24 null mutant embryos (Fig. 2O-V). Loss of zygotic paf1 resulted in reduction of foxd3 gene expression, an early lineage marker of premigratory NC, and complete loss of crestin, snai1b or sox10 in the trunks of 16 hpf mutant embryos. As observed previously, trunk NC was affected more severely than cranial NC in aln mutant embryos (Cretekos and Grunwald, 1999). Whereas trunk derivatives were not detected in the absence of zygotic paf1 function, craniofacial NC structures were present but reduced in mutants (Cretekos and Grunwald, 1999) or upon knockdown of zygotic Paf1 protein expression (Fig. 4R). In summary, premigratory NC arises normally in alnz24 mutant embryos, but zygotic paf1 function is required to maintain this NC lineage in the trunk.
Fig. 4.
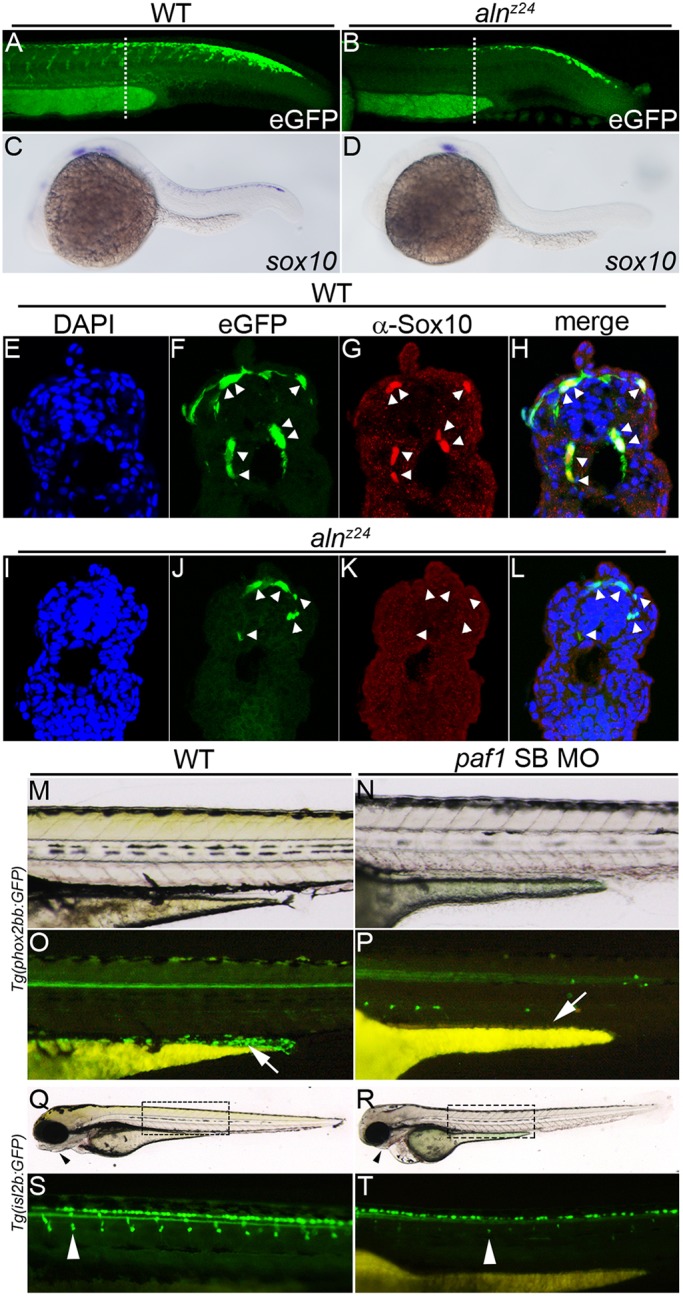
Paf1 is necessary to maintain NC gene expression, identity and the normal complement of multipotent premigratory NC. (A,B) Confocal images of eGFP expression in 26-28 hpf wild-type (A) and alnz24 mutant (B) embryos carrying the Tg(-4.9sox10:egfp)ba2 transgene. Dashed lines indicate approximate plane of section in E-L. (C,D) sox10 gene expression detected by whole-mount in situ hybridization in 24 hpf wild-type (C) and alnz24 mutant (D) embryos. (E-L) Transverse sections through the trunks of 26-28 hpf wild-type (E-H) and alnz24 mutant (I-L) transgenic embryos. In wild-type embryos, eGFP expression colocalizes with Sox10 protein expression (NC cells, indicated by arrowheads) (F-H). In contrast, all migrating GFP+ cells (arrowheads) in the trunk of alnz24 mutant embryos do not express Sox10 (J-L). (M-P) Lateral views with anterior towards the left of 72 hpf wild-type (M,O) and low dose paf1 MO-injected (N,P) Tg(phox2bb:GFP) embryos. GFP-expressing enteric neurons (arrow) are present in the gut of wild-type embryos (M) and are completely absent (arrow) in embryos with reduced paf1 function (P). GFP+ cells in paf1 MO-injected embryos are likely sympathetic neurons. (Q-T) Lateral views with anterior towards the left of 72 hpf wild-type (Q,S) and low dose paf1 MO-injected (R,T) Tg(isl2b:GFP) embryos. Reduction of paf1 function diminishes melanophore formation, and results in cardiac edema and loss of jaw structures (arrowheads) in MO-injected embryos (R) when compared with wild-type embryos (Q). GFP fluorescence marking DRG cells (arrowheads) in wild-type (S) and paf1 MO-injected embryos (T). DRG cell formation in paf1 MO-injected embryos is significantly reduced (T).
paf1 function is required cell autonomously for maintenance of NC progenitors
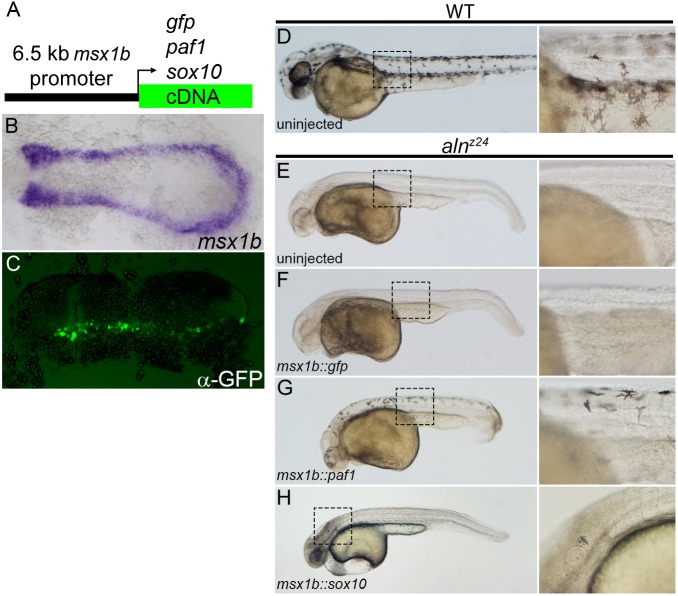
Many factors required for NC development have primary functions in the patterning of precursor tissues and only indirectly affect the generation of NC (Mayor et al., 1997; Nguyen et al., 1998; Villanueva et al., 2002). To determine whether Paf1 is required in developing NC cells, we expressed the Paf1 protein in the NC lineage of embryos otherwise devoid of zygotic paf1 product. A 6.5 kb fragment of the msx1b promoter was used to drive gene expression in the NC precursor domain (Fig. 3A,B). As injected plasmid DNA is distributed mosaically in developing embryos, injection of the msx1b::GFP plasmid into one-cell stage embryos resulted in GFP expression in subsets of the msx1b expression domain at 11-11.5 hpf (Fig. 1C), confirming the spatial and temporal specificity of the promoter fragment. Whereas expression of GFP in the msx1b domain did not rescue NC development in alnz24 embryos (Fig. 3D-F), most mutant embryos injected with the msx1b::paf1 construct (48/72) had significant numbers of melanophores present in both the cranial and trunk regions at 48 hpf (Fig. 3G). The rescued melanophores appeared normal, migrating away from the dorsal neural tube and assuming the stellate morphology of wild-type cells (Fig. 3D,G). These results demonstrate that defective NC development in aln mutants is due solely to the cell-autonomous requirement for paf1 function in developing NC cells.
Fig. 3.
Paf1 is required cell-autonomously in the NC lineage for melanophore development. (A) Schematic representation of a msx1b promoter construct used to express gfp, paf1 or sox10 cDNAs. (B) Endogenous msx1b expression in an 11.5 hpf embryo detected by whole-mount in situ hybridization. (C) GFP (detected by immunohistochemistry) is expressed mosaically, but only within the normal msx1b expression domain of an 11.5 hpf embryo injected at the one-cell stage with 50 pg msx1b::GFP plasmid DNA. (D-H) 48 hpf control and DNA-injected embryos. (D) Wild-type embryo exhibiting normal distribution and morphology of melanophores. Uninjected alnz24 mutant embryos (E) and alnz24 mutants injected with msx1b::GFP plasmid DNA (F) completely lack NC-derived melanophores. (G) alnz24 mutants injected with msx1b::paf1 plasmid DNA have widely distributed melanophores with normal stellate morphology. (H) Expression of sox10 in the msx1b expression domain fails to rescue melanophore development; abnormal pigment cells were occasionally found in the heads of plasmid-injected mutant embryos. (B,C) Dorsal views of flat-mounted embryos, with anterior towards the left. D-H are lateral views of entire embryos (anterior towards the left); boxed regions are shown at higher magnification to the right of each whole-embryo view.
The aln/paf1 mutant reveals a previously unrecognized function in NC development
We attempted to place paf1 function within the gene regulatory network that has been proposed to drive NC development (Simoes-Costa and Bronner, 2015). Although the first detectable alteration in alnz24 embryos is slightly reduced foxd3 and snai1b expression (Fig. 2K-N), forced overexpression of those genes failed to ameliorate the mutant phenotype (data not shown), suggesting paf1 provides additional functions necessary for maintaining the premigratory NC population. Mutants that lack both foxd3 and tfap2a functions are phenotypically similar to paf1-deficient embryos (Arduini et al., 2009; Wang et al., 2011). These genes are thought to promote early stages in specification, and the requirement for them can be bypassed by forced expression of SoxE family genes (Arduini et al., 2009). However overexpression of sox10 (a SoxE family member) in the msx1b expression domain of alnz24 mutants failed to rescue trunk melanophore development in this mutant background (Fig. 3H). Finally, we investigated whether aln mutant NC cells appear to transfate into neural tissue, as they do in embryos lacking both tfap2a and tfap2c (Li and Cornell, 2007). In 11-11.5 hpf wild-type embryos, the NC occupies the region between the sox2-expressing neural plate and dlx3b-expressing pre-placodal ectoderm. In embryos depleted for the tfap2a and tfap2c genes, the sox2 expression domain is expanded so it abuts directly onto the dlx3b domain. In contrast, the patterning of sox2 and dlx3b domains appears normal in alnz24 mutants, consistent with the interpretation that NC is induced normally in aln mutants (Fig. S3). In summary, paf1 provides functions that are distinct from genes whose contributions to NC development have been previously characterized, including snai1b, sox10, foxd3 or tfap2a and tfap2c.
The cells occupying the premigratory NC domain do not simply die in situ. Confirming earlier work that did not detect apoptosis in the early NC of alnz12 mutants (Cretekos and Grunwald, 1999), we could not distinguish mutant and wild-type 14 hpf embryos by virtue of staining for anti-activated caspase 3, nor were we able to rescue the alnz24 phenotype by suppressing p53-mediated cell death using a p53 MO (Robu et al., 2007) (data not shown).
As sox10 mRNA expression is unaffected at early stages of NC specification in 11.5-12 hpf alnz24 mutant embryos (Fig. 2I,J), we used a transgene [Tg(-4.9sox10:egfp)ba2] (Park et al., 2005) in which eGFP expression is driven from the sox10 promoter to trace the fates of the NC progenitors cells in alnz24 mutants (Fig. 4). At 26-28 hpf, premigratory as well as migrating NC express eGFP in the trunk of wild-type embryos (Fig. 4A,F), and sox10 mRNA and protein are detectable in these cells (Fig. 4C,G,H). Fewer eGFP+ cells are apparent in alnz24 mutant embryos of the same stage (Fig. 4B); many of those remain close to the dorsal neural tube with some that appear to be migrating from this region along normal pathways (Fig. 4J). Significantly, unlike in wild-type embryos, the mutant eGFP+ NC cells do not maintain expression of sox10 mRNA or protein (Fig. 4D,K,L). In summary, in the absence of zygotic paf1, mutant NC cells initiate the gene expression program and normal behaviors of NC, but they fail to maintain NC gene expression or identity, and fail to populate their normal destinations.
We sought to determine whether the aberrantly migrating trunk NC eventually gave rise to appropriate derivatives. Neither alnz12 nor alnz24 embryos generate pigmented melanophores during the first 2 days of development, the time at which null mutants begin to appear severely disturbed (Fig. 1) (Cretekos and Grunwald, 1999). To measure the effect of loss of paf1 on the later development of NC-derived dorsal root ganglion (DRG) cells and enteric neurons, moderate doses of paf1 SB MO were injected into eggs harboring transgenes that marked formation of these cell types. These embryos displayed intermediate phenotypes: some melanophores formed, but other characteristics of the aln phenotype were observed, including pericardial edema and reduction of the jaw (Fig. 4M,N,Q,R). The partially paf1-depleted embryos failed to generate any detectable enteric neurons (Fig. 4O,P), and possessed consistently reduced numbers of DRGs (Fig. 4S,T and Fig. S4). It is likely that NC progenitors completely lacking zygotic paf1 fail to give rise to any of the normal trunk NC derivative tissues.
paf1 is required for additional progenitor populations in the embryo
The aln mutant was identified solely on the basis of its NC phenotype; initial characterization demonstrated that loss of aln function did not have additional discernible effects on patterning or cell differentiation in the central nervous system (Cretekos and Grunwald, 1999). As loss of zygotic paf1 affected maintenance of the multipotent premigratory population of NC, we examined the development of additional progenitor cell types in aln mutant embryos. Primitive blood progenitors are first detected in the lateral mesoderm at 10-12 hpf and are marked by expression of myb and tal1. Blood progenitors appear to arise normally in alnz24 mutant embryos, as expression of myb and tal1 in both the anterior and posterior lateral mesoderm are unaffected in 12 hpf alnz24 mutant embryos (Fig. 5A-D). However, by 26 hpf, the levels of expression of myb and tal1 in the intermediate cell mass are significantly downregulated in alnz24 mutant embryos compared with controls (Fig. 5E-H), consistent with the interpretation that Paf1 is needed to maintain development of the erythroid lineage.
Fig. 5.
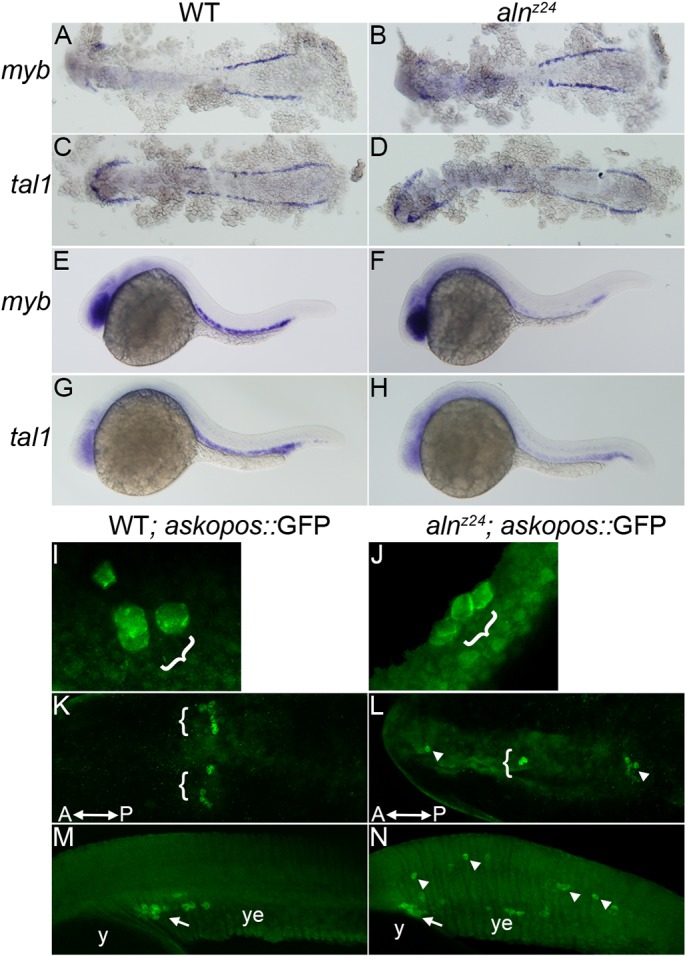
Paf1 is necessary for maintenance of blood and germline stem cell populations. (A-D) Expression of myb (A,B) or tal1 (C,D), markers of early blood precursors in the anterior and posterior lateral mesoderm at 12 hpf, is similar in alnz24 mutant and wild-type sibling embryos. (E-H) In contrast, by 26 hpf, expression of myb (E,F) and tal1 (G,H) in the intermediate cell mass is significantly reduced in alnz24 mutant embryos. (I-N) Wild-type sibling and alnz24; askopos::GFP transgenic embryos that express GFP in PGCs. Small clusters of PGCs arise similarly in wild-type and alnz24 mutant 50-60% epiboly embryos (I,J). PGCs are present in two bilateral clusters at 12 hpf (K) and coalesce at the midline above the yolk extension in 32 hpf (M) wild-type sibling embryos. In contrast, PGCs are present in ectopic locations in 12 hpf (L) and 32 hpf (N) alnz24 mutant embryos. Brackets and arrows indicate normal clusters of PGCs, whereas arrowheads indicate ectopic PGCs. y, yolk; ye, yolk extension. A-D,K,L are dorsal views with anterior towards the left; E-H,M,N, are lateral views with anterior towards the left; I,J are dorsolateral views with animal pole upwards.
The dependence of primordial germ cell (PGC) development on Paf1 function was measured in transgenic embryos that express GFP specifically in PGCs under the control of the askopos promoter (Blaser et al., 2005). Although PGCs arise normally in 5-6 hpf alnz24 mutants (Fig. 5I,J), they behave abnormally during subsequent development. During wild-type development, the initial streams of migrating PGCs arrest mid-axis and form two bilateral clusters of cells evident at 12 hpf (Fig. 5K). In alnz24 mutants of the same stage, PGCs are found scattered along the anteroposterior axis (Fig. 5L). Subsequently, in 30 hpf embryos, wild-type PGCs normally coalesce at the midline above the anterior portion of the yolk extension (Fig. 5M). In dramatic contrast, upon loss of zygotic paf1 function, PGCs wander into numerous ectopic locales, including the brain and throughout the trunk and tail (Fig. 5N). In other experiments aimed at generating maternal-zygotic paf1 mutants, we found homozygous alnz24 mutant PGCs did not survive and develop when transplanted into wild-type hosts (data not shown). In all, these experiments are consistent with the hypothesis that Paf1C is required to maintain proper PGC behavior and identity.
All Paf1C components are required for NC development
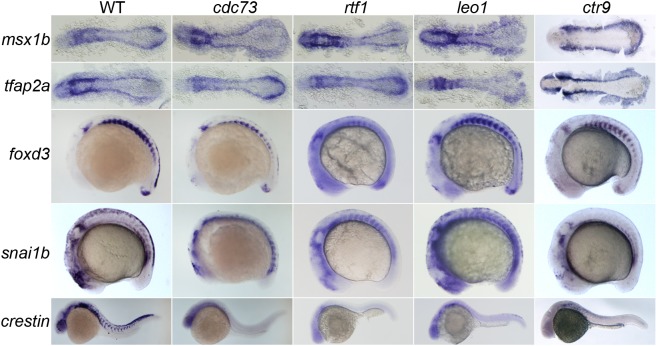
The protein product of aln/paf1 is but one of five components of the vertebrate Paf1C – a cohort of proteins, Paf1, Ctr9, Cdc73, Rtf1 and Leo1, recruited to cooperate with RNA Pol II and facilitate transcription (Jaehning, 2010; Mueller and Jaehning, 2002; Squazzo et al., 2002; Van Oss et al., 2017). Previous work showed members of the Paf1C were required in some way for NC development, as mutations in leo1 or rtf1, or inhibition of ctr9 function by MO knockdown, disrupted formation of NC-derived tissues (Akanuma et al., 2007; Nguyen et al., 2010). We hypothesized the entire intact Paf1C was required to maintain the premigratory NC. We characterized early stages of NC development of two existing Paf1C mutants known to have NC defects, leo1LA1186 and rtf1kt641, and one mutant, cdc73/sunrise, which had been analyzed previously only in the context of its role in erythroid development (Bai et al., 2010). In addition, we characterized the consequences of ctr9 loss of function after we isolated an ENU-induced null mutation (zy13) in the ctr9 gene (p.Trp580X) (Fig. S5). Similar to other Paf1C genes, ctr9 is normally ubiquitously expressed during development and is refined to the anterior neural tissue by 24 hpf (Fig. S5). However, in ctr9zy13 homozygous mutants, ctr9 transcripts are undetectable by early somite stages, indicating it is a null allele (Fig. S5).
Embryos lacking any member of the Paf1C closely resemble the aln mutant phenotype (Fig. 6). Expression of markers of NC induction and early specification, such as msx1b and tfap2a, appear unaffected in 12-12.5 hpf Paf1C mutant embryos. However, the Paf1C mutant embryos fail to maintain normal expression levels of foxd3 and snai1b in the trunk of 16-18 hpf embryos, indicating premigratory NC is not properly maintained. Despite differences in the absolute numbers of NC cells at 16-18 hpf in the different mutants, by 24 hpf all Paf1C mutant embryos are devoid of crestin expression with the exception of leo1LA1186 mutants, which have a few crestin-positive cells in the trunk. Each member of the Paf1C is needed to maintain the premigratory trunk NC population.
Fig. 6.
All members of the Paf1C are needed for maintenance of the trunk premigratory NC. Gene expression in 12-12.5 hpf (msx1b and tfap2a), 16-18 hpf (foxd3 and snai1b) and 24 hpf (crestin) wild-type, cdc73/sunrise, rtf1kt641, leo1LA1186 and ctr9zy13 mutant embryos. Expression of msx1b and tfap2a, which mark the NC precursor domain and NC lineage, respectively, is not altered in Paf1C mutant embryos, while expression of foxd3 and snai1b, which mark the premigratory NC, is severely reduced or absent in the trunk of all Paf1C mutant embryos. Paf1C mutant embryos are devoid of crestin-positive cells, except leo1LA1186 mutants, which have a few crestin-positive cells in the trunk. msx1b- and tfap2a-stained embryos are shown as dorsal views with anterior to the left. foxd3-, snai1b- and crestin-stained embryos are lateral views with anterior towards the left.
Components of the Paf1C are non-redundant. For example, overexpression of Leo1 could not rescue the alnz24 mutant phenotype, and overexpression of Paf1 could not rescue the cdc73 mutant phenotype (data not shown). We found that knockdown of either Ctr9 or Rtf1 function results in a significant reduction of Paf1 protein expression at 16-18 hpf (Fig. S6). Thus, it appears individual components are required for the stability of the entire Paf1C, explaining why loss of component genes have similar mutant phenotypes. We surmise the previously reported NC defects in these mutants arise from the cell-autonomous requirement for Paf1C function in developing NC cells.
The antagonistic functions of P-TEFb and Paf1C regulate NC progenitor development
Paf1C is thought to function as a scaffold, or ‘platform’, necessary for the recruitment of additional transcription factors and enzymes needed to modulate many aspects of transcription (Jaehning, 2010; Van Oss et al., 2017). To identify the Paf1C-associated pathway needed to maintain the NC progenitor population, we sought to identify factors that execute this Paf1C function and thus whose loss might recapitulate the aln mutant phenotype, or opposing factors whose depletion might rescue melanophore development in alnz24 mutant embryos. We used MO knockdown to determine the effects of eliminating expression of individual proteins known to interact with the Paf1C (Costa and Arndt, 2000; Jaehning, 2010; Ng et al., 2003; Rozenblatt-Rosen et al., 2009; Squazzo et al., 2002). Knockdown of Rad6a and Rad6b, E2 ubiquitin-conjugating enzymes required for histone H2B ubiquitylation, CstF3, a factor needed for 3′ end processing, or Spt16, a histone chaperone subunit of the FACT complex that facilitates transcription progression by destabilizing nucleosome structure, failed either to block pigment cell development in wild-type embryos or to rescue melanophore formation in alnz24 mutant embryos (Fig. S7). In contrast, knockdown of Cdk9, the kinase subunit of P-TEFb whose phosphorylation activity triggers release of paused RNA Pol II (Peterlin and Price, 2006), was able to rescue melanophore formation in all MO-injected alnz24 mutant embryos (n=58) (Fig. S7). These data demonstrate that among the potential roles of Paf1C, it is its function in opposing P-TEFb that is needed to maintain the NC progenitor population.
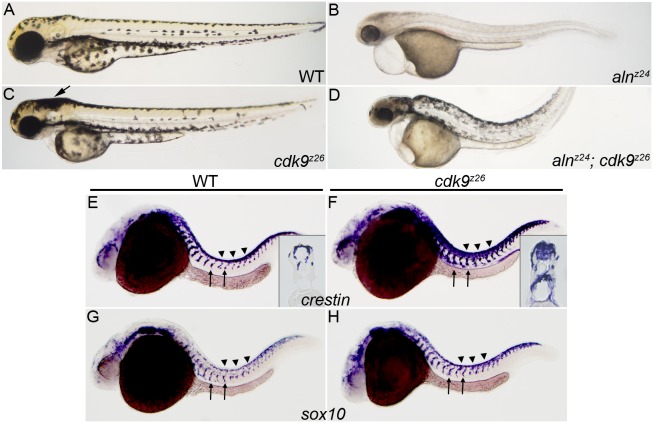
To further explore cdk9 function in NC development, we generated a cdk9 null allele, cdk9z26 (see Materials and Methods). As cdk9 transcripts are supplied maternally (data not shown), the cdk9-null phenotype reflects requirements for the zygotic product. Consistent with the results of Cdk9 knockdown, loss of cdk9 function had a strong effect on NC development (Fig. 7). First, alnz24; cdk9z26 double mutant embryos displayed significant rescue of NC and melanophore development (Fig. 7B,D), indicating that loss of P-TEFb counteracted the effects of loss of the Paf1C on NC progenitors. Second, cdk9z26 mutant embryos developed a superabundance of NC, with an evident excess of melanophores in the head of 48 hpf mutant embryos (Fig. 7A,C). Loss of cdk9 also produced expansion of the streams of crestin+ and sox10+ migrating NC precursor population, with crestin-expressing cells infiltrating the neural tube (Fig. 7E-H). Hence, loss of Paf1C and loss of Cdk9 have reciprocal effects on the NC progenitor population, and the role of Paf1C in maintaining NC progenitors is counteracted by the pause release and/or transcription elongation promoted by P-TEFb.
Fig. 7.
Loss of Cdk9 function expands both the premigratory and migratory NC populations, and suppresses loss of melanophore formation in aln/paf1 mutants. (A-D) Lateral views of 48 hpf wild-type (A), alnz24 mutant (B), cdk9z26 mutant (C) or alnz24; cdk9z26 double mutant embryos (D). (E-H) NC expression of crestin (E,F) and sox10 (G,H) in 26 hpf wild-type and cdk9z26 mutant embryos. Arrow in C indicates an increase in the number of melanophores in the head of cdk9z26 mutant embryos. Insets in E and F are cross-sections through the trunk of crestin-expressing embryos. Arrows indicate migrating NC and arrowheads indicate premigratory NC.
DISCUSSION
Paf1C is required to maintain NC progenitors
Loss of any of the five genes that encode Paf1C components results in the short-lived development of NC progenitors and the failure of these cells to give rise to NC-derived tissues in the trunk of the zebrafish embryo. By detailed examination of NC lineage development, we found the cascade of gene expression events associated with induction and generation of NC progenitors occurred with normal spatiotemporal characteristics in mutant embryos. Lineage tracing indicated that premigratory NC cells failed to maintain expression of the definitive NC marker sox10 and lost their normal developmental identity without immediately dying. Owing to the presence of maternally supplied Paf1 protein and paf1 mRNA deposited in eggs, it is likely the phenotypic changes we observe in the null mutants represent a tapering off of essential functions provided to NC cells, rather than a complete abrogation of Paf1C functions. Maternal supply may also explain different zygotic requirements for paf1 in cranial versus trunk neural crest. As a reflection of their waning specified precursor fate, mutant NC progenitors migrated a few cell diameters from their source, failing to populate their normal destinations or to transfate and join adjacent tissues.
Our studies reveal a unique role for Paf1C that has been previously unrecognized in NC development. Loss of the complex results in a novel defect: the failure to maintain established progenitors. Paf1C is required in NC cells, as expression of paf1 solely in the NC was sufficient to rescue NC development in aln mutants. The inability to maintain the progenitor population is distinct from any function predicted by gene regulatory network models that describe pathways controlling the progressive development of the NC lineage (Simoes-Costa and Bronner, 2015). It is likely the Paf1C has a permissive role as part of the machinery required to support maintenance of the progenitor state rather than an instructive one designating a particular fate. It seems unlikely that many additional genes can be mutated to uncover this function, as independent forward genetic screens repeatedly found that loss of Paf1C components was responsible for the inhibition of the development of premigratory NC in mutants.
Paf1C and P-TEFb functions are common requirements for maintenance of tissue precursors
Whereas our data highlight the important antagonistic relationship between the Paf1C and P-TEFb in maintaining multipotent cells of the NC lineage, the interacting functions of the two complexes may be essential for regulating pluripotent precursor cells in general. Two additional stem cell-like populations, primordial germ cells and erythroid progenitors, were affected in the aln/paf1 mutant embryos. In each instance, progenitor cells appeared to arise transiently, but the normal behavior of the precursors was not completed. We hypothesize that differences in the requirements for the maternally supplied Paf1 protein accounted for the variations in the progenitor phenotypes caused by loss of zygotic paf1 gene expression. Owing to our inability to completely deplete embryos of maternally supplied protein, we could not determine whether Paf1C might be required in additional pluripotent cells in the early zebrafish embryo.
Previous studies have linked the two transcription complexes to the behavior of specific precursor populations. In the zebrafish spinal cord, oligodendrocyte differentiation from an established precursor population is dependent on antagonistic Paf1C/P-TEFb interactions (Kim et al., 2012). Paf1C function has been previously associated with maintenance of a multipotent progenitor state. Paf1C is required for maintenance of embryonic stem cells (ESCs) in vitro, and its forced overexpression blocks their differentiation (Ding et al., 2009; Ponnusamy et al., 2009; Strikoudis et al., 2016). Forced expression of the complex also appears to stabilize the undifferentiated state of certain primitive hematopoietic cells (Muntean et al., 2010). Reciprocally, the function of P-TEFb, i.e. release of promoter-proximal paused Pol II, is required for ESC differentiation (Tastemel et al., 2017). In summary, the Paf1C and P-TEFb have been associated recurrently with maintenance functions in several pluripotent precursor populations.
Paf1C may contribute to transcription pausing in progenitor cells
To test whether loss of pausing might be responsible for loss of NC progenitors, we asked whether loss of P-TEFb activity, the crucial factor involved in promoting pause-release and transcription elongation (Li et al., 2018; Lu et al., 2016; Peterlin and Price, 2006; Zhou et al., 2012), might counteract the effects of loss of Paf1. Indeed null mutations in cdk9, the gene encoding the catalytic subunit of P-TEFb, strongly rescued NC progenitor fate in aln/paf1 mutants. Double mutants produced streams of crestin-expressing migrating NC cells and normal-appearing terminally differentiated melanophores, neither of which were evident in the aln/paf1 mutant. Moreover, loss of Cdk9 in zygotic cdk9z26 mutant embryos led to a superabundance of NC, results that are consistent with the interpretation that inhibiting or delaying pause release is associated with maintenance of the NC progenitor state. Cdk9 was the only Paf1C-interacting factor whose loss suppressed the NC phenotype aln mutant embryos, indicating that pause-release is a crucial function in maintaining the NC progenitor population.
As a result of the antagonistic roles that P-TEFb and the Paf1C have in maintaining the NC progenitor population, we hypothesize that Paf1C is required to achieve transcription pausing. Consistent with this interpretation, the null foggy/spt5sk8 mutant, which is deficient in a component required for transcription pausing, has a phenotype similar to the Paf1C mutants (Keegan et al., 2002; White et al., 2011). Other studies that linked Paf1C to maintenance of tissue precursor populations also indicated that Paf1C functioned to curtail transcription elongation. Paf1C mutations were found to suppress the phenotype of the zebrafish moonshine (trim33) blood mutant, which lacks TIF1γ, a factor that promotes transcription elongation in erythroid progenitors (Bai et al., 2010). In those studies, loss of any member of the Paf1C had effects similar to the spt5m806 mutation, affecting a factor known to be required for RNA Pol II pausing (Guo et al., 2000). It has also been demonstrated that disrupting RNA Pol II pausing reduces hematopoietic stem cells in zebrafish and this defect can be rescued by inhibiting P-TEFb function (Yang et al., 2016). Finally, in tissue culture cells, loss of Paf1 function has been linked to a release of proximal-promoter paused RNA Pol II and its progression onto gene bodies at many loci (Chen et al., 2015), and enhancer-bound Paf1 prevents the release of proximal-promoter paused RNA Pol II (Chen et al., 2017).
Nevertheless, we recognize that the role of Paf1C in maintaining promoter-proximal paused RNA Pol II is controversial (Hou et al., 2019; Vos et al., 2018a,b). Several studies have demonstrated that Paf1C helps promote CTD phosphorylation of paused RNA Pol II and its release into productive elongation (Vos et al., 2018a; Yu et al., 2015) and that Paf1C is a positive modulator of RNA Pol II elongation rate in vitro (Hou et al., 2019). Paf1C functions in transcription termination and processing are also well established (Nordick et al., 2008; Penheiter et al., 2005). Together, these data suggest that the primary function of Paf1C may depend on cell- and tissue-specific factors that interact with Paf1C to modulate its effect on RNA Pol II transcription (Van Oss et al., 2017).
The essential roles of Paf1C and P-TEFb in regulating transcription and their likely pleiotropic roles in development make it difficult to associate these factors with specific developmental events or states. We imagine that knowledge of the specific cells in which Paf1C or P-TEFb function are perturbed experimentally will be crucial to the interpretation of the consequences of the effects. Recent studies found that P-TEFb is required for NC specification in Xenopus embryos as a consequence of its role in the release of paused Myc transcription (Hatch et al., 2016). Expression of Myc was shown previously to be needed for early stages of NC development (Bellmeyer et al., 2003). At first glance these results might appear to contradict the current work indicating that P-TEFb promotes pause release in NC progenitors leading to their differentiation. The disparity likely arises in the different experimental approaches used to analyze the developmental functions of Cdk9. Hatch et al. used translation-blocking morpholinos, which can inhibit the functions of maternally supplied as well as zygotically expressed transcripts, to knockdown Cdk9 function in Xenopus embryos. Their studies likely uncovered the earliest role of pause release in NC development. In contrast, the present studies examined zygotic cdk9 and aln/paf1 mutants, in which maternal products are available to support early development. P-TEFb likely supplies distinct temporal functions that are required to support different stages of NC development: its function may be required early for Myc expression and NC specification, while later promoting the expression of differentiation genes. In conclusion, direct examination of RNA Pol II occupancy and transcript production in tissue progenitors will be required to definitively test the role of transcription pausing in regulating pluripotency and cell differentiation.
MATERIALS AND METHODS
Zebrafish strains
Zebrafish (Danio rerio) were maintained and all experiments were performed in accordance with approved institutional protocols at the University of Utah (IACUC). Adult zebrafish were maintained under standard conditions (Westerfield, 2000) and kept on a light-dark cycle of 14 h in light and 10 h in dark at 27°C. AB and Tu strains were used as wild-type zebrafish. The previously described alnz12 mutation (Cretekos and Grunwald, 1999) is maintained on the AB background. The c4 allele is a gamma ray-induced deletion on chromosome 15 (Fisher et al., 1997). The previously isolated Paf1C mutant lines used in this study were: sunrise/cdc73 (Bai et al., 2010); leo1LA1186 (Nguyen et al., 2010) and rtf1kt641 (Akanuma et al., 2007). The ctr9zy13 and alyronz24 mutations isolated in this study were recovered from AB animals mutagenized with ENU as described previously (Mullins et al., 1994). The ctr9zy13 mutation was recovered from a standard F2 screen and was maintained by outcrosses to wild-type WIK animals. The alyronz24 mutation was recovered in a screen for mutations that failed to complement alyronz12 and is maintained in the AB background. cdk9z26 was induced by TALEN-mediated mutagenesis of Tu zebrafish and is maintained on the Tu background. The sox10:egfp (Park et al., 2005), isl2b:GFP (Pittman et al., 2008) and phox2bb:GFP (Nechiporuk et al., 2007) transgenes have been previously described.
Zebrafish embryo culture
Embryos from natural spawnings were generated and collected as described previously (Westerfield, 2000). Live embryos were maintained at 28°C. All developmental staging was based on counting somite number and calculated based on somitogenesis beginning at 10 hpf, with the first six somites forming three per hour and all subsequent somites forming two per hour (18-somite stage is 18 hpf) (Kimmel et al., 1995).
Molecular analysis of the z12 and c4 genomic regions
Genomic DNA (gDNA) was isolated from pools of alnz12 mutant or wild-type sibling embryos. PCR amplification from the gDNA identified the following markers as absent from alnz12 chromosomes: z37, z1195, z13759, z27227, z24, z6190, z25713, z40966, z858, z846 and z5223, and the gene gap-43 (Fig. S1). To define the alyron critical region, gDNA was isolated from alnz12/c4 mutant and wild-type sibling 32 hpf embryos, and PCR amplification from the gDNA samples was used to determine marker sequences that were missing in transheterozygous genomes (Fig. S1). The critical region corresponds to ∼1-2.5cM. Z-marker primer sequences can be obtained from the Zebrafish Information Network (zfin.org). gap-43 primer sequences are: gap-43 F, 5′-GAAGAGGCCGATCAGGAGATC-3′; gap-43 R, 5′-GTTTCTGTGGAGGCGTCAGC-3′.
Genotyping of aln mutant embryos
The genotypes of mutant embryos were determined by PCR analysis of gDNA isolated from individual embryos. Homozygous alnz12 mutant embryos were identified by the absence of gap-43 sequences. The alnz24 C>A transversion at base pair 843 of the paf1 coding sequence disrupts an NspI restriction endonuclease site present in the wild-type genome. To genotype alnz24 embryos, the region of the paf1 gene spanning the alnz24 mutation was amplified from gDNA using the following primers: paf1g F, 5′-GTTCAGAGGTATGATGGATGAGG-3′ and paf1g R, 5′-GTATGCAGCTTTATGAAAACACTC-3′. The PCR product was digested with NspI (New England Biolabs) and resolved on a 2% agarose gel.
Identification of ctr9zy13
The gene responsible for the ctr9zy13 mutation was identified using a combination of restriction site-associated DNA sequencing (RADseq) mapping (Miller et al., 2007) and a candidate gene approach. Briefly, gDNA was prepared from pools of 70 mutant and 70 wild-type sibling embryos, and mapped by RADseq analysis (Floragenex). A region of ∼10 Mb on LG 7 was identified as the probable site of the genetic lesion. Analysis of the Ensemble zebrafish Zv8 assembly revealed the candidate region included ctr9, a gene encoding a component of the Paf1C. A homozygous G-to-A nucleotide substitution was detected in mutant gDNA. The mutation changes a tryptophan (W) residue at amino acid position 580 to a stop codon; the wild-type Ctr9 protein is 1160 amino acids.
Cdk9 mutant generation
TALENs were generated to exon 2 of the cdk9 coding sequence (NM_212591) and injected into Tu embryos, and an 11 bp deletion that removes nucleotides 144-154 of the coding sequence was recovered (Dahlem et al., 2012). The cdk9z26 mutation results in a premature stop codon; no Cdk9 protein is detectable in 26 hpf mutant protein extracts by immunoblot using a zebrafish-specific Cdk9 antibody (GeneTex #zf124698) (data not shown).
Cloning and analysis of zebrafish paf1 and ctr9 sequences
The zebrafish paf1 cDNA (NM_001024453) and ctr9 cDNA (NM_001083583) sequences were used to generate primers to amplify full-length cDNA from wild-type and mutant embryos. Primers paf1 F (5′-TCCTCCATGGCTCCTACCATAC-3′) and paf1 R (5′-GTGAACTATGCCTCTGCAGGAGC-3′) or ctr9 F (5′-GCTGCTGTTCATCACCACTG-3′) and ctr9 R (5′-CAGGGTGTGTTGTCCCTTT-3′) were used to PCR amplify coding sequences, which were cloned into pGEM-T Easy (Promega) or pCRII (Invitrogen), sequence verified, and subcloned into the pCS2+ vector for generation of full-length wild-type and mutant mRNAs and antisense probes.
Immunoblot analysis
Protein extracts were prepared as previously described (Jurynec and Grunwald, 2010) from groups of 30 alnz12 mutant, alnz24 mutant or wild-type sibling embryos harvested at 36 hpf or groups of 30 wild-type control, ctr9 MO-injected or rtf1 MO-injected embryos harvested at 16-18 hpf. Protein was prepared as described (Link et al., 2006) from unfertilized eggs collected from adult females. Total lysate (25 µg) was resolved by electrophoresis through 4–20% polyacrylamide gradient gels (NuSep). Blots of the gels were probed with α-pMAPK (1:10,000; SAB4504395, Sigma) or α-Paf1 (1:6500; #A300-173A, Bethyl Laboratories). Secondary antibodies were HRP-linked goat anti-mouse (1:10,000, Jackson ImmunoResearch, 115-035-174) and HRP-linked goat anti-rabbit (1:10,000, Jackson ImmunoResearch, 111-035-144). Signal was developed following incubation with ECL Plus (GE Healthcare).
Morpholino oligonucleotide injections
An antisense splice-blocking morpholino oligonucleotide targeting the paf1 exon 1 splice donor site was synthesized (Gene Tools): paf1 SB MO 5′-TATTATGGTTTATTCCTCTCTCACCGG-3′. Approximately 1 nl (4 ng) morpholino was injected into the yolk of one-cell stage wild-type embryos. paf1 SB MO is highly effective at blocking paf1 expression: RT-PCR and sequence analyses of paf1 cDNA from paf1 SB MO-injected embryos detected only unspliced RNA containing intron 1; intron inclusion introduced premature stop codons in all three open reading frames. To generate embryos with reduced Paf1 function that are viable at 72 hpf for analysis of DRG and enteric neuron formation (Fig. 4M-T), 1 ng of paf1 SB MO was injected into the yolk of one-cell stage embryos. Additional antisense MOs were used as previously described (Bai et al., 2010): cdk9, rtf1, spt16, cstF3, rad6a and rad6b. Fixed tfap2a and tfap2c MO-injected and control embryos were kindly provided by Rob Cornell (University of Iowa, Iowa City, IA, USA).
mRNA and DNA injections
Full-length wild-type and mutant paf1, wild-type and mutant ctr9, and snai1b and foxd3 mRNAs were made using the AmpliCap SP6 High Yield Message Maker Kit (Epicentre Biotechnologies). Approximately 200 pg paf1 mRNA and 50-100 pg ctr9 RNA were injected into one-cell stage embryos. Embryos were scored for the presence or absence of melanophores at 36-48 hpf. Injected embryos were genotyped by PCR. The 6.5 kb msx1b promoter upstream of the translational start site was isolated from a PAC (182o13-BUSM1 PAC library) following restriction digestion and cloned into pBluescript SK. The 6.5 kb promoter corresponds approximately to bp 49,852,799-49,859,299 on LG 1 (Ensemble zebrafish Zv9 assembly). cDNA sequences encoding eGFP, Paf1 or Sox10 were cloned downstream of the promoter. DNA constructs (50 pg) containing msx1b::GFP, msx1b::paf1 or msx1b::sox10 were injected into one-cell stage z24 intercross embryos, and embryos were scored for presence of melanophores at 48 hpf. Anti-GFP antibodies (1:1000, Thermo Fisher Scientific, A-11122) were used to detect mosaic GFP expression (Jurynec et al., 2008) at 11.5 hpf in embryos injected with msx1b::GFP DNA; anti-Sox10 antibody (1:1000) (a gift from B. Appel, University of Colorado School of Medicine, USA) was used as described by Park et al. (2005).
Immunohistochemistry and in situ hybridization
Embryos were fixed with fresh 4% paraformaldehyde in PBS at room temperature for 2 h or overnight at 4°C. Fixed embryos were dehydrated in methanol and stored at −20°C until processing for immunohistochemistry according to standard procedures (Westerfield, 2000). In brief, embryos were rehydrated into Ptw (PBS with 0.1% Tween-20) and then incubated in blocking agent (10% heat-inactivated sheep serum, 1% DMSO, 2 mg/ml BSA and 0.1% Triton X-100 in PBS) for at least 1 h at room temperature. Embryos were incubated in primary antibodies diluted in blocking agent overnight at 4°C. Primary antibodies were removed and embryos were washed extensively with PBDT (2 mg/ml BSA, 0.1% TritonX-100 and 1% DMSO in PBS). Embryos were next incubated with appropriate secondary antibodies in the dark for either 2 h at room temperature or overnight at 4°C followed by extensive washes in PBDT. Primary antibodies used were: anti-pMAPK (1:10,000; SAB4504395, Sigma), anti-Paf1 (1:6500; #A300-173A, Bethyl Laboratories), anti-GFP (1:1000, A-11122, ThermoFisher), anti-Cdk9 (1:1000, GeneTex, zf124698) and anti-Sox10 antibody (1:1000; a gift from B. Appel). Secondary antibodies used were donkey anti-mouse IgG-488 at 1:500 (Jackson ImmunoResearch, 715-545-151), or goat anti-rabbit IgG-594 at 1:500 (Jackson ImmunoResearch, 111-585-144). DAPI was used at 1:10,000 as a nuclear counterstain. Embryos were taken stepwise through a glycerol series into 75% glycerol. All fluorescence images comparing wild-type and alnz24 mutant embryos or wild-type and paf1 SB MO-injected embryos were captured using identical settings.
Whole-mount in situ hybridization was performed on embryos fixed overnight at 4°C in 4% paraformaldehyde, washed in Ptw, dehydrated stepwise into 100% methanol and maintained at −20°C until processing. Riboprobe hybridization with digoxigenin (DIG; Roche)-labeled riboprobes followed standard procedures (Jurynec et al., 2008). Embryos were rehydrated into Ptw and treated with 5 µg/ml proteinase K in Ptw for 2 min and 0.1 M triethanolamine (TEA) in Ptw twice for 5 min. Acetic anhydride was added to the TEA mixture and embryos were refixed in 3.7% formaldehyde in Ptw for 20 min followed by thorough washing. Embryos were incubated in hybridization buffer (70% formamide, 5× SSC, 1 mg/ml yeast RNA, 100 μg/ml heparin, 1× Denhardts, 0.1% Tween-20, 5 mM EDTA) at 70°C for a minimum of 16 h followed by incubation in probe (1 ng/µl) in hybridization buffer at 70°C for 1-2 days. After a series of washes and incubation with anti-digoxigenin-AP overnight, embryos were developed in NBT/BCIP (Roche) to detect alkaline phosphatase. Probes for the following genes were used for in situ hybridization: crestin, msx1b, sox10, myb, tfap2a, foxd3, snai1b, paf1, sox2, dlx3b and ctr9. Embryos were mounted in 75% glycerol. Transverse sections approximately one somite thick were collected manually using a razor blade.
Supplementary Material
Acknowledgements
This work is dedicated to our friend Chris J. Cretekos (1966-2014), who had the insight to generate and characterize the alyronz12 mutant. Rob Cornell generously provided tfap2a/c MO-depleted embryos for analysis and Bruce Appel generously provided the Sox10 antibody. We thank our colleagues Chi-Bin Chien, Rich Dorsky and Rod Stewart for scientific guidance and critical feedback. We thank the University of Utah Health Sciences Core Facilities for DNA sequencing, oligonucleotide synthesis, imaging support and zebrafish husbandry.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.J.J., D.J.G.; Methodology: M.J.J.; Validation: M.J.J.; Formal analysis: M.J.J., B.W.B., H.J., R.A.S.P., H.J.Y., D.J.G.; Investigation: M.J.J., B.W.B., H.J., R.A.S.P., H.A.G., Y.-C.S., K.H., D.J.G.; Resources: X.B., A.N., L.I.Z.; Writing - original draft: M.J.J., B.W.B., H.J.Y., D.J.G.; Writing - review & editing: M.J.J., X.B., B.W.B., H.J.Y., L.I.Z., D.J.G.; Visualization: M.J.J., B.W.B.; Supervision: D.J.G.; Project administration: D.J.G.; Funding acquisition: D.J.G.
Funding
This work was supported by grants to D.J.G. from the National Institutes of Health (NIH) (1P01HD048886 and 1R01HD081950); to L.I.Z. from the Melanoma Research Alliance and the NIH (PO1 CA163222); and to H.J.Y. from the NIH (UM1 HL098160). We acknowledge direct financial support for the research reported in this publication from the Huntsman Cancer Foundation and the Nuclear Control of Cell Growth and Differentiation Program at the Huntsman Cancer Institute. We also acknowledge support from the National Cancer Institute/National Institutes of Health (P30CA042014). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.180133.supplemental
References
- Adelman K. and Lis J. T. (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720-731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K., Kennedy M. A., Nechaev S., Gilchrist D. A., Muse G. W., Chinenov Y. and Rogatsky I. (2009). Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. USA 106, 18207-18212. 10.1073/pnas.0910177106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanuma T., Koshida S., Kawamura A., Kishimoto Y. and Takada S. (2007). Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep. 8, 858-863. 10.1038/sj.embor.7401045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduini B. L., Bosse K. M. and Henion P. D. (2009). Genetic ablation of neural crest cell diversification. Development 136, 1987-1994. 10.1242/dev.033209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrampour S. and Thor S. (2016). Ctr9, a key component of the Paf1 complex, affects proliferation and terminal differentiation in the developing Drosophila nervous system. G3 6, 3229-3239. 10.1534/g3.116.034231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A. et al. (2010). TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142, 133-143. 10.1016/j.cell.2010.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmeyer A., Krase J., Lindgren J. and LaBonne C. (2003). The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev. Cell 4, 827-839. 10.1016/S1534-5807(03)00160-6 [DOI] [PubMed] [Google Scholar]
- Blaser H., Eisenbeiss S., Neumann M., Reichman-Fried M., Thisse B., Thisse C. and Raz E. (2005). Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J. Cell Sci. 118, 4027-4038. 10.1242/jcs.02522 [DOI] [PubMed] [Google Scholar]
- Boettiger A. N. and Levine M. (2009). Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471-473. 10.1126/science.1173976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner M. E. and Simões-Costa M. (2016). The neural crest migrating into the twenty-first century. Curr. Top. Dev. Biol. 116, 115-134. 10.1016/bs.ctdb.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yamaguchi Y., Tsugeno Y., Yamamoto J., Yamada T., Nakamura M., Hisatake K. and Handa H. (2009). DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 23, 2765-2777. 10.1101/gad.1834709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. X., Woodfin A. R., Gardini A., Rickels R. A., Marshall S. A., Smith E. R., Shiekhattar R. and Shilatifard A. (2015). PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell 162, 1003-1015. 10.1016/j.cell.2015.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. X., Xie P., Collings C. K., Cao K., Aoi Y., Marshall S. A., Rendleman E. J., Ugarenko M., Ozark P. A., Zhang A. et al. (2017). PAF1 regulation of promoter-proximal pause release via enhancer activation. Science 357, 1294-1298. 10.1126/science.aan3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. X., Smith E. R. and Shilatifard A. (2018). Born to run: control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464-478. 10.1038/s41580-018-0010-5 [DOI] [PubMed] [Google Scholar]
- Costa P. J. and Arndt K. M. (2000). Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156, 535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretekos C. J. and Grunwald D. J. (1999). alyron, an insertional mutation affecting early neural crest development in zebrafish. Dev. Biol. 210, 322-338. 10.1006/dbio.1999.9287 [DOI] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., Weis A. M., Voytas D. F. and Grunwald D. J. (2012). Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Paszkowski-Rogacz M., Nitzsche A., Slabicki M. M., Heninger A.-K., de Vries I., Kittler R., Junqueira M., Shevchenko A., Schulz H. et al. (2009). A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4, 403-415. 10.1016/j.stem.2009.03.009 [DOI] [PubMed] [Google Scholar]
- Fisher S., Amacher S. L. and Halpern M. E. (1997). Loss of cerebum function ventralizes the zebrafish embryo. Development 124, 1301-1311. [DOI] [PubMed] [Google Scholar]
- Gaertner B. and Zeitlinger J. (2014). RNA polymerase II pausing during development. Development 141, 1179-1183. 10.1242/dev.088492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariglio P., Bellard M. and Chambon P. (1981). Clustering of RNA polymerase B molecules in the 5′ moiety of the adult beta-globin gene of hen erythrocytes. Nucleic Acids Res. 9, 2589-2598. 10.1093/nar/9.11.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach J. M., Furrer M., Gallant M., Birkel D., Baluapuri A., Wolf E. and Gallant P. (2017). PAF1 complex component Leo1 helps recruit Drosophila Myc to promoters. Proc. Natl. Acad. Sci. USA 114, E9224-E9232. 10.1073/pnas.1705816114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Yamaguchi Y., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H. and Rosenthal A. (2000). A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408, 366-369. 10.1038/35042590 [DOI] [PubMed] [Google Scholar]
- Hatch V. L., Marin-Barba M., Moxon S., Ford C. T., Ward N. J., Tomlinson M. L., Desanlis I., Hendry A. E., Hontelez S., van Kruijsbergen I. et al. (2016). The positive transcriptional elongation factor (P-TEFb) is required for neural crest specification. Dev. Biol. 416, 361-372. 10.1016/j.ydbio.2016.06.012 [DOI] [PubMed] [Google Scholar]
- Henriques T., Gilchrist D. A., Nechaev S., Bern M., Muse G. W., Burkholder A., Fargo D. C. and Adelman K. (2013). Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell 52, 517-528. 10.1016/j.molcel.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Wang Y., Liu Y., Zhang N., Shamovsky I., Nudler E., Tian B. and Dynlacht B. D. (2019). Paf1C regulates RNA polymerase II progression by modulating elongation rate. Proc. Natl. Acad. Sci. USA 116, 14583-14592. 10.1073/pnas.1904324116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning J. A. (2010). The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799, 379-388. 10.1016/j.bbagrm.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurynec M. J. and Grunwald D. J. (2010). SHIP2, a factor associated with diet-induced obesity and insulin sensitivity, attenuates FGF signaling in vivo. Dis. Model. Mech. 3, 733-742. 10.1242/dmm.000703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurynec M. J., Xia R., Mackrill J. J., Gunther D., Crawford T., Flanigan K. M., Abramson J. J., Howard M. T. and Grunwald D. J. (2008). Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc. Natl. Acad. Sci. USA 105, 12485-12490. 10.1073/pnas.0806015105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan B. R., Feldman J. L., Lee D. H., Koos D. S., Ho R. K., Stainier D. Y. and Yelon D. (2002). The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development 129, 1623-1632. [DOI] [PubMed] [Google Scholar]
- Kim J., Guermah M. and Roeder R. G. (2010). The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140, 491-503. 10.1016/j.cell.2009.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim J.-D., Chung A.-Y., Kim H.-S., Kim Y.-S., Kim M.-J., Koun S., Lee Y. M., Rhee M., Park H.-C. et al. (2012). Antagonistic regulation of PAF1C and p-TEFb is required for oligodendrocyte differentiation. J. Neurosci. 32, 8201-8207. 10.1523/JNEUROSCI.5344-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Langenbacher A. D., Nguyen C. T., Cavanaugh A. M., Huang J., Lu F. and Chen J.-N. (2011). The PAF1 complex differentially regulates cardiomyocyte specification. Dev. Biol. 353, 19-28. 10.1016/j.ydbio.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N. and Kalcheim C. (1999). The Neural Crest, 2nd edn. Cambridge: Cambridge University Press. [Google Scholar]
- Levine M. (2011). Paused RNA polymerase II as a developmental checkpoint. Cell 145, 502-511. 10.1016/j.cell.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. and Cornell R. A. (2007). Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev. Biol. 304, 338-354. 10.1016/j.ydbio.2006.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu M., Chen L.-F. and Chen R. (2018). P-TEFb: Finding its ways to release promoter-proximally paused RNA polymerase II. Transcription 9, 88-94. 10.1080/21541264.2017.1281864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link V., Carvalho L., Castanon I., Stockinger P., Shevchenko A. and Heisenberg C. P. (2006). Identification of regulators of germ layer morphogenesis using proteomics in zebrafish. J. Cell Sci. 119, 2073-2083. 10.1242/jcs.02928 [DOI] [PubMed] [Google Scholar]
- Lu X., Zhu X., Li Y., Liu M., Yu B., Wang Y., Rao M., Yang H., Zhou K., Wang Y. et al. (2016). Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res. 44, 6853-6867. 10.1093/nar/gkw571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Landry H. M. and Churchman L. S. (2017). Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr. Opin. Cell Biol. 46, 72-80. 10.1016/j.ceb.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R., Guerrero N. and Martínez C. (1997). Role of FGF and noggin in neural crest induction. Dev. Biol. 189, 1-12. 10.1006/dbio.1997.8634 [DOI] [PubMed] [Google Scholar]
- Miller M. R., Atwood T. S., Eames B. F., Eberhart J. K., Yan Y.-L., Postlethwait J. H. and Johnson E. A. (2007). RAD marker microarrays enable rapid mapping of zebrafish mutations. Genome Biol. 8, R105 10.1186/gb-2007-8-6-r105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min I. M., Waterfall J. J., Core L. J., Munroe R. J., Schimenti J. and Lis J. T. (2011). Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 25, 742-754. 10.1101/gad.2005511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C., Hausmann G. and Basler K. (2006). Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125, 327-341. 10.1016/j.cell.2006.01.053 [DOI] [PubMed] [Google Scholar]
- Mosimann C., Hausmann G. and Basler K. (2009). The role of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control. Mech. Dev. 126, 394-405. 10.1016/j.mod.2009.02.002 [DOI] [PubMed] [Google Scholar]
- Mueller C. L. and Jaehning J. A. (2002). Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22, 1971-1980. 10.1128/MCB.22.7.1971-1980.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins M. C., Hammerschmidt M., Haffter P. and Nüsslein-Volhard C. (1994). Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr. Biol. 4, 189-202. 10.1016/S0960-9822(00)00048-8 [DOI] [PubMed] [Google Scholar]
- Muntean A. G., Tan J., Sitwala K., Huang Y., Bronstein J., Connelly J. A., Basrur V., Elenitoba-Johnson K. S. J. and Hess J. L. (2010). The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 17, 609-621. 10.1016/j.ccr.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S. and Adelman K. (2011). Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809, 34-45. 10.1016/j.bbagrm.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A., Linbo T., Poss K. D. and Raible D. W. (2007). Specification of epibranchial placodes in zebrafish. Development 134, 611-623. 10.1242/dev.02749 [DOI] [PubMed] [Google Scholar]
- Ng H. H., Dole S. and Struhl K. (2003). The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278, 33625-33628. 10.1074/jbc.C300270200 [DOI] [PubMed] [Google Scholar]
- Nguyen V. H., Schmid B., Trout J., Connors S. A., Ekker M. and Mullins M. C. (1998). Ventral and lateral regions of the zebrafish gastrula, including the neural crest progenitors, are established by a bmp2b/swirl pathway of genes. Dev. Biol. 199, 93-110. 10.1006/dbio.1998.8927 [DOI] [PubMed] [Google Scholar]
- Nguyen C. T., Langenbacher A., Hsieh M. and Chen J.-N. (2010). The PAF1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish. Dev. Biol. 341, 167-175. 10.1016/j.ydbio.2010.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordick K., Hoffman M. G., Betz J. L. and Jaehning J. A. (2008). Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell 7, 1158-1167. 10.1128/EC.00434-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-C., Boyce J., Shin J. and Appel B. (2005). Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J. Neurosci. 25, 6836-6844. 10.1523/JNEUROSCI.0981-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter K. L., Washburn T. M., Porter S. E., Hoffman M. G. and Jaehning J. A. (2005). A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20, 213-223. 10.1016/j.molcel.2005.08.023 [DOI] [PubMed] [Google Scholar]
- Peterlin B. M. and Price D. H. (2006). Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297-305. 10.1016/j.molcel.2006.06.014 [DOI] [PubMed] [Google Scholar]
- Pittman A. J., Law M.-Y. and Chien C.-B. (2008). Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development 135, 2865-2871. 10.1242/dev.025049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy M. P., Deb S., Dey P., Chakraborty S., Rachagani S., Senapati S. and Batra S. K. (2009). RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells 27, 3001-3011. 10.1002/stem.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A. and Young R. A. (2010). c-Myc regulates transcriptional pause release. Cell 141, 432-445. 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A. and Ekker S. C. (2007). p53 activation by knockdown technologies. PLoS Genet. 3, e78 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E. and Lis J. T. (1988). The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795-804. 10.1016/S0092-8674(88)91087-2 [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O., Nagaike T., Francis J. M., Kaneko S., Glatt K. A., Hughes C. M., LaFramboise T., Manley J. L. and Meyerson M. (2009). The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl. Acad. Sci. USA 106, 755-760. 10.1073/pnas.0812023106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon K. E., Mauger D. M. and Arndt K. M. (2005). A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20, 225-236. 10.1016/j.molcel.2005.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M. and Bronner M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242-257. 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., Jennings J. L., Link A. J., Arndt K. M. and Hartzog G. A. (2002). The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21, 1764-1774. 10.1093/emboj/21.7.1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strikoudis A., Lazaris C., Trimarchi T., Galvao Neto A. L., Yang Y., Ntziachristos P., Rothbart S., Buckley S., Dolgalev I., Stadtfeld M. et al. (2016). Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat. Cell Biol. 18, 1127-1138. 10.1038/ncb3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tastemel M., Gogate A. A., Malladi V. S., Nguyen K., Mitchell C., Banaszynski L. A. and Bai X. (2017). Transcription pausing regulates mouse embryonic stem cell differentiation. Stem Cell Res. 25, 250-255. 10.1016/j.scr.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss S. B., Shirra M. K., Bataille A. R., Wier A. D., Yen K., Vinayachandran V., Byeon I.-J. L., Cucinotta C. E., Héroux A., Jeon J. et al. (2016). The histone modification domain of Paf1 complex subunit Rtf1 directly stimulates H2B ubiquitylation through an interaction with Rad6. Mol. Cell 64, 815-825. 10.1016/j.molcel.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss S. B., Cucinotta C. E. and Arndt K. M. (2017). Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem. Sci. 42, 788-798. 10.1016/j.tibs.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Lopez G. A., Cerrizuela S. and Aybar M. J. (2017). Trunk neural crest cells: formation, migration and beyond. Int. J. Dev. Biol. 61, 5-15. 10.1387/ijdb.160408gv [DOI] [PubMed] [Google Scholar]
- Villanueva S., Glavic A., Ruiz P. and Mayor R. (2002). Posteriorization by FGF, Wnt, and retinoic acid is required for neural crest induction. Dev. Biol. 241, 289-301. 10.1006/dbio.2001.0485 [DOI] [PubMed] [Google Scholar]
- Vos S. M., Farnung L., Boehning M., Wigge C., Linden A., Urlaub H. and Cramer P. (2018a). Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607-612. 10.1038/s41586-018-0440-4 [DOI] [PubMed] [Google Scholar]
- Vos S. M., Farnung L., Urlaub H. and Cramer P. (2018b). Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601-606. 10.1038/s41586-018-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-D., Melville D. B., Montero-Balaguer M., Hatzopoulos A. K. and Knapik E. W. (2011). Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population. Dev. Biol. 360, 173-185. 10.1016/j.ydbio.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. Eugene: University of Oregon Press. [Google Scholar]
- White R. M., Cech J., Ratanasirintrawoot S., Lin C. Y., Rahl P. B., Burke C. J., Langdon E., Tomlinson M. L., Mosher J., Kaufman C. et al. (2011). DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518-522. 10.1038/nature09882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Liu X., Zhou T., Cook J., Nguyen K. and Bai X. (2016). RNA polymerase II pausing modulates hematopoietic stem cell emergence in zebrafish. Blood 128, 1701-1710. 10.1182/blood-2016-02-697847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Yang W., Ni T., Tang Z., Nakadai T., Zhu J. and Roeder R. G. (2015). RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 350, 1383-1386. 10.1126/science.aad2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Li T. and Price D. H. (2012). RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119-143. 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.