Significance
Natural products form the basis for most drugs in clinical use. Advances in genome sequencing and bioinformatic tools have revealed thousands of biosynthetic gene clusters encoding these products. However, linking natural products identified by genome mining to their corresponding products in untargeted metabolomics data remains a key challenge. Here we present a platform, DeepRiPP, which integrates genomic and metabolomic data to automate the discovery of new ribosomally synthesized posttranslationally modified peptides (RiPPs), a subclass of natural products with diverse chemistry and activities. We apply DeepRiPP to discover 3 novel RiPPs.
Keywords: natural products, RiPPs, genome mining, machine learning, metabolomics
Abstract
Microbial natural products represent a rich resource of evolved chemistry that forms the basis for the majority of pharmacotherapeutics. Ribosomally synthesized and posttranslationally modified peptides (RiPPs) are a particularly interesting class of natural products noted for their unique mode of biosynthesis and biological activities. Analyses of sequenced microbial genomes have revealed an enormous number of biosynthetic loci encoding RiPPs but whose products remain cryptic. In parallel, analyses of bacterial metabolomes typically assign chemical structures to only a minority of detected metabolites. Aligning these 2 disparate sources of data could provide a comprehensive strategy for natural product discovery. Here we present DeepRiPP, an integrated genomic and metabolomic platform that employs machine learning to automate the selective discovery and isolation of novel RiPPs. DeepRiPP includes 3 modules. The first, NLPPrecursor, identifies RiPPs independent of genomic context and neighboring biosynthetic genes. The second module, BARLEY, prioritizes loci that encode novel compounds, while the third, CLAMS, automates the isolation of their corresponding products from complex bacterial extracts. DeepRiPP pinpoints target metabolites using large-scale comparative metabolomics analysis across a database of 10,498 extracts generated from 463 strains. We apply the DeepRiPP platform to expand the landscape of novel RiPPs encoded within sequenced genomes and to discover 3 novel RiPPs, whose structures are exactly as predicted by our platform. By building on advances in machine learning technologies, DeepRiPP integrates genomic and metabolomic data to guide the isolation of novel RiPPs in an automated manner.
A substantial majority of small molecule therapeutics presently in clinical use are derived from naturally occurring molecules produced by bacteria, fungi, and plants (1). The complex and diverse chemistries of these molecules have been refined over evolutionary timescales in order to provide their producing organisms with selective advantages in their natural environments, and consequently, they can be viewed as structures privileged by evolution (2). During the mid-20th century, natural products formed the backbone of industrial drug development programs. However, the extensive exploitation of biologically active molecules that are abundantly produced by organisms readily cultured in laboratory environments—the so-called “low-hanging fruit” of microbial natural products (3)—made traditional bioactivity-guided screening of microbial extracts economically infeasible by the end of the 20th century, in part due to high rediscovery rates (4). Studies of sequenced bacterial genomes indicate a vast genetically encoded resource of undiscovered natural products, many of which are likely to have biological activities of considerable pharmaceutical or industrial utility (5–7). However, leveraging genomic information toward the directed discovery of novel molecules has proven substantially less straightforward than anticipated (8).
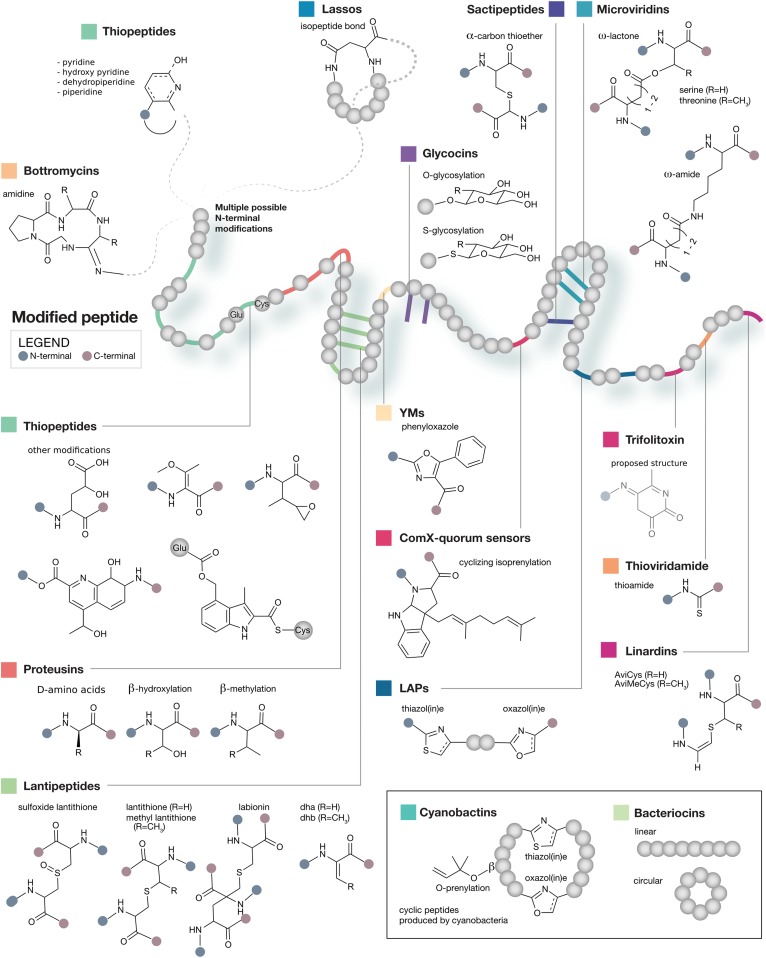
Among microbial natural products, ribosomally synthesized and posttranslationally modified peptides (RiPPs) are of particular interest due to their structural diversity (Fig. 1) and attendant biological activities (9). Biosynthesis of RiPPs initiates with direct translation of a core peptide by the ribosome, continues with decoration by tailoring reactions, and terminates with cleavage and release of the mature product (9). Thousands of putatively unknown RiPPs are encoded within sequenced bacterial genomes (10). However, the process of RiPP discovery remains a low-throughput endeavor. Several prominent obstacles exist to automating the process of genome-guided RiPP discovery. Chief among these is the enormous structural diversity of known RiPPs, which are diversified from simple precursor peptides by a vast range of enzymatic tailoring reactions (Fig. 1). Even after accounting for the structural diversity of known pathways, the problem of distinguishing between genomic loci encoding known and novel natural products with maximum accuracy remains. Further, existing approaches center around a paradigm whereby potential biosynthetic genes are only considered when they are grouped with other known biosynthetic genes; this prevents the identification of novel classes of RiPPs with divergent biosynthetic machinery and impedes the analysis of fragmented or low-quality genome assemblies. Finally, a critical challenge is linking the biosynthetic loci that are most likely to produce novel products to metabolomic data. Although important strides have been made in the semiautomated matching of genomic and metabolomic data (11, 12), existing approaches rely primarily on interpretation of tandem mass spectra, ignoring the broader spectrum of data available from sources such as isotope distributions and comparative metabolomics. An integrated pipeline that translates genomic data directly into the physical detection of novel compounds could accelerate the process of novel RiPP discovery by linking biosynthetic loci to their products.
Fig. 1.
Overview of known RiPP tailoring reactions. Posttranslational tailoring modifications are shown within a hypothetical core peptide backbone.
Here we present DeepRiPP, a modular platform designed to automate the process of novel RiPP discovery, from strain selection to compound isolation. First, we developed a bipartite algorithm adapted from natural language processing to identify precursor peptides independent of genomic context (NLPPrecursor). NLPPrecursor overcomes limitations in genome mining associated with fragmented assemblies or the presence of distantly encoded and unclustered modification enzymes, thereby capturing a wider diversity of RiPPs. The second component of DeepRiPP is Basic Alignment of Ribosomal Encoded Products Locally (BARLEY), which combines retrobiosynthetic processing of known RiPP structures with local alignment to genomic information in order to assign a novelty index to candidate RiPPs identified by genome mining and dereplicate known products. Finally, we developed Computational Library for Analysis of Mass Spectra (CLAMS), an algorithm that integrates disparate sources of mass spectral information, including isotopic distributions, intensity, exact mass, fragmentation patterns, and comparative metabolomics to pinpoint the products of identified biosynthetic loci within a database of thousands of microbial extracts. We apply DeepRiPP to analyze 65,421 sequenced bacterial genomes and identify 19,498 unique unknown RiPPs, expanding the number of RiPP natural products by a factor of 6 from previous estimates. We link a subset of these genes to their potential products in metabolomic data, facilitating the directed isolation of 3 new products in their native hosts. DeepRiPP is publicly available online as a user-friendly, interactive web application at http://deepripp.magarveylab.ca to facilitate rapid analysis of genomic and metabolomic data.
Results
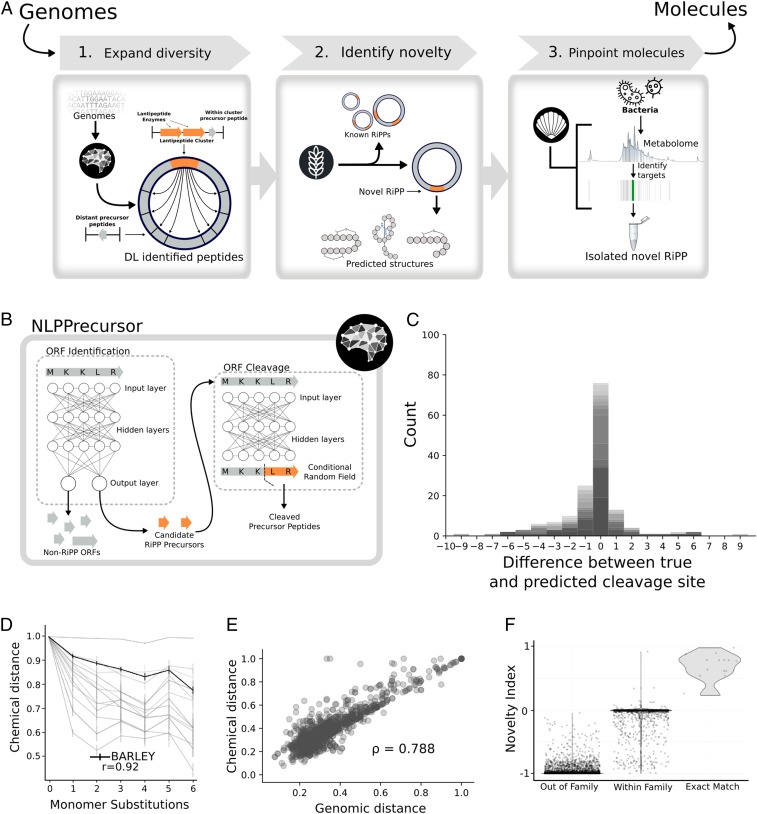
To enable the automated discovery of novel RiPPs from paired genomic and metabolomic data, we envisioned an integrated workflow to expand genomic discovery, prioritize the discovery of novel genes, and pinpoint the target gene products in crude extracts (Fig. 2A). DeepRiPP first uses a deep learning approach inspired by natural language processing, NLPPrecursor, to identify precursor peptides across the entire genome and predict their cleavage patterns. The cleaved precursor peptides identified by NLPPrecursor are then integrated into our RiPP-PRISM (10) system to enable combinatorial prediction of complete chemical structures, including complete enzymatic tailoring reaction cascades. The BARLEY algorithm employs a cheminformatic local alignment framework to match predicted RiPPs identified from genome sequence data to a chemical structure database of all previously characterized RiPPs (Datasets S1 and S2). The CLAMS algorithm applies comparative metabolomic analysis across a database containing thousands of extracts to pinpoint target products in mass spectrometry data. Collectively, these algorithms constitute the DeepRiPP workflow.
Fig. 2.
Illustration of the DeepRiPP gene to molecule workflow and performance of its genomic modules, NLPPrecursor and BARLEY. (A) The DeepRiPP workflow that guides the discovery strategy from genomes to isolated molecules. DeepRiPP consists of 3 modules. Module 1, NLPPrecursor, implements deep learning techniques inspired by natural language processing to expand the diversity of genomically detected RiPPs by including all potential precursor peptides outside putative biosynthetic gene cluster boundaries. Module 2, BARLEY, identifies novel RiPPs by aligning genomic information to a database of known RiPP chemical structures and scoring the novelty of each candidate RiPP identified by genome mining. Module 3, CLAMS, identifies putative RiPPs in metabolomics data. (B) The architecture of NLPPrecursor, highlighting the 2 components responsible for precursor identification and cleavage, respectively. (C) Histogram depicting the prediction accuracy of NLPPrecursor ORF cleavage, where the x axis is the difference between the predicted and true cleavage site in number of amino acids. Gray shading represent different families. (D) Line chart describing the relationship between increasing chemical divergence in an artificially generated, combinatorial dataset (33) of 600 compounds to chemical distance scores. BARLEY is highlighted in black, while other metrics are shown in light gray. The relationship between the number of monomer substitutions and the chemical similarity assigned by each metric is computed using the Spearman rank correlation coefficient (ρ). (E) Scatterplot representing the relationship between BARLEY chemical distances and genomic distances generated by BARLEY. The comparison was performed on a dataset of 136 known RiPP clusters which encode 161 small molecules (Dataset S3). The Spearman rank correlation coefficient (ρ) is used to quantify the relationship between genomic and chemical BARLEY distances. (F) Validation of BARLEY novelty index. A violin plot is shown with BARLEY predicted novelty index (y axis) and the true relationship type (exact match, family match, or out of family) between encoded RiPP and chemical scaffold (x axis). Using a cutoff of 0.2 on the BARLEY novelty index yields a 99.7% accuracy in classifying exact matches from other comparison types.
A Deep Learning Approach to Genome-Wide Discovery of RiPPs.
Computational approaches for identifying natural product gene clusters from genome sequence data rely on the assumption that these pathways are encoded by chromosomally adjacent genes (8). In the context of RiPP discovery, this represents a limiting assumption for at least 3 reasons. First, entirely novel classes of RiPPs may share key sequence features with known precursor peptides but diverge in their tailoring reaction cascades, such that the requirement of complete biosynthetic pathways for cluster detection limits the sensitivity of the algorithm. Second, fragmented or low-quality genome assemblies often fail to resolve complete biosynthetic gene clusters across contigs (13), potentially leading to scenarios where the precursor peptide is distant to the remainder of the biosynthetic machinery. Finally, examples of precursor peptides separated from the rest of the encoded RiPP biosynthetic machinery have been described, most notably for the prochlorosin family of lantipeptides (14). The limitations of homology-directed approaches are evident when considering the fact that among the 30,261 RiPP clusters previously identified by RiPP-PRISM (10), 5,459 did not contain a precursor peptide with homology to a known RiPP (SI Appendix, Fig. S1).
We sought to expand the framework for chemical structure prediction of genetically encoded RiPPs introduced in our previous work (10) by using a family of deep neural network-based models known as language models to systematically identify RiPP precursor peptides genome-wide and predict their likely cleavage patterns. Recent work has demonstrated that deep language models based on recurrent neural networks are not only extremely effective in natural language processing tasks (15–17) but can also be applied in biological contexts, such as regulatory genomics or protein sequence analysis (18–20). However, learning robust language models from limited training data has historically been challenging (21–23). Recent advances in unsupervised or self-supervised pretraining provide a means to train accurate models from even very small labeled training datasets (24–26). Here we designed NLPPrecursor to extend methodologies used for sentiment analysis (25) and named entity recognition (27, 28). NLPPrecursor is a 2-stage deep learning pipeline that first uses protein sequence information to classify ORFs as RiPP precursors and subsequently to predict their cleavage sites (Fig. 2B). We framed this as an annotation problem, where each amino acid within the ORF sequence must be labeled as either part of the final peptide or not. In natural language processing, several models have been developed for a similar task, labeling parts of speech within a sentence (29).The algorithm was assessed by cross-validation using a training set compiled from all RiPPs identified by RiPP-PRISM (10). The RiPP classification algorithm had a positive predictive value of 98% in discriminating true RiPP precursors from nonprecursor ORFs and a prediction accuracy of 95% in classifying RiPP precursors into their biosynthetic subfamilies (Dataset S3). Of note, this model is not biased according to ORF size, suggesting that the accuracies represented here are conservative as larger ORFs were not taken into consideration (an analysis into the confounding effects of ORF size is provided in SI Appendix, SI Results and Fig. S2). The precursor cleavage algorithm predicted N-terminal cleavage sites with 90% accuracy, when considering cleavage points ±5 amino acids from the true prediction site, a range within which all possible complete chemical structures can be elaborated in silico by combinatorial structure prediction (Fig. 2C and SI Appendix, Fig. S3 and Dataset S4). Of note, these results were obtained in a completely automated manner and validated on a dataset entirely independent of the training set, suggesting NLPPrecursor achieves excellent performance in genome-wide RiPP precursor identification and analysis. When comparing NLPPrecursor to the manually curated sequence motifs in RiPP-PRISM, we found both RiPP-PRISM and NLPPrecursor predict N-terminal cleavage sites with a median error of 0, but RiPP-PRISM predicts cleavage sites on our dataset of characterized clusters with a lower SD (1.34 vs. 3.16; SI Appendix, Fig. S4). However, NLPPrecursor generates predicted cleavage patterns for a much broader range of precursor peptides than originally designed into RiPP-PRISM, especially within all classes of lantipeptides (SI Appendix, Fig. S1), helping to extend structure prediction to the ∼18% of clusters for which the original RiPP-PRISM algorithm was unable to generate a predicted structure.
To further characterize the performance of NLPPrecursor, we additionally compared it to 2 methods designed for specific subsets of RiPPs, including RODEO (30) and RiPPMiner (31) (SI Appendix, SI Results and Figs. S5 and S6). Although NLPPrecursor is capable of processing a larger number of RiPP families than these tools, to ensure a fair comparison we limited our analysis to families predicted by both methods in each comparison. We observed that NLPPrecursor very slightly underperforms RODEO (∆AUC [area under receiver operating characteristic curve] = 0.012, P = 0.027, DeLong test; SI Appendix, Fig. S5), despite making use exclusively of information encoded within the precursor peptide sequence and not the broader genomic context, as utilized by RODEO. In contrast, we demonstrate that NLPPrecursor is substantially more accurate than RiPPMiner across a broad range of RiPP families (SI Appendix, Fig. S6).
Cheminformatic Local Alignment Prioritizes Novel Genomically Encoded RiPPs.
In order to prioritize novel RiPPs for discovery from genomic information, we next envisioned an automated method to compare biosynthetic loci to the structures of known RiPPs that would incorporate information beyond the sequence of the precursor ORF, including cleavage sites and tailoring reactions. We therefore developed BARLEY, a local alignment algorithm which extends our previous work on the global alignment of gene clusters for nonribosomal peptides and polyketides to the structures of their products, GRAPE (generalized retrobiosynthetic assembly prediction engine) and GARLIC (global alignment for natural products chemoinformatics) (32), by implementing in silico retrobiosynthesis of known RiPP tailoring reactions (Fig. 1), followed by local alignment of the precursor peptide and the identifies of the inferred tailoring reactions. BARLEY is capable of comparing chemical structures to chemical structures, genes to genes, and chemical structures to genes. This last mode is used in the DeepRiPP workflow to dereplicate putative RiPPs identified by NLPPrecursor and RiPP-PRISM with reference to a database of all known RiPP structures.
For any of the 3 types of comparisons enumerated above, BARLEY generates a relative similarity score scaled between 0 and 1, quantifying the strength of the local alignment and similarity of inferred tailoring reactions. To validate DeepRiPP in an unbiased manner, we extended our LEMONS (Library for the Enumeration of Modular Natural Structures) algorithm (33) to systematically generate modified versions of RiPP scaffolds. We compared the BARLEY chemical similarity score to 13 widely used chemical similarity metrics, using a library of 600 hypothetical RiPPs sampled based on the structure of nisin, substituting between 1 and 6 monomers from the nisin precursor peptide. Within this library of hypothetical RiPPs, the BARLEY similarity score is most strongly correlated to the number of monomer substitutions (Spearman’s ρ = −0.92), significantly more so than the next most accurate method (topological torsion fingerprints, ρ = −0.78; P < 0.01, Fisher z transformation; Fig. 2D and SI Appendix, Fig. S7A). Next, we assembled a dataset of 638 structurally characterized RiPPs and compared the ability of BARLEY and the topological torsion fingerprint to discriminate between RiPPs of different classes. BARLEY was significantly more accurate in this task (P < 0.01, DeLong test; SI Appendix, Fig. S7B), with an increase of 18% in accuracy at a fixed false-positive rate of 10%. Finally, having validated the accuracy of BARLEY, we applied its chemical similarity score to perform hierarchical clustering of all known RiPPs (SI Appendix, Fig. S8), finding that biosynthetic classes with clear defining tailoring reactions such as lantipeptides, thiopeptides, bacteriocins, and linear azole-containing peptides are well clustered, whereas classes such as cyanobactins with diverse and inconsistent tailoring reactions were not.
Because BARLEY can also assign similarity scores to pairs of clusters, we further compared BARLEY to 2 more tools designed to score the similarity of 2 RiPP clusters: BiG-SCAPE (Biosynthetic Gene Similarity Clustering And Prospecting Engine) (34) and the Tanimoto coefficient between RiPP-PRISM predicted structures (10). We compared genomic similarity scores to the Tanimoto coefficient between pairs of true products, as assessed using topological torsion fingerprints (35). To more precisely capture the ability of each method to discriminate between closely related RiPPs, we limited comparisons to pairs of RiPPs from the same class and found BARLEY is significantly more correlated with chemical similarity (Spearman’s ρ = 0.79) than either BiG-SCAPE (ρ = 0.02; P < 10−15; Methods) or RiPP-PRISM Tanimoto coefficients (ρ = 0.31; P < 10−15) (Fig. 2E and SI Appendix, Fig. S9 B and C). Both RiPP-PRISM and BARLEY’s stronger correlation to chemical scores suggest that using information from the specific precursor peptide is essential in modeling RiPP genomic diversity, which is not captured by BiG-SCAPE. These results provide additional evidence that BARLEY similarity scores accurately reflect both chemical and genomic similarity with significantly higher resolution than existing tools.
Finally, having extensively validated BARLEY similarity scores for pairs of compounds and pairs of clusters, we sought to evaluate its ability to determine the novelty of genetically encoded RiPPs by comparison to a library of known RiPP structures (Fig. 2F and SI Appendix, Fig. S10A). To this end, we designed a machine-learning framework to classify pairwise relationships between genetically encoded RiPPs and known RiPPs into 1 of 3 categories: unknown, within-family, or exact match. We compared the performance of a random forest classifier given BARLEY scores as input to the structure prediction engine within RiPP-PRISM, finding that BARLEY distinguished family-wise chemical relationships between RiPPs significantly more accurately than direct comparison of RiPP-PRISM predicted structures (AUC 0.96 vs. 0.89; P < 0.01, DeLong test) (SI Appendix, Fig. S10B). In predicting novel RiPPs, BARLEY demonstrates a 99.5% accuracy at a fixed false positive rate of 1% (AUC 0.997; Fig. 2F and SI Appendix, Fig. S10). For this task of scoring clusters according to novelty, BARLEY was trained and validated on 2 independent datasets (Dataset S1), suggesting its high accuracy is likely to extend to genome-wide analyses.
Large-Scale Analysis of Bacterial Genomes Reveals Unappreciated Diversity of Novel RiPPs.
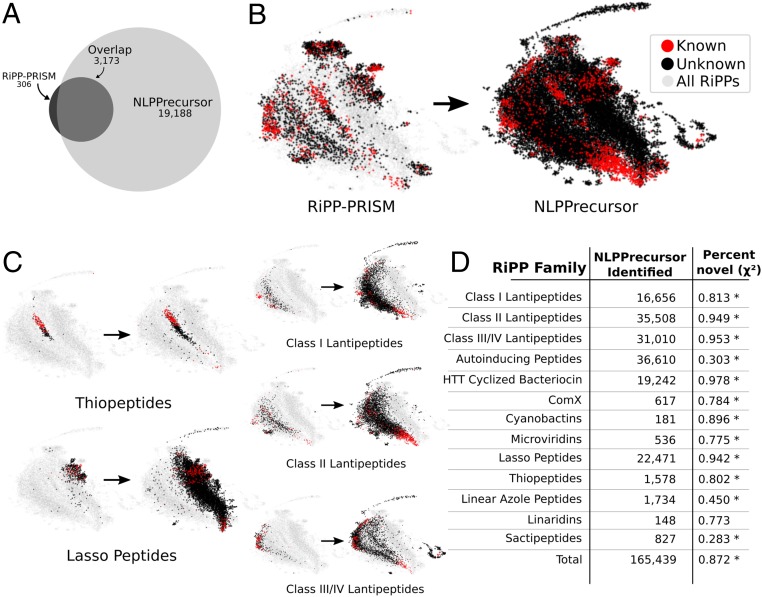
To obtain a global view of the ability of NLPPrecursor and BARLEY to capture RiPP diversity, we conducted genome-wide searches for RiPPs across a set of 65,421 prokaryotic genomes, of which 19,113 genomes encoded at least 1 RiPP cluster as revealed by RiPP-PRISM (10). In total, 165,439 RiPPs were detected, of which 25,840 represent unique precursor peptide sequences, suggesting many RiPPs are observed in duplicate or more within this dataset. NLPPrecursor identified more than 6 times as many unique RiPP precursor peptides as RiPP-PRISM (22,361 vs. 3,479; Fig. 3A), while still capturing 91.2% of RiPPs detected by RiPP-PRISM alone. To assess the overall diversity and novelty within this dataset, we used BARLEY to generate a pairwise distance matrix across all detected RiPPs and further align each detected RiPP to our library of characterized RiPP chemical scaffolds (Dataset S2). In total, 87.2% of RiPPs detected by NLPPrecursor were denoted as novel, a significant upward shift from that observed with RiPP-PRISM (45.9%, P < 10−15, χ2 test). We then used the nonlinear dimensionality reduction tool Uniform Manifold Approximation and Projection (UMAP) (36) to visualize the global distribution of RiPP chemical diversity (Fig. 3B) revealed by both RiPP-PRISM and NLPPrecursor. The resulting visualization highlights the expanded chemical diversity of RiPPs revealed by NLPPrecursor from genomic data. NLPPrecursor identifies significantly more diverse and unknown thiopeptides, lasso peptides, and lantipeptides than RiPP-PRISM, with similar trends observed for almost all RiPP families (Fig. 3C and SI Appendix, Fig. S11).
Fig. 3.
DeepRiPP expands the discovery of novel RiPPs in a reanalysis of genomes analyzed by RiPP-PRISM. (A) Venn diagram depicts the total number of unique RiPP precursor peptides identified via NLPPrecursor as compared to RiPP-PRISM. A total of 65,421 bacterial genomes were analyzed from National Center for Biotechnology Information (downloaded March 2016) through DeepRiPP. (B) Total diversity of genomically encoded RiPPs as identified by RiPP-PRISM and NLPPrecursor distributed according to BARLEY similarity and subsequently plotted using UMAP (36). Each point represents a unique RiPP as determined via BARLEY and is colored according to its novelty as determined by BARLEY. All RiPPs identified by either RiPP-PRISM or NLPPrecursor are shaded in gray within the background to visualize overall localization. (C) Depicting the family-wise increase in diversity using UMAP. (D) Number of RiPPs identified by NLPPrecursor and the percentage of these that are novel. The χ2 test is used to determine whether a significant increase in the percentage novel are observed within the NLPPrecursor set (*P < 10−10).
A critical challenge in the discovery of novel RiPPs is identifying genera or species with a high probability of producing novel compounds. BARLEY facilitates the prioritization of specific microbial taxa for the targeted discovery of divergent RiPPs. After normalizing for the number of sequenced genomes within each genus, the top 3 genera most enriched in unique novel RiPPs are Nocardiopsis, Kitasatospora, and Actinomadura, with 8.78, 6.22, and 6.09 novel unique RiPPs per genome, respectively (Dataset S5). Conversely, certain RiPPs are duplicated among a wide diversity of taxa: for example, we detected subtilosin A across 121 organisms covering 4 genera (Salinibacillus, Bacillus, Streptococcus, and Jeotgalibacillus). Taken together, these analyses highlight the utility of DeepRiPP for prioritizing microbial genera based on their capacity for production of novel RiPPs.
Integrative Analysis of Genomic and Metabolomic Data Automates Discovery of 3 Novel RiPPs.
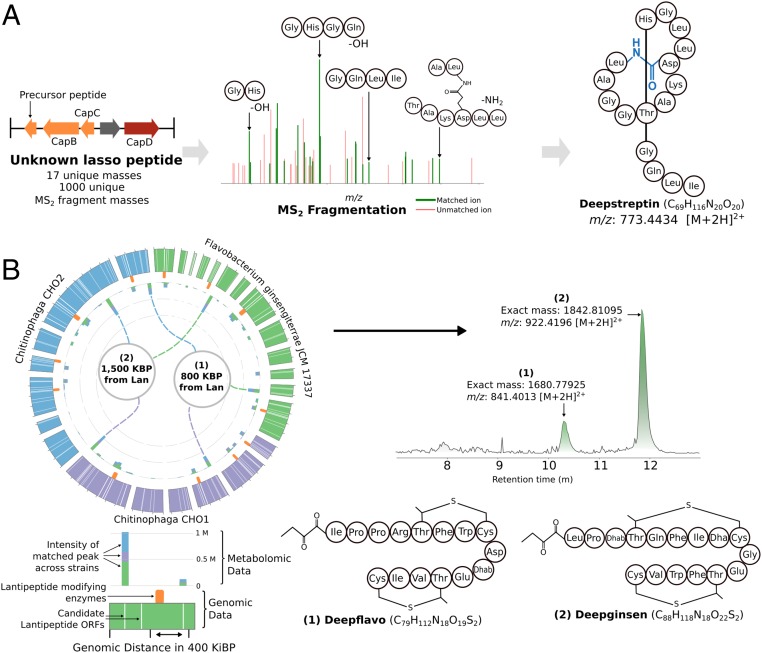
As the final module of the DeepRiPP pipeline, we envisioned a computational platform to pinpoint the products of genomically identified RiPPs within metabolomic data of crude bacterial extracts. We therefore developed CLAMS, an algorithm for mass spectral analysis that takes into account the full complement of available metabolomic information, including not only fragmentation patterns reflected in tandem mass spectra (37, 38) but also the shape and intensities of isotopic distributions of ions and cross-species comparative metabolomics data. Using CLAMS, we developed a subtractive strategy to decrease the number of candidate ions linked to a target cluster and reduce the amount of noise present in mass spectrometric datasets (SI Appendix, Fig. S12). We first compiled a large-scale metabolomic database, consisting of 10,498 extracts generated from 463 strains, each with a standard panel of media and growth conditions (described further in SI Appendix, SI Methods, Microbial Strains and Culturing). In parallel, we conducted 118 diverse blank media extractions in order to readily eliminate metabolite signatures matching known media constituents. These resources allowed us to identify peaks that were unique to strains containing a given cluster, as determined by BARLEY, and which do not share an exact mass with either, or a database of 50,317 known natural products (39) (SI Appendix, Fig. S12B). We then leveraged the increased resolution afforded by the genomic modules of the DeepRiPP workflow to automatically link individual peaks to RiPP clusters. In particular, we considered all precursor cleavage sites within ±5 amino acids of the NLPPrecursor predictions, in order to account for a certain degree of error in prediction. In combination, these strategies can generate thousands of predicted structures for a given RiPP. We therefore required that a given match between an encoded RiPP and a candidate peak be supported both by 1) a matching exact mass and 2) the presence of supporting fragmentation patterns in the tandem mass spectrum. The combination of these 5 distinct filtering steps (Fig. 4A) enables the automated matching of genomically encoded RiPPs to candidate peaks, such that a single peak can be selected among the 2,066 (SD 466.9) peaks observed per microbial extract.
Fig. 4.
DeepRiPP enables a gene to molecule strategy and leads to discovery of 3 new RiPP products. (A) A gene cluster encoding a lasso peptide in Streptomyces sp. BTA-0171 was inferred by BARLEY to represent a novel chemical scaffold. Using the comparative metabolomic reductionist workflow (SI Appendix, S12B), CLAMS correlated a single ion to this specific cluster. The MS2 fragmentation of this ion is shown where 40% of detected fragmentation ions were predicted in silico. Downstream isolation led to the discovery of a lasso peptide with the exact structure as predicted, named deepstreptin. (B) Discovery of 2 RiPP products with precursor ORFs distant from tailoring enzymes. A subset of 3 closely related strains, F. ginsengiterrae JCM 17337, Chitinophaga sp. CHO1, and Chitinophaga sp. CHO2, were found to match a high number of metabolites originating from ORFs outside of traditional boundaries and were further investigated. Notably, these precursor ORFs were only identified by NLPPrecursor. Shown are the genomes of this subset of 3 strains, the relative genomic positions of RiPP precursor ORFs, and the intensity of the matched ions. Here the outer circle represents the genomic coordinates within these 3 strains (colored according to strain). In orange are the highlighted locations of lantipeptide modifying enzymes. On the inner circular axis, bars represent the intensity of matched peaks from metabolomic data, while the colors represent the different strains in which they were identified. The 2 most abundant peaks are further highlighted in the center with their genomic distance from lantipeptide modifying enzymes. These corresponding ions for 1 and 2 are shown as an EIC within a crude extract from F. ginsengiterrae JCM 17337. The full structures for 1 and 2 were elucidated, and we named them deepflavo and deepginsen, respectively.
We validated the metabolomic component of the DeepRiPP workflow by pursuing a unique Streptomyces sp. BTA0171 lasso peptide cluster identified by BARLEY (SI Appendix, Fig. S13A), identified both by RiPP-PRISM and DeepRiPP. Local alignment to known natural products (Dataset S2) and genomically encoded RiPPs (Dataset S1) suggested the product likely to be both novel and unique, with no closely related clusters (Fig. 4A). Filtering media components and peaks that were not unique to the strain under investigation yielded a set of 1,235 candidate peaks. Of these, 5 had at least 1 MS2 fragment matching the cluster of interest, 1 of which had a similarity score of 40% based on MS2 and an exact match to the monoisotopic mass of a RiPP-PRISM predicted structure. The target ion, 773.4434 [M + 2H]2+, was therefore selected for downstream purification and structural elucidation. The structure of this compound, which we named deepstreptin, was revealed to be exactly as predicted by DeepRiPP (Fig. 4A and SI Appendix, Figs. S33–S39).
The DeepRiPP workflow enables the identification of RiPPs in a genome-wide manner, independent of their chromosomal adjacency to conventional modification enzymes. As noted above, this process led to the identification of over 6 times as many candidate RiPPs as in our previous analysis, with 49% of putative novel RiPPs detected by NLPPrecursor being lantipeptides. We sought to validate the unbiased precursor detection and cleavage module within DeepRiPP by selecting 20 bacterial strains that contained 263 unique and novel lantipeptide precursors, as predicted by DeepRiPP, for further study. We processed 200 extracts of these strains through liquid chromatography-tandem mass spectrometry (LC-MS/MS), followed by downstream DeepRiPP analysis to reveal specific metabolites corresponding to targeted RiPPs (Fig. 4B). Among the target ions matched to genomically encoded RiPPs, we selected 2 peaks to pursue further due to their independent appearance and match in 3 strains of interest, Flavobacterium ginsengiterrae JCM 17337, Chitinophaga sp. CHO1, and Chitinophaga sp. CHO2 (Fig. 4B). These precursors were of particular interest because their genomic coordinates suggested a distance of at least 1.5 Mbp and 0.8 Mbp away from the nearest lantipeptide modification enzymes (Fig. 4B) and indeed were only detectable by DeepRiPP (SI Appendix, Fig. S13 B and C), whereas RiPP-PRISM alone failed to predict any of them. The ions matching structure predictions, mass-to-charge ratio (m/z) 841.4013 [M + 2H]2+ and 922.4196 [M + 2H]2+, are shared between all 3 strains (Fig. 4B), with the highest abundance in F. ginsengiterrae JCM 17337. We purified the 2 targets, naming them deepflavo and deepginsen, and confirmed their structures to be exactly as predicted by DeepRiPP (SI Appendix, Figs. S41–S55). Taken together, these 3 identified RiPPs validate the DeepRiPP workflow for the automated discovery of novel RiPPs.
Discussion
The promise of genome-guided approaches to unlock unknown chemistry encoded within microbial genomes is considerable but, to date, has been incompletely realized. At present, a critical gap in this regard is the development of fully automated approaches to establish links between biosynthetic loci and their products. Our DeepRiPP workflow combines multiple machine learning technologies to expand the landscape of unknown RiPPs and delineate the molecules corresponding to targeted genomic loci among millions of metabolites and is available online as a comprehensive, user-friendly platform for easy access and use by the broader scientific community.
Deep Learning Captures Encoded RiPPs Beyond Cluster Boundaries.
The dominant paradigm in genomic approaches to natural product discovery consists of identifying clusters of chromosomally adjacent biosynthetic genes (8). Although this paradigm has led to advances in the study of microbial biosynthesis to date, it poses obstacles to RiPP discovery from fragmented genome assemblies or in cases where the precursor is distant to the remainder of the biosynthetic machinery. An example of the latter case is provided by the prochlorosins, in which some precursor peptides are nearly 1 Mbp distant from the ProcM tailoring enzyme (14). To identify precursor peptides in an unbiased manner regardless of their genomic context, we developed a sequence-based deep learning framework, NLPPrecursor, with a high degree of accuracy to enable precise identification of RiPPs outside the boundaries of conventional biosynthetic gene clusters. Further, we show that NLPPrecursor readily identifies precursor peptides in biosynthetic gene clusters (BGCs) where conventional homology-based approaches such as hidden Markov models fail to generate predictions. Finally, we show that our deep learning framework enables accurate prediction of cleavage sites directly from protein sequence. Whereas deep learning approaches are often thought to require extremely large training datasets (40), we achieve excellent performance using a modest dataset composed of ∼3,000 RiPPs by applying a deep learning strategy developed for natural language processing (25). The success of this strategy in the context of RiPP discovery suggests that deep learning may be more broadly applicable to natural product discovery from genome sequence information. Furthermore, the expanded landscape of unknown RiPPs revealed by our large-scale genomic analysis suggests that many novel RiPPs may be invisible to existing computational strategies.
DeepRiPP Enables Strain Selection for RiPP Discovery.
RiPP biosynthesis is widespread across bacterial genomes, but many biosynthetic pathways produce known compounds (10). Prioritizing organisms most likely to produce novel RiPPs is therefore a central challenge in RiPP discovery. To effectively prioritize strains and candidate products for discovery, we developed BARLEY, a cheminformatic local alignment algorithm that can align genomically identified RiPPs to other genomic loci, RiPP chemical structures to other chemical structures, or genomic loci to chemical structures. Importantly, this functionality makes BARLEY the only available tool capable of directly inferring the novelty of encoded RiPPs by comparison to a library of characterized natural product scaffolds. Because substantially more RiPPs with characterized chemical structures are known (638; Dataset S2) than have fully sequenced and experimentally confirmed biosynthetic gene clusters (136; Dataset S1), this allows BARLEY to consider a much larger resource of known RiPPs when assigning the likelihood that a given locus produces a novel product than other approaches, such as BiG-SCAPE (34) or RiPP-PRISM (10). Furthermore, we show that BARLEY is more accurate than previously described methods, even in comparisons involving products with known gene clusters. In combination, these properties enable BARLEY to target divergent and uncharacterized genomically encoded RiPPs for downstream isolation (SI Appendix, Fig. S13).
Uncharacterized natural products can be divided conceptually into 2 classes: those with novel chemical modifications and those with rearrangements of existing chemical modifications. Like most existing approaches to genome-guided natural product discovery, DeepRiPP is limited in its ability to predict chemical reactions catalyzed by unknown enzymes and leading to entirely novel chemical scaffolds. Instead, DeepRiPP is designed primarily in consideration of the many uncharacterized products with rearrangements of known chemical modifications. Whereas recent works have described how combining bioinformatic methods with novel experimental approaches (41, 42) can prioritize novel chemical modifications, the primary goal of DeepRiPP is not to discover new biosynthetic routes but instead to leverage a large corpus of knowledge on RiPP biosynthesis to automate the pursuit of novel chemical scaffolds using untargeted metabolomics with minimal sample preparation. However, even when uncharacterized products are modified by novel enzymatic reactions, DeepRiPP can provide substantial value by the genome-wide identification of RiPP precursors (NLPPrecursor), assessment of their novelty based on the precursor peptide and known tailoring reactions (BARLEY), and their identification via comparative metabolomics and in silico fragmentation of partially correct structures (CLAMS).
DeepRiPP Integrates Genomic and Metabolomic Data to Automate Isolation of Novel RiPPs.
Despite the maturation of approaches to identify biosynthetic loci encoded within microbial genomes at a large scale (8, 10, 30, 38, 42–52), attempts to link genomic information to metabolomic datasets remain limited. Statistical approaches have been developed to link tandem mass spectra to biosynthetic clusters in a semiautomated manner (37, 38, 53, 54), but most strategies for connecting clusters to their products are driven primarily by manual annotation (12, 55). In developing CLAMS, we sought to exploit a broader range of metabolomic information than has been considered to date by existing approaches, which rely primarily on fragmentation patterns to establish cluster–compound links (11, 53, 56–59). In contrast, CLAMS leverages large-scale metabolomic resources of empty media extractions, as well as crude extracts of hundreds of bacterial species, in order to selectively identify candidate peaks unique to strains producing a RiPP of interest. CLAMS further combines both exact mass information and in silico fragmentation to pinpoint compounds from complete combinatorial libraries containing thousands of predicted structures within metabolomic datasets. We show that expanding the sources of metabolomic information considered in multiomics analysis of natural product biosynthesis beyond tandem mass spectra permits a truly automated system for target molecule isolation, without manual intervention at any stage. As demonstrated herein, the specificity of CLAMS allows for detection of gene products in their native host, bypassing the need for heterologous gene expression (60–62). By doing so, we overcome the challenges associated with this technology, including limited availability of optimized hosts (63–65), differential codon bias (66), absence of regulatory elements (66), metabolic balance (67), and toxicity (68).
By combining 3 distinct modules—NLPPrecursor, BARLEY, and CLAMS—into a single platform, DeepRiPP represents an integrated tool for RiPP discovery. We believe automated tools such as this will be critical to advance genome-guided natural product discovery and shed light on the vast unknown universe of microbial chemistry.
Methods
Data Availability Statement.
All public genomic and chemical data used in this study are available through SI Appendix, List of Supplementary Datasets. Source code for the software presented in this manuscript can be found at https://github.com/magarveylab under the repositories NLPPrecursor and clams-release. A full protocol for all methodologies presented here is available within SI Appendix, SI Methods.
Genomic and Chemical Datasets.
In order to validate the genomic distance analysis, and to train BARLEY’s novelty index (identifying a genomically encoded RiPP as novel or previously characterized), we curated a total of 138 RiPP biosynthetic gene clusters, stored in FASTA format, and mapped to 161 chemical structures, stored in SMILES (Simplified Molecular Input Line Entry System) format (Dataset S3). To validate BARLEY’s chemical distances, we used 638 chemical structures of known RiPPs with family-level annotation but without necessarily having matching clusters, stored in SMILES format (Dataset S1).
Development of NLPPrecursor.
NLPPrecursor is composed of 2 distinct deep learning models and methodologies for the identification of precursor peptides and predicting their cleavage respectively. A full methodology is provided in SI Appendix, SI Methods. All training data are publicly available online (https://github.com/magarveylab/NLPPrecursor/tree/master/training_data/), with a step-by-step tutorial for reproducing the results presented here (https://github.com/magarveylab/nlpprecursor/). Pretrained models are available for inference through our web application (http://deepripp.magarveylab.ca/) and as raw files (https://github.com/magarveylab/nlpprecursor/releases).
Construction of BARLEY.
BARLEY is a RiPP comparison tool that can function in 3 modes (chemical–chemical, genome–genome, and genome–chemical). For processing chemical structures from SMILES format, we have updated our retrobiosynthetic algorithm, GRAPE (32), with additional biosynthetic tailoring reactions. A full description of GRAPE and BARLEY’s 3 functionalities are provided in SI Appendix, SI Methods.
Genomic Analysis of RiPP Scaffolds.
From a total of 65,421 genomes run through RiPP-PRISM, we identified 24,756 RiPP-BGCs (Dataset S5) and their resulting cleaved precursor peptides. In parallel, prodigal was used to find all ORFs between 20 and 200 AAs long within these genomes and were subsequently processed through NLPPrecursor to identify and cleave precursor peptides. These cleaved precursor peptides and associated tailoring enzymes were parsed through BARLEY, and all pairwise scores were stored in an n × n distance matrix where n represents the total number of identified and cleaved precursor peptides identified by RiPP-PRISM. Since BARLEY scores are directionally dependent, the maximum score of each side is considered for subsequent analysis. Each encoded product was also compared to library of 638 RiPP chemical structures using BARLEY to determine structural novelty.
Metabolomic Mass Spectral Analysis.
Mass spectrometry data were analyzed using CLAMS (source code available at https://github.com/magarveylab/clams-release) to format MS1 ions as individual entities, mapping to each their relative isotopic distribution, monoisotopic m/z, retention time, charge, and intensity. Precise values were obtained for each MS1 ion at their maximal intensity. Where observed, MS2 spectra containing relative intensity and m/z of each ion were associated with each MS1 ion. Each MS1 ion is then compared across our metabolomic database and matched according to RiPP structure predictions and in silico fragmentation described in SI Appendix, SI Methods, Metabolomic Mass Spectral Analysis and RiPP Structure Prediction and Peak Matching.
DeepRiPP Web Application.
The DeepRiPP web application integrates NLPPrecursor, BARLEY, and CLAMS into a single interactive platform. Using this design, a registration and login system provide users the ability to manage long-running jobs and revisit analyses completed in the past. Support for the entire DeepRiPP web application is provided at https://github.com/magarveylab/NLPPrecursor/issues. Screenshots and a description of its implementation can be found in SI Appendix, SI Methods and Figs. S19–S32).
General Experimental Procedures.
A full summary of microbial strains used in this study; their growth and metabolite extraction methods (Dataset S7); LC-MS procedures; and the structure elucidation of deepstreptin, deepflavo, and deepginsen can be found in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
W.K.M. is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) postdoctoral scholarship. N.A.M. is supported by Canada Research Chair. We thank Jeff Janso (Pfizer Inc.) for providing the bacterial strain Streptomyces sp. BTA-0171 used in this study.
Footnotes
Competing interest statement: N.A.M. is a founder of Adapsyn Bioscience. M.A.S. is a consultant to Adapsyn Bioscience. Haoxin Li and C.A.D. are employed by Adapsyn Bioscience.
This article is a PNAS Direct Submission. Hongzhe Li is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901493116/-/DCSupplemental.
References
- 1.Newman D. J., Cragg G. M., Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Li J. W.-H., Vederas J. C., Drug discovery and natural products: End of an era or an endless frontier? Science 325, 161–165 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Wright G. D., Antibiotics: A new hope. Chem. Biol. 19, 3–10 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Koehn F. E., Carter G. T., The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 4, 206–220 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Doroghazi J. R., et al. , A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimermancic P., et al. , Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crits-Christoph A., Diamond S., Butterfield C. N., Thomas B. C., Banfield J. F., Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Medema M. H., Fischbach M. A., Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnison P. G., et al. , Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skinnider M. A., et al. , Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc. Natl. Acad. Sci. U.S.A. 113, E6343–E6351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohimani H., et al. , Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 9, 1545–1551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medema M. H., et al. , Pep2Path: Automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput. Biol. 10, e1003822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein S., Beka L., Graf J., Klassen J. L., Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genomics 20, 23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B., et al. , Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 10430–10435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves A., Generating sequences with recurrent neural networks. arxiv:1308.0850 (4 August 2013).
- 16.Sutskever I., Vinyals O., Le Q. V., Sequence to sequence learning with neural networks. arxiv:1409.3215 (10 September 2014).
- 17.Bahdanau D., Cho K., Bengio Y., Neural machine translation by jointly learning to align and translate. arxiv:1409.0473 (1 September 2014).
- 18.Sønderby S. K., Winther O., Protein secondary structure prediction with long short term memory networks. arxiv:1412.7828 (25 December 2014).
- 19.Alipanahi B., Delong A., Weirauch M. T., Frey B. J., Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Angermueller C., Pärnamaa T., Parts L., Stegle O., Deep learning for computational biology. Mol. Syst. Biol. 12, 878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran P., Liu P. J., Le Q. V., Unsupervised pretraining for sequence to sequence learning. arxiv:1611.02683 (8 November 2016).
- 22.Dai A. M., Le Q. V., Semi-supervised sequence learning. arxiv:1511.01432 (4 November 2015).
- 23.Mou L., et al. , How transferable are neural networks in NLP applications? arxiv:1603.06111 (19 March 2016).
- 24.Devlin J., Chang M.-W., Lee K., Toutanova K., “BERT: Pre-training of deep bidirectional transformers for language understanding” in Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (Long and Short Papers), Burstein J., Doran C., Solorio T., Eds. (Association for Computational Linguistics, 2019), pp. 4171–4186, vol. 1. [Google Scholar]
- 25.Howard J., Ruder S., “Universal language model fine-tuning for text classification” in Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Long Papers), Gurevych I., Miyao Y., Eds. (Association for Computational Linguistics, Stroudsburg, PA, 2018), pp. 328–339, vol. 1. [Google Scholar]
- 26.Peters M., et al. , “Deep contextualized word representations” in Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (Long Papers), Walker M., Ji H., Stent A., Eds. (Association for Computational Linguistics, 2018), pp. 2227–2237, vol. 1. [Google Scholar]
- 27.Gardner M., et al. , “AllenNLP: A deep semantic natural language processing platform” in Proceedings of Workshop for NLP Open Source Software (NLP-OSS), Park E. L., Hagiwara M., Milajevs D., Tan L., Eds. (Association for Computational Linguistics, Stroudsburg, PA, 2018), pp. 1–6. [Google Scholar]
- 28.McCallum A., Li W., “Early results for named entity recognition with conditional random fields, feature induction and web-enhanced lexicons” in Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL 2003, Daelemans W., Osborne M., Eds. (Association for Computational Linguistics, Stroudsburg, PA, 2003), pp. 188–191, vol. 4. [Google Scholar]
- 29.Wang X., et al. , Institute of Automation, Chinese Academy of Sciences ; Labeling sequential data based on word representations and conditional random fields. Int. J. Mach. Learn. Comput. 5, 439–444 (2015). [Google Scholar]
- 30.Tietz J. I., et al. , A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 13, 470–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal P., Khater S., Gupta M., Sain N., Mohanty D., RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 45, W80–W88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dejong C. A., et al. , Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol. 12, 1007–1014 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Skinnider M. A., Dejong C. A., Franczak B. C., McNicholas P. D., Magarvey N. A., Comparative analysis of chemical similarity methods for modular natural products with a hypothetical structure enumeration algorithm. J. Cheminform. 9, 46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro-Muñoz J., et al. , A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. bioRxiv:10.1101/445270 (17 October 2018).
- 35.Nilakantan R., Bauman N., Dixon J. S., Venkataraghavan R., Topological torsion: A new molecular descriptor for SAR applications. Comparison with other descriptors. J. Chem. Inf. Comput. Sci. 27, 82–85 (1987). [Google Scholar]
- 36.McInnes L., Healy J., Melville J., UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arxiv:1802.03426 (9 February 2018).
- 37.Zhang Q., et al. , Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS. Proc. Natl. Acad. Sci. U.S.A. 111, 12031–12036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston C. W., et al. , An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat. Commun. 6, 8421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston C. W., et al. , Assembly and clustering of natural antibiotics guides target identification. Nat. Chem. Biol. 12, 233–239 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Zou J., et al. , A primer on deep learning in genomics. Nat. Genet. 51, 12–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox C. L., et al. , Nucleophilic 1,4-additions for natural product discovery. ACS Chem. Biol. 9, 2014–2022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwalen C. J., Hudson G. A., Kille B., Mitchell D. A., Bioinformatic expansion and discovery of thiopeptide antibiotics. J. Am. Chem. Soc. 140, 9494–9501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skinnider M. A., et al. , Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 43, 9645–9662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skinnider M. A., Merwin N. J., Johnston C. W., Magarvey N. A., PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 45, W49–W54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blin K., et al. , The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 47, D625–D630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blin K., et al. , antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansari M. Z., Yadav G., Gokhale R. S., Mohanty D., NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32, W405–W413 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J., Yi G.-S., PKMiner: A database for exploring type II polyketide synthases. BMC Microbiol. 12, 169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziemert N., et al. , The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7, e34064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway K. R., Boddy C. N., ClusterMine360: A database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 41, D402–D407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichikawa N., et al. , DoBISCUIT: A database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 41, D408–D414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M. H. T., Ung P. M. U., Zajkowski J., Garneau-Tsodikova S., Sherman D. H., Automated genome mining for natural products. BMC Bioinformatics 10, 185 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L., et al. , Exploration of nonribosomal peptide families with an automated informatic search algorithm. Chem. Biol. 22, 1259–1269 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Kersten R. D., et al. , A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 7, 794–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen D. D., et al. , MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. U.S.A. 110, E2611–E2620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ibrahim A., et al. , Dereplicating nonribosomal peptides using an informatic search algorithm for natural products (iSNAP) discovery. Proc. Natl. Acad. Sci. U.S.A. 109, 19196–19201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skinnider M. A., Johnston C. W., Zvanych R., Magarvey N. A., Automated identification of depsipeptide natural products by an informatic search algorithm. ChemBioChem 16, 223–227 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Mohimani H., et al. , NRPquest: Coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 77, 1902–1909 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohimani H., et al. , Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 13, 30–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo Y., Enghiad B., Zhao H., New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat. Prod. Rep. 33, 174–182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wenzel S. C., Müller R., Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr. Opin. Biotechnol. 16, 594–606 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Frattaruolo L., Lacret R., Cappello A. R., Truman A. W., A genomics-based approach identifies a thioviridamide-like compound with selective anticancer activity. ACS Chem. Biol. 12, 2815–2822 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Zhou M., et al. , Sequential deletion of all the polyketide synthase and nonribosomal peptide synthetase biosynthetic gene clusters and a 900-kb subtelomeric sequence of the linear chromosome of Streptomyces coelicolor. FEMS Microbiol. Lett. 333, 169–179 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Komatsu M., Uchiyama T., Omura S., Cane D. E., Ikeda H., Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y., Lin Y., Li L., Linhardt R. J., Yan Y., Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab. Eng. 29, 217–226 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Gustafsson C., Govindarajan S., Minshull J., Codon bias and heterologous protein expression. Trends Biotechnol. 22, 346–353 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Ikeda H., Kazuo S.-Y., Omura S., Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 41, 233–250 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Flinspach K., Kapitzke C., Tocchetti A., Sosio M., Apel A. K., Heterologous expression of the thiopeptide antibiotic GE2270 from Planobispora rosea ATCC 53733 in Streptomyces coelicolor requires deletion of ribosomal genes from the expression construct. PLoS One 9, e90499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All public genomic and chemical data used in this study are available through SI Appendix, List of Supplementary Datasets. Source code for the software presented in this manuscript can be found at https://github.com/magarveylab under the repositories NLPPrecursor and clams-release. A full protocol for all methodologies presented here is available within SI Appendix, SI Methods.