Members of the Tombusviridae family have nearly identical shells, and yet they package genomes that range from 4.6 kb (monopartite) to 5.3 kb (bipartite) in size. To understand how this genome flexibility occurs within a rigidly conserved shell, we determined the high-resolution cryo-electron microscopy (cryo-EM) structures of cucumber leaf spot virus and red clover necrotic mosaic virus. In response to genomic size differences, it appears that the ssRNA binding (R) domain of the capsid diverged evolutionarily in order to recognize the different genomes. The next region, the “arm,” seems to have also coevolved with the R domain to allow particle assembly via interactions at the icosahedral 3-fold axes. In addition, there are differences at the icosahedral 3-fold axes with regard to metal binding that are likely important for transmission and the viral life cycle.
KEYWORDS: Tombusviridae, electron microscopy, plant viruses, virion structure
ABSTRACT
Members of the Tombusviridae family have highly similar structures, and yet there are important differences among them in host, transmission, and capsid stabilities. Viruses in the Tombusviridae family have single-stranded RNA (ssRNA) genomes with T=3 icosahedral protein shells with a maximum diameter of ∼340 Å. Each capsid protein is comprised of three domains: R (RNA binding), S (shell), and P (protruding). Between the R domain and S domain is the “arm” region that studies have shown to play a critical role in assembly. To better understand how the details of structural differences and similarities influence the Tombusviridae viral life cycles, the structures of cucumber leaf spot virus (CLSV; genus Aureusvirus) and red clover necrotic mosaic virus (RCNMV; genus Dianthovirus) were determined to resolutions of 3.2 Å and 2.9 Å, respectively, with cryo-electron microscopy and image reconstruction methods. While the shell domains had homologous structures, the stabilizing interactions at the icosahedral 3-fold axes and the R domains differed greatly. The heterogeneity in the R domains among the members of the Tombusviridae family is likely correlated with differences in the sizes and characteristics of the corresponding genomes. We propose that the changes in the R domain/RNA interactions evolved different arm domain interactions at the β-annuli. For example, RCNMV has the largest genome and it appears to have created the necessary space in the capsid by evolving the shortest R domain. The resulting loss in RNA/R domain interactions may have been compensated for by increased intersubunit β-strand interactions at the icosahedral 3-fold axes. Therefore, the R and arm domains may have coevolved to package different genomes within the conserved and rigid shell.
IMPORTANCE Members of the Tombusviridae family have nearly identical shells, and yet they package genomes that range from 4.6 kb (monopartite) to 5.3 kb (bipartite) in size. To understand how this genome flexibility occurs within a rigidly conserved shell, we determined the high-resolution cryo-electron microscopy (cryo-EM) structures of cucumber leaf spot virus and red clover necrotic mosaic virus. In response to genomic size differences, it appears that the ssRNA binding (R) domain of the capsid diverged evolutionarily in order to recognize the different genomes. The next region, the “arm,” seems to have also coevolved with the R domain to allow particle assembly via interactions at the icosahedral 3-fold axes. In addition, there are differences at the icosahedral 3-fold axes with regard to metal binding that are likely important for transmission and the viral life cycle.
INTRODUCTION
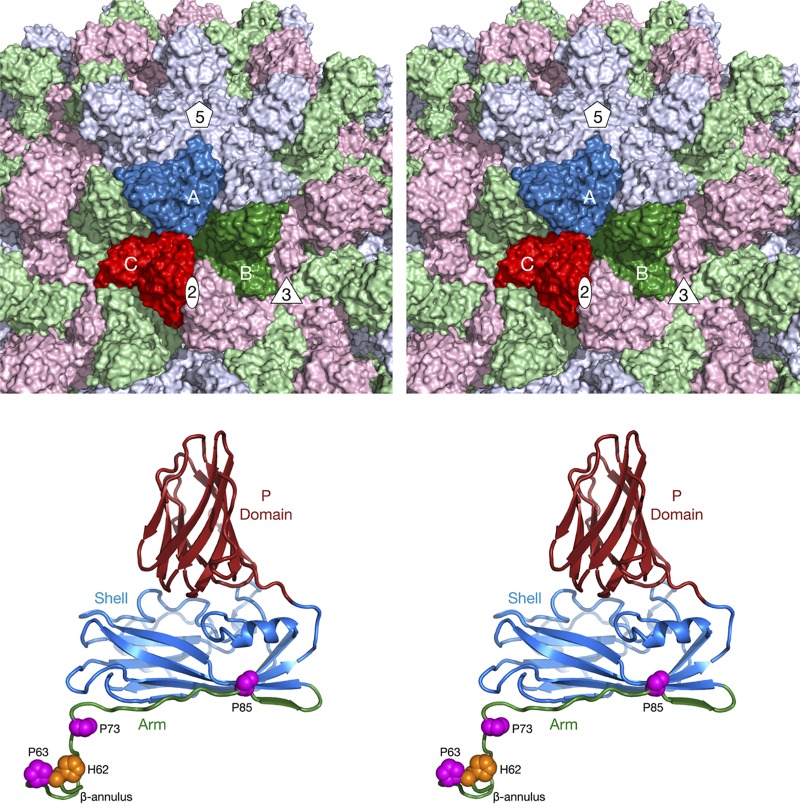
The Tombusviridae family members are single-stranded RNA (ssRNA) viruses composed of three subfamilies (International Committee on Taxonomy of Viruses [ICTV]; https://talk.ictvonline.org/ictv-reports/ictv_online_report/). The major subfamily is Procedovirinae, comprised of 13 genera, which contains one of the subjects of this study (cucumber leaf spot virus; CLSV). One of the other subfamilies is the Regressovirinae, which contains the other virus investigated in this study (red clover necrotic mottle virus; RCNMV). These two viruses have a T=3 icosahedral protein shell with a maximum diameter of ∼340 Å (Fig. 1). Each capsid protein is comprised of three domains: R (RNA binding), S (shell), and P (protruding). The residues at the N-terminal region are disordered and are called the R domain since that domain interacts with the RNA interior. Residues between the R and S domains act as a flexible “arm” connecting the two domains. The next ∼170 residues comprise the S domain, so named because it forms the tight protein shell that encapsidates the RNA genome. The final ∼110 residues form the P domains, which form protruding dimers with adjacent subunits. The A subunits encircle the icosahedral 5-fold axes, and their P domains interact with the P domains of adjacent B subunits. The C subunits form homodimers across the icosahedral 2-fold axes.
FIG 1.
Organization of the Tombusviridae capsid. The top panel shows a stereo image of a portion of the CNV capsid. The A, B, and C subunits of one icosahedral asymmetric unit are colored blue, green, and red, respectively. The icosahedrally related copies of the A, B, and C subunits are colored in light hues. Also noted are the locations of one set of 5-fold, 3-fold, and 2-fold axes. The bottom panel shows a stereo image of the C subunit of CNV. The P domain is shown in brick red, the shell in blue, and the arm in green. The locations of conserved prolines and a metal ion binding histidine, discussed in the Fig. 5 legend, are also shown.
Cucumber necrosis virus (CNV; subfamily Procedovirinae), cucumber leaf spot virus (CLSV), and red clover necrotic mosaic virus (RCNMV) belong to the Tombusvirus, Aureusvirus, and Dianthovirus genera, respectively. In CNV, the “arm” region is observed only in the C subunits of the crystal (1) and cryo-electron microscopy (cryo-EM) (2) structures and ∼17 residues of this region form β-annuli at the icosahedral 3-fold axes. The entire R and arm regions (up to approximately residue 100) are disordered in the A and B subunits. As with other members of this family, the differences in the arm structures are necessary to accommodate the angular differences between the S domain contacts in the intact icosahedral capsid structure. The S domains in the C-C subunit dimers are relatively flat, with the ordered arms lying between the C subunits, whereas the S domains in the A-B pairs are more angled, leaving no room for the arm structure.
Even though the R domain is disordered, it plays a major role in the assembly of virions. In CNV, the role of the R domain in particle formation cannot be ascribed simply to neutralization of the negative charge of the RNA core by the addition of basic residues (3). If the entire R domain or C-terminal 26 amino acids of the R-domain are removed, then nearly all of the virions are assembled into small T=1 particles (4, 5). The arm region that connects the disordered R domain to the S domain is critical for assembly since if either P73 or P85 is mutated then the particles either do not assemble or assemble poorly (3). The exception is that the P73G mutant of CNV yields the same amount of virus as the wild-type strain if propagated in Nicotiana benthamiana (3). However, these virions are less stable than the wild type and are unable to be transmitted by zoospores (2).
The R domain of RCNMV, a Dianthovirus, is at least 30 residues shorter than other members of the Tombusviridae family but is similarly critical for the viral life cycle. RCNMV exists as two different populations: an infectious form with one copy of both RNA-1 (3.8 kb) and RNA-2 (1.4 kb) and a second form containing four copies of RNA-2 (6). When more than the first 10 residues are removed from the N terminus, the resulting virus cannot form particles or move out of the initially infected leaf (7). In addition, deletion of the first 10 residues and deletions in the region consisting of residues 21 to 45 eliminated the packaging of RNA-1, showing that different regions of the R domain are responsible for selecting the genome fragment to package (7). Similarly to CNV, mutations in the arm region (K25A, K33A, and K38A) greatly reduced particle accumulation but did not affect systemic spread of the infection (8).
The structures of CNV and RCNMV have previously been determined. For CNV, the cryo-transmission electron microscopy (cryo-TEM) structures of the T=1 particles (5) and the T=3 particles (2, 5) and the X-ray structure of the wild-type virus have been determined (1). For RCNMV, the cryo-EM structure was previously determined to a resolution of 8.5 Å (9). As expected, both T=3 particles resemble the crystal structure of tomato bushy stunt virus (TBSV) (10).
There is significant internal organization in the T=3 particles of both CNV (2, 5) and RCNMV (9) but not in the CNV T=1 capsids. The general features of the internal structure for CNV are consistent with previous neutron diffraction experiments performed on Tomato bushy stunt virus (TBSV) (11, 12). The outermost shell (radii, 110 to 175 Å) represents the capsid protein shell and protruding domains and is essentially that portion of the capsid described by the X-ray structure of TBSV (10). Immediately beneath the capsid is a shell (radii, 80 to 100Å) that is composed of RNA with little or no protein. Beneath the layer of RNA, surprisingly, is a second shell of protein (radius of 50 to 80Å) that, in TBSV, contains little RNA. On the basis of a single-crystal neutron diffraction study (12), it was proposed that this second protein shell is composed of partially folded but mainly disordered R domains. At the icosahedral 3-fold axes, the ordered part of the β-annulus fades out as it extends toward the inner shell. It was also suggested that the arm domains of the A and B subunits are largely disordered. In contrast, the RCNMV neutron diffraction experiments (13) revealed similar structures for the first two layers but yielded a nearly empty region (92% solvent) where the second shell of protein is found in TBSV (10).
The structures of both CNV and RCNMV in the presence of metal chelators have been determined. For RCNMV, the removal of calcium with EGTA (pH 5.3) did not cause the particles to swell, whereas the removal of magnesium with EDTA (pH 5.3) did (9). With CNV, the particles swelled in the presence of EDTA at pH 7.6 (2). In both of the swollen structures, pores formed between the A, B, and C subunits whereas the contacts at the icosahedral 3-fold axes remained intact. Notably, the P73G mutation in the CNV arm domain disrupted the putative internal zinc-binding structure at the icosahedral 3-fold axes and the particles were less stable than those seen with the wild type and were unable to be transmitted by zoospores (2, 14).
To better understand the commonalities and differences among the members of the Tombusviridae family, we determined the structures of CLSV and RCNMV to resolutions of 3.2 Å and 2.9 Å, respectively, using cryo-electron microscopy and image reconstruction techniques. As expected, the capsids show structural homology, but there are significant differences at the icosahedral 3-fold axes. The stability of the subunit interactions has been shown to be important to CNV zoospore transmission (2) and is likely crucial for the viral life cycle in all of these viruses. A synthetic 24-residue peptide based on the TBSV β-annulus is able to self-assemble into capsid-like structures (15), demonstrating an isolated function of the arm region. From these studies, the R domain appears to be involved in RNA selection and packaging, whereas the arm region is intimately involved in capsid assembly. As such, the R domain varies among the Tombusviridae family according to the size and packaging requirements of the particular genome whereas the arm compensates for gain/loss of RNA/R domain interactions via changes at the β-annuli.
RESULTS
Comparison of CLSV and RCNMV virion structures to CNV virion structure.
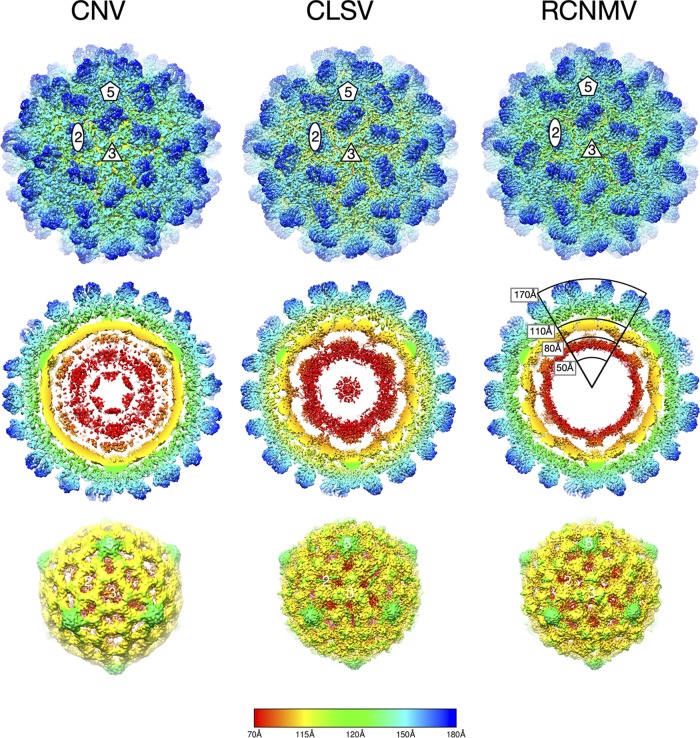
Shown in Fig. 2 are the reconstructions of CLSV and RCNMV compared to the previous 4.2-Å resolution image reconstruction of CNV (2). As expected, since all three viruses are members of the Tombusviridae family, the exterior views of all three viruses are remarkably similar (top row). The second row of images represents cross sections of the virions showing the architecture inside the capsid region. As was observed with TBSV (11), CNV (5), and RCNMV (9) at lower resolution, there are several concentric shells of density inside the capsid. From neutron scattering experiments (11, 12), it was suggested that the 110-to-175-Å shell is composed entirely of protein and corresponds to the capsid protein. The next shell down (80 to 100 Å) was found to be composed entirely of RNA, while the 50-to-80-Å shell was found to be either mostly composed of protein (in the case of TBSV and CNV) or empty (RCNMV). The data representing the density of these concentric shells are weak compared to that representing the outer capsid shell and not interpretable.
FIG 2.
Cryo-EM image reconstructions of CLSV and RCNMV at 3.2-Å resolution and 2.9-Å resolution, respectively, compared to the previously described 4.2-Å reconstruction of CNV (2). All panels are colored from red to blue with increasing radii. The top panels present exterior views of the particles looking down the icosahedral 3-fold (quasi-6-fold) axes. The locations of 2-fold, 3-fold, and 5-fold axes are denoted. The middle panels present thin sections of the particles showing the protein/RNA layers. The bottom panels present the inner RNA cores.
While the three viruses had layers of internal structures under the capsid shell, there were differences among them. Compared to CNV, RCNMV has a thin shell of weak density in this region that is consistent with the high solvent content of this region in neutron scattering experiments (13) and may be due to the truncated R domain in RCNMV. In TBSV (12), there were weak connections at the icosahedral 3-fold axes between the presumed RNA shell immediately beneath the capsid and the shell beneath. A similar connection was also observed in the 12-Å cryo-EM structure of CNV (5). In contrast, this type of connection between the RNA shell and the shell beneath appears to occur at the 2-fold axes in RCNMV and CLSV. Inspection of the core structures (bottom panels) revealed that all have protrusions at the 5-fold axes, corresponding to a convolution in the inner surface at the 5-fold axes in the capsid. The CNV core has a large hole in the density at the 3-fold axes, whereas the cores of CLSV and RCNMV have a cluster of three islands of density at this location.
Modeling of the CLSV and RCNMV CP structures.
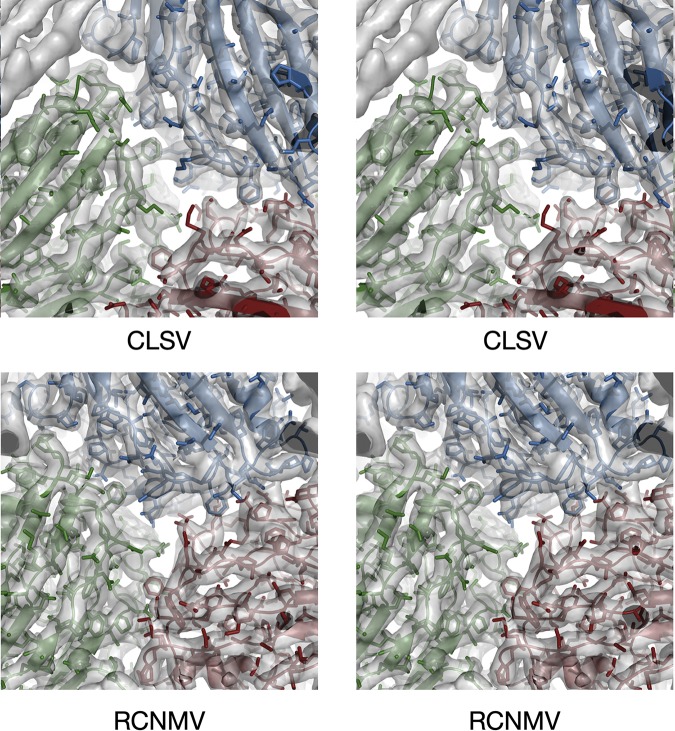
The structural models for CLSV and RCNMV were readily built using the crystal structure of CNV (1). The cryo-EM resolution values for CLSV and RCNMV were 3.2 Å and 2.9 Å, respectively. The EM density of the protein backbone was clear, but many of the side chains were truncated, and the corresponding amino acid identity was often ambiguous (Fig. 3). This is what we have found in all of our high-resolution EM reconstructions (see, e.g., reference 16), and such results are simply a function of the difference between EM and crystallographic resolution estimates. (For a thorough discussion of this observation, see a recent publication by Pintilie et al. [17]). Therefore, the model building was heavily dependent on using the bulky side chains of tryptophan and tyrosine as fiducial marks in the sequence. The models were refined in real space using PHENIX (18), and the final statistics are shown in Table 1. Residues 61 to 386 were built into the EM density for CLSV (Fig. 4), whereas residues 19 to 338 were built into the RCNMV density. However, the data representing the EM density for residues 43 to 49 in RCNMV were too weak to model those residues with any level of confidence (Fig. 4B, “A” arrows). The “B” arrows in this panel show how the N terminus of RCNMV of the C subunit markedly departs from the structures of CNV and CLSV. Also noted in RCNMV are the locations of K25, K33, and K38. When these residues were mutated to alanine, intact virions did not accumulate (8).
FIG 3.
Stereo images of portions of the EM density of CLSV and RCNMV with the refined models fitted in the density. The view here is looking down a pseudo-3-fold axis, with subunits A, B, and C colored blue, green, and red, respectively.
TABLE 1.
Summary of data collection and refinement statistics
| Parametera | Result |
|
|---|---|---|
| CLSV | RCNMV | |
| Data collection | ||
| No. of micrographs | 1,246 | 620 |
| Defocus range (μm) | 0.8–2.5 | 0.8–2.5 |
| Dose rate (e/pix/s) | ∼7 | ∼7 |
| Total dose (e/Å2) | ∼50 | ∼50 |
| No. of particles picked | 37,556 | 100,007 |
| No. of particles used | 5,908 | 3,966 |
| Refinement statistics | ||
| PDB ID | 6MRL | 6MRM |
| EMDB ID | 9204 | 9205 |
| Resolution (Å) | 3.2 | 2.9 |
| Rmodel (%) | 34 | 36 |
| Correlation coefficient (%) | 85 | 82 |
| Ramachandran favored (%) | 85 | 82 |
| Ramachandran outlier (%) | 0 | 1.2 |
| Rotamer outlier (%) | 2 | 0.3 |
| C-β outlier | 2 | 3 |
| Bond length RMSD (Å) | 0.01 | 0.01 |
| Bond angle RMSD (°) | 1.5 | 1.7 |
| EMringer | 4.5 | 3.2 |
| ClashScore | 12.1 | 6.7 |
| MolProbity | 2.2 | 2.1 |
EMDB, Electron Microscopy Data Bank; ID, identifier; pix, pixel; RMSD, root mean square deviation.
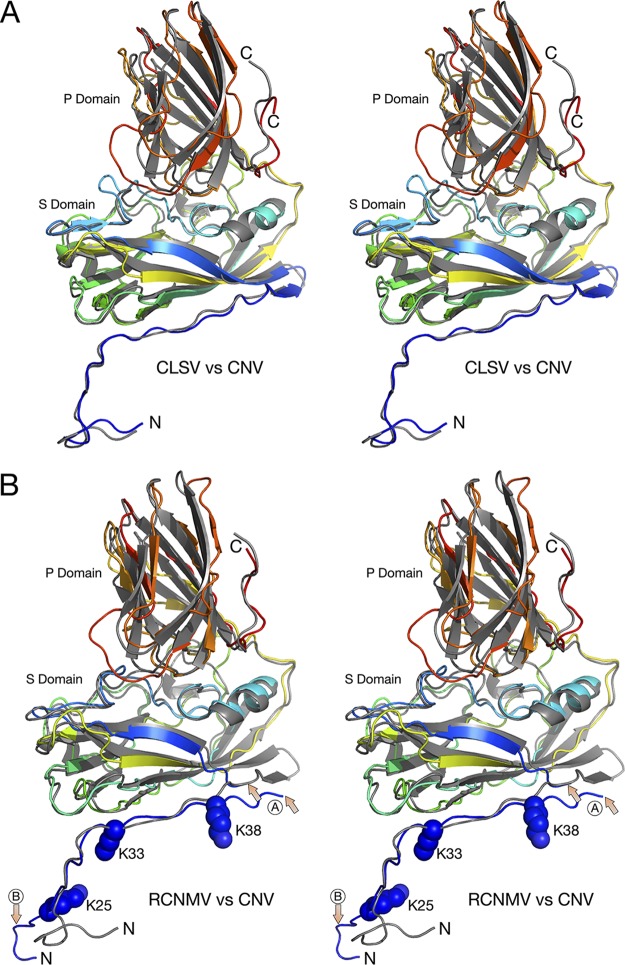
FIG 4.
Comparisons of the structures of CNV, CLSV, and RCNMV C subunits. In both panel A and panel B, the atomic structure of CNV is represented by gray ribbons whereas CLSV and RCNMV are colored from blue to red as the chain extends from the N terminus to the C terminus. In panel B, the area marked with arrows and a circled “A” shows the one region of disorder that was of insufficient quality to build a model. The other area noted with a circled “B” shows the marked differences between the structures of RCNMV and CNV. While the N terminus of CNV bends sharply to form a putative zinc ion binding site, RCNMV extends into an adjacent subunit and forms extensive β-strand interactions.
In terms of overall protein fold, the largest differences among these viruses are represented by the interactions among the N termini of the C subunits that lie at the icosahedral 3-fold axes (Fig. 5). As we previously observed with CNV (1), a metal ion sits on the icosahedral 3-fold axis and, from its interactions with three histidines, is likely a Zn2+ ion. We have also previously shown that the residues surrounding this metal binding site are also crucial, in that a P73G mutation disrupts this metal binding site, destabilizes the capsids, and blocks zoospore transmission (2). CLSV looks very similar to CNV, with P73 forming the same kink in the β-annulus. However, the histidines clustered at the 3-fold axis in CNV are replaced by the hydrophobic I63 in CLSV. In complete contrast to both CNV and CLSV, RCNMV forms a highly extended structure where the N terminus of each C subunit forms a β-sheet with the other two subunits. As with the other two viruses, P30 forms a kink in the structure so that it can turn to form these β-strand interactions. For comparison, cucumber mosaic virus (CMV), a member of the Bromoviridae family, forms a highly stable hexagonal bundle of amphipathic α-helices (19). Therefore, Fig. 5 shows increasing levels of subunit interactions. CNV is held in place by a Zn2+ ion that, when lost either by mutations or by the addition of chelating agents, can trigger a less stable form of the capsid. CLSV is very similar to CNV but, because of the presence of hydrophobic interactions rather than metal chelation, is insensitive to changes in metal concentrations at this site. RCNMV is the most stable of the three, with extensive, high-energy hydrogen bonds between the intertwining C subunit β-strands. CMV has a completely different architecture at the icosahedral 3-fold axes, with amphipathic α-helices at the N termini of the B and C subunits coming together to form a hexameric bundle (19). It is unclear why these very similar viruses evolved such different structures at the icosahedral 3-fold axes and yet the rest of the capsid structure remains so highly conserved. They likely have different stabilities and responses to environmental cues, but how do these differences translate to biological function?
FIG 5.
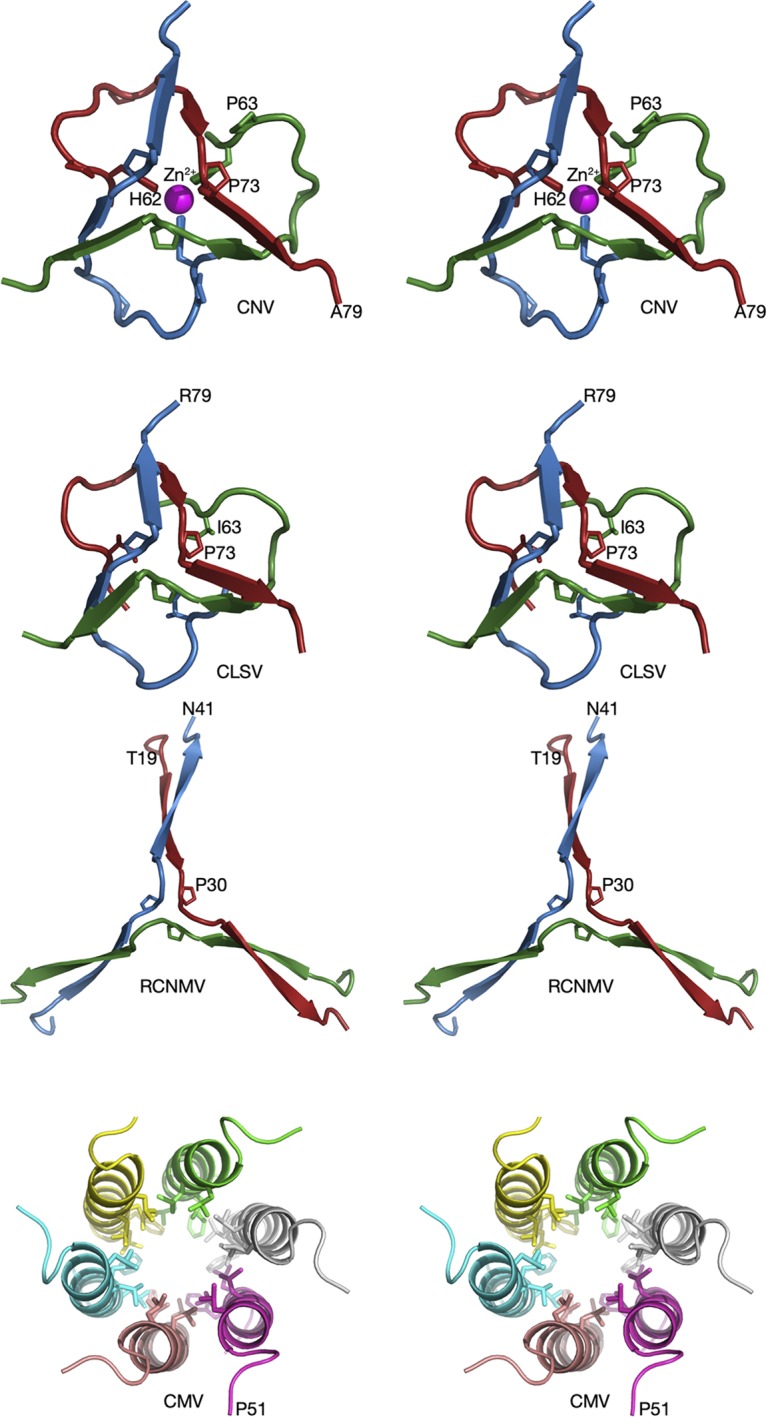
Comparisons of the β-annuli of CNV, CLSV, RCNMV, and CMV at the quasi-6-fold axis. In all panels, the different subunits at the 3-fold axis are given unique colors. Note that these colors do not correspond to subunits A, B, and C, since all three proteins are C subunits. CMV, a member of the Bromoviridae family that is transmitted by aphids, is shown for comparison.
Sequence analysis of N terminus/R domain.
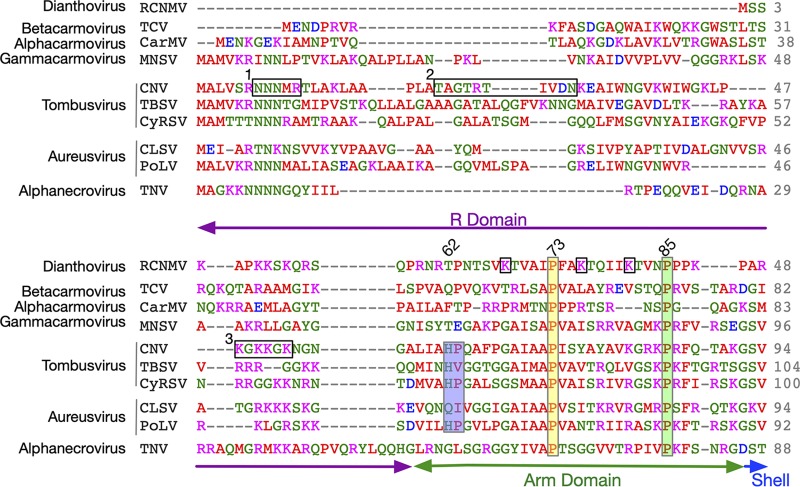
With the current and previous structural studies, along with detailed analyses on the R and arm domains in CNV, it is informative to align and compare the N-terminal sequences of several members of the Tombusviridae family. From previous mutagenesis studies on CNV (3), three areas of the R domain were found to be of interest (boxes 1 to 3 in Fig. 6). Box 1 is asparagine rich, box 2 is uncharged, and box 3 is arginine/lysine rich. Even if the entire R domain of CNV is deleted and replaced with regions 1 and 2 (R1 + 2 mutant), the virus can still form particles. However, the yield of this mutant is only ∼2.8% that of the wild type and is a mixture of T=1 and T=3 icosahedrons and only packages subgenomic fragments of CNV. It was suggested that, while regions 1 and 2 might be sufficient to form particles, lysine/arginine-rich region 3 might contain the selectivity necessary to package full-length genomic RNA (3). In the arm domain, mutating the highly conserved P85 to a glycine in CNV similarly resulted in formation of polymorphic particles that are unable to properly package the CNV genome. Indeed, when the P85G mutation was combined with the R1 + 2 mutant, all of the particles appeared to be T=1. Together, these results suggest that virion assembly/RNA packaging requires more than abrogation of the RNA charge with basic residues in the R domain and that residues in the arm domain are also critical.
FIG 6.
Clustal Omega (31) alignment of the N termini of several members of the Tombusviridae family. Shown here are the alignments of the R and arm domains of several different genera of the Tombusviridae family. Note that only the arm domain is observable in the atomic structures and that it is visible only in the C subunits around the β-annulus. The genus name is denoted on the far left. The amino acids are colored according to chemical characteristics; nonpolar, acidic, basic, and polar are colored red, mauve, blue, and green, respectively. The three horizontal black rectangles labeled 1, 2, and 3 represent regions of CNV that were analyzed in previous studies (3). The three vertical rectangles denote points of conservation with the corresponding residue number from CNV shown at the top. For RCNMV, K25, K33, and K38 in the arm region are noted with black boxes. Mutating these residues to alanine blocks capsid assembly (8). The corresponding domain designations are shown at the bottom.
In the atomic structure of CNV, three histidines (H62) come together at the β-annulus to bind a metal ion, most likely zinc (1). This histidine residue is conserved within the members of the Tombusvirus genus, including Havel river virus (sequence not shown) (20). However, only the structures of CNV and TBSV are known at this time, and a metal ion was not modeled in the TBSV structure (10). Interestingly, an Aureusvirus, Pothos latent virus (PoLV), also has the conserved histidine-proline pair at this location. However, without knowing the structure, it is not clear if it forms a zinc binding site.
Among the carmoviruses, the β-annulus in Melon necrotic spot virus (MNSV; Gammacarmovirus) looks very similar to CNV but lacks the zinc binding site (21). This part of the structure was not defined in Carnation mottle virus (CarMV; Alphacarmovirus) (22) and was truncated in Turnip crinkle virus (TCV; Betacarmovirus). As shown in Fig. 5, the β-annulus in CLSV looks very similar to that in CNV without the Zn2+ binding region whereas RCNMV is quite different and forms the extended β-strands that interlock the C subunits. Despite the differences in the β-annuli, all of the members of Tombusviridae have a conserved proline at residue 73 (CNV numbering). In CNV, P73 sits atop the zinc binding site and this proline is absolutely conserved in all members of this family. A P73G mutation disrupts the β-annulus in CNV, greatly destabilizes the particle, and prevents zoospore transmission (2, 23). Similarly, a P85G mutation generates polymorphic particles (3). It is interesting that replacing a rigid proline with a flexible glycine could disrupt the particle and its assembly to such a great extent. A major caveat in trying to understand how these residues might influence capsid stability/assembly is that we have a view only of the N termini of the C subunits, and there is no information as to how the R domain interacts with the RNA or how arm domains are structured in the A and B subunits. Certainly, the disorder at the β-annulus caused by the P73G mutation is consistent with the loss in capsid stability, but whether the effects of this and other mutations in the R/arm domains are limited to this area of the C subunits is not clear. Several studies on TBSV (11, 12) have suggested that some of the inner layers of density in the particle are composed of protein and RNA. It is not clear whether some of these mutations affect these features and play some role in disrupting assembly. Perhaps future studies on the CNV T=3 particles that package subgenomic RNA fragments could be useful to this end. Additionally, it would be interesting to try to swap these various R/arm domains to see if one could similarly swap the packaged genome.
DISCUSSION
The structures of CNV, CLSV, and RCNMV are all homologous and share a number of similar features. Like previous TBSV (12) and CNV (5) structures, CLSV and RCNMV have a multilayered internal structure. As shown by single-crystal neutron diffraction studies on TBSV, the shell immediately beneath the capsid is composed of RNA and the shell beneath that is proteinaceous (11, 12). There are connections between this layer and the one beneath at the icosahedral 3-fold axes in CNV, while they occur at the 2-fold axes in CLSV and RCNMV. Furthermore, there are large islands of density in the RNA layer at the 3-fold axes in CLSV and RCNMV, but there is a large hole in the RNA layer in CNV. It should be noted that the CNV reconstruction has been performed on several different occasions using different microscopes and that all produced the same result. It is not at all clear why CNV and CLSV are so different in this layer when the lengths and structures of the N termini at the β-annuli are so similar to each other and so different from those of RCNMV. Notably, none of this structure was observed in CNV T=1 particles that had packaged subgenomic pieces of RNA (5).
What is particularly interesting is that these viruses came up with such different solutions with respect to packing interactions at the icosahedral 3-fold axes. The arm region has intertwining β-strands in all three viruses. However, those strands in RCNMV are nearly twice as long as in CNV and CLSV. CNV is additionally stabilized by a bound metal ion, likely Zn2+, while CLSV is not. When CNV swells at neutral pH, these interactions at the 3-fold axis hold the particle together (1). Since this interaction is not mediated by calcium, the environmental response corresponding to the interactions between the subunits within the icosahedral protomers differs from that seen with the quasi-6-fold axes. CMV of the Bromoviridae family represents the most extreme version of stabilization at the 3-fold axes, with extensive interactions among the amphipathic helixes (19). It is not at all clear why these homologous viruses employ such different means of stabilization of the 3-fold axes during the swelling process.
The R domain is perhaps the least understood portion of the Tombusviridae capsid protein. Dianthoviruses have the shortest R domain and lack the asparagine-rich region found in the other Tombusviridae family members. Indeed, the R domain of RCNMV is only ∼14 residues long and contains the lysine/arginine-rich region that appears to be crucial in genome recognition in CNV (3). The R domains of carmoviruses and necroviruses are more than 40 residues in length and contain both the asparagine-rich region (region 1 in Fig. 6) and the lysine/arginine-rich region (region 3 in Fig. 6). Finally, the tombusviruses and aureusviruses have the longest R domains, with all three regions identified in CNV.
With the caveat that we do not know how the R/arm domains interact with the RNA in the A and B subunits, that of RCNMV may represent the minimal R domain required for packaging and assembly. As previously suggested (6–8), RCNMV may have evolved this relatively short R domain to create sufficient space for a genome that is significantly larger than other members of this family. To compensate for this, the virus may have evolved an extensive network of hydrogen bonds made by the interlocking arm domains around the icosahedral 3-fold axes in order to hold the particles together. Mutagenesis experiments show that the virus cannot tolerate nearly complete truncation of the R domain (7, 8). The other members of this family represent a more complicated story, and the role of the additional R domain regions is unclear. Work on CNV has shown that only small portions of the R domain are required for assembly but that the level of assembly efficiency is low and that, without the lysine/arginine-rich region, only subgenomic fragments are packaged (3). In both CNV and RCNMV, the arm plays a clear role in particle stability/assembly. In CNV, conserved prolines at 73 and 85 (CNV numbering) are required (14). With RCNMV, lysine-to-alanine mutations at arm residues 25, 33, and 38 (RCNMV numbering) block particle assembly (8). Interestingly, the arm region of RCNMV is more basic than that found in CNV and therefore may interact with the RNA more than the arm domains in the other members of the family.
We propose that, at least in part, the large differences in the β-annuli of these Tombusviridae viruses represent evolutionary solutions to the problem of packaging disparate genomes within the confines of a rigidly conserved capsid shell. As the size and type (e.g., monopartite versus bipartite) of genome changed, these viruses appear to have altered the length of the R domain rather than adjusting the size of the stable capsid formed by the shell domain. As the strength of the RNA/R domain interactions decreased, the arm domain evolved accordingly to compensate via interactions at the icosahedral 3-fold axes to help assemble and stabilize the particle. In addition to the RNA interactions, it appears that the arm domain/β-annulus may have also evolved to accommodate other changes in the viral life cycle such as vector transmission. Therefore, the members of the Tombusviridae have evolved a way to keep a highly successful capsid by making relatively small changes in its internal parts.
MATERIALS AND METHODS
Virus production and purification.
RCNMV was prepared as previously described (9). CLSV was produced and purified using a protocol similar to that used for CNV (5). CLSV was propagated in Nicotiana benthamiana plants that were at true leaf stages 5 to 7. The inoculum contained ∼50 ng/μl virus in a mixture containing 50 mM KPO4 (pH 6.8), and 20 μl of this solution was rubbed onto the leaves, which had been dusted with carborundum. The infection was allowed to proceed for 5 to 7 days, and leaves were harvested when the plants had completely wilted. The infected leaf material was stored at –80°C until processing.
Frozen leaf material (∼200 g) was ground in a chilled blender until fully homogenized in 1 to 2 volumes of ice-cold 0.1 M sodium acetate (pH 5.0) containing 10 mM β-mercaptoethanol. The material was allowed to rest on ice for 20 to 30 min, and the plant debris was removed by centrifugation at 5,000 × g for 30 min at 4°C. The supernatant was then filtered through one layer of Miracloth. The filtrate was clarified by centrifugation again at 5,000 × g for 30 min at 4°C. To the supernatant, polyethylene glycol 8000 (PEG 8000) was added to yield a final concentration of 8% (wt/vol) and stirred for 2 h at 4°C. The precipitate was collected by centrifugation at 8,000 × g for 20 min at 4°C and resuspended in a minimum amount of 10 mM sodium acetate (pH 5.0). This suspension was kept at 4°C overnight to ensure that the precipitate was fully resuspended.
The next day, the debris was removed from the suspension by centrifugation at 8,000 × g for 20 min at 4°C. The supernatant was layered onto a 10% to 40% sucrose gradient in a SW28 rotor and centrifuged for 3.75 h at 25K rpm (82,000 × g). The virus band was collected, diluted, and concentrated via centrifugation in a 50.2 Ti rotor at 40K rpm (145,000 × g) for 3 h. The virus pellet was then suspended in 10 mM sodium acetate (pH 5.0) for structural studies. The concentration of CLSV was estimated by assuming that a 1-mg/ml solution has an optical density of 4.5 at 260 nm. The purity of the virus was determined via SDS-PAGE, but the sample was first brought to a pH level between 7 and 8 prior to boiling in SDS running buffer.
Cryo-electron microscopy (cryo-EM).
Purified CLSV and RCNMV were vitrified on holey carbon film (R2x1 Quantifoil; Micro Tools GmbH, Jena, Germany) grids as previously described (2, 9). Briefly, purified and concentrated (∼1 mg/ml) suspensions of virions were applied to the holey films, blotted with filter paper, and plunged into liquid ethane. Either Vitrobot (Thermo Fisher) or EM-GP2 (Leica) automated plungers were used for plunging. Frozen grids were stored under liquid nitrogen until used for microscopy.
CLSV virions were imaged in a JEM3200 SFF microscope (JEOL Ltd., Japan) equipped with an in-column energy filter (omega type) and a field emission gun (FEG) operating at 300 keV. CLSV data were acquired using a K-2 camera (Gatan, USA) in superresolution mode under low-dose conditions. An in-column omega electron energy filter was used during imaging with zero-loss energy peak selected with 20 eV slit. Images were acquired with ∼54 electrons/Å2 dose; the image pixel size corresponded to 0.883 Å on the specimen scale. A total of 1,246 image stacks were collected manually.
RCNMV grids were imaged in a Titan Krios (Thermo Fisher) microscope. The microscope was operated at 300 keV and was equipped with a GIF (Gatan Inc.) postcolumn energy filter. The slit width was 20 eV, as was used in the omega filter in JEM 3200 sintered fiber felts (SFF) microscope. A total of 1,236 image stacks were collected with doses (∼54 electrons/Å2) similar to those used in the CLSV data collection scheme. The image pixel size was 1.07 Å on the specimen scale. Individual frames in each image stack (including both CLSV and RCNMV) were aligned using MotionCor2 script (24) with radiation damage compensation to increase the low-frequency content in the final images, facilitating subsequent particle alignment.
Image processing.
Individual virion images were boxed from the images using the E2BOXER program in the EMAN2 suite (25). EMAN2 was used for contrast transfer function (CTF) determination and correction. Subsequent processing (particle alignment, determination of orientations, and refinement) was done in IMAGIC-5 (26, 27). Approximately 37,000 individual CLSV and 10,000 individual RCNMV images were boxed out from corresponding micrographs.
Ab initio models of the viruses were obtained using the angular reconstitution technique (28). Particle sets for each virus were subdivided into halves and were processed independently. Several origin/orientation refinement cycles were performed, leading to construction of stable three-dimensional (3D) maps of the CLSV and RCNMV virions. The final 3D maps were reconstructed from 23,366 CLSV and 7,882 RCNMV virion images. The effective resolution of the CLSV map was 3.3 Å according to the 0.143 gold standard Fourier shell correlation (FSC) function criterion (29) (see Fig. S1 and S2 in the supplemental material). For RCNMV, the resolution estimate using the same criterion was 2.9 Å.
Model building and refinement.
For both CLSV and RCNMV, the crystal structure of CNV (1) was used as a starting model. COOT (30) was used to build the models into the density, and the program PHENIX (18) was used for subsequent real space refinement. While the gold standard resolutions for the reconstructions were 2.9 Å (RCNMV) and 3.2 Å (CLSV) (Fig. S2), the density values for most of the side chains were notably reduced. The backbone density was mostly interpretable, but the bulky side chains (Phe and Trp) were absolutely necessary to keep register with the sequence.
Data availability.
The EM density maps and coordinates for CLSV (PDB: 6MRL; EMD: 9204) and RCNMV (PDB: 6MRM; EMD: 9205) are available in the Protein Data Bank and Electron Microscopy Data Bank.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NIH grant to T.J.S. (1R01-AI141465). We acknowledge the support of the Sealy Center for Structural Biology at the University of Texas Medical Branch at Galveston (UTMB) and the College of Agriculture and Life Sciences at North Carolina State University (NCSU). Data were collected at NIH-supported facilities operated by Wah Chiu (1U24 GM116787-01) with support from Joanita Jakana and Jason Kalber and by Hong Zhou (1U24 GM116792-01) with support from Ivo Atanasov and Yanxiang Cui. We also acknowledge the Texas Advanced Computing Center (TACC; http://www.tacc.utexas.edu) at The University of Texas at Austin for providing high-performance-computing (HPC) resources (LONESTAR5) that contributed to the research results reported within this paper.
We do not have any conflicts of interest with the work presented here.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Li M, Kakani K, Katpally U, Johnson S, Rochon D, Smith TJ. 2013. Atomic structure of Cucumber necrosis virus and the role of the capsid in vector transmission. J Virol 87:12166–12175. doi: 10.1128/JVI.01965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherman MB, Kakani K, Rochon DA, Jiang W, Voss NR, Smith TJ. 2017. Stability of cucumber necrosis virus at the quasi-6-fold axis affects zoospore transmission. J Virol 91:e01030-17. doi: 10.1128/JVI.01030-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kakani K, Reade R, Katpally U, Smith TJ, Rochon D. 2008. Induction of particle polymorphism in cucumber necrosis virus coat protein mutants in vivo. J Virol 82:1547–1557. doi: 10.1128/JVI.01976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katpally U, Kakani K, Reade R, Rochon D, Smith TJ. 2007. Structures of T=1 and T=3 Cucumber necrosis virus particles: evidence of internal scaffolding. J Mol Biol 365:502–512. doi: 10.1016/j.jmb.2006.09.060. [DOI] [PubMed] [Google Scholar]
- 5.Katpally U, Kakani K, Reade R, Dryden K, Rochon D, Smith TJ. 2007. Structures of T=1 and T=3 Cucumber necrosis virus particles: evidence of internal scaffolding. J Mol Biol 365:502–512. doi: 10.1016/j.jmb.2006.09.060. [DOI] [PubMed] [Google Scholar]
- 6.Basnayake VR, Sit TL, Lommel SA. 2006. The genomic RNA packaging scheme of Red clover necrotic mosaic virus. Virol 345:532–539. doi: 10.1016/j.virol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Park S-H, Sit TL, Kim K-H, Lommel SA. 2013. The red clover necrotic mosaic virus capsid protein N-terminal amino acids possess specific RNA binding activity and are required for stable virion assembly. Virus Res 176:107–118. doi: 10.1016/j.virusres.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Park S-H, Sit TL, Kim K-H, Lommel SA. 2012. The Red clover necrotic mosaic virus capsid protein N-terminal lysine-rich motif is a determinant of symptomatology and virion accumulation. Mol Plant Pathol 13:744–754. doi: 10.1111/j.1364-3703.2011.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA. 2006. Removal of divalent cations induces structural transitions in Red clover necrotic mosaic virus revealing a potential mechanism for RNA release. J Virol 80:10395–10406. doi: 10.1128/JVI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. 1978. Tomato bushy stunt virus at 2.9Å resolution. Nature 276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 11.Chauvin C, Witz J, Jacrot B. 1978. Structure of the tomato bushy stunt virus: a model for protein-RNA interactions. J Mol Biol 124:641–651. doi: 10.1016/0022-2836(78)90175-4. [DOI] [PubMed] [Google Scholar]
- 12.Timmins PA, Wild D, Witz J. 1994. The three-dimensional distribution of RNA and protein in the interior of tomato bushy stunt virus: a neutron low-resolution single-crystal diffraction study. Structure 2:1191–1201. doi: 10.1016/s0969-2126(94)00121-9. [DOI] [PubMed] [Google Scholar]
- 13.Martin SL, He L, Meilleur F, Guenther RH, Sit TL, Lommel SA, Heller WT. 2013. New insight into the structure of RNA in red clover necrotic mosaic virus and the role of divalent cations revealed by small-angle neutron scattering. Arch Virol 158:1661–1669. doi: 10.1007/s00705-013-1650-6. [DOI] [PubMed] [Google Scholar]
- 14.Kakani K, Reade R, Rochon D. 2004. Evidence that vector transmission of a plant virus requires conformational change in virus particles. J Mol Biol 338:507–517. doi: 10.1016/j.jmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura K, Watanabe K, Matsuzaki T, Sakurai K, Kimizuka N. 2010. Self-assembled synthetic viral capsids from a 24-mer viral peptide fragment. Angew Chem Int Ed Engl 49:9662–9665. doi: 10.1002/anie.201004606. [DOI] [PubMed] [Google Scholar]
- 16.Sherman MB, Williams AN, Smith HQ, Nelson C, Wilen CB, Fremont DH, Virgin HW, Smith TJ. 2019. Bile salts alter the mouse norovirus capsid conformation; possible implications for cell attachment and immune evasion. J Virol 12:e00970-19. doi: 10.1128/JVI.00970-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pintilie G, Zhang K, Su Z, Li S, Schmid MF, Chiu W. 2019. Measurement of atom resolvability in CryoEM maps with Q-scores. bioRxiv 10.1101/722991. [DOI] [PMC free article] [PubMed]
- 18.Afonine PV, Grosse-Kunstleve RW, Adams PD. 2005. The Phenix refinement framework. CCP4 Newsl 42:Contribution 8. [Google Scholar]
- 19.Smith TJ, Chase E, Schmidt T, Perry KL. 2000. The structure of cucumber mosaic virus and comparison to cowpea chlorotic mottle virus. J Virol 74:7578–7586. doi: 10.1128/jvi.74.16.7578-7586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koenig R, Pfeilstetter E, Kegler H, Lesemann DE. 2004. Isolation of two strains of a new Tombusvirus (Havel river virus, HaRV) from surface waters in Germany. Eur J Plant Path 110:429–433. doi: 10.1023/B:EJPP.0000021084.38938.c3. [DOI] [Google Scholar]
- 21.Wada Y, Tanaka H, Yamashita E, Kubo C, Ichiki-Uehara T, Nakazono-Nagaoka E, Omura T, Tsukihara T. 2008. The structure of melon necrotic spot virus determined at 2.8Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun 64:8–13. doi: 10.1107/S1744309107066481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgunova EY, Dauter Z, Fry E, Stuart DI, Stel'mashchuk VY, Mikhailov AM, Wilson KS, Vainshtein BK. 1994. The atomic structure of Carnation Mottle Virus capsid protein. FEBS Lett 338:267–271. doi: 10.1016/0014-5793(94)80281-5. [DOI] [PubMed] [Google Scholar]
- 23.Kakani K, Sgro J-Y, Rochon D. 2001. Identification of specific cumber necrosis virus coat protein amino acids affecting fungus transmission and zoospore attachment. J Virol 75:5576–5583. doi: 10.1128/JVI.75.12.5576-5583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 26.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. 1996. A new generation of the IMAGIC image processing system. J Struct Biol 116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 27.van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. 2000. Single-particle electron cryo-microscopy: towards atomic resolution. Q Rev Biophys 33:307–369. doi: 10.1017/S0033583500003644. [DOI] [PubMed] [Google Scholar]
- 28.van Heel M. 1987. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21:111–124. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 29.Scheres SH, Chen S. 2012. Prevention of overfitting in cryo-EM structure determination. Nat Methods 9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EM density maps and coordinates for CLSV (PDB: 6MRL; EMD: 9204) and RCNMV (PDB: 6MRM; EMD: 9205) are available in the Protein Data Bank and Electron Microscopy Data Bank.