Abstract
Peptidoglycan (PG) is a critical component of the bacterial cell wall and is composed of a repeating β-1,4–linked disaccharide of N-acetylglucosamine and N-acetylmuramic acid appended with a highly conserved stem peptide. In Gram-negative bacteria, PG is assembled in the cytoplasm and exported into the periplasm where it undergoes considerable maturation, modification, or degradation depending on the growth phase or presence of environmental stressors. These modifications serve important functions in diverse processes, including PG turnover, cell elongation/division, and antibiotic resistance. Conventional methods for analyzing PG composition are complex and time-consuming. We present here a streamlined MS-based method that combines differential analysis with statistical 1D annotation approaches to quantitatively compare PGs produced in planktonic- and biofilm-cultured Pseudomonas aeruginosa. We identified a core assembly of PG that is present in high abundance and that does not significantly differ between the two growth states. We also identified an adaptive PG assembly that is present in smaller amounts and fluctuates considerably between growth states in response to physiological changes. Biofilm-derived adaptive PG exhibited significant changes compared with planktonic-derived PG, including amino acid substitutions of the stem peptide and modifications that indicate changes in the activity of amidases, deacetylases, and lytic transglycosylases. The results of this work also provide first evidence of de-N-acetylated muropeptides from P. aeruginosa. The method developed here offers a robust and reproducible workflow for accurately determining PG composition in samples that can be used to assess global PG fluctuations in response to changing growth conditions or external stimuli.
Keywords: peptidoglycan, Pseudomonas aeruginosa (P. aeruginosa), biofilm, microbiology, mass spectrometry (MS), bacterial cell wall, murine, peptidoglycomics, planktonic, sacculi
Introduction
Peptidoglycan (PG)3 is an important structural component of the bacterial cell wall that is found in almost all bacterial species. In Gram-negative bacteria, PG is composed of a repeating β–1,4-linked disaccharide of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) with an appended highly conserved stem peptide consisting of l-alanine, iso-d-glutamate, meso-diaminopimelic acid, followed by two terminating d-alanine residues. During PG synthesis, NAG–NAM–pentapeptide monomer subunits are produced in the cytosol, as nucleotide-linked precursors, and transferred to a membrane-bound undecaprenyl phosphate before being flipped into the periplasmic space through a well-characterized enzymatic pathway (1, 2). Within the periplasm, a collection of penicillin-binding proteins create the rigid PG structure by catalyzing linkages between the disaccharides creating a glycan chain, as well as cross-links between adjacent peptide side chains (3–6).
Once assembled, newly formed PG undergoes considerable maturation, modification, and/or degradation depending on the growth phase or presence of environmental stressors. Numerous lytic enzymes have been identified that cleave at various locations in the PG structure (7, 8). These enzymes include peptidases and amidases that cleave the peptide side chain, as well as glucosaminidases, muramidases, and lytic transglycosylases that cleave the glycan chain. They function in diverse cellular processes including PG turnover, cell elongation, cell division, and the membrane insertion of transmembrane protein complexes such as for flagella and the secretion systems (7, 9). Other well-characterized PG modifications include acetylation, deacetylation, or N-glycolylation of the PG glycan chain. These modifications are known to provide resistance to the host immune response during infection and during bacterial competition (10–12). Lastly, the replacement of the terminal d-alanine with d-serine or d-lactate has been shown to impart resistance to some antimicrobials in Gram-positive organisms (13–15).
Traditional methods for examining PG composition, such as those developed by Glauner et al. in 1988 (16), have remained virtually unchanged for more than 30 years. These methods include purifying PG sacculi from whole bacterial cells, enzymatically digesting them into individual components called muropeptides, and separating muropeptides by reverse-phase HPLC followed by identification using MS. These techniques are complex and time-consuming; specifically, the identification and correlation of individual MS ions with their corresponding muropeptide structure remains a tedious manual step. Because of this complexity, relatively few in vivo PG compositional studies have been undertaken. More recently, several groups have begun to use feature extraction software (17, 18) or isotope-labeling methods (19) to automate the identification of MS ions and to expand the known composition of PG, primarily in Gram-positive organisms.
Here, we further advance these approaches by incorporating feature extraction with a library of known Gram-negative muropeptide structures. We analyzed the resulting spectra with differential analysis approaches that are typically used to detect and analyze large proteomic data sets. This enables the automated assessment of muropeptide identity and the comparison of muropeptide modifications to quantify changes in PG composition between different growth states. In addition, we incorporated a statistical 1D annotation approach (20) that allows for the comparison of global muropeptide compositional changes.
To test this approach, we assessed Pseudomonas aeruginosa PG composition under two distinct physiological growth conditions. P. aeruginosa is an important opportunistic Gram-negative pathogen that causes significant morbidity and mortality in immune-compromised individuals, such as those with cystic fibrosis. A significant decline in the overall prognosis for cystic fibrosis patients is associated with the switch from acute (motile) to chronic (sessile) P. aeruginosa infections. Chronic infections are linked to the ability of P. aeruginosa to grow as an aggregated community of bacteria commonly referred to as a biofilm (21–23). Once established, biofilms are difficult to eradicate and demonstrate enhanced resistance to antimicrobial treatments compared with their motile “planktonic” counterparts (24). Understanding the mechanisms that drive biofilm formation and the heightened antimicrobial resistance is critical for the treatment of chronic infections. To date, only one study has examined the effects of biofilm growth on PG composition within a Gram-positive Enterococcus bacterium (19).
Our workflow was designed to highlight both large- and small-scale compositional changes that occur during this physiological transition. Based on our results, we present the most detailed compositional analysis of P. aeruginosa PG to date. We identify a core assembly of PG that is represented in high abundance but that does not significantly change between the two growth states and that likely plays a structural role within the cell wall. We also identified an adaptive assembly of PG that is present in smaller amounts but that fluctuates considerably between growth states in response to physiological changes. The PG modifications that showed significant change during biofilm growth included amino acid substitutions of the peptide side chain, as well as modifications that indicate changes in the activity of amidases, deacetylases, and lytic transglycosylases. Our methodology provides a robust and reproducible workflow for accurately determining PG composition between samples that can be used to assess global PG fluctuations in response to changing growth conditions or external stimuli.
Results and discussion
Isolation and purification of peptidoglycan from biofilm and planktonic P. aeruginosa cells
To compare muropeptide composition between biofilm and planktonic P. aeruginosa cells, cultures were processed as described in Fig. 1. We grew both types of cells to match previous proteomic studies (25–27) that showed significant shifts in proteomes at 96 h compared with earlier time points. These proteome shifts included changes in abundance for proteins related to PG synthesis and modification, such as penicillin-binding proteins and lytic transglycosylases (25–27). Planktonic and biofilm cultures were initially inoculated with 5 × 108 cfu of P. aeruginosa PAO1, and after 96 h of growth, four times more cellular biomass was obtained, which is consistent with observed reductions in growth rate in biofilm cultures (25–28). The quantity of purified sacculi obtained from the planktonic cultures (58.6 ± 5.7 mg of sacculi) when compared with biofilm cultures (13.7 ± 1.6 mg of sacculi) remained constant per weight of cellular biomass, indicating that the reduction in purified sacculi in biofilm cultures was due to reduced growth rate. In addition, biofilm cultures showed considerable blue-green pigment production compared with their planktonically grown counterparts (Fig. S1, A and B), likely because of increased pyocyanin production (25, 29, 30). Lyophilized sacculi purified from planktonic cells showed a distinct reddish pigment as compared with the overall white color of the biofilm-derived sacculi (Fig. S1, C and D). To ensure that equivalent amounts of sacculi were used for subsequent MS experiments, PG was quantified and normalized based on muramic acid content because one muramic acid represents one monomer subunit (Fig. S1E). Transmission EM was used to assess sacculi purity and integrity (Fig. S1F).
Figure 1.
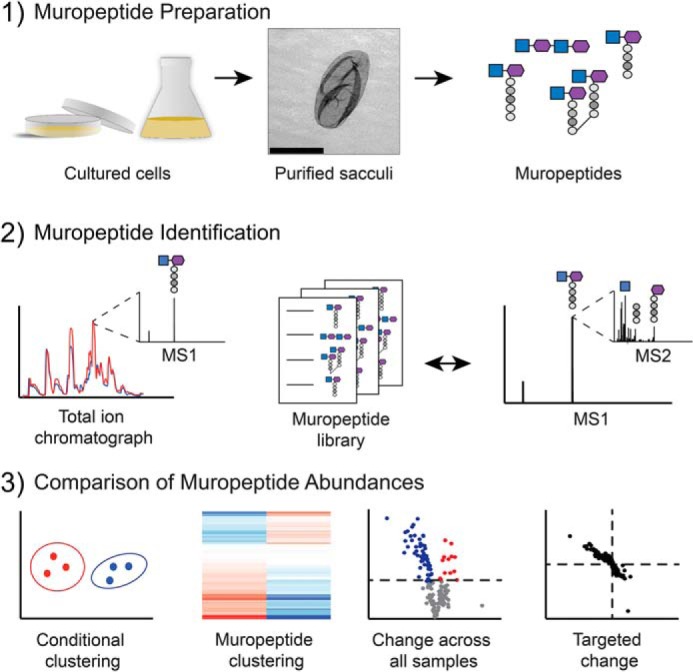
Peptidoglycomic workflow. Step 1, muropeptide preparation. Planktonic and biofilm P. aeruginosa cultures were grown in liquid medium or on 2% agar-containing medium, respectively. The peptidoglycan sacculi were extracted from all other cellular components and then digested into individual muropeptides using mutanolysin. Scale bar in middle panel, 2 μm. Step 2, muropeptide identification. Unique muropeptide m/z peaks were identified across the total ion chromatograph at all retention times and were compared with a library of predicted muropeptide m/z. MS/MS was then used to compare fragment sizes to predicted muropeptide fragmentation and repeated as necessary to confirm identity of muropeptide. Step 3, comparison of muropeptide abundances. Bioinformatic approaches were used to compare the differences in abundance of each muropeptide between culture conditions.
High-resolution muropeptide analysis
Isolated PG sacculi were digested with mutanolysin (31), and muropeptide composition was assessed using LC–Q-TOF-MS. The total ion chromatograph of single replicates of biofilm- and planktonic-derived PG showed differences in the total peak intensity at various retention times (Fig. 2A). To examine whether these differences represented genuine deviations in PG composition, we analyzed biological triplicate muropeptide samples (in technical triplicate; nine replicates in total per growth condition). Each sample was analyzed in the same sequence to decrease drift and other deviations in retention time between samples. A principal component analysis (PCA) of the data sets was performed. A PCA plot separates the data based on components inherent to the experiment (e.g. biological or technical differences). Here, a PCA of biofilm- and planktonic-derived PG muropeptides (Fig. 2B) demonstrates the largest distinction between the data sets to be the state of growth (component 1, 65.04%). Differences in replicate reproducibility is accounted for in component 2 (19.39%), but to a lesser degree, supports the importance of growth conditions to independently segregate the muropeptides.
Figure 2.
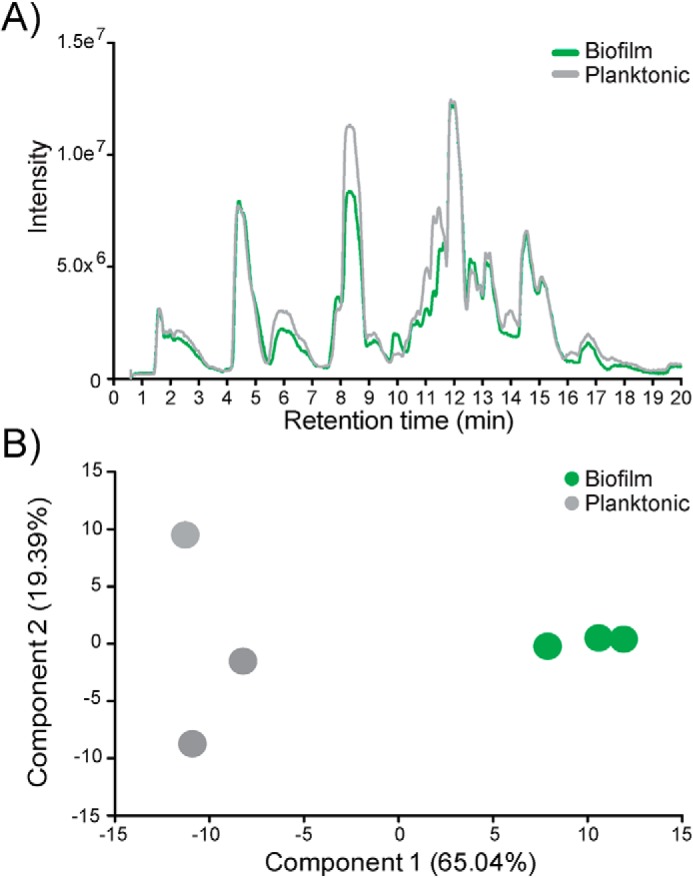
Mass spectrometry demonstrates gross compositional differences between biofilm- and planktonic-derived peptidoglycan. A, total ion chromatograph from Q-TOF-MS of biofilm-derived peptidoglycan (green) overlaid with planktonic-derived peptidoglycan (gray). B, PCA comparing biofilm-derived peptidoglycan (green) against planktonic-derived peptidoglycan (gray).
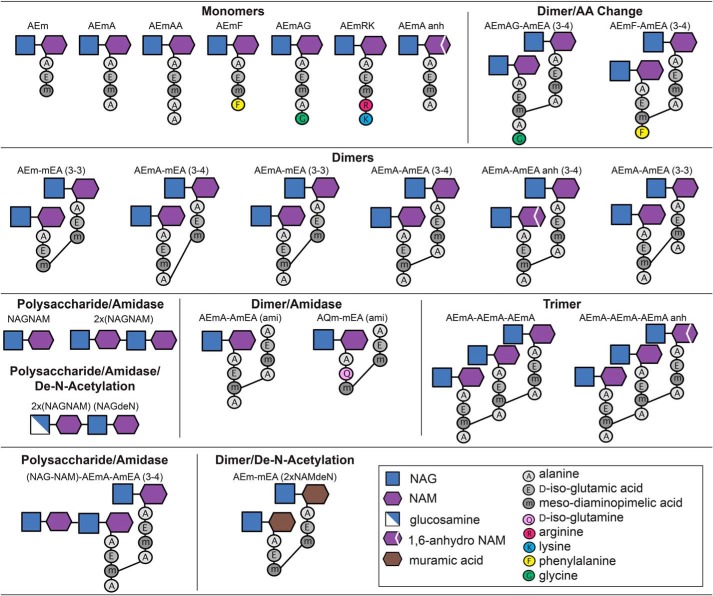
To identify individual muropeptides, the corresponding m/z was compared with an in-house prepared library comprised of >6000 potential muropeptide structures (Table S2) and confirmed using MS/MS. In total, 160 muropeptide structures were identified (Table S1), a selected subset of which is presented in Fig. 3. All of the muropeptides identified in this study were detected in both planktonic and biofilm growth conditions. Therefore, we use ion intensity as a measure of the abundance of individual muropeptides between planktonic- and biofilm-derived PG. This is not necessarily a measure of absolute molar abundance but rather an indication of muropeptide compositional changes between the growth states. However, relative abundance can be used within muropeptide groups (monomers, dimers, etc.) to compare molar stoichiometries, as described in previous studies (18). Of the 160 muropeptides identified, 106 have been assigned a structure with 68 confirmed by MS/MS (Table S1, in bold). However, because of the low abundance and uniqueness of the structures, we were unable to definitely assign structures for 54 muropeptides, which when combined represent ∼6% of the overall PG composition.
Figure 3.
Graphical representation of selected muropeptides identified in P. aeruginosa PAO1. Typical muropeptide characteristics included monomers, dimers, trimers, and tetramers. Dimers were determined to be either 3–4 or 3–3 cross-linked between muropeptide monomers. Muropeptide modifications included amino acid substitutions in the fourth and/or fifth position of the peptide side chain, as well as the resultant muropeptide products after deacetylase or amidase activity.
Growth phase-dependent changes in PG composition
A scatterplot of the MS raw intensity values showed that 10 muropeptides were the most abundant and constituted ∼70% of the overall PG composition within both growth conditions (Fig. 4A and Table S1, top 10 rows). The most abundant monomers were AEmA (which lacks the fifth alanine), AEm (which lacks the fourth and fifth alanine), and AEmAG (where the fifth alanine is substituted with a glycine) (Fig. 3). The most abundant cross-linked dimer was the muropeptide AEmA–AmEA (3–4), where (3–4) represents the formation of a peptide bond between the meso-diaminopimelic acid residue of one monomer and the alanine in the fourth position on the second monomer (Fig. 3). This dimer was also found in high abundance with both an 1,6-anhydro-NAM (anh) modification producing AEmA–AmEA (anh) (3–4), and β-1,4–linked to a NAG–NAM disaccharide, (NAG–NAM)-AEmA–AmEA (3–4) (Table S1). The second most abundant dimer in this study was the cross-linked AEmA-mEA (3–4) which has the fourth alanine on one monomer removed (Fig. 3). The trimer representing three cross-linked AEmA monomers, AEmA–AmEA–AEmA, and the anh-modified form, AEmA–AmEA–AEmA (anh), were also highly abundant in both growth conditions (Table S1). The remaining highest abundant muropeptide was AQm–mEA (ami), where the glutamic acid is changed to glutamine on one monomer, and the NAG–NAM disaccharide on one monomer was removed (Table S1).
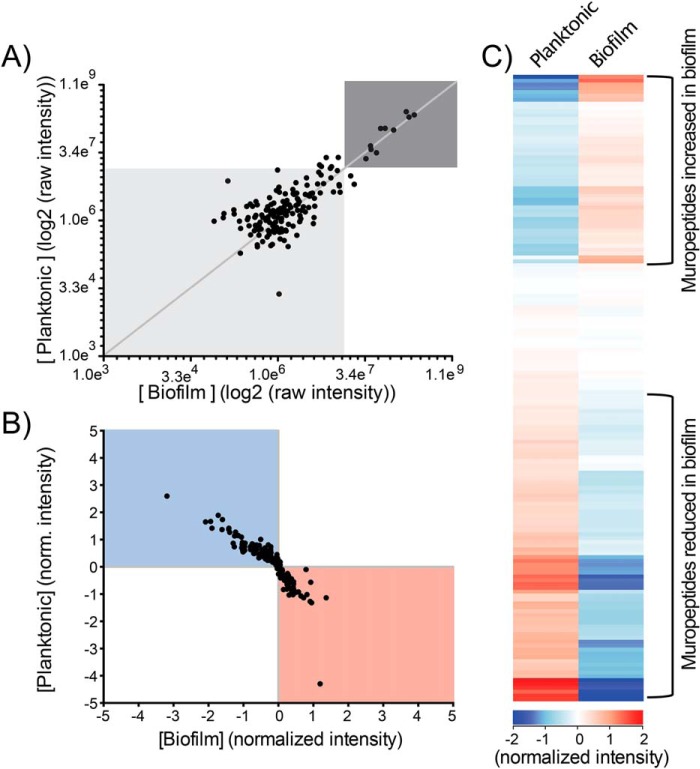
Figure 4.
Quantitative comparison of muropeptide abundance between biofilm- and planktonic-derived peptidoglycan. A, scatterplot of the raw intensity values (log2). Each dot represents the intensity of a single muropeptide recorded in both biofilm-derived (x axis) and planktonic-derived (y axis) peptidoglycan. B, scatterplot of the intensity of individual muropeptides normalized to the median intensity value across all samples. Each dot represents the normalized intensity of a single muropeptide recorded in both biofilm-derived (x axis) and planktonic-derived (y axis) peptidoglycan. C, hierarchical clustering analysis of the normalized intensity values. Each bar represents the abundance of one muropeptide in both the biofilm- and planktonic-derived peptidoglycan.
The remaining 150 muropeptides identified were present in lower abundance (Fig. 4A and Table S1). To reduce the influence of the highly abundant muropeptides, the intensity of each muropeptide was normalized to the median intensity of that muropeptide across all replicates of both growth conditions. A scatterplot of the normalized intensity displays the relative abundance of each muropeptide between the two growth conditions (Fig. 4B). Hierarchical clustering analysis of the normalized intensity demonstrates distinct sets of muropeptides that show a change in abundance in biofilm-derived compared with planktonic-derived PG (Fig. 4C). These changes in PG composition in biofilm-cultured P. aeruginosa can be clustered to indicate that ∼80 of the 160 muropeptides were decreased in abundance, whereas ∼50 muropeptides were increased in abundance in the biofilm-derived PG (Fig. 4C).
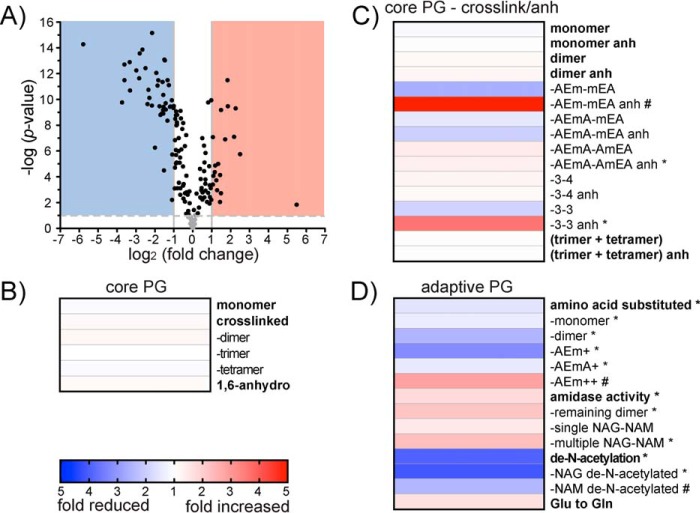
To determine whether the apparent differences in abundance were significant, a Student's t test was performed. To account for the rate of type I errors in null hypothesis testing when conducting multiple comparisons, we used a Benjamini–Hochberg false discovery rate (FDR) correction for the performed t tests (32). The corrected p value was plotted on a volcano plot against the fold change in abundance between the two growth conditions (Fig. 5A). Twenty-seven muropeptides showed no significant change in abundance between the two conditions (Fig. 5A, gray circles). In contrast, 133 muropeptides showed a significant change in abundance (Fig. 5A, black circles), where 19 muropeptides demonstrated a >2-fold increase (Fig. 5A, red area), and 43 demonstrated a >2-fold decrease in abundance (Fig. 5A, blue area) in biofilm-derived PG (Table S1).
Figure 5.
Changes in composition for biofilm- and planktonic-derived peptidoglycan. A, volcano plot where each dot represents an individual muropeptide plotted against the significance (Student's t test p value <0.05, FDR < 0.05, s0 = 1) and fold change (log2) in the biofilm-derived samples compared with the planktonic-derived samples. Lines represent the p value = 0.05 (horizontal lines) and a 2-fold change (two vertical lines +2 and −2 fold change). The red area indicates muropeptides that demonstrate a 2-fold or greater significant increase in biofilm-derived peptidoglycan. The blue area indicates muropeptides that demonstrate a 2-fold or greater significant decrease in biofilm-derived peptidoglycan. B, heat map of fold-change differences between biofilm- and planktonic-derived PG of core muropeptide characteristics. C, heat map of fold-change differences between biofilm- and planktonic-derived PG of detailed core muropeptide characteristics including variations in cross-linkages and variations in anh production. D, heat map of fold-change differences between biofilm- and planktonic-derived PG of adaptive muropeptide characteristics. * represents muropeptide characteristics enriched following 1D annotation (Student's t test p value < 0.05; FDR < 0.05, s0 = 1). # represents muropeptide characteristics with only one or two muropeptides within the category and could not be analyzed for significance using 1D annotation.
To determine which muropeptides were differentially represented in biofilm cells, muropeptides that showed a significant change in abundance between the two growth conditions were compiled into categories based on their structural characteristics (as described in Fig. 3) and assessed for enrichment using 1D annotation (Table 1 and Fig. 5, B and C). 1D annotation enrichment tests for every annotation term (e.g. muropeptide category) if the corresponding numerical values (e.g. intensity) have a preference to be systematically larger or smaller than the global distribution of the values for all muropeptides (20). The 1D annotation score represents the direction in which the muropeptide tends to deviate from the overall distribution of all muropeptides (i.e. a positive or negative enrichment of the muropeptide category).
Table 1.
Abundance differences of core and adaptive muropeptide characteristics identified in P. aeruginosa PAO1 between biofilm- and planktonic-derived PG
ND indicates categories with only one or two representative muropeptides, 1D annotation could not be performed.
| Muropeptide category | Biofilm | Planktonic | Difference | Fold change | 1D score |
|---|---|---|---|---|---|
| % | % | % | |||
| Core | |||||
| Monomer | 28.08 ± 1.32 | 30.31 ± 2.75 | −2.23 | 1.08 | −0.25 |
| Cross-linked | 47.65 ± 0.77 | 42.85 ± 4.55 | 4.80 | 1.11 | 0.10 |
| Dimer | 38.57 ± 0.47 | 33.63 ± 3.86 | 4.94 | 1.15 | 0.12 |
| Trimer | 8.65 ± 0.32 | 8.74 ± 0.68 | −0.09 | 1.01 | 0.01 |
| Tetramer | 0.43 ± 0.01 | 0.47 ± 0.03 | −0.04 | 1.09 | 0.10 |
| Glycan chain | 10.86 ± 0.99 | 7.03 ± 1.78 | 3.83 | 1.54 | 0.17 |
| 1,6-Anhydro | 14.94 ± 0.52 | 13.54 ± 1.48 | 1.40 | 1.11 | 0.20 |
| Detailed core | |||||
| Monomer | 27.80 ± 1.31 | 30.03 ± 2.70 | −2.23 | 1.08 | −0.28 |
| Monomer anh | 0.27 ± 0.01 | 0.27 ± 0.06 | 0.00 | 1.00 | ND |
| Dimer | 28.48 ± 0.35 | 24.94 ± 2.84 | 3.54 | 1.14 | −0.02 |
| Dimer anh | 10.09 ± 0.37 | 8.69 ± 1.04 | 1.40 | 1.16 | 0.29 |
| AEm–mEA | 0.45 ± 0.10 | 1.16 ± 0.13 | −0.71 | 2.58 | −0.51 |
| AEm–mEA anh | 0.31 ± 0.04 | 0.05 ± 0.02 | 0.26 | 4.81 | ND |
| AEmA–mEA | 5.07 ± 0.49 | 7.49 ± 0.78 | −2.42 | 1.48 | −0.35 |
| AEmA–mEA anh | 0.91 ± 0.10 | 1.77 ± 0.16 | −0.86 | 1.95 | −0.23 |
| AEmA–AmEA | 22.92 ± 0.57 | 16.27 ± 2.06 | 6.65 | 1.41 | 0.49 |
| AEmA–AmEA anh | 8.87 ± 0.26 | 6.87 ± 0.87 | 2.00 | 1.29 | 0.71a |
| 3–4-linked | 27.46 ± 0.33 | 23.24 ± 2.71 | 4.22 | 1.18 | 0.23 |
| 3–4-linked anh | 9.21 ± 0.32 | 8.40 ± 0.97 | 0.81 | 1.10 | 0.07 |
| 3–3-linked | 0.84 ± 0.10 | 1.58 ± 0.14 | −0.74 | 1.89 | −0.35 |
| 3–3-linked anh | 1.05 ± 0.10 | 0.29 ± 0.09 | 0.76 | 3.57 | 0.88a |
| Trimer | 4.42 ± 0.16 | 4.55 ± 0.31 | −0.13 | 1.03 | −0.08 |
| Trimer anh | 4.23 ± 0.18 | 4.20 ± 0.37 | 0.03 | 1.01 | 0.09 |
| Tetramer | 0.08 ± 0.01 | 0.09 ± 0.01 | −0.01 | 1.33 | ND |
| Tetramer anh | 0.35 ± 0.01 | 0.38 ± 0.03 | −0.03 | 1.09 | 0.10 |
| Adaptive | |||||
| Amino acid substitution | 8.78 ± 0.43 | 13.60 ± 1.20 | −4.83 | 1.55 | −0.46a |
| Monomer | 7.42 ± 0.33 | 10.27 ± 0.93 | −2.85 | 1.39 | −0.39a |
| Dimer | 1.36 ± 0.11 | 3.33 ± 0.29 | −1.97 | 2.45 | −0.43a |
| AEm+ | 1.03 ± 0.12 | 3.50 ± 0.51 | −2.47 | 3.40 | −0.51a |
| AEmA+ | 6.76 ± 0.31 | 9.73 ± 0.87 | −2.97 | 1.44 | −0.44a |
| AEm++ | 0.99 ± 0.05 | 0.36 ± 0.06 | 0.63 | 2.76 | ND |
| Amidase activity | 15.87 ± 1.47 | 9.29 ± 1.96 | 6.58 | 1.71 | −0.26a |
| Remaining dimer | 4.69 ± 0.39 | 2.26 ± 0.21 | 2.43 | 2.07 | −0.77a |
| Single NAG–NAM | 7.74 ± 0.57 | 5.38 ± 1.07 | 2.36 | 1.44 | −0.01 |
| Longer NAG–NAM chains | 3.60 ± 0.53 | 1.62 ± 0.73 | 1.98 | 2.23 | −0.54a |
| De-N-acetylation of NAG or NAM | 0.16 ± 0.01 | 0.65 ± 0.10 | −0.49 | 3.96 | 0.26a |
| NAG de-N-acetylation | 0.15 ± 0.01 | 0.61 ± 0.10 | −0.46 | 4.13 | −0.82a |
| NAM de-N-acetylation | 0.02 ± 0.00 | 0.04 ± 0.01 | −0.02 | 2.94 | ND |
| Glu changed to Gln at amino acid position 2 | 3.10 ± 0.22 | 1.95 ± 0.13 | 1.15 | 1.59 | 0.30 |
| Overall | |||||
| Muropeptides >1% of PG composition (10 total) | 73.61 ± 1.93 | 70.88 ± 6.81 | 2.73 | 1.04 | 0.06 |
| Below 1% | 21.60 ± 0.46 | 23.43 ± 2.88 | −1.83 | 1.08 | −0.10 |
| Unknown or to be determined | 5.76 ± 0.20 | 6.22 ± 0.83 | −0.46 | 1.08 | 0.07 |
a Student's t test p value < 0.05, FDR < 0.05, s0 = 1.
Based on these analyses, P. aeruginosa muropeptides could be divided into two distinct groups. The first group consisted of the most common muropeptides identified to date (16, 33) and were generally found in high abundance with only minor fluctuations between the growth conditions (Fig. 5, B and C). Therefore, we refer to these muropeptides as being part of the “core PG.” The second group consisted of muropeptides with less well-characterized modifications and were present in lower abundance with a higher degree of variability between growth conditions (Fig. 5D). We refer to these muropeptides as being part of the “adaptive PG.”
Muropeptide composition of the core PG
The core PG constitutes the major proportion of the Gram-negative sacculi and consists of muropeptides that have roles in PG synthesis or turnover (16, 33). These included modifications indicative of carboxypeptidase reactions producing the AEm and AEmA; d,d-transpeptidase and l,d-transpeptidase reactions producing the 3–4 and 3–3 cross-links, respectively, as well as lytic transglycosylase activity producing the NAM anh that represent terminal ends of the PG glycan chain (34). Based on fold changes and 1D annotation, we did not observe enrichment of these core muropeptide characteristics between the two growth conditions (Fig. 5B). The relatively low variability of these high-abundance muropeptides suggests that they are involved in maintaining the structural viability of the overall PG sacculi.
To delve deeper into the core PG composition, we reassessed the core muropeptides to group them by differences in cross-linking (including 3–4 and 3–3), as well as variations in anh modification (Fig. 5C). When separated by these parameters, higher-fold change differences were seen overall; however, only two categories demonstrated a significant change between the growth conditions. These categories were enriched in biofilm-grown cells and consisted of the AEmA–AmEA (anh) dimer and 3–3 linked (anh), which were enriched 1.3- and 3.6-fold, respectively (Table 1 and Fig. 5C). It is possible the anh-modified 3–3-linked AEmA–AmEA was responsible for the enrichment of the AEmA–AmEA (anh) muropeptide category (Table 1). We could not individually test the enrichment of 3–4- and 3–3-linked AEmA–AmEA using 1D annotation because of the limited number of muropeptides with these characteristics. However, the abundance of the single muropeptide, AEmA–AmEA (anh) (3–4), did not change between the two growth conditions (Table S1). Lytic transglycosylases form the anh product when cleaving the glycan chains to loosen the overall rigid PG structure to carry out numerous biological processes (7, 34, 35). There are six families of lytic transglycosylases based on sequence similarity (34, 36, 37), with each family potentially participating in distinct cellular functions (38). These include cellular processes, such as elongation, division septum formation, PG turnover, and the incorporation of macromolecular complexes, such as flagella and secretion systems (34, 38). In addition, the resulting anh muropeptides, such as AEm (anh), when removed from the PG during turnover, are well-known signal molecules that regulate the activity of various resistance mechanisms (39, 40). Also, the loss of lytic transglycosylase activity has been shown to reduce biofilm formation in a number of bacterial species (41–43). Therefore, the relevance of the anh formation on the 3–3 cross-link produced by l,d-transpeptidases that are a known bypass resistance mechanism for β-lactam antimicrobials (44, 45) may be of interest.
Muropeptide composition of the adaptive PG
The adaptive PG constitutes a smaller proportion of the overall PG; however, these muropeptides undergo significant fluctuations in abundance between the different culture conditions. It is estimated that Escherichia coli has ∼3.5 × 106 muropeptides/cell (46). Therefore, an individual muropeptide comprising 1.0% of the overall PG (Fig. 4A, light gray box) would amount to ∼35,000 copies/cell. Even a muropeptide comprising 0.01% of the total PG composition, which represents the lowest abundant muropeptide detected (Table S1), would still equal ∼350 copies/cell. The low-abundance adaptive muropeptides exhibited the highest fold difference between the biofilm and planktonic cells (Fig. 5D). It is the variability of this dynamic pool of diverse muropeptides that could allow for flexibility in response to growth or stimuli, whereas their low abundance would not affect overall structural viability. In P. aeruginosa, the adaptive muropeptide 1D annotation categories that showed distinct abundance differences between the two growth conditions were the amino acid substitutions of the peptide side chain, amidase activity that separates the NAM from the peptide side chain, and de-N-acetylation of the glycan chain (Fig. 5D).
The majority of Gram-negative muropeptides consist of peptide side chains with d-alanine as the terminal amino acid in positions 4 and 5 (Fig. 3). However, other amino acid substitutions can occur, with glycine as the most well-known (16, 47–49). In this study, we identified glycine, arginine, cysteine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, tyrosine, and valine incorporated into the peptide side chain at either the fourth and/or fifth position (Table S1). Glycine at position five (AEmAG) was the most abundant amino acid–substituted muropeptide. AEmAG was one of the 10 most abundant muropeptides (Table S1) and decreased in abundance 1.4-fold in biofilm-derived PG. Glycine was also found substituted in position four (AEmG) in very low abundance but demonstrated a much higher 4.0-fold decrease in biofilm-derived PG (Table S1). Overall, amino acid substitutions were reduced 1.5-fold in biofilm-derived PG compared with planktonic-derived PG. Amino acid substitutions on monomers were depleted 1.4-fold (Table 1 and Fig. 5D), whereas dimers containing one amino acid–substituted monomer were depleted 2.4-fold in biofilm-derived PG (Table 1 and Fig. 5D). We did not identify any trimers or tetramers with amino acid substitutions. However, we did identify numerous, as-yet-unknown, m/z peaks that were consistent with potential multimers, some of which may have amino acid substitutions (Table S1). We next determined whether there was a preference for the position on the peptide chain that was substituted. Substitution of the fifth amino acid (AEmA+) was approximately three times more prevalent than substitution of the fourth position (AEm+) (Table 1). However, it was the lower abundant AEm+ that had the greatest variation between the two growth conditions, with 3.4-fold depletion compared with 1.4-fold depletion of the AEmA+ (Table 1 and Fig. 5D). Only two muropeptides were identified with substitution of both the fourth and fifth amino acid (AEm++), which was AEmKR, found as both a monomer and as part of a dimer.
The incorporation of alternate amino acids into PG is generally dependent on their concentration within the growth medium (47, 50). Biofilms are known to form internal gradients and contain nutrient-limited zones (51–53). Therefore, the observed reduction of amino acid–substituted muropeptides in the biofilm could be a result of limited availability of these compounds within the biofilm. Additionally, exogenous application of d-amino acids can incorporate unique amino acids into muropeptides (50, 54, 55) and have been shown to reduce biofilm formation or disperse preformed biofilms (54, 56–58). Exogenous amino acids can also act synergistically with antimicrobials, producing an increased killing effect (56, 59). Furthermore, some bacteria (including P. aeruginosa) excrete d-amino acids into the medium in stationary phase (55). The amino acids excreted by P. aeruginosa into the media (55) correlates well with the amino acid–substituted muropeptides that we identified as increased in abundance in the planktonic-grown culture. Using the MS-based methods presented here, we are unable to determine whether the amino acid–substituted muropeptides contain d- or l-conformations. However, it is possible that the addition of nonstandard amino acids into PG decreases its stability, in turn increasing the susceptibility to antibiotics. This may, in part, contribute to the differences in antibiotic susceptibility observed between planktonic cells and biofilms. Both penicillin-binding proteins and l,d-transpeptidases (responsible for the production of 3–3 cross-links) have been shown to incorporate d-amino acids into PG (55, 60–62). In Vibrio cholera, l,d-transpeptidases catalyzed the incorporation of d-amino acids preferentially into the fourth position of the muropeptide side chain with other enzymes responsible for incorporation into the fifth position (61). The ortholog of the putative l,d-transpeptidase, PA2854, in the P. aeruginosa strain PA14 (PA14_27180) was shown to function in the incorporation of d-amino acids into the PG (61). In addition, the abundance of PA2854 increased in the planktonic-cultured P. aeruginosa strain PAO1 proteome at the 96-h time point (25–27). Therefore, the production of the 3–3 cross-links and the amino acid substitutions may be linked by the activity of a single enzyme, possibly PA2854.
Removal of the entire peptide side chain from the NAM is the result of enzymatic cleavage by periplasmic-localized amidase enzymes (7, 63). Amidase activity on a dimer results in both a NAG–NAM disaccharide and the remaining cross-linked peptide side chain, e.g. AEmA–AmEA (ami), as residual products (Fig. 3). Previous studies have only detected the amidase-cleaved dimer in PAO1 (64), which highlights the sensitivity of our methodology. Within P. aeruginosa PG, muropeptides containing a single NAG–NAM disaccharide did not show a significant change when growing as a biofilm (Fig. 5D). However, muropeptides containing longer NAG–NAM glycan chains of two, three, and four repeats and the amidase residual dimers showed a significant 2.2- and 2.1-fold enrichment, respectively, in biofilm-derived PG (Table 1 and Fig. 5D). Amidases function in combination with other hydrolases to break down the rigid structure of the PG for incorporation of transmembrane complexes, such as the type VI secretion system (9). The type VI secretion system is up-regulated in biofilm and are important for biofilm antimicrobial resistance (65, 66). Accordingly, an increase in macromolecular complexes within the biofilm culture could account for the increased PG amidase activity. However, PG amidase activity has been shown to be important for cell division (67), as well as peptidoglycan turnover (68–70), and amidase abundance is up-regulated by the CpxA–CpxR two component system in response to cell envelope stress in P. aeruginosa (71).
Changes to the acetylation state of the glycan chain are common modifications to Gram-negative PG and generally occurs as either an addition of an acetyl group to the C6 hydroxyl (O-acetylation) or a loss of the acetyl group on the C2 amine (de-N-acetylation) (10, 11). P. aeruginosa does not contain the genes necessary for adding the O-acetyl group (72, 73). However, PAO1 does possess the pa1517 gene, which is a homolog (26% amino acid identity) of the Helicobacter pylori enzyme responsible for the de-N-acetylation of the NAG (74). In P. aeruginosa, the de-N-acetylation of the glycan chain was depleted 4.0-fold in the biofilm-derived PG (Table 1 and Fig. 5D). NAG de-N-acetylation occurred more frequently, constituting a 4.1-fold decrease in the biofilm-derived PG and was found exclusively on the NAG–NAM disaccharide product of amidase activity (Fig. 3). This could indicate that the P. aeruginosa deacetylase may have an active site that limits substrate binding when the NAM is appended to a peptide side chain. Only a single muropeptide was identified containing a de-N-acetylation modification of the NAM residue, i.e. AEm-mEA (2xNAMdeN) (Fig. 3), which decreased 2.9-fold in biofilm-derived PG. Additional as-yet-unknown muropeptides have m/z values that indicate potential de-N-acetylation modifications, some of which may be NAM de-N-acetylated; however, none were confirmed by MS/MS. Changes to the acetylation of the glycan chain is known to provide protection from hydrolytic enzymes, such as lysozyme, produced by the host immune system during host–pathogen interactions (10, 75, 76). It is possible that planktonic-grown P. aeruginosa increases the content of the de-N-acetylation in preparation for environmental conditions where host immune stressors and/or bacterial competition may be higher.
Identification of multimeric and unique muropeptides
The high sensitivity of our experimental approach led to the identification of numerous muropeptide trimers and tetramers. The abundance of trimeric muropeptides detected here was ∼2-fold higher than reported previously for PAO1 (∼4%) (3, 77) but was not enriched in either culture condition (Fig. 5B). The fact that we detected not only tetramers but also two anh-containing forms is a testament to the sensitivity of this methodology. These multimeric muropeptides indicate connection points between several glycan chains and therefore may have significant structural importance. It was also interesting that tetramer formation occurred three times more often at an anh glycan chain termination. Whereas ∼50% of trimers, ∼25% of dimers, and only ∼1% of monomers occurred with an anh modification. The preference for glycan chain termination at a cross-linked muropeptide is also a feature found in H. pylori PG (78) and the PG-derived from stationary phase growth in E. coli (16). Further work into the effects these multimer muropeptides have on the overall ordered structure of PG will be needed.
When examining the abundance of muropeptides grouped by amino acid substitutions, several muropeptides, particularly AEmK, AEmKR, and AQmT-mQA (Fig. 3), were increased in abundance in biofilm-derived PG (Table S1), whereas all other muropeptides containing an amino acid substitution were depleted as mentioned above (Fig. 5C). AEmK increased in abundance 2.8-fold, AEmKR increased 2.8-fold, and AQmT-mQA increased 2.1-fold in the biofilm-derived PG (Table S1). The uniqueness of these muropeptides could indicate a divergent role for them within the biofilm-grown cultures compared with the remaining amino acid substituted muropeptides. AEmKR is the well-characterized attachment site of PG to the Braun's lipoprotein homolog OprI in P. aeruginosa (16, 79, 80). The attachment of the PG to the outer membrane through a lipoprotein is thought to increase the stability of the overall cell envelope (81). Therefore, the increased abundance of AEmKR indicates enhanced outer membrane attachment of PG and possibly a more stable cell envelope within biofilm-grown cells, which may increase the overall resistance of the bacteria within a biofilm to antimicrobial treatment.
Conclusions
Coupling high-resolution MS with software capable of handling complex data processing makes high-throughput analysis of PG composition more readily accessible and has been coined “peptidoglycomics” (82, 83). However, the use of this technology for the quantitative comparison of PG composition is, as of yet, highly limited (17, 18, 84). More specifically, we can now use the semiautomated pipeline described here to compare the detailed makeup of PG under a variety of growth or stress conditions. Using 1D annotation, we can assess the enrichment of muropeptide characteristics to make inferences concerning enzymatic activity. This will potentially provide novel insight as to how physiological and environmental conditions shape the enzymatic pathways involved in the synthesis and modification of a critical cell envelope component.
Gram-negative PG is thought to be relatively consistent in composition between species (33, 77, 85). We have shown that the Gram-negative bacterium P. aeruginosa PAO1 has a more varied PG composition than previously demonstrated (3, 64, 77, 85). We identified 160 unique muropeptides (Table S1), which represents eight times more muropeptides than previously shown for P. aeruginosa (3, 64, 77, 85) and double the number characterized for E. coli (16). This is the first report of de-N-acetylated muropeptides isolated from P. aeruginosa. In addition, this is the first demonstration of the high diversity and significant quantity of amino acid substituted muropeptides in Gram-negative PG. Therefore, to our knowledge, this approach has produced the most detailed PG compositional analyses of Pseudomonas or any Gram-negative bacterium to date.
Experimental procedures
Bacterial culture
Starter cultures of P. aeruginosa PAO1 were grown in tryptic soy broth (TSB) at 37 °C for 6 h to an approximate optical density at 600 nm of 2.0. The starter culture was used to inoculate 1 liter of TSB for planktonic cells, and 250 ml of TSB + 2% agar (TSA), each with 5 × 108 cfu. TSA cultures were allowed to dry briefly after inoculation. Planktonic cultures were grown with shaking at 200 rpm, whereas biofilm cultures were grown static. Both cultures were incubated for 96 h at 37 °C. For cell collection, planktonic cultures were centrifuged at 7,000 × g, whereas biofilm cultures were scraped off the surface of the TSA and collected into a 50-ml conical tube. Cell pellets were weighed and stored at −20 °C until further use. All samples were cultured in triplicate.
Isolation of peptidoglycan sacculi
PG sacculi were isolated from planktonic- and biofilm-grown cultures following the method of Glauner (86). Frozen cell pellets were suspended in 250 ml of cold (4 °C) 20 mm sodium phosphate buffer, pH 7.5. The suspension was added dropwise to an equal volume of boiling 8% SDS in 20 mm sodium phosphate buffer, pH 7.5, to a final volume of 500 ml in a round-bottomed flask with a water-cooled condenser. The samples were boiled with stirring for 3 h and allowed to cool overnight. The SDS was removed by repeated ultracentrifugation at 70,000 × g for 40 min and washing in 20 mm sodium phosphate buffer, pH 7.5. The concentration of SDS in the supernatant was determined using Stains-All (Sigma–Aldrich), prepared as per manufacturer's instructions; absorbance was read at 510 nm (87) and compared with an SDS standard curve. Washing continued until the concentration of SDS in the supernatant was ∼0.001%. Washed sacculi were suspended in 20 mm sodium phosphate buffer, pH 7.5, and incubated with 50 μg/ml each of amylase, DNase, and RNase for 1 h at 37 °C. Next, 100 μg/ml Pronase and ∼0.02% sodium azide were added and incubated for an additional 18 h at 37 °C with nutation. Samples were then incubated in 2% SDS at ∼100 °C for 1 h followed by removal of SDS by repeated ultracentrifugation at 70,000 × g for 40 min and washing with H2O until SDS concentration was ∼0.001%. Washed sacculi were suspended in H2O and lyophilized. The lyophilized PG sacculi were weighed and stored at room temperature until prepared for quantification and MS.
Quantification of NAM
The quantity of isolated PG was determined for both planktonic and biofilm samples using the method described by Clarke (88). Briefly, 10 mg/ml of purified sacculi were acid hydrolyzed with 6 m HCl for 2 h at 95 °C under vacuum in hand-pulled glass ampules. Ampules were opened and dried by heating under vacuum suspended over solid NaOH to neutralize acid. Hydrolyzed PG components were suspended in 10 mm NaOH and injected onto a CarboPac PA-20 column (Dionex Corp., Sunnyvale, CA), equilibrated with 100 mm NaOH for 10 min, and then separated by application of a 0 to 150 mm NaOAc linear gradient over 25 min, with detection using a pulsed-amperometric electrochemical sensor. The concentration of NAM in all samples was determined by peak integration using Chromelean version 7.2.0.3765 (Dionex Corp.) and compared with a muramic acid standard (Sigma–Aldrich).
Preparation of muropeptides for MS
Lyophilized PG sacculi were suspended to 10 mg/ml in H2O and sonicated for 20 s at 30% amplitude to disperse clumps. To produce muropeptides, 8 mg/ml of isolated sacculi were digested with 100 μg/ml mutanolysin in 100 mm ammonium acetate, pH 5.5, 50 mm MgCl2 for 18 h at 37 °C. Next, an equal volume of 0.5 m borate buffer, pH 9.0, with ∼10 mg/ml sodium borohydride was added and reacted for 20 min at room temperature. Finally, pH was adjusted to <4 with diluted (1:5) phosphoric acid and filtered using Nanasep MF 0.2-μm microcentrifuge filters (PALL Ltd., Mississauga, Canada).
Mass spectrometry
LC-MS analyses were performed on an Agilent 1200 HPLC liquid chromatograph interfaced with an Agilent UHD 6540 Q-TOF mass spectrometer at the Mass Spectrometry Facility of the Advanced Analysis Centre, University of Guelph. A C18 column (Agilent AdvanceBio Peptide Map, 50 mm × 2.1 mm × 2.7 μm) was used for chromatographic separation with the following solvents; water with 0.1% formic acid for A; and acetonitrile with 0.1% formic acid for B. The mobile phase gradient was as follows: initial conditions 2% B for 2 min increasing to 15% B in 13 min and then to 50% B for an additional 10 min followed by column wash at 98% B and 10 min re-equilibration. The first 2 and last 5 min of the gradient were sent to waste and not the spectrometer. The flow rate was maintained at 0.2 ml/min. The mass spectrometer electrospray capillary voltage was maintained at 4.0 kV, and the drying gas temperature was set at 350 °C with a flow rate of 13 liters/min. Nebulizer pressure was 40 p.s.i., and the fragmentor was set to 150 V. Nitrogen was used as both nebulizing and drying gas and collision-induced gas. The mass-to-charge (m/z) ratio was scanned across the m/z range of 200–2000 m/z in 4GHz (extended dynamic range positive-ion auto MS/MS mode. Three precursor ions per cycle were selected for fragmentation. The instrument was externally calibrated with the ESI TuneMix (Agilent Technologies Inc., Santa Clara, CA). The sample injection volume was 10 μl. Triplicate technical replicates were performed for all biological replicates. Selected samples were reinjected for further characterization of ambiguous muropeptides using the same chromatography and general MS conditions as above; however, we incorporated a preferred list of targets for MS/MS at specific m/z values and retention times. These targets were fragmented sequentially with 10, 15, 20, and 25 V collision energy. Raw MS and MS/MS spectra have been deposited into the publicly accessible repository Figshare under number 10277909.
Muropeptide library production
Peptidoglycan muropeptide characteristics selected for incorporation into the library were based on: 1) P. aeruginosa muropeptides published previously (64); 2) known PG compositions in Gram-negative bacteria (11, 16, 33, 64); 3) all possible amino acid substitutions on the stem peptide; and 4) combinations of muropeptide modifications thereof. Structures were produced using ChemDraw Prime version 16.0.1.4 (PerkinElmer Life Science), and the molecular formula for each was manually entered into the MassHunter Personal Compound Database and Library version B.07.00 (Agilent Technologies Inc.). In total >6000 different muropeptide structures were included (Fig. S2).
Data analysis
Spectra processing was performed using batch untargeted recursive muropeptide feature extraction in MassHunter Profinder version B08.00 (Agilent Technologies Inc.). Profinder recursive feature extraction involves an initial naïve feature finding algorithm, molecular feature extraction, which combines coeluting related ions such as adducts or different charge states into one compound. This list of compounds is then verified in a second round of feature finding with the find-by-formula algorithm, which uses the ion m/z values and isotope ratios found by molecular feature extraction to reinterrogate the data. In this way, the Profinder software combines related m/z into single compounds, verifies the extracted compounds, and reduces false positives and negatives. The initial m/z threshold set for feature detection was 300 counts and extraction window of 40 ppm using the Molecular Feature Extraction algorithm. After isotope grouping using the peptide isotope model, the compound threshold was set to 3000 counts in at least two-thirds of samples in one group. For the recursive portion of the feature detection, a list of consensus muropeptides determined from all samples was used to reassess the raw data using the find-by-ion algorithm using a 50 ppm extracted ion chromatograph extraction window. Statistical analysis was then preformed in Mass Profiler Professional (MPP) version 14.9.1 (Agilent Technologies, Inc.). During import of the Profinder prealigned data, coeluting m/z values representing in-source loss of NAG were merged into a single compound. Prior to merging the compounds, MS/MS spectra were assessed to confirm the loss of the NAG. The MPP data were normalized using the NAM quantification and baselined to the median intensity of each individual muropeptide across samples. Significantly different (p value < 0.05) muropeptides between growth conditions were identified using a moderated t test and corrected using Benjamini–Hochberg FDR. Unsupervised hierarchical clustering of the normalized intensity values was used to group the data. Muropeptides were identified using MPP IDBrowser Identification combined with the muropeptide MassHunter Personal Compound Database and Library and confirmed using the MS/MS spectra (Fig. S2). A list of muropeptides that could not be identified were exported for preferred MS/MS fragmentation and then verified manually. For 1D annotations, the normalized intensity values from MPP of all muropeptides that had a significant FDR < 0.05 were uploaded into Perseus version 1.6.2.2 (89) and assigned into categories based on the muropeptide characteristics. Each characteristic category was assessed for significance (Student's t test, p value < 0.05, FDR < 0.05, s0 = 1) in Perseus using 1D annotation (20). All graphs were produced in Prism 5.0f (GraphPad Software Inc., San Diego, CA).
Transmission EM of peptidoglycan sacculi
PG sacculi were processed for transmission EM as previously described (90). Sacculi were adhered to 200-mesh carbon-coated copper grids and washed three times with H2O, wicking away liquid between washes with #1 Whatman filter paper. The grids were incubated with 0.5% BSA for 5 min before washing again with H2O as before. Sacculi were stained with 1% uranyl acetate, air-dried, and viewed with a single-tilt holder on an FEI Tecnai G2 F20 transmission electron microscope operating at an accelerating voltage of 200 kV and equipped with a bottom-mount Gatan 4k charge-coupled device camera.
Author contributions
E. M. A., D. B., and C. M. K. conceptualization; E. M. A., D. S., D. B., A. J. C., J. G.-M., and C. M. K. data curation; E. M. A., D. B., A. J. C., J. G.-M., and C. M. K. formal analysis; A. J. C., and C. M. K. supervision; E. M. A., A. J. C., and C. M. K. funding acquisition; E. M. A., A. J. C., J. G.-M., and C. M. K. validation; E. M. A., D. S., D. B., A. J. C., and J. G.-M. investigation; E. M. A., D. B., J. G.-M., and C. M. K. visualization; E. M. A., D. S., D. B., A. J. C., J. G.-M., and C. M. K. methodology; E. M. A., D. B. and C. M. K. writing-original draft; E. M. A. and C. M. K. project administration; E. M. A., D. S., D. B., A. J. C., J. G.-M., and C. M. K. writing-review and editing.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Operating Grants PJT 156111 (to C. M. K.) and PJT 156353 (to A. J. C.) and a scholarship from the Alexander Graham Bell Canada Graduate Scholarships–Doctoral Program of the Natural Sciences and Engineering Research Council of Canada (to E. M. A.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1 and S2.
- PG
- peptidoglycan
- NAG
- N-acetylglucosamine
- NAM
- N-acetylmuramic acid
- FDR
- false discovery rate
- PCA
- principal component analysis
- anh
- 1,6-anhydro-NAM
- TSB
- tryptic soy broth
- MPP
- Mass Profiler Professional
- Ami
- dimer product of amidase activity.
References
- 1. Typas A., Banzhaf M., Gross C. A., and Vollmer W. (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Teo A. C., and Roper D. I. (2015) Core steps of membrane-bound peptidoglycan biosynthesis: Recent advances, insight and opportunities. Antibiotics 4, 495–520 10.3390/antibiotics4040495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ropy A., Cabot G., Sánchez-Diener I., Aguilera C., Moya B., Ayala J. A., and Oliver A. (2015) Role of Pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, β-lactam resistance, and peptidoglycan structure. Antimicrob. Agents Chemother. 59, 3925–3934 10.1128/AAC.05150-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liao X., and Hancock R. E. (1997) Identification of a penicillin-binding protein 3 homolog, PBP3x, in Pseudomonas aeruginosa: gene cloning and growth phase-dependent expression. J. Bacteriol. 179, 1490–1496 10.1128/jb.179.5.1490-1496.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Legaree B. A., Daniels K., Weadge J. T., Cockburn D., and Clarke A. J. (2007) Function of penicillin-binding protein 2 in viability and morphology of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 59, 411–424 10.1093/jac/dkl536 [DOI] [PubMed] [Google Scholar]
- 6. Liao X., and Hancock R. E. (1995) Cloning and characterization of the Pseudomonas aeruginosa pbpB gene encoding penicillin-binding protein 3. Antimicrob. Agents Chemother. 39, 1871–1874 10.1128/AAC.39.8.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vollmer W., Joris B., Charlier P., and Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 10.1111/j.1574-6976.2007.00099.x [DOI] [PubMed] [Google Scholar]
- 8. Johnson J. W., Fisher J. F., and Mobashery S. (2013) Bacterial cell-wall recycling. Ann. N.Y. Acad. Sci. 1277, 54–75 10.1111/j.1749-6632.2012.06813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scheurwater E. M., and Burrows L. L. (2011) Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett. 318, 1–9 10.1111/j.1574-6968.2011.02228.x [DOI] [PubMed] [Google Scholar]
- 10. Yadav A. K., Espaillat A., and Cava F. (2018) Bacterial strategies to preserve cell wall integrity against environmental threats. Front. Microbiol. 9, 2064 10.3389/fmicb.2018.02064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vollmer W. (2008) Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32, 287–306 10.1111/j.1574-6976.2007.00088.x [DOI] [PubMed] [Google Scholar]
- 12. Brott A. S., and Clarke A. J. (2019) Peptidoglycan O-acetylation as a virulence factor: its effect on lysozyme in the innate immune system. Antibiotics. 8, E94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grohs P., Gutmann L., Legrand R., Schoot B., and Mainardi J. L. (2000) Vancomycin resistance is associated with serine-containing peptidoglycan in Enterococcus gallinarum. J. Bacteriol. 182, 6228–6232 10.1128/JB.182.21.6228-6232.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Jonge B. L., Gage D., and Xu N. (2002) The carboxyl terminus of peptidoglycan stem peptides is a determinant for methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46, 3151–3155 10.1128/AAC.46.10.3151-3155.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Putty S., Vemula H., Bobba S., and Gutheil W. G. (2013) A liquid chromatography–tandem mass spectrometry assay for d-Ala-d-Lac: a key intermediate for vancomycin resistance in vancomycin-resistant Enterococci. Anal. Biochem. 442, 166–171 10.1016/j.ab.2013.07.045 [DOI] [PubMed] [Google Scholar]
- 16. Glauner B., Höltje J. V., and Schwarz U. (1988) The composition of the murein of Escherichia coli. J. Biol. Chem. 263, 10088–10095 [PubMed] [Google Scholar]
- 17. van der Aart L. T., Spijksma G. K., Harms A., Vollmer W., Hankemeier T., and van Wezel G. P. (2018) High-resolution analysis of the peptidoglycan composition in Streptomyces coelicolor. J. Bacteriol. 200, e00290–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bern M., Beniston R., and Mesnage S. (2017) Towards an automated analysis of bacterial peptidoglycan structure. Anal. Bioanal. Chem. 409, 551–560 10.1007/s00216-016-9857-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang J. D., Wallace A. G., Foster E. E., and Kim S. J. (2018) Peptidoglycan compositional analysis of Enterococcus faecalis biofilm by stable isotope labeling by amino acids in a bacterial culture. Biochemistry 57, 1274–1283 10.1021/acs.biochem.7b01207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox J., and Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12 10.1186/1471-2105-13-S16-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh P. K., Schaefer A. L., Parsek M. R., Moninger T. O., Welsh M. J., and Greenberg E. P. (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764 10.1038/35037627 [DOI] [PubMed] [Google Scholar]
- 22. Garcia-Medina R., Dunne W. M., Singh P. K., and Brody S. L. (2005) Pseudomonas aeruginosa acquires biofilm-like properties within airway epithelial cells. Infect. Immun. 73, 8298–8305 10.1128/IAI.73.12.8298-8305.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreau-Marquis S., Stanton B. A., and O'Toole G. A. (2008) Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21, 595–599 10.1016/j.pupt.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stewart P. S. (2015) Antimicrobial tolerance in biofilms. Microbiol. Spectr. 3, MB-0010-2014 10.1128/microbiolspec.MB-0010-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park A. J., Murphy K., Surette M. D., Bandoro C., Krieger J. R., Taylor P., and Khursigara C. M. (2015) Tracking the dynamic relationship between cellular systems and extracellular subproteomes in Pseudomonas aeruginosa biofilms. J. Proteome Res. 14, 4524–4537 10.1021/acs.jproteome.5b00262 [DOI] [PubMed] [Google Scholar]
- 26. Park A. J., Murphy K., Krieger J. R., Brewer D., Taylor P., Habash M., and Khursigara C. M. (2014) A temporal examination of the planktonic and biofilm proteome of whole cell Pseudomonas aeruginosa PAO1 using quantitative mass spectrometry. Mol. Cell. Proteomics 13, 1095–1105 10.1074/mcp.M113.033985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park A. J., Surette M. D., and Khursigara C. M. (2014) Antimicrobial targets localize to the extracellular vesicle-associated proteome of Pseudomonas aeruginosa grown in a biofilm. Front. Microbiol. 5, 464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilbert P., Collier P. J., and Brown M. R. W. (1990) Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob. Agents Chemother. 34, 1865–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramos I., Dietrich L. E., Price-Whelan A., and Newman D. K. (2010) Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 161, 187–191 10.1016/j.resmic.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meirelles L. A., and Newman D. K. (2018) Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol. Microbiol. 110, 995–1010 10.1111/mmi.14132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schaub R. E., and Dillard J. P. (2017) Digestion of peptidoglycan and analysis of soluble fragments. Bio-protocol 7, e2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. Methodol. 57, 289–300 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 33. Vollmer W., Blanot D., and de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 10.1111/j.1574-6976.2007.00094.x [DOI] [PubMed] [Google Scholar]
- 34. Dik D. A., Marous D. R., Fisher J. F., and Mobashery S. (2017) Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Crit. Rev. Biochem. Mol. Biol. 52, 503–542 10.1080/10409238.2017.1337705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee M., Hesek D., Dik D. A., Fishovitz J., Lastochkin E., Boggess B., Fisher J. F., and Mobashery S. (2017) From genome to proteome to elucidation of reactions for all eleven known lytic transglycosylases from Pseudomonas aeruginosa. Angew. Chem. Int. Ed. Engl. 56, 2735–2739 10.1002/anie.201611279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blackburn N. T., and Clarke A. J. (2001) Identification of four families of peptidoglycan lytic transglycosylases. J. Mol. Evol. 52, 78–84 10.1007/s002390010136 [DOI] [PubMed] [Google Scholar]
- 37. Herlihey F. A., and Clarke A. J. (2017) Controlling autolysis during flagella insertion in Gram-negative bacteria. In Advances in Experimental Medicine and Biology: Protein Reviews (Zouhairratassi M., ed) pp. 41–56, Springer Nature, Singapore: [DOI] [PubMed] [Google Scholar]
- 38. Scheurwater E., Reid C. W., and Clarke A. J. (2008) Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40, 586–591 10.1016/j.biocel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 39. Fisher J. F., and Mobashery S. (2014) The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative enterobacteriaceae and Pseudomonas aeruginosa. Bioorg. Chem. 56, 41–48 10.1016/j.bioorg.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dhar S., Kumari H., Balasubramanian D., and Mathee K. (2018) Cell-wall recycling and synthesis in Escherichia coli and Pseudomonas aeruginosa: their role in the development of resistance. J. Med. Microbiol. 67, 1–21 10.1099/jmm.0.000636 [DOI] [PubMed] [Google Scholar]
- 41. Crépin S., Ottosen E. N., Peters K., Smith S. N., Himpsl S. D., Vollmer W., and Mobley H. L. T. (2018) The lytic transglycosylase MltB connects membrane homeostasis and in vivo fitness of Acinetobacter baumannii. Mol. Microbiol. 109, 745–762 10.1111/mmi.14000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Neudorf K. D., and Yost C. K. (2017) An uncharacterized gene coding a conserved lytic transglycosylase domain (RL4716) is required for proper cell envelope function in Rhizobium leguminosarum. FEMS Microbiol. Lett. 364, fxn035 10.1093/femsle/fnx035 [DOI] [PubMed] [Google Scholar]
- 43. Monteiro C., Fang X., Ahmad I., Gomelsky M., and Römling U. (2011) Regulation of biofilm components in Salmonella enterica serovar typhimurium by lytic transglycosylases involved in cell wall turnover. J. Bacteriol. 193, 6443–6451 10.1128/JB.00425-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mainardi J.-L., Fourgeaud M., Hugonnet J.-E., Dubost L., Brouard J.-P., Ouazzani J., Rice L. B., Gutmann L., and Arthur M. (2005) A novel peptidoglycan cross-linking enzyme for β-lactam-resistant transpeptidation pathway. J. Biol. Chem. 280, 38146–38152 10.1074/jbc.M507384200 [DOI] [PubMed] [Google Scholar]
- 45. Hugonnet J. E., Mengin-Lecreulx D., Monton A., den Blaauwen T., Carbonnelle E., Veckerlé C., Brun Y. V., van Nieuwenhze M., Bouchier C., Tu K., Rice L. B., and Arthur M. (2016) Factors essential for l,d-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. Elife 5, e19469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wientjes F. B., Woldringh C. L., and Nanninga N. (1991) Amount of peptidoglycan in cell walls of Gram-negative bacteria. J. Bacteriol. 173, 7684–7691 10.1128/jb.173.23.7684-7691.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takacs C. N., Hocking J., Cabeen M. T., Bui N. K., Poggio S., Vollmer W., and Jacobs-Wagner C. (2013) Growth medium-dependent glycine incorporation into the peptidoglycan of Caulobacter crescentus. PLoS One 8, e57579 10.1371/journal.pone.0057579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hammes W., Schleifer K. H., and Kandler O. (1973) Mode of action of glycine on the biosynthesis of peptidoglycan. J. Bacteriol. 116, 1029–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Jonge B. L., Chang Y.-S., Xu N., and Gage D. (1996) Effect of exogenous glycine on peptidoglycan composition and resistance in a methicillin-resistant Staphylococcus aureus strain. Antimicrob. Agents Chemother. 40, 1498–1503 10.1128/AAC.40.6.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Caparrós M., Pisabarro A. G., and de Pedro M. A. (1992) Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J. Bacteriol. 174, 5549–5559 10.1128/jb.174.17.5549-5559.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tolker-Nielsen T. (2015) Biofilm development. Microbiol. Spectr. 3, MB-0001-2014 [DOI] [PubMed] [Google Scholar]
- 52. Pamp S. J., Gjermansen M., Johansen H. K., and Tolker-Nielsen T. (2008) Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68, 233–240 10.1111/j.1365-2958.2008.06152.x [DOI] [PubMed] [Google Scholar]
- 53. Werner E., Roe F., Bugnicourt A., Franklin M. J., Heydorn A., Molin S., Pitts B., and Stewart P. S. (2004) Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70, 6188–6196 10.1128/AEM.70.10.6188-6196.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kolodkin-Gal I., Romero D., Cao S., Clardy J., Kolter R., and Losick R. (2010) d-Amino acids trigger biofilm disassembly. Science 328, 627–629 10.1126/science.1188628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lam H., Oh D. C., Cava F., Takacs C. N., Clardy J., de Pedro M. A., and Waldor M. K. (2009) d- Amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555 10.1126/science.1178123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sanchez C. J. Jr., Akers K. S., Romano D. R., Woodbury R. L., Hardy S. K., Murray C. K., and Wenke J. C. (2014) d-Amino acids enhance the activity of antimicrobials against biofilms of clinical wound isolates of Staphylococcus aureus and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 58, 4353–4361 10.1128/AAC.02468-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ramón-Peréz M. L., Diaz-Cedillo F., Ibarra J. A., Torales-Cardeña A., Rodríguez-Martínez S., Jan-Roblero J., Cancino-Diaz M. E., and Cancino-Diaz J. C. (2014) d-Amino acids inhibit biofilm formation in Staphylococcus epidermidis strains from ocular infections. J. Med. Microbiol. 63, 1369–1376 10.1099/jmm.0.075796-0 [DOI] [PubMed] [Google Scholar]
- 58. Hochbaum A. I., Kolodkin-Gal I., Foulston L., Kolter R., Aizenberg J., and Losick R. (2011) Inhibitory effects of d-amino acids on Staphylococcus aureus biofilm development. J. Bacteriol. 193, 5616–5622 10.1128/JB.05534-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. She P., Chen L., Liu H., Zou Y., Luo Z., Koronfel A., and Wu Y. (2015) The effects of d-tyrosine combined with amikacin on the biofilms of Pseudomonas aeruginosa. Microb. Pathog. 86, 38–44 10.1016/j.micpath.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 60. Lupoli T. J., Tsukamoto H., Doud E. H., Wang T.-S., Walker S., and Kahne D. (2011) Transpeptidase-mediated incorporation of d-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 133, 10748–10751 10.1021/ja2040656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cava F., de Pedro M. A., Lam H., Davis B. M., and Waldor M. K. (2011) Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J. 30, 3442–3453 10.1038/emboj.2011.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Peters K., Pazos M., Edoo Z., Hugonnet J. E., Martorana A. M., Polissi A., VanNieuwenhze M. S., Arthur M., and Vollmer W. (2018) Copper inhibits peptidoglycan l,d-transpeptidases suppressing β-lactam resistance due to bypass of penicillin-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 115, 10786–10791 10.1073/pnas.1809285115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vermassen A., Leroy S., Talon R., Provot C., Popowska M., and Desvaux M. (2019) Cell wall hydrolases in bacteria: Insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 10, 331 10.3389/fmicb.2019.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee M., Dhar S., De Benedetti S., Hesek D., Boggess B., Blázquez B., Mathee K., and Mobashery S. (2016) Muropeptides in Pseudomonas aeruginosa and their role as elicitors of β-lactam-antibiotic resistance. Angew. Chem. Int. Ed. Engl. 55, 6882–6886 10.1002/anie.201601693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bernard C. S., Brunet Y. R., Gueguen E., and Cascales E. (2010) Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192, 3850–3860 10.1128/JB.00370-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang L., Hinz A. J., Nadeau J.-P., and Mah T.-F. (2011) Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J. Bacteriol. 193, 5510–5513 10.1128/JB.00268-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yakhnina A. A., McManus H. R., and Bernhardt T. G. (2015) The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance, and viability. Mol. Microbiol. 97, 957–973 10.1111/mmi.13077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Moya B., Juan C., Albertí S., Pérez J. L., and Oliver A. (2008) Benefit of having multiple ampD genes for acquiring β-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52, 3694–3700 10.1128/AAC.00172-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang W., Lee M., Hesek D., Lastochkin E., Boggess B., and Mobashery S. (2013) Reactions of the three AmpD enzymes of Pseudomonas aeruginosa. J. Am. Chem. Soc. 135, 4950–4953 10.1021/ja400970n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rivera I., Molina R., Lee M., Mobashery S., and Hermoso J. A. (2016) Orthologous and paralogous AmpD peptidoglycan amidases from Gram-negative bacteria. Microb. Drug Resist. 22, 470–476 10.1089/mdr.2016.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Weatherspoon-Griffin N., Zhao G., Kong W., Kong Y., Morigen Andrews-Polymenis H., McClelland M., and Shi Y. (2011) The CpxR/CpxA two-component system up-regulates two tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J. Biol. Chem. 286, 5529–5539 10.1074/jbc.M110.200352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Clarke C. A., Scheurwater E. M., and Clarke A. J. (2010) The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J. Biol. Chem. 285, 14843–14847 10.1074/jbc.C110.120931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Clarke A. J. (1993) Extent of peptidoglycan O-acetylation in the tribe Proteeae. J. Bacteriol. 175, 4550–4553 10.1128/jb.175.14.4550-4553.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang G., Olczak A., Forsberg L. S., and Maier R. J. (2009) Oxidative stress-induced peptidoglycan deacetylase in Helicobacter pylori. J. Biol. Chem. 284, 6790–6800 10.1074/jbc.M808071200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sychantha D., Brott A. S., Jones C. S., and Clarke A. J. (2018) Mechanistic pathways for peptidoglycan O-acetylation and de-O-acetylation. Front. Microbiol. 9, 2332 10.3389/fmicb.2018.02332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang G., Maier S. E., Lo L. F., Maier G., Dosi S., and Maier R. J. (2010) Peptidoglycan deacetylation in Helicobacter pylori contributes to bacterial survival by mitigating host immune responses. Infect. Immun. 78, 4660–4666 10.1128/IAI.00307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Quintela J. C., Caparrós M., and de Pedro M. A. (1995) Variability of peptidoglycan structural parameters in Gram-negative bacteria. FEMS Microbiol. Lett. 125, 95–100 10.1111/j.1574-6968.1995.tb07341.x [DOI] [PubMed] [Google Scholar]
- 78. Costa K., Bacher G., Allmaier G., Dominguez-Bello M. G., Engstrand L., Falk P., de Pedro M. A., and García-del Portillo F. (1999) The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J. Bacteriol. 181, 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Braun V., and Bosch V. (1972) Sequence of the murein·lipoprotein and the attachment site of the lipid. Eur. J. Biochem. 28, 51–69 10.1111/j.1432-1033.1972.tb01883.x [DOI] [PubMed] [Google Scholar]
- 80. Duchêne M., Barron C., Schweizer A., von Specht B. U., and Domdey H. (1989) Pseudomonas aeruginosa outer membrane lipoprotein I gene: Molecular cloning, sequence, and expression in Escherichia coli. J. Bacteriol. 171, 4130–4137 10.1128/jb.171.8.4130-4137.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Asmar A. T., and Collet J.-F. (2018) Lpp, the Braun lipoprotein, turns 50: major achievements and remaining issues. FEMS Microbiol. Lett. 365, 10.1093/femsle/fny199 [DOI] [PubMed] [Google Scholar]
- 82. Wheeler R., Chevalier G., Eberl G., and Gomperts Boneca I. (2014) The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell. Microbiol. 16, 1014–1023 10.1111/cmi.12304 [DOI] [PubMed] [Google Scholar]
- 83. Pomin V. H. (2012) Current glycomics' approaches: subprojects and journals. J. Glycomics Lipidomics 2, 1000e102 10.4172/2153-0637.1000e102 [DOI] [Google Scholar]
- 84. Espaillat A., Forsmo O., El Biari K., Björk R., Lemaitre B., Trygg J., Cañada F. J., de Pedro M. A., and Cava F. (2016) Chemometric analysis of bacterial peptidoglycan reveals atypical modifications that empower the cell wall against predatory enzymes and fly innate immunity. J. Am. Chem. Soc. 138, 9193–9204 10.1021/jacs.6b04430 [DOI] [PubMed] [Google Scholar]
- 85. Torrens G., Escobar-Salom M., Pol-Pol E., Camps-Munar C., Cabot G., López-Causapé C., Rojo-Molinero E., Oliver A., and Juan C. (2019) Comparative analysis of peptidoglycans from Pseudomonas aeruginosa isolates recovered from chronic and acute infections. Front. Microbiol. 10, 1868 10.3389/fmicb.2019.01868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Glauner B. (1988) Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172, 451–464 10.1016/0003-2697(88)90468-X [DOI] [PubMed] [Google Scholar]
- 87. Rusconi F., Valton E., Nguyen R., and Dufourc E. (2001) Quantification of sodium dodecyl sulfate in microliter-volume biochemical samples by visible light spectroscopy. Anal. Biochem. 295, 31–37 10.1006/abio.2001.5164 [DOI] [PubMed] [Google Scholar]
- 88. Clarke A. J. (1993) Compositional analysis of peptidoglycan by high-performance anion-exchange chromatography. Anal. Biochem. 212, 344–350 10.1006/abio.1993.1339 [DOI] [PubMed] [Google Scholar]
- 89. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 90. Priyadarshini R., de Pedro M. A., and Young K. D. (2007) Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J. Bacteriol. 189, 5334–5347 10.1128/JB.00415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.