Abstract
Caloric restriction has been associated with increased life span and reduced aging-related disorders and reduces fibrosis in several diseases. Fibrosis is characterized by deposition of excess fibrous material in tissues and organs and is caused by aging, chronic stress, injury, or disease. Myofibroblasts are fibroblast-like cells that secrete high levels of extracellular matrix proteins, resulting in fibrosis. Histological studies have identified many-fold increases of myofibroblasts in aged organs where myofibroblasts are constantly generated from resident tissue fibroblasts and other cell types. However, it remains unclear how aging increases the generation of myofibroblasts. Here, using mouse models and biochemical assays, we show that sirtuin 6 (SIRT6) deficiency plays a major role in aging-associated transformation of fibroblasts to myofibroblasts, resulting in tissue fibrosis. Our findings suggest that SIRT6-deficient fibroblasts transform spontaneously to myofibroblasts through hyperactivation of transforming growth factor β (TGF-β) signaling in a cell-autonomous manner. Importantly, we noted that SIRT6 haploinsufficiency is sufficient for enhancing myofibroblast generation, leading to multiorgan fibrosis and cardiac dysfunction in mice during aging. Mechanistically, SIRT6 bound to and repressed the expression of key TGF-β signaling genes by deacetylating SMAD family member 3 (SMAD3) and Lys-9 and Lys-56 in histone 3. SIRT6 binding to the promoters of genes in the TGF-β signaling pathway decreased significantly with age and was accompanied by increased binding of SMAD3 to these promoters. Our findings reveal that SIRT6 may be a potential candidate for modulating TGF-β signaling to reduce multiorgan fibrosis during aging and fibrosis-associated diseases.
Keywords: sirtuin, transforming growth factor beta (TGF-beta), fibrosis, aging, SMAD transcription factor, aging, aging-associated fibrosis, SIRT6 deacetylase, TGF-beta signaling, SMAD3, caloric restriction, cardiac disease, extracellular matrix (ECM)
Introduction
Calorie restriction has been suggested as a strategy to increase life span and reduce aging-related disorders, including heart failure (1). Interestingly, calorie restriction reduces fibrosis in several disease models (2–4). Calorie restriction is believed to activate sirtuins, a family of class III histone deacetylase proteins (5). Sirtuins remove acetyl moieties from target Lys residues, thereby regulating the function of targets including histone and nonhistone proteins. There are seven sirtuin isoforms in humans, which are distributed in different subcellular compartments (5). Among the seven isoforms, SIRT6 is chromatin-associated and deacetylates histone 3 at Lys-9, Lys-18, and Lys-56 (5–7). In addition, SIRT6 regulates the activity of proteins by post-translational modifications, such as deacetylation, ADP-ribosylation, and demyristoylation. SIRT6 is also known to repress transcription factors HIF-1α, NF-κB, c-Myc, and c-Jun (5, 8). Interestingly, SIRT6 deficiency induces accelerated aging in mice, whereas overexpression extends lifespan in male mice (9, 10). Recent reports indicate that SIRT6 activation prevents several aging-related diseases, including metabolic diseases, cancer, and inflammation (5, 11).
One of the major contributors of tissue fibrosis is activation of TGF-β7 signaling (12). The TGF-β superfamily consists of three ligands, TGF-β1, -β2, and -β3, which are synthesized as latent precursors. Activated TGF-β1 binds to membrane receptors and initiates a series of phosphorylation-dependent signaling cascades, which finally culminate into activation of SMAD family of transcription factors (13). Activation of TGF-β is essential for proper embryogenesis, organ morphogenesis, cell polarity, and wound healing (13, 14, 16). However, chronic hyperactivation of TGF-β signaling causes aging-related fibrotic diseases (17, 18). Studies indicate that aging results in activation of TGF-β signaling in the brain (19), corneal epithelium (20), heart (21), lung (22), kidney (23), and liver (24). Moreover, the mRNA levels of different regulators of the TGF-β pathway, including TGFβRI, TGFβRII, SMAD3, and SMAD4 are significantly higher in senescent fibroblasts derived from old human donors compared with those from young donors (25). Based on several experimental studies, inhibition of TGF-β signaling was suggested to be a therapeutic target for the treatment of pathologic fibrosis (26). It is worth mentioning that neutralization of TGF-β by specific antibodies was considered to be one of the strategies for curing fibrotic diseases (27–30). However, TGF-β signaling inhibitors failed in clinical trials as anti-fibrotic drugs owing to severe adverse effects probably because of cross-talk between multiple signaling pathways (18, 26). To date, there is no definitive therapy available for treatment of fibrotic diseases.
Our previous findings indicate that activation of SIRT6 specifically in cardiomyocytes prevents the development of pressure overload–induced cardiac hypertrophy by repressing IGF/Akt signaling (8). Recently, we found that SIRT6 is a critical regulator of global protein synthesis (31). In this work, we investigated the role of SIRT6 in regulating TGF-β signaling and development of multiorgan fibrosis during aging using in vitro cell culture and mouse models. Our data suggest that SIRT6 transcriptionally regulates TGF-β signaling pathway genes, thus functioning as guardian to protect against adverse tissue remodeling during aging.
Results
SIRT6 deficiency activates TGF-β signaling in fibroblasts
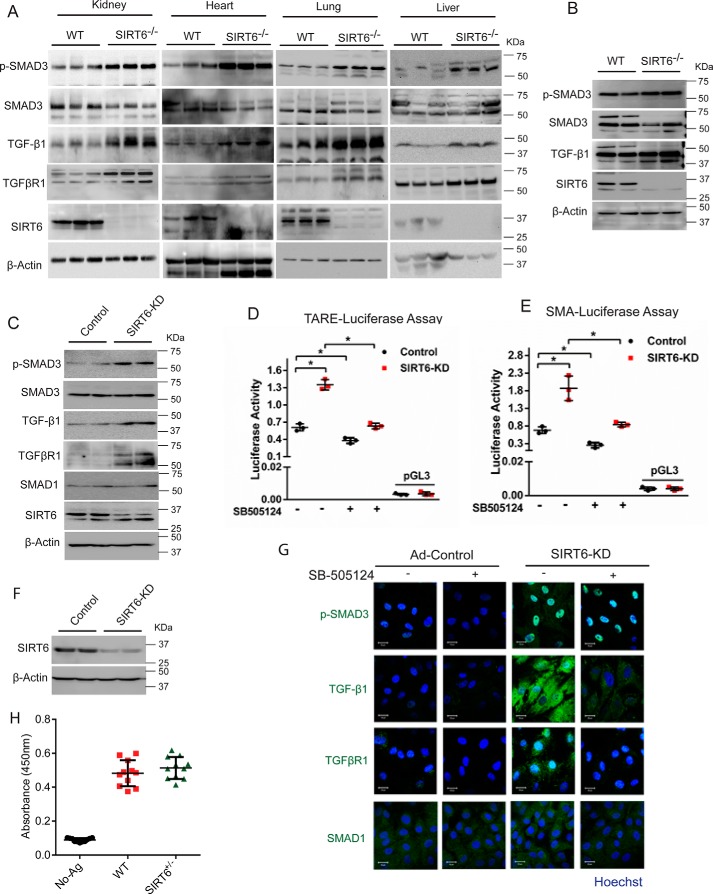
SIRT6-deficient mice develop accelerated aging and aging-associated diseases, such as heart failure, metabolic syndrome, and inflammation (7). In our previous work, we observed increased fibrosis in the hearts of SIRT6-deficient mice (8). Because TGF-β signaling is a well-established driver of fibrosis (17, 18), we tested TGF-β signaling in various organs of SIRT6−/− mice and cardiac fibroblasts isolated from SIRT6−/− mice. Western blot analysis of kidney, heart, lung, and the liver tissues of SIRT6-deficient mice revealed increased phosphorylation of SMAD3, suggesting activated TGF-β signaling (Fig. 1A and Fig. S1A). Further, we observed a marked up-regulation of TGF-β1 and TGFβRI protein levels in these tissues (Fig. 1A and Fig. S1A). To further validate our findings, we assessed the TGF-β signaling in SIRT6-deficient cardiac fibroblasts. Consistent with in vivo findings, we observed increased phosphorylation of SMAD3 and enhanced levels of TGF-β1 in SIRT6-deficient cardiac fibroblasts, indicating activation of TGF-β signaling as shown in Fig. 1B. Further, to rule out any confounding effects due to genomic instability and age-related degeneration observed in SIRT6−/− mice, we transiently depleted SIRT6 using SIRT6-specific siRNA in cardiac fibroblasts isolated from WT mice and assessed the activation of TGF-β signaling. We observed that depletion of SIRT6 spontaneously activated TGF-β signaling, as evidenced by increased phosphorylation of SMAD3 and the enhanced expression levels of TGF-β1 and TGFβRI (Fig. 1C). However, we did not see any marked changes in the expression of SMAD1 in SIRT6-depleted cardiac fibroblasts (Fig. 1C).
Figure 1.
A, Western blot analysis of kidney, heart, lung, and liver lysates from 25-day-old WT and SIRT6-KO mouse littermates for p-SMAD3, SMAD3, TGF-β1, TGFβR1, SIRT6, and β-actin. n = 3 mice/group. B, Western blot analysis of lysates from cardiac fibroblasts, isolated from WT and SIRT6-KO littermate pups for the indicated proteins. C, Western blot analysis of the indicated proteins from control or SIRT6-depleted (SIRT6-KD) cardiac fibroblasts. D and E, luciferase activity assay for TGF-β1/activin-response elements (TARE-Luciferase (D)) and α-SMA-reporter luciferase (E). Luciferase activity was measured with or without treatment with SB-505124 (2.5 μg/ml) for 24 h, with the help of a luminometer. pGL3 basic vector was used as a negative control. n = 3. Data are presented as mean ± S.D. (error bars). *, p < 0.05. One-way ANOVA was used to calculate p values. F, Western blot analysis to confirm knockdown of SIRT6 in cardiac fibroblasts upon transfection with SIRT6-siRNA. G, representative confocal images of control or SIRT6-KD cardiac fibroblasts, with or without SB-505124 treatment (2.5 μg/ml) for 24 h, for the indicated proteins. Scale bar, 20 μm. H, quantitative ELISA for estimating the levels of TGF-β1 in WT or SIRT6 heterozygous (SIRT6+/−) mice at 12 months of age. n = 11 mice/group.
Next, we checked whether SB-505124, a TGF-β signaling pathway inhibitor treatment, reduces spontaneous activation of TGF-β signaling in SIRT6-depleted fibroblasts. SB-505124 treatment indeed significantly reduced the activity of TGF-β signaling in control and SIRT6-depleted fibroblasts as measured by the TGF-β/activin-response element (TARE)-containing luciferase reporter plasmid (Fig. 1D). In addition, we performed luciferase reporter assays to test the transcriptional activity of α-smooth muscle actin (α-SMA) promoter, a well-characterized fibrosis-associated gene activated by TGF-β signaling. Notably, SIRT6-depleted cells showed increased luciferase reporter activity for the α-SMA promoter, which was attenuated by SB-505124 treatment (Fig. 1E). In line with this, inhibition of TGF-β signaling markedly reduced the phosphorylation of SMAD3 and the protein levels of TGF-β1 and TGFβRI levels in SIRT6-depleted fibroblasts (Fig. 1 (F and G) and Fig. S1B). These findings suggest that hyperactivation of TGF-β signaling might be a contributing factor for the increased protein levels of TGF-β1 and TGFβRI levels in SIRT6-depleted fibroblasts, and there could be a positive feedback loop in the activation of TGF-β signaling in SIRT6-deficient cells. Because SIRT6-deficient mice show enhanced TGF-β signaling due to increased TGF-β1 and TGFβRI levels, we tested serum TGF-β1 levels by sandwich ELISA. Surprisingly, our results suggest that TGF-β1 levels were not increased in the serum of SIRT6+/− mice (Fig. 1H), which could possibly be due to failure in detectable levels of serum TGF-β1, because TGF-β1 works mostly as a paracrine growth factor (32).
SIRT6 overexpression attenuates the activation of TGF-β signaling
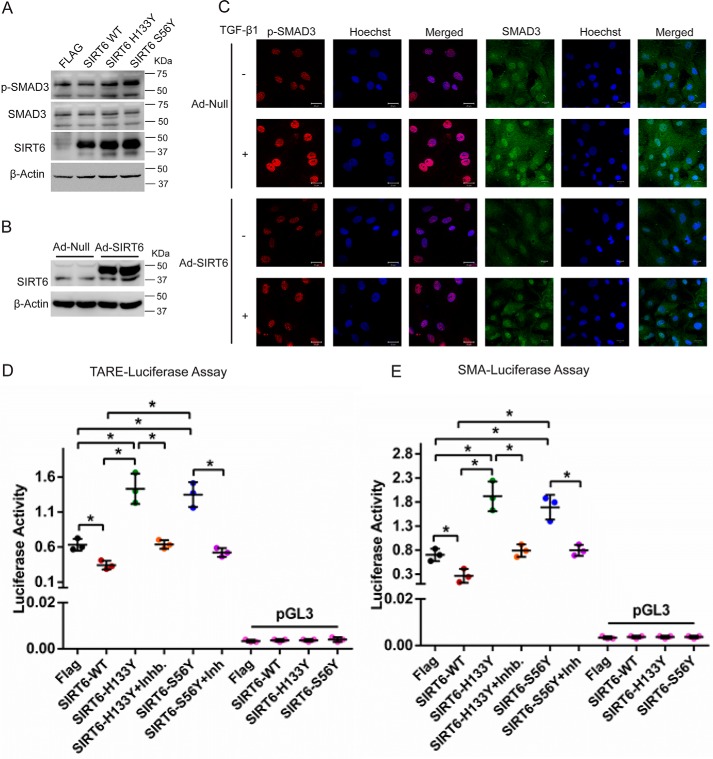
Because SIRT6 deficiency spontaneously activates TGF-β signaling, we tested the effect of transient overexpression of SIRT6 on TGF-β signaling. Overexpression of SIRT6-WT, but not SIRT6-H133Y or SIRT6-S56Y, the catalytic mutants of SIRT6, attenuated TGF-β signaling (Fig. 2A). Further immunostaining results suggest that SIRT6 overexpression attenuates the basal and TGF-β1–induced phosphorylation of SMAD3 in cardiac fibroblasts (Fig. 2 (B and C) and Fig. S1C). Consistent with our observations, TARE reporter activity was inhibited by overexpression of SIRT6. Moreover, expression of SIRT6-H133Y or SIRT6-S56Y markedly enhanced the TARE reporter activity over basal levels, indicating a dominant negative effect of these mutants. Interestingly, SB-505124 treatment reduced the enhanced TGF-β signaling in SIRT6-H133Y– and SIRT6-S56Y–expressing cells (Fig. 2D). In addition, we observed similar results with the promoter activity of α-SMA (Fig. 2E). These findings suggest that the catalytic activity of SIRT6 is required for the inhibition of TGF-β signaling, and the hyperactivation of TGF-β signaling might be a contributing factor for the enhanced SMAD3 activity.
Figure 2.
A, Western blot analysis of cardiac fibroblasts transfected with FLAG-control, FLAG-SIRT6-WT, FLAG-SIRT6-H133Y, or FLAG-SIRT6-S56Y-SIRT6. n = 3. B, Western blot analysis to confirm overexpression of SIRT6 in cardiac fibroblasts, upon infection with Ad-Null or Ad-SIRT6 for 24 h. C, representative confocal images of cardiac fibroblasts infected with either Ad-Null or Ad-SIRT6, with or without TGF-β1 treatment (10 ng/ml) for 24 h, for the indicated proteins. Scale bar, 20 μm. D and E, luciferase activity assay for TGF-β1/activin-response elements (TARE-Luciferase (D)) and α-SMA-reporter luciferase (E). Luciferase plasmid was transfected in cardiac fibroblasts, along with FLAG-control, FLAG-SIRT6-WT, FLAG-SIRT6-H133Y, or FLAG-SIRT6-S56Y using Lipofectamine 2000 for 48 h. Luciferase activity was measured with or without the treatment of SB-505124 (2.5 μg/ml) for 24 h. pGL3 basic vector was used as a negative control. n = 3. Data are presented as mean ± S.D. (error bars). *, p < 0.05. One-way ANOVA test was used to calculate p values.
Deficiency of SIRT6 transcriptionally activates TGF-β signaling and induces fibrosis in mice
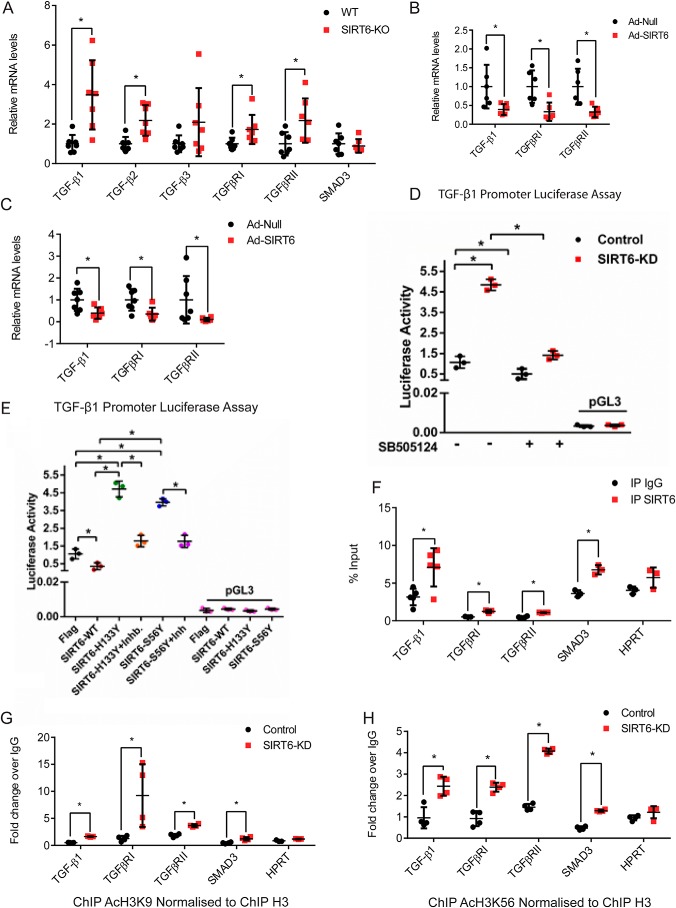
Recent studies have shown association of H3K9 acetylation with myofibroblasts differentiation and extracellular matrix accumulation in nasal polyposis (33). SIRT6 is a chromatin-associated histone deacetylase known to transcriptionally regulate IGF/Akt signaling by regulating acetylation of histone 3 at lysine 9 (8). Therefore, we hypothesized that SIRT6 could transcriptionally modulate the expression of TGF-β signaling genes. To test this hypothesis, we quantified mRNA levels of key TGF-β signaling genes by real-time qPCR in SIRT6−/− mouse hearts. The mRNA levels of TGF-β1, TGF-β2, TGFβRI, and TGFβRII were significantly up-regulated (Fig. 3A). However, we did not observe significant changes in mRNA levels of TGF-β3 and SMAD3 in SIRT6−/− mouse hearts (Fig. 3A). These results are consistent with an increase in protein levels of TGF-β1 and TGFβRI (Fig. 1, A–C). On the other hand, mRNA levels of TGF-β1, TGFβRI, and TGFβRII were significantly down-regulated in SIRT6-overexpressing cardiomyocytes and fibroblasts (Fig. 3, B and C). To test whether increased protein and mRNA levels of TGF-β1 in SIRT6-depleted cells are associated with increased transcriptional activation, we performed luciferase reporter assays with the construct containing the promoter region of TGF-β1. Our results suggest that SIRT6 deficiency increased TGF-β1 promoter activity, which was significantly inhibited following treatment with SB-505124 (Fig. 3D). On the other hand, transient overexpression of SIRT6, but not SIRT6-H133Y or SIRT6-S56Y, repressed the activity of TGF-β1 promoter (Fig. 3E). Further, expression of SIRT6-H133Y and SIRT6-S56Y markedly enhanced the TGF-β1 promoter reporter activity over basal levels, suggesting that inhibition of SIRT6 by dominant negative SIRT6-H133Y or SIRT6-S56Y activates TGF-β1 promoter activity (Fig. 3E). These findings reveal that SIRT6 might transcriptionally control expression of TGF-β signaling genes.
Figure 3.
A–C, scatterplot representing real-time qPCR analysis of heart tissue samples from WT and SIRT6-KO mice (A), cardiomyocytes infected with Ad-Null or Ad-SIRT6 for 24 h (B), and cardiac fibroblasts (C). GAPDH was used to normalize the mRNA levels. Data are presented as mean ± S.D. (error bars), n = 6–7. *, p < 0.05. Student's t test was used to calculate p values. D and E, luciferase activity assay for TGF-β promoter-luciferase. TGF-β-luciferase plasmid was transfected in control and SIRT6-KD cardiac fibroblasts with or without SB-505124 (2.5 μg/ml) treatment (D) or after transfection with either FLAG-control, FLAG-SIRT6-WT, FLAG-SIRT6-H133Y, or FLAG-SIRT6-S56Y, with or without SB-505124 treatment (E). pGL3 basic vector was used as a negative control. n = 3. Data are presented as mean ± S.D. *, p < 0.05. Student's t test was used to calculate p values. F, chromatin Immunoprecipitation analysis to check for the binding of SIRT6 to the promoters of the indicated genes. HPRT was used as the negative control. n = 3–5. Data are presented as mean ± S.D. *, p < 0.05. Student's t test was used to calculate p values. G and H, ChIP analysis for the indicated genes in control or SIRT6-depleted (SIRT6-KD) mouse cardiac fibroblasts. Immunoprecipitation was performed with anti-H3–specific and anti-acetylated H3K9 and H3K56 antibodies. Acetylated H3K9 and H3K56 levels are shown relative to total H3 levels. HPRT was used as the negative control. n = 3–5. Data are presented as mean ± S.D. *, p < 0.05. Student's t test was used to calculate p values.
We next tested whether SIRT6 binds to the promoters of TGF-β signaling genes by ChIP and found that SIRT6 binds significantly to TGF-β1, TGFBRI, TGFBRII, and SMAD3 promoters in cardiac fibroblasts (Fig. 3F). Similarly, we found significant enrichment of SIRT6 at the promoters of TGF-β signaling genes in fibroblasts (Fig. S2A). Further, for the validation of anti-SIRT6 antibody used for ChIP, we tested the occupancy of SIRT6 at the TGF-β1 gene promotor in control and SIRT6-depleted cells. We find that SIRT6 displays significant binding only in control cells and not in SIRT6-depleted cells (Fig. S2B), indicating the specificity of the antibody. Therefore, these data suggest that SIRT6 might regulate TGF-β signaling by binding to the promoters of TGF-β signaling genes. Because SIRT6 is known to deacetylate histone 3 at Lys-9 and Lys-56 and control gene expression at the chromatin level (5, 6), we tested the levels of acetylated H3K9 and H3K56 on promoters of TGF-β signaling genes. We found increased acetylated H3K9 and H3K56 at the promoters of TGF-β1, TGFβRI, TGFβRII, and SMAD3 genes in SIRT6-depleted cardiac fibroblasts (Fig. 3, G and H). Similarly, we observed increased H3K9 acetylation at the promoters of TGF-β signaling genes in SIRT6-depleted 293T fibroblasts (Fig. S2C). These data demonstrate that SIRT6 may repress the expression of key TGF-β signaling genes by deacetylating H3K9 and H3K56. Collectively, our findings suggest that SIRT6 transcriptionally regulates expression of key TGF-β signaling pathway genes through deacetylation of histone 3 at Lys-9 and Lys-56.
SIRT6 suppresses transcriptional activity of SMAD3 transcription factor
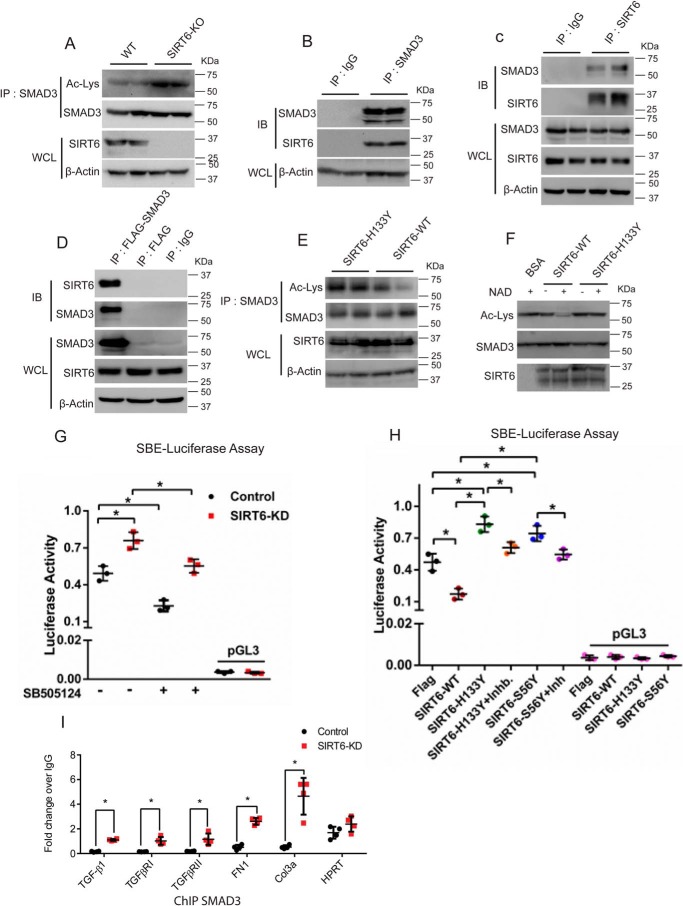
We performed in silico analysis of TGF-β signaling genes to find the transcription factors that could be targeted by SIRT6. We identified conserved binding sites for SMAD3 in the promoters of TGF-β1, TGF-β2, TGFBRI, TGFBRII, and SMAD3, but not in TGF-β3 (File S1). Earlier studies indicate that autocrine TGF-β/SMAD3 transcription factor signal is necessary for TGF-β1 expression in dendritic cells (34). Thus, we hypothesized that SIRT6 might regulate SMAD3 transcription factor to control TGF-β signaling genes. SMAD3 has been shown to be an acetylated protein (35), and SIRT6 deacetylates proteins such as PKM2 (36). Thus, we tested whether SMAD3 acetylation is regulated by SIRT6. We immunoprecipitated SMAD3 from SIRT6−/− mouse hearts and found increased acetylation of SMAD3 (Fig. 4A). In addition, SIRT6 was found to interact with both endogenous as well as ectopically overexpressed SMAD3 (Fig. 4, B–D). Further, we tested whether catalytic activity of SIRT6 plays a role in SMAD3 deacetylation. Our data suggest that SMAD3 can be deacetylated by SIRT6-WT, but not SIRT6-H133Y (Fig. 4E). Further, in vitro deacetylation assay suggests that SIRT6 regulates acetylation of SMAD3 in an NAD+ dependent manner (Fig. 4F). Hence, SIRT6-mediated deacetylation might regulate the transcriptional activity of SMAD3. To further verify our findings, we assessed the interplay between SIRT6 and SMAD3 by using a reporter construct carrying SMAD3-binding elements (SBE). The luciferase reporter assay suggested an increase in transcriptional activity of SMAD3 in SIRT6-depleted cells, which was abrogated by SB-505124 treatment (Fig. 4G). On the other hand, SIRT6, but not SIRT6-H133Y or SIRT6-S56Y, was found to attenuate the transcriptional activity of SMAD3 (Fig. 4H). Interestingly, overexpression of SIRT6-H133Y and SIRT6-S56Y significantly up-regulated the transcriptional activity of SMAD3 under basal conditions (Fig. 4H). These findings suggest that SIRT6 regulates the transcriptional activity of SMAD3 in a deacetylation-dependent manner. To test whether SIRT6-dependent deacetylation regulates the occupancy of SMAD3 on the promoters of TGF-β1, TGF-β receptor I (TGFβRI), TGFβRII, fibronectin 1, and collagen 3a, which are targets of SMAD3, we performed ChIP experiments on SIRT6-depleted cardiac fibroblasts. Our results suggest that the occupancy of SMAD3 was significantly increased on the promoters of TGF-β1, TGFβRI, TGFβRII, fibronectin 1, and collagen 3a in SIRT6-depleted cardiac fibroblasts (Fig. 4I). Further, we also observed similar results in SIRT6-depleted 293T fibroblasts (Fig. S3). These data suggest that the deficiency of SIRT6 may promote activation of TGF-β signaling by increase in SMAD3 binding to key TGF-β signaling genes.
Figure 4.
A, Smad3 was immunoprecipitated to assess its acetylation status in heart samples of 25-day-old WT and SIRT6-KO littermate mice using anti-SMAD3 antibody. Western blotting was performed to detect SMAD3 acetylation using anti-acetyl-lysine antibody. Whole-cell lysates (WCL) were probed for SIRT6 and actin by Western blotting. B, co-immunoprecipitation analysis to assess the interaction between SIRT6 and SMAD3 in heart tissue lysates of WT mice. Western blotting was performed for SMAD3 and SIRT6. IgG was used as the control. WCL were probed for actin. C, co-immunoprecipitation analysis to assess the interaction between SIRT6 and SMAD3 in heart tissue lysates of WT mice. Western blotting was performed for SMAD3 and SIRT6 using specific antibodies. IgG was used as the control. WCL were probed for the indicated proteins. D, assessment of the interaction in vitro between SIRT6 and SMAD3 in FLAG-control or FLAG-SMAD3 plasmid–transfected cells. IgG was used as the control. WCL were probed for SMAD3, SIRT6, and actin by Western blotting. E, Western blot analysis showing changes in the acetylation levels of SMAD3 upon SIRT6 overexpression. Cells were transfected with FLAG-SIRT6 H133Y (catalytically inactive mutant) or FLAG-SIRT6 (WT). SMAD3 was immunoprecipitated from the cell lysate using anti-SMAD3 antibody. Western blotting was performed to detect SMAD3 acetylation using anti-acetyl-lysine antibody. WCL were probed for SIRT6 and actin by Western blotting. F, in vitro deacetylation assay showing SIRT6 as SMAD3 deacetylase. SMAD3 was overexpressed by transfection of the plasmid pcDNA3-HA-SMAD3. SMAD3 was immunoprecipitated and then incubated with FLAG-tagged SIRT6 or FLAG-SIRT6-H133Y. Deacetylation reaction was carried out either in the presence or absence of NAD+ in HDAC buffer. BSA was used as control. Anti-acetyl-lysine antibody was used to analyze SMAD3 acetylation. G and H, luciferase activity assay for SMAD-binding element (SBE-luciferase). SBE-luciferase plasmid was transfected in control and SIRT6-KD cardiac fibroblasts, with or without SB-505124 (2.5 μg/ml) treatment (G) or after transfection with either FLAG-control or FLAG-SIRT6-WT, FLAG-SIRT6-H133Y, or FLAG-SIRT6-S56Y, with or without SB-505124 treatment (H). pGL3 basic vector was used as a negative control. n = 3. Data are presented as mean ± S.D. (error bars). *, p < 0.05. One-way ANOVA test was used to calculate p values. I, ChIP analysis of SMAD3 binding to the promoters of the indicated genes in cardiac fibroblasts. HPRT was used as the negative control. n = 4. Data are presented as mean ± S.D. *, p < 0.05. Student's t test was used to calculate the p values.
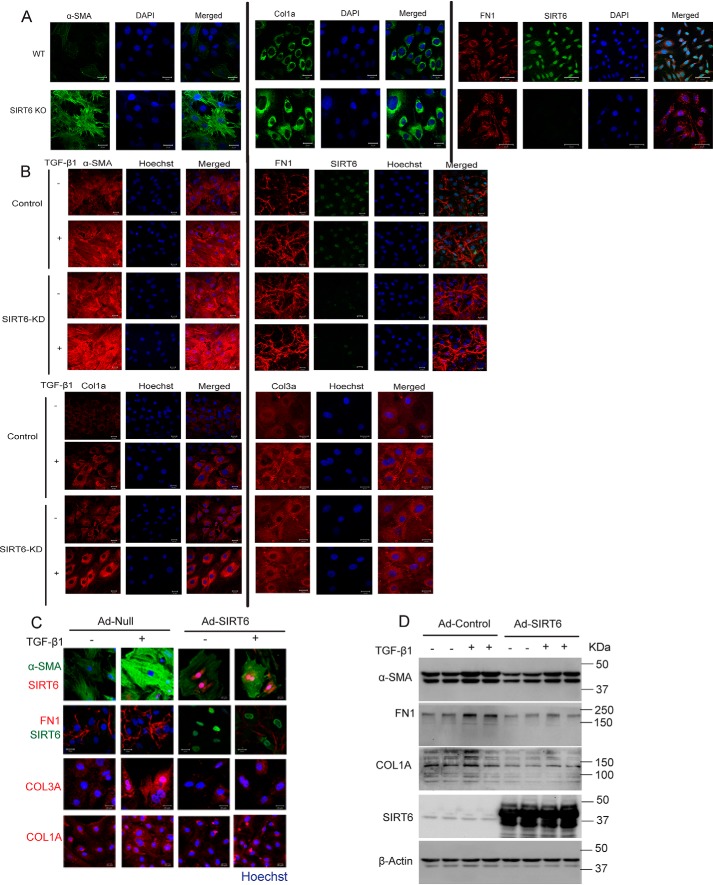
SIRT6 deficiency increases expression of fibrosis-associated markers in myofibroblasts
TGF-β signaling is involved in transformation of fibroblasts to myofibroblasts as well as expression of fibrosis-associated markers, such as α-SMA, collagen 1a (Col1a), collagen 3a (Col3a), and fibronectin 1 (FN1) (14–16). Because SIRT6 deficiency was found to augment the TGF-β signaling, we assessed whether SIRT6 deficiency affects fibrotic markers such as α-SMA, Col1a, and FN1 in fibroblasts. Immunostaining of SIRT6−/− fibroblasts suggested increased expression of fibrotic markers (Fig. 5A and Fig. S4A). Similarly, depletion of SIRT6 in cardiac fibroblasts enhanced the expression of fibrosis-associated myofibroblast markers under basal conditions, which was further augmented by the treatment with TGF-β1 (Fig. 5B and Fig. S4B). These findings indicate that SIRT6 deficiency promotes spontaneous and TGF-β1–induced transformation of fibroblasts to myofibroblasts. To test whether SIRT6 overexpression could inhibit transformation of fibroblasts, we overexpressed SIRT6 in cardiac fibroblasts and analyzed the expression of fibrosis-related myofibroblast markers. Our results suggest that TGF-β1 treatment increases the expression of fibrotic markers in control, but not in SIRT6-overexpressing, cardiac fibroblasts (Fig. 5 (C and D) and Fig. S4C). Together, these findings reveal SIRT6 to be an endogenous negative regulator of transformation of fibroblasts to myofibroblasts.
Figure 5.
A, representative confocal images of cardiac fibroblasts isolated from WT or SIRT6-KO littermate mouse pups, for α-SMA, Col1a, FN1, and SIRT6. Scale bar, 20 μm. n = 3. B, representative confocal images of control or SIRT6-KD cardiac fibroblasts, with or without TGF-β1 treatment (10 ng/ml) for 24 h. Scale bar, 50 μm. n = 3. C, representative confocal images of cardiac fibroblasts infected with Ad-Null or Ad-SIRT6, with or without TGF-β1 treatment (10 ng/ml) for 24 h. Scale bar, 50 μm. n = 3. D, Western blot analysis of cardiac fibroblasts infected with Ad-Null or Ad-SIRT6 for 24 h, with or without TGF-β1 treatment (10 ng/ml) for 24 h.
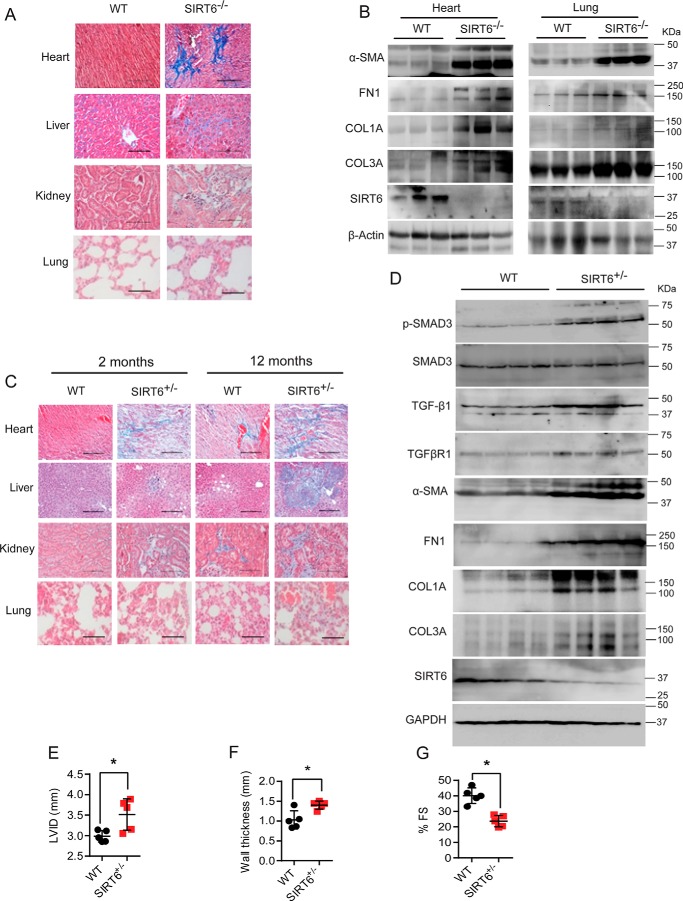
SIRT6 deficiency induces aging-dependent multiorgan fibrosis in mice
To verify the in vitro results of increased expression of fibroblast differentiation markers, we assessed fibrosis in multiple organs of SIRT6−/− mice by histology. Masson's trichrome staining suggested increased fibrosis in heart, liver, kidney, and lungs of SIRT6−/− mice (Fig. 6A and Fig. 5A). Further, Western blot analysis suggested increased expression of α-SMA, FN1, Col1a, and Col3a in heart and lung tissues of SIRT6−/− mice (Fig. 6B and Fig. S5B). Because the life span of whole-body SIRT6−/− mice is only 4 weeks, we further verified our findings in SIRT6+/− mice at 2 and 12 months of age. Whereas 2-month-old SIRT6+/− mice showed mild fibrosis, we observed severe fibrosis in heart, liver, kidney, and lung of 12-month-old SIRT6+/− mice (Fig. 6C and Fig. S5C). Along the same lines, we found significantly increased expression of α-SMA, FN1, Col1a, and Col3a in the heart samples of SIRT6+/− mice at 12 months of age as measured by Western blotting (Fig. 6D and Fig. S5D). Moreover, the phosphorylation of SMAD3 and the protein levels of TGF-β1 and TGFβRI were significantly high in 12-month-old SIRT6+/− mice (Fig. 6D and Fig. S5D), suggesting that augmented TGF-β/SMAD3 signaling might be a contributing factor for multiorgan fibrosis found in SIRT6+/− mice. These findings were consistent with our observations on SIRT6−/− mice (Fig. 1A). Overall, our findings suggest that haploinsufficiency of SIRT6 is sufficient to activate TGF-β signaling, transformation of fibroblasts, and tissue fibrosis in an age-dependent manner.
Figure 6.
A and C, histology of heart, liver, kidney, and lung tissue samples from 25-day-old WT and SIRT6-KO littermate mice (A) and 2-month-and 12-month-old WT and SIRT6+/− mice (C) showing tissue fibrosis. Scale bar, 50 μm. n = 3. B, Western blot analysis of heart and lung lysates from 25-day-old WT and SIRT6-KO mouse littermates for the indicated proteins. n = 3. D, Western blot analysis of heart lysates from 12-month-old WT and SIRT6+/− mouse littermates for the indicated proteins. n = 4 mice/group. E–G, scatterplot showing the left-ventricular internal diameter (LVID), left-ventricular posterior wall thickness, and cardiac contractile functions, as measured by fractional shortening of 12-month-old WT and SIRT6+/− mice, n = 5; data are presented as mean ± S.D. (error bars). *, p < 0.05. Student's t test was used to calculate the p values.
Organ fibrosis is generally associated with impaired functioning; hence, we tested whether fibrosis observed in 12-month-old SIRT6+/− mouse hearts impairs cardiac function. We performed transthoracic echocardiography to assess the structure and contractile functions of heart in 12-month-old SIRT6+/− mice. We found that SIRT6+/− mouse hearts show increased left-ventricular chamber diameter and ventricular wall thickness, indicating cardiac hypertrophy (Fig. 6, E and F). Moreover, SIRT6+/− mouse hearts showed contractile dysfunction, as assessed by reduction in fractional shortening (Fig. 6G). These data suggest that SIRT6 deficiency augments aging-associated fibrosis and causes contractile dysfunction in mouse hearts.
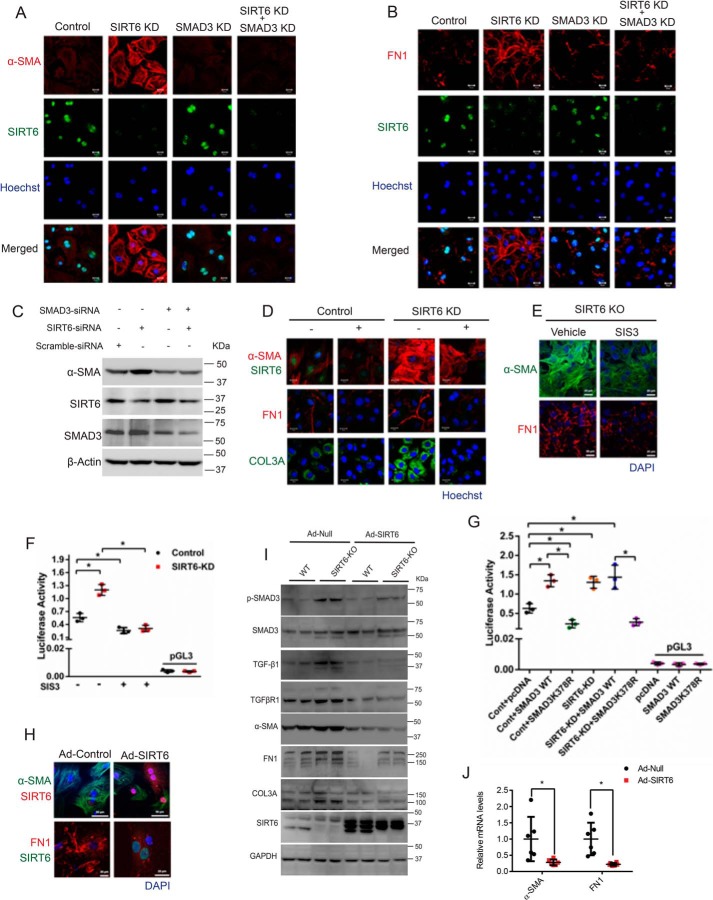
Inhibition of SMAD3 rescues fibrosis phenotype in SIRT6−/− fibroblasts
To test whether SMAD3 is responsible for the increase in expression of fibrosis-related markers observed in SIRT6-depleted cells, we transiently depleted SMAD3 in SIRT6-depleted cells and assessed their phenotype. Our results suggest that depletion of SMAD3 markedly reduces the expression of α-SMA and fibronectin-1 in SIRT6-depleted fibroblasts (Fig. 7 (A–C) and Fig. S6 (A and B). To further confirm our findings, we inhibited TGF-β/SMAD3 signaling in SIRT6-depleted fibroblasts with SB-505124 and tested the phenotype. Our findings reveal that treatment of SB-505124 reduces the expression of α-SMA, FN1, and Col3a in SIRT6-depleted fibroblasts (Fig. 7D and Fig. S6C). We further verified these findings with SIS3, a small-molecule inhibitor of SMAD3 (37). Our findings show that SIS3 reduces expression of α-SMA and fibronectin-1 in SIRT6-knockout (KO) fibroblasts as well as causes attenuation of SMAD3 activation, as reflected in its reduced binding to the SMAD-binding element (Fig. 7 (E and F) and Fig. S6D). These findings suggest that hyperactive TGF-β/SMAD3 signaling is responsible for the expression of fibrosis-related myofibroblast markers in SIRT6-depleted fibroblasts. Previous studies suggest that acetylation of SMAD3 regulates its activity (35). In line with this, our data suggest that K378R, which mimics the deacetylated state of SMAD3 and reduces SMAD3 activity, failed to show enhanced activity in SIRT6-KD fibroblasts (Fig. 7G). This finding indicates that SIRT6 might target Lys-378 of SMAD3. However, future work is required for confirming this observation. Next, we tested whether SIRT6 reconstitution could rescue the fibrotic phenotype observed in SIRT6−/− fibroblasts. We therefore cultured SIRT6−/− fibroblasts and then infected them with adenovirus expressing SIRT6. We found that reconstitution of SIRT6 in SIRT6−/− fibroblasts markedly reduces the expression of α-SMA and FN1 (Fig. 7H and Fig. S6E). Further, Western blot analysis revealed that reconstitution of SIRT6 into the SIRT6−/− myofibroblasts markedly reduced the phosphorylation of SMAD3 as well as the expression of TGF-β1, TGFβRI, α-SMA, FN1, and Col3a in SIRT6−/− fibroblasts (Fig. 7I). Furthermore, reconstitution of SIRT6 into SIRT6−/− myofibroblasts reduced the mRNA expression of α-SMA and FN1 (Fig. 7J). These data suggest that SIRT6 is sufficient to reverse the fibrotic phenotype in SIRT6−/− myofibroblasts by attenuation of TGF-β/SMAD3 signaling.
Figure 7.
A and B, representative confocal images of control, SIRT6-depleted (SIRT6-KD), SMAD3-depleted (SMAD3-KD), or both SIRT6- and SMAD3-depleted (SIRT6-KD + SMAD3-KD) cardiac fibroblasts for α-SMA and FN1. Scale bar, 20 μm. n = 3. C, Western blot analysis of control, SIRT6-KD, SMAD3-KD, or SIRT6-KD + SMAD3-KD cardiac fibroblasts for the indicated proteins. D, representative confocal images of control or SIRT6-KD cardiac fibroblasts, with or without SB-505124 treatment (2.5 μg/ml). Scale bar, 20 μm. n = 3. E, representative confocal images of control or SIS3-treated cardiac fibroblasts isolated from SIRT6-KO mouse pups. Scale bar, 20 μm. n = 3. F and G, scatter plots showing a luciferase activity assay for SBE (SBE-Luciferase). SBE-luciferase plasmid was transfected in control and SIRT6-KD cardiac fibroblasts, with or without SIS3 treatment (F) or after transfection with either pcDNA, pcDNA-SMAD3-WT, or pcDNA-SMAD3-K378R (G). H, representative confocal images of cardiac fibroblasts infected with Ad-Null or Ad- SIRT6 for the indicated proteins. Scale bar, 50 μm (for α-SMA) or 20 μm (for FN1). n = 3. I, Western blot analysis of cardiac fibroblasts isolated from WT or SIRT6-KO littermate mouse pups, infected with Ad-Null or Ad-SIRT6 for the indicated proteins. J, scatter plot representing real-time qPCR analysis of α-SMA and FN1 from fibroblasts isolated from SIRT6-KO mice and then infected with either control or SIRT6-overexpressing adenovirus. GAPDH was used to normalize the mRNA levels. Data are presented as mean ± S.D. (error bars), n = 5; *, p < 0.05. Student's t test was used to calculate the p values.
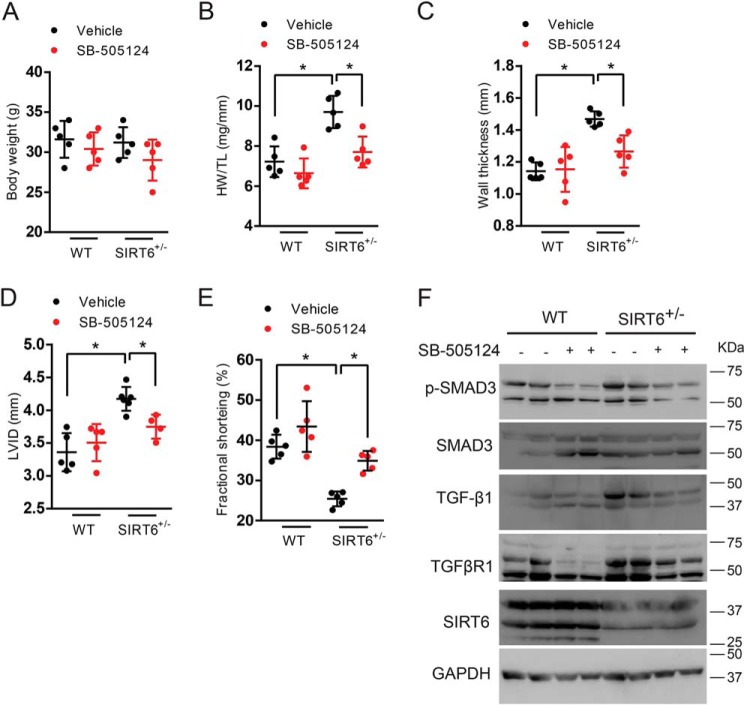
Inhibition of TGF-β/SMAD3 signaling improves cardiac function in SIRT6+/− mice
Because we found fibrosis in 12-month-old SIRT6+/− mouse hearts where their cardiac structural and functional parameters were compromised, we treated SIRT6+/− mice with TGF-β/SMAD3 signaling inhibitor. SIRT6+/− mice were injected intraperitoneally with either vehicle or SB-505124 at a dose of 10 mg/kg at every alternate day for 15 days. SB-505124 treatment did not significantly change the body weight, but it rescued the cardiac hypertrophy observed in 12-month-old SIRT6+/− mice, as depicted by heart weight/tibia length ratio (HW/TL), left-ventricular wall thickness, and left-ventricular internal diameter (Fig. 8, A–D). Moreover, cardiac contractile function, as assessed by fractional shortening, improved after SB-505124 treatment in 12-month-old SIRT6+/− mice (Fig. 8E). Further, Western blot analysis suggested that TGF-β/SMAD3 signaling, as assessed by the phosphorylation of SMAD3, was attenuated following SB-505124 treatment in 12-month-old SIRT6+/− mice. In addition, the levels of TGF-β1 and TGFβRI were reduced in hearts of 12-month-old SIRT6+/− mice following SB-505124 treatment (Fig. 8F). These data suggest that inhibition of TGF-β/SMAD3 signaling rescues cardiac function in 12-month-old SIRT6+/− mouse hearts.
Figure 8.
A–E, scatter plot depicting body weight, heart weight to tibia length ratio (HW/TL), left-ventricular posterior wall thickness, left-ventricular internal diameter (LVID), and cardiac contractile functions, respectively, of WT and SIRT6+/− mice, before and after the treatment with SB-505124. SB-505124 was injected intraperitoneally at a concentration of 10 mg/kg every alternate day for 15 days. Sterile peanut oil was used as the vehicle. n = 5. *, p < 0.05. One-way ANOVA test was used to calculate the p values. F, Western blot analysis of heart lysates from WT and SIRT6+/− mice, before and after the treatment with SB-505124 (10 mg/kg). Error bars, S.D.
Reduced SIRT6 levels might cause aging-related activation of TGF-β/SMAD3 signaling
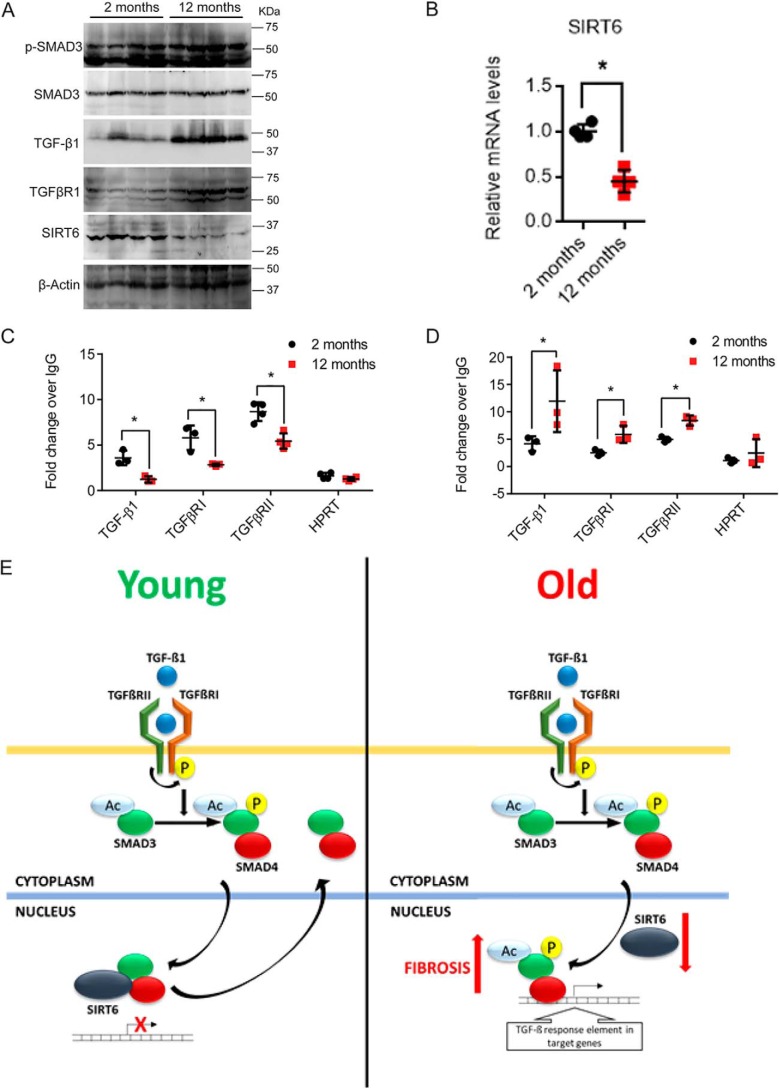
Aging activates TGF-β signaling in the brain (19), corneal epithelium (20), heart (21), lung (22), kidney (23), and liver (24). We previously found low SIRT6 levels in the tissues of heart failure patients (8). Hence, we tested whether SIRT6, which is considered to be longevity-promoting sirtuin, has a role in fibrosis associated with physiological aging. Western blotting results suggest increased phosphorylation of SMAD3 and increased levels of TGF-β1 and TGFβRI in hearts of 12-month-old mice as compared with 2-month-old mice, whereas the levels of SIRT6 were markedly reduced in the 12-month-old mice (Fig. 9A and Fig. S7). Further, we observed that the mRNA levels of SIRT6 were also significantly reduced in 12-month-old mouse hearts (Fig. 9B). Because SIRT6 levels were reduced in heart samples of 12-month-old mice, we tested the occupancy of SIRT6 on the promoters of TGF-β/SMAD3 signaling genes in young and old mice. Our ChIP assay results suggest reduced binding of SIRT6 to the promoters of TGF-β1, TGFβRI, and TGFβRII in heart samples of 12-month-old mice, when compared with 2-month-old mice (Fig. 9C). Because our findings suggest that SIRT6 represses SMAD3, we tested whether SMAD3 occupancy increased 12-month-old mouse hearts. Our results suggest increased binding of SMAD3 to promoters of TGF-β1, TGFβRI, and TGFβRII in heart tissues of 12-month-old mice as compared with 2-month-old mice (Fig. 9D). These data indicate that the binding of SIRT6 deacetylase to TGF-β signaling pathway genes significantly reduced with age, which corresponds to the increased binding of SMAD3 to these promoters. As a result, SIRT6-deficient fibroblasts express increased levels of TGF-β1, TGFβRI, and TGFβRII and activate TGF-β signaling in a cell-autonomous manner. Enhanced TGF-β signaling transforms fibroblasts to myofibroblasts and causes deposition of fibrous material in aged organs, leading to organ dysfunction (Fig. 9E).
Figure 9.
A, Western blot analysis of heart lysates from 2- and 12-month-old WT mice for the indicated proteins. n = 4. B, scatter plot representing real-time qPCR analysis of SIRT6 in heart samples of 2-month-old and 12-month-old mice. GAPDH was used to normalize the mRNA levels. Data are presented as mean ± S.D. (error bars), n = 4; *, p < 0.05. Student's t test was used to calculate the p values. C and D, chromatin immunoprecipitation analysis of heart tissue samples to check for the binding of SMAD3 and SIRT6 to the promoters of the indicated genes in 2-month-old and 12-month-old mice. HPRT was used as the negative control. n = 3–7 mice/group. Data are presented as mean ± S.D. *, p < 0.05. Student's t test was used to calculate the p values. E, representative model depicting the findings of the study. Reduction in levels of SIRT6 with aging results in increased binding of SMAD3 to TGF-β signaling genes, hence leading to enhanced transformation of fibroblasts to myofibroblasts and development of multiorgan fibrosis. SIRT6 acts as a transcriptional repressor of TGF-β/SMAD signaling to regulate the aging-related fibrosis.
Discussion
Whole-body SIRT6−/− mice show features of accelerated aging and age-related fibrosis (10). However, the exact mechanism of SIRT6 in the development of fibrosis is not well-elucidated. Our work provides evidence that SIRT6 regulates fibrosis via TGF-β/SMAD3 signaling. We have shown that mRNA levels of TGF-β1, TGF-β2, and TGFβRI are up-regulated in SIRT6-deficient hearts, and SIRT6 transcriptionally regulates these genes by deacetylation of SMAD3 and histone 3 at Lys-9 and Lys-56.
Previous studies have shown that TGF-β1 and TGFβRI activation is sufficient to induce TGF-β signaling almost in any cell type (13, 14, 16). Our findings suggest a similar phenomenon in SIRT6-deficient fibroblasts, which show increased levels of TGF-β1 and TGFβRI and transformation of fibroblasts to myofibroblasts in a cell-autonomous manner. Recently, SIRT6 was shown to inhibit cardiac fibrosis by inhibiting the transcriptional activity of NF-κB (39). Furthermore, SIRT6 inhibits inflammation and fibrosis in liver by c-Jun and H3K9 deacetylation (40). Reduced SIRT6 levels, leading to hyperacetylation of H3K9 and H3K56, are also implicated in early neurodegenerative events in diabetic retinopathy (41). Similarly, SIRT6 inhibits TGF-β–induced epithelial senescence, a key feature associated with idiopathic pulmonary fibrosis in human bronchial epithelial cells (42). SIRT6 silencing in human dermal fibroblasts activates NF-κB signaling, leading to decreased collagen 1 expression (43). In contrast, SIRT6 was found to promote TGF-β signaling in systemic sclerosis (44) and in cancer (45). SIRT6 expression along with that of SMAD2, SMAD5, and other contractile markers, increased when vascular smooth muscle cells were stimulated with cyclic strain (46). Thus, it is possible that SIRT6 controls TGF-β signaling as well as fibrosis in a cell- and tissue-specific manner.
Loss of heart elasticity is one of the major causes for contractile dysfunction in cardiovascular diseases such as hypertension, myocardial infarction, diabetic cardiomyopathy, and drug-induced cardiotoxicity (47–49). It is well-proven that loss of elasticity occurs due to severe interstitial fibrosis in cardiac tissue (50, 51). During cardiac fibrosis, the resident cardiac fibroblasts transform into myofibroblasts and secrete several fibrosis-associated proteins, such as collagen, fibronectin, and laminin, resulting in remodeling of extracellular matrix (48). Although several signal transduction pathways are believed to be associated with fibroblast transformation and fibrosis, one of the well-proven mechanisms involved in fibrosis is TGF-β signaling (52). In the present study, we show that SIRT6 regulates TGF-β signaling to control the process of fibrosis in mice. Similarly, other sirtuin isoforms are known to modulate TGF-β signaling and fibrosis. Studies indicate that SIRT1 inhibits renal fibrosis by inhibiting TGF-β/SMAD3 signaling, and resveratrol, a SIRT1 activator, inhibits extracellular matrix deposition by decreasing the acetylation of SMAD3 (53). Similarly, activation of SIRT1 has been shown to attenuate doxorubicin-induced cardiac fibrosis by down-regulating the TGF-β/SMAD3 pathway (54). On the other hand, inhibition of SIRT2 suppresses hepatic fibrosis (55). Our previous work demonstrated that SIRT3 inhibits age-related tissue fibrosis (56). Similarly, SIRT5 deficiency induces cardiac fibrosis (57). In contrast, SIRT4 accelerates angiotensin II–induced cardiac fibrosis by inhibiting SIRT3-mediated deacetylation of MnSOD (58). SIRT7 deficiency down-regulates TGF-β signaling and impairs wound healing (59). These findings indicate that SIRT6 may not be the only deacetylase regulating the TGF-β signaling and fibrosis. Hence, from our work, we show evidence that SIRT6 deficiency is sufficient to cause transformation of fibroblasts and fibrosis, and the mechanism is distinct from other sirtuins. Because TGF-β signaling is a master regulator of fibrosis and warrants multilayer control, sirtuins may regulate TGF-β signaling at multiple levels to control its activation.
Earlier studies indicate that transcription factor Sp1 is essential for basal expression of TGF-β1 (60). Interestingly, we recently reported that SIRT6 deficiency up-regulates the expression of Sp1 target genes (31). Moreover, AP-1 sites are found in the promoter of TGF-β1, and transcription factors c-Jun and c-Fos are required for the induction of TGF-β1 (61, 62). Consistent with the previous findings, we found that SMAD3 binds to and activates the promoters of key TGF-β signaling genes, such as TGF-β1, TGF-β2, and FN1 (34). Our previous work demonstrated that SIRT6 represses the activity of c-Jun (8). SMAD3 forms a complex with either c-Jun/c-Fos (61) or Sp1 (64) to increase the transcription of TGF-β signaling target genes involved in the fibrotic program. Because the SMAD3/c-Jun complex may be the functional unit, inhibition of SMAD3 might inactivate the complex and rescue fibrosis in SIRT6-deficient cells.
We show here that SMAD3 inhibition can rescue expression of fibrosis markers in SIRT6-depleted cells. Published works suggest that SMAD3−/− mice are protected from fibrosis and suppression of SMAD3 is sufficient to attenuate the process of fibrosis (65, 66). SMAD3 is an acetylated protein, and p300/CBP-mediated acetylation positively regulates its transcriptional activity (35). In line with the previous findings, our work confirms that prevention of acetylation of SMAD3 at Lys-378 reduces its transcriptional activity in SIRT6-depleted cells. ERK5-induced increase in acetylation of SMAD3 augments pulmonary fibrosis (67). Studies also indicate that SIRT1 deacetylates and represses SMAD3 transcription factor to protect mice from renal and cardiac fibrosis (53, 54). Our results suggest that SIRT6 deacetylates SMAD3 and H3K9 to control SMAD3 activation.
Our previous work shows that SIRT6 levels were reduced in heart tissues of patients with heart failure (8). Cardiac-specific deletion of SIRT6 causes cardiac hypertrophy and fibrosis, leading to heart failure, whereas cardiac-specific overexpression of SIRT6 protects mice from hypertrophy and fibrosis (8). SIRT6−/− mouse hearts exhibit increased TGF-β1 and TGFBRI receptor expression. Similarly, SIRT6 deficiency leads to increased expression of TGF-β1 in fibroblasts. Further, we noted that SIRT6 overexpression suppresses the expression of TGF-β1, TGFβRI, and TGFβRII in cardiomyocytes as well as fibroblasts. It is possible that SIRT6 might regulate TGF-β signaling in both cardiomyocytes and fibroblasts. Previous studies corroborate these findings, emphasizing the importance of TGF-β signaling in cardiomyocyte hypertrophy (68).
Our work suggests that inhibition of TGF-β signaling by specific small-molecule inhibitors can reverse fibrosis markers in SIRT6-deficient fibroblasts. Upon inhibition of TGF-β signaling, the expression of TGF-β1 and TGFβRI was observed to be reduced in SIRT6-depleted fibroblasts, possibly due to prevention of SMAD3 occupancy at the gene promoters of TGF-β1 and TGFβRI and thus inhibition of a feed-forward activation loop for TGF-β signaling. Our work elucidates a novel feed-forward loop involving transcriptional regulation of TGF-β signaling. SIRT6 deacetylase plays a major role in antagonizing this feed-forward loop in resting fibroblasts under basal conditions. However, loss of SIRT6 in fibroblasts during aging amplifies this feed-forward loop to hyperactivate TGF-β signaling to favor the fibroblast transformation. However, further experiments are needed to support the hypothesis that TGF-β1 depletion or antagonizing TGFβRI can affect SIRT6/SMAD3 association with chromatin at these gene promoters.
Our finding of transcriptional up-regulation of the TGF-β pathway in a mouse model mirrors the observations noted in aged humans. We found that increased TGF-β signaling is associated with reduced levels and occupancy of SIRT6 at the promoters of TGF-β signaling genes. Our earlier work revealed that SIRT6 levels were decreased in the tissue samples of heart failure patients (8). Similarly, SIRT6 levels were significantly low in human dermal fibroblasts isolated from older human subjects (69), in senescent 2BS cells, and in liver, spleen, and kidney tissues of 18-month-old BALB/c mice (70). We believe that SIRT6 plays a critical role in maintenance of tissue architecture by regulating a fine balance of fibroblasts and myofibroblasts. Loss of SIRT6 protein could be major determining factor causing multiple-organ fibrosis. We believe that SIRT6 can be a potential therapeutic target for the treatment of aging-related fibrotic diseases.
Experimental procedures
Animal experiments and cell culture
WT and SIRT6 heterozygous (SIRT6+/−) mice were purchased from Jackson Laboratories. All animal protocols were carried out with the approval of the institutional animal ethics committee of the Indian Institute of Science, Bengaluru. Animal experiments were carried out in strict accordance with the recommendations of the Committee for Control and Supervision of Experiments on Animals (CPCSEA), Government of India. The animals were housed under a 12-h light/dark cycle in individually ventilated cages in the clean air facility of the Indian Institute of Science, with food and water ad libitum. Male mice were used for in vivo experiments. Mice were sacrificed with CO2 gas inhalation from a compressed CO2 gas cylinder. Neonatal mouse pups were anesthetized with 1–2% isoflurane. In vitro experiments were carried out in primary neonatal cardiac fibroblasts isolated from WT and SIRT6−/− mouse pups. Cardiac fibroblasts were then isolated using standard protocols (63). Third-passage cardiac fibroblasts were used for experiments. The cells were cultured in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Animal echocardiography
Transthoracic echocardiography was performed by a Vevo 1100 imaging system (FUJIFILM Visual Sonics Inc.) as per a previously published protocol (38). Briefly, 1–2% isoflurane was used for anesthetizing mice. A heated imaging platform was used to maintain the temperature of mice throughout the procedure. Chest hair was removed prior to recording. Images were captured at both the long and short axis at parasternal projection. M-mode recordings were captured for both of the views at midventricular level. All of the echocardiography readings were taken for at least three beats from each projection. An in-built software was used for data analysis of left-ventricular wall thickness, left-ventricular cavity diameter, and fractional shortening.
Histology
Age-matched WT, SIRT6+/−, and SIRT6−/− mouse heart, liver, lung, muscle, and kidney were collected in 10% neutral buffered formalin. Tissues were then processed with an automated tissue processer (Leica). Paraffin-embedded 4-μm-thick tissue sections were prepared and stained with Masson's trichrome stain (Sigma) as per the protocols described in our previous publication (8).
Chromatin immunoprecipitation assay
The detailed protocol for ChIP from young and old mice is described in our previous publication (8). The protocol for the ChIP assay from 293 cells and cardiac fibroblasts was adapted from previously published work (15). The SimpleChIP® enzymatic chromatin IP kit (CST, Magnetic Beads) was used for this assay. The hypoxanthine phosphoribosyltransferase (HPRT) gene promoter was used as the negative control. Primers used for ChIP were as follows: human TGF-β1, forward (5′-GARGGCACAGTGGTCAAGAG-3′) and reverse (5′-AGGATGGAAGGGTCAGGAG-3′); human TGF-β2, forward (5′-CATCCAGGAACAAACTGAG-3′) and reverse (5′-TGCCAGCAGATAACATCAC-3′); human TGF-β3, forward (5′-CCGAGGTGCTGGTGACCCTG-3′) and reverse (5′-CCAGTGAGTAGGTGGGGAGA-3′); human TGFβRI, forward (5′-CATAGTGGAAACTTGAACG-3′) and reverse (5′-GCTTCCTTCTTCTCACACA-3′); human TGFβRII, forward (5′-ATCCCACCGCACGTTCAGAAGT-3′) and reverse (5′-GACTGTCAAGCGCAGCGGAGA-3′); human SMAD3, forward (5′-GCGTGTGTGTGAGAGTGG-3′) and reverse (5′-AGGTGTGGAAGCCAGAGT-3′); human HPRT, forward (5′-GCCACAGGTAGTGCAAGGTCTT-3′) and reverse (5′-TTCATFFCFFCCFTAAAC-3′); mouse TGF-β1, forward (5′-CACGCAGATACCATCTACAGC-3′) and reverse (5′-ACCCATGAGAAATACACGCTT-3′); mouse TGFβRI, forward (5′-TCTGGAGCTCATTTTGGCGT-3′) and reverse (5′-AGCGGGAGCAGTCATAGGTA-3′); mouse TGFβRII, forward (5′-TGAGCAGGAGGCTGGGGCTC-3′) and reverse (5′-TCTCGGCGCCCGGGTAAAGT-3′); mouse SMAD3, forward (5′-ATGACTTGTTCCTGTCCTTC-3′) and reverse (5′-GCTAGGCAGAGTTCCCAGAA-3′); and mouse HPRT, forward (5′-TGTGAAAGAACTGGGCCTAAA-3′) and reverse (5′-TCCCAGAGGATTCCCAGATA-3′).
Real-time qPCR
Real-time qPCR analysis was performed as described previously (8). Briefly, total RNA was isolated from tissues using TRIzol reagent as per the manufacturer's instructions (Invitrogen). Corresponding cDNA was synthesized from 1 μg of RNA using the RevertAid first-strand cDNA synthesis kit (Thermo Scientific). The real-time PCR was then carried out for TGF-β1, TGF-β2, TGF-β3, TGFβRI, TGFβRII, SMAD3, and HPRT. GAPDH was used as the housekeeping reference gene. The primer sequences used were as follows: TGF-β1, forward (TTGCTTCAGCTCCACAGAGA) and reverse (TGGTTGTAGAGGGCAAGGAC); TGF-β2, forward (CCGCATCTCCTGCTAATGTTG) and reverse (AATAGGCGGCATCCAAAGC); TGF-β3, forward (AAGAGAAGCAAGGGACAGAAG) and reverse (GCTGGGATGAGGGATTATGTAC); TGFβRI, forward (TGCCATAACCGCACTGTCA) and reverse (AATGAAAGGGCGATCTAGTGATG); TGFβRII, forward (CCGCTGCATATCGTCCTGTG) and reverse (AGTGGATGGATGGTCCTATTACA); SMAD3, forward (GGAGGGGAGGTCTTTGCG) and reverse (GCTCGGGGAACCCATCTG); and GAPDH, forward (TATGTCGTGGAGTCTACTGGT) and reverse (GAGTTGTCATATTTCTCGTGG).
Western blot analysis
Western blotting was performed as per a previously published protocol (8). The cell lysates or tissue lysates were prepared in lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Nonidet P-40, 1% SDS, and 0.5% sodium deoxycholate, 10 mm sodium fluoride, 2.5 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture (Roche Applied Science). Protein concentration was determined by a Bradford assay. Cell or tissue lysates were mixed with 2× Laemmli buffer (Bio-Rad). Samples were mixed and boiled for 5 min at 96 °C. The proteins were resolved by SDS-PAGE, and Western blotting was performed to transfer resolved proteins onto polyvinylidene difluoride membrane. Membrane was blocked with a solution of 5% nonfat dried milk in TBST buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Tween 20) for 1 h at room temperature. Blots were probed overnight with primary antibodies at 4 °C. The membrane was washed three times with TBST and HRP-conjugated secondary antibody probed at room temperature for 1 h. The antibodies used were phosphorylated SMAD3 (p-SMAD3) (Abcam), SMAD3 (Abcam), α-SMA (Sigma), fibronectin-1 (Santa Cruz Biotechnology, Inc.), collagen-1a (Millipore), collagen-3a (Santa Cruz Biotechnology), and SIRT6 (Cell Signaling). GAPDH (Santa Cruz Biotechnology) and β-actin (Cell Signaling) were used as loading control. The blots were developed with Clarity ECL Western blotting substrate (Bio-Rad) or SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) in a chemiluminescence imager (Chemidoc Touch, Bio-Rad).
Co-immunoprecipitation experiments
The immunoprecipitation assay was performed as per a previously published protocol (38). Tissues were homogenized in modified radioimmune precipitation assay lysis buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 10 mm sodium fluoride, 2.5 mm sodium pyrophosphate, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture (Roche Applied Science)). Cells were transfected with pcDNA FLAG-SMAD3 plasmid vector. pcDNA-FLAG plasmid served as control. Cells were lysed, and proteins were harvested in radioimmune precipitation assay buffer. Cell or tissue homogenates were centrifuged at 12,000 rpm at 4 °C for 10 min, and the supernatant was collected. 500–1000 μg of protein was incubated with 2 μg of SMAD3 or SIRT6 or control IgG antibodies. Immune complex was incubated with protein A–conjugated agarose beads for 2 h. Beads were washed, and the immunoprecipitated proteins were subjected to SDS-PAGE. Western blotting was performed to detect the protein-protein interactions.
Confocal microscopy
For confocal microscopy, cells were washed twice with 1× PBS followed by fixing with 3.7% formaldehyde for 15 min at room temperature. Fixed cells were then washed three times with 1× PBS and then permeabilized with 0.2% Triton X-100 prepared in PBS. The cells were washed three times with 1× PBS and blocked with 5% BSA prepared in PBST (PBS containing 1% Tween 20). Blocked cells were incubated overnight with specific primary antibodies prepared in 1% BSA in PBST at 4 °C. The cells were then incubated with the respective secondary antibodies conjugated with Alexa Fluor 488 and/or Alexa Fluor 546 prepared in 1% BSA in PBST for 1 h at room temperature. The cells were washed three times with 1× PBS followed by incubation with Hoechst 33342 prepared in PBS for 10 min at room temperature for staining the nucleus. The cells were finally washed three times with 1× PBS before mounting with Fluoromount G on a clean glass slide. Images were captured with a Zeiss LSM 710 or 880 confocal microscope.
Luciferase assay
The luciferase reporter assay was performed using TARE-luc, TGF-β1 promoter-luc, SBE-luc, and α-SMA-luc reporter plasmid constructs as described previously (56). Cardiac fibroblasts were transfected with the above plasmid constructs. Renilla luciferase was used for normalizing the luciferase signal, and pGL3 vector was used as nonresponding negative control reporter. Luminescence was measured using a luminometer (Pharmingen Moonlight 3010, BD Biosciences).
SMAD3 deacetylation assay
The detailed protocol for the deacetylation assay is adapted from our previous publication (38). FLAG-tagged SIRT6-WT or SIRT6-H133Y was transfected to 293 cells by Lipofectamine using the manufacturer's instructions. Cells were harvested in lysis buffer, the homogenate was centrifuged at 12,000 rpm at 4 °C for 10 min, and the supernatant was collected in fresh microcentrifuge tubes. Recombinant SIRT6 was purified using agarose beads conjugated to FLAG antibody. Beads were washed, and the protein was eluted. Recombinant SMAD3 protein overexpressed in 293 was incubated with eluted SIRT6-WT or SIRT6-H133Y protein in deacetylation buffer (250 mm Tris-Cl, pH 9.0, 20 mm MgCl2, 250 mm NaCl, 2.5 mm DTT, 5 mm NAD+, 2.5 μm trichostatin A) for 2 h at 30 °C. SMAD3 acetylation was tested by Western blotting using acetyl-lysine–specific antibody.
ELISA
Sandwich ELISA was performed as per a standard protocol. Briefly, a 96-well ELISA plate was coated with 1 μg of TGF-β antibody (Thermo MAI-21479) overnight at room temperature in 100 μl of PBS. The plate was then blocked with 0.5% gelatin in PBS for 1 h. 1 μg of antigen was added and incubated for 2 h; the plate was washed three times with PBST, followed by three washings with PBS and incubation with 1 μg of TGF-β antibody (Sc-146, Santa Cruz Biotechnology) for 1 h. The plate was again washed and incubated with 100 μl of HRP-conjugated rabbit anti-mouse secondary antibody (1:2000) in PBS for 45 min. The plate was washed, and 100 μl of substrate, 0.03% H2O2 in citrate phosphate buffer, pH 5.5, and chromogen 3,3′,5,5′-tetramethylbenzidine (60 μg/ml) was added to detect the HRP activity. The reaction was quenched by the addition of 50 μl of 1 m H2SO4, and absorbance was measured at 450 nm using an ELISA microplate reader.
Statistics
Student's t test was performed for pairwise comparison. One-way ANOVA or two-way ANOVA was used to calculate the statistical significance for multiple comparisons between experimental groups using GraphPad Prism software version 6.04. Western blots were processed using Image Lab software (Bio-Rad). Densitometric analyses of Western blots were performed by ImageJ. Confocal image processing was performed with ZEN-Black software, and signal was quantified using ImageJ. The promoter sequences were retrieved using the Genomatix Software Suite, and sequence alignment was performed using Clustal Omega from EMBL.
Author contributions
S. M., J. M., M. S., S. K., V. R., A. J., D. K., and N. R. S. resources; S. M., J. M., M. S., S. K., F. A., K. M. S., A. J., D. K., B. P. A., and P. A. D. data curation; S. M., J. M., M. S., S. K., F. A., K. M. S., V. R., A. J., D. K., B. P. A., P. A. D., and N. R. S. formal analysis; S. M., J. M., M. S., S. K., F. A., K. M. S., A. J., D. K., and P. A. D. validation; S. M., J. M., M. S., S. K., F. A., K. M. S., V. R., A. J., D. K., P. A. D., and N. R. S. investigation; S. M., J. M., M. S., S. K., F. A., K. M. S., V. R., A. J., D. K., and P. A. D. methodology; S. M. and J. M. writing-original draft; V. R. and N. R. S. writing-review and editing; N. R. S. conceptualization; N. R. S. supervision; N. R. S. funding acquisition; N. R. S. project administration.
Supplementary Material
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains File S1 and Figs. S1–S7.
- TGF-β1
- transforming growth factor β1
- TARE
- TGF-β/activin-response element
- α-SMA
- α-smooth muscle actin
- H3K9 and H3K56
- histone H3 Lys-9 and Lys-56, respectively
- IGF
- insulin-like growth factor
- qPCR
- quantitative PCR
- SBE
- SMAD3-binding element(s)
- Col1a and Col3a
- collagen 1a and 3a, respectively
- FN1
- fibronectin 1
- HPRT
- hypoxanthine phosphoribosyltransferase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HRP
- horseradish peroxidase
- ANOVA
- analysis of variance
- KO
- knockout
- p-SMAD3
- phosphorylated SMAD3
- WCL
- whole-cell lysate(s)
- CBP
- CREB-binding protein.
References
- 1. Ravussin E., Redman L. M., Rochon J., Das S. K., Fontana L., Kraus W. E., Romashkan S., Williamson D. A., Meydani S. N., Villareal D. T., Smith S. R., Stein R. I., Scott T. M., Stewart T. M., Saltzman E., et al. (2015) A 2-year randomized controlled trial of human caloric restriction: feasibility and effects on predictors of health span and longevity. J. Gerontol. A Biol. Sci. Med. Sci. 70, 1097–1104 10.1093/gerona/glv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dolinsky V. W., and Dyck J. R. B. (2011) Calorie restriction and resveratrol in cardiovascular health and disease. Biochim. Biophys. Acta 1812, 1477–1489 10.1016/j.bbadis.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 3. Yan L., Gao S., Ho D., Park M., Ge H., Wang C., Tian Y., Lai L., De Lorenzo M. S., Vatner D. E., and Vatner S. F. (2013) Calorie restriction can reverse, as well as prevent, aging cardiomyopathy. AGE 35, 2177–2182 10.1007/s11357-012-9508-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harvey A. E., Lashinger L. M., Hays D., Harrison L. M., Lewis K., Fischer S. M., and Hursting S. D. (2014) Calorie restriction decreases murine and human pancreatic tumor cell growth, nuclear factor-κB activation, and inflammation-related gene expression in an insulin-like growth factor-1-dependent manner. PLoS ONE 9, e94151 10.1371/journal.pone.0094151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chalkiadaki A., and Guarente L. (2015) The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer 15, 608–624 10.1038/nrc3985 [DOI] [PubMed] [Google Scholar]
- 6. Tasselli L., Xi Y., Zheng W., Tennen R. I., Odrowaz Z., Simeoni F., Li W., and Chua K. F. (2016) SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 23, 434–440 10.1038/nsmb.3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tasselli L., Zheng W., and Chua K. F. (2017) SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 28, 168–185 10.1016/j.tem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sundaresan N. R., Vasudevan P., Zhong L., Kim G., Samant S., Parekh V., Pillai V. B., Ravindra P. V., Gupta M., Jeevanandam V., Cunningham J. M., Deng C.-X., Lombard D. B., Mostoslavsky R., and Gupta M. P. (2012) The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 18, 1643–1650 10.1038/nm.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., and Cohen H. Y. (2012) The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 10.1038/nature10815 [DOI] [PubMed] [Google Scholar]
- 10. Mostoslavsky R., Chua K. F., Lombard D. B., Pang W. W., Fischer M. R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M. M., Mills K. D., Patel P., Hsu J. T., Hong A. L., Ford E., et al. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 10.1016/j.cell.2005.11.044 [DOI] [PubMed] [Google Scholar]
- 11. Bonkowski M. S., and Sinclair D. A. (2016) Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679–690 10.1038/nrm.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgley A. J., Krum H., and Kelly D. J. (2012) Targeting fibrosis for the treatment of heart failure: a role for transforming growth factor-β. Cardiovasc. Ther. 30, e30–e40 10.1111/j.1755-5922.2010.00228.x [DOI] [PubMed] [Google Scholar]
- 13. Heldin C. H., Miyazono K., and ten Dijke P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471 10.1038/37284 [DOI] [PubMed] [Google Scholar]
- 14. Shi Y., and Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- 15. Gomes N. P., Bjerke G., Llorente B., Szostek S. A., Emerson B. M., and Espinosa J. M. (2006) Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20, 601–612 10.1101/gad.1398206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Massagué J., and Chen Y. G. (2000) Controlling TGF-β signaling. Genes Dev. 14, 627–644 [PubMed] [Google Scholar]
- 17. Leask A., and Abraham D. J. (2004) TGF-β signaling and the fibrotic response. FASEB J. 18, 816–827 10.1096/fj.03-1273rev [DOI] [PubMed] [Google Scholar]
- 18. Meng X. M., Nikolic-Paterson D. J., and Lan H. Y. (2016) TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338 10.1038/nrneph.2016.48 [DOI] [PubMed] [Google Scholar]
- 19. Doyle K. P., Cekanaviciute E., Mamer L. E., and Buckwalter M. S. (2010) TGFβ signaling in the brain increases with aging and signals to astrocytes and innate immune cells in the weeks after stroke. J. Neuroinflammation 7, 62 10.1186/1742-2094-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Z. Y., Chen Z. L., Zhang T., Wei C., and Shi W. Y. (2016) TGF-β and NF-κB signaling pathway crosstalk potentiates corneal epithelial senescence through an RNA stress response. Aging 8, 2337–2354 10.18632/aging.101050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biernacka A., and Frangogiannis N. G. (2011) Aging and cardiac fibrosis. Aging and disease 2, 158–173 [PMC free article] [PubMed] [Google Scholar]
- 22. Neveu W. A., Mills S. T., Staitieh B. S., and Sueblinvong V. (2015) TGF-β1 epigenetically modifies Thy-1 expression in primary lung fibroblasts. Am. J. Physiol. Cell Physiol. 309, C616–C626 10.1152/ajpcell.00086.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruiz-Torres M. P., Bosch R. J., O'Valle F., Del Moral R. G., Ramírez C., Masseroli M., Pérez-Caballero C., Iglesias M. C., Puyol M., and Rodríguez-Puyol D. (1998) Age-related increase in expression of TGF-β1 in the rat kidney: relationship to morphologic changes. J. Am. Soc. Nephrol. 9, 782–791 [DOI] [PubMed] [Google Scholar]
- 24. Kim I. H., Xu J., Liu X., Koyama Y., Ma H. Y., Diggle K., You Y. H., Schilling J. M., Jeste D., Sharma K., Brenner D. A., and Kisseleva T. (2016) Aging increases the susceptibility of hepatic inflammation, liver fibrosis and aging in response to high-fat diet in mice. Age 38, 291–302 10.1007/s11357-016-9938-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rapisarda V., Borghesan M., Miguela V., Encheva V., Snijders A. P., Lujambio A., and O'Loghlen A. (2017) Integrin β3 regulates cellular senescence by activating the TGF-β pathway. Cell Rep. 18, 2480–2493 10.1016/j.celrep.2017.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edgley A. J., Krum H., and Kelly D. J. (2012) Targeting fibrosis for the treatment of heart failure: a role for transforming growth factor-β. Cardiovasc. Ther. 30, e30–e40 10.1111/j.1755-5922.2010.00228.x [DOI] [PubMed] [Google Scholar]
- 27. Laurent G. J., Coker R. K., and McAnulty R. J. (1993) TGF-β antibodies: a novel treatment for pulmonary fibrosis? Thorax 48, 953–954 10.1136/thx.48.10.953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamamoto T., Takagawa S., Katayama I., and Nishioka K. (1999) Anti-sclerotic effect of transforming growth factor-β antibody in a mouse model of bleomycin-induced scleroderma. Clin. Immunol. 92, 6–13 10.1006/clim.1999.4720 [DOI] [PubMed] [Google Scholar]
- 29. Giri S. N., Hyde D. M., and Hollinger M. A. (1993) Effect of antibody to transforming growth factor β on bleomycin induced accumulation of lung collagen in mice. Thorax 48, 959–966 10.1136/thx.48.10.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akhurst R. J., and Hata A. (2012) Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 10.1038/nrd3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ravi V., Jain A., Khan D., Ahamed F., Mishra S., Giri M., Inbaraj M., Krishna S., Sarikhani M., Maity S., Kumar S., Shah R. A., Dave P., Pandit A. S., Rajendran R., et al. (2019) SIRT6 transcriptionally regulates global protein synthesis through transcription factor Sp1 independent of its deacetylase activity. Nucleic Acids Res. 47, 9115–9131 10.1093/nar/gkz648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khan S. A., Joyce J., and Tsuda T. (2012) Quantification of active and total transforming growth factor-β levels in serum and solid organ tissues by bioassay. BMC Res. Notes 5, 636 10.1186/1756-0500-5-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho J. S., Moon Y. M., Park I. H., Um J. Y., Moon J. H., Park S. J., Lee S. H., Kang H. J., and Lee H. M. (2012) Epigenetic regulation of myofibroblast differentiation and extracellular matrix production in nasal polyp-derived fibroblasts. Clin. Exp. Allergy 42, 872–882 10.1111/j.1365-2222.2011.03931.x [DOI] [PubMed] [Google Scholar]
- 34. Kashiwagi I., Morita R., Schichita T., Komai K., Saeki K., Matsumoto M., Takeda K., Nomura M., Hayashi A., Kanai T., and Yoshimura A. (2015) Smad2 and Smad3 inversely regulate TGF-β autoinduction in Clostridium butyricum-activated dendritic cells. Immunity 43, 65–79 10.1016/j.immuni.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 35. Inoue Y., Itoh Y., Abe K., Okamoto T., Daitoku H., Fukamizu A., Onozaki K., and Hayashi H. (2007) Smad3 is acetylated by p300/CBP to regulate its transactivation activity. Oncogene 26, 500–508 10.1038/sj.onc.1209826 [DOI] [PubMed] [Google Scholar]
- 36. Bhardwaj A., and Das S. (2016) SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc. Natl. Acad. Sci. U.S.A. 113, E538–E547 10.1073/pnas.1520045113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jinnin M., Ihn H., and Tamaki K. (2006) Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-β1-induced extracellular matrix expression. Mol. Pharmacol. 69, 597–607 10.1124/mol.105.017483 [DOI] [PubMed] [Google Scholar]
- 38. Sarikhani M., Mishra S., Maity S., Kotyada C., Wolfgeher D., Gupta M. P., Singh M., and Sundaresan N. R. (2018) SIRT2 deacetylase regulates the activity of GSK3 isoforms independent of inhibitory phosphorylation. eLife 7, e32952 10.7554/eLife.32952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian K., Liu Z., Wang J., Xu S., You T., and Liu P. (2015) Sirtuin-6 inhibits cardiac fibroblasts differentiation into myofibroblasts via inactivation of nuclear factor κB signaling. Transl. Res. 165, 374–386 10.1016/j.trsl.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 40. Xiao C., Wang R.-H., Lahusen T. J., Park O., Bertola A., Maruyama T., Reynolds D., Chen Q., Xu X., Young H. A., Chen W.-J., Gao B., and Deng C.-X. (2012) Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in Sirt6 mutant mice. J. Biol. Chem. 287, 41903–41913 10.1074/jbc.M112.415182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zorrilla-Zubilete M. A., Yeste A., Quintana F. J., Toiber D., Mostoslavsky R., and Silberman D. M. (2018) Epigenetic control of early neurodegenerative events in diabetic retinopathy by the histone deacetylase SIRT6. J. Neurochem. 144, 128–138 10.1111/jnc.14243 [DOI] [PubMed] [Google Scholar]
- 42. Minagawa S., Araya J., Numata T., Nojiri S., Hara H., Yumino Y., Kawaishi M., Odaka M., Morikawa T., Nishimura S. L., Nakayama K., and Kuwano K. (2011) Accelerated epithelial cell senescence in IPF and the inhibitory role of SIRT6 in TGF-β-induced senescence of human bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 300, L391–L401 10.1152/ajplung.00097.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baohua Y., and Li L. (2012) Effects of SIRT6 silencing on collagen metabolism in human dermal fibroblasts. Cell Biol. Int. 36, 105–108 10.1042/CBI20110268 [DOI] [PubMed] [Google Scholar]
- 44. Zerr P., Palumbo-Zerr K., Huang J., Tomcik M., Sumova B., Distler O., Schett G., and Distler J. H. W. (2014) Sirt1 regulates canonical TGF-β signalling to control fibroblast activation and tissue fibrosis. Ann. Rheum. Dis. 75, 226–233 10.1136/annrheumdis-2014-205740 [DOI] [PubMed] [Google Scholar]
- 45. Feng X.-X., Luo J., Liu M., Yan W., Zhou Z.-Z., Xia Y.-J., Tu W., Li P.-Y., Feng Z.-H., and Tian D.-A. (2015) Sirtuin 6 promotes transforming growth factor-β1/H2O2/HOCl-mediated enhancement of hepatocellular carcinoma cell tumorigenicity by suppressing cellular senescence. Cancer Sci. 106, 559–566 10.1111/cas.12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yao Q. P., Zhang P., Qi Y. X., Chen S. G., Shen B. R., Han Y., Yan Z. Q., and Jiang Z. L. (2014) The role of SIRT6 in the differentiation of vascular smooth muscle cells in response to cyclic strain. Int. J. Biochem. Cell Biol. 49, 98–104 10.1016/j.biocel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- 47. Knaapen P., Götte M. J. W., Paulus W. J., Zwanenburg J. J. M., Dijkmans P. A., Boellaard R., Marcus J. T., Twisk J. W. R., Visser C. A., van Rossum A. C., Lammertsma A. A., and Visser F. C. (2006) Does myocardial fibrosis hinder contractile function and perfusion in idiopathic dilated cardiomyopathy? PET and MR imaging study. Radiology 240, 380–388 10.1148/radiol.2402051038 [DOI] [PubMed] [Google Scholar]
- 48. Krenning G., Zeisberg E. M., and Kalluri R. (2010) The origin of fibroblasts and mechanism of cardiac fibrosis. J. Cell. Physiol. 225, 631–637 10.1002/jcp.22322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mann D. L., and Bristow M. R. (2005) Mechanisms and models in heart failure: the biomechanical model and beyond. Circulation 111, 2837–2849 10.1161/CIRCULATIONAHA.104.500546 [DOI] [PubMed] [Google Scholar]
- 50. Díez J. (2007) Mechanisms of cardiac fibrosis in hypertension. J. Clin. Hypertens. 9, 546–550 10.1111/j.1524-6175.2007.06626.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Creemers E. E., and Pinto Y. M. (2010) Molecular mechanisms that control interstitial fibrosis in the pressure-overloaded heart. Cardiovascular research 89, 265–272 10.1093/cvr/cvq308 [DOI] [PubMed] [Google Scholar]
- 52. Davis J., and Molkentin J. D. (2014) Myofibroblasts: trust your heart and let fate decide. J. Mol. Cell. Cardiol. 70, 9–18 10.1016/j.yjmcc.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang X.-Z., Wen D., Zhang M., Xie Q., Ma L., Guan Y., Ren Y., Chen J., and Hao C.-M. (2014) Sirt1 activation ameliorates renal fibrosis by inhibiting the TGF-β/Smad3 pathway. J. Cell. Biochem. 115, 996–1005 10.1002/jcb.24748 [DOI] [PubMed] [Google Scholar]
- 54. Cappetta D., Esposito G., Piegari E., Russo R., Ciuffreda L. P., Rivellino A., Berrino L., Rossi F., De Angelis A., and Urbanek K. (2016) SIRT1 activation attenuates diastolic dysfunction by reducing cardiac fibrosis in a model of anthracycline cardiomyopathy. Int. J. Cardiol. 205, 99–110 10.1016/j.ijcard.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 55. Arteaga M., Shang N., Ding X., Yong S., Cotler S. J., Denning M. F., Shimamura T., Breslin P., Lüscher B., and Qiu W. (2016) Inhibition of SIRT2 suppresses hepatic fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G1155–G1168 10.1152/ajpgi.00271.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sundaresan N. R., Bindu S., Pillai V. B., Samant S., Pan Y., Huang J.-Y., Gupta M., Nagalingam R. S., Wolfgeher D., Verdin E., and Gupta M. P. (2015) SIRT3 blocks aging-associated tissue fibrosis in mice by deacetylating and activating GSK3β. Mol. Cell Biol. 36, 678–692 10.1128/MCB.00586-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sadhukhan S., Liu X., Ryu D., Nelson O. D., Stupinski J. A., Li Z., Chen W., Zhang S., Weiss R. S., Locasale J. W., Auwerx J., and Lin H. (2016) Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc. Natl. Acad. Sci. U.S.A. 113, 4320–4325 10.1073/pnas.1519858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo Y.-X., Tang X., An X.-Z., Xie X.-M., Chen X.-F., Zhao X., Hao D.-L., Chen H.-Z., and Liu D.-P. (2016) Sirt4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur. Heart J. 38, 1389–1398 10.1093/eurheartj/ehw138 [DOI] [PubMed] [Google Scholar]
- 59. Araki S., Izumiya Y., Rokutanda T., Ianni A., Hanatani S., Kimura Y., Onoue Y., Senokuchi T., Yoshizawa T., Yasuda O., Koitabashi N., Kurabayashi M., Braun T., Bober E., Yamagata K., and Ogawa H. (2015) Sirt7 contributes to myocardial tissue repair by maintaining TGF-β signaling pathway. Circulation 132, 1081–1093 10.1161/CIRCULATIONAHA.114.014821 [DOI] [PubMed] [Google Scholar]
- 60. Zhang X., Yang J., Li Y., and Liu Y. (2005) Both Sp1 and Smad participate in mediating TGF-β1-induced HGF receptor expression in renal epithelial cells. Am. J. Physiol. Renal Physiol. 288, F16–F26 10.1152/ajprenal.00318.2003 [DOI] [PubMed] [Google Scholar]
- 61. Verrecchia F., Vindevoghel L., Lechleider R. J., Uitto J., Roberts A. B., and Mauviel A. (2001) Smad3/AP-1 interactions control transcriptional responses to TGF-β in a promoter-specific manner. Oncogene 20, 3332–3340 10.1038/sj.onc.1204448 [DOI] [PubMed] [Google Scholar]
- 62. Weigert C., Sauer U., Brodbeck K., Pfeiffer A., Häring H. U., and Schleicher E. D. (2000) AP-1 proteins mediate hyperglycemia-induced activation of the human TGF-β1 promoter in mesangial cells. J. Am. Soc. Nephrol. 11, 2007–2016 [DOI] [PubMed] [Google Scholar]
- 63. Li R., Gong K., and Zhang Z. (2017) [Isolation, purification and primary culture of adult mouse cardiac fibroblasts]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 33, 67–71 [PubMed] [Google Scholar]
- 64. Poncelet A. C., and Schnaper H. W. (2001) Sp1 and Smad proteins cooperate to mediate transforming growth factor-β1-induced α2(I) collagen expression in human glomerular mesangial cells. J. Biol. Chem. 276, 6983–6992 10.1074/jbc.M006442200 [DOI] [PubMed] [Google Scholar]
- 65. Wang W., Huang X. R., Canlas E., Oka K., Truong L. D., Deng C., Bhowmick N. A., Ju W., Bottinger E. P., and Lan H. Y. (2006) Essential role of Smad3 in angiotensin II–induced vascular fibrosis. Circ. Res. 98, 1032–1039 10.1161/01.RES.0000218782.52610.dc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang X. R., Chung A. C. K., Yang F., Yue W., Deng C., Lau C. P., Tse H. F., and Lan H. Y. (2010) Smad3 mediates cardiac inflammation and fibrosis in angiotensin II–induced hypertensive cardiac remodeling. Hypertension 55, 1165–1171 10.1161/HYPERTENSIONAHA.109.147611 [DOI] [PubMed] [Google Scholar]
- 67. Kim S., Lim J. H., and Woo C.-H. (2013) ERK5 inhibition ameliorates pulmonary fibrosis via regulating Smad3 acetylation. Am. J. Pathol. 183, 1758–1768 10.1016/j.ajpath.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 68. Schultz Jel J., Witt S. A., Glascock B. J., Nieman M. L., Reiser P. J., Nix S. L., Kimball T. R., and Doetschman T. (2002) TGF-β1 mediates the hypertrophic cardiomyocyte growth induced by angiotensin II. J. Clin. Invest. 109, 787–796 10.1172/JCI0214190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sharma A., Diecke S., Zhang W. Y., Lan F., He C., Mordwinkin N. M., Chua K. F., and Wu J. C. (2013) The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J. Biol. Chem. 288, 18439–18447 10.1074/jbc.M112.405928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao G., Wang H., Xu C., Wang P., Chen J., Wang P., Sun Z., Su Y., Wang Z., Han L., and Tong T. (2016) SIRT6 delays cellular senescence by promoting p27Kip1 ubiquitin-proteasome degradation. Aging 8, 2308–2323 10.18632/aging.101038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.