Abstract
Cyclophyllidean tapeworms obligatorily parasitize numerous mammalian species, including herbivores, domestic animals and humans, of which, the genera Taenia and Mesocestoides are well characterized. However, little is known about these parasitic infections in wild animals. This study aims to investigate the prevalence and distribution of Taenia sp. and Mesocestoides sp. in wild carnivores in Mongolia by identifying tapeworm species based on mtDNA analysis. The field survey was carried out in 2012–2013 in 19 provinces located in different ecological regions. A total of 405 fecal samples from wild carnivores were collected. Specific DNA markers in fecal samples was detected via copro-DNA analysis and tapeworm species were identified by DNA sequencing. From 27.7% (112/405) of samples, cox1 and 12S rRNA genes of tapeworms were amplified. Further, Taenia hydatigena (50.0%, 56/112) and two Mesocestoides species, including Mesocestoides sp.-1 (36.6%, 41/112) and Mesocestoides sp.-2 (13.4%, 15/112) were identified by DNA sequencing. The prevalence of T. hydatigena was 19.9% (27/136), 13.8% (23/167), 4.8% (3/62), and 7.5% (3/40) in wolves, red foxes, corsac foxes, and snow leopards, respectively. The prevalence of Mesocestoides sp.-1 was 14.7% (20/136), 9% (15/167), 9.7% (6/62) in wolves, red foxes, and corsac foxes, while the prevalence of Mesocestoides sp.-2 was 4.4% (6/136), 1.8% (3/167), 3.2% (2/62), and 10.0% (4/40) in wolves, red foxes, corsac foxes, and snow leopards, respectively. T. hydatigena was found throughout all ecological regions, while Mesocestoides sp.-1 was in the mountain taiga, forest-steppe, steppe, desert-steppe, and desert, and Mesocestoides sp.-2 in the alpine, forest-steppe, steppe, and desert-steppe ecoregions. This study revealed the prevalence and distribution of cyclophyllidean tapeworms in wild carnivores in Mongolia; while also confirming that wolves, red foxes, corsac foxes, and snow leopards serve as definitive hosts for unidentified Mesocestoides species.
Keywords: Taenia hydatigena, Mesocestoides sp., cox1 gene, 12S rRNA gene, Wild carnivores, Mongolia
Highlights
-
•
Taenia hydatigena and two Mesocestoides species were first identified by copro-DNA analysis in wild carnivores in Mongolia.
-
•
The persistent infections by T. hydatigena and Mesocestoides species in these animals were shown.
-
•
T. hydatigena was detected in all ecoregions in Mongolia.
-
•
Mesocestoides sp.-1 was found in wild carnivores inhabiting in all ecoregion unless in the alpine, and Mesocestoides sp.-2 in the alpine, forest-steppe, steppe, and desert-steppe ecoregions.
-
•
Wolf, red fox, corsac fox, and snow leopard were confirmed as definitive hosts for Mesocestoides species.
1. Introduction
Mongolia is a land locked country located in Central and East Asia bordering with the Russian Federation in the north and the People's Republic of China in the west, south, and east, and consists of 21 provinces. Great diversities characterize the geography of the country. Currently, from north to south, it can be divided into four regions (Western, Khangai, Central, and Eastern), and six ecological regions: alpine, mountain taiga, forest-steppe, steppe, desert-steppe, and desert (Table 2, Table 3, Fig. 1). The total population of Mongolia is 3.238.479 (NSO, 2019); of which nearly 40% of the rural population is nomadic or semi-nomadic herdsmen (Myadagsuren et al., 2007).
Table 2.
Prevalence of T. hydatigena and Mesocestoides sp. by region and province in Mongolia.
| Region | Province | Number and prevalence (%) |
||
|---|---|---|---|---|
| T. hydatigena | Mesocestoides sp.-1 | Mesocestoides sp.-2 | ||
| Western | ||||
| Bayan-Ulgii | 2/16 (12.5) | 0/16 (0.0) | 2/16 (12.5) | |
| Govi-Altai | 0/16 (0.0) | 1/16 (6.3) | 2/16 (12.5) | |
| Zavkhan | 3/19 (15.8) | 6/19 (31.6) | 3/19 (15.8) | |
| Uvs | 4/21 (19.0) | 2/21 (9.5) | 1/21 (4.8) | |
| Khovd | 2/16 (12.5) | 0/16 (0.0) | 2/16 (12.5) | |
| Khangai | ||||
| Arkhangai | 3/26 (11.5) | 2/26 (7.7) | 0/26 (0.0) | |
| Bayankhongor | 2/21 (9.5) | 2/21 (9.5) | 1/21 (4.8) | |
| Bulgan | 5/23 (21.7) | 5/23 (21.7) | 0/23 (0.0) | |
| Uvurkhangai | 3/14 (21.4) | 2/14 (14.3) | 1/14 (7.1) | |
| Khuvsgul | 0/8 (0.0) | 1/8 (12.5) | 0/8 (0.0) | |
| Orkhon-Uul | 5/28 (17.9) | 1/28 (3.6) | 0/28 (0.0) | |
| Central | ||||
| Gobi-Sumber | ||||
| Dornogobi | 2/49 (4.1) | 2/49 (4.1) | 1/49 (2.0) | |
| Dundgobi | 5/28 (17.9) | 3/28 (10.7) | 0/28 (0.0) | |
| Umnu-Gobi | 2/25 (8.0) | 2/25 (8.0) | 2/25 (8.0) | |
| Selenge | 2/23 (8.7) | 4/23 (17.4) | 0/23 (0.0) | |
| Tuv | 7/25 (28.0) | 1/25 (4.0) | 0/25 (0.0) | |
| Darkhan-Uul | 3/12 (25.0) | 2/12 (16.7) | 0/12 (0.0) | |
| Eastern | ||||
| Dornod | ||||
| Sukhbaatar | 4/22 (18.2) | 4/22 (18.2) | 0/22 (0.0) | |
| Khentii | 2/13 (15.4) | 1/13 (7.7) | 0/13 (0.0) | |
Table 3.
Prevalence of T. hydatigena and Mesocestoides sp. infections in wild carnivores by ecoregions in Mongolia.
| Ecoregion/Wild carnivore | No.of fecal sample | Number and prevalence (%) |
||
|---|---|---|---|---|
| T. hydatigena | Mesocestoides sp.-1 | Mesocestoides sp.-2 | ||
| Alpine (n = 38) | ||||
| Wolf | 10 | 1/10 (10.0) | 0/10 (0.0) | 0/10 (0.0) |
| Snow leopard | 28 | 3/28 (10.7) | 0/28 (0.0) | 4/28 (14.3) |
| Mountain taiga (n = 26) | ||||
| Wolf | 14 | 2/14 (14.3) | 3/14 (21.4) | 0/14 (0.0) |
| Snow leopard | 12 | 0/12 (0.0) | 0/12 (0.0) | 0/12 (0.0) |
| Forest-steppe (n = 113) | ||||
| Wolf | 41 | 10/41(24.4) | 7/41 (17.1) | 0/41 (0.0) |
| Red fox | 49 | 7/49 (14.3) | 3/49 (6.1) | 1/49 (2.0) |
| Corsac fox | 23 | 0/23 (0.0) | 2/23 (8.7) | 0/23 0.0) |
| Steppe (n = 89) | ||||
| Wolf | 40 | 8/40 (20.0) | 2/40 (5.0) | 6/40 (15.0) |
| Red fox | 36 | 5/36 (13.9) | 5/36 (13.9) | 2/36 (5.6) |
| Corsac fox | 13 | 0/13 (0.0) | 4/13 (30.8) | 0/13 (0.0) |
| Desert-steppe (n = 101) | ||||
| Wolf | 23 | 5/23 (21.7) | 6/23 (26.1) | 0/23 (0.0) |
| Red fox | 59 | 11/59 (18.6) | 0/59 (0.0) | 0/59 (0.0) |
| Corsac fox | 19 | 1/19 (5.3) | 0/19 (0.0) | 2/19 (10.5) |
| Desert (n = 38) | ||||
| Wolf | 8 | 1/8 (12.5) | 2/8 (25.0) | 0/8 (0.0) |
| Red fox | 23 | 0/23 (0.0) | 7/23 (30.4) | 0/23 (0.0) |
| Corsac fox | 7 | 2/7 (28.6) | 0/7 (0.0) | 0/7 (0.0) |
Fig. 1.
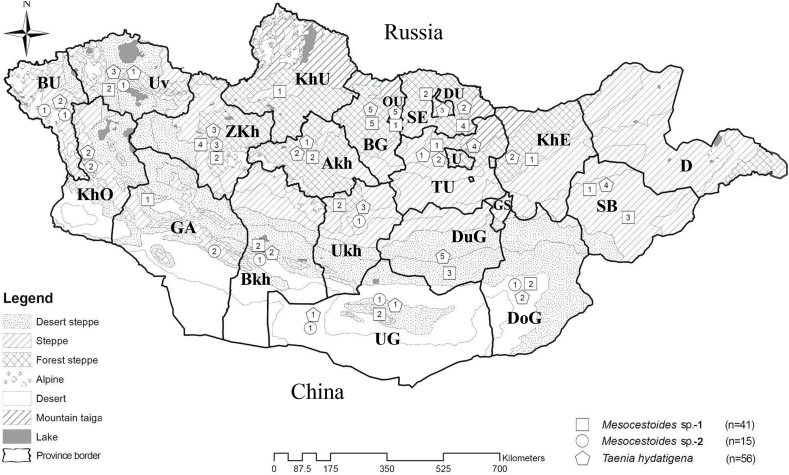
Map of Mongolia showing the distribution of Taenia hydatigena (pentangle), Mesocestoides sp.-1 (square), and Mesocestoides sp.-2 (circle) by province detected by molecular identification of fecal samples from wild carnivores. Mongolia consists of 21 provinces: Arkhangai (Akh), Bayankhongor (Bkh), Bayan-Ulgii (BU), Bulgan (BG), Darkhan-Uul (DU), Dornogobi (DoG), Dundgobi (DuG), Govi-Altai (GA), Khentii (KhE), Khovd (KhO), Khuvsgul (KhU), Orkhon-Uul (OU), Selenge (SE), Sukhbaatar (SB), Tuv (TU), Umnu-Gobi (UG), Uvs (Uv), Uvurkhangai (Ukh), Zavkhan (ZKh), Dornod (D), and Gobi-Sumber (GS). The field survey was conducted in all provinces, unless Dornod (D), and GS (Gobi-Sumber). Ulaanbaatar (U) is the capital city of Mongolia. The field survey was conducted in all provinces, unless Dornod (D), and GS (Gobi-Sumber). Ulaanbaatar (U) is the capital city of Mongolia located in Tuv Province.
Tapeworms of the genus Taenia include over 100 species (Gonzalez et al, 2018) that affect dogs, cats, goats, sheep, cattle, pigs, and other livestock, domestic and wild animals as well as humans. The lifecycle of the Taenia species relies on a vertebrate intermediate host in which the infective larvae develop, as well as on a definitive host that ingests the uncooked flesh of the intermediate host (Gonzalez et al, 2018). Taenia sp. is distributed worldwide, whereby abundance and incidence in different regions depend on each particular species (Rostami et al., 2015; Poglayen et al., 2017; Kubečka et al., 2018).
T. hydatigena is an omnipresent tapeworm found in domestic animals worldwide (Nguyen et al., 2016; Miran, 2017), exerting a significant constraint on the development of the livestock industry in developing countries. Furthermore, in Mongolia it has been reported that carnivores such as gray wolves, red fox, corsac fox, and dogs are the definitive hosts for T. hydatigena (Dubinin and Dubinina, 1951; Danzan, 1978; Tinnin et al., 2002); whereas, the metacestodes found in sheep, goat, cattle, argali, ibex deer, and roe deer, serve as intermediate hosts (Danzan, 1978; Sharkhuu, 2001; Sharhuu and Sharkhuu, 2004). These intermediate hosts may become infected by environments that are contaminated by infected wild carnivores or dogs, primarily wolves, which become infected following consumption of infected wild and domestic livestock. In fact, within dogs in Mongolia, the prevalence of T. hydatigena was reported as 61.3% (Danzan, 1978). Intermediate hosts containing T. hydatigena cysticerci in the migratory phase, present with hemorrhaging in the liver parenchyma and beneath the liver surface (Blazek et al., 1985). Moreover, cysticerci are permitted to develop in hosts when immature immune systems are incapable of defeating the parasite. Mass infection of T. hydatigena cysticerci may cause the death of infected animals during the migration of cysticerci (Scala et al., 2016; Sgroi et al., 2019).
T. hydatigena accounts for one of the most prevalent tapeworm infections in intermediate hosts, causing significant negative impacts to the health of infected animals (Getaw et al., 2010; Dumitri et al., 2011; Oryan et al., 2012; Debas and Ibrahim, 2013), and significantly impacting the livestock industry in developing countries such as Mongolia (Jenkins et al., 2014; Nguyen et al., 2016; Miran, 2017). Specifically, T. hydatigena infected livestock can result in meat condemnation (Oryan et al., 2012; Debas and Ibrahim, 2013; Rashid et al., 2019). Since animal husbandry accounts for approximately 20–30% of Mongolia's GDP (MoFALI, 2018), tapeworm infections represent a significant economic burden on herders who practice a nomadic lifestyle. However, T. hydatigena is not only detrimental to developing countries, but has also been reported to cause significant economic losses, reaching 330,000 euros, in developed nations (Scala et al., 2015).
Mesocestoides sp. exhibit unique characteristics compared to other groups of cyclophyllideans (Cho et al., 2013). For instance, the life cycle of Mesocestoides is complex, requiring two or three intermediate hosts to complete its development (Zalesny and Hildebrand, 2012; McAllister et al., 2013; Poglayen et al., 2017). The oncosphere Mesocestoides develops into a second-stage larva in the first intermediate host (arthropod); while in the second intermediate host, such as small mammals (Loos-Frank, 1991) and reptiles, the larva develops into third-stage (tetrathyridium). Among the Mesocestoides isolates, M. litteratus and M. lineatus were commonly isolated from red foxes, dogs, coyotes and wolves, which served as definitive hosts (Hrčkova et al., 2011; Zalesny and Hildebrand, 2012). Alternatively, in Mongolia domestic or wild animals such as sheep, goats, cattle, argali, ibex deer, and roe deer serve as the second intermediate hosts (Danzan, 1978; Sharhuu and Sharkhuu, 2004). Although there are few studies regarding the identity of definitive hosts in Mongolia, it has been reported that definitive host becomes infected after eating meat contaminated with tetrathyridia (CDC, 2017). Once infected, the intermediate host may experience hemorrhagia within the liver (Blazek et al., 1985), while the definitive hosts have their small intestines colonized by Mesocestoides sp., which can prove to be very dangerous (Jabbar et al., 2012) for rare species such as the snow leopard.
Cestodes represent a significant risk to the health of both humans and their livestock. However, little is known about these tapeworm infections in wild animals. Within Mongolia, soil-transmitted cestode infections are increasing in prevalence, making the need for accurate molecular characterization, and mapping of the geographic distribution of the parasites, of utmost importance if efficient prevention and control options are to be designed (Ebright et al., 2003; McFadden et al., 2016).
This study aims to investigate the prevalence and distribution of Taenia sp. and Mesocestoides sp. in wild carnivores in Mongolia by identifying tapeworm species based on copro-DNA analysis.
2. Materials and methods
2.1. Collection of canid and felid fecal samples
A total of 405 fecal samples were randomly collected from 167, 136, 62, and 40 wolves, red foxes, corsac foxes, and snow leopards, respectively, from 19 of the 21 Mongolian provinces, save for Dornod and Gobi-Sumber, representing all ecological regions of Mongolia (Fig. 1, Table 1). Each fecal sample weighed 18 g. The feces were collected using standard techniques (Lawrence and Brown, 1967; Strachan et al., 1996). The species for each feces sample was visually identified based on color, shape, location, pugmarks, scrapes, and the nearby remains of prey species, by biologists, ecologists and local hunters (Oli et al., 1993; Bachi and Mishra, 2006). Each sample was stored in individually labeled zip-lock plastic bags to prevent contamination, and were stored at −80 °C until processing.
Table 1.
Number of PCR amplified in 405 fecal samples and prevalence of T. hydatigena and Mesocestoides sp. infections by wild carnivores in Mongolia.
| PCR amplified/Infecting parasite | Number and prevalence (%) |
|||
|---|---|---|---|---|
| Wild carnivore | ||||
| Wolf (n = 136) | Red fox (n = 167) | Corsac fox (n = 62) | Snow leopard (n = 40) | |
| PCR amplified | 53/112 (47.3) | 41/112 (36.6) | 11/112 (9.8) | 7/112 (6.3) |
| Infecting parasite | ||||
| T. hydatigena | 27/136 (19.9) | 23/167 (13.8) | 3/62 (4.8) | 3/40 (7.5) |
| Mesocestoides sp.-1 | 20/136 (14.7) | 15/167 (9.0) | 6/62 (9.7) | 0/40 (0.0) |
| Mesocestoides sp.-2 | 6/136 (4.4) | 3/167 (1.8) | 2/62 (3.2) | 4/40 (10.0) |
2.2. DNA analysis
The copro-DNA samples were extracted from frozen feces using the QIAamp DNA Stool Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The cytochrome c oxidase subunit 1 (cox1) gene and 12S rRNA was amplified by polymerase chain reaction (PCR) using previously published primers (Bowles and McManus,1993; von Nickisch-Rosenegk et al., 1999). Amplicons were sequenced in both directions on a 3100-Advant Genetic Analyzer (ABI PRISM, Applied Biosystems, Hitachi, Japan).
2.3. Phylogenetic analysis
Sequence data were assembled and edited using BioEdit (version 7.2.5) and were compared to the reference sequences in GenBank® by using BLAST. Phylogenetic trees were constructed based on the partial nucleotide sequences of cox1 and 12S rRNA, together with reference sequences available in GenBank and using the MEGA software, version 7.0 (http://www.megasoftware.net/mega7). The evolutionary distances were computed using the maximum likelihood estimation method (HKY + G + I substitution model) and neighbor joining (TN93 + G substitution model) was presented as the number of base substitutions per site (Kumar et al., 2016).
2.4. Data analysis
Infection prevalence was determined for the study years. A comparison of infection prevalence was conducted using the Pearson's chi-squared test. All analyses were performed using SPSS v.22 (IBM). Arlequin 3.1 (Excoffier et al., 2005) was used to calculate population diversity indices (haplotype (h) and nucleotide (π) diversities), neutrality indices, Tajima's D (Tajima, 1989) and Fu's Fs (Fu, 1997). A p-value < 0.05 was considered statistically significant.
This study was approved by the Research Ethics Committee of the Mongolian National University of Medical Sciences, Ulaanbaatar, Mongolia (Number 2012/3–4, dated November, 24).
3. Results
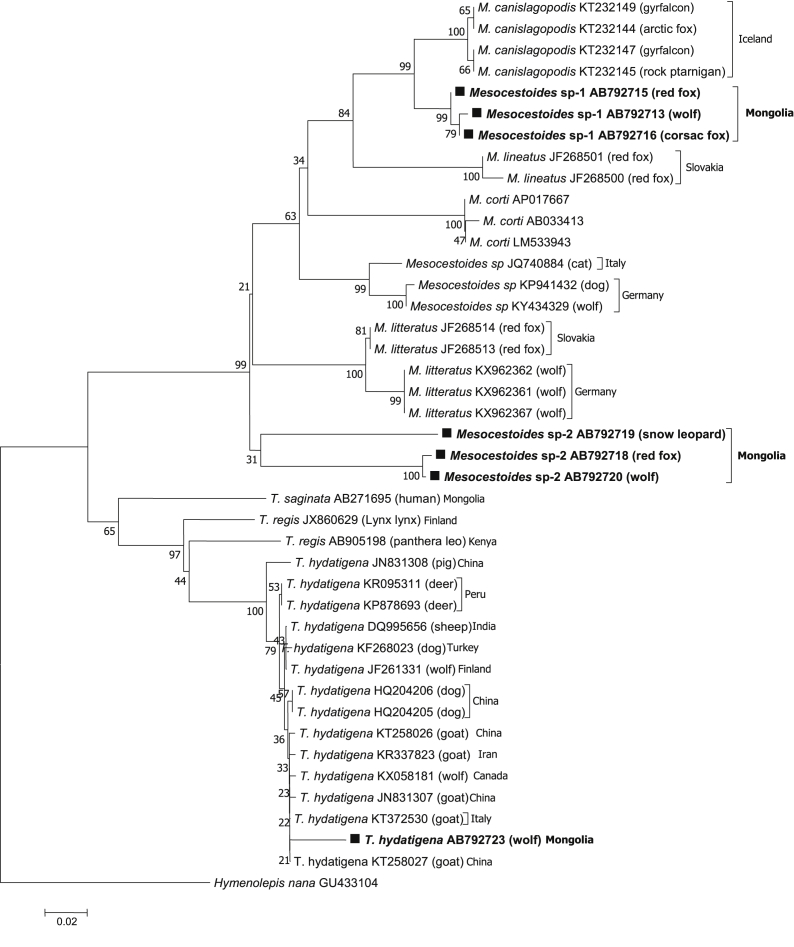
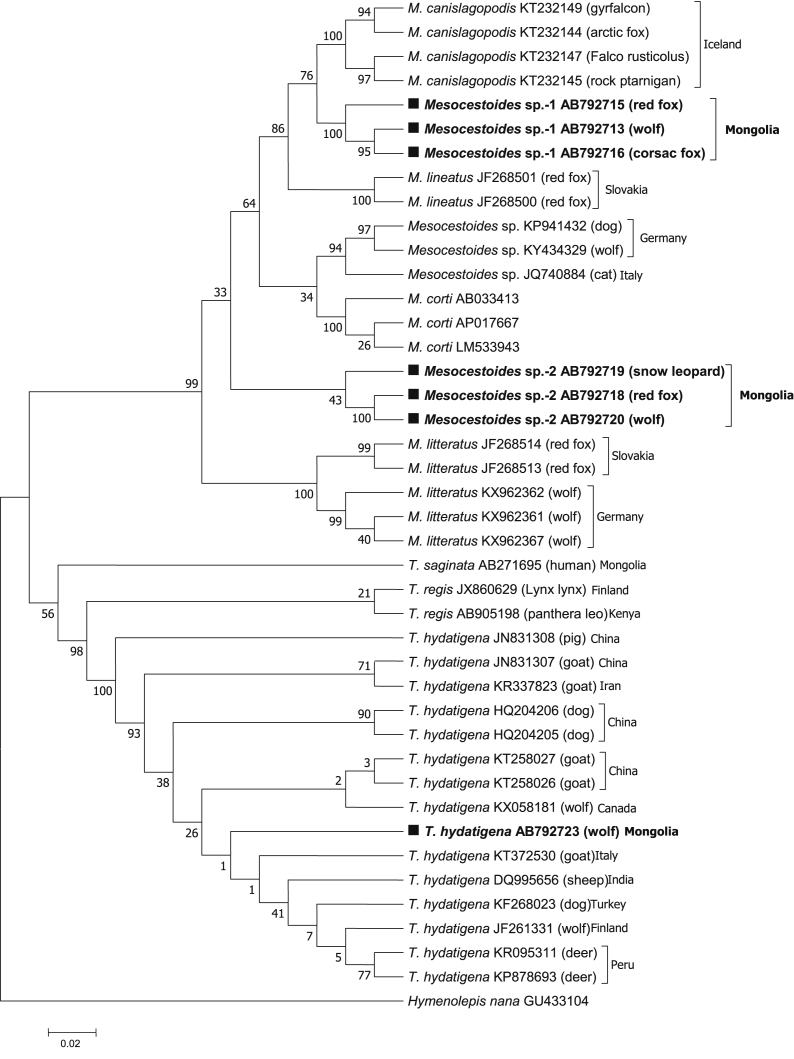
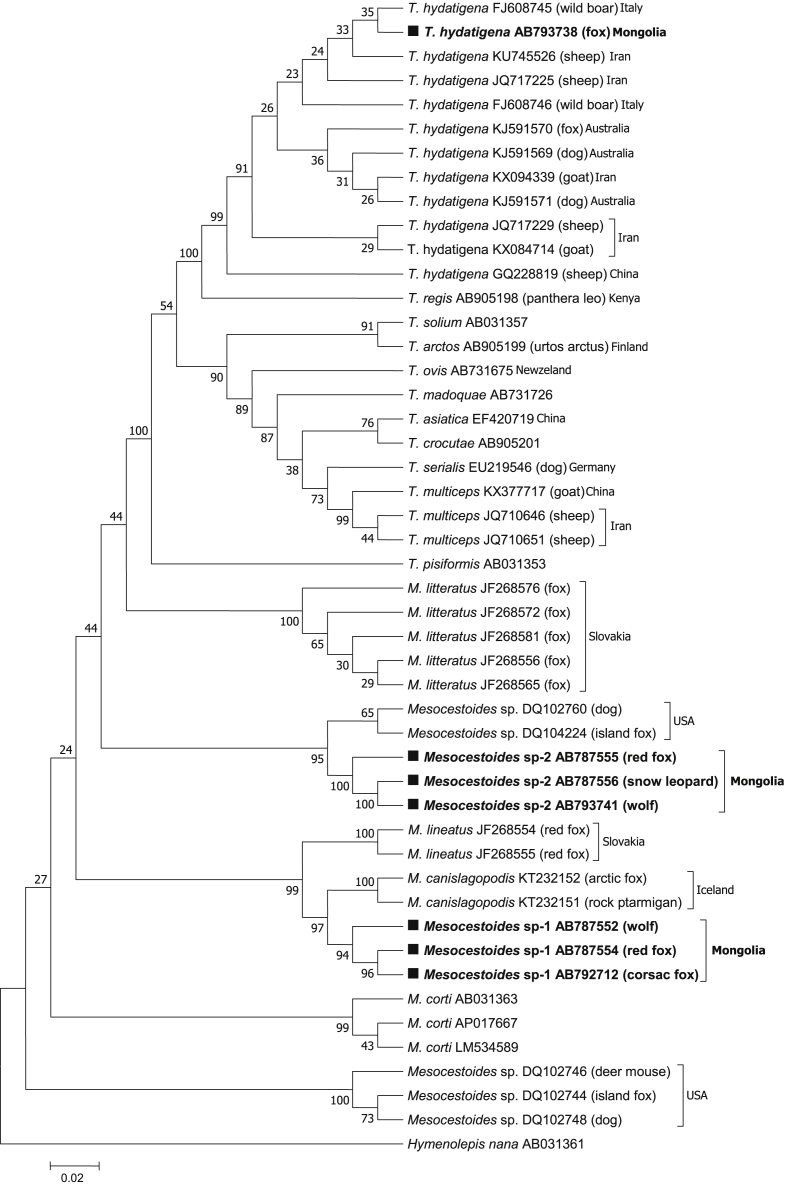
Table 1 shows results from PCR amplification of 405 wild carnivores fecal samples, which indicate that 27.7% (112/405) of samples were amplified with molecular sizes of 400 bp and 314 bp for cox1 and 12S rRNA, respectively. Tapeworm DNA was detected in wolves (47.3%, 53/112), red foxes (36.6%, 41/112), corsac foxes (9.8%, 11/112), and snow leopards (6.3%, 7/112). GenBank® accession numbers for cyclophyllidean tapeworms identified in Mongolia are presented in Table 6, along with a breakdown by DNA marker, parasite species/isolate code, wild carnivore, and coordinate. Of the 112 amplified samples, 54 and 56 different sequences were detected in the cox1 and 12S rRNA genes, respectively, and were classified as T. hydatigena (50.0%, 56/112), Mesocestoides sp.-1 (36.6%, 41/112), and Mesocestoides sp.-2 (13.4%, 15/112) via the sequence homology search. Phylogenetic tree analysis revealed that T. hydatigena from Mongolia was included in the clade composed of T. hydatigena from other countries (Table 4, Fig. 2, Fig. 3b).
Table 6.
GenBank® accession numbers for cyclophyllidean tapeworms identified in Mongolia.
| DNA marker | Parasite species/isolate code | Wild carnivore | Accession numbera |
|---|---|---|---|
| cox1 | |||
| Mesocestoides sp.-1 | |||
| Isolate Ml01 | Wolf | AB792713 | |
| Isolate Ml03 | Red fox | AB792715 | |
| Isolate Ml04 | Corsac fox | AB792716 | |
| Mesocestoides sp.-2 | |||
| Isolate Msp04 | Wolf | AB792720 | |
| Isolate Msp02 | Red fox | AB792718 | |
| Isolate Msp03 | Snow leopard | AB792719 | |
| Taenia hydatigena | |||
| Isolate Th03a | Wolf | AB792723 | |
| 12S rRNA | |||
| Mesocestoides sp.-1 | |||
| Isolate Ml01 | Wolf | AB787552 | |
| Isolate Ml03 | Red fox | AB787554 | |
| Isolate Msp04 | Corsac fox | AB792712 | |
| Mesocestoides sp.-2 | |||
| Isolate Msp03 | Wolf | AB793741 | |
| Isolate Msp01 | Red fox | AB787555 | |
| Isolate Msp02 | Snow leopard | AB787556 | |
| Taenia hydatigena | |||
| Isolate Th03b | Red fox | AB793738 | |
Accession numbers reported in this study.
Table 4.
Pairwise genetic distance in cox1 gene among Mesocestoides isolates from Mongolia and related taxa.
| Mesocestoides isolates | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mesocestoides sp.-1 AB792715 (red fox) Mongolia | |||||||||||||||||||||||
| 2 | Mesocestoides sp.-1 AB792713 (wolf) Mongolia | 0.005 | ||||||||||||||||||||||
| 3 | Mesocestoides sp.-1 AB792716 (corsac fox) Mongolia | 0.003 | 0.003 | |||||||||||||||||||||
| 4 | Mesocestoides sp.-2 AB792718 (red fox) Mongolia | 0.160 | 0.163 | 0.160 | ||||||||||||||||||||
| 5 | Mesocestoides sp.-2 AB792720 (wolf) Mongolia | 0.160 | 0.164 | 0.160 | 0.013 | |||||||||||||||||||
| 6 | Mesocestoides sp.-2 AB792719 (snow leopard) Mongolia | 0.192 | 0.196 | 0.192 | 0.174 | 0.165 | ||||||||||||||||||
| 7 | Mesocestoides sp. KP941432 (dog) Germany | 0.125 | 0.133 | 0.129 | 0.166 | 0.170 | 0.171 | |||||||||||||||||
| 8 | Mesocestoides sp. KY434329 (wolf) Germany | 0.121 | 0.129 | 0.125 | 0.162 | 0.166 | 0.167 | 0.003 | ||||||||||||||||
| 9 | Mesocestoides sp. JQ740884 (cat) Italy | 0.137 | 0.145 | 0.141 | 0.180 | 0.174 | 0.181 | 0.037 | 0.034 | |||||||||||||||
| 10 | M. lineatus JF268501 (red fox) Slovakia | 0.125 | 0.132 | 0.129 | 0.203 | 0.189 | 0.229 | 0.155 | 0.151 | 0.170 | ||||||||||||||
| 11 | M. lineatus JF268500 (red fox) Slovakia | 0.131 | 0.139 | 0.135 | 0.211 | 0.196 | 0.242 | 0.162 | 0.158 | 0.177 | 0.008 | |||||||||||||
| 12 | M. litteratus KX962362 (wolf) Germany | 0.164 | 0.172 | 0.168 | 0.156 | 0.156 | 0.164 | 0.157 | 0.154 | 0.153 | 0.166 | 0.178 | ||||||||||||
| 13 | M. litteratus KX962361 (wolf) Germany | 0.164 | 0.172 | 0.168 | 0.156 | 0.156 | 0.164 | 0.157 | 0.154 | 0.153 | 0.166 | 0.178 | 0.000 | |||||||||||
| 14 | M. litteratus KX962367 (wolf) Germany | 0.164 | 0.172 | 0.168 | 0.156 | 0.156 | 0.164 | 0.157 | 0.154 | 0.153 | 0.166 | 0.178 | 0.000 | 0.000 | ||||||||||
| 15 | M. litteratus JF268514 (red fox) Slovakia | 0.173 | 0.181 | 0.176 | 0.161 | 0.161 | 0.162 | 0.157 | 0.154 | 0.153 | 0.175 | 0.187 | 0.022 | 0.022 | 0.022 | |||||||||
| 16 | M. litteratus JF268513 (red fox) Slovakia | 0.172 | 0.180 | 0.176 | 0.160 | 0.160 | 0.161 | 0.154 | 0.150 | 0.149 | 0.175 | 0.187 | 0.019 | 0.019 | 0.019 | 0.003 | ||||||||
| 17 | M. canislagopodis KT232149 (gyrfalcon) Iceland | 0.049 | 0.056 | 0.053 | 0.172 | 0.172 | 0.183 | 0.114 | 0.110 | 0.125 | 0.131 | 0.142 | 0.155 | 0.155 | 0.155 | 0.167 | 0.163 | |||||||
| 18 | M. canislagopodis KT232147 (gyrfalcon) Iceland | 0.049 | 0.056 | 0.053 | 0.172 | 0.172 | 0.183 | 0.114 | 0.110 | 0.125 | 0.131 | 0.142 | 0.155 | 0.155 | 0.155 | 0.167 | 0.163 | 0.005 | ||||||
| 19 | M. canislagopodis KT232145 (rock ptarnigan) Iceland | 0.049 | 0.056 | 0.053 | 0.172 | 0.172 | 0.183 | 0.114 | 0.110 | 0.125 | 0.131 | 0.142 | 0.155 | 0.155 | 0.155 | 0.167 | 0.163 | 0.005 | 0.000 | |||||
| 20 | M. canislagopodis KT232144 (arctic fox) Iceland | 0.049 | 0.056 | 0.053 | 0.172 | 0.172 | 0.183 | 0.114 | 0.110 | 0.125 | 0.131 | 0.142 | 0.155 | 0.155 | 0.155 | 0.167 | 0.163 | 0.000 | 0.005 | 0.005 | ||||
| 21 | M. corti AP017667 | 0.156 | 0.164 | 0.161 | 0.195 | 0.185 | 0.188 | 0.126 | 0.123 | 0.133 | 0.168 | 0.175 | 0.150 | 0.150 | 0.150 | 0.149 | 0.145 | 0.162 | 0.162 | 0.162 | 0.162 | |||
| 22 | M. corti AB033413 | 0.157 | 0.165 | 0.161 | 0.194 | 0.194 | 0.197 | 0.125 | 0.122 | 0.142 | 0.169 | 0.176 | 0.158 | 0.158 | 0.158 | 0.157 | 0.154 | 0.170 | 0.170 | 0.170 | 0.170 | 0.005 | ||
| 23 | M. corti LM533943 | 0.156 | 0.164 | 0.161 | 0.195 | 0.185 | 0.188 | 0.126 | 0.123 | 0.133 | 0.168 | 0.175 | 0.150 | 0.150 | 0.150 | 0.149 | 0.145 | 0.162 | 0.162 | 0.162 | 0.162 | 0.000 | 0.005 | |
Fig. 2.
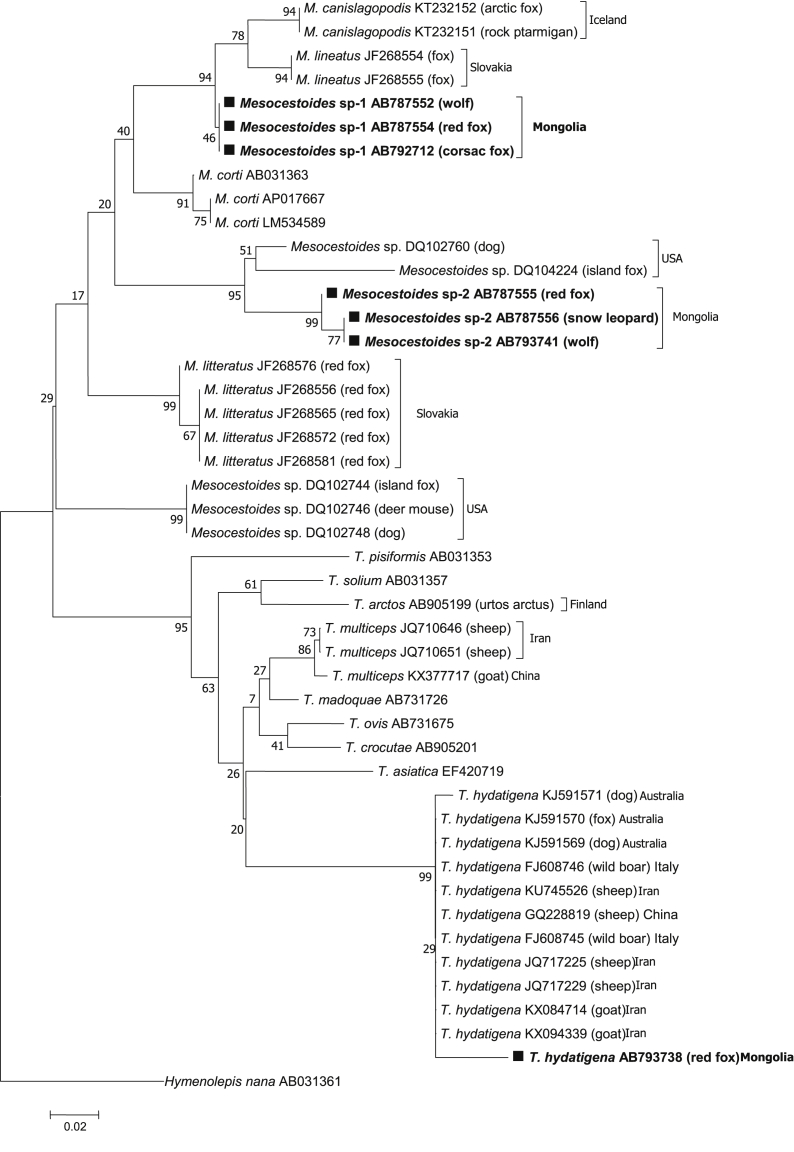
Phylogenetic tree based on a partial sequence of tapeworms obtained by the neighbor joining method was conducted using the TN93 + I nucleotide substitution model. Numbers above branches are percent bootstrap values based on 1,000 replicates. Bootstrap value >70% are shown.
(a) Phylogenetic tree based on the cox1 sequences of T. hydatigenaand Mesocestoides sp. isolates available in the GenBank® database were included. Hymenolepis nana served as an out-group.
(b) Phylogenetic analysis of the 12SrRNA partial sequence of T. hydatigena, Mesocestoides sp., and M. lineatus inferred using the sequence distance method and maximum likelihood. Hymenolepis nana was used as an out-group.
Fig. 3.
Phylogenetic tree based on a partial sequence of tapeworms obtained by the maximum likelihood method was conducted using the HKY + G + I nucleotide substitution model. Numbers above branches are percent bootstrap values based on 1,000 replicates. Bootstrap value >70% are shown.
(a) Phylogenetic tree based on the cox1 sequences of T. hydatigenaand Mesocestoides sp. isolates available in the GenBank® database were included. Hymenolepis nana served as an out-group.
(b) Phylogenetic analysis of the 12SrRNA partial sequence of T. hydatigena, Mesocestoides sp., and M. lineatus inferred using the sequence distance method and maximum likelihood. Hymenolepis nana was used as an out-group.
T. hydatigena infection in wild carnivores was detected throughout all regions, 19.6% (11/56), 32.1% (18/56), 37.5% (21/56), and 10.7% (6/56) in Western, Khangai, Central, and Eastern, respectively (Table 2, Fig. 1). T. hydatigena infection in wild carnivores was detected in 17 provinces. The highest prevalence was observed in Tuv (28.0%, 7/25), while the lowest was in Dornogobi (4.1%, 2/49). No T. hydatigena was isolated in samples from Govi-Altai and Khuvsgul (Table 2). Furthermore, the prevalence in wolf samples from Tuv was significantly higher than in other provinces (12.9%, 49/380) (p = 0.034).
The prevalence of T. hydatigena infection in wild carnivores was 19.9% (27/136), 13.8% (23/167), 4.8 (3/62), and 7.5% (3/40) in wolves, red foxes, corsac foxes, and snow leopards, respectively (Table 1). Prevalence in wolves was significantly higher compared to other wild carnivores (10.7%, 29/269) (p = 0.012).
The prevalence of T. hydatigena infection in wolves was 10.0% (1/10), 8.3% (2/24), 24.4% (10/41), 20.0% (8/40), 21.7% (5/23), and 12.5% (1/8) in alpine, mountain taiga, forest-steppe, steppe, desert-steppe, and desert, respectively. The prevalence in red foxes was 14.3% (7/49), 13.9% (5/36), and 18.6% (11/59) in forest-steppe, steppe, and desert-steppe, and the prevalence in corsac foxes was 5.3% (1/19) and 28.6% (2/7) in desert-steppe and desert, respectively. The prevalence of T. hydatigena infection in the snow leopards was 10.7% (3/28) in alpine region (Table 3).
Using the cox1 and 12S rRNA sequence data we observed that Mesocestoides species from Mongolia formed two genetically distant clades designated Mesocestoides sp.-1 and Mesocestoides sp.-2, which are not similar to the reference sequences present in GenBank (Fig. 2, Fig. 3b). Moreover, the genetic distances (d value) of the cox1 gene between Mesocestoides sp.-1 and M. lineatus from Slovakia were as large as 0.107–0.120 (Table 4). These results were also supported by the 12S rRNA sequence analysis and genetic distance data (Table 5). Haplotype diversity high compared to the nucleotide diversity was observed for cox1 and 12S rRNA gene. Due to Tajima’D was >0 for the cox1 and 12S rRNA sequences, rare alleles scarce in the population. The statistically significant Fu and Li's D and F values observed for the cox1 sequences showed P < 0.02, Fu and Li's D value observed for the 12S rRNA sequence showed P < 0.02 and Fu and Li's F value showed P < 0.05 (Table 7).
Table 5.
Pairwise genetic distance in 12S rRNA gene among Mesocestoides isolates from Mongolia and related taxa.
| Mesocestoides isolates | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mesocestoides sp-1 AB792712 (corsac fox) Mongolia | |||||||||||||||||||||||
| 2 | Mesocestoides sp-1 AB787554 (red fox) Mongolia | 0.004 | ||||||||||||||||||||||
| 3 | Mesocestoides sp-1 AB787552 (wolf) Mongolia | 0.008 | 0.004 | |||||||||||||||||||||
| 4 | Mesocestoides sp-2 AB787555 (red fox) Mongolia | 0.174 | 0.168 | 0.163 | ||||||||||||||||||||
| 5 | Mesocestoides sp-2 AB787556 (snow leopard) Mongolia | 0.192 | 0.186 | 0.180 | 0.012 | |||||||||||||||||||
| 6 | Mesocestoides sp-2 AB793741 (wolf) Mongolia | 0.198 | 0.192 | 0.186 | 0.016 | 0.004 | ||||||||||||||||||
| 7 | Mesocestoides sp. DQ102748 (dog) USA | 0.161 | 0.156 | 0.151 | 0.154 | 0.172 | 0.178 | |||||||||||||||||
| 8 | Mesocestoides sp. DQ102760 (dog) USA | 0.190 | 0.184 | 0.178 | 0.067 | 0.081 | 0.086 | 0.173 | ||||||||||||||||
| 9 | Mesocestoides sp. DQ102746 (deer mouse) USA | 0.155 | 0.150 | 0.145 | 0.148 | 0.166 | 0.171 | 0.004 | 0.166 | |||||||||||||||
| 10 | Mesocestoides sp. DQ104224 (island fox) USA | 0.212 | 0.207 | 0.201 | 0.104 | 0.120 | 0.125 | 0.224 | 0.072 | 0.232 | ||||||||||||||
| 11 | Mesocestoides sp. DQ102744 (island fox) USA | 0.155 | 0.150 | 0.145 | 0.161 | 0.179 | 0.185 | 0.004 | 0.173 | 0.008 | 0.224 | |||||||||||||
| 12 | M. lineatus JF268554 (red fox) Slovakia | 0.052 | 0.047 | 0.043 | 0.169 | 0.187 | 0.193 | 0.150 | 0.183 | 0.144 | 0.213 | 0.144 | ||||||||||||
| 13 | M. lineatus JF268555 (red fox) Slovakia | 0.052 | 0.047 | 0.043 | 0.169 | 0.187 | 0.193 | 0.150 | 0.183 | 0.144 | 0.213 | 0.144 | 0.000 | |||||||||||
| 14 | M. canislagopodis KT232152 (arctic fox) Iceland | 0.037 | 0.033 | 0.029 | 0.169 | 0.187 | 0.193 | 0.150 | 0.190 | 0.145 | 0.214 | 0.157 | 0.056 | 0.056 | ||||||||||
| 15 | M. canislagopodis KT232151 (rock ptarmigan) Iceland | 0.037 | 0.033 | 0.029 | 0.169 | 0.187 | 0.193 | 0.150 | 0.190 | 0.145 | 0.214 | 0.157 | 0.056 | 0.056 | 0.000 | |||||||||
| 16 | M. litteratus JF268556 (red fox) Slovakia | 0.159 | 0.154 | 0.148 | 0.131 | 0.148 | 0.153 | 0.155 | 0.137 | 0.155 | 0.188 | 0.155 | 0.174 | 0.174 | 0.174 | 0.174 | ||||||||
| 17 | M. litteratus JF268565 (red fox) Slovakia | 0.159 | 0.154 | 0.148 | 0.131 | 0.148 | 0.153 | 0.155 | 0.137 | 0.155 | 0.188 | 0.155 | 0.174 | 0.174 | 0.174 | 0.174 | 0.000 | |||||||
| 18 | M. litteratus JF268572 (red fox) Slovakia | 0.159 | 0.154 | 0.148 | 0.131 | 0.148 | 0.153 | 0.155 | 0.137 | 0.155 | 0.188 | 0.155 | 0.174 | 0.174 | 0.174 | 0.174 | 0.000 | 0.000 | ||||||
| 19 | M. litteratus JF268576 (red fox) Slovakia | 0.154 | 0.148 | 0.143 | 0.131 | 0.148 | 0.154 | 0.149 | 0.137 | 0.149 | 0.188 | 0.149 | 0.168 | 0.168 | 0.167 | 0.167 | 0.004 | 0.004 | 0.004 | |||||
| 20 | M. litteratus JF268581 (red fox) Slovakia | 0.159 | 0.154 | 0.148 | 0.131 | 0.148 | 0.153 | 0.155 | 0.137 | 0.155 | 0.188 | 0.155 | 0.174 | 0.174 | 0.174 | 0.174 | 0.000 | 0.000 | 0.000 | 0.004 | ||||
| 21 | M. corti AP017667 | 0.112 | 0.107 | 0.102 | 0.116 | 0.132 | 0.138 | 0.077 | 0.131 | 0.082 | 0.156 | 0.077 | 0.124 | 0.124 | 0.118 | 0.118 | 0.116 | 0.116 | 0.116 | 0.121 | 0.116 | |||
| 22 | M. corti LM534589 | 0.112 | 0.107 | 0.102 | 0.116 | 0.132 | 0.138 | 0.077 | 0.131 | 0.082 | 0.156 | 0.077 | 0.124 | 0.124 | 0.118 | 0.118 | 0.116 | 0.116 | 0.116 | 0.121 | 0.116 | 0.000 | ||
| 23 | M. corti AB031363 | 0.106 | 0.101 | 0.096 | 0.121 | 0.137 | 0.143 | 0.082 | 0.137 | 0.087 | 0.162 | 0.082 | 0.130 | 0.130 | 0.124 | 0.124 | 0.110 | 0.110 | 0.110 | 0.116 | 0.110 | 0.004 | 0.004 | |
Table 7.
Diversity and neutrality analysis for Mesocestoides isolates from Mongolia.
| Genes | n | Hn | Hd | VarHd | nd | SD of Hd | TajimaD | SigD | FuLiD* | SigD | FuLiF* | SigF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12S rRNAa | 56 | 6 | 0.775 | 0.00098 | 0.23468 | 0.031 | 0.3935 | Not | 2.28560 | ** | 1.85576 | * |
| cox1b | 56 | 6 | 0.775 | 0.00098 | 0.23975 | 0.031 | 1.6513 | Not | 2.28452 | ** | 2.44219 | ** |
12S rRNAa sequences of Mesocestoides from canids in Mongolia.
cox1b sequences of Mesocestoides from canids in Mongolia.
Abbreviation: n-number of isolate; H-haplotype; Hd-haplotype diversity; nd-nucleotide diversity; SD−standard deviation; SigD-statistical significance D; FuLiD- Fu and Li's D test statistic; FuLiF- Fu and Li's F test statistic; SigF- statistical significance F12S rRNA gene for Tajima's D test statistic: Not significant, P > 0.10; Fu and Li's D* test statistic: Significance **, P < 0.02; Fu and Li's F* test statistic: Significance: *, P < 0.05.
cox1 gene for Tajima's D test statistic: Not significant, P > 0.10; Fu and Li's D and F* test statistic: Significance: **, P < 0.02.
Mesocestoides sp.-2 isolated from snow leopards differed slightly from those isolated from red foxes and wolves. However, it did not form any clades with known Mesocestoides species in cox1 genes (Fig. 2, Fig. 3b), and the d values between Mesocestoides sp.-2 and European isolates were as large as 0.149–0.164 (Table 4). Similarly, Mesocestoides sp.-2 did not form any clades with known Mesocestoides species, and the d values were also large (Table 5).
Mesocestoides sp.-1 infection in wild carnivores was detected in all regions, 22.0% (9/41), 31.7% (13/41), 34.1% (14/41), and 12.2% (5/41) in Western, Khangai, Central, and Eastern, respectively (Table 2, Fig. 1). The prevalence of Mesocestoides sp.-1 infection in wild carnivores was 14.7% (20/136), 9.0% (15/167), and 9.7% (6/62) in wolves, red foxes, and corsac foxes, respectively. However, no Mesocestoides sp.-1 infection was observed in the snow leopard (Table 3). Additionally, Mesocestoides sp.-1 infection was detected in wild carnivores from 17 of the 21 provinces (Table 2). The highest prevalence was observed in Zavkhan (31.6%, 6/19), while the lowest was in Dornogobi (4.1%, 2/49). No Mesocestoides sp.-1 infection was identified in Bayan-Ulgii or Khovd (Table 2).
The prevalence of Mesocestoides sp.-1 infection in wolves was 21.4% (3/14), 17.0% (7/41), 5.0% (2/40), 26.1% (6/23), and 25.0% (2/8) in mountain taiga, forest-steppe, steppe, desert-steppe, and desert (Table 3). The prevalence in red foxes was 6.1% (3/49), 13.9% (5/36), and 30.4% (7/23) in forest-steppe, steppe, and desert, and the prevalence in corsac foxes was 8.7% (2/23) and 30.8% (4/13) in forest-steppe and steppe, respectively. There was no Mesocestoides sp.-1 infection observed in snow leopards (Table 3).
Mesocestoides sp.-2 infection in wild carnivores was detected in three regions, 66.7% (10/15), 13.3% (2/15), and 20.0% (3/15) in Western, Khangai, and Central, respectively (Table 2, Fig. 1). The prevalence of Mesocestoides sp.-2 infection in wild carnivores was 15.0% (6/40), 3.5% (3/85), 10.5% (2/19), and 14.3% (4/28) in wolves, red foxes, corsac foxes and snow leopards, respectively (Table 3).
The prevalence of Mesocestoides sp.-2 infection was 10.5% (4/38), 0.9% (1/113), 9.0% (8/89), and 2.0% (2/101) in alpine, forest-steppe, steppe, and desert-steppe. The prevalence in wolves was 15.0% (6/40) in steppe. The prevlence in red foxes was 2.0% (1/49) and 5.6% (2/36) in forest-steppe and steppe. The prevalence in corsac foxes was 10.5% (2/19) in desert-steppe, and the prevlence in snow leopards was 14.3% (4/28) in alpine (Table 3).
Mesocestoides sp.-2 infection in wild carnivores was detected in nine provinces. The prevalence of which is shown in Table 2. The highest was in Zavkhan (15.8%, 3/19), and the lowest in Dornogobi (2.0%, 1/49) (Table 2).
4. Discussion
Ecological regions, the existence of wild carnivores and the behavior of local herders all serve to create favorable conditions for parasites to complete their life cycle in Mongolia. The steppe region represents a typical broad grassland that supports stable herds of larger vertebrates. Currently, more than 180.000 herders (34%) in Mongolia live a nomadic or semi-nomadic live style (NSO, 2019).
Animal husbandry is essential for Mongolia's economy. The number of livestock has reached 66.5 million consisting primarily of goats and sheep, followed by horses, cattle, and camels. (MoFALI, 2018). Most herders in rural areas of Mongolia own livestock, which are kept in common open pastures (Myadagsuren et al., 2007). The majority of the 32 million head of livestock are located within the grassland in the steppe ecoregion, which accounts for about 70% of the country's territory. However, herein, we did not investigate T. hydatigena infection in livestock, which serve as the intermediate host for these tapeworm species, but rather this study is the first to investigate the prevalence and distribution of Taenia sp. and Mesocestoides sp. using specific molecular tools in wild carnivores in 19 provinces of Mongolia, covering all ecological regions. The tapeworm eggs in the fecal samples were not examined microscopically due to the difficulty of morphological identification.
This study demonstrated that parasitic infections were identified in wild carnivores from all ecosystems in Mongolia. Geographically these animals were distributed from western to eastern regions in all ecoregions and provinces, save for Dornod and Gobi-Sumber, from which there was no available. The infections were primarily detected in wolves (47.3%) followed by red foxes (36.6%), corsac foxes (9.8%), and snow leopards (6.3%) (Table 2).
Phylogenetic tree analysis revealed that T. hydatigena from Mongolia was included in the clade composed of T. hydatigena from other countries (Table 4, Fig. 2, Fig. 3). Moreover, wild carnivores infected with this parasite were detected in 17 provinces, with the highest prevalence observed in Tuv province (p < 0.05), where the capital city of Mongolia, Ulaanbaatar, is located. The population of Tuv province is approximately 1.2 million (37.5%) (NSO, 2019), and this province provides suitable conditions that support nomadic pastoralism. Additionally, we show that the prevalence of T. hydatigena infection is most common in wolves (p < 0.05) (Table 1). Similar results were reported in Iran (Nabavi et al., 2014), Italy and Serbia (Ćírovíč et al., 2015), where wild and domestic hoofed animals were reported to serve as intermediate hosts. Alternatively, no T. hydatigena infection was observed in the Govi-Altai and Khuvsgul provinces of Mongolia (Table 2).
In contrast, Mesocestoides sp.-1, and Mesocestoides sp.-2 isolated from wild carnivores in Mongolia were not found to be similar to reference sequences in GenBank, nor with known Mesocestoides species (Fig. 2, Fig. 3), suggesting that Mesocestoides sp.-1 is a unique species from M. lineatus. These results were also supported by 12S rRNA sequence analysis and genetic distance data (Table 5). Phylogenetic analysis of T. hydatigena, Mesocestoides based on cox1 and 12S rRNA was performed with a 410 bp long aligment comprising 48 sequences. Phylogenetic trees obtained by the use two tree-bulding methods have the same topology (Fig. 2, Fig. 3b). Further, Mesocestoides sp.-2 isolated from snow leopards differed slightly from red fox and wolf isolates, and did not form clades with known Mesocestoides species in cox1 genes (Fig. 2, Fig. 3a). In fact, the d values between Mesocestoides sp.-2 and European isolates were as large as 0.149–0.164 (Table 4). Similarly, Mesocestoides sp.-2 did not form any clades with known Mesocestoides species, and the d values were also quite high (Table 5), indicating that these two unidentified Mesocestoides species from Mongolia likely represent new species or species unregistered in GenBank. Using two mitochondrial genes, the cox1 and the 12S rRNA we molecularly confirmed the presence of Mesocestoides in canids from Mongolia. The overall high haplotype, low nucleotide diversities (Table 7) and the mainly significant negative neutrality indices detected for both cox1 and 12S rRNA sequences for Mongolian Mesocestoides sp-1 and Mesocestoides sp-2 from wolf, red fox, corsac fox, snow leopard from different ecological regions. This is similar to results reported for Mesocestoides from many regions worldwide (Foronda et al., 2007; Hrčkova et al., 2011; Zaleśny and Hildebrand, 2012; Skirnisson et al., 2016; Varcasia et al., 2018; Montalbano et al., 2018).
Based on the study results, Mesocestoides sp.-1 infection in wild carnivores was detected primarily (34.1%) in the Central region, with the highest prevalence observed in Zavkhan (31.6%, 6/19), and the lowest in Dornogobi (4.1%, 2/49) (Table 2). Moreover, the highest prevalence was in corsac foxes (30.8%), followed by red foxes (30.4%), and wolves (26.1%) in steppe, desert, and desert-steppe, respectively. There was no Mesocestoides sp.-1 infection seen in the snow leopard (Table 3).
Mesocestoides sp.-2 infection in wild carnivores was detected primarily in the Western (66.7%) region where the highest prevalence was again in Zavkhan (15.8%) and the lowest in Dornogobi (2.0%) (Table 2). Mesocestoides sp.-2 infection was found to be more prevalent in wolves (15.0%) and snow leopards (14.3%) in steppe and alpine ecoregions, respectively (Table 3).
The distribution of Mesocestoides sp.-1 and Mesocestoides sp.-2 infections in wild carnivores may be related to the natural habitat of these wild carnivores in Mongolia. The prey of these wild animals primarily comprise large herbivorous hoofstock including blue sheep, wild sheep, mountain goat, deer, Siberian ibex, and domestic animals such as goats, sheep, cattle, and horses (McCarthy, 2000; McCarthy et al., 2005; Shehzad et al., 2012). However, they also hunt birds, marmots, rodents, and other small mammals (Shehzad et al., 2012). We, therefore, assumed that the relationship between wolves, corsac foxes, and snow leopards and livestock has a central role in the lifecycle of Mesocestoides sp.-1, Mesocestoides sp.-2 in Mongolia. Hence, although felids and canids often act as definitive hosts for tapeworm species, they may also serve as secondary intermediate hosts (Venco et al., 2005; Elini et al., 2007; Jabbar et al., 2012).
The increase in the prevalence of parasitic diseases in wildlife poses a significant challenge to endangered species conservation (Aguirre, 2009; Pedersen and Greives, 2008; Smith et al., 2009). Therefore, it is imperative that appropriate measures be taken to prevent the spread of potentially fatal organisms, such as T. hydatgena and Mesocestoides sp., to the endangered snow leopard. The snow leopard is included on the International Union for Conservation of Nature's (IUCN) red list and the Mongolian Red List of Mammals (Clark et al., 2006) due to its endangered status. Approximately 20% (over 1000 animals) of the world's population of snow leopards are found in Mongolia (Fox ., 1994; McCarthy et al, 2010).
Moreover, as pasture lands are likely extensively contaminated with the feces of wild animals infected with T. hydatigena, Mesocestoides sp.-1 and Mesocestoides sp.-2, feral herbivores, livestock, herders, and the local people are at risk of acquiring these tapeworm infections. Thus, preventive measures that limit the spread of these parasites are necessary. The results of this study serve as a baseline with the potential inform the development of improved practices to better control the spread of parasitic infections in domestic and wild animals, particularly those considered to be endangered.
The limitation of this study, PCR-positive products were detected in only 112 samples (27.7%). This low isolation rate may have been caused by improper preservation conditions of feces (old and dry) or the presence of inhibitors. In addition, DNA extraction of mtDNA from fecal samples serves to preserve fragments less than 200 bp and are, therefore, more reliably amplified compared to fragments longer than 300 bp (Frantzen et al., 1998). Moreover, these DNA products employ short mtDNA markers that ensure effective and accurate genotyping.
This study reports the prevalence of T. hydatigena and Mesocestoides species in wild carnivores based on copro-DNA analysis. The most salient finding is that Mesocestoides sp.-1, and Mesocestoides sp.-2 from Mongolia likely account for new species, or species that are currently unregistered on GenBank. Furthermore, T. hydatigena was detected throughout all ecoregions was Mesocestoides sp.-1, save for in the alpine; while Mesocestoides sp.-2 was isolated from samples in the alpine, forest-steppe, steppe, and desert-steppe regions. Finally, wolf, red fox, corsac fox, and snow leopard were confirmed as new definitive hosts for Mesocestoides sp.-1 and Mesocestoides sp.-2.
Funding
This study was partially supported by the Fellowship Awards 2012 from the Japanese Association of University Women to MN.
Declaration of competing interest
The authors have no conflict of interest regarding the contents of this manuscript.
Acknowledgments
We are grateful for the support of the Mongolian National University of Medical Sciences and staff of the Laboratory of Helminthology, Department of Parasitology, National Institute of Infectious Diseases for their kind cooperation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2019.12.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aguirre A.A. Wild canids as sentinels of ecological health: a conservation medicine perspective. Parasites Vectors. 2009;2(Suppl):S7. doi: 10.1186/1756-3305-2-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi S., Mishra C. Living with large carnivores: predation on livestock by the snow leopard (Uncia uncia) J. Zool. 2006;268:217–224. [Google Scholar]
- Blazek K., Schramlová J., Hulínská D. Pathology of the migration phase of Taenia hydatigena (Pallas, 1766) larvae. Folia Parasitol. (Praha) 1985;32:127–137. [PubMed] [Google Scholar]
- Bowles J., McManus D.P. Rapid discrimination of Echinococcus species and strains using a polymerase chain reaction-based RFLP method. Mol. Biochem. Parasitol. 1993;57:231–239. doi: 10.1016/0166-6851(93)90199-8. [DOI] [PubMed] [Google Scholar]
- CDC (Centers for Disease Control and Prevention) 2017. Mesocestoidiasis.https://www.cdc.gov/dpdx/mesocestoidiasis/index.html [Google Scholar]
- Cho S.H., Kim T.S., Kong Y., Na B.K., Sohn W.M. Tetrathyridia of Mesocestoides lineatus in Chinese snakes and their adults recovered from experimental animals. Korean J. Parasitol. 2013;51:531–536. doi: 10.3347/kjp.2013.51.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ćirović D., Pavlović I., Penezić A. Intestinal helminth parasites of the grey wolf (Canis lupus L.) in Serbia. Acta Vet. Hung. 2015;63:189–198. doi: 10.1556/AVet.2015.016. [DOI] [PubMed] [Google Scholar]
- Clark E.L., Munkhbat J., Dulamtseren S., Baillie J.E.M., Batsaikhan N., Samiya R., Stubbe M. Zoological Society of London; London: 2006. Mongolian Red List of Mammals. Regional Red List Series 1. [Google Scholar]
- Danzan G. VIGIS; Moscow: 1978. Helminths of the Wild Mammals in Mongolia. D.Sc. Thesis. (in Russian) [Google Scholar]
- Debas E., Ibrahim N. Prevalence and economic importance of hydatidosis in cattle slaughtered at North Gonder Elfora abattoir. Eur. J. Appl. Sci. 2013;5:29–35. 10.1007/s00436-011-2598-7. Epub 2011 Aug 17. [Google Scholar]
- Dubinin V.B., Dubinina M.N. Parasite fauna of mammals in Daurian steppe. Materialy k poznaniyu fauny i flory SSSR (MOIP) Novaya Seriya. 1951;22:98–156. (in Russian) [Google Scholar]
- Dumitru I.M., Dumitru E., Rugina S. Role of epidemiologic data in management of hydatidosis in Constanta County, Romania. Ther. Pharmacol. Clin. Toxicol. 2011;15:132–138. [Google Scholar]
- Ebright J.R., Altantsetseg T., Oyungerel R. Emerging infectious diseases in Mongolia. Emerg. Infect. Dis. 2003;9:1509–1515. doi: 10.3201/eid0912.020520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleni C., Scaramozzino P., Busi M., Ingrosso S., D'Amelio S., De Liberato C. Proliferative peritoneal and pleural cestodiasis in a cat caused by metacestodes of Mesocestoides sp. anatomohistopathological findings and genetic identification. Parasite. 2007;14:71–76. doi: 10.1051/parasite/2007141071. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinfor Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Foronda P.В., Pérez Rivero A., Santana Morales M.A.В., Kabdur A., González A.C., Quispe Ricalde M.A., Feliu C., Valladares B. First larval record of Mesocestoides in carnivora of tenerife (canary islands) J Parasitol. Feb. 2007;93(1):138–142. doi: 10.1645/GE-932R1.1. [DOI] [PubMed] [Google Scholar]
- Fox J.L. Snow leopard conservation in the wild-a comprehensive perspective on a low density and highly fragmented population. In: Fox J.L., Jizeng D., editors. Proceedings of the Seventh International Snow Leopard Symposium. Xining; Qinghhai, China: 1994. pp. 3–15. [Google Scholar]
- Frantzen M.A.J., Silk, Ferguson J.W.H., Wayne P.K., Kohn M.H. Empirical evaluation of preservation methods for faecal DNA. Mol. Ecol. 1998;7:1423–1428. doi: 10.1046/j.1365-294x.1998.00449.x. [DOI] [PubMed] [Google Scholar]
- Fu Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getaw A., Beyene D., Ayana D., Megersa B., Abunna F. Hydatidosis: prevalence and its economic importance in ruminants slaughtered at Adama municipal abattoir, Central Oromia, Ethiopia. Acta Trop. 2010;113:221–225. doi: 10.1016/j.actatropica.2009.10.019. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Thomas L. Taenia spp. In: Rose J.B., Jiménez-Cisneros B., editors. Global Water Phathogens Project. UNESCO; E. Lansing, MI: 2018. http://www.water.pathogens.orghttp://www.waterpathogens.org/book/taenia Michigan State University Robertson, L (eds) Part 4 Helminths. [Google Scholar]
- Hrčkova G., Miterpáková M., O'Connor A., Šnábel V., Olson P.D. Molecular and morphological circumscription of Mesocestoides tapeworms from red foxes (Vulpes vulpes) in central Europe. Parasitology. 2011;138(5):638–647. doi: 10.1017/S0031182011000047. April 2011. [DOI] [PubMed] [Google Scholar]
- Jabbar A., Papini R., Ferrini N., Gasser R.B. Use of a molecular approach for the definitive diagnosis of proliferative larval mesocestoidiasis in a cat. Infect. Genet. Evol. 2012;12:1377–1380. doi: 10.1016/j.meegid.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Jenkins D.J., Urwin N.A., Williams T.M., Mitchell K.L., Lievaart J.J., Armua-Fernandez M.T. Red foxes (Vulpes vulpes) and wild dogs (dingoes (Canis lupus dingo) and dingo/domestic dog hybrids), as sylvatic host for Australian Taenia hydatigena and Taenia ovis. Int. J. Parasitol. Parasites Wildl. 2014;3:75–80. doi: 10.1016/j.ijppaw.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubečka B.W., Traub N.J., Tkach V.V., Shirley T.R., Rollins D., Fedynich A. Mesocestoides sp. in wild northern bobwhite (Colinus virginianus) and scaled quail (Callipepla squamata) J. Wildl. Dis. 2018;54:612–616. doi: 10.7589/2017-11-275. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.J., Brown R.W. Blandford; London: 1967. Mammals of Britain, Their Tracks, Trails and Signs. [Google Scholar]
- Loos-Frank B. One or two intermediate hosts in the life cycle of Mesocestoides (Cyclophyllidea, Mesocestoididae) Parasitol. Res. 1991;77(Issue 8):726–728. doi: 10.1007/BF00928692. [DOI] [PubMed] [Google Scholar]
- McAllister C.T., Trauth S.E., Plummer M.V. A new host record for Mesocestoides sp. (Cestoidea: cyclophyllidea: Mesocestoididae) from a rough green snake Opheodrys aestivus (Ophidia: colubridae) in Arkansas. U.S.A. Comp. Parasitol. 2013;80:130–133. [Google Scholar]
- McCarthy T. University of Massachusett Amherst; 2000. Ecology and Conservation of Snow Leopards, Gobi Brown Bears and Wild Bactrain Camels in Mongolia.http://www.snowleopardnetwork.org/bibliography/McCarthy_Ecology_2000.pdf Ph.D. Thesis. [Google Scholar]
- McCarthy T., Murray K., Sharma K., Johansson O. Preliminary results of along-term study of snow leopards in South Gobi, Mongolia. IUCN Cat News. 2010;5:15–19. [Google Scholar]
- McCarthy T.M., Fuller T.K., Munkhtsog B. Movements and activities of snow leopards in Southwestern Mongolia. Biol. Conserv. 2005;124:527–537. [Google Scholar]
- McFadden A.M., Muellner P., Baljinnyam Z., Vink D., Wilson N. Use of multicriteria risk ranking of zoonotic diseases in a developing country: case study of Mongolia. Zoonoses Public Health. 2016;63:138–151. doi: 10.1111/zph.12214. [DOI] [PubMed] [Google Scholar]
- Miran M.B., Kasuku A.A., Swai E.S. Prevalence of echinococcosis and Taenia hydatigena cysticercosis in slaughtered small ruminants at the livestock-wildlife interface areas of Ngorongoro, Tanzania. Vet. World. 2017;10:411–417. doi: 10.14202/vetworld.2017.411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MoFALI (Ministry of Food, Agriculture and Light Industry) 2018. National Report on the Rangeland Health of Mongolia.http://greenmongolia.mn/upload/news_files/ee55b0b158c70450d3a2ac13ee10c3e7.pdf [Google Scholar]
- Montalbano, Di Filippo M., Meoli R., Cavallero S., Eleni C., De Liberato C., Berrilli F. Molecular identification of Mesocestoides sp. metacestodes in a captive gold-handed tamarin (Saguinus midas) Infect Genet Evol.Nov. 2018;65:399–405. doi: 10.1016/j.meegid.2018.08.008. Epub 2018 Aug 12. [DOI] [PubMed] [Google Scholar]
- Myadagsuren N., Davaajav A., Wandra T., Sandar T., Ichinkhorloo P., Yamasaki H., Sako Y., Nakao M., Sato M.O., Nakaya K., Ito A. Taeniasis in Mongolia, 2002–2006. Am. J. Trop. Med. Hyg. 2007;77:342–346. [PubMed] [Google Scholar]
- Nabavi R., Manouchehri Naeini K., Zebardast N., Hashemi H. Epidemiological study of gastrointestinal helminthes of canids in Chaharmahal and Bakhtiari Province of Iran. Iran. J. Parasitol. 2014;9:276–281. [PMC free article] [PubMed] [Google Scholar]
- Nguyen M.T., Gabriël S., Abatih E.N., Dorny P. A systematic review on the global occurrence of Taenia hydatigena in pigs and cattle. Vet. Parasitol. 2016;226:97–103. doi: 10.1016/j.vetpar.2016.06.034. [DOI] [PubMed] [Google Scholar]
- NSO (National Statistics Office of Mongolia) 2019. http://www.en.nso.mn
- Oli M.K., Taylor I.R., Rogers M.E. Diet of the snow leopard (Panthera uncia) in the annapurna conservation area, Nepal. J. Zool., London. 1993;231:365–370. [Google Scholar]
- Oryan A., Goorgipour S., Moazeni M., Shirian S. Abattoir prevalence, organ distribution, public health and economic importance of major metacestodes in sheep, goats and cattle in Fars, southern Iran. Trop. Biomed. 2012;29:349–359. [PubMed] [Google Scholar]
- Pedersen A.B., Greives T.J. The interaction of parasites and resources cause crashes in a wild mouse population. J. Anim. Ecol. 2008;77:370–377. doi: 10.1111/j.1365-2656.2007.01321.x. [DOI] [PubMed] [Google Scholar]
- Poglayen G., Gori F., Morandi B., Galuppi R., Fabbri E., Caniglia R., Milanesi P., Galaverni M., Randi E., Marchesi B., Deplazes P. Italian wolves (Canis lupus italicus alberto, 1921) and molecular detection of taeniids in the foreste casentinesi national park, northern Italian apennines. Int. J. Parasites Wildl. 2017;6:1–7. doi: 10.1016/j.ijppaw.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M., Rashid M.I., Akbar H., Ahmad L., Hassan M.A., Ashraf K., Saeed K., Gharbi M. A systematic review on modelling approaches for economic losses studies caused by parasites and their associated diseases in cattle. Parasitology. 2019;146:129–141. doi: 10.1017/S0031182018001282. Epub 2018 Aug 2. [DOI] [PubMed] [Google Scholar]
- Rostami S., Salavati R., Beech R.N., Babaei Z., Sharbatkhori M., Baneshi M.R., Hajialilo E., Shad H., Harandi M.F. Molecular and morphological characterization of the tapeworm Taenia hydatigena (Pallas, 1766) in sheep from Iran. J. Helminthol. 2015;89:150–157. doi: 10.1017/S0022149X13000667. Epub 2013 Oct 8. [DOI] [PubMed] [Google Scholar]
- Scala A., Pipia A.P., Dore F., Sanna G., Tamponi C., Marrosu R., Bandino E., Carmona C., Boufana B., Varcasia A. Epidemiological updates and economic losses due to Taenia hydatigena in sheep from Sardinia. Italy. 2015;114(8):3137–3143. doi: 10.1007/s00436-015-4532-x. [DOI] [PubMed] [Google Scholar]
- Scala A., Urrai G., Varcasia A., Nicolussi P., Mulas M., Goddi L., Pipia A.P., Sanna G., Genchi M., Bandino E. vol. 90. 2016. pp. 113–116. (Acute Visceral Cysticercosis by Taenia Hydatigena in Lambs and Treatment with Praziquantel). 1. [DOI] [PubMed] [Google Scholar]
- Sgroi G., Varcasia A., Dessì G., D'Alessio N., Pacifico L., Buono F., Neola B., Fusco G., Santoro M., Toscano V., Fioretti A., Veneziano V. 2019. Massive Taenia Hydatigena Cysticercosis in a Wild Boar (Sus scrofa) from Italy. [DOI] [PubMed] [Google Scholar]
- Sharhuu G., Sharkhuu T. The helminth fauna of wild and domestic ruminants in Mongolia – a review. Eur. J. Wildl. Res. 2004;50:150–156. [Google Scholar]
- Sharkhuu T. Helminths of goats in Mongolia. Vet. Parasitol. 2001;101:161–169. doi: 10.1016/s0304-4017(01)00508-8. [DOI] [PubMed] [Google Scholar]
- Shehzad W., McCarthy T.M., Pompanon F., Purevjav L., Coissac E., Riaz T., Taberlet P. Prey preference of snow leopard (Panthera uncia) in South Gobi, Mongolia. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirnisson K., Jouet D., Ferté H., Nielsen O.K. Occurrence of Mesocestoides canislagopodis (Rudolphi, 1810) (Krabbe, 1865) in mammals and birds in Iceland and its molecular discrimination within the Mesocestoides species complex. Parasitol. Res. 2016 doi: 10.1007/s00436-016-5006-5. [DOI] [PubMed] [Google Scholar]
- Smith K.F., Acevedo-Whitehouse K., Pedersen A.B. The role of infectious diseases in biological conservation. Anim. Conserv. 2009;12:1–12. [Google Scholar]
- Strachan R., Jefferies D.J., Chanin P.R.F. Joint Nature Conservation Committee; Peterborough: 1996. Pine Marten Survey of England and Wales 1987±1988. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA 484 polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinnin D.S., Dunnum J.L., Salazar-Bravo J.A., Batsaikhan N., Burt M.S., Gardner S.L., Yates T.L. Faculty Publications from the Harold W. Manter. Laboratory of Parasitology; 2002. Contributions to the Mammalogy of Mongolia, with a Checklist of the Species for the Country.http://digitalcommons.unl.edu/parasitologyfacpubs/36 Paper 36. [Google Scholar]
- Varcasia A., Sanna D., Casu M., Lahmar S., Dessì G., Pipia A.P., Tamponi C., Gaglio G., Hrčková G., Otranto D., Scala A. Species delimitation based on mtDNA genes suggests the occurrence of new species of Mesocestoides in the Mediterranean region. Parasites Vectors. 2018;11(1):619. doi: 10.1186/s13071-018-3185-x. Dec 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venco L., Kramer L., Pagliaro L., Genchi C. Ultrasonographic features of peritoneal cestodiasis caused by Mesocestoides sp. in a dog and a cat. Vet. Radiol. Ultrasound. 2005;5:417–422. doi: 10.1111/j.1740-8261.2005.00076.x. [DOI] [PubMed] [Google Scholar]
- von Nickisch-Rosenegk M., Silva-Gonzalez R., Lucius R. Modification of universal 12S rDNA primers for specific amplification of contaminated Taenia sp. (Cestoda) gDNA enabling phylogenetic studies. Parasitol. Res. 1999;85:819–825. doi: 10.1007/s004360050638. [DOI] [PubMed] [Google Scholar]
- Zaleśny G., Hildebrand J. Molecular identification of Mesocestoides spp. from intermediate hosts (rodents) in central Europe (Poland) Parasitol. Res. 2012;110:1055–1061. doi: 10.1007/s00436-011-2598-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.