Abstract
Background and Purpose
Epilepsy is a common chronic neurological disorder. About one third of epilepsy patients will suffer from drug resistance after rational selection of antiepileptic drug treatment. The formation of drug-resistant epilepsy has quite a few causes of which genetic factors are considered to be the most important. Previous studies have suggested that the aquaporin 4(AQP4) and inward rectifier potassium ion channel Kir4.1 (encoded by gene KCNJ10) may act in concert to adjust water homeostasis and concentration of potassium ions in extracellular spaces of the central nervous system. Therefore, these two molecules would play a major role in the regulation of the excitability of neurons. In order to explore the potential mechanism of genetic factors related to AQP4 and Kir4.1, we conducted a study to analyze the effects of the AQP4 and KCNJ10 genes' single nucleotide polymorphisms (SNPs) on epileptic drug resistance and seizure susceptibility in a group of Chinese Han patients with focal epilepsy.
Materials and Methods
In total, 510 patients with focal-onset seizures and 206 healthy controls were recruited. Among the patients, 222 were drug resistant and 288 were responsive. The selection of tag SNPs was based on the Hapmap database and Haploview software. Genotyping of three loci of the AQP4 gene (rs1058424, rs3763043 and rs35931) and nine loci of the KCNJ10 gene (rs12122979, rs1186685, rs6690889, rs2486253, rs1186675, rs12402969, rs12729701, rs1890532 and rs3795339) was conducted on the Sequenom MassARRAY iPLEX platform.
Results
The distribution of genotype and allele frequencies of selected SNP loci of AQP4 and KCNJ10 genes showed no significant difference between the drug-resistant and drug-responsive groups (p>0.05), and no significant difference between all the idiopathic focal epilepsy patients and healthy controls either.
Conclusion
AQP4 and KCNJ10 genetic polymorphisms may not be associated with drug resistance or seizure susceptibility of focal epilepsy in the Chinese Han population.
Keywords: AQP4, KCNJ10, focal epilepsy, polymorphism, Chinese Han population
Introduction
Epilepsy is one of the common chronic disorders of the nervous system, with an increasing prevalence in the global population.1 Although there are multiple antiepileptic drugs (AEDs) of different mechanisms available for clinicians, one third of the patients with epilepsy cannot be controlled effectively and would suffer from drug resistance after a sufficient amount and reasonable use of AEDs.2 The formation of drug-resistant epilepsy has quite a few causes of which the genetic factor is considered to be significant.3 Since the single nucleotide polymorphism (SNP) of ABCB1 gene C3435T (rs1045642) was first confirmed to be associated with epileptic drug resistance in a Caucasian population, several other SNP loci such as rs717620 of the ABCC2 gene, and rs2706110 of the NFE2L2 gene were also found to be correlated with epileptic drug resistance in researches of different ethnic groups.4–8 These pharmacogenomic results identified novel polymorphism loci that increase the vulnerability to drug resistance of epilepsy, which provides a new idea for the future clinical therapeutic target.
Aquaporin 4(AQP4) is widely found in the central nervous system (CNS), especially in astrocytes, which is of great significance to the water regulation balance in the microenvironment of the CNS. Neuronal excitability is closely related to extracellular space (ECS) volume and potassium ion concentration in the ECS. Dysfunction of AQP4 can lead to a reduction of ECS volume and therefore an increase of extracellular potassium ion concentration, resulting in increased neuronal excitability and abnormal highly synchronized electrophysiological activities in epileptic foci. Meanwhile, AQP4 may be involved in glial cell scar formation in CNS after the occurrence of epilepsy, which may play a critical role in the occurrence of epilepsy and the formation of drug resistance.9 In addition, the inward rectifying potassium channel Kir4.1, which is encoded by gene KCNJ10 and mostly expressed in the end-feet of astrocytes, is believed to act in concert with AQP4 to regulate water homeostasis and concentration of potassium ions in the ECS of the CNS. The main function of Kir4.1 is to carry potassium ions into cells accompanied by water entry through AQP4, which could reduce the excitability of neurons.10,11 In epilepsy, water homeostasis and ion concentration regulation imbalance of ECS may also lead to reduced sensitivity to AEDs and generate drug resistance. AQP4 was found to be significantly increased in protein levels in surgical resection from patients with refractory temporal lobe epilepsy (TLE), suggesting that AQP4 may be associated with epileptic drug resistance.12 Meanwhile, Kir4.1 expression was obviously reduced in astrocytes in hippocampi from patients with drug-resistance.13 These findings above reveal that AQP4 and Kir4.1 may be crucial determinants of epileptic drug resistance and seizure susceptibility.
So far, there has been no report regarding the exact correlation between drug-resistant epilepsy and SNPs of these two genes. Previous genetic polymorphism studies chiefly focused on validating its increasing susceptibility to seizures. Seven SNP loci of the KCNJ10 gene, including rs1053074, rs12729701, rs1186685, rs4656873, rs946429, rs2820585 and rs6656873, were found to be correlated with temporal lobe epilepsy with febrile seizure (TLE-FS) in Norwegian patients, suggesting that KCNJ10 had an essential influence on epilepsy susceptibility. Moreover, five SNP loci of the AQP4 gene (ss119336753, ss119336756, rs151244, ss119336763 and rs9951307) were found to be coupled to TLE-FS in the same population, indicating that AQP4 and KCNJ10 have potentially an obligatory correlation with TLE-FS.14 However, this study did not discuss the association between SNPs of these two genes and epileptic drug resistance. A better understanding of the influence on epilepsy of these two genes may develop much needed new therapeutic strategies for epilepsy individuals. Therefore, in our present study, we aimed to make a further investigation to find out whether AQP4 and KCNJ10 gene polymorphisms might be risk factors for drug resistance or seizure attacks of focal epilepsy.
Materials and Methods
Participants Recruitment and Classification
In total, 510 patients with focal-onset seizures and 206 healthy controls without neurological disorders were recruited. The participants of our study were uncorrelated Chinese Hans randomly collected from the Neurology Clinic, Xiangya Hospital, Central South University, Changsha, People's Republic of China. Upon the approval of the Ethics Committee, all participants had signed an informed consent. On the basis of the operational classification of seizure types by the International League Against Epilepsy (ILAE) in 2017,15 the 510 patients were diagnosed and classified. The types of seizures and AED applications of each focal epilepsy patient were evaluated and checked by well-trained neurologists in our department. Patients with a history of drug abuse or alcoholism, severe systemic illness, poor drug compliance and uncertain seizure history were excluded. The drug resistance was accepted as incapable of complete seizure freedom after at least two appropriately used AEDs with adequate dosage and duration, meanwhile, drug responsiveness was accepted as complete seizure freedom for more than 12 months or three times the longest interseizure interval prior to treatment.16 After screening, in all participants recruited, 222 patients and the other 288 were classified into the drug resistant and responsive groups respectively.
Selection of SNP Loci and Genotyping
The detailed information of SNP loci of the AQP4 and KCNJ10 genes in the Chinese Han population was acquired from the database of the International HapMap Project (HapMap Data Rel 24/Phase II Nov08, on NCBI B36 assembly, dbSNP b126). The selection criteria for tag SNP include minor allele frequency more than the value of 0.05 and an r2 threshold of 0.80. By doing so, SNP loci rs1058424, rs3763043, rs35931 of AQP4 and SNP loci rs12122979, rs1186685, rs6690889, rs2486253, rs1186675, rs12402969, rs12729701, rs1890532, rs3795339 of KCNJ10 were selected respectively. Peripheral whole blood samples (5 mL) from each participant were acquired with EDTA through venipuncture. We used QIAamp DNA Blood Mini Kit (QIAGEN, Germany) to extract genomic DNA from peripheral blood samples. Following the commendatory protocols with measurement of charge-to-mass ratio, the matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) was used for genotyping on the Sequenom MassARRAY iPLEX (Sequenom, USA) platform. TYPER 4.0 software was then applied to output the genotyping results.
Statistical Analysis
The statistical analysis was carried out by using SPSS software (version 21.0). We used the binary logistic regression to analyze the difference of genotype distribution and frequency of seizures among groups. Independent samples t-test was used to evaluate the differences of duration time, number of AEDs, age of life and onset between drug-resistant and responsive groups. Allele distribution, sex distribution and the Hardy-Weinberg equilibrium were assessed by the Pearson chi-square test. A p-value less than 0.05 was accepted to be statistically significant. Accompanied by the 95% confidence intervals (CI), the strengths of the associations were described as odds ratios (OR). Bonferroni’s method for correction of multiple testing was used in our study.17
Results
The clinical features of all participants are summarized in Table 1. In general, compared to the patients in the responsive group, the drug-resistant patients had more frequent seizures, earlier onset age, longer duration of illness and had taken more AEDs. While there was no significant difference in age and sex between the two groups. The call rate of genotyping, including AQP4 and KCNJ10 genes, were over 98% for each tag SNPs (Figure 1A–C, Figure 2A–I). Genotypes or allele frequencies of the three tag SNPs of AQP4 or nine of KCNJ10 have no significance between the drug-responsive and resistant groups (Tables 2 and 3). Among all the recruited focal epilepsy patients, 393 idiopathic ones were further stratified to compare with the healthy controls for genetic predisposition analysis. However, no clear correlation was found between SNPs and epileptic seizure susceptibility with regards to AQP4 and KCNJ10 (Tables 4 and 5). The distributions of genotypes for all selected SNP loci both in the patients and healthy controls are in alignment with the Hardy-Weinberg equilibrium.
Table 1.
Clinical Features of Enrolled Focal Epilepsy Patients
| Parameters | Total Patients | AED Resistant N=222 | AED Responsive N=288 | p | Healthy Controls | |
|---|---|---|---|---|---|---|
| Age of life (years) | 26.64 11.91 11.91 |
27.42 10.63 10.63 |
26.03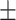 12.79 12.79 |
0.193 | 42.87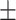 12.31 12.31 |
|
| Sex | Male | 334 (65.5%) | 150 (67.6%) | 184 (63.9%) | 0.386 | 116 (56.3%) |
| Female | 176 (34.5%) | 72 (32.4%) | 104 (36.1%) | 90 (43.7%) | ||
| Age of onset (years) | 17.11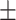 11.41 11.41 |
15.56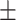 9.77 9.77 |
18.30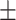 12.42 12.42 |
0.007 | – | |
| Duration time (years) | 9.44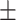 8.17 8.17 |
11.74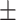 8.20 8.20 |
7.67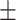 7.71 7.71 |
<0.001 | – | |
| Number of AEDs | 1.90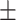 0.98 0.98 |
2.40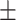 0.94 0.94 |
1.52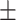 0.82 0.82 |
<0.001 | – | |
| Frequency of seizure before therapy | <once half a year | 34 (6.7%) | 8 (3.6%) | 26 (9.0%) | 0.003 | – |
| <once a month | 166 (32.5%) | 49 (22.0%) | 117 (40.6%) | <0.001 | – | |
| ≥once a month | 251 (49.2%) | 132 (59.4%) | 119 (41.3%) | 0.643 | – | |
| Daily | 59 (11.6%) | 33 (14.9%) | 26 (9.0%) | – | – | |
| Seizure types | FOA | 40 (7.8%) | 14 (6.3%) | 26 (9.0%) | – | – |
| FOIA | 163 (32.0%) | 88 (39.6%) | 75 (26.0%) | – | – | |
| FO to BTC | 307 (60.2%) | 120 (54.0%) | 187 (65.0%) | – | – | |
Abbreviations: FOA/FOIA, focal onset aware/impaired awareness; BTC, bilateral tonic-clonic.
Figure 1.
Polar plot of genotyping results of AQP4 gene rs1058424 (A), rs3763043 (B) and rs335931 (C) for all participants by MALDI TOF. The call rate of genotyping was over 98% for each tag SNPs.
Figure 2.
Polar plot of genotyping results of KCNJ10 gene rs12122979 (A), rs1186685 (B), rs6690889 (C), rs2486253 (D), rs1186675 (E), rs12402969 (F), rs12729701 (G), rs1890532 (H) and rs3795339 (I) for all participants by MALDI TOF. The call rate of genotyping was over 98% for each tag SNPs.
Table 2.
Genotype and Allele Frequency of AQP4 Between Drug Resistant and Responsive Patients
| SNP Locus | Genotype and Allele | AED Resistant N=222 | AED Responsive N=288 | OR (95% CI) | p |
|---|---|---|---|---|---|
| rs1058424 | TT | 67 (30.92%) | 107 (38.1%) | Reference | – |
| TA | 110 (50.7%) | 124 (44.1%) | 1.413 (0.947–2.109) | 0.090 | |
| AA | 40 (18.4%) | 50 (17.8%) | 1.245 (0.741–2.091) | 0.408 | |
| T | 244 (56.2%) | 338 (60.1%) | 1.175 (0.911–1.515) | 0.213 | |
| A | 190 (43.8%) | 224 (39.9%) | |||
| rs3763043 | GG | 72 (33.3%) | 114 (40.6%) | Reference | – |
| GA | 105 (48.6%) | 118 (42.0%) | 1.394 (0.938–2.072) | 0.101 | |
| AA | 39 (18.1%) | 49 (17.4%) | 1.222 (0.729–2.049) | 0.446 | |
| G | 249 (57.6%) | 346 (61.6%) | 1.177 (0.912–1.520) | 0.211 | |
| A | 183 (42.4%) | 216 (38.4%) | |||
| rs335931 | AA | 73 (33.5%) | 86 (30.7%) | Reference | – |
| CG | 112 (51.5%) | 140 (50.0%) | 0.960 (0.643–1.433) | 0.841 | |
| GG | 33 (15.1%) | 54 (19.3%) | 0.718 (0.421–1.227) | 0.226 | |
| A | 258 (59.2%) | 312 (55.7%) | 1.152 (0.894–1.484) | 0.273 | |
| G | 178 (40.8%) | 248 (44.3%) |
Notes: After multiple testing correction by using Bonferroni’s method, p<0.017 is accepted to be significant.
Table 3.
Genotype and Allele Frequency of KCNJ10 Between Drug Resistant and Responsive Patients
| SNP Locus | Genotype and Allele | AED Resistant N=222 | AED Responsive N=288 | OR (95% CI) | p |
|---|---|---|---|---|---|
| rs12122979 | GG | 105 (47.9%) | 148 (52.5%) | Reference | – |
| GA | 90 (41.4%) | 95 (33.7%) | 1.334 (0.910–1.957) | 0.140 | |
| AA | 24 (11.0%) | 39 (13.8%) | 0.832 (0.470–1.473) | 0.528 | |
| G | 300 (68.5%) | 391 (69.3%) | 1.039 (0.794–1.361) | 0.777 | |
| A | 138 (31.5%) | 173 (30.7%) | |||
| rs1186685 | TT | 126 (57.5%) | 176 61.5%) | Reference | – |
| TC | 77 (35.2%) | 88 (30.8%) | 1.125 (0.828–1.782) | 0.320 | |
| CC | 16 (7.3%) | 22 (7.7%) | 0.975 (0.480–1.908) | 0.900 | |
| T | 329 (75.1%) | 440 (76.9%) | 1.104 (0.825–1.478) | 0.504 | |
| C | 109 (24.9%) | 132 (23.1%) | |||
| rs6690889 | TT | 110 (50.2%) | 162 (57.7%) | Reference | – |
| TC | 90 (41.1%) | 92 (32.7%) | 1.428 (0.977–2.087) | 0.066 | |
| CC | 19 (8.7%) | 27 (9.6%) | 0.981 (0.515–1.868) | 0.954 | |
| T | 310 (70.8%) | 416 (74.0%) | 1.176 (0.899–1.555) | 0.254 | |
| C | 128 (29.2%) | 146 (26.0%) | |||
| rs2486253 | GG | 213 96.8%) | 272 (95.8%) | Reference | – |
| GT | 7 (3.2%) | 12 (4.2%) | 0.823 (0567–1.195) | 0.305 | |
| GG | 0 (0.0%) | 0 (0.0%) | – | – | |
| G | 433 (98.4%) | 556 (97.9%) | 1.335 (0.521–3.420) | 0.545 | |
| T | 7 (1.6%) | 12 (2.1%) | |||
| rs1186675 | CC | 197 (89.1%) | 258 (90.5%) | Reference | – |
| CT | 23 (10.4%) | 24 (9.1%) | 1.150 (0635–2.083) | 0.645 | |
| TT | 1 (0.5%) | 1 (0.4%) | 1.280 (0.079–20.676) | 0.862 | |
| C | 417 (94.3%) | 542 (95.1) | 0.861 (0.495–1.500) | 0.276 | |
| T | 25 (5.7%) | 28 (4.9%) | |||
| rs12402969 | TT | 194 (88.2%) | 242 (85.5%) | Reference | – |
| TC | 24 (10.9%) | 38 (13.4%) | 0.796 (0.460–1.475) | 0.413 | |
| CC | 2 (0.9%) | 3 (1.1%) | 0.835 (0.137–5.077) | 0.844 | |
| T | 412 (93.6%) | 522 (92.2%) | 0.806 (0.493–1.318) | 0.389 | |
| C | 28 (6.4%) | 44 (7.8%) | |||
| rs12729701 | AA | 127 (58.0%) | 163 (57.6) | Reference | – |
| AG | 80 (36.5%) | 106(37.5%) | 0.949(0.653–1.378) | 0.782 | |
| GG | 12 (5.5%) | 14 (4.9%) | 1.151 (0.513–2.584) | 0.734 | |
| A | 334 (76.3%) | 432 (76.3%) | 0.996 (0.743–1.336) | 0.980 | |
| G | 104 (23.7%) | 134 (23.7%) | |||
| rs1890532 | CC | 137 (62.3%) | 182 (64.5%) | Reference | – |
| CG | 65 (29.3%) | 86 (30.5%) | 1.007 (0.860–1.419) | 0.972 | |
| GG | 18 (8.2%) | 14 (5.0%) | 1.706 (0.819–3.555) | 0.154 | |
| C | 339 (77.0%) | 450 (79.8%) | 0.850 (0.628–1.150) | 0.293 | |
| G | 101 (23.0%) | 114 (20.2%) | |||
| rs3795339 | GG | 217 (98.6%) | 274 (96.5%) | Reference | – |
| GA | 3 (1.4%) | 10 (3.5%) | 0.388 (0.105–1.433) | 0.155 | |
| AA | 0 (0.0%) | 0 (0.0%) | – | – | |
| G | 437 (99.3%) | 558 (98.2%) | 2.611 (0.714–9.543) | 0.132 | |
| A | 3 (0.7%) | 10 (1.8%) |
Notes: After multiple testing correction by using Bonferroni’s method, p<0.006 is accepted to be significant.
Table 4.
Genotype and Allele Frequency of AQP4 Between All Idiopathic Focal Epilepsy Patients and Healthy Controls
| SNP Loci | Genotype and Allele | Patients N=393 | Healthy Controls N=206 | OR (95% CI) | p |
|---|---|---|---|---|---|
| rs1058424 | TT | 135 (35.0%) | 82 (40%) | Reference | – |
| TA | 185 (47.9%) | 88 (42.9%) | 1.362 (0.869–2.135) | 0.178 | |
| AA | 66 (17.1%) | 35 (17.1%) | 1.224 (0.680–2.204) | 0.500 | |
| T | 455 (58.9%) | 252 (61.5%) | 0.900 (0.704–1.150) | 0.399 | |
| A | 317 (41.4%) | 158 (38.5%) | |||
| rs3763043 | GG | 143 (37.1%) | 83 (40.5%) | Reference | – |
| GA | 178 (46.2%) | 87 (42.4%) | 1.338 (0.855–2.093) | 0.203 | |
| AA | 64 (16.6%) | 35 (17.1%) | 1.188 (0.660–2.139) | 0.565 | |
| G | 464 (60.2%) | 253 (61.7%) | 0.941 (0.736–1.203) | 0.628 | |
| A | 306 (39.8%) | 157 (38.3%) | |||
| rs335931 | AA | 122 (31.8%) | 68 (33.2%) | Reference | – |
| CG | 196 (51.0%) | 102 (49.8%) | 0.992 (0.627–1.569) | 0.972 | |
| GG | 66 (17.2%) | 35 (17.1%) | 1.110 (0.606–2.033) | 0.735 | |
| A | 440 (57.3%) | 238 (58.0%) | 0.969 (0.761–1.236) | 0.802 | |
| G | 328 (42.7%) | 172 (42.0%) |
Notes: After multiple testing correction by using Bonferroni’s method, p<0.017 is accepted to be significant.
Table 5.
Genotype and Allele Frequency of KCNJ10 Between All Idiopathic Focal Epilepsy Patients and Healthy Controls
| SNP Loci | Genotype and Allele | Patients N=393 | Healthy Controls N=206 | OR (95% CI) | p |
|---|---|---|---|---|---|
| rs12122979 | GG | 187 (48.4%) | 96 (46.8%) | Reference | – |
| GA | 150 (38.9%) | 84 (41.0%) | 0.824 (0.543–1.272) | 0.383 | |
| AA | 49 (12.7%) | 25 (12.2%) | 1.018 (0.519–2.000) | 0.958 | |
| G | 524 (67.8%) | 276 (67.3%) | 1.026 (0.794–1.325) | 0.845 | |
| A | 248 (32.2%) | 134 (32.7%) | |||
| rs1186685 | TT | 224 (57.6%) | 121 (58.7%) | Reference | – |
| TC | 138 (35.5%) | 70 (34.0%) | 1.040 (0.674–1.602) | 0.860 | |
| CC | 27 (6.9%) | 15 (7.3%) | 1.366 (0.562–3.075) | 0.512 | |
| T | 586 (75.3%) | 312 (75.7%) | 0.978 (0.741–1.292) | 0.877 | |
| C | 192 (24.7%) | 100 (24.3%) | |||
| rs6690889 | TT | 202 (52.5%) | 104 (50.7%) | Reference | – |
| TC | 148 (38.4%) | 87 (42.4%) | 0.794 (0.519–1.126) | 0.289 | |
| CC | 35 (9.1%) | 14 (6.8%) | 1.678 (0.718–3.924) | 0.232 | |
| T | 552 (71.7%) | 295 (72.0%) | 0.987 (0.756–1.289) | 0.924 | |
| C | 218 (28.3%) | 115 (28.0%) | |||
| rs2486253 | GG | 371 (95.6%) | 191 (93.2%) | Reference | – |
| GT | 17 (4.4%) | 14 (6.8%) | 0.610 (0.255–1.459) | 0.267 | |
| GG | 0 (0.0%) | 0 (0.0%) | – | – | |
| G | 759 (97.8%) | 396 (96.6%) | 1.578 (0.770–3.235) | 0.209 | |
| T | 17 (2.2%) | 14 (3.4%) | |||
| rs1186675 | CC | 349 (89.7%) | 187 (91.7%) | Reference | – |
| CT | 38 (9.8%) | 17 (8.3%) | 1.204 (0.584–2.841) | 0.615 | |
| TT | 2 (0.5%) | 0 (0.0%) | – | – | |
| C | 736 (94.6%) | 391 (95.8%) | 0.762 (0.428–1.356) | 0.354 | |
| T | 42 (5.4%) | 17 (4.2%) | |||
| rs12402969 | TT | 333 (85.8%) | 169 (86.8%) | Reference | – |
| TC | 51 (13.1%) | 33 (16.2%) | 0.615 (0.345–1.094) | 0.098 | |
| CC | 4 (1.0%) | 2 (1.0%) | 0.399 (0.053–2.156) | 0.252 | |
| T | 717 (92.4%) | 371 (90.9%) | 1.212 (0.789–1.863) | 0.380 | |
| C | 59 (7.6%) | 37 (9.1%) | |||
| rs12729701 | AA | 226 (58.4%) | 117 (57.1%) | Reference | – |
| AG | 144 (37.2%) | 74 (36.1%) | 1.120 (0.728–1.723) | 0.607 | |
| GG | 17 (4.4%) | 14 (1.8%) | 0.445 (0.187–1.063) | 0.068 | |
| A | 596 (77.0%) | 308 (75.1%) | 1.109 (0.838–1.467) | 0.469 | |
| G | 178 (23.0%) | 102 (24.9%) | |||
| rs1890532 | CC | 248 (64.2%) | 120 (58.5%) | Reference | – |
| CG | 111 (28.8%) | 70 (34.3%) | 0.636 (0.409–0.989) | 0.045 | |
| GG | 27 (7.0%) | 14 (6.9%) | 1.243 (0.541–2.853) | 0.608 | |
| C | 607 (78.6%) | 310 (76.0%) | 1.163 (0.875–1.546) | 0.299 | |
| G | 165 (21.4%) | 98 (34.0%) | |||
| rs3795339 | GG | 378 (97.4%) | 200 (97.1%) | Reference | – |
| GA | 10 (2.6%) | 5 (2.4%) | 0.626 (0.179–2.191) | 0.463 | |
| AA | 0 (0.0%) | 0 0.0%) | – | – | |
| G | 766 (98.8%) | 405 (98.3%) | 0.946 (0.321–2.785) | 0.919 | |
| A | 10 (1.2%) | 5 (0.7%) |
Notes: After multiple testing correction by using Bonferroni’s method, p<0.006 is accepted to be significant.
Discussion
This study is so far the first to explore the correlation between AQP4 and KCNJ10 gene polymorphisms and focal epilepsy drug resistance. As for clinical features, the most significant parameters associated with drug resistance of epilepsy patients include number of AEDs, age of onset, frequency of seizures before therapy and illness duration time, which would be considered as predictive factors of drug-resistant epileptogenesis probably. The genotypes of drug-resistant and drug-responsive patients, as well as healthy controls were all in alignment with the Hardy-Weinberg equilibrium, manifesting that a representative population sample was used. The allele and genotype frequencies were basically consistent with the Hapmap database. We tried to find the exact relationship between SNPs of these two genes and drug-resistant of focal epilepsy. It is helpful to increase our knowledge about how AQP4 and KCNJ10 genes affect drug resistance in focal epilepsy. Meanwhile, we took one step further to test the effects of selected SNPs on seizure susceptibility. However, according to our results, there is no association between AQP4 and KCNJ10 gene polymorphisms and seizure susceptibility or drug resistance in the participants we recruited.
AQP4 is mainly expressed in the end-feet around the blood vessels of astrocytes and the processes around the synapses of the neural fiber network, which is very beneficial to the bidirectional transport of water molecules in ECS and blood, affecting the volume and osmotic pressure of ECS. Thus, AQP4 plays a very significant role in regulating the water and example homeostasis in the extracellular microenvironment of neurons.9 The migration and self-healing rate of astrocytes were also significantly reduced after inhibiting AQP4 RNA, suggesting that AQP4 may be involved in the activation and migration of astrocytes and the formation of glial scars.18 The scars can result in abnormal neural circuits, leading to increased excitability of neural networks and a blocking effect on AEDs, which is one of the possible causes of drug resistance in epilepsy.19,20 We heretofore were capable of finding some previous studies on AQP4 gene polymorphisms and formation of epilepsy. The former reports have presented that AQP4 expression is altered in several epilepsy models.21 Some studies suggest perivascular upregulation of AQP4 whereas others revealed downregulation.22 In intrahippocampal kainic acid model of TLE, expression of AQP4 protein was acutely downregulated at 1 day post status epilepticus and was followed by a return to baseline levels gradually at the 30 days of post status epilepticus, which suggested AQP4 may express differently on early or later duration in epileptogenesis.23 The mean duration of patients we recruited are 11.71 and 7.67 years in the drug-resistant and drug-responsive groups respectively. Most of our patients were in the chronic period of epilepsy. According to the research findings on animal models, the expression of AQP4 protein would return to the baseline level at this period, speculating that equal quantities of AQP4 may be found in chronic epilepsy patients and healthy controls, which may be an explanation for our results.
Kir4.1 and AQP4 are involved tightly with the regulation of water, potassium redistribution and volume of ECS in the CNS,24 and play an important role in regulating the excitability of neurons. In the CA1 region of the hippocampus of mice knocked out by the AQP4 gene, potassium clearance was delayed, indicating that AQP4 and Kir4.1 not only co-expressed in the same region, but were also closely related in function.25 By using conditional knockout mice, KCNJ10 gene deletion can affect the excitatory transmitter of astrocytes to release glutamate and regulate the excitability of the neural network in the hippocampus.26 It is therefore likely that astrocytic Kir4.1 genes potentially affect drug resistance in epilepsy.27 Evidence that gene polymorphisms of KCNJ10 may be closely involved with the onset and development of epileptogenesis is accumulated. In previous studies, for the locus rs1130183, T allele of KCNJ10 was significantly higher in healthy people than in idiopathic generalized epilepsy patients in a German population.28 Meanwhile, in a pediatric epilepsy patients group in Japan, the GT genotype of KCNJ10 gene rs2486253 polymorphism also significantly increased the risk for development of idiopathic generalized epilepsy of children.29 Among a Chinese Han population, Hapmap database shows that rs1130183 are all CC genotypes, with extremely low frequency of T allele, which is not suitable for our SNPs study. Previously, rs6690889 TC and TT genotypes were found to have lower frequencies in genetic generalized epilepsy patients than in healthy controls recruited from southern China by the research of Guo et al. Hence, they speculated that potential SNP loci of KCNJ10 may contribute to seizure susceptibility and antiepileptic drug resistance.30 Compared to the former study, our design is different in two ways. Firstly, different types of epilepsy were chosen in our study, which probably have differences in their underlying etiopathogenesis. Secondly, a drug-responsive group was not set in Guo et al’s research. Therefore, in the former study, it is a limitation to expound whether SNPs of KCNJ10 affect epileptic drug resistance or not.
As a result of few relevant studies and the complicated function of AQP4 and Kir4.1, the explicit evidence that these two molecules are involved with antiepileptic drug resistance is still lacking. Considering the results of absence of correlation between AQP4 and KCNJ10 genetic polymorphisms and epileptic drug resistance on the population level, it is possible that these two genes may not be directly involved in drug-resistant epileptogenesis of focal epilepsy. The altered expression of AQP4 and Kir4.1 in epilepsy patients and animal models may be the result of frequent seizures attacks, rather than one of the causes of drug resistance to epilepsy. Various AEDs primarily act on neural components, such as voltage-gated sodium ion channels (eg, phenytoin and lamotrigine), GABAA receptors (eg, phenobarbital and diazepam) and voltage-gated calcium ion channel (gabapentin), not glial cell directly.26 This may be another reason that SNPs of these two genes do not contribute to AEDs resistance. Regarding focal epilepsy, our study is the first to analyze the effects of AQP4 and KCNJ10 gene SNPs on seizure susceptibility in the Chinese Han population. The frequency of rs1890532 CT genotype of KCNJ10 in healthy controls was higher than that in focal epilepsy patients (OR = 0.636, 95% CI (0.409 0.989), p = 0.045), prompting heterozygous CT genotype groups to have a lower risk for focal epilepsy. Regretfully, there is no significant difference between these two groups after multiple testing is corrected by Bonferroni method. The rs1890532 of KCNJ10 gene is located in the intron region. The locus may not play a direct role in encoding Kir4.1 protein, but it may be essential for accurate alternative splicing as well as efficient translation of KCNJ10 mRNA.31 The specific role of the locus rs1890532 may need to be further explored in the future.
The limitation of this study is that the sample size is relatively small, so it is impossible to evaluate the effects of SNP loci with low allele frequency but which play a crucial genetic role. Diverse regions and ethnic groups also would lead to significant differences in allele frequency distribution at the same SNP loci so that our results are different from the previous studies. Besides that, the mechanism of epileptogenesis for drug resistance and the clinical classification of epilepsy are very complicated. The diagnosis and treatment strategies of different types of epilepsy are also different. Therefore, it is necessary to repeat verification studies on the correlation between SNPs of these two genes and susceptibility or drug resistance of epilepsy in a larger sample size, different regions and ethnic groups in the future, so as to provide new ideas for clinical individualized treatment of epilepsy.
Conclusion
AQP4 and KCNJ10 genetic polymorphisms may not be associated with drug resistance or seizure susceptibility of focal epilepsy in the Chinese Han population.
Acknowledgments
We would like to thank all the focal epilepsy patients and healthy subjects for their cooperation in the study. This work was supported by the following funding: National Natural Science Foundation of China (Grant No. 81601139 and No. 81671299); and Natural Science Foundation of Hunan Province (Grant No. 2017JJ3500).
Ethical Approval and Consent to Participate
This study was approved by the Ethics Committee of the Xiangya Hospital of Central South University of Medical Science, Approval No. 201303120. This study was conducted in accordance with the 1964 declaration of Helsinki and its later amendments or comparable ethical standards. The subjects were assured of the following: All participants were voluntary, and could withdraw at any time without facing any negative consequences. All participants provided their written informed consent.
Consent for Publication
Informed consent for publication has been obtained from all participants. All patients have signed for agreement.
Author Contributions
LZ and BX conceived and designed the experiments. LZ, BX and HYZ performed the experiments. All authors contributed to data analysis, drafting or revising the article, gave the final approval of the version to be published and agreed to be accountable for all aspects of work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yuen A, Keezer MR, Sander JW. Epilepsy is a neurological and a systemic disorder. Epilepsy Behav. 2018;78:57–61. doi: 10.1016/j.yebeh.2017.10.010 [DOI] [PubMed] [Google Scholar]
- 2.Xiong J, Mao D, Liu L. Research progress on the role of ABC transporters in the drug resistance mechanism of intractable epilepsy. Biomed Res Int. 2015;2015:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng J, Pang N, Wang Y, et al. Next-generation sequencing improves treatment efficacy and reduces hospitalization in children with drug-resistant epilepsy. CNS Neurosci Ther. 2019;25(1):14–20. doi: 10.1111/cns.2019.25.issue-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Yin X, Liu L, et al. Association of KEAP1 and NFE2L2 polymorphisms with temporal lobe epilepsy and drug resistant epilepsy. Gene. 2015;571(2):231–236. doi: 10.1016/j.gene.2015.06.055 [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui A, Kerb R, Weale ME, et al. Association of multidrug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. N Engl J Med. 2003;348(15):1442–1448. doi: 10.1056/NEJMoa021986 [DOI] [PubMed] [Google Scholar]
- 6.Ufer M, von Stulpnagel C, Muhle H, et al. Impact of ABCC2 genotype on antiepileptic drug response in Caucasian patients with childhood epilepsy. Pharmacogenet Genomics. 2011;21(10):624–630. doi: 10.1097/FPC.0b013e3283498131 [DOI] [PubMed] [Google Scholar]
- 7.Qu J, Zhou BT, Yin JY, et al. ABCC2 polymorphisms and haplotype are associated with drug resistance in Chinese epileptic patients. CNS Neurosci Ther. 2012;18(8):647–651. doi: 10.1111/j.1755-5949.2012.00336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian L, Fang S, Yan Y, Zeng S, Xu Z, Gong Z. The ABCC2 c.-24C>T polymorphism increases the risk of resistance to antiepileptic drugs: a meta-analysis. J Clin Neurosci. 2017;37:6–14. doi: 10.1016/j.jocn.2016.10.014 [DOI] [PubMed] [Google Scholar]
- 9.Binder DK, Nagelhus EA, Ottersen OP. Aquaporin-4 and epilepsy. Glia. 2012;60(8):1203–1214. doi: 10.1002/glia.22317 [DOI] [PubMed] [Google Scholar]
- 10.Devinsky O, Vezzani A, Najjar S, De Lanerolle NC, Rogawski MA. Glia and epilepsy: excitability and inflammation. Trends Neurosci. 2013;36(3):174–184. doi: 10.1016/j.tins.2012.11.008 [DOI] [PubMed] [Google Scholar]
- 11.Kinboshi M, Shimizu S, Mashimo T, et al. Down-regulation of astrocytic Kir4.1 channels during the audiogenic epileptogenesis in Leucine-Rich Glioma-Inactivated 1 (Lgi1) mutant rats. Int J Mol Sci. 2019;20(5):1013. doi: 10.3390/ijms20051013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A, Wallace GT, Holmes C, et al. Hippocampal tissue of patients with refractory temporal lobe epilepsy is associated with astrocyte activation, inflammation, and altered expression of channels and receptors. Neuroscience. 2012;220:237–246. doi: 10.1016/j.neuroscience.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heuser K, Eid T, Lauritzen F, et al. Loss of perivascular Kir4.1 potassium channels in the sclerotic hippocampus of patients with mesial temporal lobe epilepsy. J Neuropathol Exp Neurol. 2012;71(9):814–825. doi: 10.1097/NEN.0b013e318267b5af [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuser K, Nagelhus EA, Tauboll E, et al. Variants of the genes encoding AQP4 and Kir4.1 are associated with subgroups of patients with temporal lobe epilepsy. Epilepsy Res. 2010;88(1):55–64. doi: 10.1016/j.eplepsyres.2009.09.023 [DOI] [PubMed] [Google Scholar]
- 15.Fisher RS, Cross JH, French JA, et al. Operational classification of seizure types by the international league against epilepsy: position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58(4):522–530. doi: 10.1111/epi.13670 [DOI] [PubMed] [Google Scholar]
- 16.Kwan P, Arzimanoglou A, Berg AT, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE commission on therapeutic strategies. Epilepsia. 2010;51(6):1069–1077. doi: 10.1111/j.1528-1167.2009.02397.x [DOI] [PubMed] [Google Scholar]
- 17.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310(6973):170. doi: 10.1136/bmj.310.6973.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saadoun S, Papadopoulos MC, Watanabe H, Yan D, Manley GT, Verkman AS. Involvement of aquaporin-4 in astroglial cell migration and glial scar formation. J Cell Sci. 2005;118(Pt 24):5691–5698. doi: 10.1242/jcs.02680 [DOI] [PubMed] [Google Scholar]
- 19.Pecorelli A, Natrella F, Belmonte G, et al. NADPH oxidase activation and 4-hydroxy-2-nonenal/aquaporin-4 adducts as possible new players in oxidative neuronal damage presents in drug-resistant epilepsy. Biochim Biophys Acta. 2015;1852(3):507–519. doi: 10.1016/j.bbadis.2014.11.016 [DOI] [PubMed] [Google Scholar]
- 20.Verkhratsky A, Rodriguez JJ, Parpura V. Astroglia in neurological diseases. Future Neurol. 2013;8(2):149–158. doi: 10.2217/fnl.12.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubbard JA, Szu JI, Binder DK. The role of aquaporin-4 in synaptic plasticity, memory and disease. Brain Res Bull. 2018;136:118–129. doi: 10.1016/j.brainresbull.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 22.Bebek N, Ozdemir O, Sayitoglu M, et al. Expression analysis and clinical correlation of aquaporin 1 and 4 genes in human hippocampal sclerosis. J Clin Neurosci. 2013;20(11):1564–1570. doi: 10.1016/j.jocn.2012.12.023 [DOI] [PubMed] [Google Scholar]
- 23.Hubbard JA, Szu JI, Yonan JM, Binder DK. Regulation of astrocyte glutamate transporter-1 (GLT1) and aquaporin-4 (AQP4) expression in a model of epilepsy. Exp Neurol. 2016;283(Pt A):85–96. doi: 10.1016/j.expneurol.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Short B, Kozek L, Harmsen H, et al. Cerebral aquaporin-4 expression is independent of seizures in tuberous sclerosis complex. Neurobiol Dis. 2019;129:93–101. doi: 10.1016/j.nbd.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 25.Nagelhus EA, Mathiisen TM, Ottersen OP. Aquaporin-4 in the central nervous system: cellular and subcellular distribution and coexpression with KIR4.1. Neuroscience. 2004;129(4):905–913. doi: 10.1016/j.neuroscience.2004.08.053 [DOI] [PubMed] [Google Scholar]
- 26.Djukic B, Casper KB, Philpot BD, Chin LS, McCarthy KD. Conditional knock-out of Kir4.1 leads to glial membrane depolarization, inhibition of potassium and glutamate uptake, and enhanced short-term synaptic potentiation. J Neurosci. 2007;27(42):11354–11365. doi: 10.1523/JNEUROSCI.0723-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno Y, Kinboshi M, Shimizu S. Inwardly rectifying potassium channel Kir4.1 as a novel modulator of BDNF expression in astrocytes. Int J Mol Sci. 2018;19(11):3313. doi: 10.3390/ijms19113313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenzen KP, Heils A, Lorenz S, et al. Supportive evidence for an allelic association of the human KCNJ10 potassium channel gene with idiopathic generalized epilepsy. Epilepsy Res. 2005;63(2–3):113–118. doi: 10.1016/j.eplepsyres.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 29.Dai AI, Akcali A, Koska S, Oztuzcu S, Cengiz B, Demiryurek AT. Contribution of KCNJ10 gene polymorphisms in childhood epilepsy. J Child Neurol. 2015;30(3):296–300. doi: 10.1177/0883073814539560 [DOI] [PubMed] [Google Scholar]
- 30.Guo Y, Yan KP, Qu Q, et al. Common variants of KCNJ10 are associated with susceptibility and anti-epileptic drug resistance in Chinese genetic generalized epilepsies. PLoS ONE. 2015;10(4):e124896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathieu O, Bouche N. Interplay between chromatin and RNA processing. Curr Opin Plant Biol. 2014;18:60–65. doi: 10.1016/j.pbi.2014.02.006 [DOI] [PubMed] [Google Scholar]