Abstract
Lycian salamanders (genus Lyciasalamandra) constitute an exceptional case of micro-endemism of an amphibian species on the Asian Minor mainland. These viviparous salamanders are confined to karstic limestone formations along the southern Anatolian coast and some islands. We here study the genetic differentiation within and among 118 populations of all seven Lyciasalamandra species across the entire genus’ distribution. Based on circa 900 base pairs of fragments of the mitochondrial 16SrDNA and ATPase genes, we analysed the spatial haplotype distribution as well as the genetic structure and demographic history of populations. We used 253 geo-referenced populations and CHELSA climate data to infer species distribution models which we projected on climatic conditions of the Last Glacial Maximum (LGM). Within all but one species, distinct phyloclades were identified, which only in parts matched current taxonomy. Most haplotypes (78%) were private to single populations. Sometimes population genetic parameters showed contradicting results, although in several cases they indicated recent population expansion of phyloclades. Climatic suitability of localities currently inhabited by salamanders was significantly lower during the LGM compared to recent climate. All data indicated a strong degree of isolation among Lyciasalamandra populations, even within phyloclades. Given the sometimes high degree of haplotype differentiation between adjacent populations, they must have survived periods of deteriorated climates during the Quaternary on the spot. However, the alternative explanation of male biased dispersal combined with a pronounced female philopatry can only be excluded if independent nuclear data confirm this result.
Introduction
Small-range endemism at both species and higher taxonomic level is a common phenomenon among all three amphibian orders. Various mechanisms exist leading to small ranges and isolation (e.g., [1, 2]). We here put light on Lycian salamanders (Lyciasalamandra), an old clade of the Salamandridae, which has evolved high diversity of allopatric micro-endemic lineages [3].
Lycian salamanders occur in the western Taurus Mountains along the Mediterranean coast of Turkey and on the Greek Karpathos Archipelago. Seven species, some with various subspecies, have been suggested. Each is endemic to an isolated mountain ridge of its own, with almost no signs of admixture among each other even when they live in close proximity [4]. Lyciasalamandra is the sister genus of Salamandra and evolved circa 9.29 million years ago (mya) (95% confidence interval (CI): 6.12–12.8 mya; [5]). The final emergence of the mid-Aegean trench 10.2–12.3 mya probably initiated species evolution within Lyciasalamandra [6]. Phylogenetic studies consistently support a scenario of largely simultaneous emergence of seven major lineages [7, 8], and the hypothesis of a hard polytomy could in fact not be rejected [6].
Intraspecific evolution within Lyciasalamandra species can be explained by the Messinian Salinity Crisis (5.3–5.6 mya) as well as repeated climatic alterations since the Late Pliocene and throughout the Pleistocene [5, 6]. Given this high evolutionary age of Lyciasalamandra species and their intraspecific lineages [6] in combination with a low level of genetic diversity of local populations and a high amount of private local mitochondrial DNA haplotypes [8], it is sound to assume that these lineages, once having evolved, were not able to substantially disperse out of their mountains. Repeated bottlenecks associated with an increasing effect of genetic drift may have contributed to or even reinforced this pattern of low degrees of local genetic variability. And if eventually small-scale dispersal may have occurred, such founder events again should have reduced local genetic variability. All this may have led to today’s small-scale differences of pattern and colour among populations, forming the basis for the description of numerous new taxa within the last years (e.g., [9–16]). Unfortunately, up to know data supporting such a scenario solely stem from organelle data, so male-biased dispersal, which may erode local population differentiation, can only be detected by also analysing nuclear DNA.
But how could these salamanders survive such long periods ‘on the spot’, especially with regard to the tremendous climatic alterations which repeatedly occurred during the last 2.4 my [17]? Especially, since that time, cold periods (stitials) and warm (interstitials) periods regularly alternated [18]. This is known to have drastically affected the spatial distribution of plants and animals in the entire Western Palearctic (e.g., [19, 20]). Although ice ages along the Mediterranean coast may not have been as severe as in the interior land masses [21], it is suggested that the increasingly dry conditions during glacial periods had an impact on amphibian populations [22, 23].
The ecological niche of the enigmatic Lycian salamanders might be the key to understand this unique situation. Almost all known populations live on karstic limestone. Throughout the entire Mediterranean, such rock formations are associated with an increased biological diversity because they offer a variety of micro-habitats [24]. Especially at the slopes of south-facing coastal mountains karstic limestone provides a sufficient moisture gradient to organisms due to the opportunity to deeply hide inside crevices of rock formations and boulder fields. As major pre-requisite of this life-style, the Lycian salamanders had evolved a viviparous mode of reproduction, with females giving birth to only two juveniles per year.
Relying on stable micro-climatic conditions through vertical movements in order to constantly follow suitable micro-habitat conditions within short distances (e.g., [25]) may also have constrained Lycian salamanders to evolve a high degree of ecological plasticity. Rödder et al. [26] demonstrated that the climate niche of six Lyciasalamandra species is similar, with merely the single south-east Aegean Sea Island (i.e. non-coastal) species, L. helverseni, showing a deviating climate niche. Further support for intra-generic climate niche conservatism comes from a study on demographic life-history parameters. Sinsch and co-authors [27] found that detectable differences in life history traits among populations of different species and subspecies are mainly due to variation in the period of surface activity rather than being the result of fundamental differences in their ecological adaptation.
On the one hand, the highly specialised ecological and reproductive adaptations of Lyciasalamandra species allowed them to survive in primarily hostile environments. On the other hand, this may have ‘trapped’ them on suitable habitat spots, with restricted options to disperse and to cross areas lacking suitable micro-habitats. Such rare colonization events would result in low levels of within and high levels of among population genetic differentiation (e.g., [8]). Up to now, evidence for restricted gene flow among Lyciasalamandra populations comes from studies that were mainly conducted to solve their inter- and intra-specific phylogenetic relation (e.g., [6–8]). Accordingly, sample designs were not elaborated to infer evolutionary processes at smaller geographic scales.
We here apply a population sampling across all species to test the hypothesis that most Lyciasalamandra populations are strongly isolated from each other. In the absence of strong male-biased dispersal, this would be shown by the existence of numerous local haplotypes and by low levels of genetic (haplotype) diversity within and high levels of haplotype diversity among populations. This prediction is based on the assumption that the pronounced adaptation of Lycian salamanders to a largely subterranean life style, coupled with viviparity, has continuously trapped them inside once colonised mountains. By applying species distribution models (SDMs) and using current and past climate scenarios, we therefore test a second hypothesis that these salamanders do not need to horizontally move out of trouble in times of drastically changed climatic conditions (as reflected by the Last Glacial Maximum (LGM)), which we use as a proxy for the numerous climatic alterations that repeatedly occurred during the Pleistocene; rather were they able to survive on the spot during periods of cold and dry climate.
Materials and methods
Population sampling
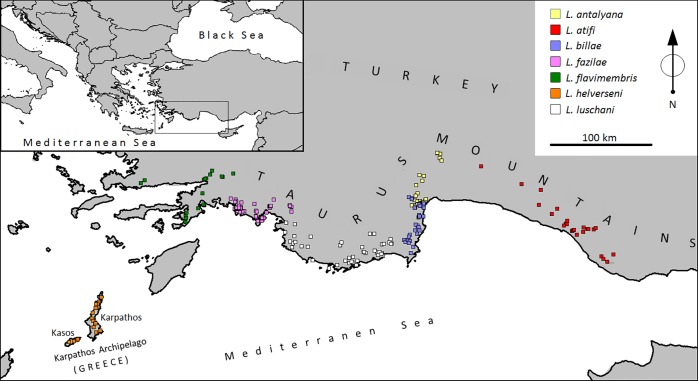
We analysed 559 specimens of Lyciasalamandra from 118 populations (population average 4.7 specimens) by either using sequences stored in GenBank or by analysing new tissue samples (Fig 1, S1 File and S1 Table). They cover all currently described species and subspecies as well as their entire ranges. Turkish samples were collected under license according to the ethical permission of the Ege University Animal Experiments Ethics Committee (2014#001) and special permission (2014#62406) for field studies from the Republic of Turkey, Ministry of Forestry and Water Affairs. Sampling in Greece took place according to the Hellenic National Law (Presidential Decree 67/81) and under a special permit (#107439/758) issued by the Ministry of the Environment. Salamanders were collected by turning stones in the respective habitats; tissue samples (toe clips) were immediately stored in absolute ethanol.
Fig 1. Localities along the Western Taurus Mountains and the Karpathos Archipelago in the eastern Mediterranean (see inset) of seven Lyciasalamandra species, used for Species Distribution Models (SDMs) and genetic analyses.
For details see S1 File.
Due to the large number of species and populations studied, we throughout this study consistently use the following terms: first three digits (###) of a species’ name are followed by phyloclade (###-<Roman numeral>), population (###-p<Arabic numeral>) and haplotype (###-h<Arabic numeral>) codes.
DNA extraction and sequencing
DNA was isolated using the Qiagen Blood and Tissue Kit following the manufacturer’s protocol. We sequenced fractions of two mitochondrial genes: 16SrDNA (primers 16SAL and 16SBH of [28]; initial melting for 120 s at 94°C, 33 cycles of denaturation for 30 s at 94°C, primer annealing for 30 s at 51°C, extension for 60 s at 65°C, final step at 65°C for 10 min) and ATPase (primers L-LYS-ML and H-COIII-ML of [8], covering fractions of subunits 8 and 6; initial melting for 10 s at 94°C, 30 cycles of denaturation for 30 s at 98°C, primer annealing for 30 s at 67°C, extension for 30 s at 72°C, final step at 72°C for 1 min). PCR reactions were prepared using either 5Prime Master Mix (16S) or the Phusion Flash Master Mix of Thermo Science (ATPase). PCR products were purified using the High Pure PCR Product Purification Kit of Roche. Sanger reactions for all genes were run using the Big Dye Terminator (ABI) with initial melting for 60 s at 96°C, 25 cycles of denaturation for 10 s at 96°C, primer annealing for 5 s at 50°C, extension for 240 s at 60°C. We sequenced single stranded fragments from both directions each on an ABI 3500 Genetic Analyzer Serie 2 automatic sequencer using standard protocols.
Sequence alignment
We automatically aligned the single gene datasets (16S, ATP6 and ATP8) with MAFFT (version 7, [29]) using the iterative refinement [30, 31] and the Needleman-Wunsch algorithm [32]. We examined our datasets for stop codons and excluded them where necessary. Between base pair positions 244–251 we found an AT tandem repeat in several species; we had to exclude this section from further analyses since it produces additional local variation which could mask the true haplotype genealogy. All ambiguous areas in the alignment were excluded using GBlocks (version 0.91b, [33]). We used the package ape [34] and the function cbind in R (version 3.3.1, [35]) for sequence concatenation. We removed redundant sequences by collapsing them into haplotypes using FaBox [36].
Partition Finder (version 2.0.0, [37]) was used to select the best-fitting partitioning schemes and substitution models from a predefined number of subsets (one subset for 16S and one for each codon position in ATP6 and ATP8), implementing the greedy algorithm and the Akaike Information Criterion (AIC). We compared the Partition Finder runs with linked and unlinked branch length and selected the model with linked branch length across partitions according to the AIC value.
Analyses of demographic population history
Nucleotide diversity (π) and haplotype diversity (hd) were calculated to estimate the genetic diversity of distinct phyloclades using DnaSP v. 6.10 [38] (for phyloclade delineation see below). To gain information about the demographic history of phyloclades we calculated further statistics: pairwise mismatch distribution [39], with observed distributions tested against expected distributions under a constant growth and a growth-decline model, respectively; Tajima’s D [40]; and Fu’s Fs [41]; all analyses done with DnaSP (see S1 Fig for mismatch distribution). The mismatch distribution describes the frequency of pairwise substitutional differences among individuals of a given group and is expected to be unimodal in populations that underwent recent bottleneck and rapid expansion [39]. Tajima’s D statistic tests for a departure from neutrality as measured by the difference between the number of segregating sites (h) and the average number of pairwise nucleotide differences (p). In the absence of balancing or purifying selection, population expansion can cause significant negative departures of Tajima’s D from zero, while a population bottleneck can cause a significant positive departure from zero [40]. Under the assumption of neutrality, Fu’s Fs statistic provides a test for population growth as well by identifying an excess of rare haplotypes in an expanding population when compared with the number of expected haplotypes in a stationary population (Fs<0; [41]). Conversely, Fs>0 would indicate a recent population bottleneck. Fs-values should be regarded significant if p<0.02 [41]. As additional test statistics for detecting patterns of population growth, we calculated R2 of [42], which has shown by these authors to be superior to the raggedness statistic rg of [43]. Statistical significance of R2 was tested using coalescent simulations in DnaSP. In addition, the demographic population history was reconstructed for each phyloclade using the Bayesian Skyline Plots (BSP) [44] method in Beast (version 2.5.0, [45]). We used the selected partitions of Partition Finder as predefined subsets and bModeltest for calculating the substitution models and the phylogeny in one single analysis [46]. As estimation for the substitution rate we inferred the mean rate of [6] (9.70E-3 sites per Mya) as strict clock rate. Coalescent Bayesian Skyline was used as tree prior and MCMC tests were run for 10 million steps each and sampled every 1,000th generation. BSP can be found in S2 Fig.
Analysis of molecular variance (AMOVA)
Ordered mitochondrial haplotypes (based on a p-distance matrix) were used to test for the degree of population fragmentation since, depending on their phylogenetic background, they may harbour information about their own history [47]. A hierarchical AMOVA [48] (ARLEQUIN software of [49]) partitioned the total genetic variance into variance components within populations (VIP), among populations within phyloclades (VPC) and among phyloclades within species (VCS). We also calculated the respective inbreeding coefficients (F-values) according to Weir and Cockerham [50]. Exact probabilities were calculated using the Markov-chain method [51], with 1023 recombinations and replicates each.
Phylogenetic tree and haplotype networks
A phylogenetic tree of all haplotypes was calculated using Maximum Likelihood (ML) and Bayesian Inference (BI). For hierarchical outgroup rooting we added homologous gene fragments from complete mitochondrial genomes of Salamandra salamandra (EU880331), Chioglossa lusitanica (EU880308), Mertensiella caucasica (EU880319), Pleurodeles poireti (EU880329), Pleurodeles waltl (EU880330), Euproctus platycephalus (EU880317) and Euproctus montanus (EU880316). The ML tree was calculated with RAxML (Stamatakis 2014) using rapid bootstrapping and the greedy algorithm, running 2,000 bootstrap replicates. The BI tree was calculated with MrBayes (version 3.2.6, [52, 53, 54]) implementing two runs with four independent chains; each run with 10 million generations and sampled every 1,000th generation with a burn-in of 20% (see S3 Fig for the BI tree). Afterwards we checked if the tree likelihood had converged. The results of Partition Finder were used as a priori configurations in MrBayes (see S2 Table). Since we could only use one substitution model for all partitions in RAxML [55], we decided to use the GTR+G model for all partitions.
We generated species specific population level genealogies using TCS [56]. TCS is widely used to calculate haplotype networks based on statistical parsimony. We set a probability cut-off value of 90% that defined the maximum number of mutational connections between pairs of sequences and thus delineated unconnected haplotype networks as distinct intraspecific phyloclades.
Modelling of current and past species distributions
Grid-based correlative Species Distribution Models (SDMs; [57]) were used to predict the current potential distributions of Lyciasalamandra species within the general region of the genus’ known range. For this purpose, we compiled 253 geo-referenced localities of all species based on literature and on own fieldwork data (S1 File). The minimum number of records per species was 18 (details below) after elimination of duplicates in the same grid cell, applying the grid system of SDM climate data (see below). Although SDM building is delicate when the number of records is low (cf. [58]), it can be feasible in narrow-ranged species [59], which is why we continued to compute SDMs for all seven species.
As ecological predictors we used current high resolution climate data for the period 1979–2013 from the CHELSA project ([60]; version 1.2; data available at: http://chelsa-climate.org/, accessed 10 May 2018). CHELSA operates on monthly means and is based on a quasi-mechanistic statistical downscaling of the ERA interim global circulation model (GCM) with a GPCC bias correction [60]. The CHELSA website provides ‘bioclim’ variables (cf. [61]). From the 19 variables available, six were selected via pair-wise Pearson correlation analyses to avoid effects of multicollinearity, which is important when projecting SDMs into new space and time [62]. Of highly correlated variables (|r| > 0.7), we excluded the less informative one, based on a priori assumptions on biological importance to our target organisms: bio3, isothermality (BIO2/BIO7) (* 100); bio11, mean temperature of coldest quarter; bio15, precipitation seasonality (coefficient of variation); bio18, precipitation of warmest quarter; bio19, precipitation of coldest quarter.
CHELSA climate data are available at grid resolution 30 arc sec, thus reflecting macro- or mesoclimate. Like multiple authors before (cf. [57]), we consider these data as proxies to micro-climatic conditions. This is a crucial point to mention, especially with regard to Lyciasalamandra species, because part of their life these amphibians exploit deep-reaching systems of crevices in karstic limestone systems [3, 27, 63]. We defend our approach, as micro-climatic data for the focal taxa are not available and Rödder et al. [26] have shown that macro-climate can be effectively used for the computation of Lyciasalamandra SDMs. Along with these authors, we expect that CHELSA climate data show where the climate in general is favorable to our study species.
With the goal to predict the potential geographic ranges of the target species during the LGM (ca. 21 K BP), SDMs based on the current climate were projected to LGM data available from the CHELSA website (i.e., bioclim variables, downloaded 10 May 2018). They are based on the implementation of the CHELSA algorithm on PMIP3 (Paleoclimate Modelling Intercomparison Project Phase III) data using the widely used and well performing Community Climate System Model 4 global circulation model [64] as a test dataset and a paleo-digital elevation model with resolution 30 arc sec [60].
Maxent 3.4.1 [65, 66] was used for SDM building (https://biodiversityinformatics.amnh.org/open_source/maxent/, accessed 10 May 2018). This presence-only/background method operates with a machine-learning algorithm following the principle of maximum entropy. It makes predictions on the potential geographic range of a taxon by taking environmental (here: climatic) data from geo-referenced species records and random background data [65, 67]. In this way, it contrasts the environmental conditions at species’ presences against those at the background to fit a function to estimate the relative suitability to the species [68]. Maxent is a widely used SDM tool and often performs better than other SDM methods [69, 70]. It offers various settings for SDM building allowing fine-tuning [65, 70]. This requires some caution, however, as with these, the output can be dramatically altered when uncritically used [67, 68, 71, 72]. Therefore, it is important to explore settings and to adapt them to the available data [66, 70, 73].
In our final model runs, we employed Maxent with specifications for SDMs based on small sample sizes with hinge features. In three species (L. antalyana, L. flavimembris, L. helverseni), with 29, 18 and 23 records, respectively, the cross-validate approach was chosen. For all other species (with 33 or more records), the subsample approach was used with each 25% of the records randomly set aside as test data. The number of replicates was equal to the number of records in the cross-validate and 100 in the subsample approach. Extrapolation was used, but no clamping, and response curves were explored. One background was chosen for all species as a window enclosing all Lyciasalamandra records. The number of background points was 100,000. All other settings were default [66, 68, 70, 74]. A Multivariate Environmental Similarity Surfaces (MESS) analysis [71] revealed that there were no conditions in the general region of the genus’ known range (i.e. coastal mountain ranges and coastal islands) potentially leading to unrealistic extrapolations of response curves (S4 Fig).
Maxent calculates the area under the receiver operating characteristic curve (AUC) as a measure of predictive accuracy [65]. Following the classification of [75], AUC values range between 0.5 for models with no predictive ability and 1.0 for models giving perfect predictions; values > 0.9 describe ‘very good’, > 0.8 ‘good’, > 0.7 ‘useable’ models. Although criticized (e.g., [67]), the AUC is informative as it mirrors the model’s ability to distinguish between species records and background points, i.e. showing how general or restricted a distribution is along the range of the variables in the studied region [68]. To account for AUC critics, we also calculated True Skill Statistic (TSS) as sensitivity + specificity– 1 [76].
The Maxent default ClogLog output format (ranging 0–1) was chosen for processing the resulting SDM maps in a GIS approach, and the ‘maximum training sensitivity plus specificity ClogLog threshold’ was used to distinguish potential presence from absence, as this threshold might not overestimate distributions [66]. Because of the almost exclusive association of Lyciasalamandra species with karstic limestone formations (see above), areas identified with SDMs climatically suitable but outside these geological conditions were cut off using a geology shapefile of the Central Energy Resource Team of the US Geological Survey (http://energy.cr.usgs.gov/oilgas/wep/, accessed 10 May 2018). In addition, in LGM maps a sea level layer was overlaid to only show areas beyond today’s coast line that indeed were accessible continental shelf. It was obtained from the LGM Vegetation Download Page (http://anthro.unige.ch/lgmvegetation/download_page_js.htm, accessed 10 May 2018).
We prepared boxplots of Maxent ClogLog values at species records for current and LGM SDMs. These were tested using a Wilcoxon signed rank test.
Results
Molecular diversity and population demography
Altogether, we found 153 combined 16S and ATPase haplotypes, with the fewest in L. flavimembris and the most in L. billae (Table 1). The majority of haplotypes were private to a single population (78.4%). Only one haplotype occurred in a maximum of seven populations (ant-h1 of L. antalyana). Consequently, haplotype diversity was high in all phyloclades, with hd almost always approaching a maximum value of 1.0. In contrast, nucleotide diversity π was mostly below 1%.
Table 1. Genetic diversity, neutrality tests and mismatch goodness-of-fit tests for Lyciasalamandra phyloclades.
| Phyloclade | np | ns |
nh (nph) %ph |
hd
(s.d.) |
π
(s.d.) |
Neutrality tests | Goodness-of-fit tests | |
|---|---|---|---|---|---|---|---|---|
| Tajima’s D
(P-value) |
Fu’s Fs
(P-value) |
R2 (P-value) |
||||||
| L. atifi-I (ati-I) | 3 | 18 | 4 (3) 75% |
1.000 (0.177) |
0.0028 (0.0009) |
-0.79684 (> 0.10) |
-1.514 (0.180) |
0.2179 (0.164) |
| L. atifi-II (ati-II) | 10 | 52 | 14 (10) 71% |
1.000 (0.027) |
0.0067 (0.0006) |
-0.16290 (> 0.10) |
-9.421 (< 0.001) |
0.1299 (0.374) |
| L. atifi-III (ati-III) | 3 | 14 | 3 (2) 67% |
1.000 (0.272) |
0.0028 (0.0009) |
n.a. | n.a. | n.a. |
| L. antalyana-I (ant-I) | 8 | 36 | 11 (8) 72% |
0.964 (0.051) |
0.00431 (0.0042) |
0.11338 (> 0.10) |
-3.679 (0.021) |
0.1413 (0.251) |
| L. antalyana-II (ant-II) | 8 | 30 | 3 (2) 67% |
1.000 (0.272) |
0.0022 (0.0008) |
n.a. | n.a. | n.a. |
| L. billae-I (bil-I) | 7 | 29 | 8 (6) 75% |
1.000 (0.063) |
0.0653 (0.0009) |
0.43162 (> 0.10) |
-3.463 (0.030) |
0.1665 (0.348) |
| L. billae-II (bil-II) | 1 | 13 | 2 (2) 100% |
1.000 (0.500). |
0.0011 (0.0006) |
n.a. | n.a. | n.a. |
| L. billae-III (bil-III) | 9 | 35 | 14 (10) 71% |
1.000 (0.027) |
0.0064’ (0.0007) |
0.41361 (> 0.10) |
-9.990 (< 0.001) |
0.1555 (0.451) |
| L. billae-IV (bil-IV) | 9 | 30 | 3 (0) 0% |
1.000 (0.272) |
0.0015 (0.0005) |
n.a. | n.a. | n.a. |
| L. luschani-I (lus-I) | 4 | 14 | 4 (3) 75% |
1.000 (0.177) |
0.0048 (0.0013) |
1.32331 (> 0.10) |
-0.715 (0.328) |
0.2486 (0.342) |
| L. luschani-II(lus-II) | 2 | 4 | 2 (2) 100% |
1.000 (0.500) |
0.0022 (0.0011) |
n.a. | n.a. | n.a. |
| L. luschani-III (lus-III) | 1 | 6 | 3 (3) 100% |
1.000 (0.272) |
0.0015 (0.0005) |
n.a. | n.a. | n.a. |
| L. luschani-IV (lus-IV) | 2 | 8 | 4 (4) 100% |
1.000 (0.177) |
0.0019 (0.0005) |
0.00186 (> 0.10) |
-2.181 (0.101) |
0.2003 (0.059) |
| L. luschani-V (lus-V) | 9c | 48 | 13 (11) 85% |
1.000 (0.030 |
0.0055 (0.0006) |
-0.18976 (> 0.10) |
-9.501 (< 0.001) |
0.1242 (0.199) |
| L. luschani-VI (lus-VI) | 7 | 30 | 7 (6) 86% |
1.000 (0.076) |
0.0021 (0.0003) |
-1.12898 (> 0.10) |
-5.925 (0.003) |
0.0851 (<0.001) |
| L. fazilae-I (faz-I) | 9 | 34 | 8 (5) 63% |
1.000 (0.063 |
0.0054 (0.0009) |
0.21018 (> 0.10) |
-4.022 (0.018) |
0.1698 (0.352) |
| L. fazilae-II (faz-II) | 1 | 1 | 1 (1) 100% |
n.a. | n.a. | n.a. | n.a. | n.a. |
| L. fazilae-III (faz-III) | 2 | 14 | 5 (5) 100% |
1.000 (0.126) |
0.0031 (0.0007) |
-0.19092 (> 0.10) |
-2.371 (0.085) |
0.1599 (0.056) |
| L. fazilae-IV (faz-IV) | 6 | 27 | 10 (9) 90% |
1.000 (0.045) |
0.0030 (0.0007) |
-1.62991 (> 0.05) |
-8.843 (< 0.001) |
0.0710’ (< 0.001) |
| L. fazilae-V (faz-V) | 2 | 8 | 3 (2) 67% |
1.000 (0.272) |
0.0015 (0.0005) |
n.a. | n.a. | n.a. |
| L. flavimembris (fla-I) | 8 | 39 | 10 (7) 70% |
1.000 (0.045 |
0.0047 (0.0005) |
-0.36945 (> 0.10) |
-6.650 (0.001) |
0.1192 (0.046) |
| L. helverseni-I (hel-I) | 12 | 55 | 17 (15) 88% |
0.985 (0.025) |
0.0044 (0.0006) |
-1.86018 (< 0.05) |
-10.682 (< 0.001) |
0.0675 (<0.001) |
| L. helverseni-II (hel-II) | 1 | 4 | 3 (3) 100% |
1.000 (0.272) |
0.0030 (0.0011) |
n.a. | n.a. | n.a. |
| L. helverseni-III (hel-III) | 1 | 5 | 1 (1) 100% |
n.a | n.a. | n.a. | n.a. | n.a. |
np, number of populations; ns, number of samples; nh (nph) %ph, number of (private) haplotypes and percentage of private haplotypes; π (s.d.), nucleotide diversity with standard deviation; hd (s.d.), haplotype diversity with standard deviation; R2, Ramos-Onsins’ and Rozas’ [42] raggedness index; n.a., not applied.
Analyses of population demography show in parts contradicting results within and among phyloclades (Fig 2, Table 1 and S1 Fig). Bayesian skyline plots indicate a recent increase in effective population size for ant-I, ant-II, bil-III, lus-VI, faz-IV and fla-I, while a recent decrease is indicated for ati-III, bil-I, lus-V, faz-III and, most pronounced, for hel-I (S2 Fig). Interestingly, for the latter significant negative values of Tajima’s D and Fu’s Fs indicate possible recent population expansion. In all other phyloclades, according to Bayesian skyline plots population sizes remained more or less constant (S2 Fig), although in several cases significantly negative values of Fu’s Fs indicate possible recent population expansion, while Tajima’s D did not significantly deviate from zero. In contrast, significantly negative values of Fu’s Fs (p<0.02) indicate possible recent population expansion in ati-II, bil-III, lus-V, lus-VI, faz-I, faz-IV, fla-I and hel-I, while the same is indicated by a significantly negative value of Tajima’s D only for hel-I. The graphic representation of the mismatch distribution tentatively fits an expected growth-decline model only in phyloclades ant-II, lus-IV, lus-VI, and hel-I. In all other phyloclades, the observed mismatch distribution seems to fit neither a growth decline nor a constant growth model (S1 Fig). R2 values significantly different from a neutral model were only found for lus-VI, faz-IV, fla-I and hel-I (Table 1). For all other phyloclades, multimodal mismatch distributions indicate a pattern of recurrent population bottlenecks and expansions.
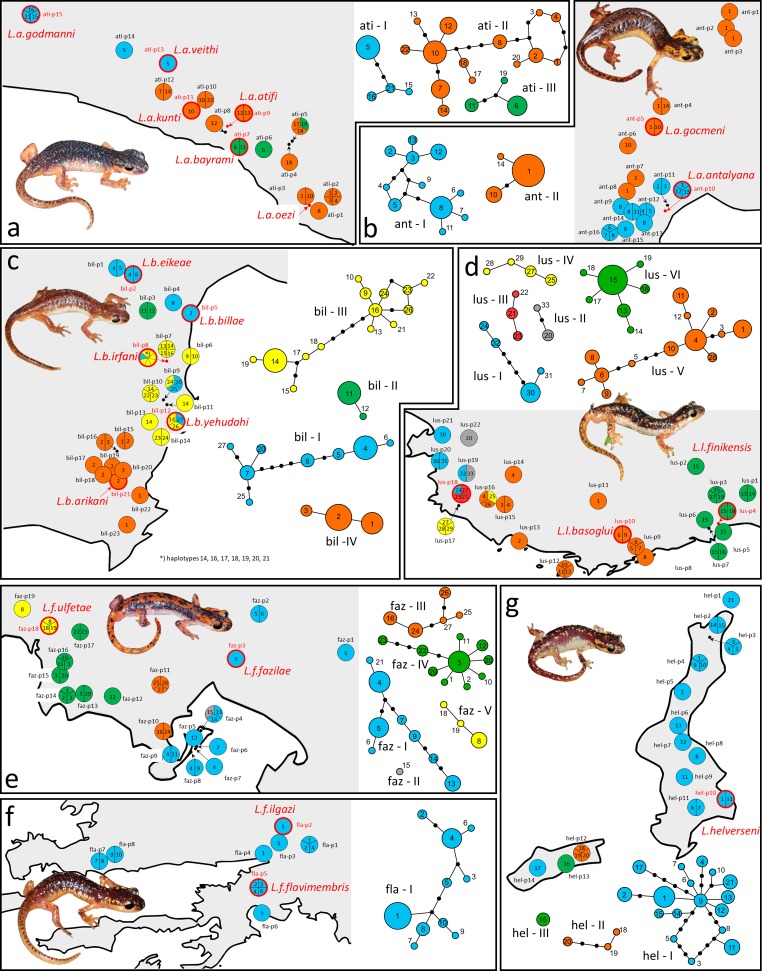
Fig 2. Geographic haplotype distribution and unrooted haplotype networks of Lycian salamander phyloclades.
From East to West (see also Fig 1 for the distribution of all species): a) L. atifi phyloclades ati-I to -III, b) L. antalyana phyloclades ant-I and -II, c) L. billae phyloclades bil-I to -IV, d) L. luschani phyloclades lus-I to -VI, e) L. fazilae phyloclades faz-I to V, f) L. flavimembris (fla-I) and g) L. helverseni phyloclades hel-I to -III. Names and type localities of currently recognized subspecies are indicated in red in the distribution maps. Numbers in circles refer to found haplotypes, with circle size being proportional to haplotype abundance; black dots indicate haplotypes not found. Each line in between two haplotypes accounts for one mutational difference.
Total molecular variance was lowest in L. atifi (6.9%) and highest in L. fazilae 19.8 (Table 2). Molecular variance distribution shows a consistent pattern across species. Within-population variance was extremely low, ranging from 1.50% in L. luschani to 7.89% in L. billae. The respective fixation indices FIP were close to one, indicating almost fixed molecular differences among populations. Among phyloclade molecular variance made up most of the entire molecular variance, with a maximum of 91.5% in L. fazilae (a comparable three-level AMOVA could not be calculated for L. flavimembris since only one phyloclade was found in this species).
Table 2. Intraspecific distribution of molecular haplotype variance (AMOVA).
| among phyloclades | among populations within phyloclades | within populations | total variance | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| species | df | VCS | %VCS | FCS | p | df | VPC | %VPC | FPC | p | df | VIP | %VIP | FIP | p | df | VT |
| L. atifi | 2 | 4.810 | 69.56 | 0.696 | < 0.001 | 13 | 1.748 | 25.80 | 0.847 | < 0.001 | 68 | 0.321 | 4.64 | 0.953 | < 0.001 | 83 | 6.915 |
| L. antalyana | 1 | 6.270 | 83.59 | 0.836 | < 0.001 | 14 | 1.002 | 13.36 | 0.814 | < 0.001 | 52 | 0.229 | 3.05 | 0.969 | < 0.001 | 67 | 7.051 |
| L. billae | 3 | 6.651 | 79.17 | 0.791 | < 0.001 | 22 | 1.087 | 12.94 | 0.621 | < 0.001 | 81 | 0.663 | 7.89 | 0.921 | < 0.001 | 106 | 8.401 |
| L. luschani | 5 | 16.447 | 91.18 | 0.912 | < 0.001 | 19 | 1.321 | 7.32 | 0.830 | < 0.001 | 87 | 0.271 | 1.50 | 0.985 | < 0.001 | 111 | 18.039 |
| L. fazilae | 4 | 18.090 | 91.50 | 0.915 | < 0.001 | 15 | 1.341 | 6.78 | 0.798 | < 0.001 | 64 | 0.340 | 1.72 | 0.983 | < 0.001 | 83 | 19.771 |
| L. helverseni | 2 | 15.548 | 87.26 | 0.873 | < 0.001 | 11 | 1.703 | 9.56 | 0.750 | < 0.001 | 63 | 0.567 | 3.18 | 0.968 | < 0.001 | 63 | 17.818 |
Among phyloclades (VCS), among populations within phyloclades (VPC), within populations (VIP); the respective fixation indices FCS, FPC and FIP are given. Not calculated for L. flavimembris due to the lack of an intraspecific phyloclade sub-structure.
Haplotype networks and spatial haplotype distribution
Two to six distinct phyloclades appeared in all species but L. flavimembris (only one phyloclade; Fig 2A–2G). Most phyloclades correspond to distinct geographic clusters. Exceptions are ati-II (interspersed by ati-III), bil-I (some haplotypes also occur throughout the range of bil-III) and hel-I (it occurs on all islands of the Karpathos Archipelago, but with hel-II and–III being found in between its populations). Within most phyloclades there seems to exist a fine-scale geographic differentiation that is largely mirrored by haplotype positions within the networks, e.g. in L. atifi (ati-I, ati-III), L. antalyana (ant-I, ant-II), L. billae (buil-I, bil-IV), L. luschani, L. fazilae and L. flavimembris. Discordant network and geographic positions of haplotypes or groups of haplotypes are obvious in L. atifi (ati-II), L. billae (bil-III) and, to a lower degree, in L. helverseni (hel-I). Widespread haplotypes spanning almost the entire range of a phyloclade only occur in L. antalyana (ant-h1), L. billae (bil-h1) and L. luschani (lus-h15). Haplotypes of two phyloclades co-occur in L. atifi (ati-p5), L. billae (bil-p8, bil-p9, bil-p12), L. luschani (lus-p16, lus-p18, lus-p19) and L. fazilae (faz-p4)
Modelled current and past potential species distributions
AUC values of SDMs ranged from 0.854 to 0.9997, suggesting ‘good’ to ‘very good’ model performance. This was also supported by TSS values ranging 0.762 to 0.879. Acceptable model accuracy was also indicated by the markedly low standard deviation of single model runs (ClogLog ≤ 0.2). The modelled potential distributions of the species under current climate (S5 Fig) well explained most known species’ presences and identified more or less continuous areas as suitable within the proximity of the species’ records and beyond. However, in L. atifi, L. billae, L. fazilae and L. luschani some records remained unexplained under the ‘maximum training sensitivity plus specificity ClogLog threshold.
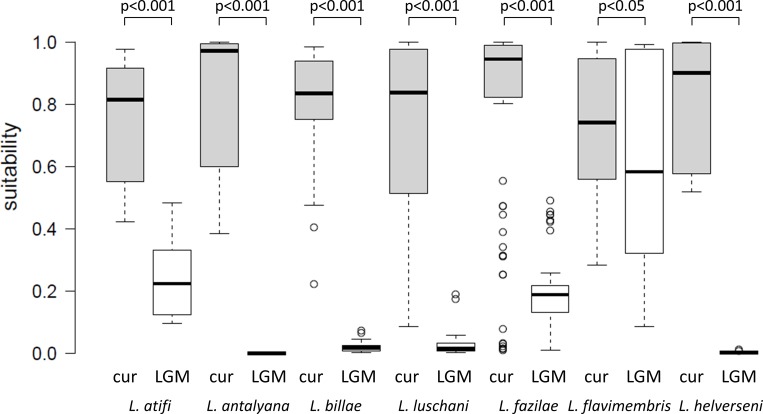
When projecting SDMs into climatic conditions of the LGM, species’ potential distributions markedly shifted (L. fazilae, L. flavimembris, L. helverseni), decreased (L. atifi, L. luschani) or even no suitable area could be identified (L. antalyana, L. billae) (S5 Fig). In all taxa, it became evident that all or almost all of today’s known records had been in unsuitable areas during the LGM. The suitability alteration in terms of Maxent ClogLog values at species records is illustrated in Fig 3. The median was highly significantly different in all species (p < 0.001), except in L. flavimembris (p = 0.041), when comparing current and LGM climates.
Fig 3. Box-Whisker plots for Maxent ClogLog values at known records of Lyciasalamandra species under current (grey) and LGM (white) climatic conditions.
Values range from 0 (no suitability) to 1 (perfect suitability).
Discussion
Phylogeographic structure of species
Lycian salamanders have inhabited the Anatolian Mediterranean coast and the Karpathos Archipelago for more than 10 mya [6]. Subsequent diversification resulted in the seven species known today, with a varying number of distinct intraspecific lineages (i.e. some suggested to be subspecies). Several authors had already emphasized that Lyciasalamandra populations often harbour exclusive haplotypes, indicating a high degree of isolation among populations (e.g., [6, 8, 77]); however, their sampling was geographically more limited. Our dense spatial sampling of 118 populations across the entire known geographic range of the genus, combined with new analytic approaches, now offers the opportunity to evaluate the degree of possible isolation among populations as well as the processes that shaped this unique pattern in this enigmatic amphibian clade.
All but one (L. flavimembris) species show a pattern of deep haplotype differentiation in the mtDNA with pronounced geographic structure, indicating possible long-term isolation and/or limited dispersal and gene flow capabilities [78; currently, the alternative hypothesis of male-biased gene flow, which would erode population differentiation, cannot be ruled out due to the fact that bi-parentally inherited nuclear genes have not yet been investigated]. These highly divergent allopatric phyloclades show almost no sign of geographic overlap, with the shortest distance between two phyloclades being 2.5 km in L. helverseni and the longest being 22 km in L. luschani. Within most phyloclades, further well-differentiated haplogroups exist, which again commonly are allopatric (shallow haplotype trees showing geographic structure; [78]). Given the high degree of differentiation among phyloclades and their small-scale geographic occurrence, we consider this one of the most exceptional cases of fine-scale genetic differentiation in mtDNA within an amphibian. Of course, comparative levels of intraspecific differentiation at mitochondrial loci were described also in other amphibian species. European examples include Rana temporaria [79, 80], Rana iberica [81], Hyla orientalis [82], Lissotriton boscai [83], Lissotriton helveticus [84] and Salamandra algira [85]. However, samples of these species came from ranges of up to two orders of magnitude larger than those inhabited by Lyciasalamandra species (maximum distance between two samples of a species range from 40 km in L. flavimembris to 160 km in L. atifi, respectively). In addition, admixture of haplotypes of neighbouring phyloclades as observed in other species (e.g. R. temporaria; [80]), a pattern that often mirrors geographic introgression after secondary contact of lineages (in Europe most often after re-expansion from glacial refugia; [86, 87]), is found only in eight out of the 118 populations studied. Pronounced intraspecific phylogenetic discontinuity visible through high mitochondrial haplotype differentiation is typical for amphibians, presumably because their relative dispersal ability is low [88]. A complex population differentiation comparable to what we found in Lyciasalamandra, is also observed in some tropical species, mainly driven by forest habitats and topographic heterogeneity (e.g., [89]). However, in such cases haplotypes are often shared among populations (e.g., [90]).
Phylogeographic structure within phyloclades
In all Lyciasalamandra species, even geographically nearby populations harbour private haplotypes, although, from a human point of view, the habitat in-between seems to be appropriate (most often karstic limestone with pine forests [3, 63]). This is most obvious in phyloclades ati-I of L. atifi, ant-I of L. antalyana, bil-I of L. billae, lus-I and lus-II of L. luschani, faz-II of L. fazilae and hel-I of L. helverseni (own field observation). From a geographic point of view, an extreme example is the island of Kasos, where all three phyloclades of L. helverseni occur within only 7.3 km (populations hel-p12, -13 and -14 in Fig 2F). Given the large mutational difference of such neighbouring haplotypes, a pattern like this is most easily be explained by strict isolation. Widespread haplotypes, covering a maximum area of 10 to 15 km in Lyciasalamandra, are rare and indicate either moderate recent female dispersal or incomplete lineage sorting. The most common haplotype in terms of populations (ant-h1 was found in seven populations) covers a distance of more than 50 km.
Widespread haplotypes are possibly indicative of geographic expansion, a process that is usually associated with demographic expansion (e.g., [88, 91]). Therefore, one would expect respective population genetic signals within phyloclades. This is in fact the case in ant-II and lus-VI, both having wide-spread haplotypes and both showing a signal of recent demographic expansion in the Bayesian skyline plots (S4 Fig). On the other hand, also some phyloclades with no widespread haplotypes showed signs of recent demographic expansion after a bottleneck (Table 1: significantly negative values of Fu’s Fs; alternatively, a selective sweep of mtDNA haplotypes may also produce such a pattern; however, this invokes pronounced female dispersal, which contradicts current knowledge of the ecology of Lyciasalamandra [4]). Only in hel-I this was corroborated by a significantly negative Tajima’s D (hel-I). Bayesian skyline plots in most cases showed only moderate signs of change in effective population sizes towards the recent (again hel-I is an exception). We therefore consider the calculated population genetic parameters to be less conclusive, which might be attributed to the fact that most phylogroups harboured predominantly haplotypes private to single populations.
Klewen [63] suggested that rivers and rock formations other than limestone act as strong barriers to dispersal and hence gene flow among Lyciasalamandra populations. However, in between such river systems, dispersal should have easily been possible, given that most often karstic limestone connects even adjacent populations. In fact, star-like haplotype networks with locally occurring descendent haplotypes or haplogroups branching off from a central one occur in several Lyciasalamandra phylogroups. Such patterns are typical of range expansions, followed by local differentiation and subsequent isolation [88, 91]. One reason why in most cases this pattern is not identified by population demographic parameters may be that such star-like structures almost always form only part of the respective haplotype network, with longer branches also connecting more differentiated haplotypes to the central haplotype. This inevitably increases the raggedness of mismatch distributions and increases deviation from a pattern expected under a simple growth-decline model. Therefore, and apart from the idea of strong isolation of populations, we conclude that in fact range expansion occasionally must have occurred regularly in the past. Future research including bi-parentally inherited genes is needed to test this assumption.
Interestingly, within phyloclade hel-I of L. helverseni, haplotype hel-h1 occurs at two localities, Olympos village (hel-p4) and the city of Pigadia (hel-p10) (see Fig 2F) at the almost respective ends of the island of Karpathos, ca. 27 km away from each other. Pigadia is the only harbour connecting Karpathos Island with the Greek mainland. We therefore assume that hel-h1 may be a case of anthropogenic translocation, in this case from Olympos village to the city of Pigadia (see review on distribution patterns of Aegean herpetofauna and examples of human mediated translocations therein [20]). Admittedly, incomplete lineage sorting, which may also produce a pattern of disjunctive haplotype distribution throughout a species’ distribution range by preserving ancestral polymorphisms cannot completely be ruled out as an alternative explanation, although in none of the other Lyciasalamandra phyloclades a similar pattern was observed.
Potential range dynamics during the Quaternary
To explain the pattern of haplotype differentiation and distribution observed today, there must have been geological episodes when female dispersal was possible. According to [6], the Messinian Salinity Crisis (MSC) was most likely responsible for the onset of intraspecific evolution in L. fazilae, L. helverseni and L. luschani. In L. atifi, L. antalyana, L. billae and L. flavimembris, intraspecific diversification seems to have started in the Quaternary, a period when recurrent and dramatic climatic alterations [18, 22, 92] seem to have shaped the evolution of western Palearctic biota (e.g., [93, 94]). Unfortunately, the number of stitials and interstitials that occurred in the Quaternary is high [22]; so, given the uncertainty associated with split time estimation based on the molecular clock it is not possible to affiliate any split within this period to a single climatic event.
During the LGM, sites where nowadays Lyciasalamandra species occur were largely not explained (S5 Fig). At these sites, Maxent ClogLog values were significantly lower compared to current climate (Fig 3), strongly supporting that unsuitable conditions for salamander survival prevailed except for L. fazilae and L. flavimembris. In some species, SDM projections into the LGM revealed suitable area not too far from nowadays occurrences, all in all including areas which contain karstic limestone, too (S5 Fig). In part, e.g. in L. helverseni, this included the shelf area of the Mediterranean Sea which was about 100 m lower than today (e.g., [95]). In consequence, one may argue that range shifts in order to following suitable habitats could have occurred. There exist numerous examples of European plant and animal species for the validity of the so-called expansion-contraction (EC) model, a demographic scenario in temperate species whereby glacial cycles result in the contraction in size and shift toward lower latitudes into refugial areas during periods of cooling, followed by population growth and re-colonisation during postglacial warming (e.g., [86, 96]). Interestingly, numerous L. fazilae populations, especially those living far from the Mediterranean coast, appear to live under suboptimal current macroclimatic conditions (Fig 3, S5 Fig). This can be interpreted as an indication that horizontal range shifts due to climate change do not always allow theses salamanders to quickly adapt their geographic ranges to the climate.
Also not in line with such a past range shift scenario is our observation that almost no suitable area for four species, L. antalyana, L. billae, L. luschani and L. helverseni, could not be identified during the LGM (Fig 3, S5 Fig). This casts some doubt on potential range shift scenarios. Also, range shifts are typical bottleneck events and should leave characteristic imprints in the genetic signature of populations [87, 88, 93]. In the species where this has been observed, they led to an erosion of genetic diversity within and among populations, and after re-expansion genetically uniform populations would inhabited large areas [88, 93]. Species with a low dispersal capacity were ultimately doomed to extinction through loss of suitable habitat([97], unless they could shift their ranges at small scales along altitudinal gradients [93]. Alternatively, they might have survived in so-called refugial sanctuaries [84]. Any recurrent range shift, either on larger or on smaller geographic scales, would produce the above mentioned pattern of genetically similar and at the same time depauperate populations. We should observe such a pattern if Lyciasalamandra populations had followed shifting suitable habitats between stitials and interstadials and vice versa. Our results of numerous local haplotypes across most phyloclades’ ranges contrast with this expectation. Hence, micro-refugial survival in glacial sanctuaries best explains this unique pattern of fine-scale geographic allopatry of mitochondrial haplotypes within Lyciasalamandra (see [23, 98] for a further examples of cryptic diversity that evolved in the same area during Plio-Pleistocene glaciations).
Based on demographic and growth pattern analyses, [27] recently showed that the mainly subterranean life-style of Lycian salamanders in deep-reaching systems of crevices allows them to survive both cold winter and dry and hot summer. They observed a very small age- and size-related life-history variation across populations and species and concluded that this hints at a pronounced niche conservatism in Lyciasalamandra. Transferring this into a temporal dimension, this highly specialised and at the same time conserved ecological niche in combination with viviparity provides a sufficient explanation why many populations could have survived repeated climatic deteriorations ‘on the spot’. A pronounced female philopatry, which is considered advantageous when living in harsh environments as it can help females to acquire and defend an appropriate shelter (see [99] for the viviparous alpine salamander), may have enforced this niche conservatism.
Did our mitochondrial markers capture the whole story?
Mitochondrial markers as studied by us are inherited almost exclusively by females [100], so gene flow mediated by males would remain undetected when studying organelle genes. Slight male-biased dispersal was suggested by [4] to explain discordant patterns of mitochondrial versus nuclear genes in a hybrid zone of L. antalyana and L. billae. Such gene flow, if strong enough, should admix nuclear alleles among populations, while population specific mitochondrial haplotypes would persist. However, several authors mentioned that neighbouring populations often show divergent, but within populations seemingly stable, phenotypes in terms of coloration and pattern (e.g., [3, 63]). Unfortunately, such studies neither were based on statistically solid sample sizes, nor did they appreciate the potentially high environmental and ontogenetic plasticity in amphibian colouration (e.g., [101, 102]). Genes for coloration and pattern are coded by the nuclear genome, and unless strong natural selection would locally stabilize distinct colour and pattern morphs, even occasional male-mediated gene flow would homogenize populations in terms of nuclear genes. Therefore, our observed pattern of strong isolation among populations does not seem to be an artefact of a merely mitochondrial based analysis. However, analyses of bi-parentally inherited nuclear genes are needed to test this hypothesis.
Taxonomic implications
Within Lyciasalamandra, numerous new taxa have been described during the last few years, however, almost always based on morphology alone (see above). Our phyloclade structure and subspecies designation only match perfectly in L. antalyana, which is also supported by the phylogenetic tree. In L. fazilae and L. helverseni, phyloclades diversity is higher than current taxonomic diversity. In the phylogenetic tree, the five phyloclades of L. fazilae form three well supported clusters ((faz-I + faz-II), faz-II and (faz-IV + faz-V)), while in L. helverseni, phyloclade structure perfectly matches the three clusters supported by the pylogenetic tree.
In L. atifi, L. billae and L. flavimembris several subspecies have been described even within phyloclades that have formed in our analysis. Most extremely, in L. billae populations morphologically and geographically assigned to three subspecies, L. b. billae (bil-p6), L. b. irfani (bil-p7, -8) and L. b. yehudahi (bil-p9 to -14), respectively, even harbour haplotypes of the same phyloclades. Such patterns may be due to incomplete lineage sorting or secondary contact. L. billae, phyloclades bil-I, bil-II and bil-III form a common cluster, however, without three clearly delineated sub-clusters. The situation is similar in L. atifi: although ati-I and ati-III are well supported by the phylogenetic tree (S3 Fig), phyloclade differentiation is not mirrored by cluster delineation. L. flavimembris does not show any significant intraspecific differentiation, although two subspecies have been discriminated based on morphology alone. In L. luschani, clusters formed by the phylogenetic tree perfectly match current subspecies taxonomy, while the four phyloclades identified within L. l. luschani do not transfer to an unambiguous cluster pattern in the phylogenetic tree.
Variance distribution among phyloclades is more than twice as high in L. luschani, L. fazilae and L. helverseni compared to the other species (Table 2), indicating that also taxonomic differentiation within these species might be more justified. Almost all taxa newly described during the last decade were delineated exclusively on the basis of colour and pattern polymorphisms (e.g., [10–13]) and allopatric distribution. While the latter criterion is concordant with the definition of subspecies [103], the discriminant power of the morphological characters used for the delimitation of recently described taxa within Lyciasalamandra has never been proven. Nevertheless, and although we doubt the validity of the current taxonomy within Lyciasalamandra, we abstain from a taxonomic revision since our result are based on mitochondrial data alone.
Supporting information
Comparison of observed mismatch distributions (obs) of Lyciasalamandra phyloclades with distributions expected under a growth-decline (exp (G-D)) and a constant growth (exp (cG)) model; X-axis: number of base substitutions between two haplotypes; Y-axis: frequency of the number of base substitutions between two haplotypes.
(JPG)
Bayesian Skyline Plots (BSP) for phyloclades of Lycian salamanders (a-f): a) L. atifi BSPs for phyloclades ati-I, ati-II and ati-III, b) L. antalyana BSPs for phyloclades ant-I and ant-II, c) L. billae BSPs for phyloclades bil-I and bil-III, d) L. luschani BSPs for phyloclades lus-I, lus-IV, lus-V and lus-VI, e) L. fazilae BSPs for phyloclades faz-I, faz-III and faz-IV, f) L. flavimembris BSP for phyloclade fla-I, and g) L. helverseni BSP for phyloclade hel-I. Phyloclades were coloured according to the colours of the haplotype networks in Fig 2.
(TIF)
Outgroups were excluded from the figure. We only represent Bayesian Posterior Probabilities ≥0.95 (see numbers at nodes). Phyloclades of each species were colour marked according to Fig 2 in the main text. Due to the low resolution of the ML tree, we only represent the BI tree.
(TIF)
(TIF)
Mapped Maxent SDMs for all Lyciasalamandra species under current (left) and LGM (right) climatic conditions. Note that under the LGM land masses exceeded those of today due to sea level change. Suitability to species increases with higher ClogLog values, indicated by warmer colours. Known species records are indicated by blue dots (cf. S1 File). For details of modelling and mapping see Materials and Methods.
(TIF)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
We thank the Management Agency of Karpathos-Saria for facilitating samplings at northern Karpathos and Saria islands. Thanks to Sabine Naber for help with DNA sequence analyses.
Data Availability
All sequences are available from the GenBank database, as listed in the Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Buckley LB, Jetz W. Environmental and historical constraints on global patterns of amphibian richness. Proc Roy Soc B. 2007; 274: 1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeisset I, Beebee TJC. Amphibian phylogeography: a model for understanding historical aspects of species distributions. Heredity. 2008; 101: 109–119. 10.1038/hdy.2008.30 [DOI] [PubMed] [Google Scholar]
- 3.Veith M, Baran I, Godmann O, Kiefer A, Öz M, Tunc MR. A revision of population designation and geographic distribution of Mertensiella luschani (Steindachner, 1891). Zool Middle East. 2001; 22: 67–82. [Google Scholar]
- 4.Johannesen J, Johannesen B, Baran I, Rizvan-Tunc M, Kiefer A, Veith M. Distortion of symmetrical introgression in a hybrid zone: evidence for locus-specific selection and uni-directional range expansion. J Evol Biol. 2006; 19: 705–716. 10.1111/j.1420-9101.2005.01064.x [DOI] [PubMed] [Google Scholar]
- 5.Ehl S, Vences M, Veith M. Reconstructing evolution at the community level: a case study on Mediterranean amphibians. Mol Phyl Evol. Re-submitted. [DOI] [PubMed] [Google Scholar]
- 6.Veith M, Göçmen B, Sotiropoulo K, Kieren S, Godmann O, Steinfartz S. Seven at one blow: The origin of major lineages of the viviparous Lycian salamanders (Lyciasalamandra) was triggered by a single paleo-historic event. Amph-Rept. 2016; 37: 373–387. [Google Scholar]
- 7.Weisrock DW, Macey JR, Ugurtas IH, Larson A, Papenfuss TJ. Molecular phylogenetics and historical biogeography among Salamandrids of the ‘‘true” salamander clade: rapid branching of numerous highly divergent lineages in Mertensiella luschani associated with the rise of Anatolia. Mol Phyl Evol. 2001; 18: 434–448. [DOI] [PubMed] [Google Scholar]
- 8.Veith M, Lipscher E, Öz M, Kiefer A, Baran I, Polymeni RM. et al. Cracking the nut: Geographical adjacency of sister taxa supports vicariance in a polytomic salamander clade in the absence of node support. Mol Phyl Evol. 2008; 47: 916–931 [DOI] [PubMed] [Google Scholar]
- 9.Akman B, Godmann O. A new subspecies of Lyciasalamandra antalyana (Amphibia: Salamandridae) from the Lycian Coast, Turkey. Salamandra 2014; 50: 125–132. [Google Scholar]
- 10.Göçmen B, Arikan H, Yalçinkaya D. A new Lycian salamander, threatened with extinction, from the Göynük Canyon (Antalya, Anatolia), Lyciasalamandra irfani n. sp. (Urodela: Salamandridae). NW J Zool. 2011; 7: 151–160. [Google Scholar]
- 11.Göçmen B, Akman B, Lyciasalamandra arikani n. sp. & L. yehudahi n. sp. (Amphibia: Salamandridae), two new Lycian salamanders from Southwestern Anatolia. NW J Zool. 2012; 8: 181–194. [Google Scholar]
- 12.Üzüm N, Avci A, Bozkurt E, Olgun K. A new subspecies of Lyciasalamandra flavimembris (Urodela: Salamandridae) from Muğla, southwestern Turkey. Turk J Zool. 2015; 39: 328–334. [Google Scholar]
- 13.Yildiz MZ, Akman B. A new subspecies of Atif’s Lycian Salamander, Lyciasalamandra atifi (Başoğlu, 1967), from Alanya (Antalya, Turkey). Herpetozoa. 2015; 8: 3–13. [Google Scholar]
- 14.Oguz MA, Göcmen B, Yalcinkaya D. Comparison of Lyciasalamandra atifi (BAŞOĞLU, 1967) (Urodela: Salamandridae) populations with description of three new subspecies from Antalya province. SW J Hortic Biol Environ. 2016; 7: 61–113. [Google Scholar]
- 15.Tok CV, Afsar M, Yakın BY. A new subspecies, Lyciasalamandra atifi oezi n. ssp. (Urodela: Salamandridae) from Gazipaşa (Antalya, Turkey). Ecol Montenegrina. 2016; 9: 38–45. [Google Scholar]
- 16.Göçmen B, Ehl S, Karış M, Thiesmeier B, Kordges T. Molecular and morphological investigations on the Fazıla’s Lycian salamander Lyciasalamandra fazilae (Amphibia: Salamandridae) populations and description of a new subspecies. Zool Middle East. 2018; 64: 304–314. [Google Scholar]
- 17.Hays JD, Imbrie J, Shackleton NJ. Variation in the Earth’s orbit: pacemaker of the ice age. Science. 1976; 194: 1121–1132. 10.1126/science.194.4270.1121 [DOI] [PubMed] [Google Scholar]
- 18.Andersen BG, Borns HW. The ice age world Oslo: Scandinavia University Press; 1994. [Google Scholar]
- 19.Blondel J, Aronson J, Bodiou J-Y, Boeuf G. The Mediterranean region: Biological diversity in space and time 2nd edition Oxford/ New York: Oxford University Press; 2010. [Google Scholar]
- 20.Lymberakis P, Poulakakis N. Three continents claiming an archipelago: The evolution of Aegean’s herpetofaunal diversity. Diversity. 2010; 2: 233–255. [Google Scholar]
- 21.Vanderplank SE, Moreira-Muñoz A, Hobohm C, Pils G, Noroozi J, Clark VR et al. Endemism in Mainland Regions–Case Studies In: Hobohm C, editor. Endemism in Vascular Plants. Dordrecht: Springer; 2014. pp. 205–308. [Google Scholar]
- 22.Wilson RCL, Drury SA, Chapman JL. The Great Ice Age. 1st ed. London, New York: The Open University; 1999. [Google Scholar]
- 23.Veith M, Schmidtler FJ, Kosuch J, Baran I, Seitz A. Paleoclimatic changes explain Anatolian mountain frogs: evolution: a test for alternating vicariance and dispersal events. Mol Ecol. 2003; 12: 185–199. 10.1046/j.1365-294x.2003.01714.x [DOI] [PubMed] [Google Scholar]
- 24.Atalay I, Efe R, Öztürk M. Effects of topography and climate on the ecology of Taurus Mountains in the Mediterranean region of Turkey. Procedia–Soc Behav Sci. 2014; 120: 142–156. [Google Scholar]
- 25.Coope GR. The response of insect faunas to glacial-interglacial climatic fluctuations. Phil Trans R Soc London B. 1994; 344: 19–26. [Google Scholar]
- 26.Rödder D, Lötters S, Öz M, Bogaerts S, Eleftherakos K, Veith M. A novel method to calculate climatic niche similarity among species with restricted ranges–the case of terrestrial Lycian salamanders. Org Div Evol. 2011; 11: 409–423. [Google Scholar]
- 27.Sinsch U, Böcking H, Leskovar C, Öz M, Veith M. Demography and lifetime growth patterns in Lyciasalamandra spp.: Living underground attenuates among-species variation. Zool Anz. 2017; 269: 48–56. [Google Scholar]
- 28.Palumbi S, Martin A, Romano S, McMillan OW, Stice L, Grabowski G. The Simple Fool’s Guide to PCR. Version 2. Honolulu:,HI: Department of Zoology and Kewalo Marine Laboratory, University of Hawai; 2002. [Google Scholar]
- 29.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013; 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger MP, Munson PJ. A novel randomized iterative strategy for aligning multiple protein sequences. Bioinformatics 1991; 7: 479–484. [DOI] [PubMed] [Google Scholar]
- 31.Gotoh O. Optimal alignment between groups of sequences and its application to multiple sequence alignment. Computer applications in the biosciences. CABIOS. 1993; 9: 361–370. 10.1093/bioinformatics/9.3.361 [DOI] [PubMed] [Google Scholar]
- 32.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970; 48 443–453. 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- 33.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000; 17: 564–577. [DOI] [PubMed] [Google Scholar]
- 34.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004; 20: 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 35.R Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria; 2016. [Google Scholar]
- 36.Villesen P. FaBox: an online toolbox for fasta sequences. Mol Ecol Notes. 2007; 7: 965–968. [Google Scholar]
- 37.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2017. 34: 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- 38.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE. et al. DnaSP v.6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol. 2017; 34: 3299–3302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 39.Rogers AR, Harpending HC. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992; 9:552–69. 10.1093/oxfordjournals.molbev.a040727 [DOI] [PubMed] [Google Scholar]
- 40.Tajima F. Statistical method for testing the neutral mutation hypothesis. Genetics. 1989; 123: 585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997; 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002; 19: 2092–2100. 10.1093/oxfordjournals.molbev.a004034 [DOI] [PubMed] [Google Scholar]
- 43.Harpending HC, Sherry ST, Rogers AR, Stoneking M. Genetic structure of ancient human populations. Current Anthr. 1993; 34: 483–496. [Google Scholar]
- 44.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005; 22: 1185–1192. 10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- 45.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PloS Comput Biol. 2014; 10: e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouckaert RR, Drummond AJ. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol. 2017; 17: 42 10.1186/s12862-017-0890-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pons O, Petit RJ. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996; 144: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Excoffier L, Smousse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992; 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Excoffier L, Laval G, Schneider S. Arelquin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005; 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 50.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984; 38: 1358–1370. 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- 51.Guo SW, Thompson EA. Performing the exact test of Hardy–Weinberg proportions for multiple alleles. Biometrics. 1992; 48: 361–371. [PubMed] [Google Scholar]
- 52.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 53.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012, 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wertheim JO, Sanderson MJ, Worobey M, Bjork A. Relaxed molecular clocks, the bias–variance trade-off, and the quality of phylogenetic inference. Syst Biol. 2010, 59: 1–8. 10.1093/sysbio/syp072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000; 9: 1657–1659. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- 57.Franklin J. Mapping species distributions. Spatial inference and prediction Cambridge/New York: Cambridge University Press; 2010. [Google Scholar]
- 58.Wisz MS, Hijmans RJ, Li J, Peterson AT, Graham CH, Guisan A. Effects of sample size on the performance of species distribution models. Div Dist. 2008; 2008: 14: 763–773. [Google Scholar]
- 59.Proosdij ASJ van Sosef MSM, Wieringa JJ, Raes N. Minimum required number of specimen records to develop accurate species distribution models. Ecography. 2016; 38: 542–552. 10.1002/hed.23929 [DOI] [Google Scholar]
- 60.Karger DN, Conrad O, Böhner J, Kawohl T, Kreft H, Soria-Auza RW, et al. Climatologies at high resolution for the earth’s land surface areas. Sci Data. 2017; 4: 170122 10.1038/sdata.2017.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Booth TH, Nix HA, Busby JR, Hutchinson MF, Franklin J. bioclim. The first species distribution modelling package, its early applications and relevance to most current MaxEnt studies. Div Dist. 2014; 20: 1–9. [Google Scholar]
- 62.Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, et al. Collinearity. A review of methods to deal with it and a simulation study evaluating their performance. Ecography, 2013, 36: 27–46. [Google Scholar]
- 63.Klewen R. Die Landsalamander Europas. 2nd edition Wittenberg Lutherstadt: Die Neue Brehm-Bücherei; 584 1991. [Google Scholar]
- 64.Varela S, Lima-Ribeiro KS, Terribile LC. A short guide to the climatic variables of the Last Glacial Maximum for biogeographers. PLoS ONE. 2015; 10(6): e0129037 10.1371/journal.pone.0129037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips SJ, Anderson RP,Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006; 190: 231–259. [Google Scholar]
- 66.Phillips SJ, Anderson RP, Dudík M, Schapire RE, Blair ME. Opening the black box: an open‐source release of Maxent. Ecography. 2017; 40: 887–893. [Google Scholar]
- 67.Yackulic CB, Chandler R, Zipkin EF, Royle J, Nichols JD, Campbell EH et al. Presence-only modelling using Maxent: when can we trust the inferences? Methods Ecol Evol. 2013; 4: 236–243. [Google Scholar]
- 68.Merow C, Smith MJ, Silander JA. A practical guide to MaxEnt for modeling species’ distributions. What it does, and why inputs and settings matter. Ecography. 2013; 36: 1058–1069. [Google Scholar]
- 69.Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006; 29: 129–151. [Google Scholar]
- 70.Phillips SJ, Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008; 31: 161–175. [Google Scholar]
- 71.Elith J, Kearney M, Phillips S. The art of modelling range-shifting species. Meth Ecol Evol. 2010; 1: 330–342. [Google Scholar]
- 72.Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ. A statistical explanation of MaxEnt for ecologists. Div Dist. 2011; 17: 43–57. [Google Scholar]
- 73.Warton D, Aarts G. Advancing our thinking in presence-only and used-available analysis. J Anim Ecol. 2013; 82: 1125–1134. 10.1111/1365-2656.12071 [DOI] [PubMed] [Google Scholar]
- 74.Shcheglovitova M, Anderson RP. Estimating optimal complexity for ecological niche models: A jackknife approach for species with small sample sizes. Ecol Model. 2013; 269: 9–17. [Google Scholar]
- 75.Swets J. Measuring the accuracy of diagnostic systems. Science. 1988; 240: 1285–1293. 10.1126/science.3287615 [DOI] [PubMed] [Google Scholar]
- 76.Allouche O, Tsoar A, Kadmon R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J Appl Ecol. 2006; 43: 1223–1232. [Google Scholar]
- 77.Eleftherakos K, Sotiropoulos K, Polymeni RM. Conservation units in the insular endemic salamander Lyciasalamandra helverseni (Urodela, Salamandridae). Ann Zool Fenn. 2007; 44:387–399. [Google Scholar]
- 78.Avise JC, Arnoldt J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987; 18:489–522. [Google Scholar]
- 79.Vences M, Hauswaldt JS, Steinfartz S, Rupp O, Goesmann A, Künzel S et al. Radically different phylogeographies and patterns of genetic variation in two European brown frogs, genus Rana. Mol Phyl Evol. 2013; 68: 657–670. [DOI] [PubMed] [Google Scholar]
- 80.Vences M, Sarasola-Puente V, Sanchez E, Amat F, Hauswaldt JS. Diversity and distribution of deep mitochondrial lineages of the common frog, Rana temporaria, in northern Spain. Salamandra. 2017; 53: 25–33. [Google Scholar]
- 81.Teixeira J, Goncalves H, Ferrand N, García-París M, Recuero E. Mitochondrial phylogeography of the Iberian endemic frog Rana iberica, with implications for its conservation. Current Zool. 2018; 64: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dufresnes CM, Litvinchuk S, Leuenberger J, Ghali K, Zinenko O, Stöck M, et al. Evolutionary melting pots: A biodiversity hotspot shaped by ring diversifications around the Black Sea in the Eastern tree frog (Hyla orientalis). Mol Ecol. 2016; 25: 4285–4300. 10.1111/mec.13706 [DOI] [PubMed] [Google Scholar]
- 83.Martinez-Solana I, Teixeira J, Buckley D., García-Paris M. Mitochondrial DNA phylogeography of Lissotriton boscai (Caudata, Salamandridae): Evidence for old, multiple refugia in an Iberian endemic. Mol Ecol. 2006; 15: 3375–3388. 10.1111/j.1365-294X.2006.03013.x [DOI] [PubMed] [Google Scholar]
- 84.Recuero E, García-París M. Evolutionary history of Lissotriton helveticus, multilocus assessment of ancestral vs. recent colonization of the Iberian Peninsula. Mol Phyl Evol. 2011; 60: 170–182. [DOI] [PubMed] [Google Scholar]
- 85.Dinis M, Merabet K, Martínez-Freiría F, Steinfartz S, Vences M, Burgon JD, et al. Allopatric diversification and melting pot in a North African Palearctic relict: the biogeographic history of Salamandra algira. Mol Phyl Ecol. 2019; 130: 81–91. [DOI] [PubMed] [Google Scholar]
- 86.Taberlet P, Fumagalli L, Wust–Saucy AG, Cosson, JF. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 1998; 7: 453–464. 10.1046/j.1365-294x.1998.00289.x [DOI] [PubMed] [Google Scholar]
- 87.Hewitt GM. The structure of biodiversity—insights from molecular phylogeography. Front Zool. 2004; 1: 1–22. 10.1186/1742-9994-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Avise JC. Phylogeography Cambridge/London: Harvard University Press; 2000. [Google Scholar]
- 89.Rodríguez A, Börner M, Pabijan M, Gehara M, Haddad CFB, Vences M. Genetic divergence in tropical anurans: deeper phylogeographic structure in forest specialists and in topographically complex regions. Evol Ecol. 2015; 29: 765–785. [Google Scholar]
- 90.Crottini A, Brown JL, Mercurio V, Glaw F, Vences M, Andreone F. Phylogeography of the poison frog Mantella viridis (Amphibia: Mantellidae) reveals chromatic and genetic differentiation across ecotones in northern Madagascar. J Zool Syst Evol Res. 2012; 50: 305–314. [Google Scholar]
- 91.Templeton AR, Routman E, Phillips CA. Separating population structure from population history: a cladistic analysis of thegeographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics. 1995; 140: 767–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Müller C. Late Miocene to recent Mediterranean biostratigraphy and paleoenvironments based on calcareous nannoplankton In: Stanley DJ, Wezel F-C, editors. Geological Evolution of the Mediterranean Basin. New York: Springer; 1985. pp. 458–471. [Google Scholar]
- 93.Hewitt G. The genetic legacy of the quaternary ice ages. Nature. 2000; 405: 907–913. 10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- 94.Beheregaray LB. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol Ecol. 2008; 17: 3754–3774. 10.1111/j.1365-294X.2008.03857.x [DOI] [PubMed] [Google Scholar]
- 95.Mazza PPA, Lovari S, Masini F, Masseti M, Rustioni M. A multidisciplinary approach to the analysis of multifactorial land mammal colonization of islands: BioScience. 2013; 63: 939–951. [Google Scholar]
- 96.Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol. 2008; 23: 564–571. 10.1016/j.tree.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 97.Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC et al. Extinction risk from climate change. Nature. 2004; 427: 145–148. 10.1038/nature02121 [DOI] [PubMed] [Google Scholar]
- 98.Skourtanioti E, Kapli P, Ilgaz Ç, Kumlutaş Y, Avcı A, Ahmadzadeh F, Crnobrnja-Isailović J, Gherghel I, Lymberakis P, Poulakakis N. A reinvestigation of phylogeny and divergence times of the Ablepharus kitaibelii species complex (Sauria, Scincidae) based on mtDNA and nuDNA genes. Mol Phyl Evol. 2016; 103: 199–214. [DOI] [PubMed] [Google Scholar]
- 99.Helfer V, Broquet T, Fumagali L. Sex-specific estimates of dispersal show female philopatry and male dispersal in a promiscuous amphibian, the alpine salamander (Salamandra atra). Mol Ecol. 2012; 21: 4706–4720. 10.1111/j.1365-294X.2012.05742.x [DOI] [PubMed] [Google Scholar]
- 100.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004; 13: 729–744. 10.1046/j.1365-294x.2003.02063.x [DOI] [PubMed] [Google Scholar]
- 101.Alho JS, Herczeg G, Söderman F, Laurila A, Jönsson KI, Merilä J. Increasing melanism along a latitudinal gradient in a widespread amphibian: local adaptation, ontogenic or environmental plasticity? BMC Evol Biol. 2010; 10: 317 10.1186/1471-2148-10-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Urban MC, Richardson JL, Freidenfels NA. Plasticity and genetic adaptation mediate amphibian and reptile responses to climate change. Evol Applic. 2014; 7: 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mayr E. Systematics and the origin of species New York: Columbia University Press; 1942. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of observed mismatch distributions (obs) of Lyciasalamandra phyloclades with distributions expected under a growth-decline (exp (G-D)) and a constant growth (exp (cG)) model; X-axis: number of base substitutions between two haplotypes; Y-axis: frequency of the number of base substitutions between two haplotypes.
(JPG)
Bayesian Skyline Plots (BSP) for phyloclades of Lycian salamanders (a-f): a) L. atifi BSPs for phyloclades ati-I, ati-II and ati-III, b) L. antalyana BSPs for phyloclades ant-I and ant-II, c) L. billae BSPs for phyloclades bil-I and bil-III, d) L. luschani BSPs for phyloclades lus-I, lus-IV, lus-V and lus-VI, e) L. fazilae BSPs for phyloclades faz-I, faz-III and faz-IV, f) L. flavimembris BSP for phyloclade fla-I, and g) L. helverseni BSP for phyloclade hel-I. Phyloclades were coloured according to the colours of the haplotype networks in Fig 2.
(TIF)
Outgroups were excluded from the figure. We only represent Bayesian Posterior Probabilities ≥0.95 (see numbers at nodes). Phyloclades of each species were colour marked according to Fig 2 in the main text. Due to the low resolution of the ML tree, we only represent the BI tree.
(TIF)
(TIF)
Mapped Maxent SDMs for all Lyciasalamandra species under current (left) and LGM (right) climatic conditions. Note that under the LGM land masses exceeded those of today due to sea level change. Suitability to species increases with higher ClogLog values, indicated by warmer colours. Known species records are indicated by blue dots (cf. S1 File). For details of modelling and mapping see Materials and Methods.
(TIF)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All sequences are available from the GenBank database, as listed in the Supporting Information files.