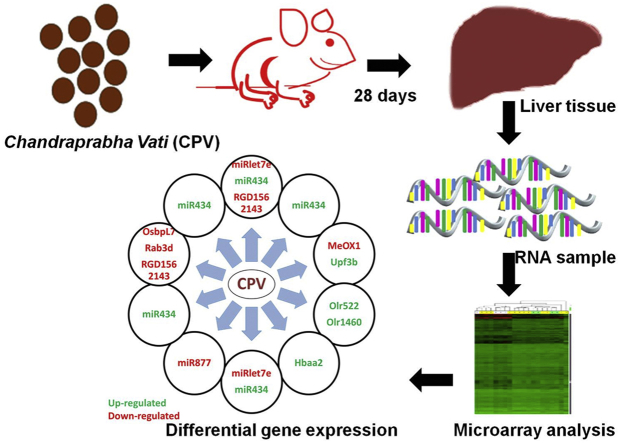
Abstract
Background
Traditional medicinal preparations have not received global acceptance, and their therapeutic benefits remain disputed due to lack of scientific evidence on their mechanism of action. Microarray analysis has emerged as a powerful technique that can aid in understanding the complex signaling networks activated by these formulations and thereby assess their beneficial as well as adverse effects.
Aim
The present work aims to investigate the differential influence of ChandraprabhaVati, Ayurvedic formulation used in the treatment of diabetes, anemia, urinary, respiratory, skin and liver disorders.
Materials and methods
The RNA from the liver of rats treated with different doses of ChandraprabhaVati for 28 days was isolated and studied for the genome-wide changes in the expression.
Results
The results revealed several molecular targets that could contribute to the therapeutic effects of ChandraprabhaVati. Several genes have been differentially expressed, among those miRNAs miR-434, miR877, and miRlet7e contribute to the anti-diabetic, anti-fibrotic and anti-inflammatory of CPV. The rejuvenative activity of CPV may be due to the MeOX1 and Upf3b genes. Up-regulation of Hbaa2 gene facilitates the anti-anemic effect. Interestingly gender-specific differential expressions of genes were also observed. Rab3d were found to be altered in female when compared to male animals.
Conclusion
Thus the microarray data for the CPV treated animals has revealed molecular targets that may be responsible for the various known therapeutic effects and also identified new beneficial effects of CPV.
Keywords: ChandraprabhaVati, Ayurveda, Microarray analysis
Graphical abstract
Abbrevations
- CPV
ChandraprabhaVati
- miRNA
microRNAs
- ONECUT1
One cut homeobox 1
- PRDM1
PR domain zinc finger protein
- XBP-1
X-box binding protein
- SEPT1
septin
- TAGLN
Transgelin
- MeOX1 gene
mesenchyme homeobox gene 1
- Osbpl7
Oxysterol binding protein like-7
- FXR
Farsenoid-X-receptor
- PXR
pregnane-X-receptor
1. Introduction
Traditional medicinal formulations that are widely used in India are polyherbal and herbo-mineral preparations whose molecular mechanisms of action have not been thoroughly deciphered due to the numerous constituents that are present in the formulations. However, recent evidence points out that the multi-component nature of such formulations can exert multi-target activity and hence may have superior properties when compared with the individual constituents.1 Indeed, many of these formulations have been used to treat more than one type of ailments by traditional medicine practitioners. Controversies on the safety of these formulations persist that have limited their global market. However, with the emergence of new lifestyle diseases and multi-drug resistance, there is renewed interest to explore phytoformulations to treat the modern age maladies. In this context, it becomes essential to confirm the safety of these formulations apart from deciphering their molecular targets. In recent years, microarray analysis has emerged as a powerful tool to understand the molecular targets of an individual molecule or a formulation.2 Also, it can provide insights into possible therapeutic or toxic effects of the drug sample being investigated at a molecular level.3
Microarray analysis has been employed to investigate several traditional Chinese preparations to identify possible molecular mechanisms, discover novel therapeutic action and confirm the safety of the preparations.4 Microarray studies on the breast cancer cells treated with the Chinese traditional medicine preparation Si-Wu-Tang revealed that the formulation functions as a phytoestrogen and influences the genes involved in the Nrf2 (Nuclear factor erythroid 2-Related Factor 2) pathway thereby indicating its potential as a chemopreventive and anti-cancer agent.5 Traditional use of this formulation was primarily restricted to antibacterial and estrogen-related disorders. But microarray analysis had revealed new therapeutic benefits for this formulation. Microarray analysis of DNA from MG-63 osteoblast cells treated with the Chinese herbal preparation DangguiBuxue Tang revealed that the decoction acted upon several unique targets that were not affected by the individual constituents thereby emphasizing the importance of poly-herbal preparations in therapy.6 Based on the microarray analysis, it was concluded that the preparation might also be beneficial for the treatment of osteoporosis in addition to the traditional use. The molecular mechanism of action of an Indo-Tibetan traditional medicinal preparation Padma-28 was deciphered using microanalysis of DNA isolated from hepatoma cells treated with the formulation.7 The formulation was found to alter 19 inter-related signaling networks that contributed to its beneficial effects against oxidative stress. Molecular mechanisms for extracts from a wide variety of plants such as Anemarrhenaasphodeloides, Centellaasiatica, Coptidis rhizome, Tripterygiumhypoglaucum, Ginkgo biloba,etc. have been unraveled using DNA microarray analysis.8 In the context of Indian traditional medicine, an Ayurvedic preparation, Medhyarasayana whose primary herbal ingredient is Clitoriaternatea, was found to act upon pathways influencing axon guidance and memory potentiation apart from autophagy regulation using DNA microarray analysis of animals treated with the formulation thereby revealing the molecular targets contributing to its memory enhancing effects.9
ChandraprabhaVati (CPV), a camphor-basedAyurvedicpreparation comprising 31 herbal and 6 mineral ingredients, has been widely used to treat a diverse range of ailments that include diabetes, urinary tract disorders, urinary calculi, abdominal colic, liver cirrhosis, anorexia, anemia, constipation, digestive disorders, anal fistula, menorrhagia, cancer and skin disorders.10 It is generally administered with water or honey.11Clinical studies on patients treated with CPV did not reveal any significant adverse effects indicating its safe nature.12 Earlier studies have shown that methanolic extract of CPV was found to effectively scavenge superoxide radicals13 and exhibited anti-inflammatory effects in a rat model with paw edema14.In vivo studies have also revealed that CPV exerts an anti-inflammatory action against urinary tract infections in mice.15 Several groups have reported the anti-diabetic activity of CPV.10 Our group had earlier demonstrated using in silico and in vitro studies that CPV exerts its anti-diabetic activity through down-regulation of pro-inflammatory cytokines and activation of the pregnane-X-receptor mediated triggering of the PPAR-γ-GLUT4 pathway.16 However, molecular mechanisms for the diverse range of therapeutic benefits are yet to be deciphered for this formulation. The present work aims to address this lacuna by performing a DNA microarray analysis from the liver samples obtained from animals treated with different doses of CPV for 28 days.
2. Materials & methods
2.1. Materials
CPV tablets (IMPCOPS, Chennai) were procured from a local Ayurvedic medical store at Thanjavur. Honey was procured fromKhadi Co-operative Stores, Thanjavur. Microarray Gene chip - Rat Gene 2.0 ST, wt plus kit, Gene chip Hybridization wash and stain kit was procured from Affymetrix, USA.
2.2. Methods
Wistar rats (Rattusnorvegicus) of either gender aged 6–8 weeks were chosen for the study. Animals were housed individually in autoclaved standard polypropylene rat cages. Sieved and sterilized paddy husk was used as the bedding material. The bedding material,cages, grills and water bottles were changed twice a week. The temperature and relative humidity of the test room was maintained between 22 ± 3 °C and 50–70% during the experimental period. The experimental room was provided with a 12 h light and 12 h dark lighting condition using an automatic timer. Standard rodent pellet feed (Altromin) supplied by M/s. ATNT Laboratories, Mumbai, India, and water were provided to the animals ad libitum. All animal experimentswereconductedat the CentralAnimalFacilityof SASTRA Deemed University after obtaining approval from the Institutional Animal Ethics Committee (IAEC Approval Number: 355/SASTRA/IAEC/RPP).
2.3. Treatment with CPV
The therapeutic dose of CPVin humans is 500 mg for an adult.17 Assuming the average adult weight as 60 kg, the human dose of CPVis 8.33 mg/kg. The animal dose conversion factor was calculated from the following equation:
where the Km refers to conversion factor associated with body surface area. The rat Km and human Km values used from the standard literature were 6 and 37 respectively.18 Using the animal dose conversion factor, the rat dose for CPVwas calculated as 50 mg/kg. This was considered as the therapeutic dose (Low dose) for the rats. Four times (4X) and sixteen times (16X) the therapeutic (Low dose) were assigned as medium and high dose respectively. Oral administration of the drugs was chosen for the study as they are intended for oral use in humans. The test substances were suspended in honey:water(2:3) and were administered daily through oral gavage attached by a disposable syringe (3 mL) for 28 days. The animals were randomly divided into five groups comprising ten animals each with equal numbers of either gender. The observation period of the study was 28 days.
After the study period, the animals were euthanized following standard protocols and the livers of the animals from different groups were collected for extraction of RNA.
2.4. RNA extraction
Liver Tissues collected from the animals were homogenized and total RNA were extracted using RNeasy Mini Kit (QIAGEN, California), following the manufacturer's protocol. The RNA concentrations were measured by Nanodrop (Thermo).The RNA samples were stored at −80 °C before further processing for microarray analysis.
2.5. Microarray analysis
The RNA was converted to single-stranded cDNA with T7 promoter sequence at the 5′ end and then converted to double-stranded cDNA, which acts as a template for in vitro transcription for cRNA synthesis. The sense-strand of cDNA was synthesized by the reverse transcription of cRNA using 2nd- Cycle Primers.Thesynthesizedsingle stranded cDNA was purified and from that 5.5 μg was taken for fragmentation and labelling.Sense-strand cDNA was fragmented by uracil-DNA glycosylase (UDG) and apurinic/apyrimidinic endonuclease 1 (APE 1) and labeled by terminal deoxynucleotidyltransferase (TdT) using the Affymetrix proprietary DNA Labeling Reagent that is covalently linked to biotin. The fragmented and labeled cDNA was added to the hybridization mixture comprising salts, blocking agents, and spike-in controls from the bacterial RNA. This cocktail was then injected into the AffymetrixGeneChip array and hybridized at 45 °C in the hybridization chamber for 16 h. Immediately after hybridization, the GeneChip array was subjected to a several washing and staining steps in the Fluidics Station that involves a biotin binding step by fluorescent molecule and a signal amplification step. The array was then scanned using Affymetrix GeneChip3000 7G scanner, a confocal laser scanner and the fluorescence signal pattern on the array was recorded. The Affymetrix microarray software suite defines the probe cells and computes intensity for each cell, which was analyzed. Transcriptome Analysis Console (TAC) Software was used to identify the differentially expressed genes between groups that are statistically significant.
2.6. qPCR analysis
To validate the microarray results, qPCR were done to validate the similar expression of genes in therapeutic dose CPV treated vs normal animals.One microgram of total RNA was reverse-transcribed using iScriptcDNA Synthesis Kit (BioRad, USA). cDNA was amplified by primer pairs specific for MeOX1 and Rab3d and quantification was done using SYBR green (Bio Rad, USA).The GAPDH gene was used as a normalizing control. The primer sequences of MeOX1, Rab3d and GAPDH are as follows: MeOX1F-GCCCATGAGACGGAGAAGAA, MeOX1R-GAACCACACTTTGACCTGCC; Rab3dF- GTGCGGGCCATAGCAACTTC, Rab3dR- CCATCTTGGTGGGATCTCGG; GAPDHF-TTGTGCAGTGCCAGCCTCGT, GAPDHR-GCCACTGCAAATGGCAGCCC. Relative gene expression (ΔΔct) was calculated and the graph was represented as fold change.
3. Results and discussion
No mortality of the animals in any group was observed during the study period. The animals did not display any abnormality in the feeding pattern or behavior during the study period. Biochemical, hematological and histopathological studies did not reveal any significant toxicity in all groups treated with different doses of CPV (data not shown) confirming the non-toxic nature of the formulation.
Microarray analysis was performed using RNA isolated from the livers of animals treated with different doses of CPV-administered with honey, and the results were compared with the control animals and honey-administered animals. The changes in the gene expressions were analyzed based on the magnitude of variations in expression levels. Expression levels of +2 and higher were considered up-regulated and −2 and lower were considered to be down-regulated.
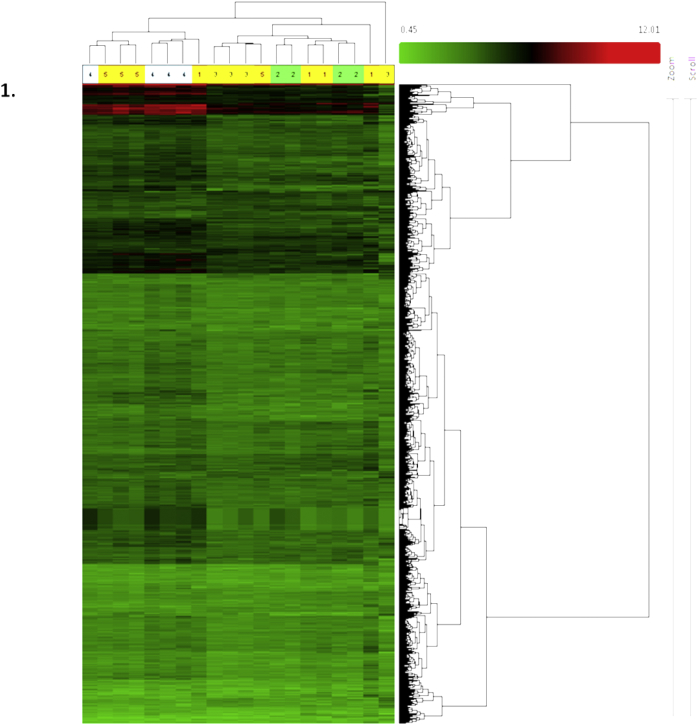
Hierarchical cluster analysis was generated in Transcriptome Analysis Console (TAC) Software and a cluster image map constructed using the microarray data (Fig. 1). The four replicates in each treatment group were evaluated. The colored bar above the heat map (horizontal dimension) indicates the grouping variable: 1-Normal; 2-Honey; 3–50 mg/kg; 4–200 mg/kg; 5–800 mg/kg. The normalized expression value of each gene is colour-coded, the under-expressed genes weregiven green and over expressed genes were given red and genes with no change in expression are represented in black.
Fig. 1.
Hierarchical clustering of gene expression levels in the liver samples of animals from different groups treated with CPV.
Fig. 1 shows the hierarchical clustering of the gene expression levels in the liver samples of control, honey treated, CPV (therapeutic dose), CPV (medium dose) and CPV (high dose) treated animals.
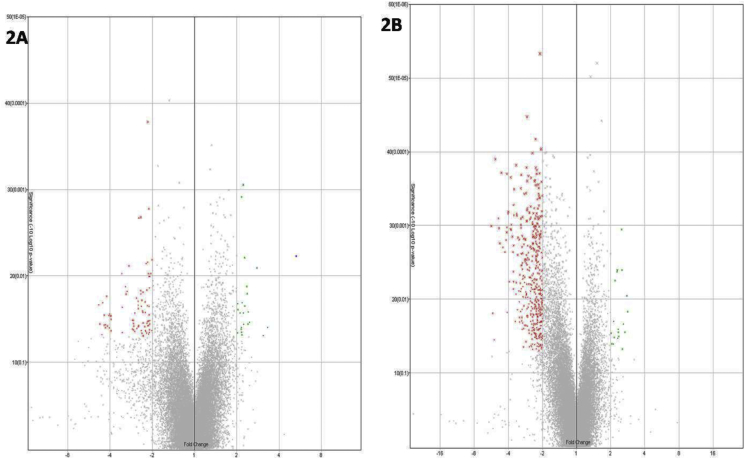
Out of 30,429 genes, a total of 930 genes were found to be differentially expressed. Honey administered groups showed up-regulation of 30 genes, while14 genes were found to be down-regulated when compared to control animals. The therapeutic dose of CPV up-regulated 27 genes while 14 genes were down-regulated when compared to the control group. Treatment with a medium dose of CPV showed an up-regulation of 26 genes while a total of 108 genes were down-regulated when compared with the control group. High dose treatment of CPV, when compared with the control group, shows up-regulation of 52 genes and down-regulation of 17 genes. Comparison with the honey administered groups reveal an up-regulation of 20, 19 and 21 genes in the therapeutic dose, medium dose, and high dose CPV treated samples respectively. Down-regulation of 21, 484 and 116 genes was recorded for a therapeutic dose, medium dose and high dose treated samples respectively when compared with the honey-treated group. When the different CPV treated groups were compared with each other, up-regulation of 18 genes, and down-regulation of 388 genes were observed when therapeutic dose CPV administered animal samples were compared with the medium dose CPV treated group. The comparison of therapeutic dose CPV treated animals with high dose CPV administered animals revealed up-regulation of 7 genes while 36 genes were down-regulated. 14 genes were up-regulated and 5 genes were down-regulated when medium dose CPV administered liver samples were compared with the high dose of CPV treated samples. Fig. 2 shows the volcano plots for control versus a medium dose of CPV treated groups (A) and honey administered versus medium dose of CPV treated samples (B).
Fig. 2.
Volcano plots for control versus medium dose of CPV.
[A] Control versus medium dose of CPV; [B] Honey-administered versus medium dose of CPV.
Comparison of the gene expression levels of animals treated with only honey and medium dose of CPV showed a significant up-regulation of miR-434, Olr522 and Olr1460. miR-434 belongs to a class of microRNAs (miRNA) that regulate the expression levels of other genes.19 miR-434 is a target that is yet to be completely explored. A recent work has found that there are several miRNAsthat are differentially expressed in rat liver. These include miR-434, and it has been suggested that it may play a positive role in regulating the morphology, growth, proliferation, development and signaling pathways of the cells.20 miR-434 has also been found to be down-regulated in diabetic animals with beta cell failure, and hence, it's up-regulation has been suggested to be a good sign to abrogate diabetic symptoms.20 Interestingly, traditional literature, as well as our earlier work on CPV, has also confirmed the anti-diabetic effects of CPV.16 We had reported that CPV activates PXR optimally that results in the up-regulation of Glut-4 and PPAR-gamma that may contribute to its anti-diabetic activity.16 The microarray analysis has suggested that the anti-diabetic effect of CPV may be potentiated by up-regulation of miR-434.
Further, it has been reported that miR-434 may regulate the functions of the genes ONECUT1, PRDM1, CNTN4, SEPT3, TAGLN, XBP1.21 The gene ONECUT1 (One cut homeobox 1) is expressed in liver and has been reported to play a major role in glucose metabolism and cell cycle regulation. It is also found to antagonize the action of glucocorticoid-stimulated gene transcription that leads to gluconeogenesis.22 PRDM1 (PR domain zinc finger protein) gene is associated with the expression of proteins regulating the innate and adaptive immune response.23 Similarly, CNTN4 (Contactin-4) and XBP-1 (X-box binding protein) genes have also been associated with immune response.24 CPV has been reported to exhibit anti-inflammatory effects by earlier groups,25 and it is likely that miR-434 may influence this property of CPV through regulation of the PRDM1 activity. SEPT1 (septin) gene regulated by miR-434, on the other hand has been implicated in the cytokinesis process. TAGLN (Transgelin) is another gene whose dysregulation has been associated with fibrosis26 and liver cirrhosis.27 CPV has been reported in the traditional literature to be useful in the treatment of liver fibrosis and liver complications, and miR-434 may be a key player in the therapeutic action of CPV for these disorders. Another recent work has indicated the up-regulation of miR-434 expression in skeletal muscles for the restoration of function in animals with spinal cord injury19 thereby opening up new therapeutic vistas for this molecular target and CPV in the treatment of other disorders.
Olr522 and Olr1460 genes encode for olfactory receptors, which are G-protein coupled receptors. These receptors have also been found to be expressed in non-olfactory tissues and have been implicated in chemosensitive response leading to activation of signaling pathways.28 Several reports have implicated the role of these olfactory receptors in tissue regeneration, repair and angiogenesis.29 Therefore, the microarray analysis has indicated new targets that may help decipher newer molecular targets for the multi-component formulation.
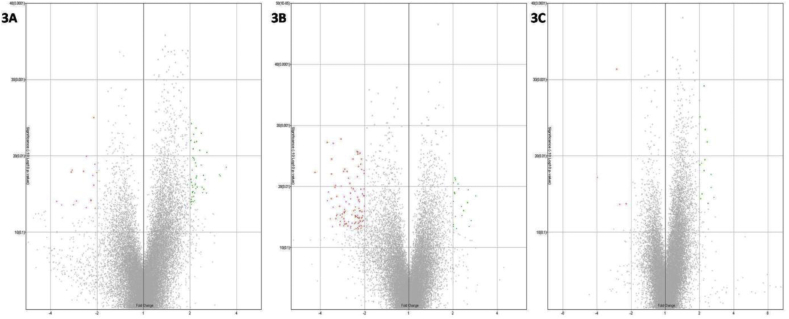
The analysis of the genes differentially regulated in the livers of animals administered with the therapeutic dose of CPV when compared with livers from normal animals reveals that the mi-RNA miR877 was down-regulated in the CPV treated animals. It has been recently identified that down-regulation of miR877 plays an important role in inhibiting the development of pulmonary fibrosis and enhancing the levels of the inhibitory protein Smad7.30 This target may be explored further as a possible molecular target that may be responsible for the ability of CPV to treat respiratory disorders. Further, CPV treatment was also found to up-regulate the Upf3b gene, an important target that is involved in the formation of post-splicing multi-protein complex and facilitating the nuclear export of mRNA.31 It also aids in promoting non-sense mediated RNA degradation (NMD) pathway to reduce the expression of aberrant mRNA.31 Another interesting gene target that has been found to be down-regulated in the CPV treated animals (therapeutic dose) is the MeOX1 gene (mesenchyme homeobox gene 1) that has been recently discovered to induce cell cycle arrest and endothelial cell senescence by activating p21 and p16.32 As CPV treatment has been found to reduce this gene, it may lead to improved cell survival and exhibit rejuvenative effects that have been traditionally reported for CPV. The therapeutic dose equivalent of CPV also was found to reduce the expression levels of the gene Osbpl7 (Oxysterol binding proteinlike-7). This gene has been implicated in the bile acid synthesis, and its levels have been reported to be up-regulated in several biliary complications including cholangiocarcinoma.33 The levels of Osbp proteins have also been suggested to be regulated by the Farsenoid-X-receptor (FXR) based pathways.33 Our earlier work on CPV has demonstrated that it activates the pregnane-X-receptor (PXR) optimally that may have implications on its therapeutic effects. The microarray data suggests that further exploring these targets can lead to more an understanding of the underlying pathways and mechanisms regulated by CPV, especially for its action against liver disorders.Fig. 3 shows the volcano plots for normal versus high dose of CPV treated group (A), honey versus high dose of CPV treated samples (B), and medium dose versus high dose of CPV treated groups (C).
Fig. 3.
Volcano plots for
[A] normal versus high dose of CPV treated group, [B] honey versus high dose of CPV treated samples, [C] medium dose versus high dose of CPV treated groups.
It is found on comparison of the medium dose of CPV treated sample with the high dose CPV treated sample that the gene miRlet7e was down-regulated in both groups with greater down-regulation in the higher dose when compared with the medium dose. High expression levels of miRlet7e have been reported to be associated with negative regulation of the anti-inflammatory cytokine IL-10 and have been implicated in Hashimoto's disease, an autoimmune disorder.34 In yet another report, miRlet7e has been found to be up-regulated in samples of liver fibrosis.35 Interestingly, CPV has been reported for treatment of liver disorders. Therefore, microarray analysis results reveal that the ability of CPV to down-regulate miRlet7e may be responsible for the therapeutic effect of CPV against liver disorders.
Comparison of the expression data of honey treated samples with high dose CPV treated samples shows up-regulation of the Hbaa2 gene that encodes for the hemoglobin alpha a2 chain. One of the ingredients of CPV is Lauhabhasma, an iron-based Ayurvedic preparation. Our earlier work with Lauhabhasma has demonstrated its ability to treat anemia (Krishnamacharyet al., unpublished data). The micro-array data suggests that CPV may have a direct role in improving hemoglobin levels through up-regulation of associated genes such as Hbaa2. Another interesting target that has emerged from the microarray analysis is the down-regulation of RGD1562143 gene in livers of groups treated with high dose of CPV. Though very less information is present in the literature on the role of this gene, its action has been cited to be similar to the Ctps gene that encodes for CTP synthase. The CTP synthase has been associated with enhanced lymphocyte proliferation and activation leading to inflammation and tumorigenesis.36 Therefore, down-regulation of this gene and similar targets may also contribute to the anti-inflammatory and anti-cancer properties of CPV.
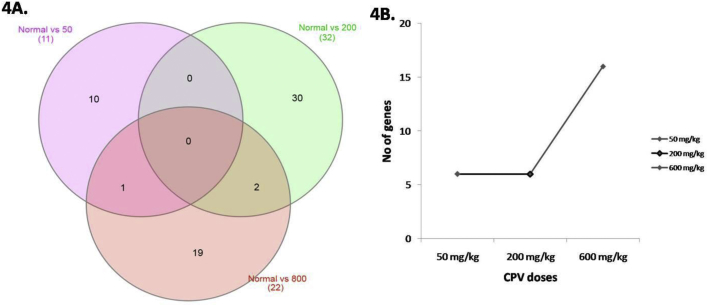
Fig. 4Ashows the Venn diagram of the various treated groups and the overlapping genes for different groups have also been illustrated.Cytochrome P450, family 3, subfamily ‘a'was found to be down-regulated in both therapeutic and high dose CPV treated groups. Many herbal supplements can activate pregnane X receptor (PXR) leading to an increased expression of CYP450 enzymes and can affect the metabolism of other xenobiotics. The induction of CYP450 enzymes may result in hepatotoxicty by increasing the metabolism of less toxic parent compounds to much more toxic daughter compounds.37Another gene that has been down-regulated in medium and higher dose CPV treated animals is the miRlet7e whose down-regulation can be beneficial in the treatment of liver fibrosis.With increase in dose of CPV number of down regulated genes were also increased (Fig. 4B). Similar observations were earlier reported for other phytoformulations.38 However, in case of multi-component formulations, it is difficult to identify dose-dependency in differential expression of a specific gene. This is because the multiple components in the formulation may exert different effects on multiple targets and hence exhibit a narrow concentration range where their therapeutic efficiency is observed, and therefore, deviations from this range can result in reversal or loss of this activity.16
Fig. 4.
; Differential gene expression of therapeutic, medium and high dose treated groups. A. Venn diagram for treated groups:50 mg/kg; 200 mg/kg; 800 mg/kg; B. Number of genes down regulated.
Differences due to gender were also observed in the expression profiles of genes compared between male and female animals. The Rab3d oncogene was found to be down-regulated in female animals administered with the therapeutic dose of CPV when compared to control animals and animals treated with medium and high doses of CPV. The Rab3d gene that encodes for a small Rho-GTPase protein has been reported to be involved in exocytosis and also influences the secretion of amylase and other enzymes from the granules in secretory cells.39 Interestingly, this gene was not altered in male animals treated with CPV indicating strong gender-based differences in response to CPV treatment. A scan of literature reveals that concerted efforts have been directed to understand the gender-based differences to therapeutic regimens but the progress has been minimal. In the context of the secretory cells, it has been identified that females produce an insulin-like small 6 kDa protein relaxin that reduces the levels of enzymes such as amylase and lipase secreted from cells.40 Our data suggest that alteration in the expression levels of Rab3d gene in female animals may be mediated through such gender-specific proteins like relaxin. In the context of therapy, over-expression of Rab3d, an oncogene has been implicated as a possible cause of tumorigenesis41 and therefore, down-regulation of this gene indicates a positive role for CPV in reducing the risk of cancer.
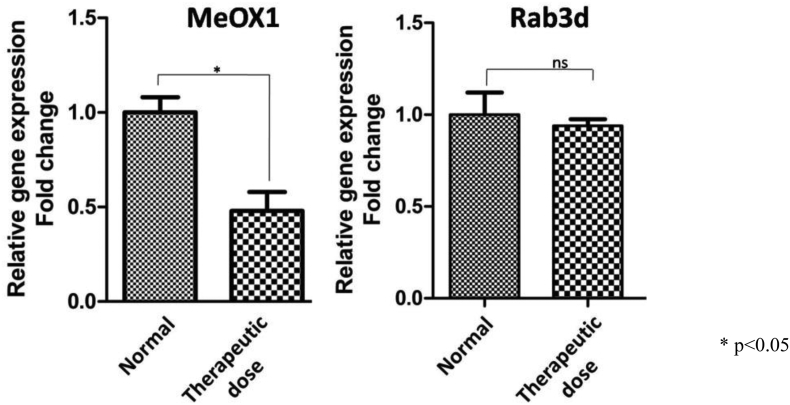
The validation of two genes MeOX1 and Rab3d that were found altered in the therapeutic group with respect to normal group using microarray analysis was performed using PCR (Fig. 5). The MeOX1 gene that is involved in cell cycle arrest was found to be down-regulated in the therapeutic dose treated group when compared to that of normal concurring with the microarray data. Thus it may be concluded that CPV treatment increases cell survival. The Rab3d which is involved in tumorigenesis was slightly down-regulated in the therapeutic dose treated female animals when compared to normal animals.
Fig. 5.
Gene expression analysis using RT-PCR. The relative gene expressions was depicted in fold change.
*p < 0.05.
No KEGG pathways were significantly enriched with the differentially expressed genes in the 50 mg/kg treated group and only 3 relative pathways were enriched from 200 mg/kg CPV treated group. 4 pathways were found to be involved in high dose CPV treated group.The pathways derived from DAVID (KEGG pathways) with p valuesare listed in Table 2.
Table 1.
Group assignment for sub-acute toxicity studyofChandraprabhavati
| Group No. | No. ofAnimals | Description |
|---|---|---|
| G1 | 10 (5 male +5 Female) | Control |
| G2 | 10 (5 male +5 Female) | Vehicle control (2:3 Honey and water) |
| G3 | 10 (5 male +5 Female) | Low dose of CPV (50 mg/kg body wt.) |
| G4 | 10 (5 male +5 Female) | Medium dose of CPV (200 mg/kg body wt.) |
| G5 | 10 (5 male +5 Female) | High dose of CPV (800 mg/kg body wt.) |
Table 2.
Pathway analysis for the CPV treated groups.
| Signaling pathways | p-value |
| Normal vs 50 mg/kg | |
| Olfactory transduction | 2.9E−1 |
| Normal vs 200 mg/kg | |
| Olfactory transduction | 4.8 E-7 |
| MAPK signaling pathway | 1. E0 |
| Metabolic pathways | 1. E0 |
| Normal vs 800 mg/kg | |
| Bile secretion | 6.2E−2 |
| Metabolic pathways | 3.3 E-1 |
| ABC transporters | 1.0 E0 |
| PPAR signaling pathway | 1.0 E0 |
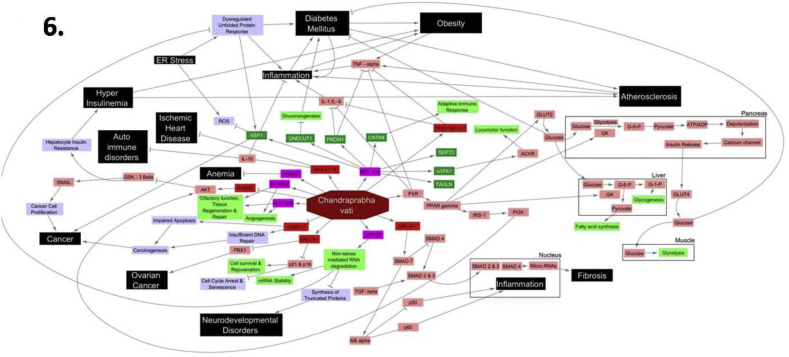
The key pathways altered by CPV have been depicted in the Fig. 6. The microarray data also reveals that the treatment with CPV does not alter the expression levels of many genes especially those that contribute to the toxic effects. This indicates that administration of CPV even at sixteen times the therapeutic dose does not cause any toxic manifestation even in the gene level. This is in line with our safety studies carried out in rats where no significant adverse effects were observed in the hematological and biochemical investigations.
Fig. 6.
Pathways altered by ChandraprabhaVati (CPV).
4. Concluding remarks
The present work is the first report of microarray analysis of animals administered with the Ayurvedic formulation ChandraprabhaVati (CPV)at different doses ranging from the therapeutic dose to sixteen times the therapeutic dose. No adverse effects were discernible in the animals both at a macroscopic scale as well as the genetic level indicating the safety of this formulation. The microarray data analysis has revealed several molecular targets that could be responsible for the reported therapeutic effects of CPV. These include the regulation of the miRNAsmiR-434, miR877, andmiRlet7e that may play a major role in the anti-diabetic, anti-fibrotic and anti-inflammatory activities respectively of CPV. The gene targets MeOX1 and Upf3b may contribute to the rejuvenative properties of CPV while the Osbpl7can be a major player in the therapeutic effect of CPV against hepatic disorders. The analysis also reveals that CPV up-regulates the expression of Hbaa2 gene that presents as a molecular target for the anti-anemic effect of CPV. Interestingly, the microarray data also revealed a gender-specific regulation of the genes with the female animals showing a marked alteration in Rab3d gene when compared male animals, which may have implications in anti-tumor properties for the formulation. It is evident from the microarray data that CPV regulates several molecular targets that have implications in the known therapeutic action and also some new beneficial effects for this formulation that can be explored in future.
Acknowledgement
The authors acknowledge the financial support from the Department of AYUSH (Sanction No. Z.15015/1/2010-COE (AYUSH)) and SASTRA Deemed University for financial and infrastructural support.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtcme.2019.08.001.
Author disclosure statement
No competing financial interests exist.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Tarkang P.A., Appiah-Opong R., Ofori M.F., Ayong L.S., Nyarko A.K. Application of multi-target phytotherapeutic concept in malaria drug discovery: a systems biology approach in biomarker identification. Biomark Res. 2016;4(1):25. doi: 10.1186/s40364-016-0077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C.-C., Lo H.-Y., Hsiang C.-Y., Ho T.-Y. DNA microarray analysis as a tool to investigate the therapeutic mechanisms and drug development of Chinese medicinal herbs. Biomedicine. 2012;2(1):10–16. [Google Scholar]
- 3.Waring J.F., Gum R., Morfitt D. Identifying toxic mechanisms using DNA microarrays: evidence that an experimental inhibitor of cell adhesion molecule expression signals through the aryl hydrocarbon nuclear receptor. Toxicology. 2002;181–182:537–550. doi: 10.1016/S0300-483X(02)00477-8. [DOI] [PubMed] [Google Scholar]
- 4.KiyamaR DNA microarray-based screening and characterization of traditional Chinese medicine. Microarrays. 2017;6(1):4. [Google Scholar]
- 5.Wen Z., Wang Z., Wang S. Discovery of molecular mechanisms of traditional Chinese medicinal formula Si-Wu-Tang using gene expression Microarray and Connectivity Map. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0018278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi R.C.Y., Gao Q.T., Cheung A.W.H. A Chinese herbal decoction, danggui buxue Tang, stimulates proliferation, differentiation and gene expression of cultured osteosarcoma cells: genomic approach to reveal specific gene activation. Evidence-Based Complement Altern Med. 2011;2011:1–13. doi: 10.1093/ecam/nen085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein A., Wrulich O.A., Jenny M. Pathway-focused bioassays and transcriptome analysis contribute to a better activity monitoring of complex herbal remedies. BMC Genomics. 2013;14(1):1–14. doi: 10.1186/1471-2164-14-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavan P., Joshi K., Patwardhan B. DNA microarrays in herbal drug research. Evid Based Complement Altern Med. 2006;3(4):447–457. doi: 10.1093/ecam/nel075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghu K.S., Shamprasad B.R., Kabekkodu S.P. Age dependent neuroprotective effects of medhya rasayana prepared from Clitoria ternatea Linn. in stress induced rat brain. J Ethnopharmacol. 2017;197:173–183. doi: 10.1016/j.jep.2016.07.068. [DOI] [PubMed] [Google Scholar]
- 10.Wanjari M.M., Mishra S., Dey Y.N., Sharma D., Gaidhani S.N., Jadhav A.D. Antidiabetic activity of Chandraprabha vati - a classical Ayurvedic formulation. J Ayurveda Integr Med. 2016;7(3):144—150. doi: 10.1016/j.jaim.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamadagni P.S., Jamadagni S.B., Singh A. Toxicity study of swarna bhasma, an ayurvedic medicine containing gold, in wistar rats. Toxicol Int (Formerly Indian J Toxicol. 2015;22(3):11. [Google Scholar]
- 12.Tripathi A.K., Pandey S.D., Sastry J.L.N., Vedula S. Observations on clinical safety of chandraprabha. Vati- An Ayurvedic Metallo-Herbo Mineral Formulation. 2016;1:17–23. [Google Scholar]
- 13.Bagul M.S., Kanaki N.S., Rajani M. Evaluation of free radical scavenging properties of two classical polyherbal formulations. Indian J Exp Biol. 2005;43(8):732–736. [PubMed] [Google Scholar]
- 14.Rajani M., Bagul M., Srinivasa H., Kanaki N. Antiinflammatory activity of two Ayurvedic formulations containing guggul. Indian J Pharmacol. 2005;37(6):399. [Google Scholar]
- 15.Christa S.S., Swetha A., Christina E., Ganesh R.N., Viswanathan P. Modulatory effect of chandraprabha vati on antimicrobial peptides and inflammatory markers in kidneys of mice with urinary tract infection. Iran J Kidney Dis. 2013;7(5):390–398. [PubMed] [Google Scholar]
- 16.Selvaraj S., Ramanathan R., Vasudevaraja V. vol. 4. 2014. (Transcriptional Regulation of the Pregnane-X Receptor by the Ayurvedic Formulation Chandraprabha Vati). [Google Scholar]
- 17.Ministry for Health and Family Welfare, Ministry fo Health and Family Welfare, Ministry for Health and Family Welfare The ayurvedic pharmacopoeia of India. The Ayurvedic Pharmacopoeia of India. 2010;2:171. [Google Scholar]
- 18.Reagan-Shaw S., Nihal M., Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2007;22(3):659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 19.Shang F.F., Xia Q.J., Liu W. MiR-434-3p and DNA hypomethylation co-regulate eIF5A1 to increase AChRs and to improve plasticity in SCT rat skeletal muscle. Sci Rep. 2016;6 doi: 10.1038/srep22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delic D., Eisele C., Schmid R., Luippold G., Mayoux E., Grempler R. Characterization of micro-RNA changes during the progression of type 2 diabetes in Zucker diabetic fatty rats. Int J Mol Sci. 2016;17(5):1–16. doi: 10.3390/ijms17050665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwekel J.C., Vijay V., Han T., Moland C.L., Desai V.G., Fuscoe J.C. Sex and age differences in the expression of liver microRNAs during the life span of F344 rats. Biol Sex Differ. 2017;8(1):1–21. doi: 10.1186/s13293-017-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K., Holterman A.X. Pathophysiologic role of hepatocyte nuclear factor 6. Cell Signal. 2012;24(1):9–16. doi: 10.1016/j.cellsig.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Smith M.A., Maurin M., Cho H Il. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J Immunol. 2010;185(10) doi: 10.4049/jimmunol.1001682. 10.4049/jimmunol.1001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olivera A., Kitamura Y., Wright L.D. Sphingosine-1-phosphate can promote mast cell hyper-reactivity through regulation of contactin-4 expression. J Leukoc Biol. 2013;94(5):1013–1024. doi: 10.1189/jlb.0313163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumbre R.K. Effect of Chandraprabha Vati in Experimental Prostatic Hyperplasia and Inflammation in Rats. 2012;5(430):5302–5304. [Google Scholar]
- 26.Yu H., Königshoff M., Jayachandran A. Transgelin is a direct target of TGF-β/Smad3-dependent epithelial cell migration in lung fibrosis. FASEB J. 2008;22(6):1778–1789. doi: 10.1096/fj.07-083857. [DOI] [PubMed] [Google Scholar]
- 27.Mölleken C., Sitek B., Henkel C. Detection of novel biomarkers of liver cirrhosis by proteomic analysis. Hepatology. 2009;49(4):1257–1266. doi: 10.1002/hep.22764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrer I., Garcia-Esparcia P., Carmona M. Olfactory receptors in non-chemosensory organs: the nervous system in health and disease. Front Aging Neurosci. 2016;8(JUN) doi: 10.3389/fnagi.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abaffy T. Human olfactory receptors expression and their role in non-olfactory tissues – a mini-review. J Pharmacogenomics Pharmacoproteomics. 2015;06(04) [Google Scholar]
- 30.Wang C., Gu S., Cao H. MiR-877-3p targets Smad7 and is associated with myofibroblast differentiation and bleomycin-induced lung fibrosis. Sci Rep. 2016;6 doi: 10.1038/srep30122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shum E.Y., Jones S.H., Shao A. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell. 2016;165(2):382–395. doi: 10.1016/j.cell.2016.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douville J.M., Cheung D.Y.C., Herbert K.L., Moffatt T., Wigle J.T. Mechanisms of MEOX1 and MEOX2 regulation of the cyclin dependent kinase inhibitors p21CIP1/WAF1 and p16INK4a in vascular endothelial cells. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0029099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuver R. Mechanism of oxystero-induced disease: insights from the biliary system. Clin Lipidol. 2012;7(5):537–548. doi: 10.2217/clp.12.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagawa T., Watanabe M., Inoue N. Increases of microRNA let-7e in peripheral blood mononuclear cells in Hashimoto's disease. Endocr J. 2016;63(4):375–380. doi: 10.1507/endocrj.EJ15-0577. [DOI] [PubMed] [Google Scholar]
- 35.Murakami Y., Toyoda H., Tanaka M. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin E., Palmic N., Sanquer S. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature. 2014;510(7504):288–292. doi: 10.1038/nature13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brewer C.T. Hepatotoxicity of Herbal Supplements Mediated by Modulation of Cytochrome P450. 2017;DILI:1–28. doi: 10.3390/ijms18112353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Du Q., Wang F. 2004. Microarray Analysis of Gene Expression on Herbal Glycoside Recipes Improving Deficient Ability of Spatial Learning Memory in Ischemic Mice; pp. 1406–1415. [DOI] [PubMed] [Google Scholar]
- 39.Millar A.L., Pavlos N.J., Xu J., Zheng M.H. Rab3D: a regulator of exocytosis in non-neuronal cells. Histol Histopathol. 2002;17(3):929–936. doi: 10.14670/HH-17.929. [DOI] [PubMed] [Google Scholar]
- 40.Lazarus G.M. Marianne J. BT - Principles of Gender-specific Medicine. second ed. Academic Press; San Diego: 2010. Introduction A2 - legato; pp. 1–2. [DOI] [Google Scholar]
- 41.Yang J., Liu W., Lu X., Fu Y., Li L., Luo Y. High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget. 2015;6(13) doi: 10.18632/oncotarget.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.