Abstract
Receptor-interacting serine-threonine kinase 3 (RIPk3) is a key signaling molecule in the regulation of cell apoptosis and necroptosis, it plays an important role in the pathophysiological changes of many hematologic diseases. However, the regulatory role of RIPk3 in programmed cell death (PCD) is not fully known. In this study, bone marrow-specific RIPk3 gene knockout homozygotes (RIPk3-/- mice) were established by homologous recombination. The physiological index of peripheral blood, the morphology and structure of the bone marrow, the bone marrow nucleated cells (BMNCs), the hemopoietic stem cells (HSCs), interleukin-6 (IL-6) level and the colony formation capacity of bone marrow hematopoietic progenitor cells were compared between RIPk3-/- mice and wild-type mice. The results showed that, the cell death rate of BMNCs in RIPk3-/- mice was significantly higher than that in control mice, indicated that RIPk3 gene knockout may cause damage to bone marrow cells to some extent. However, the bone marrow had normal structure and morphology in the bone marrow-specific RIPk3-knockout mice, and there were not significantly different between the two mice in most of the blood physiological indicators, and colony yields of hemopoietic stem/progenitor cells. Further study found that the bone marrow IL-6 level of the RIPk3-/- mice increased significantly, besides, the number of BMNCs and HSCs in the bone marrow of the RIPk3-/- mice increased considerably as compared with the control mice. The findings implies that bone marrow RIPk3 gene knockout may lead to the increase of BMNCs cell death, however, increased secretion of hematopoietic cytokines such as IL-6 may promote the proliferation of hematopoietic stem/progenitor cells and thus maintain the stability of bone marrow hematopoiesis. This hypothesis and the detailed mechanisms remain to be further investigated.
Keywords: Bone marrow, RIPK3, gene knockout, hematopoiesis, mouse
Introduction
Programmed cell death (PCD) is a physiological cell death process involved in the selective elimination of unwanted cells. This process is closely connected with the activation, expression and regulation of multiple genes and plays an important role in maintaining homeostasis and normal cell functions. Bone marrow is the major site of hematopoiesis. The balance between the proliferation and death of bone marrow cells is the basis of normal hematopoiesis. When the regulatory mechanism of cell death is in disorder, bone marrow hematopoiesis may be affected by excessive proliferation or death of bone marrow cells caused by PCD abnormalities. This may further lead to various hematologic diseases such as aplastic anemia, leukemia and myeloproliferative diseases [1-5]. Understanding the regulatory mechanism of bone marrow PCD can facilitate hunting for pathogenesis of hematologic diseases and provide scientific clues for the diagnosis and new therapy exploration of hematologic diseases.
PCD mainly consists of apoptosis and necroptosis, which are crucial for the development and homeostasis in organisms. Recent studies have shown that RIPk3 is a key signaling molecule in the regulation of cell apoptosis and necroptosis [6,7]. It plays an important role in the pathophysiological changes of many diseases [8-12]. According to recent studies, RIPk3 is involved in the occurrence and progression of many hematologic diseases, including aplastic anemia (AA) [13], chronic lymphocytic leukemia (CLL) [14] and acute myeloid leukemia (AML) [15]. The role of RIPk3 in the pathogenesis of diseases has drawn wide-spread attention. RIPk3 can act as an important target in gene therapy [16-20]. However, the regulatory role of RIPk3 in PCD is not fully known. Establishing the bone marrow RIPk3 knockout mice may facilitate the understanding on the molecular mechanism of RIPk3-mediated PCD of bone marrows and the pathogenesis of related hematologic diseases. In this study, bone marrow-specific RIP3 knockout in the mouse model was induced by homologous recombination and the influence of bone marrow-specific RIPk3 gene knockout on the bone marrow hemopoietic function was observed. The purpose was to provide experimental data for understanding about the pathological role of RIPk3 in hematologic diseases.
Materials and methods
The study protocol was approved by the ethics committee of the Institute of Tumor, Medical College of Taizhou University and conformed to the Guide for the Care and Use of Laboratory Animals published by the Chinese National Institutes of Health.
Reagents
Bacterial Artificial Chromosome (BAC) containing RIPk3 was purchased from BAC/PAC Resources Center (Children’s Hospital Oakland Research Institute, Oakland, CA, USA). pL-451 Plasmid and EL-350 Strain were provided by Pengtao Liu (Welcome Trust Sanger Institute, Cambridge, UK). pSC101-BAD-γβα-A-tet was provided by Youming Zhang (Gene Bridges GmbH, Germany). pBR322-2S and pDTA were provided by Shanghai Research Center For Model Organisms. Restriction enzymes, T4 DNA ligase, T4 DNA polymerase, Taq enzyme and PCR kit were purchased from TaKaRa Biotechnology (Dalian, China) and NEB company (Carlsbad, CA, USA). Alexa Fluor 488 Annexin V and a PI kit were purchased from KeyGEN BioTECH (JiangXu, China). IL-6 ELISA kits, IMDM substratum, erythropoietin (EPO), thrombopoietin (TPO), granulocyte-macrophage colony stimulating factor (GM-CSF), fluorescent antibody PEc-kit and FITC-CD45 were all purchased from the Boster Company (Wuhan, China).
Construction and identification of the targeting plasmid
The loxp sequence was inserted to the intron 3 and 9 in the RIPk3 gene, respectively. Using pGK-Neo gene as the positive selection marker (flanked by frt sequence, with the deletion of pGK-Neo gene using the flpe enzyme) and TK gene in the targeting vector as the negative selection marker, the targeting vector for exon 4-9 knockout was constructed (by reacting with the Cre recombinase).
ES cell targeting, blastocyst microinjection and transplantation
The linearized targeting vector was transferred to the ES cells by electroporation. After that, the ES cells were screened with 200 μg/mL G418. Resistant ES cell clones were subjected to further culture. Genomic DNA was extracted from the resistance ES cells, and the positive clones were screened by long fragment PCR. The 5’ homology arm was identified using the forward primer 5’-GGCAGGCTGGTTTCTGAGTTTG-3’, and the reverse primer 5’-GGCCTACCCGCTTCCATTGCTC-3; the 3’ homology arm was identified using the forward primer 5’-CCGTGCCTTCCTTGACCCTGG-3’, and the reverse primer 5’-CATGGGCAGGCAACAGTCACA-3’. Blastocytes were harvested from female C57BL/6J mice aged 3-4 weeks, the positive ES cells were used for blastocyst microinjection after check and transplanted to the mice during spurious pregnancy.
Mice of F1 generation and genotyping
Coat color chimerism was determined in newborn mice from the female C57BL/6J mice transplanted with the blastocysts. Those with high degree of chimerism were chosen for breeding with the FLP mice to obtain the F1 generation. At 10-12 d after birth, the tail tip was harvested, and 200 μl Tail buffer and 10 μl protease K (20 μg/ml) were added. After digestion at 55°C overnight, protease K was deactivated at 95°C for 15 min and centrifuged. The supernatant was collected, amplified by PCR and sequenced. Identification of 5’ homology arm was performed using the forward primer 5’-GGCAGGCTGGTTTCTGAGTTTG-3’, and the reverse primer 5’-ATTCATCTCCTGAGCCCATTCCA-3’; identification of 3’ homology arm was performed using 5’-CCCTCCACAGACTAAGACATCCCTAA-3’, and the reverse primer 5’-CATGGGCAGGCAACAGTCACA-3’. PCR reactions were performed: 94°C for 2 min, then 98°C for 20 sec, 66°C for 20 sec and 68°C for 2.5 min, a total of 34 cycles; 68°C for 5 min.
Breeding
In theory, the breeding between RIPk3 loxp/+ and lyzcre/+ mice can lead to individuals that contain loxp sites and express Cre recombinase (i.e., loxp/+_cre/+). Similarly, the breeding between RIPk3 loxp/loxp and loxp/+_cre/+ mice can lead to loxp/loxp_cre/+ individuals. Study has shown that the binding of the loxp site to Cre recombinase will result in gene knockout [21]. Therefore, the loxp/+_cre/+ individuals will be heterozygotes with bone marrow-specific knockout of RIPk3 gene (lyz-/+), while loxp/loxp_cre/+ individuals will be homozygotes.
Acquisition and identification of homozygotes
Tails were cut off from the mice reaching 2 weeks. DNA was extracted conventionally and subjected to PCR and electrophoresis. The loxp site was detected using the forward primer 5’-CTCCTTACCAGACGCCCTTCT-3’ and the wild-type genomic site was detected using the forward primer 5’-CAGCGACACCTTGTGATCTCC-3’. The homozygous loxp site corresponded to a 629 bp band, and the heterozygous loxp site was split into two bands, which were 629 bp and 476 bp, respectively. The wild-type gene containing no loxp site corresponded to one 476 bp band. That is, loxp/loxp = 629 bp; wt/wt = 476 bp; wt/loxp = 476 bp + 629 bp. Cre recombinase gene was detected using the forward prime 5’-GACACGGCACTCCTTGGTAT-3’. If Cre recombinase gene was inserted into the genome, a 335 bp band would be produced; otherwise, there would be no band.
Peripheral blood and bone marrow examination
Bone marrow-specific RIPk3 gene knockout homozygotes (RIPk3-/- mice) were obtained and wild-type mice were taken as control. Routine blood examination was conducted, then three mice of each group were sacrificed, and the femurs of each mouse were taken. One femur was surgically dissected, the BMNCs suspension was prepared and number of BMNCs was counted by flow cytometer.
60 μl of the BMNCs suspension was centrifuged, then washed with PBS and stained twice with PE-c-kit and FITC-CD45, bone marrow HSCs were counted by flow cytometer. 100 μl BMNCs suspension was used for cell death rate evaluation by annexin V and PI staining, and 100 μl suspension was used in ELISA for the measurement of levels of IL-6. All operations step by kits instructions.
Morphological evaluation of the bone marrow
Another femur were dissected and fixed in 10% formalin solution for histopathological evaluation. The femur was further decalcified in 5% nitric acid solution for 7~12 h. Then, paraffin-embedded sections were prepared routinely for hematoxylin and eosin (HE) staining and histopathological evaluation.
Hemopoietic progenitor cell culture in vitro
BMNCs were adjusted to a concentration of 5×105 cells/ml, cells were cultured at 37°C with 5% CO2 and saturated humidity. Three days later, CFU-E was counted under the microscope, and 7 days later, BFU-E, CFU-Meg and CFU-GM were counted.
Statistical analysis
All data are expressed as the mean ± standard deviation (SD). The t-test was used to evaluate the difference between groups. A P value less than 0.05 was considered statistically significant.
Results
Identification of the homologous recombination vector using restriction enzyme cutting
The map of restriction enzyme analysis for the homologous recombination vector is shown in Figure 1A. The fragments obtained by restriction enzyme digestion were of the same size as expected, DNA sequencing also verified that the vector was successfully constructed (see Supplementary File 1).
Figure 1.
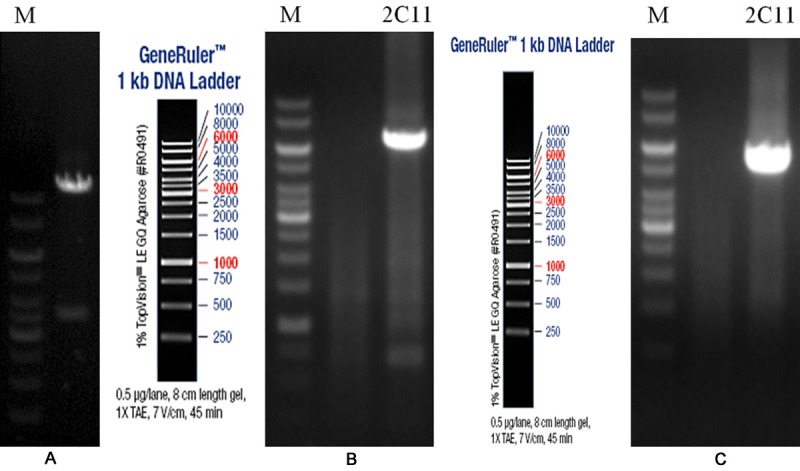
Analysis of the vector and ES cell clones positive for homologous recombination. A. Map of restriction enzyme analysis for the homologous recombination vector (For the digestion with SalI, the theoretical band was 3.7 kb and 14.2 kb in length, respectively; M: 1 kb DNA ladder). B, C. Electrophoretogram of PCR products for ES cell clones positive for homologous recombination (2-C11, positive clone; M, DNA marker; B. Identification result of 5’ homology arm; C. Identification result of 3’ homology arm).
PCR identification of ES cells positive for homologous recombination
After targeting the ES cells, screening with G418 and further culture were performed. Thus a total of 96 G418-resistant ES cell clones were obtained. Genomic DNA extraction was performed for these positive clones, which were further identified using long fragment PCR and sequenced (see Supplementary File 2). One ES cell clone positive for homologous recombination was obtained (Figure 1B and 1C).
Identification of mice of F1 generation with Neo deletion
Breeding between the male chimeras and FLP mice yielded F1 generation. After PCR identification (Figure 2) and sequencing (see Supplementary File 3), four positive heterozygous mice with Neo deletion (RIPk3 loxp/+) were obtained. They were numbered as 7, 8, 10 and 11, respectively.
Figure 2.
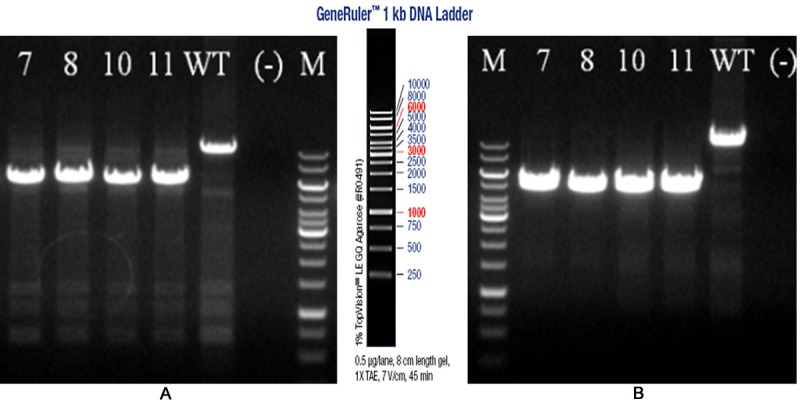
Electrophoretogram of PCR products for the 5’ and 3’ homology arm of mice of F1 generation with Neo deletion. Number, serial number of mice of F1 generation with Neo deletion; wt, wild-type control; M, DNA marker, 1 kb DNA ladder. A. Identification result of 5’ homology arm; B. Identification result of 3’ homology arm.
Acquisition and identification of homozygotes
Breeding between RIPk3 loxp/loxp and loxp/+_cre/+ mice yielded two different individuals homozygous for loxp, namely, individuals containing only homozygous loxp sites (RIPk3 loxp/loxp) and individuals containing homozygous loxp sites and expressing Cre recombinase (loxp/loxp_cre/+), and the latters are bone marrow-specific RIPk3 gene knockout homozygotes (RIPk3-/-) (Figure 3).
Figure 3.
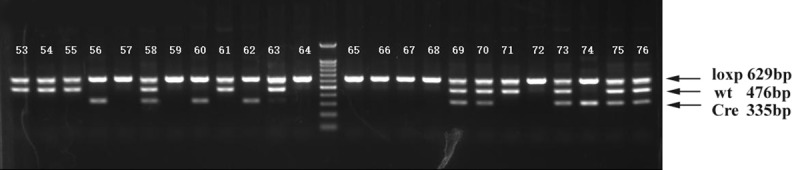
Electrophoretogram of PCR products of Rip3 floxed mice. (Number, serial number of mouse) Mice numbered 57, 59, 64, 65, 66, 67 and 68 were homozygous for loxp; those numbered 56, 60 and 62 were homozygotes with bone marrow-specific knockout of the RIP 3 gene (RIPk3-/-). A total of 6 homozygous were obtained after breeding.
Peripheral blood and bone marrow examination
Detection of peripheral blood indicated that RET increased considerably in the RIPk3 knockout mice, while other peripheral blood indicators were not significantly different from those of the control mice (Table 1). Compared with the control mice, the counts of BMNCs, HSCs and bone marrow IL-6 level in the knockout mice increased considerably. In addition, flow cytometer showed that the cell death rate of BMNCs in RIPk3-/- mice was significantly higher than that in control mice (Figure 4).
Table 1.
Peripheral blood and bone marrow examination (x̅ ± s)
| Items | RIPk3-/- | Control |
|---|---|---|
| WBC (×109/l) | 10.40±2.02 | 10.92±2.17 |
| RBC (×1012/l) | 10.33±0.59 | 10.24±0.48 |
| HGB (g/l) | 161.67±6.28 | 159.33±5.75 |
| HCT (%) | 50.50±2.42 | 48.70±1.41 |
| MCV (fl) | 48.90±0.68 | 47.60±0.93 |
| MCH (pg) | 15.67±0.29 | 15.57±0.19 |
| MCHC (g/l) | 320.33±3.61 | 327.00±3.58 |
| PLT (×109/l) | 1385.00±72.21 | 1377.00±121.00 |
| RDW-SD (fl) | 30.10±0.50 | 29.37±0.36 |
| RDW-CV (%) | 20.47±0.81 | 20.77±0.85 |
| PDW (fl) | 7.40±0.24 | 7.23±0.26 |
| MPV (fl) | 6.77±0.23 | 6.73±0.29 |
| P-LCR (%) | 4.73±0.81 | 4.67±1.56 |
| PCT (%) | 0.94±0.81 | 0.93±0.12 |
| NEUT (×109/l) | 1.20±0.15 | 1.36±0.32 |
| LYMPH (×109/l) | 8.95±2.09 | 9.26±1.91 |
| MONO (×109/l) | 0.08±0.03 | 0.11±0.06 |
| EO (×109/l) | 0.17±0.01 | 0.18±0.01 |
| BASO (×109/l) | 0.00±0.00 | 0.01±0.01 |
| RET (×109/l) | 781.37±184.51* | 624.50±31.70 |
P < 0.05, compared with control group.
Figure 4.
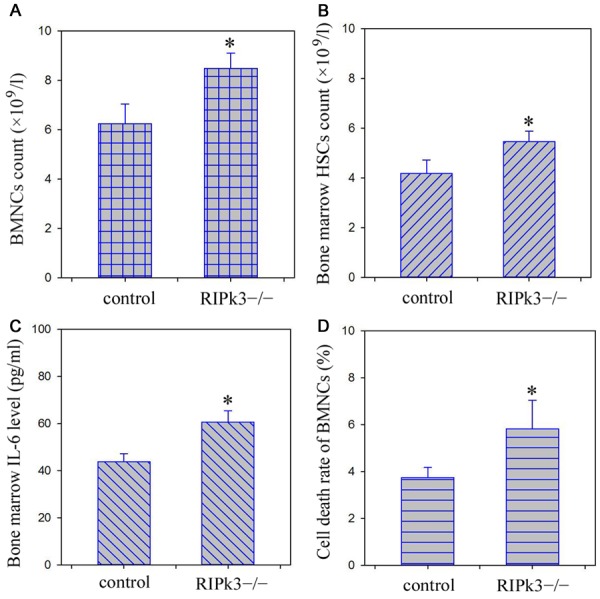
Bone marrow BMNCs and HSCs count, IL-6 level and cell death rate analysis. A. BMNCs count; B. HSCs count; C. IL-6 level; D. Cell death rate. *P < 0.05, compared with control group.
Morphological evaluation of the bone marrow
The bone marrow had normal structure and morphology in the bone marrow-specific RIPk3-knockout mice. Bone marrow cells proliferated actively in mice of two different phenotypes. A large number of hematopoietic cells were observed, megakaryocytes were easy to be seen (Figure 5). There was no sinusoidal dilatation and the sinus wall was intact.
Figure 5.
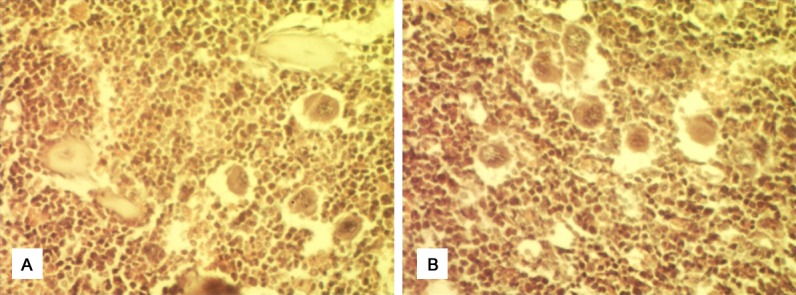
Histological analysis of bone marrow tissue in mice (100×), stained by HE. A. RIPk3-/- mice; B. Control.
Colony yields of CFU-E, BFU-E, CFU-GM and CFU-Meg
In vitro cell culture indicated no significant difference between RIPk3-/- mice and control mice in the colony yields of CFU-E, BFU-E, CFU-GM and CFU-Meg (Figure 6).
Figure 6.
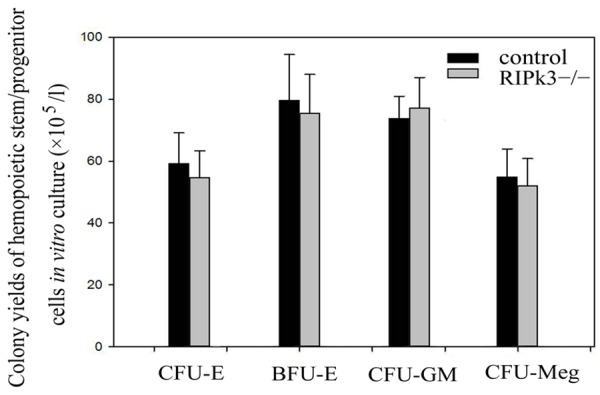
Colony yields of hemopoietic stem/progenitor cells in vitro culture.
Discussion
RIPk3 is a member of the receptor-interacting protein family and exhibits specific serine/threonine kinase activity [22]. RIPk3 has been found to be an important molecular switch for PCD signaling. Excessive RIPk3 expression can induce Caspase-mediated apoptosis of cells. When Caspase activity is inhibited, overexpression of RIPk3 can promote the conversion from cell apoptosis to non-Caspase-medicated PCD [23]. RIPk3 is involved in the pathogenesis of many hematologic diseases [24,25]. Through preliminary experiments, it was found that RIPk3-medicated PCD was involved in the cyclophosphamide- and busulfan-induced aplastic anemia in mice [4]. Xiao et al. reported that RIPk3-medicated PCD was related to the death regulation of hematopoietic stem/progenitor cells in Tak1-knockout mice. Moreover, it played an important role in the progression of bone marrow failure syndrome of Tak1(-/-) mice [26]. According to a recent study, defect in the RIPk3-mediated PCD is related to the pathogenesis of CLL and AML, and it is an important reason for resistance to chemotherapy [14,15]. Thus RIPk3 is a valuable therapeutic target for hematologic diseases.
Flox modification of the RIPk3 gene was performed by targeting ES cells using homology recombination. The Cre/Loxp system was used for specific knockout of RIPk3 gene in the mouse bone marrow. Cre recombinase is derived from bacteriophage P1. Loxp is a 34 bp palindromic sequence. Cre recombinase can specifically recognize the Loxp sequence, causing recombination between 2 Loxp sequences and deletion of the sequence between them. DNA sequences in the important functional domain of the target gene are usually flanked by 1 Loxp on each side. The DNA sequence in the important functional domain is marked by Loxp in the target gene of the knockout mice. Breeding between such knockout mice and the transgenic mice expressing Cre recombinase in specific cells and tissues may lead to Cre recombinase-mediated deletion of the target gene in the cells expressing the Cre recombinase. However, since Cre recombinase is not expressed in other tissues or cells, DNA deletion will not affect these tissues and cells. Thus the target gene remains normal in the offspring. This method can prevent embryonic death or severe developmental disorder at an early developmental stage due to systemic knockout in all cells and tissues [27-30].
Many reports have been published on the specific knockout by the breeding between mice with specific Cre recombinase expression in the bone marrow and Loxp transgenic mice. The target gene is specifically knocked out in the bone marrow of the offspring [21,31]. The bone marrow-specific RIPk3 knock-out mouse model was successfully established using this method. However, this does not mean that the target gene is removed from all bone marrow cells. For example, the knockout rate is only 83%-98% in the mature macrophages, and knockout may be not limited to bone marrow cells. Clausen et al. reported a 16% knockout rate in a type of dendritic cells in the spleen [21]. More researches are needed for the genotyping of bone marrow-specific homozygous and heterozygous RIPk3 knockout mice obtained in this study.
Bone marrow is an important organ involved in hematopoiesis and immune regulation, the healthy bone marrow tissue is the structural basis for maintaining normal hematopoiesis. Our results showed that bone marrow had normal structure and morphology for bone marrow-specific RIPk3-knockout homozygotes. However, flow cytometer showed that the cell death rate of BMNCs in RIPk3-/- mice was significantly higher than that in control mice, indicated that bone marrow RIPk3 gene knockout may lead to the increase of BMNCs cell death.
Physiological value of blood can reflect the hematopoietic function of bone marrow to a certain extent and also serves as an indicator of the health status and genetic stability of animals. It is also an important basis for clinical diagnosis. Our results showed that, compared with the control mice, RET in peripheral blood increased significantly in the homozygotes, while other physiological indicators of peripheral blood did not change significantly. All primitive blood cells are derived from bone marrow hemopoietic stem/progenitor cells. Colony yields of hemopoietic stem/progenitor cells in vitro culture can reflect the proliferative capacity of hemopoietic cells. Our results of in vitro culture showed that there was no significant difference in the colony yields of hemopoietic stem/progenitor cells of three lineages between the two mice. This findings indicated that the stability of the bone marrow hemopoiesis can be maintained under RIPk3 gene knockout.
IL-6 is a kind of cytokine with a wide range of biological activities, it is involved not only in regulation of proliferation and differentiation of early hemopoietic stem/progenitor, but also in the progress of the stress reaction, autoimmune and neoplastic diseases of the body [32,33]. In the present study, we found that the bone marrow IL-6 level in RIPk3-/- mice was increased significantly than that in control mice. In addition, flow cytometry revealed that the number of BMNCs and HSCs in the bone marrow of the RIPk3-/- mice increased considerably, indicated that the positive hematopoietic factors such as IL-6, may involved in the promotion of proliferation, differentiation, and maturation of HSCs in RIPk3-/- mice.
Taken together, the findings of our study implies that bone marrow RIPk3 gene knockout may lead to the increase of BMNCs cell death, however, increased secretion of hematopoietic factors such as IL-6 may promote the proliferation of hematopoietic stem/progenitor cells and thus maintain the stability of bone marrow hematopoiesis. This hypothesis and the detailed mechanisms remain to be further investigated.
Acknowledgements
This research was funded by the Public Welfare Technology Application Research Project of Zhejiang Province under Grant No. 2015C37122, Zhejiang, China. National natural science foundation of China under Grant no. 81373139.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Caraballo JM, Acosta JC, Cortés MA, Albajar M, Gómez-Casares MT, Batlle-López A, Cuadrado MA, Onaindia A, Bretones G, Llorca J, Piris MA, Colomer D, León J. High p27 protein levels in chronic lymphocytic leukemia are associated to low Myc and Skp2 expression, confer resistance to apoptosis and antagonize Myc effects on cell cycle. Oncotarget. 2014;5:4694–4708. doi: 10.18632/oncotarget.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Zou Z, Wu Z, Zhao Z, Luo X, Xie C, Liang Y. TNF-α-induced programmed cell death in the pathogenesis of acquired aplastic anemia. Expert Rev Hematol. 2015;8:515–526. doi: 10.1586/17474086.2015.1049593. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Lu J, van den Berghe J, Lee SH. Increased incidence of spontaneous apoptosis in the bone marrow of hyperdiploid childhood acute lymphoblastic leukemia. Exp Hematol. 2002;30:333–339. doi: 10.1016/s0301-472x(02)00771-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen YF, Liu H, Luo XJ, Zhao Z, Zou ZY, Li J, Lin XJ, Liang Y. The roles of reactive oxygen species (ROS) and autophagy in the survival and death of leukemia cells. Crit Rev Oncol Hematol. 2017;112:21–30. doi: 10.1016/j.critrevonc.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Xia G, Chen B, Ding J, Gao C, Lu H, Shao Z, Gao F, Wang X. Effect of magnetic Fe3O4 nanoparticles with 2-methoxyestradiol on the cell-cycle progression and apoptosis of myelodysplastic syndrome cells. Int J Nanomedicine. 2011;6:1921–1927. doi: 10.2147/IJN.S24078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriwaki K, Balaji S, McQuade T, Malhotra N, Kang J, Chan FK. The necroptosis adaptor RIPK3 promotes injury-induced cytokine expression and tissue repair. Immunity. 2014;41:567–578. doi: 10.1016/j.immuni.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton K, Dugger DL, Wickliffe KE, Kapoor N, de Almagro MC, Vucic D, Komuves L, Ferrando RE, French DM, Webster J, Roose-Girma M, Warming S, Dixit VM. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 8.Roychowdhury S, McMullen MR, Pisano SG, Liu X, Nagy LE. Absence of receptor interacting protein kinase 3 prevents ethanol-induced liver injury. Hepatology. 2013;57:1773–1783. doi: 10.1002/hep.26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Liu Z, Ren J, Morgan S, Assa C, Liu B. Receptor-interacting protein kinase 3 contributes to abdominal aortic aneurysms via smooth muscle cell necrosis and inflammation. Circ Res. 2015;116:600–611. doi: 10.1161/CIRCRESAHA.116.304899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng L, Jin W, Wang Y, Huang H, Li J, Zhang C. RIP3-dependent necrosis induced inflammation exacerbates atherosclerosis. Biochem Biophys Res Commun. 2016;473:497–502. doi: 10.1016/j.bbrc.2016.03.059. [DOI] [PubMed] [Google Scholar]
- 11.Afonso MB, Rodrigues PM, Carvalho T, Caridade M, Borralho P, Cortez-Pinto H, Castro RE, Rodrigues CM. Necroptosis is a key pathogenic event in human and experimental murine models of non-alcoholic steatohepatitis. Clin Sci (Lond) 2015;129:721–739. doi: 10.1042/CS20140732. [DOI] [PubMed] [Google Scholar]
- 12.Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, Kitada T, Glass K, Owen CA, Mahmood A, Washko GR, Hashimoto S, Ryter SW, Choi AM. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest. 2014;124:3987–4003. doi: 10.1172/JCI74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YF, Zhao ZQ, Wu ZM, Zou ZY, Luo XJ, Li J, Xie C, Liang Y. The role of RIP1 and RIP3 in the development of aplastic anemia induced by cyclophosphamide and busulphan in mice. Int J Clin Exp Pathol. 2014;7:8411–8420. [PMC free article] [PubMed] [Google Scholar]
- 14.Liu P, Xu B, Shen W, Zhu H, Wu W, Fu Y, Chen H, Dong H, Zhu Y, Miao K, Xu W, Li J. Dysregulation of TNFα-induced necroptotic signaling in chronic lymphocytic leukemia: suppression of CYLD gene by LEF1. Leukemia. 2012;26:1293–1300. doi: 10.1038/leu.2011.357. [DOI] [PubMed] [Google Scholar]
- 15.Nugues AL, El Bouazzati H, Hétuin D, Berthon C, Loyens A, Bertrand E, Jouy N, Idziorek T, Quesnel B. RIP3 is downregulated in human myeloid leukemia cells and modulates apoptosis and caspase-mediated p65/RelA cleavage. Cell Death Dis. 2014;5:e1384. doi: 10.1038/cddis.2014.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bozec D, Iuga AC, Roda G, Dahan S, Yeretssian G. Critical function of the necroptosis adaptor RIPK3 in protecting from intestinal tumorigenesis. Oncotarget. 2016;7:46384–46400. doi: 10.18632/oncotarget.10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höckendorf U, Yabal M, Herold T, Munkhbaatar E, Rott S, Jilg S, Kauschinger J, Magnani G, Reisinger F, Heuser M, Kreipe H, Sotlar K, Engleitner T, Rad R, Weichert W, Peschel C, Ruland J, Heikenwalder M, Spiekermann K, Slotta-Huspenina J, Groß O, Jost PJ. RIPK3 restricts myeloid leukemogenesis by promoting cell death and differentiation of leukemia initiating cells. Cancer Cell. 2016;30:75–91. doi: 10.1016/j.ccell.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Lin Z, Zhao N, Zhou L, Liu F, Cichacz Z, Zhang L, Zhan Q, Zhao X. Receptor interactive protein kinase 3 promotes cisplatin-triggered necrosis in apoptosis-resistant esophageal squamous cell carcinoma cells. PLoS One. 2014;9:e100127. doi: 10.1371/journal.pone.0100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng X, Song Q, Yu A, Tang H, Peng Z, Wang X. Receptor-interacting protein kinase 3 is a predictor of survival and plays a tumor suppressive role in colorectal cancer. Neoplasma. 2015;62:592–601. doi: 10.4149/neo_2015_071. [DOI] [PubMed] [Google Scholar]
- 20.Xin J, You D, Breslin P, Li J, Zhang J, Wei W, Cannova J, Volk A, Gutierrez R, Xiao Y, Ni A, Ng G, Schmidt R, Xia Z, Pan J, Chen H, Patel MM, Kuo PC, Nand S, Kini AR, Zhang J, Chen J, Zhu J, Zhang J. Sensitizing acute myeloid leukemia cells to induced differentiation by inhibiting the RIP1/RIP3 pathway. Leukemia. 2017;31:1154–1165. doi: 10.1038/leu.2016.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 22.Park SM, Yoon JB, Lee TH. Receptor interacting protein is ubiquitinated by cellular inhibitor of apoptosis proteins (c-IAP1 and c-IAP2) in vitro. FEBS Lett. 2004;566:151–6. doi: 10.1016/j.febslet.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 23.Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 24.Laubach JP, Fu P, Jiang X, Salter KH, Potti A, Arcasoy MO. Polycythemia vera erythroid precursors exhibit increased proliferation and apoptosis resistance associated with abnormal RAS and PI3K pathway activation. Exp Hematol. 2009;37:1411–1422. doi: 10.1016/j.exphem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JP, Zheng CL, Han ZC. Abnormal immunity and stem/progenitor cells in acquired aplastic anemia. Crit Rev Oncol Hematol. 2010;75:79–93. doi: 10.1016/j.critrevonc.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Li H, Zhang J, Volk A, Zhang S, Wei W, Zhang S, Breslin P, Zhang J. TNF-α/Fas-RIP-1-induced cell death signaling separates murine hematopoietic stem cells/progenitors into 2 distinct populations. Blood. 2011;118:6057–6067. doi: 10.1182/blood-2011-06-359448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gui YS, Wang L, Tian X, Feng R, Ma A, Cai B, Zhang H, Xu KF. SPC-Cre-ERT2 transgenic mouse for temporal gene deletion in alveolar epithelial cells. PLoS One. 2012;7:e46076. doi: 10.1371/journal.pone.0046076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsuno T, Umeda K, Matsui T, Hata M, Tamura A, Itoh M, Takeuchi K, Fujimori T, Nabeshima Y, Noda T, Tsukita S, Tsukita S. Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol Biol Cell. 2008;19:2465–2475. doi: 10.1091/mbc.E07-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang SM, Hou ZH, Yang G, Zhang JS, Hu YY, Sun JH, Guo WW, He DZ, Han DY, Young WY, Yang X. Chondrocyte-specific Smad4 gene conditional knockout results in hearing loss and inner ear malformation in mice. Dev Dyn. 2009;238:1897–1908. doi: 10.1002/dvdy.22014. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Li YP, Fu XY, Deng CX. Anterior visceral endoderm SMAD4 signaling specifies anterior embryonic patterning and head induction in mice. Int J Biol Sci. 2010;6:569–583. doi: 10.7150/ijbs.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinclair GB, Jevon G, Colobong KE, Randall DR, Choy FY, Clarke LA. Generation of a conditional knockout of murine glucocerebrosidase: utility for the study of Gaucher disease. Mol Genet Metab. 2007;90:148–156. doi: 10.1016/j.ymgme.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Möbius-Winkler S, Hilberg T, Menzel K, Golla E, Burman A, Schuler G, Adams V. Time-dependent mobilization of circulating progenitor cells during strenuous exercise in healthy individuals. J Appl Physiol. 2009;107:1943–1950. doi: 10.1152/japplphysiol.00532.2009. [DOI] [PubMed] [Google Scholar]
- 33.Nishimoto N, Kishimoto T. Interleukin 6: from bench to bedside. Nat Clin Pract Rheumatol. 2006;2:619–626. doi: 10.1038/ncprheum0338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


