Abstract
Osteoporosis has become a major disease that threatened post-menopausal women and elder people. Circulating micorRNAs (miRNA) could provide useful information for diagnosis and therapeutics. The study employed RT-real time PCR to detect the circulating miRNAs between osteoporotic patients and healthy controls. Human and mouse osteoblast cell lines were used to test the differential induction effects by miRNAs. Alkaline phosphatase activity and Alizarin red staining were examined after miRNA mimics stimulation. The authors found 14 of 150 tested miRNAs were significantly aberrant expressed between patients and healthy controls. Results showed miR-328-3p, let-7g-5p, miR-133b, miR-22-3p, miR-2861, miR-518 miR-100 were down-regulated osteoporotic patient, while miR-10b-5p, miR-21, miR-125b and miR-127 were up-regulated. MiR-10b-3p, miR-328-3p, miR-100 and let-7 showed tight association with Wnt pathway. MiR-10b-5p increased ALP activity and mineral deposition in human and mouse osteoblast cells, indicating miR-10b-3p promoted osteoblast cell differentiation. MiR-328-3p and let-7g-5p decreased ALP activity and suppressed mineral deposition in both cell lines. Conclusively, miR-10b-5p promoted osteoblast cells differentiation; miR-328-3p, miR-100 and let-7 inhibited osteoblast cells differentiation.
Keywords: Osteoporosis, circulating miRNAs, osteoblast, differentiation, cell lines
Introduction
Osteoporosis is characterized by systemic ske-letal disorder associated with deterioration of bone mass and microarchitecture [1]. Osteoblasts and osteoclasts make dynamic equilibrium of bone homeostasis. Cytokines deficiency becomes a major risk factor in osteoporosis in postmenopausal women, causing diminished osteoblast differentiation and maturation. Additionally, other investigators reminded that aberrant microRNAs (miRNAs) signatures might not be specific to the pathophysiology in bone, and indicated that more work remains to be done until the robust diagnostic biomarkers were established [2]. Despite these promising primary data, many studies demonstrated that miRNAs participated in bone formation, indicating the therapeutic roles of miRNAs in bone related diseases [3].
MiRNAs are a class of endogenous approximately 22 nucleotides, single chain and non-coding RNAs. MiRNAs participate in bone homeostasis through regulation of specific gene expression, such as Runx2 [4], and FGF2 [5]. It was reported that Osterix and miRNAs are involved in bone formation and Osterix-controlled osteogenesis [6]. Circulating miRNAs are regarded as special transporting miRNAs, which might have its specific function to nearby or long distance target cells. It was reported that osteoclasts-derived exosomes contain miRNAs, which target and inhibit osteoblasts activity and maturation [7]. New discoveries insight into the circulating miRNAs indicated circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation [8]. Data implied circulating miRNAs can be taken up by cells and thereby influence the recipient cell’s behavior in the context of diverse biological functions [9]. In the aspect of biomarkers using miRNAs, measurability, validation and utility are realized as the fundamental bases of valuated biomarker for diagnosis. Because of noninvasive and easy sampling, circulating miRNAs have great potential to serve as biomarkers for osteoporosis [10]. Many therapeutic trials on osteoporosis and other bone related diseases using miRNAs have started in animal, indicating the age of miRNA therapy [11].
Many studies demonstrate that miRNAs can promote osteoblasts differentiation and participate in the regulation of differentiation [12]. A miRNAs cluster miR-23a~27a~24-2 regulated osteoblast differentiation, and connecting Runx2 and SATB2 in osteogenic cells [13]. In addition, miRNA also affect BMMSCs [14] and MSCs [15] in bone mass and density, providing a new strategy and insight for the treatment of osteoporosis. We realized miRNAs signature might not only serve as biomarkers, but also great potential as therapeutics drugs targeting osteoporosis. In the present study, we hypothesized that osteoporotic-specific miRNAs might be presented in serum as circulating biomarkers. These miRNAs was regarded as potential diagnostic biomarkers, however, it might be also used as promising drug targets. According to the above idea, we quantified the levels of 150 circulating miRNAs in serum samples from osteoporotic patients, and age-matched health controls from the affiliated hospital. This study aims to find the early diagnosis marker for osteoporosis using circulating miRNAs, as well as to explore the miRNA drug for osteoporosis therapeutics.
Materials and methods
Osteoporotic patients and healthy donor
Recruited osteoporotic patients were diagnosed in the university affiliated hospital, and agreed to join the current study. Patients were diagnosed medicine doctor under osteoporosis diagnostic BMD, CT results and other serological results. Healthy donors are persons who had routine physical examine in the hospital, and age- and sex-matched with patients. Included controls were without any obvious diseases, such as cold, chronic disease etc. Serum samples (9 patients and 9 controls) were collected and stored at -20 degree. This study was approved by The Ethical Committee of Southern Medical University (China), Referenced No.201605339.
RNA isolation
RNAs were extracted from cells using Trizol (Life sciences)-chloroform extraction and isopropanol precipitation method. 1 ml serum was mixed with 4 ml Trizol reagent and carried out exaction procedures following the manufactory’s instructions. For good quality and safety, glycogen as a carrier at 1 μg/μl was also used (Thermo Scientific). Incubate the mixture for 5 minutes at room temperature, and add 0.8 mL of chloroform and incubate for 3 minutes at room temperature. Centrifuge the sample at 12,000 × g for 15 minutes at 4°C. Collect the RNA phase by angling the tube at 45° and pipetting the solution out. After twice wash with 70% EtOH, RNA pellets were air dried and resuspended in 50 μL of enzyme-free water. RNA concentration was analyzed by NanoDrop 2000 at 260 nm (Thermo Fisher). The RNA quality and purity were determined by 260/230 nm and 260/280 nm respectively.
Circulating microRNAs qPCR
Real-time PCR (RT-qPCR) quantification of circulating microRNAs was performed followed previous reported experiment [16]. Briefly, serum total RNA from RNA extraction step was used as template. MiRNAs real-time PCR reactions were performed using 96-well plate (Thermo Scientific). Firstly, 1 µg of total RNA was reverse transcribed using 20 μl reactions system by commercial kit (Thermo Scientific). After reversed transcription reaction, 2 μl PCR product was added for subsequent miRNA quantification in 10 μl reaction buffer system (Applied Biosystem). The reverse transcript primers and the real time PCR primers of four miRNAs were listed in Supplementary Table 1. U6 RNA and its primers were used as control. MiRNA PCR was carried out at 94°C for 30 s, 55°C for 30 s and 72°C for 45 s for 30 cycles, using ABI prism 7700 sequence detector and software to analyze the data (Applied Biosystems, CA).
Cell culture
Human osteoblast cells hFOB1.19 (cat: CRL-11372TM) and mouse osteoblast cells MC3T3-E1 were purchased from American Type Culture Collection (ATCC). hFOB1.19 cells were cultured with complete growth medium, for quick growth setting the temperature at 33.5°C, for differentiation and maturation analysis setting the culture temperature at 39.5°C, with 5% CO2 in humidity air incubator. The complete growth culture medium contains 10% fetal bovine serum (Gibco), 1 ng/ml recombinant human basic fibroblast growth factor (human FGF, Peprotech) and 4 mM L-glutamine in HAM’s F-12 medium (Gibco, Thermo). MC3T3-E1 was cultured with Alpha Minimum Essential Medium supplement with 2 mM L-glutamine and 1 mM sodium pyruvate, and 10% fetal bovine serum. Cell culture condition was 37°C, with 5% CO2 in humidity air. Cells were passaged when the density was up to 85% at a split ratio of 1:3.
Alkaline phosphatase assay
In order to quantify the activity of alkaline phosphatase (ALP), culture medium was aspirated and cells were lysed in 100 μL ALP lysis buffer (0.25% v/v Triton X-100 in 0.5 M 2-amino-2-methyl-1-propanol, 2.0 mM magnesium chloride (Sigma-Aldrich). Seven days after treatment of osteogenesis induction and miRNAs treatment. After centrifuge with 13,000 × g for 10 min, collect the supernatant, and add 50 μL ALP Buffer A, and incubate for 20 min at room temperature. Finally, stop reaction by 50 μL NaOH (0.2 M). The absorption at 405 nm was measured, setting reference absorbance at 620 nm [8]. Quantification of ALP activity was performed using human and mouse osteoblast cell lines and 4 independent replicate wells each.
Alizarin red staining
Alizarin red staining experiment was carried out to show the efficacy of calcium mineralization. MiRNA transfected cells were fixed in 70% ethanol for 1 h, followed by wash with PBS twice. Cells were stained with 50 mM Alizarin Red solution for 20 min (Sigma). Wash the cells until unbound dye was removed. Observe and count the positive nudes under microscopy.
Statistical analysis
MiRNA expression level comparison in paired patients and healthy controls was used U-test method. In ALP activity and alizarin red staining experiments, U-test method was applied. In four miRNAs ROC dichotomization, t-test was used as default method. Data are presented as mean ± standard derivation, unless stated otherwise. All the statistical comparisons were used SPSS 13.0 software. ROC curve analysis was performed by Prism 5.0 software, and its statistical analysis method. Statistical significance was achieved when the p value is <0.05.
Results
Biostatistical analysis identified 14 differentially expressed circulating miRNAs in osteoporotic patients with age-matched healthy controls
The study was approved by the Xiamen University Ethic Committee Conference. All patients and age-matched healthy controls agreed with study content and signed the agreement form. Detail information was listed in Table 1. We performed RT-real time PCR to detect the circulating miRNAs between osteoporotic patients and healthy controls, and clustered the differential miRNAs against patients and controls. P value of paired t-test was used as y-axis, and Log2 (-2Δct) as x-axis (Figure 1A). Among 150 tested miRNAs, 14 of them were significantly aberrant expressed. as showed in volcanic chart (Figure 1B). Results showed down-regulated miRNAs in osteoporotic patient: let-7g-5p, miR-133b, miR-328-3p, miR-22-3p, miR-2861, miR518, and up-regulated miR-10b-5p, miR-21, miR-125b, miR-23 and miR-100.
Table 1.
Summary of patients and controls characteristics
| Osteoporotic | SD | Control | SD | |
|---|---|---|---|---|
| Number of samples | 9 | - | 9 | - |
| Age (years) | 69.2 | ±2.7 | 67.1 | ±2.6 |
| BMI (kg/m2) | 31.3 | ±1.5 | 33.4 | ±1.9 |
| Vitamin D (μg/L) | 21.2 | ±5.3 | 14.8 | ±3.6 |
| PTH (ng/L) | 49.4 | ±4.1 | 45.2 | ±5.8 |
Figure 1.
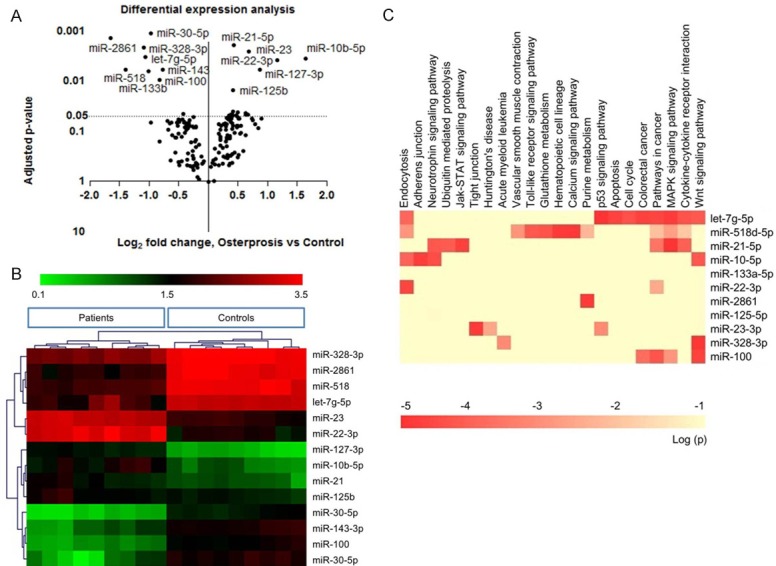
A: Volcanic chart showed aberrant expressed miRNAs among 150 tested circulating miRNAs. P value of paired t-test was used as y-axis, and Log2 (-2Δct) as x-axis. B: Real time PCR results showed down-regulated miRNAs in osteoporotic patient: let-7g-5p, miR-133b, miR-328-3p, miR-22-3p, miR-2861, miR518, and up-regulated miR-10b-5p, miR-21, miR-125b, miR-23 and miR-100. C: Clustering of these miRNAs correctly classify osteoporotic patients and healthy controls. Heatmap of KEGG pathways enriched from 14 miRNAs target genes was plotted. The 14 miRNAs were involved in multiple pathways, especially Wnt pathway, bone metabolic, cell growth and differentiation pathway.
Identification of selected miRNAs in osteoblast differentiation
We predicted potential targets of each miRNA using online software (www.targetscan.org/vert_71/). Predicted targets were listed in Supplementary Table 2. We put the miRNA targets for KEGG pathway analysis one by one, for the purpose of miRNA involved signal pathway. Heatmap of KEGG pathways enriched from 14 miRNA target genes was plotted. The 14 miRNAs were involved in multiple pathways, especially Wnt pathway, bone metabolic, cell growth and differentiation pathway (Figure 1C). Additionally, miR-10b-3p, miR-328-3p, miR-100 and let-7 showed tight association with Wnt pathway (Table 2).
Table 2.
Detail KEGG pathway analysis of 4 miRNAs
| miRNA | Term | Count | p Value | Genes |
|---|---|---|---|---|
| mir-10b | hsa04310:Wnt signaling pathway | 3 | 0.0018 | CTNNBIP1, MAP3K7, CAMK2B |
| hsa04520:Adherens junction | 3 | 0.0058 | MAP3K7, ACTG1, SSX2IP | |
| hsa04722:Neurotrophin signaling pathway | 3 | 0.0131 | BDNF, PIK3CA, CAMK2B | |
| hsa04144:Endocytosis | 3 | 0.0242 | SMAP1, TFRC, NEDD4 | |
| let-7 | hsa04115:p53 signaling pathway | 4 | 0.0053 | CCND2, TP53, FAS, MDM4 |
| hsa04010:MAPK signaling pathway | 5 | 0.005 | TGFBR1, DUSP16, TP53, FAS, NGF | |
| hsa04310:Wnt signaling pathway | 3 | 0.0191 | CCND2, TP53, FZD3 | |
| hsa05210:Colorectal cancer | 3 | 0.0073 | TGFBR1, TP53, FZD3 | |
| hsa04210:Apoptosis | 3 | 0.0077 | TP53, FAS, NGF | |
| hsa05200:Pathways in cancer | 5 | 0.0092 | TGFBR1, TP53, FZD3, ZBTB16, FAS | |
| hsa04110:Cell cycle | 3 | 0.0141 | CCND2, TP53, CDC25A | |
| hsa04060:Cytokine-cytokine receptor interaction | 4 | 0.016 | TGFBR1, FAS, TNFSF9, CCL7 | |
| hsa04144:Endocytosis | 3 | 0.0256 | PARD6B, ADRB2, TGFBR1 | |
| miR-328 | hsa04310:Wnt signaling pathway | 3 | 0.0212 | CTNNBIP1, CHP2, TCF7L2 |
| hsa05221:Acute myeloid leukemia | 3 | 0.0043 | EIF4EBP1, PIM1, TCF7L2 | |
| hsa05412:Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 3 | 0.0069 | ITGA9, ITGA5, TCF7L2 | |
| hsa04810:Regulation of actin cytoskeleton | 3 | 0.0349 | ITGA9, ITGA5, WASF2 | |
| miR-100 | hsa05200:Pathways in cancer | 5 | 0.0017 | IGF1R, FZD8, FGFR3, MTOR, FZD5 |
| hsa05210:Colorectal cancer | 3 | 0.0028 | IGF1R, FZD8, FZD5 | |
| hsa04114:Oocyte meiosis | 3 | 0.0046 | IGF1R, PPP3CA, PPP1CB | |
| hsa04010:MAPK signaling pathway | 4 | 0.0048 | FGFR3, RASGRP3, TAOK1, PPP3CA | |
| hsa00534:Heparan sulfate biosynthesis | 2 | 0.0079 | HS3ST2, HS3ST3B1 | |
| hsa04310:Wnt signaling | 3 | 0.008 | FZD8, PPP3CA, FZD5 |
ROC curve analysis of four miRNAs in osteoporosis diagnosis
In order to explore the sensitivity and specificity of these four miRNAs in prediction of osteoporosis, we use ROC curve method to analyze the data using Prism software 5.0. Results showed that the sensitivity and specificity of miRNAs were quite high, specially, ROC area of let-7g-3p was up to 0.8944, with 95% confidence from 0.8146 to 0.9742 (Figure 2). Therefore, we could draw a conclusion that these four miRNAs are very potential for osteoporosis diagnosis, and further study is required to enlarge the sample size.
Figure 2.
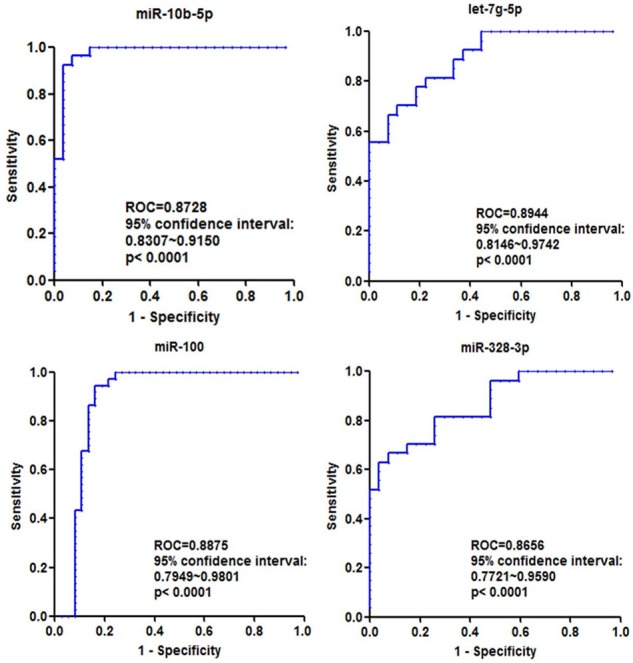
ROC curves analysis of each miRNAs to dichotomize healthy control and osteoporosis patient. The ROC area of patient related miRNAs are over 0.85, which are quite a sensitive and specific marker for osteoporosis diagnosis (all comparison are P<0.001).
miRNA mimics increase ALP activity and Alizarin red staining
We found miR-10b-5p which was up-regulated in osteoporotic patients could increase ALP activity in human and mouse osteoblast cells, indicating miR-10b-3p promoted osteoblast cell differentiation (Figure 3). miR-328-3p and let-7g-5p decreased ALP activity in both cell lines. The above miRNAs are involved in Wnt pathway, but it seems that their functions in cell differentiation are different. In order to make clear the role of three miRNAs in osteoblast differentiation, we explore if they can increase Ca2+ deposition in osteoblast cell which is a maturation marker of osteoblast cell. We used Alizarin red to stain the positive cell. Results showed miR-10b-5p increased over three fold positive staining compared with control miRNA (Figure 4A and 4B). miR-328-3p and let-7g-5p groups showed decreased Alizarin red staining (Figure 4A and 4B).
Figure 3.
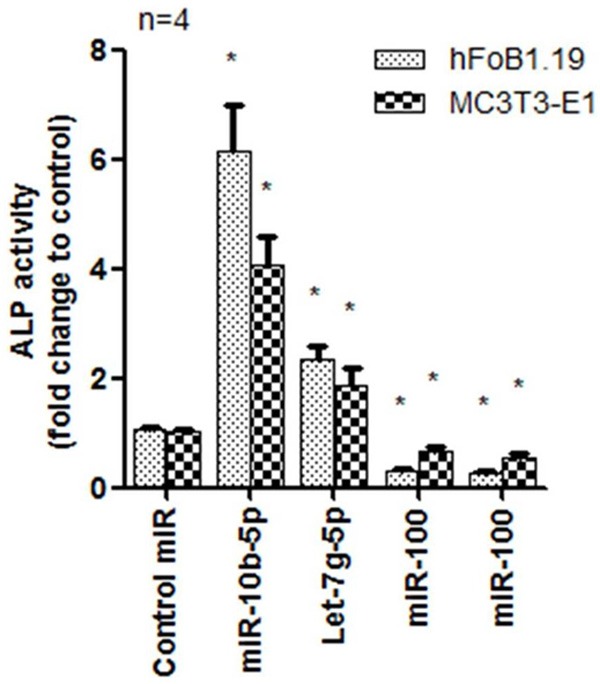
Detection of ALP activity in human and mouse osteoblast cells using miR-10b-3p, miR-328-3p and let-7g-5p. Data showed that miR-10b-3p promoted cell.
Figure 4.
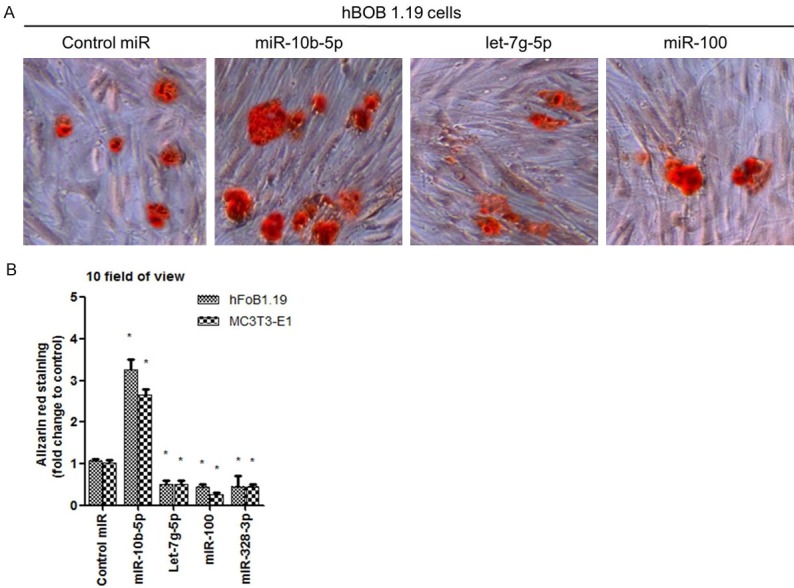
A: MiR-10b-5p increased osteoblast cell Ca2+ deposition. B: Shown in red staining), with over three fold increase when compared with control non-specific miRNA treatment. A and B: MiR-328-3p and let-7g-5p groups showed decreased Alizarin red staining.
Discussion
Many studies focused on circulating miRNAs in the aims of diagnostic biomarkers development [17,18]. Our findings showed the unique circulating miRNAs based on peers’ studies and further validated the function of interested miRNAs. In agreement with Weilner’s study [8] of let-7g-5p, miR-328, miR-127 et al., they are differentially expressed as potential biomarkers, but we showed miR-10b-5p, miR-22, and miR-100 which are reversed expressed, meet with Trompeter’ s study [19], and Zeng’s study [20]. In this aspect, we think osteoporotic patients need further detailed classification using statistic-based criteria. This study indicated the important role of aberrant miRNAs which could discriminate patients and healthy controls. The four miRNAs are associated with Wnt pathway which is vital in osteoblasts differentiation and maturation, especially bone homeostasis [21,22].
The newest study revealed that secreted miR-214 inhibited osteoblast activity [7], and showed that circulating miRNAs were capsulated in exosomes, which were a double layer lipid membrane particles. The study implies that miRNAs act as a message to deliver inhibitory signal form osteoclasts to osteoblasts, and such miRNAs was also a promising therapeutic target for osteoporosis [23]. Similarly, another study found mouse MC3T3-E1 cells secreted exosomes promoted bone marrow stromal cell (ST2) to differentiate to osteoblast [24]. Study showed that 1,25 D effect on human HOB cells was not restricted to classical VDR-mediated transcriptional responses but also involved in miRNA-directed posttranscriptional mechanisms [25]. But the study has not revealed if the osteoblast-secreted exosomes contained miRNAs. Study revealed a new cell-cell communication form using exosome materials. The secreted exosomes might benefit or damage bone mass based on the current point of view. Only have we made clear what was contained in the exosomes and the specific targets, the diagnostic and therapeutic miRNAs might come real. We found the diagnostic role of circulating miRNA, miR-10b-5p, miR-328, miR-100 and let-7 in osteoporotic patients, and characterized their function in osteoblast cell maturation. Increased miR-10b-5p stimulated human and mouse pre-osteoblast cells’ maturation. Therefore, we hypothesize that these circulating miRNAs might be also capsulated in exosomes secreted by osteoblast or osteoclasts. The idea needs further exploration and validation. Limited by the current findings, we did not have direct evidences to validate this idea. In future study, electron microscopy should be employed to visualize these miRNA-containing exosomes in serum, and to identify the specific exosomes secreted by osteoblasts or osteoclast.
Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) analysis automatically showed the related pathway of input genes. In this study, KEGG pathway was performed after potential regulated genes of miRNAs were predicted. Although the results from KEGG analysis were just possible data, it was meaningful and indicative for the subsequent study. Four miRNAs, miR-10b-5p, miR-328, miR-100 and let-7, showed significant association with Wnt pathway (P<0.001), which was regarded playing vital role in osteoblast maturation and bone mass formation [26]. The results remind us that circulating miRNAs have their specific function and targets. Therefore, we carried out the ALP activity assay and Ca2+ mineralization experiments in study, because these experiments indicate osteoblast maturation and bone mass formation. In peers’ studies, Thorfve and colleagues used KEGG analysis to identify Wnt pathway markers in Osteoarthritic Cartilage [27]; Wei et al. reported that they identified 33 risk pathways and enriched GWAS variants using KEGG analysis [28]; Gong et al. revealed Satb2-induced osteogenic differentiation finally, based on differentially regulated miRNAs using KEGG analysis [29]. KEGG pathway analysis, specially, for miRNAs potential targets, helps to achieve maximum results with little effort in the study.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Kanis JA, McCloskey EV, Johansson H, Cooper C, Rizzoli R, Reginster JY Scientific Advisory Board of the European Society for Clinical and Economic Aspects of Osteoporosis and Osteoarthritis (ESCEO) and the Committee of Scientific Advisors of the International Osteoporosis Foundation (IOF) European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int. 2013;24:23–57. doi: 10.1007/s00198-012-2074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hackl M, Heilmeier U, Weilner S, Grillari J. Circulating microRNAs as novel biomarkers for bone diseases-complex signatures for multifactorial diseases? Mol Cell Endocrinol. 2016;432:83–95. doi: 10.1016/j.mce.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Papaioannou G, Mirzamohammadi F, Kobayashi T. MicroRNAs involved in bone formation. Cell Mol Life Sci. 2014;71:4747–4761. doi: 10.1007/s00018-014-1700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res. 2011;26:72–78. doi: 10.1002/jbmr.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, Liu W, Sinha KM, Yasuda H, de Crombrugghe B. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS One. 2013;8:e58104. doi: 10.1371/journal.pone.0058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W, Zhao C, Li Y, Wang L, Nie G, Peng J, Wang A, Zhang P, Tian W, Li Q, Song J, Wang C, Xu X, Tian Y, Zhao D, Xu Z, Zhong G, Han B, Ling S, Chang YZ, Li Y. Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity. Cell Discov. 2016;2:16015. doi: 10.1038/celldisc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weilner S, Skalicky S, Salzer B, Keider V, Wagner M, Hildner F, Gabriel C, Dovjak P, Pietschmann P, Grillari-Voglauer R, Grillari J, Hackl M. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone. 2015;79:43–51. doi: 10.1016/j.bone.2015.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H, Fan GC. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis. 2011;1:138–149. [PMC free article] [PubMed] [Google Scholar]
- 10.Seeliger C, Karpinski K, Haug AT, Vester H, Schmitt A, Bauer JS, van Griensven M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res. 2014;29:1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 11.Nakasa T, Yoshizuka M, Andry Usman M, Elbadry Mahmoud E, Ochi M. MicroRNAs and bone regeneration. Curr Genomics. 2015;16:441–452. doi: 10.2174/1389202916666150817213630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Sun Z, Wang Y, Hu Z, Zhou H, Zhang L, Hong B, Zhang S, Cao X. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci Rep. 2016;6:23170. doi: 10.1038/srep23170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Meng H, Wang X, Zhao C, Peng J, Wang Y. Differentiation of bone marrow mesenchymal stem cells in osteoblasts and adipocytes and its role in treatment of osteoporosis. Med Sci Monit. 2016;22:226–233. doi: 10.12659/MSM.897044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Wei G, Gu Q, Wang Q, Tao S, Xu L. Proliferation and differentiation of rat osteoporosis mesenchymal stem cells (MSCs) after telomerase reverse transcriptase (TERT) transfection. Med Sci Monit. 2015;21:845–854. doi: 10.12659/MSM.893144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gimondi S, Dugo M, Vendramin A, Bermema A, Biancon G, Cavane A, Corradini P, Carniti C. Circulating miRNA panel for prediction of acute graft-versus-host disease in lymphoma patients undergoing matched unrelated hematopoietic stem cell transplantation. Exp Hematol. 2016;44:624–634.e1. doi: 10.1016/j.exphem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Garnero P. New developments in biological markers of bone metabolism in osteoporosis. Bone. 2014;66:46–55. doi: 10.1016/j.bone.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao D, Chen G, Li D, Wang X, Cao H, Xie Y, Liang Z. A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res Treat. 2015;154:423–434. doi: 10.1007/s10549-015-3591-0. [DOI] [PubMed] [Google Scholar]
- 19.Trompeter HI, Dreesen J, Hermann E, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Wernet P. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics. 2013;14:111. doi: 10.1186/1471-2164-14-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Y, Qu X, Li H, Huang S, Wang S, Xu Q, Lin R, Han Q, Li J, Zhao RC. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett. 2012;586:2375–2381. doi: 10.1016/j.febslet.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 21.Rauner M, Rachner TD, Hofbauer LC. Bone formation and the Wnt signaling pathway. N Engl J Med. 2016;375:1902. doi: 10.1056/NEJMc1609768. [DOI] [PubMed] [Google Scholar]
- 22.Xie W, Ji L, Zhao T, Gao P. Identification of transcriptional factors and key genes in primary osteoporosis by DNA microarray. Med Sci Monit. 2015;21:1333–1344. doi: 10.12659/MSM.894111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Zhao J, Liu Q, Xiong X, Zhang Z, Jiao Y, Li X, Liu B, Li Y, Lu Y. MicroRNA-124 promotes hepatic triglyceride accumulation through targeting tribbles homolog 3. Sci Rep. 2016;6:37170. doi: 10.1038/srep37170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Y, Luan J, Li H, Zhou X, Han J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016;590:185–192. doi: 10.1002/1873-3468.12024. [DOI] [PubMed] [Google Scholar]
- 25.Lisse TS, Chun RF, Rieger S, Adams JS, Hewison M. Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J Bone Miner Res. 2013;28:1478–1488. doi: 10.1002/jbmr.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karner CM, Long F. Wnt signaling and cellular metabolism in osteoblasts. Cell Mol Life Sci. 2017;74:1649–1657. doi: 10.1007/s00018-016-2425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorfve A, Dehne T, Lindahl A, Brittberg M, Pruss A, Ringe J, Sittinger M, Karlsson C. Characteristic markers of the WNT signaling pathways are differentially expressed in osteoarthritic cartilage. Cartilage. 2012;3:43–57. doi: 10.1177/1947603511414178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei J, Li M, Gao F, Zeng R, Liu G, Li K. Multiple analyses of large-scale genome-wide association study highlight new risk pathways in lumbar spine bone mineral density. Oncotarget. 2016;7:31429–31439. doi: 10.18632/oncotarget.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong Y, Xu F, Zhang L, Qian Y, Chen J, Huang H, Yu Y. MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol Cell Biochem. 2014;387:227–239. doi: 10.1007/s11010-013-1888-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


