Abstract
As high fetal hemoglobin levels ameliorate the underlying pathophysiological defects in sickle cell anemia and beta (β)-thalassemia, understanding the mechanisms that enforce silencing of fetal hemoglobin postnatally offers the promise of effective molecular therapy. Depletion of the Nucleosome Remodeling and Deacetylase complex member MBD2 causes a 10-20-fold increase in γ-globin gene expression in adult β-globin locus yeast artificial chromosome transgenic mice. To determine the effect of MBD2 depletion in human erythroid cells, genome editing technology was utilized to knockout MBD2 in Human Umbilical cord Derived Erythroid Progenitor-2 cells resulting in γ/γ+β mRNA levels of approximately 50% and approximately 40% fetal hemoglobin by high performance liquid chromatography. In contrast, MBD3 knockout had no appreciable effect on γ-globin expression. Knockdown of MBD2 in primary adult erythroid cells consistently increased γ/γ+β mRNA ratios by approximately 10-fold resulting in approximately 30-40% γ/γ+β mRNA levels and a corresponding increase in γ-globin protein. MBD2 exerts its repressive effects through recruitment of the chromatin remodeler CHD4 via a coiled-coil domain, and the histone deacetylase core complex via an intrinsically disordered region. Enforced expression of wild-type MBD2 in MBD2 knockout cells caused a 5-fold decrease in γ-globin mRNA while neither the coiled-coil mutant nor the intrinsically disordered region mutant MBD2 proteins had an inhibitory effect. Co-immunoprecipitation assays showed that the coiled-coil and intrinsically disorder region mutations disrupt complex formation by dissociating the CHD4 and the histone deacetylase core complex components, respectively. These results establish the MBD2 Nucleosome Remodeling and Deacetylase complex as a major silencer of fetal hemoglobin in human erythroid cells and point to the coiled-coil and intrinsically disordered region of MBD2 as potential therapeutic targets.
Introduction
Both sickle cell disease (SCD) and beta (β)-thalassemia result from genetic defects in β-globin production. SCD, which results from a single glutamic acid to valine substitution in the β-globin chain, is the most common inherited blood disorder in the US, affecting approximately 100,000 Americans, as well as millions of people worldwide, most of whom live in underdeveloped nations.1,2 The vascular sequelae of SCD lead to a shortened and reduced quality of life. Current treatments for SCD are primarily supportive. Hydroxyurea and L-glutamine are the only standard agents available that reduce the frequency of sickle cell crises. β-thalassemia major resulting from insufficient β-globin production has a high prevalence worldwide3 and has limited treatment options, with most patients remaining transfusion-dependent throughout life. The only curative treatment for either SCD or β-thalassemia is stem cell transplantation,4 which carries significant risks and is not readily accessible in developing nations. Thus new treatment options are needed. Importantly, sufficient levels of fetal hemoglobin (HbF) ameliorate the underlying pathophysiological defects in β-thalassemia5,6 and SCD.1,7 Studies aimed at a full understanding of the mechanisms that enforce silencing of HbF expression in adult erythroid cells offer the promise of effective targeted molecular therapy.
During development, humans undergo a progressive switch from embryonic ε (Hb Gower-1, Hb Gower-2) to fetal γ (HbF) and finally to adult β (HbA) and δ (HbA2) type globin production. By adulthood, γ -globin typically makes up approximately 1-2% of total β-like globin chains in hemoglobin.8 Numerous transcriptional and epigenetic regulators of γ-globin expression have been shown to mediate γ-globin gene silencing, including BCL11A, KLF1/EKLF, LRF/Pokemon, MBD2-NuRD, and LSD-1, among others.9–16 The zinc finger transcription factors BCL11A and LRF have been shown to independently exert especially strong silencing of the γ-globin gene in an immortalized Human Umbilical cord Derived Erythroid Progenitor-2 (HUDEP-2) cell line that displays an adult erythroid phenotype.13,17
In addition to transcription factors, epigenetic mechanisms, including DNA methylation and histone modifications,12,18–23 are of importance in developmental globin gene regulation. MBD2, a member of the methyl-CpG binding domain (MBD) protein family that includes MeCP2, MBD1, MBD2, MBD3, and MBD4, binds to DNA containing methylated CpG rich sequences with high affinity and recruits other members of the Nucleosome Remodeling and Deacetylase (NuRD) co-repressor complex through specific protein-protein interactions.24–28 The NuRD co-repressor complex, classically made up of one or more of at least six core proteins, including MBD2/3, CHD3/4, HDAC1/2, MTA1/2/3, RBBP4/7, and GATAD2A/B is unique in containing both an ATPase chromatin remodeling complex and a histone deacetylase complex (HDCC).29–31 Previous work by our group has shown that depletion of MBD2 or disruption of NuRD complex components abrogates silencing of fetal hemoglobin in multiple mammalian erythroid model systems.9,27,32
MBD2 interacts with GATAD2A and in turn CHD4 through a C-terminal coiled-coil (CC) motif and enforced expression of a GATAD2A CC domain inhibitory peptide abrogates the interaction of MBD2 with GATAD2A/CHD4 and partially relieves γ-globin gene silencing in β-YAC bearing murine CID cells.27 More recently we have shown the functional importance of an intrinsically disordered region (IDR) within MBD2 for recruitment of the HDAC core of the NuRD complex to silence a highly methylated tumor suppressor gene in breast cancer cells.25
Unlike MBD2, MBD3 shows greatly reduced selectivity for methylated DNA. Additionally, MBD2 and MBD3 are mutually exclusive within the same NuRD complex.33 Previous to this report, the precise role of MBD3-NuRD on γ-globin gene repression had been less clearly defined.34,35
Here we show that MBD2 is among the strongest repressors of HbF expression in the adult erythroid phenotype HUDEP-2 cell line, as knockout (KO) of MBD2 results in ~50% γ/γ+β mRNA compared to <1% in controls, while KO of MBD3 in HUDEP-2 cells has no appreciable effect on γ-globin gene expression. We also establish the functional importance of the CC domain and the IDR of MBD2 for γ-globin silencing in human erythroid cells. Together these data solidify MBD2’s role as a major silencer of HbF and suggest novel strategies for targeting MBD2-NuRD in the β-type hemoglobinopathies.
Methods
Isolation and maturation of human CD34+ cells
Human CD34+ cells were purified from deidentified apheresis units discarded by the VCU Bone Marrow Transplant Unit, and therefore Institutional Review Board exempt. CD34+ cells were isolated using the EasySep Human CD34 Positive Selection Kit (StemCell Technologies Inc.) as described previously.9,27 Erythroid differentiation and maturation were monitored by measuring expression of the erythroid lineage markers CD235a and CD71 via flow cytometry after expansion and differentiation (Online Supplementary Methods).
HUDEP-2 cell culture and erythroid differentiation
HUDEP-2, an immortalized human erythroid progenitor cell line, was a kind gift from Dr. Yukio Nakamura.17 Expansion and differentiation protocols for HUDEP-2 cells have been previously described17 and are detailed in the Online Supplementary Methods.
Genome editing-mediated depletion of MBD2/MBD3 in HUDEP-2 cells
sgRNA sequences targeting MBD2 or MBD3 were cloned into a LentiCRISPR-AcGFP backbone, packaged, and transduced (MOI=40) into HUDEP-2 cells. sgRNA sequences and cloning protocols are detailed in the Online Supplementary Methods. For stable depletion of MBD2 or MBD3 in HUDEP-2 cells, single cell colonies were isolated by limiting dilution and screened by western blotting. After three weeks of clonal expansion, three bi-allelic MBD2KO clones and five bi-allelic MBD3KO clones were expanded and analyzed individually as well as in pools.
Lentiviral-mediated “Add-back” of MBD2 in MBD2 null cells
pLV203 vectors containing sequences encoding MBD2sgR, IDRmutsgR, or CCmutsgR were packaged as described previously25,27 and used to infect MBD2KO HUDEP-2 cells. MBD2 mutant sequences are provided (Online Supplementary Appendix and Online Supplementary Figures S7-S10). Translationally silent mutations were introduced into the MBD2 expression constructs to confer resistance to MBD2 shRNA and CRISPR/Cas9 sgRNA. The assessment of exogenous MBD2 expression in HUDEP-2 MBD2KO cells was carried out by western immunoblotting five days post lentiviral infection as described.27
Hemoglobin high performance liquid chromatography
Hemolysates were prepared from scramble sgRNA or MBD2KO HUDEP-2 cells (4 × 107 cells) on day 7 of differentiation. High performance liquid chromatography (HPLC) analysis was conducted in the VCU Health System clinical lab using a Clinical Laboratory Improvement Amendments certified protocol with standard controls.
Nuclear magnetic resonance
Uniform 13C, 15N labeled wild-type and mutant (R286E/L287A) MBD2 IDR were expressed and purified as described previously.25 The purified proteins were buffer exchanged into nuclear magnetic resonance (NMR) buffer (10 mM NaPO4, pH 6.0, 0.02% sodium azide, 1 mM dithiothreitol, and 10% 2H2O, 0.5-1 mM protein) and spectra collected on a Bruker Avance III 700 MHz instrument at 25°C. Assignments for the wild-type (WT) protein at pH 6.5, reported previously,25 were extended to the WT and mutant samples at pH 6.0 using standard double and triple resonance experiments [15N-HSQC, HNCO, HNCACB, HBHA(CO)NH, 15N-NOESY-HSQC], which were processed and analyzed with NMRPipe36 and CcpNmr,37 respectively.
Statistical analysis and data sharing
All experiments were carried out in at least three independent biological repeats. Statistical significance between groups within experiments were determined as described in the figure legends. Sequencing data are available at the NCBI Gene Expression Omnibus (GEO accession number: GSE121992).
Results
Depletion of MBD2 greatly increases levels of fetal hemoglobin production in HUDEP-2 cells
The effect of MBD2 KO on γ-globin gene expression has previously been described in human β-YAC transgenic mice;32 however, mice differ considerably from humans in developmental regulation of the β-type globin genes in that they lack a direct homolog to human fetal γ-globin. Recently, a human immortalized HUDEP-2 cell line was generated through doxycycline-inducible expression of HPV E6/E7.17 With a β/γ-globin expression profile of <1% γ-globin and >95% β-globin, very similar to adult erythroid cells, the HUDEP-2 line has become a useful model system for studying globin switching.11,13,38
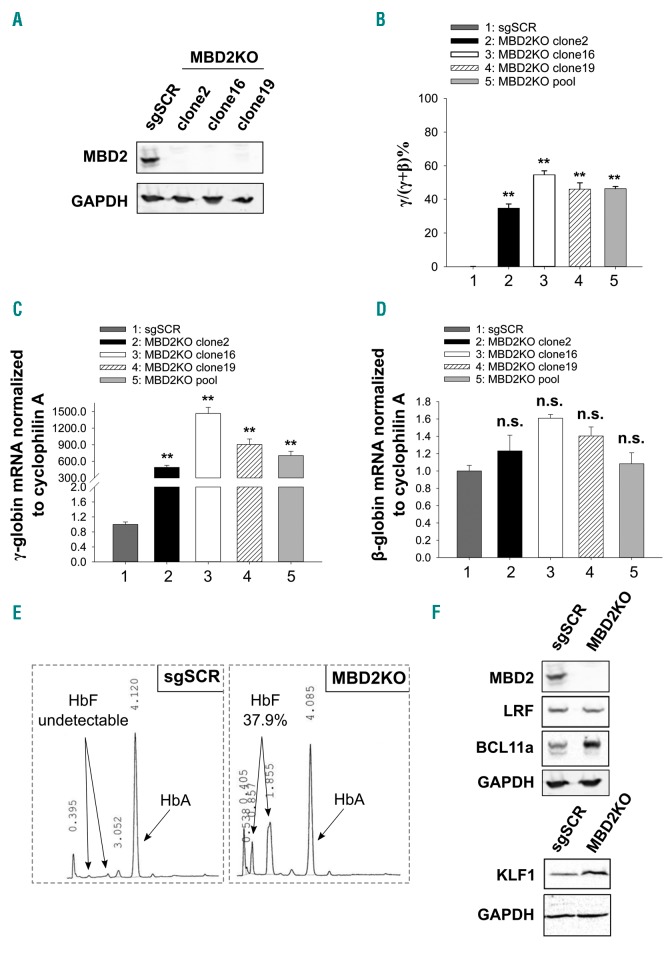
Based on the 10-20-fold increase in γ-globin gene expression in β-YAC mice lacking MBD2,32 we hypothesized that there would be a similar effect in HUDEP-2 cells. To test this and to compare the effect of MBD2 KO to that observed with other strong γ-globin silencers, we utilized CRISPR/Cas9 genome editing to biallelically knock out MBD2 in HUDEP-2 cells using two independent MBD2 sgRNA sequences and a scrambled control sgRNA guide. Three independent MBD2 KO clones were generated and absence of MBD2 was confirmed by western blot (Figure 1A). These MBD2KO clones were then analyzed individually and as pools to control for off-target CRISPR effects. Knockout (KO) of MBD2 in HUDEP-2 cells resulted in 40-55% γ/γ+β mRNA compared to approximately 0.15% γ/γ+β mRNA in the scramble sgRNA controls (Figure 1B), and high relative γ-globin mRNA levels (Figure 1C) comparable to those seen in BCL11A and LRF KO HUDEP-2 cells.13 None of the MBD2KO clones or the pooled population showed significantly different expression of β-globin mRNA (Figure 1D). We observed greatly increased γ-globin protein expression with little change in β-globin, consistent with their respective RNA levels for each individual clone (Online Supplementary Figure S1A). MBD2KO HUDEP-2 cells made approximately 38% HbF protein compared to undetectable levels of HbF in the scramble control as measured by HPLC (Figure 1E). We wished to investigate whether knockout of MBD2 had any deleterious effects on the ability of HUDEP-2 cells to differentiate, such as a differentiation block or a shift to an earlier stage of erythroid differentiation. To address this question, we performed RNA-sequencing of MBD2KO and scrambled sgRNA control HUDEP-2 cells before and after erythroid differentiation and compared the differential expression of 15 marker genes of erythroid differentiation, as described in the Online Supplementary Methods. Interestingly, MBD2KO cells demonstrate a pattern of gene expression consistent with a later stage of erythroid differentiation compared to scramble control cells, with significantly higher levels of GYPA, SLC4A1, ALAS2, EPB42, SPTA1, FECH, EPOR, and UROS, and significantly lower levels of CD44 (Online Supplementary Tables S2 and S3 and Online Supplementary Figure S2). Additionally, MBD2KO cells have an unaltered morphological appearance after differentiation compared to controls (Online Supplementary Figures S3 and S4). To determine whether MBD2 silences γ-globin by regulating expression of known potent silencing factors, we measured protein levels of LRF, BCL11A, and KLF1 in MBD2KO HUDEP-2 cells. KO of MBD2 did not change expression of LRF, and actually increased expression of both BCL11A and KLF1 (Figure 1F), demonstrating that MBD2 is not silencing γ-globin through regulation of these factors. Together these results indicate that MBD2 is among the most potent known repressors of γ-globin in HUDEP-2 cells.
Figure 1.
CRISPR Cas9 mediated knockout of MBD2 in HUDEP-2 cells results in approximately 50% γ/γ+β RNA expression and proportionally increased the fold effect on fetal hemoglobin (HbF) by high performance liquid chromatography (HPLC). (A) Western blot showing complete depletion of MBD2 protein in three independent clonal MBD2KO HUDEP-2 cell lines. (B-D) Levels of globin gene mRNA expression by qualitative polymerase chain reaction in the three clonal MBD2KO and pooled MBD2KO HUDEP-2 cell lines compared to scrambled guide RNA (sgSCR) controls. (B) γ-globin mRNA as a percentage of total globin (γ/γ+β%) mRNA. (C) Relative γ-globin expression compared to sgSCR. (D) Relative β-globin expression compared to sgSCR (E) Approximately 40-fold increase in HbF as measured by HPLC in MBD2KO HUDEP-2 cells compared to scramble. Note that a monoallelic polymorphism in the γ-globin gene of HUDEP-2 cells results in two distinct HbF HPLC peaks, consistent with published data.17 (F) Western blot showing protein levels of established γ-globin gene silencers (LRF, BCL11A and KLF1) in MBD2KO HUDEP-2 cells compared to sgSCR control cells. Error bars represent ± Standard Deviation of three biological repeats. *P<0.05; **P<0.01; n.s.: P>0.05 between sample and scramble. Statistical testing was performed using analysis of variance followed by the Tukey’s honestly significant difference procedure post-hoc test.
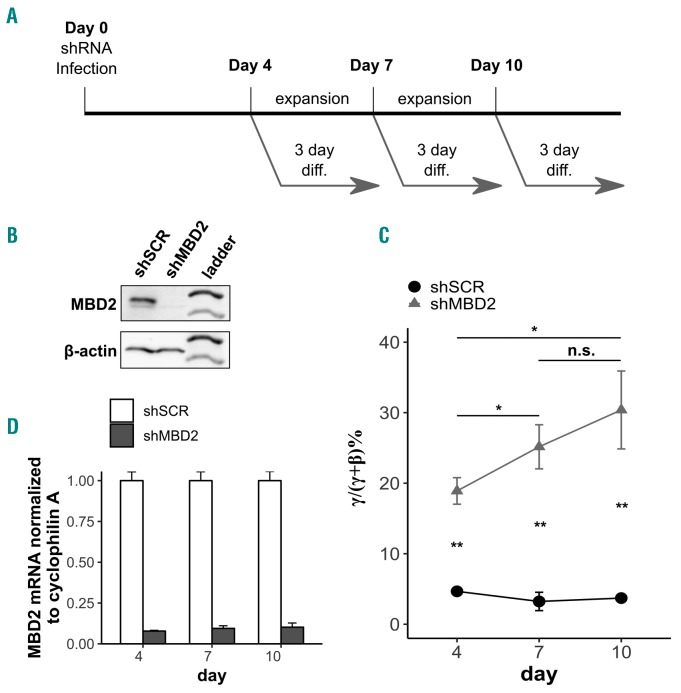
To test the effect of partial depletion of MBD2 in this model, we performed lentiviral shRNA Knockdown (Kd) in HUDEP-2 cells, and quantified the levels of β and γ-globin expression by quantitative polymerase chain reaction (qPCR). Cells were first transduced with shRNA lentiviral constructs and then expanded for 4, 7, or 10 days prior to the erythroid differentiation protocol to investigate whether there is a time-dependent response to MBD2 depletion (Figure 2A). Levels of MBD2 RNA knockdown were approximately 80-90% compared to scramble control cells across all three expansion periods with a comparable response at the protein level at day 4 of expansion (Figure 2B and D). The %γ/γ+β was significantly higher in the MBD2 Kd samples compared to scramble controls across all samples. Interestingly, in the day 10 MBD2 Kd sample, γ-globin gene induction (30% γ/γ+β) was significantly higher than the day 4 MBD2 Kd sample (19% γ/γ+β), and the level in the day 7 sample was intermediate at approximately 25% γ/γ+β, compared to no change in γ-globin gene expression in the scramble shRNA controls across the three time points (Figure 2C). There was a stepwise increase in relative γ-globin mRNA expression from day 4 to day 10 of expansion (Online Supplementary Figure S5A) with no significant change in β-globin mRNA (Online Supplementary Figure S5B).
Figure 2.
Lentiviral knockdown of MBD2 in HUDEP-2 cells results in progressively increased γ-globin gene expression. (A) Schema of HUDEP-2 cell expansion for four, seven, or ten days after MBD2 shRNA lentiviral transduction, prior to a 3-day differentiation period. mRNA and protein were harvested after the expansion and differentiation period for all samples. (B) Western blot showing qualitative degree of MBD2 protein knockdown at the day-4 time point. (C) Time-course plot of γ-globin mRNA as a percentage of total globin mRNA by qRT-PCR at the indicated time points. (D) Relative MBD2 mRNA expression normalized by cyclophilin A compared to the shSCR sample by quantitative real-time polymerase chain reaction at four, seven, and ten-day time points. Error bars represent ± Standard Deviation of three biological repeats. *P<0.05; **P<0.01; n.s.: P>0.05 between sample and scramble. Statistical testing was performed using the Student’s t-test.
MBD3-NuRD does not mediate γ-globin gene silencing in HUDEP-2 cells
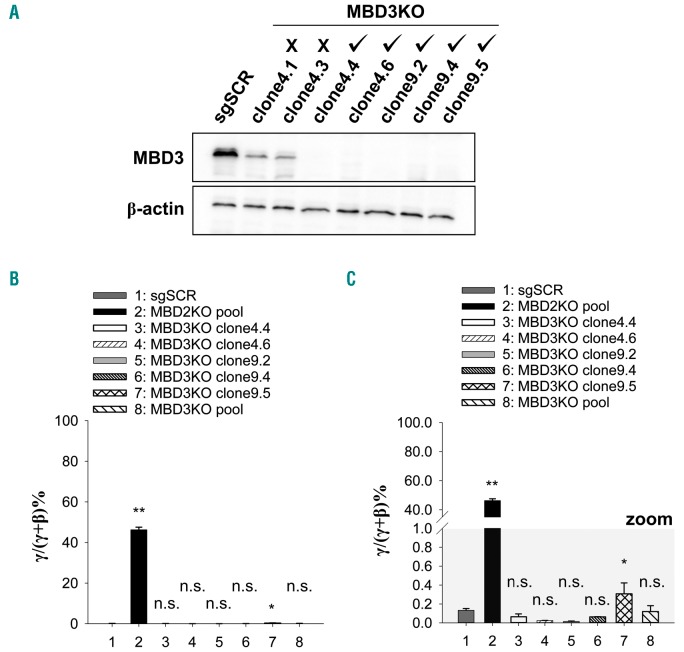
MBD3-NuRD has been biochemically associated with some γ-globin gene silencers.39,40 MBD3-NuRD was also associated with γ-globin gene silencing in β-YAC transgenic mice in some studies but not others.34,35 We previously observed no effect on γ-globin gene expression after approximately 75% siRNA Kd of MBD3 in CID-dependent β-YAC containing murine bone marrow progenitor cells.27 As we could not achieve sufficient Kd of MBD3 in human erythroid cell model systems, we utilized CRISPR/Cas9 genomic editing to genetically knock out MBD3 in HUDEP-2 cells using two independent guide RNA targeting exon 3 and exon 5 of human MBD3, isolating five independent clones with complete knockout of MBD3 as confirmed by western blot (Figure 3A). These five clones were then pooled to control for off-target effects. In stark contrast to KO of MBD2, four out of five MBD3KO clones and the pooled MBD3KO HUDEP-2 line showed no increase in γ-globin mRNA as a percent of total globin mRNA compared to scrambled guide RNA control cells (Figure 3B and C) and no difference in relative γ-globin mRNA (Online Supplementary Figure S6A). MBD3KO clone9.5 showed a minimal but statistically significant increase to 0.31% γ/γ+β compared to 0.13% in the sgSCR population with a correlative increase in relative γ-globin mRNA level (Figure 3C and Online Supplementary Figure S6A); however, these effects were very small and inconsistent compared to those for MBD2KO cells. Importantly, we observed no detectable increase in gglo-bin protein by western blot compared to scramble for any MBD3KO clone, while the MBD2KO pooled line showed robust γ-globin protein expression (Online Supplementary Figure S1B). Relative β-globin mRNA levels trended slightly higher in MBD3KO cells compared to scramble controls (Online Supplementary Figure S6B); this was reflected at the protein level (Online Supplementary Figure S1B). These results provide strong evidence that MBD3-NuRD is not an important mediator of γ-globin gene silencing in human erythroid cells.
Figure 3.
CRISPR/Cas9 mediated knockout of MBD3 in HUDEP-2 cells has no effect on γ-globin gene expression. (A) Western blot showing complete depletion of MBD3 protein in 5 out of 7 independent clonal MBD3KO HUDEP-2 cell lines, (B and C) Plots of γ-globin mRNA as a percentage of total globin mRNA by qRT-PCR in the pooled MBD2KO, five clonal MBD3KO, and pooled MBD3KO HUDEP-2 cell lines compared to scrambled guide RNA (sgSCR) controls. (B) y-axis is continuous from 0 to 100% γ/γ+β. (C) The same data with a break in the y-axis and zoomed in to more clearly show sgSCR and MBD3KO data points. Error bars represent ± Standard Deviation of three biological repeats. *P<0.05; **P<0.01; n.s.: P>0.05 between sample and scramble. Statistical testing was performed using analysis of variance followed by the Tukey’s honestly significant difference procedure post-hoc test.
Amino acid substitutions in the intrinsically disordered region and C terminal coiled-coil domains of MBD2 that disrupt MBD2-NuRD component interactions prevent γ-globin gene repression in HUDEP-2 cells
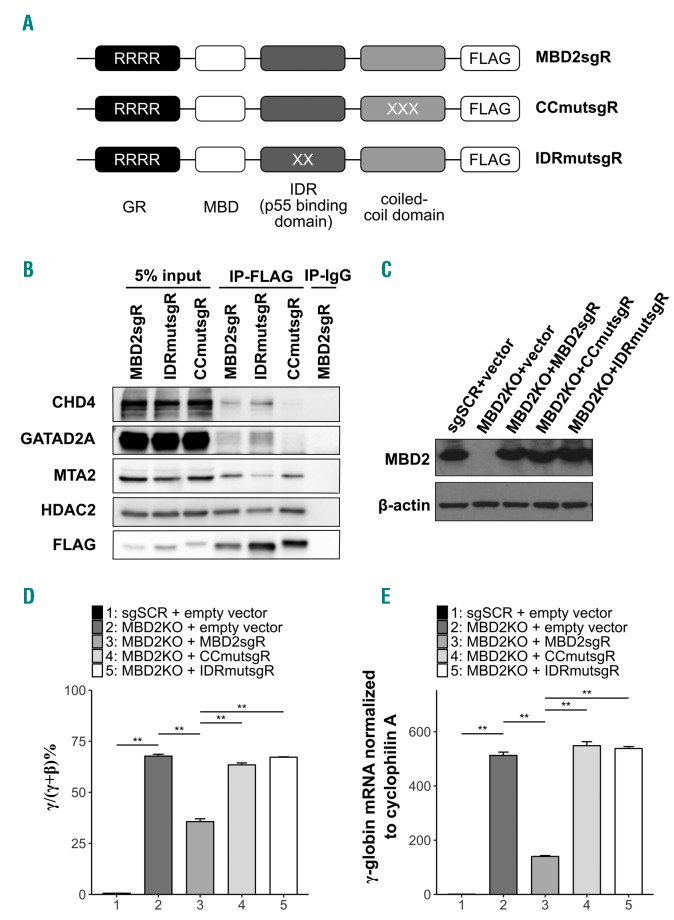
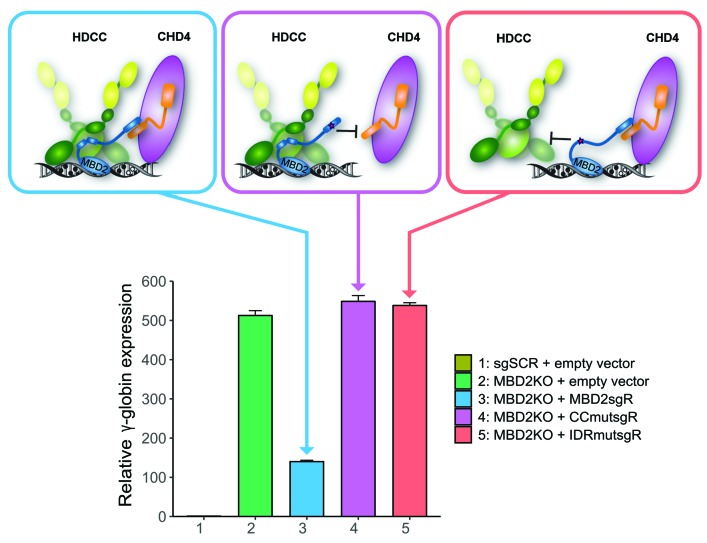
The coiled-coil (CC) domains of MBD2 and GATAD2A interact to form a stable heterodimeric complex, and this interaction is necessary for the recruitment of GATAD2A and CHD4 to the MBD2-NuRD complex.27 By determining the structure and binding dynamics between MBD2 and GATAD2A, we identified three critical charged residues in the CC region of MBD2 that when mutated disrupt binding to GATAD2A (D366R/R375E/R380E).41 We have previously demonstrated that two sequential amino acid substitutions (R286E/L287A) in the intrinsically disordered region (IDR) of MBD2 are sufficient to disrupt the ability of MBD2 to pull down the HDAC core complex (HDCC) consisting of MTA2, RBBP4/7, and HDAC2, collectively in 293T cells.25 In order to study the functional importance of these domains and interactions in human erythroid cells, we tested the effect of enforced expression of WT or mutant MBD2 in MBD2KO HUDEP-2 cells. For these studies, three lentiviral MBD2 expression vectors were engineered. ‘MBD2sgR’ is a WT MBD2 construct with translationally silent mutations in the GR domain designed to convey resistance to cleavage by Cas9 (denoted as the sgR mutation). The ‘CCmutsgR’ construct contains the sgR mutation along with three amino acid substitutions (D366R/R375E/R380E) in the CC domain of MBD2. ‘IDRmutsgR’ contains both the sgR mutation and two sequential amino acid substitutions (R286E/L287A) in the IDR of MBD2 (Figure 4A and Online Supplementary Figures S7-S10). MBD2KO HUDEP-2 cells were infected with each of these constructs. The level of exogenously expressed MBD2 proteins was closely matched with endogenous MBD2 expression seen in the scrambled guide (sgSCR + empty vector) control cells (Figure 4C). When WT MBD2 (MBD2sgR) was added back to the MBD2KO HUDEP-2 cells, there was a 5-fold reduction in relative γ-globin expression and a decrease in γ/γ+β mRNA level compared to the MBD2KO + empty vector control (Figure 4D and E). This corresponds to a significant rescue of the WT MBD2 phenotype compared to the empty vector control. In contrast, neither the CC or IDR mutant restore γ-globin gene silencing. In order to determine whether the CC mutation and IDR mutation selectively cause dissociation of the CHD4 and HDCC subcomponents, or disrupt the entire complex, we tested the ability of these mutants to pull down NuRD components by performing co-immunoprecipitation in 293T cells. The CCmutsgR construct pulled down almost no GATAD2A or CHD4, but did pull down similar amounts of MTA2 and HDAC2 compared to the MBD2sgR construct. Conversely, the IDRmutsgR construct pulled down less MTA2 and HDAC2 compared to the MBD2sgR construct, but did pull down similar levels of CHD4 and GATAD2A (Figure 4B), consistent with our structural predictions. Together these data provide strong evidence that perturbation of either of these two interaction domains is sufficient to functionally diminish MBD2-NuRD mediated γ-globin gene silencing by independently decoupling one of the NuRD subcomplexes.
Figure 4.
Enforced expression of wild-type (WT) MBD2 (MBD2sgR) but not MBD2 containing mutations in its IDR or coiled-coil domain suppresses gamma globin RNA expression in MBD2 knockout HUDEP-2 cells. (A) Schematic depicting domains mutated in MBD2 lentiviral expression vectors. Silent mutations in the GR domain (sgR) convey resistance to CRISPR/Cas9 cleavage. (B) Co-IP of exogenously expressed FLAG-tagged MBD2 mutant contructs in 293T cells using anti-FLAG shows differing abilities to pull down NuRD members CHD4, GATAD2A, MTA2, and HDAC2. (C) Western blot showing enforced expression levels of WT MBD2sgR, and CCmutsgR, and IDRmutsgR MBD2 mutants in MBD2KO HUDEP-2 cells compared to scramble control cell levels of MBD2. (D and E) MBD2 knockout HUDEP-2 cells with enforced expression of WT MBD2 (MBD2KO+MBD2sgR) but not IDR-mutant (IDRmutsgR) or CC-mutant (CCmutsgR), causes decreased γ/γ+β and relative γ-globin mRNA. Error bars represent ± Standard Deviation of three biological repeats. *P<0.05; **P<0.01; n.s.: P>0.05. Statistical testing was performed using analysis of variance followed by the Tukey’s honestly significant difference procedure post-hoc test.
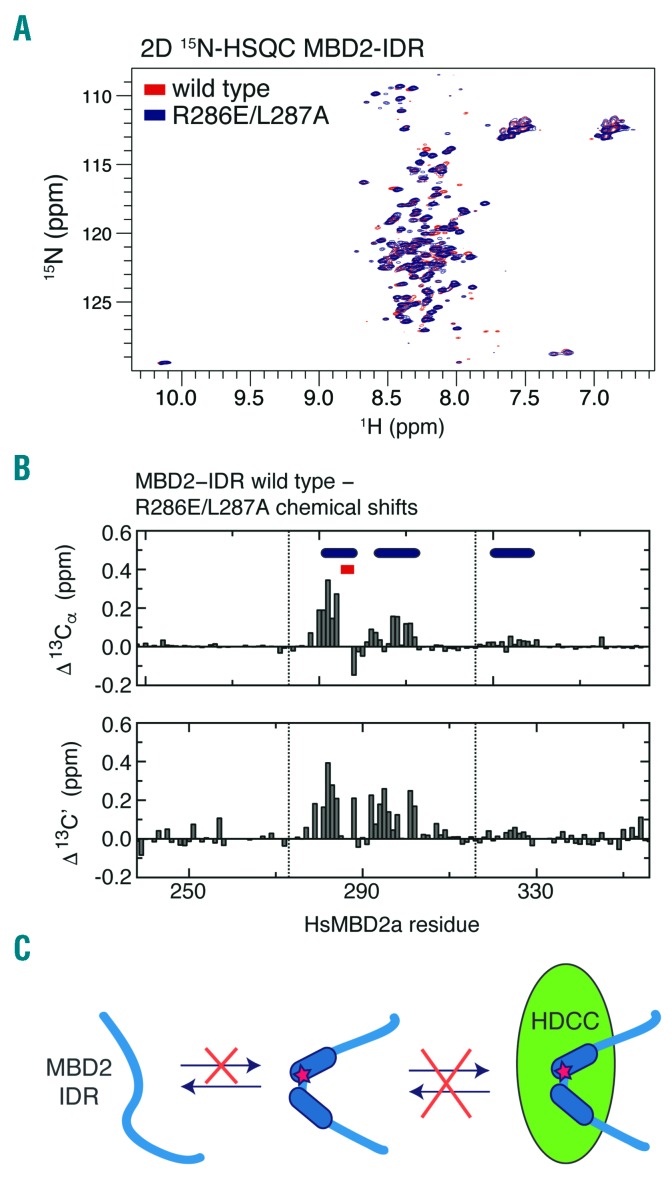
The R286E/L287 mutation of the MBD2 intrinsically disordered region disrupts helical propensity
Based on previous structural analyses,27 the CC mutations involve residues that form critical interactions with GATAD2A. However, the structure of the IDR bound to the HDCC has yet to be determined, and it is unclear whether the IDR mutations involve residues that make direct contact with HDCC components or if they reduce the structural propensity of the IDR and thereby indirectly disrupt binding. We previously demonstrated, based on NMR chemical shift analyses, that the IDR contains three regions with inherent helical propensity.25 The double mutation occurs within the first of these regions, as highlighted in Figure 5B. To test whether this mutation disrupts the helical propensity of the IDR, we assigned the NMR resonances for the mutant domain and compared 13Ca and 13C’ chemical shifts with the WT IDR. The 15N-HSQC spectrum of the mutant IDR is nearly identical to that of the WT (Figure 5A). Differences in 13Ca and 13C’ chemical shifts between WT and mutant IDR (effectively the difference in chemical shift index) are plotted in Figure 5B. This analysis reveals a positive deviation in chemical shifts (wild-type – mutant) throughout the first two helical regions, showing that the R286E/L287A mutation disrupts the structural propensity of the IDR, thereby reducing its ability to bind the HDCC (Figure 5C).
Figure 5.
The R286E/L287A double mutation reduces the helical propensity of the intrinsically disordered region (IDR). (A) Wild-type (WT) and mutant spectra. An overlay of the 2D 15N-HSQC spectra is shown for the WT (red) and R286E/L287A (blue) MBD2-IDR. (B) 13C chemical shift changes. Bar graphs depict the differences in chemical shift of the carbonyl (C’) and for a-carbons (Cα) between WT and R286E/L287A MBD2-IDR (WT – mutant in ppm). Three regions that show helical propensity are indicated with blue ellipses and the site of mutation indicated with red squares. Positive chemical shift changes indicate that the mutant MBD2-IDR shows less helical propensity in the region surrounding the site of mutation. (C) The results suggest that the R286E/L287A mutation disrupts binding to the histone deacetylase core of NuRD by reducing inherent helical propensity.
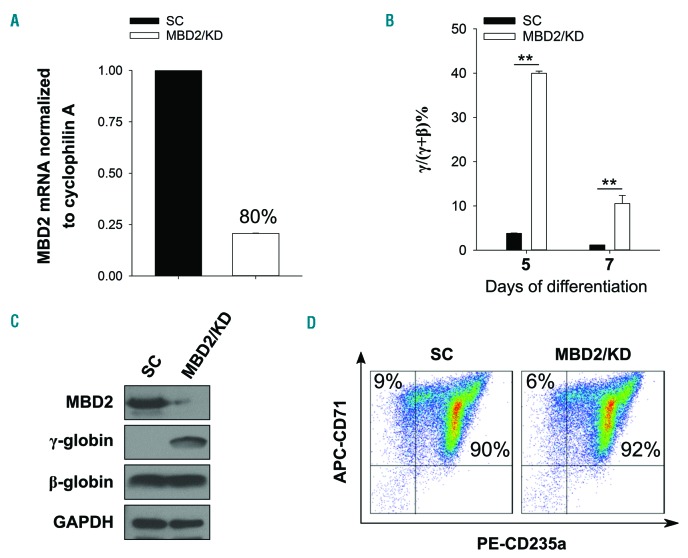
Knockdown of MBD2 in primary human CD34+ erythroid progenitor cells strongly up-regulates γ-globin expression across different levels of erythroid differentiation
Comparison of the relative effects of depleting γ-globin gene silencers in primary CD34+ progenitor-derived erythroblasts on γ/γ+β globin mRNA levels is confounded by assay conditions at inconsistent stages of differentiation.9,13–15,27 To address this, we carried out shRNA MBD2 knockdown in primary CD34+ progenitor-derived erythroblasts, and harvested mRNA at day 5 and day 7 of erythroid differentiation. This resulted in a consistent approximately 10-fold increase in γ/γ+β mRNA, and a level of 40% γ/γ+β compared to 4% in scramble controls after five days of differentiation (Figure 6A and B).
Figure 6.
Lentiviral shRNA knockdown (Kd) of MBD2 in CD34 progenitor-derived primary human erythroid cells results in high level γ/γ+β RNA expression and γ-globin protein without affecting erythroid differentiation. (A) Relative Kd of MBD2 mRNA’. (B) approximately 10-fold increase in γ/γ+β mRNA in MBD2 kd primary erythroid cells, across two different levels of differentiation, compared to scrambled shRNA controls. (C) Increase in γ-globin protein without change in β-globin protein as measured by western blot using anti γ-globin and anti β-globin antibody. (D) Flow cytometry analysis showing equivalent erythroid differentiation profiles of scramble control (sc) and MBD2 Kd CD34+ cells at day 7. Error bars represent ± Standard Deviation of three biological repeats. *P<0.05; **P< 0.01; n.s.: P>0.05. Statistical testing was performed using the Student’s t-test.
We observed a robust induction of γ-globin protein with no change in β-globin protein level assayed by western blot (Figure 6C), and no aberration in the differentiation profile, as assayed by flow cytometric analysis of erythroid markers CD71 and CD235a (Figure 6D), in MBD2 Kd cells. Thus MBD2 depletion (approx. 80%) does not impede the overall developmental stage or differentiation state of primary erythroblasts, independently validating the results in HUDEP-2 cells.
Discussion
Here we demonstrate that CRISPR/Cas9 mediated KO of MBD2 results in markedly increased levels of γ-globin mRNA and protein as well as HbF in HUDEP-2 cells. The approximately 50% γ/γ+β mRNA level of g induction in MBD2KO HUDEP-2 cells is comparable to the effect seen with KO of BCL11A or LRF, two of the strongest γ-globin gene silencers reported.13 Similarly knockdown of MBD2 in CD34+ progenitor-derived primary human erythroid cell results in a consistent approximately 10-fold increase in % γ/γ+β compared to scramble controls across multiple days of differentiation. This results in up to 40% γ/γ+β mRNA levels, compared to 4-5% in controls. Given the ≥5% level of HbF in most sickle cell patients, these results support the therapeutic potential of disruption of MBD2-NuRD-mediated silencing. Importantly, MBD2 knockdown in primary human erythroid cells does not affect erythroid differentiation.
In contrast, CRISPR/Cas9 mediated KO of MBD3 in HUDEP-2 cells does not appreciably increase γ/γ+β or relative γ-globin mRNA or protein expression compared to scramble sgRNA controls, consistent with our published observation of siRNA Kd of MBD3 in human β-YAC bearing CID cells.27 Other studies have demonstrated that MBD3-NuRD interacts with the TR2/TR4 co-repressor complex which binds the embryonic β-type globin promoter39 and BCL11A in murine MEL erythroid cells.40 While MBD3 may indeed associate with these or other complexes in vivo, it does not appear to be essential in the context of γ-globin gene silencing in adult phenotype human erythroid cells.
Genomic engineering technologies have been shown to recapitulate hereditary persistence of fetal hemoglobin (HPFH) mutations,38 and edit the β-globin gene sickle mutation.42 However, significant technological and safety barriers remain, and the vast majority of the worldwide sickle cell burden lies in underdeveloped nations, where small molecule therapeutics will likely be more feasible than cell-based therapy in the foreseeable future. We have pursued a structure-function guided approach to identify heretofore “undruggable” small molecule targets for disruption of the MBD2-NuRD gene silencing effects.26 We show that enforced expression of mutant MBD2-CCmutsgR (D366R/R375E/R380E) fails to suppress γ-globin in MBD2KO HUDEP-2 cells while expression of WT MBD2 partially rescues the MBD2KO phenotype consistent with published data showing that enforced expression of the CC domain of GATAD2A competitively disrupts the MBD2-GATAD2A interaction and mimics the effect of MBD2 Kd in murine CID cells bearing a human β-YAC.27
We previously identified two critical residues in the intrinsically disordered region of MBD2 that are necessary and sufficient for mediating recruitment of the HDAC core complex (HDCC) and silencing of a methylated tumor suppressor gene in breast cancer cells.25 Enforced expression of MBD2-IDRmutsgR (R286E/L287A) also fails to suppress γ-globin in MBD2KO HUDEP-2 cells. Moreover, this mutation disrupts the inherent helical propensity of the unstructured domain. While this observation does not exclude the possibility that R286 or L287 directly interact with components of the HDCC, it indicates that these two residues contribute to the structural propensity of the IDR. The inherent structural propensity of intrinsically disordered regions can be critical for their high-affinity association with binding partners.43–45 Hence, our results support a model in which the helical propensity of the IDR is necessary for binding to the HDCC (Figure 5C), raising the possibility that inhibiting this structural propensity with a small molecule ligand could disrupt formation of a functional NuRD complex. To our knowledge, these results show for the first time the functional effects of disrupting small protein interaction domains within the MBD2-NuRD complex on γ-globin gene silencing in human erythroid cells. We infer from this that small molecules or peptides which specifically bind to and disrupt the IDR or CC domains of MBD2 respectively would be potential candidates for the treatment of the β-hemoglo-binopathies. Intrinsically disordered regions have recently been implicated as potential drug targets and novel screening strategies have been utilized to target them.46–48
While the precise mechanism(s) by which MBD2 enforces silencing of HbF remains incomplete, the work presented here demonstrates several insights into its function, most crucially that recruitment of specific components of the NuRD corepressor complex via the IDR and CC domains of MBD2 is necessary for silencing of γ-globin by MBD2. One observation that remains perplexing is the fact that there are no CpG rich regions in the proximal γ-globin promoter, and in fact the entire β-globin locus is CpG poor. Previous work has shown that MBD2 does not bind directly to sequences in the β-globin gene locus in β-YAC transgenic mice;32 this is expected since there are no CpG rich regions in the locus. This suggests that MBD2-NuRD may exert its effect through regulation of other silencers. Here we investigated whether MBD2 depletion changes the expression of known γ-globin silencers, and found that MBD2KO does not affect the expression of LRF and actually increases expression of BCL11A and KLF1. It remains possible that BCL11A interacts functionally with MBD2-NuRD in human erythroid cells, as depletion of either results in high levels of HbF without impairing differentiation. Current studies to identify targets and interaction partners of MBD2 through which γ-globin silencing is mediated are ongoing.
The data presented here firmly establish that MBD2-NuRD is a potent repressor of HbF expression in adult human erythroid cells, while MBD3-NuRD is not. Specific mutations in the intrinsically disordered region and CC domains of MBD2 necessary for association of other NuRD components that recruit HDAC and chromatin remodeling sub-complexes abrogate the silencing effect of NuRD on the γ-globin gene (Figure 7). Finally, we re-enforce the finding that MBD2 knockdown in primary human erythroid cells results in an 8-10-fold increase in %γ/γ+β mRNA expression without affecting erythroid differentiation. The fact that MBD2 null mice show only minor phenotypic abnormalities (mild deficits in maternal nurturing, a lower than normal body weight, and altered B-cell differentiation) but are otherwise fully viable and fertile49,50 suggests that therapies targeting MBD2, especially in somatic tissues, may have acceptable side effects.
Figure 7.
Working model of the functional importance of MBD2-NuRD interacting domains in fetal hemoglobin (HbF) regulation. Previous work by our group9,25,27 and the findings presented here support a model in which the HDAC core complex (HDCC) members HDAC1/2, RBBP4-7, and MTA1/2 are recruited to MBD2-NuRD through an intrinsically disordered region of MBD2, while GATAD2A/B and CHD4 are recruited through a c-terminal coiled-coil motif of MBD2; independently decoupling either subcomplex results in an abrogation of MBD2-NuRD-mediated HbF silencing.
Acknowledgments
The authors wish to thank Amy Jones and Lindsey Paisley for expert assistance in preparation of this manuscript.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/12/2361
Funding
This work was supported by National Institutes of Health grants; R56 DK29902 (GDG) and R01 DK115563 (GDG and DCW) from the National Institute of Diabetes, Digestive and Kidney Diseases, R01 GM098264 (DCW) from the National Institute of General Medical Sciences, and P30 CA16059 (VCU Massey Cancer Center Core Grant) from the National Cancer Institute.
References
- 1.Lettre G, Bauer DE. Fetal haemoglobin in sickle-cell disease: from genetic epidemiology to new therapeutic strategies. Lancet. 2016;387(10037):2554–2564. [DOI] [PubMed] [Google Scholar]
- 2.Modell B, Darlison M. global epidemiology of haemoglobin disorders and derived service indicators. Bull World Health Organ. 2008;86(6):480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vichinsky EP. Changing patterns of thalassemia worldwide. Ann N Y Acad Sci. 2005;1054:18–24. [DOI] [PubMed] [Google Scholar]
- 4.Gaziev J, Lucarelli G. Stem cell transplantation for hemoglobinopathies. Curr Opin Pediatr. 2003;15(1):24–31. [DOI] [PubMed] [Google Scholar]
- 5.Olivieri NF. Reactivation of fetal hemoglobin in patients with beta-thalassemia. Semin Hematol. 1996;33(1):24–42. [PubMed] [Google Scholar]
- 6.Wood WG, Clegg JB, Weatherall DJ. Hereditary persistence of fetal haemoglobin (HPFH) and delta beta thalassaemia. Br J Haematol. 1979;43(4):509–520. [DOI] [PubMed] [Google Scholar]
- 7.Platt OS, Brambilla DJ, Rosse WF, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330(23):1639–1644. [DOI] [PubMed] [Google Scholar]
- 8.Sankaran VG, Orkin SH. The switch from fetal to adult hemoglobin. Cold Spring Harb Perspect Med. 2013;3(1):a011643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaya M, Desai M, Gnanapragasam MN, et al. Mi2beta-mediated silencing of the fetal gamma-globin gene in adult erythroid cells. Blood. 2013;121(17):3493–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borg J, Patrinos GP, Felice AE, Philipsen S. Erythroid phenotypes associated with KLF1 mutations. Haematologica. 2011; 96(5):635–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginder GD. Epigenetic regulation of fetal globin gene expression in adult erythroid cells. Transl Res. 2015;165(1):115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda T, Wang X, Maeda M, et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351(6270):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Cui S, Engel JD, Tanabe O. Lysine-specific demethylase 1 is a therapeutic target for fetal hemoglobin induction. Nat Med. 2013;19(3):291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet. 2010;42(9):742–744. [DOI] [PubMed] [Google Scholar]
- 17.Kurita R, Suda N, Sudo K, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8(3):e59890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charache S, Dover G, Smith K, Talbot CC, Jr, Moyer M, Boyer S. Treatment of sickle cell anemia with 5-azacytidine results in increased fetal hemoglobin production and is associated with nonrandom hypomethylation of DNA around the gamma-delta-beta-globin gene complex. Proc Natl Acad Sci U S A. 1983;80(15):4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSimone J, Heller P, Hall L, Zwiers D. 5-Azacytidine stimulates fetal hemoglobin synthesis in anemic baboons. Proc Natl Acad Sci U S A. 1982;79(14):4428–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginder GD, Whitters MJ, Pohlman JK. Activation of a chicken embryonic globin gene in adult erythroid cells by 5-azacytidine and sodium butyrate. Proc Natl Acad Sci U S A. 1984;81(13):3954–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley TJ, DeSimone J, Anagnou NP, et al. 5-azacytidine selectively increases gamma-globin synthesis in a patient with beta+ thalassemia. N Engl J Med. 1982;307(24): 1469–1475. [DOI] [PubMed] [Google Scholar]
- 22.Perrine SP, Ginder GD, Faller DV, et al. A short-term trial of butyrate to stimulate fetal-globin-gene expression in the beta-globin disorders. N Engl J Med. 1993; 328(2):81–86. [DOI] [PubMed] [Google Scholar]
- 23.Witt O, Monkemeyer S, Ronndahl G, et al. Induction of fetal hemoglobin expression by the histone deacetylase inhibitor apicidin. Blood. 2003;101(5):2001–2007. [DOI] [PubMed] [Google Scholar]
- 24.Cramer JM, Scarsdale JN, Walavalkar NM, Buchwald WA, Ginder GD, Williams DC., Jr Probing the dynamic distribution of bound states for methylcytosine-binding domains on DNA. J Biol Chem. 2014;289(3):1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desai MA, Webb HD, Sinanan LM, et al. An intrinsically disordered region of methyl-CpG binding domain protein 2 (MBD2) recruits the histone deacetylase core of the NuRD complex. Nucleic Acids Res. 2015;43(6):3100–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginder GD, Williams DC., Jr Readers of DNA methylation, the MBD family as potential therapeutic targets. Pharmacol Ther. 2018;184:98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gnanapragasam MN, Scarsdale JN, Amaya ML, et al. p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc Natl Acad Sci U S A. 2011;108(18):7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez J, Dege C, Kutateladze TG, Hagman J. MBD2 and multiple domains of CHD4 are required for transcriptional repression by Mi-2/NuRD complexes. Mol Cell Biol. 2012;32(24):5078–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15(7):827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng HH, Zhang Y, Hendrich B, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23(1):58–61. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13(15):1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupon JW, Wang SZ, Gaensler K, Lloyd J, Ginder GD. Methyl binding domain protein 2 mediates gamma-globin gene silencing in adult human betaYAC transgenic mice. Proc Natl Acad Sci U S A. 2006; 103(17):6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Guezennec X, Vermeulen M, Brinkman AB, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26(3):843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harju-Baker S, Costa FC, Fedosyuk H, Neades R, Peterson KR. Silencing of Agamma-globin gene expression during adult definitive erythropoiesis mediated by GATA-1-FOG-1-Mi2 complex binding at the -566 GATA site. Mol Cell Biol. 2008;28(10):3101–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miccio A, Blobel GA. Role of the GATA-1/FOG-1/NuRD pathway in the expression of human beta-like globin genes. Mol Cell Biol. 2010;30(14):3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. [DOI] [PubMed] [Google Scholar]
- 37.Vranken WF, Boucher W, Stevens TJ, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59(4):687–696. [DOI] [PubMed] [Google Scholar]
- 38.Traxler EA, Yao Y, Wang YD, et al. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med. 2016;22(9):987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui S, Kolodziej KE, Obara N, et al. Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic beta-type globin promoters in differentiated adult erythroid cells. Mol Cell Biol. 2011;31(16):3298–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Bauer DE, Kerenyi MA, et al. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci U S A. 2013;110(16):6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walavalkar NM, Gordon N, Williams DC., Jr Unique features of the anti-parallel, heterodimeric coiled-coil interaction between methyl-cytosine binding domain 2 (MBD2) homologues and GATA zinc finger domain containing 2A (GATAD2A/p66alpha). J Biol Chem. 2013;288(5):3419–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dever DP, Bak RO, Reinisch A, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539(7629):384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. [DOI] [PubMed] [Google Scholar]
- 44.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004; 338(5): 1015–1026. [DOI] [PubMed] [Google Scholar]
- 45.Shammas SL, Crabtree MD, Dahal L, Wicky BI, Clarke J. Insights into Coupled Folding and Binding Mechanisms from Kinetic Studies. J Biol Chem. 2016;291(13): 6689–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14(4):481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezaei-Ghaleh N, Blackledge M, Zweckstetter M. Intrinsically disordered proteins: from sequence and conformational properties toward drug discovery. Chembiochem. 2012;13(7):930–950. [DOI] [PubMed] [Google Scholar]
- 48.Yu C, Niu X, Jin F, Liu Z, Jin C, Lai L. Structure-based Inhibitor Design for the Intrinsically Disordered Protein c-Myc. Sci Rep. 2016;6:22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15(6):710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood KH, Zhou Z. Emerging Molecular and Biological Functions of MBD2, a Reader of DNA Methylation. Front Genet. 2016;7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]