Abstract
Fucosylation is a post-translational modification that attaches fucose residues to protein- or lipid-bound oligosaccharides. Certain fucosylation pathway genes are aberrantly expressed in several types of cancer, including non-small cell lung cancer (NSCLC), and this aberrant expression is associated with poor prognosis in patients with cancer. However, the molecular mechanism by which these fucosylation pathway genes promote tumor progression has not been well-characterized. The present study analyzed public microarray data obtained from NSCLC samples. Multivariate analysis revealed that altered expression of fucosylation pathway genes, including fucosyltransferase 1 (FUT1), FUT2, FUT3, FUT6, FUT8 and GDP-L-fucose synthase (TSTA3), correlated with poor survival in patients with NSCLC. Inhibition of FUTs by 2F-peracetyl-fucose (2F-PAF) suppressed transforming growth factor β (TGFβ)-mediated Smad3 phosphorylation and nuclear translocation in NSCLC cells. In addition, wound-healing and Transwell migration assays demonstrated that 2F-PAF inhibited TGFβ-induced NSCLC cell migration and invasion. Furthermore, in vivo bioluminescence imaging analysis revealed that 2F-PAF attenuated the metastatic capacity of NSCLC cells. These results may help characterize the oncogenic role of fucosylation in NSCLC biology and highlight its potential for developing cancer therapeutics.
Keywords: fucosylation, fucosyltransferase, transforming growth factor β, non-small cell lung cancer, tumor metastasis
Introduction
Lung cancer is the leading cause of cancer-related death in the world (1). Non-small cell lung cancer (NSCLC) accounts for >85% of all lung cancer cases (2). Patients are usually diagnosed with NSCLC at an advanced or metastatic stage with poor prognosis (3). However, the molecular mechanisms driving NSCLC development and progression remain poorly understood.
Fucosylation is one of the most common types of mammalian glycosylation, in which fucose is conjugated to protein- or lipid-bound oligosaccharides (4). Cell surface fucosylation serves a crucial role in fine-tuning biological recognition processes that mediate cell adhesion and migration (4). Aberrant fucosylation has been observed in multiple types of cancer (5-7) and is associated with tumor development and metastatic capability (8-11). Therefore, fucosylation has gained attention as a promising target for the development of novel cancer therapeutics.
The fucosylation pathway consists of GDP-fucose synthesis in the cytosol, GDP-fucose transport into the Golgi, and fucose transfer to acceptor substrates inside the Golgi (12). GDP-fucose is synthesized by the de novo or salvage enzymatic pathways involving GDP-mannose 4,6-dehydratase (GMDS), GDP-L-fucose synthase (TSTA3), L-fucose kinase (FUK) and fucose-1-phosphate guanylyltransferase (FPGT) (4,13). GDP-fucose is transported into the Golgi lumen by GDP-fucose transporter 1 (SLC35C1) (13). A fucose residue from GDP-fucose is transferred to the sugar moieties of glycoconjugates or the serine/threonine residues on substrate proteins by fucosyltransferases (FUTs) (14,15). FUTs catalyze α-1,2 (by FUT1 and 2), α-1,3 (by FUT3-7 and 9-11), α-1,4 (by FUT3 and 5) and α-1,6 (by FUT8) glycosidic bond formation or protein O-fucosylation (by POFUT1 and 2) (4,16,17). Certain FUT isotypes are aberrantly expressed in several types of cancer, including NSCLC (18-26), and this aberrant expression is associated with poor prognosis in patients with cancer (21,26,27). However, the molecular mechanism by which the fucosylation pathway genes promote tumor progression has not been well-characterized.
The present study aimed to analyze microarray data obtained from NSCLC samples. The findings of this study may help characterize the oncogenic role of fucosylation in NSCLC biology and highlight its potential for therapeutic targeting.
Materials and methods
Data collection
Microarray data of samples from patients with lung adenocarcinoma were obtained from the Gene Expression Omnibus (GEO) database under the accession number GSE31210 (28) as the discovery dataset. The GSE31210 data were obtained from 20 normal and 226 NSCLC samples [68 epidermal growth factor receptor (EGFR)/KRAS/echinoderm microtubule-associated protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) fusion-negative (triple-negative), 127 EGFR mutation-positive, 20 KRAS mutation-positive and 11 EML4-ALK fusion-positive samples]. The replication datasets were GSE30219 (29), GSE29013 (30), GSE37745 (31) and GSE50081 (32). RNA expression values of TCGA LUAD dataset were obtained from the RNA-Seq V2 RSEM data of cBioPortal (http://www.cbioportal.org). Survival data of patients from TCGA LUAD dataset were collected using TCGA-assembler 2.0.5 with R software (http://www.r-projects.org) (33). Baseline characteristics of the survival datasets were summarized in Table SI.
Computational analysis
The microarray datasets were subjected to stringent quality control tests to filter out low-quality microarray samples (34). The criteria used were as previously described (35,36). Of the 246 samples, 235 passed the quality control test (Table SII). These samples were normalized by the MAS5.0 algorithm in Expression Console (Build 1.3.1.187, Affymetrix; Thermo Fisher Scientific, Inc.), and 18 normal, 66 triple-negative, 121 EGFR mutation-positive, 19 KRAS mutation-positive and 11 EML4-ALK fusion-positive samples were used for further analysis. From the microarray data, probes corresponding to the fucosylation pathway genes (FUT1, FUT2, FUT3, FUT4, FUT5, FUT6, FUT7, FUT8, FUT9, FUT10, FUT11, POFUT1, POFUT2, GMDS, TSTA3, FUK, FPGT and SLC35C1) were extracted. When multiple probes were matched with one gene, the probe with the highest expression was selected as the representative value of gene expression. The fucosylation pathway gene (FUT1, FUT2, FUT3, FUT4, FUT5, FUT6, FUT7, FUT8, FUT9, FUT10, FUT11, POFUT1, POFUT2, GMDS, TSTA3, FUK, FPGT and SLC35C1) expression values from tumor samples were then divided by those from normal samples to calculate relative expression levels. Correlation analysis was performed for the gene expression levels between the TGFβ receptor complex pathway and the fucosylation pathway to examine the association between the pathways. The 'TGFβ receptor complex pathway gene set’ was created by collecting genes listed in the gene signatures, such as 'TGFβ receptor signaling activates SMADs’, 'TGFβ receptor signaling in EMT’ and 'transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer’ in Reactome pathway database version 65 (https://reactome.org) (37). Fucosylation pathway genes and TGFβ receptor complex pathway genes were presented as scatterplots, and correlations were expressed using a linear regression model.
Survival analysis
Survival analysis was performed using microarray datasets. For each fucosylation pathway gene, the patient samples were divided into four groups according to their gene expression pattern. Kaplan-Meier survival curves, log-rank test, univariate and multivariate Cox proportional hazards regression models were used to determine the association between gene expression and patient survival in the fourth quartile of expression of fucosylation pathway genes in terms of relapse-free survival rate (RFS) or overall survival rate (OS). A multivariate Cox proportional hazard regression model was used to calculate hazard ratios (HRs) and 95% confidence intervals (CIs) of the expression on RFS or OS of NSCLC adjusting for age, sex, smoking status and pathological stage. Among the 514 TCGA samples, 396 patients with stage I and II were analyzed, as microarray datasets only consisted of patients with tumor stage I or II (Table SI).
Cell culture and reagents
NCI-H3122 cells were kindly provided by Professor Pasi A. Janne (Dana Faber Cancer Institute, Boston, MA, USA). Calu-1 cells were supplied by the American Type Culture Collection. Cells with passage number <20 were used in the experiments. All cell culture reagents were obtained from Gibco; Thermo Fisher Scientific, Inc. or HyClone; GE Healthcare Life Sciences. The FUT inhibitor 2F-peracetyl-fucose (2F-PAF) was purchased from Merck KGaA. 2F-PAF was dissolved in DMSO and stored at -20˚C. Prior to treatment with TGFβ1 (R&D Systems, Inc.), cells were cultured in RPMI-1640 medium (HyClone Laboratories, Inc.) containing 0.2% FBS (HyClone Laboratories, Inc.) and/or 2F-PAF at the indicated concentrations (25-200 µM) for 24 h. All other cell culture reagents were supplied by Sigma-Aldrich.
Cell viability assay
NCI-H3122 and Calu-1 cells (3x104) were treated with 2F-PAF (0-200 µM) for 72 h in 12-well culture plates prior to MTT assay using thiazolyl blue tetrazolium bromide (Sigma-Aldrich; Merck KGaA). The purple formazan was dissolved in DMSO and quantified by measuring the absorbance at 570 nm on a BioTek Synergy MX microplate spectrophotometer (BioTek Instruments, Inc.).
Luciferase assay
NCI-H3122 cells and Calu-1 cells (3x104) were transfected with pGL2-3TP-luciferase (Addgene, Inc.) and pCMV-β-galactosidase gene constructs (Takara Biotechnology Co., Ltd.) (38) using FuGENE 6 (Roche Diagnostics) according to the manufacturer's instructions. After 24 h of transfection, cells were treated with 2F-PAF for 24 h and further incubated with TGFβ for 24 h. Cells were harvested and assayed for luciferase or β-galactosidase activity using a Luciferase Assay System kit (Promega Corporation). Luciferase activity was normalized to β-galactosidase activity as previously described (38).
Western blot analysis. NCI-H3122 or Calu-1 cells (2x105) were seeded on 60 mm dishes. After 24 h, cells were treated with the conditions indicated in figure legends (1 or 5 ng/ml TGFβ and 25-200 µM 2F-PAF). The crude extracts were prepared by incubation with RIPA buffer containing protease and phosphatase inhibitor cocktails (Merck KGaA). The protein concentration was determined by bicinchoninic acid assay. The samples were resolved using 6 or 10% SDS-PAGE and then transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk in TBS + 0.1% Tween-20 for 1 h at room temperature and probed with the indicated antibodies. The signals were determined using a SuperSignal West chemiluminescent substrate (Thermo Fisher Scientific, Inc.). ImageJ software (ImageJ bundled with 64-bit Java 1.8.0_112; National Institutes of Health) was used to quantify band intensity. The data were representative of at least three independent experiments. Antibody information and experimental conditions are presented in Table SIII.
Confocal microscopy
NCI-H3122 cells (2x104) were seeded on glass coverslips in 12-well plates. At 24 h, cells were treated with the conditions indicated in figure legends (1 ng/ml TGFβ and 50-200 µM 2F-PAF). Cells were fixed with 4% formaldehyde in PBS for 5 min, permeabilized with 0.3% Triton X-100 for 7 min, and blocked with 3% normal goat serum in PBS for 1 h at room temperature. Subsequently, cells were probed with an anti-Smad3 antibody (1:200; cat. no. 9523; Cell signaling Technology, Inc.) overnight at 4˚C and stained with FITC-conjugated anti-rabbit IgG antibody (1:1,000; cat. no. A21441; Invitrogen; Thermo Fisher Scientific, Inc.) and DAPI (Roche Diagnostics GmbH). The stained cells were imaged with a FluoView 1000 confocal microscope (x40 magnification; Olympus Corporation).
Wound-healing assay
Wound-healing assays were used to assess cell migration as previously described (39). NCI-H3122 and Calu-1 cells were cultured with 2F-PAF in 6-well plates for 24 h. A scratch was made on the cell monolayer using a sterile 10 µl pipette tip, and then cells were treated with TGFβ, 2F-PAF or TGFβ + 2F-PAF in RPMI-1640 medium containing 0.2% FBS for 24 h. Migrated cells within the scratch area were counted in five random fields using a Nikon Eclipse TS100 phase-contrast microscope (Nikon Instruments, Inc.).
Transwell migration assay
Transwell migration assays were performed in 24-well chambers with 8 µm pore size Transwell inserts (Corning, Inc.) as previously described (40,41). The inserts were coated with 0.1 mg/ml collagen for 1 h. NCI-H3122 cells were trypsinized, and single-cell suspensions were placed into the upper chamber (1×105 cells/well) in 100 µl serum-free medium. TGFβ (5 ng/ml) in serum-free medium (800 µl) was placed in the lower chamber as a chemoattractant. After 48-h incubation, the cells from the upper surface of the chamber were removed using a cotton swab. The migrated cells were fixed with 4% formaldehyde in PBS for 7 min and stained with 0.5% crystal violet for 20 min at room temperature, imaged and counted using a phase-contrast microscope (×100). The number of migrated cells was counted in five random fields.
In vivo metastasis assay
Calu-1-Luc cells were established by infection with RediFect Red-FLuc-Puromycin Lentiviral Particles (PerkinElmer, Inc.) for 24 h at 37˚C and puromycin selection for 2 weeks with a final concentration of 2 µg/ml. Calu-1-Luc cells were treated with 20 µg/ml 2F-PAF for 96 h. BALB/c-nude mice (male, 8 weeks old, n=5 per group) were housed in a specific pathogen-free environment at 22±2˚C and 55±5% relative humidity with light. BALB/c-nude mice injected intravenously with 1x106 2F-PAF-treated Calu-1-Luc cells and analyzed 2 weeks later. For in vivo bioluminescence imaging (BLI), mice were injected intraperitoneally with D-Luciferin (150 mg/kg, 200 µl; PerkinElmer, Inc.) under gas anesthesia [1% (w/v) isoflurane in 2 l oxygen] and imaged 10 min later using the IVIS spectrum system (PerkinElmer, Inc.). BLI intensity was measured using region of interest analysis. All experiments were conducted under protocols approved by the Institutional Animal Care and Use Committee of the Asan Institute for Life Sciences at the Asan Medical Center (approval no. 2019-14-201).
Statistical analysis
GraphPad Prism 7.04 (GraphPad Software, Inc.) was used for statistical analysis. Data are expressed as the mean ± SEM. Comparison of mean values among experimental groups was performed using one-way ANOVA followed by a Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of fucosylation pathway genes is altered in NSCLC
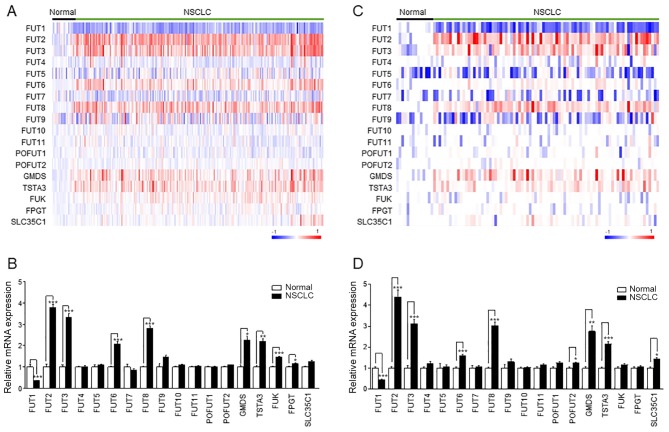
To determine which fucosylation pathway genes were differentially expressed between normal lung and NSCLC tissue samples, 235 samples (18 normal and 217 NSCLC tissues) from the public microarray dataset GSE31210 were analyzed. A heatmap demonstrated distinct expression patterns of fucosylation pathway genes (Fig. 1A). The expression levels of FUT2, FUT3, FUT6, FUT8, GMDS, TSTA3, FUK and FPGT were increased in NSCLC, whereas FUT1 expression decreased in NSCLC (Fig. 1B). This result was confirmed in the replication datasets GSE30219 (Fig. 1C and D) and GSE19188 (Fig. S3A and B). The expression levels of FUT1, FUT2, FUT3, FUT6, FUT8, GMDS and TSTA3 were commonly altered in NSCLC in the three datasets (Figs. 1 and S3).
Figure 1.
Expression of fucosylation pathway genes in NSCLC. (A and C) The heatmap of the log2-transformed expression level of fucosylation pathway genes in each microarray sample. The heatmap was generated using GenePattern HeatMapImage. (B and D) The expression levels of fucosylation pathway genes in patients with NSCLC were normalized to those in the control groups. Left panel, GSE31210; right panel, GSE30219. Data are expressed as the mean ± SEM and compared using an unpaired Student's t-test. *P<0.05, **P<0.01, ***P<0.005. NSCLC, non-small cell lung cancer; FUT, fucosyltransferase; TSTA3, GDP-L-fucose synthase; GMDS, GDP-mannose 4,6-dehydratase; FUK, L-fucose kinase; FPGT, fucose-1-phosphate guanylyltransferase; SLC35C1, GDP-fucose transporter 1; POFUT, protein O-fucosyltransferase.
The altered expression of these genes compared with normal lung tissue was observed in NSCLC independently of the triple-negative, EGFR mutation-positive, KRAS mutation-positive, and EML4-ALK fusion-positive status. The analysis results demonstrated that the gene expression levels of FUT1, FUT2, FUT3, FUT6, FUT8, GMDS, TSTA3 and FUK were commonly altered in all four NSCLC subtypes compared with those in normal tissues (Fig. S4 and Table SIV). However, the change was most prominent in the EML4-ALK fusion-positive subtype of NSCLC.
Altered expression of fucosylation pathway genes is associated with poor prognosis in patients with NSCLC
Kaplan-Meier survival analysis with log-rank test demonstrated that the altered expression of 7 out of 18 fucosylation pathway genes was associated with poor OS and RFS (Figs. S5 and S6). Patients with NSCLC with high expression of FUT2, FUT3, FUT6, FUT8 and TSTA3 and low expression of FUT1 and FUT5 exhibited significantly worse RFS, even after adjusting for epidemiological and clinicopathological factors (Table I). To validate the results of the survival analysis, OS analysis was performed using two different datasets (Table SV). These results demonstrated that patients with altered expression of FUT1 (downregulated) and FUT8 (upregulated) exhibited significantly worse survival rates. This result of the replication dataset was consistent with the result of the discovery dataset (Tables I and SV).
Table I.
HRs for relapse-free survival based on the expression of fucosylation pathway genes of 217 patients with non-small cell lung cancer.
| Gene | Expression quartile | Patients (n) | Death (n) | Crude HR (95% CI) | Adjusted HRa (95% CI) |
|---|---|---|---|---|---|
| FUT2 | Q1 (0-25%) | 54 | 11 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 15 | 1.299 (0.601-2.809) | 1.458 (0.673-3.161) | |
| Q3 (50-75%) | 54 | 12 | 1.023 (0.459-2.277) | 1.255 (0.559-2.817) | |
| Q4 (75-100%) | 54 | 22 | 2.275 (1.125-4.601) | 2.527 (1.228-5.203) | |
| FUT3 | Q1 (0-25%) | 54 | 13 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 17 | 1.364 (0.663-2.809) | 2.207 (1.042-4.679) | |
| Q3 (50-75%) | 54 | 10 | 0.736 (0.323-1.679) | 1.027 (0.440-2.395) | |
| Q4 (75-100%) | 54 | 20 | 1.737 (0.864-3.493) | 2.190 (1.073-4.470) | |
| FUT4 | Q1 (0-25%) | 54 | 13 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 14 | 1.144 (0.537-2.434) | 0.868 (0.400-1.880) | |
| Q3 (50-75%) | 54 | 11 | 0.843 (0.378-1.883) | 0.869 (0.386-1.954) | |
| Q4 (75-100%) | 54 | 22 | 2.020 (1.016-4.018) | 1.605 (0.791-3.258) | |
| FUT6 | Q1 (0-25%) | 54 | 14 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 11 | 0.614 (0.276-1.367) | 0.754 (0.336-1.690) | |
| Q3 (50-75%) | 54 | 12 | 0.803 (0.376-1.715) | 0.834 (0.385-1.805) | |
| Q4 (75-100%) | 54 | 23 | 1.765 (0.920-3.385) | 1.962 (1.018-3.780) | |
| FUT8 | Q1 (0-25%) | 54 | 8 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 13 | 1.869 (0.774-4.510) | 1.666 (0.685-4.052) | |
| Q3 (50-75%) | 54 | 18 | 2.511 (1.092-5.777) | 2.489 (1.078-5.748) | |
| Q4 (75-100%) | 54 | 21 | 3.096 (1.371-6.990) | 2.530 (1.112-5.760) | |
| FUT9 | Q1 (0-25%) | 54 | 13 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 9 | 0.702 (0.300-1.641) | 0.637 (0.271-1.497) | |
| Q3 (50-75%) | 54 | 17 | 1.535 (0.745-3.162) | 1.496 (0.721-3.106) | |
| Q4 (75-100%) | 54 | 21 | 1.854 (0.928-3.706) | 1.424 (0.698-2.906) | |
| FUT10 | Q1 (0-25%) | 54 | 16 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 15 | 0.958 (0.473-1.938) | 0.988 (0.479-2.040) | |
| Q3 (50-75%) | 54 | 13 | 0.840 (0.404-1.745) | 0.947 (0.448-2.000) | |
| Q4 (75-100%) | 54 | 16 | 1.031 (0.516-2.063) | 1.188 (0.578-2.444) | |
| FUT11 | Q1 (0-25%) | 54 | 11 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 11 | 1.070 (0.464-2.468) | 1.058 (0.457-2.453) | |
| Q3 (50-75%) | 54 | 16 | 1.629 (0.756-3.511) | 1.394 (0.639-3.039) | |
| Q4 (75-100%) | 54 | 22 | 2.411 (1.168-4.977) | 1.834 (0.868-3.876) | |
| POFUT1 | Q1 (0-25%) | 54 | 11 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 13 | 1.266 (0.567-2.827) | 1.030 (0.455-2.331) | |
| Q3 (50-75%) | 54 | 16 | 1.551 (0.720-3.342) | 1.159 (0.527-2.548) | |
| Q4 (75-100%) | 54 | 20 | 2.273 (1.088-4.747) | 1.638 (0.760-3.530) | |
| GMDS | Q1 (0-25%) | 54 | 7 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 19 | 3.180 (1.337-7.567) | 2.620 (1.092-6.291) | |
| Q3 (50-75%) | 54 | 18 | 2.916 (1.218-6.983) | 2.656 (1.103-6.396) | |
| Q4 (75-100%) | 54 | 16 | 2.649 (1.089-6.442) | 2.160 (0.881-5.296) | |
| TSTA3 | Q1 (0-25%) | 54 | 9 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 16 | 2.069 (0.922-4.645) | 1.847 (0.817-4.175) | |
| Q3 (50-75%) | 54 | 15 | 1.689 (0.731-3.902) | 1.475 (0.632-3.442) | |
| Q4 (75-100%) | 54 | 20 | 2.563 (1.167-5.629) | 2.464 (1.118-5.431) | |
| FUK | Q1 (0-25%) | 54 | 18 | 1 (Reference) | 1 (Reference) |
| Q2 (25-50%) | 55 | 12 | 0.620 (0.299-1.288) | 0.669 (0.318-1.410) | |
| Q3 (50-75%) | 54 | 10 | 0.497 (0.229-1.077) | 0.633 (0.281-1.426) | |
| Q4 (75-100%) | 54 | 20 | 1.062 (0.562-2.008) | 1.052 (0.547-2.024) | |
| FUT1 | Q1 (75-100%) | 54 | 26 | 1 (Reference) | 1 (Reference) |
| Q2 (50-75%) | 55 | 12 | 3.839 (1.406-10.483) | 3.030 (1.094-8.397) | |
| Q3 (25-50%) | 54 | 17 | 2.636 (0.929-7.485) | 1.838 (0.626-5.398) | |
| Q4 (0-25%) | 54 | 5 | 6.768 (2.604-17.591) | 4.469 (1.657-12.051) | |
| FUT5 | Q1 (75-100%) | 54 | 26 | 1 (Reference) | 1 (Reference) |
| Q2 (50-75%) | 55 | 11 | 1.104 (0.487-2.502) | 1.064 (0.467-2.425) | |
| Q3 (25-50%) | 54 | 12 | 1.036 (0.449-2.390) | 1.099 (0.469-2.577) | |
| Q4 (0-25%) | 54 | 11 | 2.882 (1.423-5.836) | 2.619 (1.286-5.332) | |
| FUT7 | Q1 (75-100%) | 54 | 16 | 1 (Reference) | 1 (Reference) |
| Q2 (50-75%) | 55 | 9 | 1.616 (0.822-3.177) | 1.593 (0.809-3.137) | |
| Q3 (25-50%) | 54 | 21 | 0.623 (0.270-1.439) | 0.657 (0.281-1.534) | |
| Q4 (0-25%) | 54 | 14 | 1.133 (0.552-2.321) | 1.130 (0.538-2.371) | |
| POFUT2 | Q1 (75-100%) | 54 | 19 | 1 (Reference) | 1 (Reference) |
| Q2 (50-75%) | 55 | 17 | 1.168 (0.523-2.607) | 1.275 (0.569-2.857) | |
| Q3 (25-50%) | 54 | 13 | 1.510 (0.707-3.223) | 1.634 (0.759-3.520) | |
| Q4 (0-25%) | 54 | 11 | 1.874 (0.891-3.939) | 1.570 (0.741-3.327) | |
| FPGT | Q1 (75-100%) | 54 | 17 | 1 (Reference) | 1 (Reference) |
| Q2 (50-75%) | 55 | 19 | 0.615 (0.266-1.422) | 0.646 (0.278-1.503) | |
| Q3 (25-50%) | 54 | 10 | 1.424 (0.714-2.842) | 1.291 (0.645-2.587) | |
| Q4 (0-25%) | 54 | 14 | 1.309 (0.651-2.633) | 0.958 (0.466-1.971) | |
| SLC35C1 | Q1 (75-100%) | 54 | 14 | 1 (Reference) | 1 (Reference) |
| Q2 (50-75%) | 55 | 17 | 1.825 (0.862-3.866) | 1.759 (0.829-3.733) | |
| Q3 (25-50%) | 54 | 18 | 1.613 (0.755-3.443) | 1.802 (0.835-3.892) | |
| Q4 (0-25%) | 54 | 11 | 1.294 (0.587-2.852) | 1.528 (0.688-3.391) |
HR adjusted for age, sex, smoking status, and pathological stage. HR, hazard ratio; CI, confidence interval.
Inhibition of FUTs attenuates TGFβ-induced cell migration and tumor metastasis
Fucosylation of the TGFβ receptor has previously been demonstrated to enhance TGFβ signaling (42,43). To determine the association between TGFβ signaling and fucosylation in patients with lung cancer, the correlation of gene expression between the fucosylation pathway and the TGFβ receptor complex pathway was first analyzed. The TGFβ receptor complex pathway gene set was constructed using the Reactome database, and the expression levels of these genes were extracted from GSE31210. The hierarchical clustering results demonstrated that the expression of TGFβ receptor complex pathway genes such as cbl proto-oncogene, cadherin1, desmoplakin, E2F transcription factor 4 (E2F4), histone deacetylase 1, matrix metallopepti-dase 9, partitioning defective 6 homolog α, poly (ADP-ribose) polymerase 1, protein phosphatase 1A, protein phosphatase 1 catalytic subunit α, ras homolog family member A, SKI-like proto oncogene, SMAD3, SRY-box transcription factor 9, Sp1 transcription factor, zinc finger E-box-binding homeobox 1, TGFβR2, tight junction protein-1, ubiquitin-conjugating enzyme E2, WW domain containing transcription regulator 1, zinc finger E-box binding homeobox 2 and zinc finger FYVE-type containing 9 exhibited strong correlations with the expression of FUT1, FUT2, FUT3, FUT4, FUT6, FUT7, FUT8, FUT11, POFUT2, GMDS, FPGT, FUK and TSTA3 in patients with NSCLC (Fig. S7). The expression of E2F4 exhibited a fair correlation with the expression of FUT4 and GMDS (R=0.53 and R=0.57, respectively) 30191186. In the linear regression model, E2F4-FUT4 (R2=0.28) and E2F4-GDMS (R2=0.32) had explanatory power (Fig. S8).
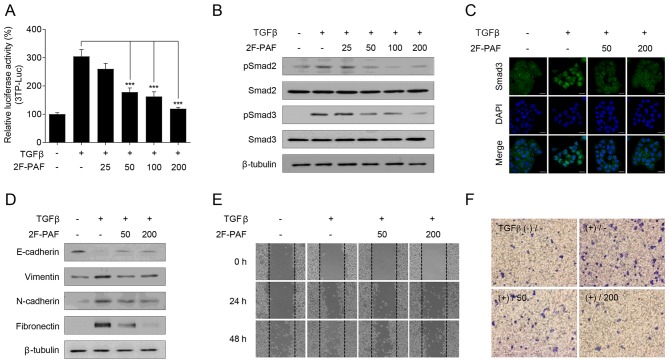
To confirm the association between TGFβ signaling and fucosylation, the FUT inhibitor 2F-PAF was used. Since altered expression of fucosylation pathway genes is most prominent in the EML4-ALK fusion-positive subtype of NSCLC, NCI-H3122, which is an NSCLC cell line that expresses the EML4-ALK fusion, was selected for the experiments. 2F-PAF suppressed TGFβ-induced Smad activation in a dose-dependent manner (Fig. 2A). In addition, 2F-PAF attenuated the TGFβ-mediated phosphorylation of Smad2/3 (Fig. 2B) and translocation of Smad3 into the nucleus (Fig. 2C). Additionally, 2F-PAF inhibited TGFβ signaling in NCI-H3122 cells without exhibiting any cytotoxicity effects (Fig. S9A). The effects of 2F-PAF on TGFβ-mediated phenotypes in NCI-H3122 cells were further examined; 2F-PAF suppressed TGFβ-induced downregulation of E-cadherin and upregulation of vimentin, N-cadherin and fibronectin (Fig. 2D), suggesting that 2F-PAF inhibited TGFβ-mediated epithelial-mesenchymal transition (EMT). In addition, 2F-PAF suppressed TGFβ-induced cell migration and invasion in a wound-healing (Figs. 2E and S10A-C) and Transwell invasion (Figs. 2F and S11) assays, respectively. These results demonstrated that 2F-PAF inhibited TGFβ-induced EMT, migration and invasion of NSCLC cells.
Figure 2.
Inhibition of FUTs suppresses TGFβ-induced cell migration and invasion in NCI-H3122 cells. (A) NCI-H3122 cells were transfected with a 3TP-Luc reporter construct for 24 h and incubated with TGFβ (1 ng/ml) and 2F-PAF at the indicated concentrations (0, 25, 50, 100, and 200 µM) for 24 h. The luciferase activity was expressed as a relative value compared with that of the untreated cells. Data are expressed as the mean ± SEM (n=3). ***P<0.005. (B and C) Cells were treated with TGFβ (1 ng/ml) and 2F-PAF at the indicated concentrations for 1 h prior to (B) western blot analysis and (C) confocal microscopy. The localization of Smad3 was assessed using an anti-Smad3 antibody and a FITC-conjugated IgG antibody. DAPI was used to visualize the nucleus. Scale bar, 20 µm. (D) Cells were treated with TGFβ (5 ng/ml) and/or 2F-PAF for 48 h prior to western blot analysis. (E) In a wound-healing assay, cells were treated with TGFβ (5 ng/ml) and/or 2F-PAF at 50 and 200 µM for the indicated times after the wound was created. (F) The invasive cells in the Transwell assay were fixed with 4% formaldehyde, stained with 0.5% crystal violet, imaged and counted using a phase-contrast microscope. FUT, fucosyltransferase; TGFβ, transforming growth factor β; 2F-PAF, 2F-peracetyl-fucose; p, phosphorylated; Luc, luciferase.
Inhibition of FUTs attenuates NSCLC metastasis
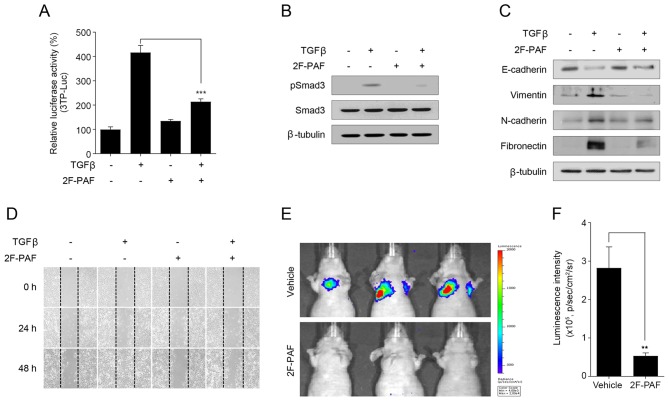
In addition to in vitro experiments, further experiments were performed to examine whether 2F-PAF may attenuate tumor metastasis in vivo. The anti-metastatic activity of 2F-PAF was examined using highly metastatic Calu-1 NSCLC cells. 2F-PAF inhibited TGFβ signaling in Calu-1 cells (Fig. 3A and B) without exhibiting any cytotoxicity effects 27683099 (Fig. S9B). The expression levels of EMT marker proteins in Calu-1 cells were also analyzed. Similar to NCI-H3122 cells (Fig. 2D), TGFβ decreased the expression level of E-cadherin and increased those of vimentin, N-cadherin, and fibronectin, and these effects were reversed by 2F-PAF (Fig. 3C). In addition, wound-healing assay demonstrated that 2F-PAF decreased TGFβ-induced Calu-1 cell migration (Fig. 3D and Fig. S10D-F), which was similar to the results observed in NCI-H3122 cells (Fig. 2E).
Figure 3.
Inhibition of FUTs suppresses tumor metastasis. (A) Calu-1 cells were transfected with a 3TP-Luc reporter construct for 24 h and then incubated with TGFβ (1 ng/ml) and 2F-PAF (200 µM) for 24 h. The luciferase activity was expressed as a relative value compared with that of the untreated cells. Data are expressed as the mean ± SEM (n=3). ***P<0.005. (B) Cells were treated with TGFβ (1 ng/ml) and 2F-PAF at 200 µM for 1 h prior to western blot analysis. (C) Cells were treated with TGFβ (5 ng/ml) and/or 2F-PAF for 48 h prior to western blot analysis. (D) Cells were treated with TGFβ (5 ng/ml) and/or 2F-PAF 200 µM for the indicated time after the wound was created. (E) Representative in vivo bioluminescence images were obtained 10 min after intraperitoneal injection of D-Luciferin (n=4). (F) Bioluminescence intensity was quantified for each mouse, and the mean was calculated for each experimental group. The radiance unit of photons/sec/cm2/sr represents the number of photons per second that leave a square centimeter of tissue and radiate into a solid angle of one sr. FUT, fucosyltransferase; TGFβ, transforming growth factor β; 2F-PAF, 2F-peracetyl-fucose; p, phosphorylated; Luc, luciferase; sr, steradian.
At 200 µM, 2F-PAF completely suppressed TGFβ-induced reporter gene activity and Smad phosphorylation (Figs. 2A and B, and 3A and B), but partially rescued TGFβ-induced EMT markers (Figs. 2D and 3C) in the two NSCLC cell lines. At 800 µM 2F-PAF fully rescued TGFβ-induced changes in EMT markers (data not shown). However, to minimize the adverse drug response, 200 µM 2F-PAF in the in vivo studies. Bioluminescence imaging analysis of a mouse NSCLC metastasis model revealed that 2F-PAF inhibited the colonizing ability of Calu-1-Luc (Fig. 3E and F), a luciferase-expressing Calu-1 cell line that has the ability to colonize to the lung following intravenous injection (44). These results indicated that inhibition of FUTs attenuated the in vivo metastatic capacity of Calu-1 cells.
Discussion
Fucosylation of cell surface receptors serves a crucial role in fine-tuning cellular responses to extracellular stimuli (13). Previous studies have reported that cellular fucosylation patterns are altered during cancer development and progression (21,45). The results of the present study demonstrated that altered expression of fucosylation pathway genes is associated with poor prognosis in patients with NSCLC. In addition, inhibition of FUTs suppressed TGFβ signaling and tumor metastasis.
TGFβ serves a crucial role in cancer metastasis by affecting various cellular processes, including cell migration (46,47). The results of the present study demonstrated that FUTs were aberrantly expressed in NSCLC and that 2F-PAF inhibited TGFβ signaling and cell migration. These results suggested that the altered expression of FUTs may stimulate cancer metastasis by potentiating TGFβ signaling in NSCLC. In addition, these results indicated that FUT inhibitors, including 2F-PAF, may be promising agents against metastasis of NSCLC. A previous study reported the feasibility of FUT inhibitors as anti-metastatic agents in prostate cancer (45). However, to corroborate the clinical significance of the present study, further studies will be required to measure the expression levels of FUTs and fucosylation on their substrates from a mouse model and patient samples. In addition, it is important to investigate how long the effect of 2F-PAF lasts in the experimental conditions and the possible secondary adverse drug effects.
The effect of each fucosylation pathway gene on TGFβ signaling was assessed in the present study using siRNAs against FUT2, FUT3, FUT6, FUT8 and TSTA3; however, siRNA-mediated silencing of any of these gene alone did not recapitulate the effects of 2F-PAF on inhibiting TGFβ signaling in NCI-H3122 cells (data not shown). These results suggested that simultaneous inhibition of all FUTs, and thus multiple types of fucosylation, may be effective in inhibiting TGFβ signaling and metastasis.
The results of the present study demonstrated that FUT1 was downregulated and associated with poor prognosis in patients with NSCLC. By contrast, the expression levels of FUT1 were elevated in samples from patient with prostate cancer (48). These studies provide insight into the cancer type-specific roles of FUT isotypes. However, further studies are needed to investigate how the expression levels of fucosylation pathway genes vary across a wide range of cancer types.
The results of the present study indicated that the change in the expression levels of fucosylation pathway genes is most prominent in the EML4-ALK fusion-positive subtype of NSCLC. These findings suggested that fucosylation may exert differing roles in different NSCLC subtypes. However, due to the small sample size in our dataset, no statistically significant differences were observed in patient prognosis across the different molecular subtypes of NSCLC.
One limitation of the present study was that patients were recruited from different datasets, and could therefore possess different clinicopathological characteristics that affect lung adenocarcinoma survival. In the discovery set (GSE31210), the expression levels of FUT1, FUT2, FUT3, FUT6, FUT8 and TSTA3 correlated with poor RFS. However, in the replication dataset, only FUT1 and FUT8 were associated with poor OS. The different results between the discovery and replication datasets result from individual heterogeneity, including the distribution of sex and smoking status in the datasets. In addition, OS analysis was performed in the replication datasets as there are not enough publicly accessible RFS datasets. Despite these limitations, the strength of the present study is that the target genes extracted from large amounts of disease tran-scriptome data were verified through survival analysis, in vitro and in vivo experiments.
In conclusion, the present study applied a data-driven approach to increase the understanding of the role of fucosylation pathway genes in NSCLC and to assess the clinical relevance of fucosylation pathway genes. In addition, the results of the present study demonstrated that inhibition of FUTs attenuated TGFβ signaling and tumor metastasis. These results suggested that targeting fucosylation may represent a promising strategy for the development of novel NSCLC therapeutics.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was supported by a grant of the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (grant no. A110057).
Availability of data and materials
All data generated or analyzed during this study are including in this published article.
Authors' contributions
All authors contributed to the acquisition, analysis, or interpretation of data for this work. TMK, DWK, SYK, IS, HGK, JYC and JHJ made substantial contributions to the conception or design of the work. SP, JML, JNC, JYC and JHJ contributed to drafting the manuscript. SHL, TMK, DWK, DJB, SMB, SYK, IS and HGK revised the manuscript. All authors approved the final version to be published and agreed to be accountable for all aspects of the work.
Ethics approval and consent to participate
All animal experiments were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Asan Institute for Life Sciences at the Asan Medical Center (2019-14-201).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chansky K, Sculier JP, Crowley JJ, et al. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 4.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 5.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta. 2012;1820:1347–1353. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Miyoshi E, Moriwaki K, Terao N, Tan CC, Terao M, Nakagawa T, Matsumoto H, Shinzaki S, Kamada Y. Fucosylation is a promising target for cancer diagnosis and therapy. Biomolecules. 2012;2:34–45. doi: 10.3390/biom2010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuccillo FM, de Laurentiis A, Palmieri C, Fiume G, Bonelli P, Borrelli A, Tassone P, Scala I, Buonaguro FM, Quinto I, et al. Aberrant glycosylation as biomarker for cancer: Focus on CD43. BioMed Res Int. 2014;2014:742831. doi: 10.1155/2014/742831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 9.Kim HJ, Kim SC, Ju W, Kim YH, Yin SY, Kim HJ. Aberrant sialylation and fucosylation of intracellular proteins in cervical tissue are critical markers of cervical carcinogenesis. Oncol Rep. 2014;31:1417–1422. doi: 10.3892/or.2013.2938. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Wang Y, Yu Y, et al. Aberrant fucosylation of glycosphin-golipids in human hepatocellular carcinoma tissues. Liver Int. 2014;34:147–160. doi: 10.1111/liv.12265. [DOI] [PubMed] [Google Scholar]
- 11.Munkley J, Mills IG, Elliott DJ. The role of glycans in the development and progression of prostate cancer. Nat Rev Urol. 2016;13:324–333. doi: 10.1038/nrurol.2016.65. [DOI] [PubMed] [Google Scholar]
- 12.Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proc Natl Acad Sci USA. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker DJ, Lowe JB. Fucose: Biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 14.Oriol R, Mollicone R, Cailleau A, Balanzino L, Breton C. Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology. 1999;9:323–334. doi: 10.1093/glycob/9.4.323. [DOI] [PubMed] [Google Scholar]
- 15.Tu Z, Lin YN, Lin CH. Development of fucosyltransferase and fucosidase inhibitors. Chem Soc Rev. 2013;42:4459–4475. doi: 10.1039/c3cs60056d. [DOI] [PubMed] [Google Scholar]
- 16.Merino P, Tejero T, Delso I, Hurtado-Guerrero R, Gómez-SanJuan A, Sádaba D. Recent progress on fucosyltransferase inhibitors. Mini Rev Med Chem. 2012;12:1455–1464. doi: 10.2174/138955712803832753. [DOI] [PubMed] [Google Scholar]
- 17.Mollicone R, Moore SE, Bovin N, Garcia-Rosasco M, Candelier JJ, Martinez-Duncker I, Oriol R. Activity, splice variants, conserved peptide motifs, and phylogeny of two new alpha1,3-fucosyltransferase families (FUT10 and FUT11) J Biol Chem. 2009;284:4723–4738. doi: 10.1074/jbc.M809312200. [DOI] [PubMed] [Google Scholar]
- 18.Guo Q, Guo B, Wang Y, Wu J, Jiang W, Zhao S, Qiao S, Wu Y. Functional analysis of α1,3/4-fucosyltransferase VI in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2012;417:311–317. doi: 10.1016/j.bbrc.2011.11.106. [DOI] [PubMed] [Google Scholar]
- 19.Barthel SR, Wiese GK, Cho J, Opperman MJ, Hays DL, Siddiqui J, Pienta KJ, Furie B, Dimitroff CJ. Alpha 1,3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc Natl Acad Sci USA. 2009;106:19491–19496. doi: 10.1073/pnas.0906074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muinelo-Romay L, Vázquez-Martín C, Villar-Portela S, Cuevas E, Gil-Martín E, Fernández-Briera A. Expression and enzyme activity of alpha(1,6)fucosyltransferase in human colorectal cancer. Int J Cancer. 2008;123:641–646. doi: 10.1002/ijc.23521. [DOI] [PubMed] [Google Scholar]
- 21.Chen CY, Jan YH, Juan YH, Yang CJ, Huang MS, Yu CJ, Yang PC, Hsiao M, Hsu TL, Wong CH. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc Natl Acad Sci USA. 2013;110:630–635. doi: 10.1073/pnas.1220425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho AS, Harduin-Lepers A, Magalhães A, Machado E, Mendes N, Costa LT, Matthiesen R, Almeida R, Costa J, Reis CA. Differential expression of alpha-2,3-sialyltransferases and alpha-1,3/4-fucosyltransferases regulates the levels of sialyl Lewis a and sialyl Lewis x in gastrointestinal carcinoma cells. Int J Biochem Cell Biol. 2010;42:80–89. doi: 10.1016/j.biocel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Zhang W, Luo J, Cao A, Zhang Y, Huang D, Sheng W, Cai S, Li J. Alpha1,3 fucosyltransferase VII plays a role in colorectal carcinoma metastases by promoting the carbohy-dration of glycoprotein CD24. Oncol Rep. 2010;23:1609–1617. doi: 10.3892/or_00000802. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Chen J, Li QK, Peskoe SB, Zhang B, Choi C, Platz EA, Zhang H. Overexpression of α (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology. 2014;24:935–944. doi: 10.1093/glycob/cwu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Liu S, Yan Q. Role of fucosyltransferase IV in epithelial-mesenchymal transition in breast cancer cells. Cell Death Dis. 2013;4:e735. doi: 10.1038/cddis.2013.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honma R, Kinoshita I, Miyoshi E, Tomaru U, Matsuno Y, Shimizu Y, Takeuchi S, Kobayashi Y, Kaga K, Taniguchi N, et al. Expression of fucosyltransferase 8 is associated with an unfavorable clinical outcome in non-small cell lung cancers. Oncology. 2015;88:298–308. doi: 10.1159/000369495. [DOI] [PubMed] [Google Scholar]
- 27.Ma L, Dong P, Liu L, Gao Q, Duan M, Zhang S, Chen S, Xue R, Wang X. Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway. Biochem Biophys Res Commun. 2016;473:503–510. doi: 10.1016/j.bbrc.2016.03.062. [DOI] [PubMed] [Google Scholar]
- 28.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 29.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY, Lantuejoul S, Hainaut P, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Y, Xiao G, Coombes KR, Behrens C, Solis LM, Raso G, Girard L, Erickson HS, Roth J, Heymach JV, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. 2011;17:5705–5714. doi: 10.1158/1078-0432.CCR-11-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et al. Biomarker discovery in non-small cell lung cancer: Integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 32.Der SD, Sykes J, Pintilie M, Zhu CQ, Strumpf D, Liu N, Jurisica I, Shepherd FA, Tsao MS. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9:59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Qiu P, Ji Y. TCGA-assembler: Open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11:599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eijssen LM, Jaillard M, Adriaens ME, Gaj S, de Groot PJ, Müller M, Evelo CT. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis.org. Nucleic Acids Res. 2013;41(W1):W71–6. doi: 10.1093/nar/gkt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, Chun JN, Kim SH, So I, Jeon JH. Icilin inhibits E2F1-mediated cell cycle regulatory programs in prostate cancer. Biochem Biophys Res Commun. 2013;441:1005–1010. doi: 10.1016/j.bbrc.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Lee S, Park YR, Kim SH, Park EJ, Kang MJ, So I, Chun JN, Jeon JH. Geraniol suppresses prostate cancer growth through down-regulation of E2F8. Cancer Med. 2016;5:2899–2908. doi: 10.1002/cam4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabregat A, Korninger F, Viteri G, Sidiropoulos K, Marin-Garcia P, Ping P, Wu G, Stein L, D'Eustachio P, Hermjakob H. Reactome graph database: Efficient access to complex pathway data. PLOS Comput Biol. 2018;14:e1005968. doi: 10.1371/journal.pcbi.1005968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park EJ, Chun JN, Kim SH, Kim CY, Lee HJ, Kim HK, Park JK, Lee SW, So I, Jeon JH. Schisandrin B suppresses TGFβ1 signaling by inhibiting Smad2/3 and MAPK pathways. Biochem Pharmacol. 2012;83:378–384. doi: 10.1016/j.bcp.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Chun JN, Kim SY, Park EJ, Kwon EJ, Bae DJ, Kim IS, Kim HK, Park JK, Lee SW, Park HH, et al. Schisandrin B suppresses TGFβ1-induced stress fiber formation by inhibiting myosin light chain phosphorylation. J Ethnopharmacol. 2014;152:364–371. doi: 10.1016/j.jep.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Kim JS, Kim JG, Moon MY, Jeon CY, Won HY, Kim HJ, Jeon YJ, Seo JY, Kim JI, Kim J, et al. Transforming growth factor-beta1 regulates macrophage migration via RhoA. Blood. 2006;108:1821–1829. doi: 10.1182/blood-2005-10-009191. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Hu G, Chen D, Gong AY, Soori GS, Dobleman TJ, Chen XM. Suppression of SCARA5 by Snail1 is essential for EMT-associated cell migration of A549 cells. Oncogenesis. 2013;2:e73. doi: 10.1038/oncsis.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirakawa M, Takimoto R, Tamura F, Yoshida M, Ono M, Murase K, Sato Y, Osuga T, Sato T, Iyama S, et al. Fucosylated TGF-β receptors transduces a signal for epithelial-mesenchymal transition in colorectal cancer cells. Br J Cancer. 2014;110:156–163. doi: 10.1038/bjc.2013.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin H, Wang D, Wu T, Dong C, Shen N, Sun Y, Sun Y, Xie H, Wang N, Shan L. Blocking core fucosylation of TGF-β1 receptors downregulates their functions and attenuates the epithelial-mesenchymal transition of renal tubular cells. Am J Physiol Renal Physiol. 2011;300:F1017–F1025. doi: 10.1152/ajprenal.00426.2010. [DOI] [PubMed] [Google Scholar]
- 44.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK, Sato M, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–3235. doi: 10.1172/JCI76725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Guillebon AD, Hsu JW, Barthel SR, Dimitroff CJ, Lee YF, King MR. Human fucosyltransferase 6 enables prostate cancer metastasis to bone. Br J Cancer. 2013;109:3014–3022. doi: 10.1038/bjc.2013.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padua D, Massagué J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 47.Drabsch Y, ten Dijke P. TGF-β signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012;31:553–568. doi: 10.1007/s10555-012-9375-7. [DOI] [PubMed] [Google Scholar]
- 48.Fukushima K, Satoh T, Baba S, Yamashita K. alpha1,2-Fucosylated and beta-N-acetylgalactosaminylated prostate-specific antigen as an efficient marker of prostatic cancer. Glycobiology. 2010;20:452–460. doi: 10.1093/glycob/cwp197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are including in this published article.