Abstract
Formation of a specialized wound epidermis is required to initiate salamander limb regeneration. Yet little is known about the roles of the early wound epidermis during the initiation of regeneration and the mechanisms governing its development into the apical epithelial cap (AEC), a signaling structure necessary for outgrowth and patterning of the regenerate. Here, we elucidate the functions of the early wound epidermis, and further reveal midkine (mk) as a dual regulator of both AEC development and inflammation during the initiation of axolotl limb regeneration. Through loss- and gain-of-function experiments, we demonstrate that mk acts as both a critical survival signal to control the expansion and function of the early wound epidermis and an anti-inflammatory cytokine to resolve early injury-induced inflammation. Altogether, these findings unveil one of the first identified regulators of AEC development and provide fundamental insights into early wound epidermis function, development, and the initiation of limb regeneration.
Research organism: Other
Introduction
Salamanders possess the impressive ability to regenerate their limbs with high fidelity throughout their lifespan (Tanaka, 2016; Flowers et al., 2017). While most mammals respond to traumatic limb injuries including amputation with a fibrotic scarring response, salamanders instead form a blastema, a transient cellular structure comprised of progenitors derived from different non-epithelial tissues (muscle, connective tissue etc.) that eventually reconstructs the new limb (Kragl et al., 2009; Sandoval-Guzmán et al., 2014; Fei et al., 2017; Gerber et al., 2018). Determining the mechanisms required for blastema formation has the potential to provide valuable insights into improving repair and regenerative outcomes in less regenerative-species including humans.
Classical salamander limb regenerative experiments determined that initiation of blastema formation requires the presence of a wound epidermis for both formation and maintenance of the blastema (Goss, 1956a; Thornton, 1957; Thornton, 1958; Mescher, 1976; Tassava and Garling, 1979). Immediately post-amputation, epithelial cells from the intact peripheral skin migrate radially over the exposed stump tissues and form a thin wound epithelium covering the amputation plane within 12 hours (Hay and Fischman, 1961). Unlike normal full thickness skin that contains two layers (epidermis and dermis), the wound epidermis only contains an epithelial layer comprised primarily of keratinocytes, allowing for direct communication between the wound epidermis and underlying stump tissues (Repesh and Oberpriller, 1978). This thin wound epidermis persists through the initial wound healing stages of regeneration. As the blastema forms during later stages of regeneration, the early wound epithelium gradually expands into a stratified epidermis commonly called the apical epithelial cap (AEC), which is a signaling structure that maintains blastemal outgrowth and directs patterning (Boilly and Albert, 1990; Christensen and Tassava, 2000; Han et al., 2001; Christensen et al., 2002). This expansion occurs primarily via proliferation and subsequent distal migration of epithelial cells in the peripheral skin (Hay and Fischman, 1961; Campbell and Crews, 2008). Classical studies showed that inhibiting early wound epidermis formation by either suturing or grafting full thickness skin over the amputation plane (Mescher, 1976; Tassava and Garling, 1979) or immediately inserting the amputated limb into the body cavity (Goss, 1956a) blocked blastema formation. However, if the amputated limb was inserted into the body cavity after the wound epidermis formed, regeneration proceeded normally, demonstrating an early need for the wound epithelium (Goss, 1956b). Furthermore, preventing AEC development by either continuous removal of the wound epidermis (Thornton, 1957) or daily irradiation to inhibit proliferative expansion also hindered limb regeneration (Thornton, 1958). Together, these findings established the necessity for a wound epidermis/AEC throughout different stages of limb regeneration.
The nature of wound epidermis dependence has been more well explored during later blastemal stages of regeneration. The AEC expresses mitogens and patterning factors that help to maintain blastemal cell proliferation and pattern the regenerate (Boilly and Albert, 1990; Campbell and Crews, 2008; Ghosh et al., 2008; Satoh et al., 2008). However, the roles of the wound epidermis during early stages of regeneration are less clear. Moreover, the early wound epidermis is likely to be functionally distinct from the AEC as the wound healing micro-environment differs substantially from that of the blastema. Preventing early wound epidermis formation was shown to affect the maintenance, but not the onset of proliferation of progenitors (Mescher, 1976; Tassava and Garling, 1979; Johnson et al., 2018), though it is worth noting that the early wound epidermis does express molecules capable of inducing proliferation (Sugiura et al., 2016). Additionally, some dedifferentiation of muscle, bone and cartilage tissue was still observed when wound epidermis formation was prevented, although not to the same extent as normal regenerating limbs. Altogether these studies indicated that the early wound epidermis is involved in maintaining progenitor cell cycling and may also play a role in tissue histolysis.
More recently, bulk and single cell transcriptional limb regeneration studies have further characterized gene expression in the early wound epidermis and AEC (Campbell et al., 2011; Leigh et al., 2018; Tsai et al., 2019). Yet, it remains unknown what gene expression programs in regenerating tissues depend on the presence of the early wound epidermis. The initiation of blastema formation is a complex event with inflammation, tissue histolysis, extracellular matrix (ECM) remodeling, and cell cycle re-entry of progenitors all transpiring concurrently (McCusker et al., 2015). Aside from the induction of proliferation, whether and to what extent the early wound epidermis may regulate these other processes remains relatively unexplored. Furthermore, little is known about the molecular mechanisms controlling the transition from the early wound epithelium into the AEC, a major milestone for successful limb regeneration.
To investigate these questions, we sought to deconvolute the functional roles of the wound epidermis from the rest of the regenerating limb tissues by revisiting the classical full skin flap surgical model to inhibit wound epidermis formation during limb regeneration. Using modern-day transcriptional approaches, we illuminate the roles of the early wound epidermis and identify distinct wound epidermis-dependent transcriptional programs in dividing progenitors and the surrounding regenerating tissues. In addition to the discovery of putative regulators of wound epidermis formation and function, our approach enabled us to identify midkine (mk) as a regulator of both AEC development and early inflammation, key events during the initiation of blastema formation. Altogether, these findings provide important insight into the molecular basis for wound epidermis dependence and further advance our understanding of wound epithelium/AEC biology during limb regeneration.
Results
The wound epithelium is a major regulator of inflammation, ECM remodeling, and tissue histolysis during early stages of regeneration
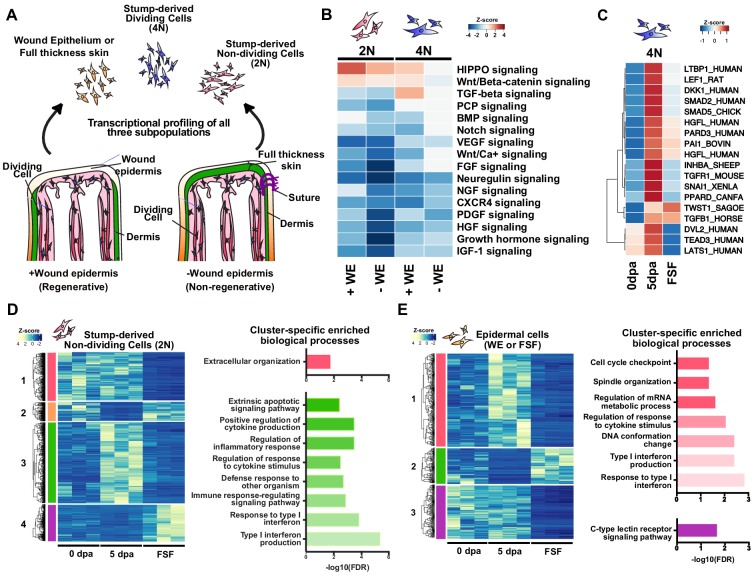
We previously developed a method leveraging differential DNA content during the cell cycle to enrich for dividing progenitors during early stages of regeneration (Tsai et al., 2019), facilitating analysis of the distinct transcriptional programs active in blastemal progenitors apart from those of the surrounding tissues (non-dividing cells in stump tissues and the wound epidermis). To examine the functional roles of the early wound epidermis, we asked how the transcriptional programs of these three populations changed when wound epidermis formation was prevented. To do this, we utilized the classical full skin flap (FSF) surgical model pioneered by Anthony Mescher (Mescher, 1976). We performed full skin flap surgeries on regenerating axolotl limbs to prevent epithelial cell migration and contact with exposed stump tissues, inhibiting both wound epidermis formation and limb regeneration (Figure 1—figure supplement 1A–B). Since preventing early wound epidermis formation via full skin flap surgery does not affect induction of proliferation (Mescher, 1976; Tassava and Garling, 1979; Johnson et al., 2018), we applied our method here to profile blastemal progenitors and the surrounding regenerating tissues in full skin flap sutured limbs at 5 days post-amputation (dpa), around the onset of proliferation. By leveraging the data generated from our previous analysis (Tsai et al., 2019), we were able to compare the gene expression programs in full skin flap sutured regenerating limbs to both intact limbs and normal regenerating limbs from the same time point (Figure 1A, Figure 1—figure supplement 1C). This approach allowed us to identify wound epidermis-dependent gene expression.
Figure 1. The early wound epidermis modulates inflammation, ECM remodeling, and tissue histolysis.
(A) Design of transcriptional profiling experiment. (B) Heatmap of Ingenuity Pathway Analysis (IPA) Z-score predictions for changes in signaling pathways in injured non-epithelial stump tissues (2N and 4N) in regenerative and full skin flap conditions. (C) Heatmap of normalized transcript levels of components/regulators of the canonical Wnt, TGF-beta, and HIPPO signaling pathways in dividing cells (4N). (D–E) Heatmaps and cluster-specific enriched biological processes of differentially expressed annotated transcripts in stump-derived non-dividing cells (2N) (D) and epithelial cells of the full skin flap (E) in regenerating vs. full skin flap conditions reveals global dysregulation of inflammation, ECM regulation, and tissue histolysis. Differentially expressed transcripts can be found in Supplementary files 1–3. Representative histological images of full skin flap sutured vs. normal regenerating limbs, PCA analysis of the samples, as well as enriched biological processes and related differentially expressed transcripts are shown in Figure 1—figure supplement 1. Z-score predictions shown in B from the IPA analysis are available in Figure 1—source data 1.
Figure 1—figure supplement 1. Sequencing of full skin flap sutured limbs reveals global dysregulation of ECM regulation, inflammation, and tissue histolysis.
Preventing wound epidermis formation led to aberrant growth factor signaling pathway activation/repression in regenerating stump tissues (Figure 1B). Pathway analysis revealed that while non-dividing cells exhibited no major differences, dividing cells (i.e. enriching for progenitors) exhibited a loss in predicted activation of canonical Wnt, HIPPO, and TGF-beta signaling. Many components of these pathways were also expressed at lower levels (Figure 1C), indicating that the wound epidermis induces transcriptional activation of these pathways in dividing progenitors. Interestingly, some components of these pathways including tgfb1 and twist1 were not transcriptionally affected, suggesting that their expression in progenitors is wound epidermis-independent. Notably, the overall transcriptional profiles of dividing progenitors in both cases were relatively similar (596 differentially expressed transcripts, 313 annotated) (Supplementary file 1), signifying that while the wound epidermis activates key signaling pathways, the overall gene expression programs in early progenitors are largely wound epidermis-independent. This lends transcriptional evidence to reinforce the notion that the earliest transcriptional programs of progenitors are likely driven in response to the injury itself (Tassava and Loyd, 1977; Wagner et al., 2017; Johnson et al., 2018).
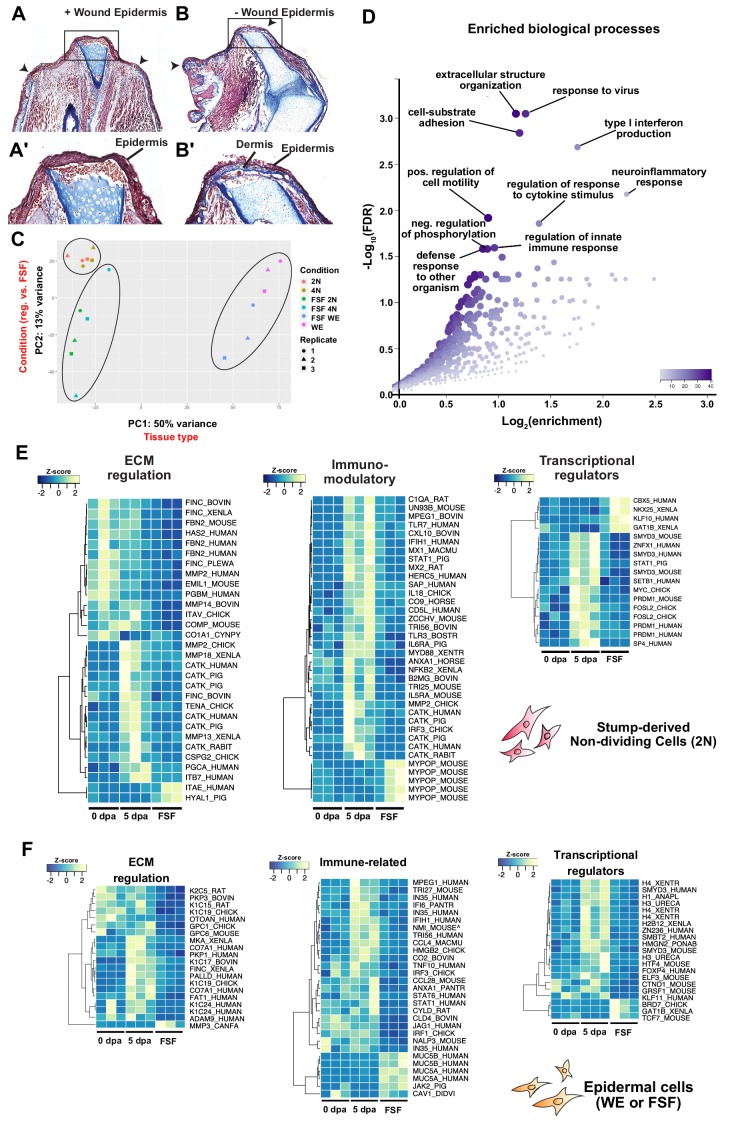
In contrast to the dividing progenitors, the transcriptional programs of the surrounding tissues diverged considerably. We found that 3911 transcripts (982 annotated) were differentially expressed in non-dividing cells in regenerating stump tissues (Figure 1D, Figure 1—figure supplement 1D–E, Supplementary file 2). The majority of these transcripts pertained to genes involved in ECM regulation, inflammation, and tissue histolysis. Many ECM-components and regulators that maintained expression in intact and normal regenerating tissues were aberrantly down-regulated (e.g. fbn2, finc, has2), and regenerative ECM components as well as degrading enzymes (e.g. tenascin, mmp2, and mmp13) failed to be induced, indicating defects in ECM remodeling. Furthermore, there was a high representation of mis-regulated immune-related transcripts, both pertaining to pro- and anti-inflammatory processes. In particular, components of pro-inflammatory signaling pathways including toll-like receptor (TLR) signaling and interferon α/β/γ signaling, as well as anti-inflammatory pathways including PPARα/RXRα signaling and anxa1 signaling were dysregulated (Figure 1—figure supplement 1D–E). Enzymes involved in tissue histolysis were mis-expressed as well. Notably, expression of the major bone matrix degrading enzyme catk failed to be induced, while the major bone and cartilage matrix component col1a1 maintained expression, indicating potential defects in bone resorption, a process that begins during the first week of regeneration (Fischman and Hay, 1962). Altogether, these data suggest that the wound epidermis is a major regulator of inflammation, ECM remodeling, and tissue histolysis in regenerating stump tissues during early stages of limb regeneration.
The presence of the dermis in full skin flap sutured limbs additionally presented a crude, yet effective opportunity to identify epithelial transcriptional programs dependent on direct contact between the wound epidermis and regenerating stump tissues. We found that 1060 transcripts (480 annotated) were differentially expressed (Figure 1E, Figure 1—figure supplement 1F, Supplementary file 3). Similar to the non-dividing cells in regenerating stump tissues, many transcripts involved in ECM regulation and inflammatory signaling (both pro- and anti-inflammatory) were also dysregulated in full skin flap epithelial cells (Figure 1—figure supplement 1F). Components of interferon and cytokine signaling failed to be induced in full skin flap derived epithelial cells, while anti-pathogenic mucins including muc5a and muc5b were overexpressed, suggesting dysregulation of inflammatory processes. Interestingly, mpeg1 commonly failed to be induced in the full skin flap epithelium as well as non-dividing cells in regenerating stump tissues, potentially indicating a defect in macrophage infiltration during early regeneration. Furthermore, ECM components expressed in intact skin (e.g. otoan, k1c15, k1c19, k2c5) lost expression in full skin flap epithelia, and other ECM regulators including finc, palld, and mmp3 were also differentially expressed. These data strongly suggest that direct communication between regenerating mesenchymal tissues and epithelial cells is crucial for proper modulation of early inflammatory responses and ECM remodeling.
Midkine is highly expressed in the basal layers of the wound epidermis/AEC and blastemal progenitors
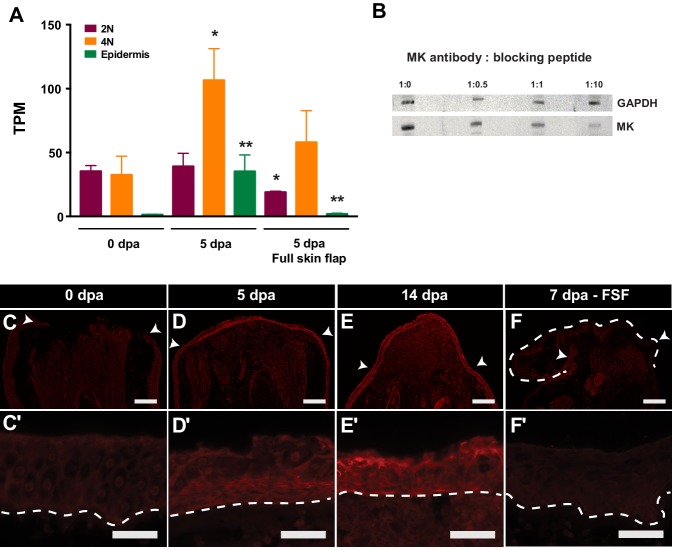
Midkine (mk) is a pleiotropic growth factor cytokine that has been shown to regulate a variety of processes including development, tumorigenesis, inflammation, and tissue repair (Muramatsu, 2010; Weckbach et al., 2011; Gramage et al., 2015; Karaman and Alitalo, 2017). Interestingly, we noticed that mk expression was induced in the wound epidermis and highly upregulated in early regenerating stump tissues. In full skin flap sutured limbs, mk expression decreased across all transcriptionally profiled subpopulations (Figure 2—figure supplement 1A). Specifically, while mk expression in regenerating stump tissues was lower, mk failed to be transcriptionally induced in full skin flap epithelial cells. These data suggested that mk may modulate wound epidermis development/function. Hence, we chose to further explore the role of mk during limb regeneration.
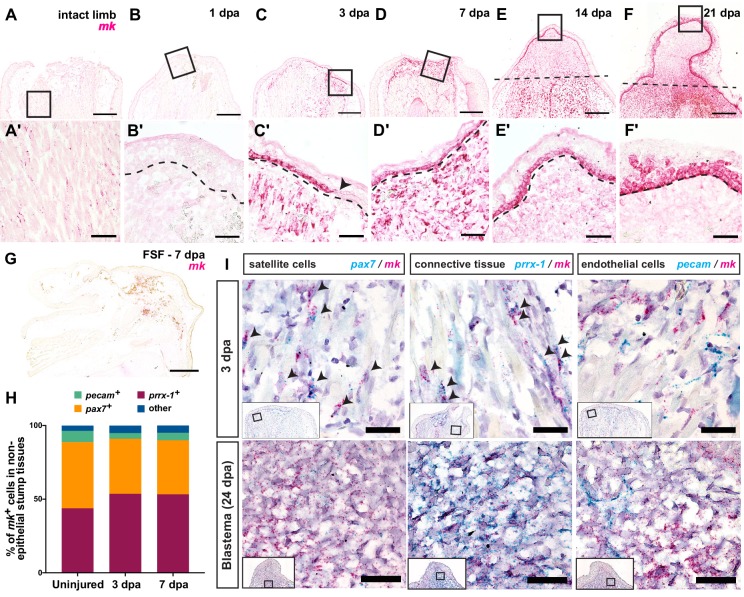
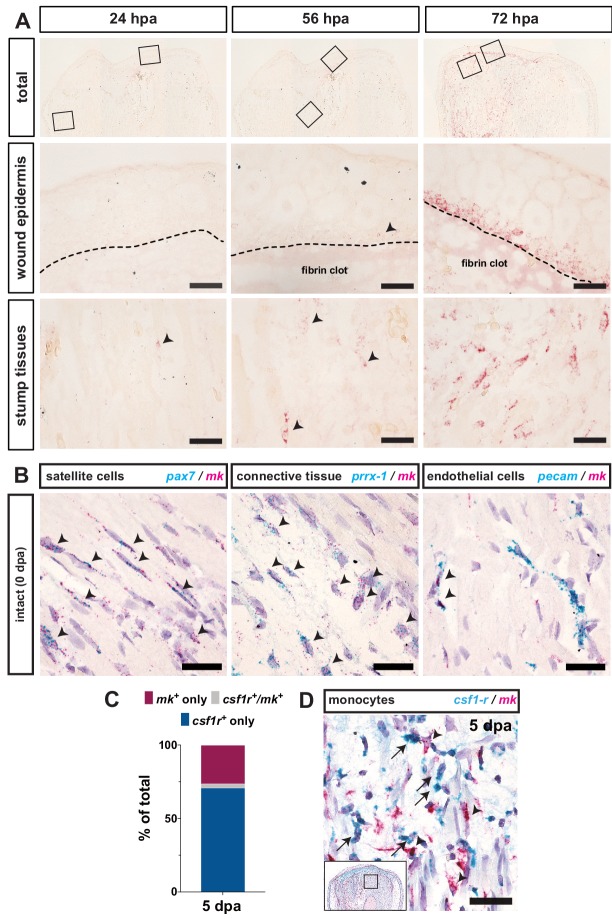
To determine when and in which cell types mk is expressed during limb regeneration, we performed a series of single and double in situ hybridizations. Time course in situ hybridization of mk confirmed that it is highly expressed during limb regeneration, but not in full skin flap sutured regenerating limbs (Figure 2A–G). In non-regenerating intact limbs, mk was lowly expressed in cells interspersed throughout the muscle and dermis (Figure 2A–A’). During regeneration, mk became strongly expressed in both the basal layers of the wound epidermis and mesenchymal cells within regenerating stump tissues beginning at 3 dpa (Figure 2B–C’). To determine whether mk expression was first up-regulated in regenerating stump tissues or the wound epidermis, we performed a more detailed analysis of the onset of mk expression prior to 3 dpa. In situ hybridization of mk at 24 and 56 hours post-amputation (hpa) revealed a small population of mk+ cells interspersed throughout regenerating stump tissues with little to no expression of mk in the basal layers of the wound epidermis until 72 hpa (Figure 2—figure supplement 2A). Together, these data indicate that mk is first expressed in regenerating stump tissues prior to the induction of expression in the wound epidermis. During later stages of regeneration, mk expression persisted at high levels in the basal layers of the AEC and lower levels in the blastema (Figure 2D–F’). Interestingly, we also noticed relatively strong mk expression within the non-regenerating tissue at the base of the blastema (Figure F-F’), suggesting mk may play a role in differentiation during later stages of regeneration. Immunostaining of axolotl MK protein further revealed the presence of MK protein throughout the AEC and blastema, providing evidence that it is indeed secreted (Figure 2—figure supplement 1B–F’).
Figure 2. Midkine (mk) is highly expressed in the basal layers of the wound epidermis/AEC and blastemal progenitors.
(A–F’) Timecourse RNAscope in situ hybridization of midkine at 0 (intact), 1, 3, 7, 14, and 21 dpa. Insets in A-F are shown in A’-F’ at higher magnification. Arrowhead in C’ denotes the beginning of the wound epidermis. Dotted line marks amputation plane in E and F or wound epidermis/AEC boundary in B’-F’. (G) In situ hybridization of mk in full skin flap sutured limbs at 7 dpa. Axolotl MK protein expression can be found in Figure 2—figure supplement 1. (H) Breakdown of the percentages of mk+ cells that are pax7+, prrx-1+, and pecam+ in regenerating stump tissues during early stages of regeneration. At 3 dpa, a total N of 1579, 444, and 1180 cells were counted for pax7, prrx-1, and pecam double in situs, respectively. At 7 dpa, a total N of 456, 1043, and 274 cells were counted for pax7, prrx-1, and pecam double in situs, respectively (Figure 2—source data 1). (I) Representative images of RNAscope double in situ hybridization of mk with pax7 (left), prrx-1 (middle), or pecam (right) at early (3 dpa) and later blastema (24 dpa) stages. Insets depict where higher magnification images were taken. Black arrowheads mark double positive cells. More detailed analysis of the onset of mk expression in early stages of regeneration, representative images of double in situ hybridization of mk with cell type-specific markers in uninjured tissue, as well as the analysis of mk co-expression with the monocyte marker csf1r can be found in Figure 2—figure supplement 2. Scale bars, A-G: 500 µm, A’-F’: 100 µm, I: 50 µm. dpa, days post-amputation, FSF, full skin flap.
Figure 2—figure supplement 1. MK protein is found throughout the wound epidermis/AEC.
Figure 2—figure supplement 2. Mk is expressed in satellite cells, connective tissue, and endothelial cells in intact tissues.
Next, to identify which cell types expressed mk in regenerating stump tissues, we performed double in situ hybridization of mk with various cell type-specific markers in non-regenerating and regenerating limbs. Because we had observed mk+ cells interspersed in the muscle and dermis of intact tissues, we surmised that mk may be expressed at low homeostatic levels in subsets of connective tissue, muscle, or endothelial cells. To examine this possibility, we performed double in situ hybridization of mk with prrx-1, pax7, and pecam, which are markers for connective tissue cells (Gerber et al., 2018), muscle satellite cells (Sandoval-Guzmán et al., 2014), and endothelial cells (Whited et al., 2013), respectively. Indeed, our analysis revealed that approximately 96.4% of mk+ expressing cells in non-regenerating tissues were comprised of prrx-1+ connective tissue cells (~43.8% of mk+ cells), pax7+ satellite cells (~44.9% of mk+ cells), and pecam+ endothelial cells (~7.7% of mk+ cells) (Figure 2H, Figure 2—figure supplement 2B).
Interestingly, we observed a relatively similar breakdown of mk+ cells across all three cell types during early stages limb regeneration (Figure 2H–I). At both 3 and 7 dpa, mk-expressing cells could be divided into connective tissue cells (~53–54%), satellite cells (~36–37%), and a small percentage of endothelial cells (4–5%). Given heavy immune cell infiltration during wound healing stages of regeneration, we also investigated whether mk was expressed in monocytes by performing double in situ hybridization with csf1-r, a monocyte marker (Grayfer et al., 2014). We found that only a small percentage (~9%) of mk+ cells were csf1-r+ and only 3.7% of csf1-r+ monocytes were mk+ at 5 dpa (Figure 2—figure supplement 2C–D), indicating mk is not predominantly expressed in monocytes. In addition, while the densely packed cells within the blastema at 24 dpa precluded the accurate quantification of double positive cells, we did observe overlapping expression patterns between mk and both prrx-1 and pax7, but not pecam (Figure 2I), indicating mk expression persists in connective tissue and muscle-derived blastemal cells. Altogether, these data strongly indicated that mk expression is highly up-regulated during limb regeneration in the majority of blastemal cells, since satellite and connective tissue cells comprise most of the blastema (Fei et al., 2017; Gerber et al., 2018). As communication between blastemal cells and the basal layers of the wound epidermis/AEC is crucial for blastema formation (Stocum and Dearlove, 1972), the unique expression pattern of mk in both cell types suggested that mk may play an important role in facilitating signaling between the two tissues.
Chemical and genetic perturbation of mk impairs limb regeneration
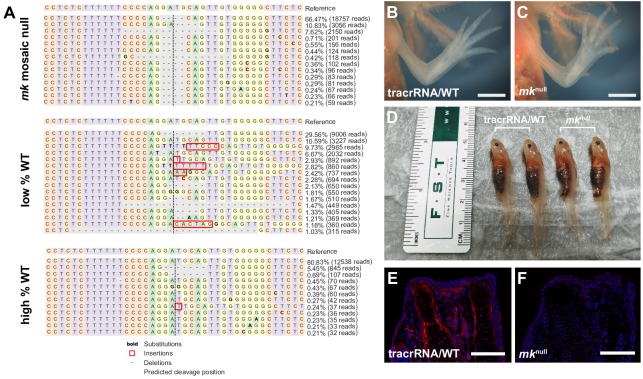
We next examined whether mk loss-of-function affected limb regeneration using both chemical and genetic methods. We first performed drug treatments with a small molecule inhibitor of MK, iMDK (Masui et al., 2016) (Figure 3). Treatment with iMDK beginning at either 0 or 7 dpa completely inhibited limb regeneration (Figure 3A–B). Alcian blue staining of limbs at 60 dpa confirmed that no skeletal structures had regenerated in iMDK-treated limbs (Figure 3C–D). We also generated F0 mk CRISPR mutants to perturb mk genetically (Figure 3E). The slow development of axolotl embryos permits high penetrance of mutations allowing phenotyping to occur at the F0 stage (Fei et al., 2018). Mk F0 mutants generated by targeting the start codon with CRISPR technology (see Materials and methods) developed with no visible abnormalities and anatomically normal limbs (Figure 3—figure supplement 1). Many of these mutants had mosaic null genotypes. Upon amputation, mk F0 mutants exhibited delayed blastema formation and regeneration (Figure 3E–F). Notably, measurements of blastema length at 14 dpa revealed that the severity of delay in regeneration segregated based on genotype, with mk mosaic null animals (>99% mutant alleles) exhibiting the most severe delay compared to control animals (Figure 3G–I). These data demonstrate that both chemical and genetic perturbations of mk impair limb regeneration.
Figure 3. Chemical and genetic perturbations of mk impair limb regeneration.
(A) Experimental design. (B) Brightfield images of DMSO- or iMDK-treated limbs (N = 4/4 iMDK-treated in each condition did not regenerate). (C–D) Alcian blue staining of DMSO- or iMDK-treated limbs at 60 dpa. (E) CRISPR strategy to target the start codon of mk to generate mutants. Control animals were generated using a non-targeted tracrRNA/cas9 complex and were unmodified at the mk locus. (F) Brightfield images of regenerating limbs from control animals or mk F0 mutants. (G) Quantification of blastema length at 14 dpa in control or mk mutant limbs. The severity of the delay in regeneration segregated based on genotype of the animal as either control (mkWT), mosaic null (mknull), or partially modified (low or high % WT alleles) (N = 14 control mkWT, 5 mknull, 8 low % WT, 4 high % WT). Graph is mean ± SD. (H–I) Representative images of picro-mallory stained sections of regenerating limbs in control animals or mk null mutants. Example genotyping analyses and mk null mutant immunofluorescence validation can be found in Figure 3—figure supplement 1. **p<0.005, two-tailed unpaired Student’s t-test was employed. Each N represents one limb from a different animal. White dotted lines mark amputation plane. Scale bars, B-D, H-I: 500 µm, F: 1 mm. WT, wildtype, dpa, days post-amputation.
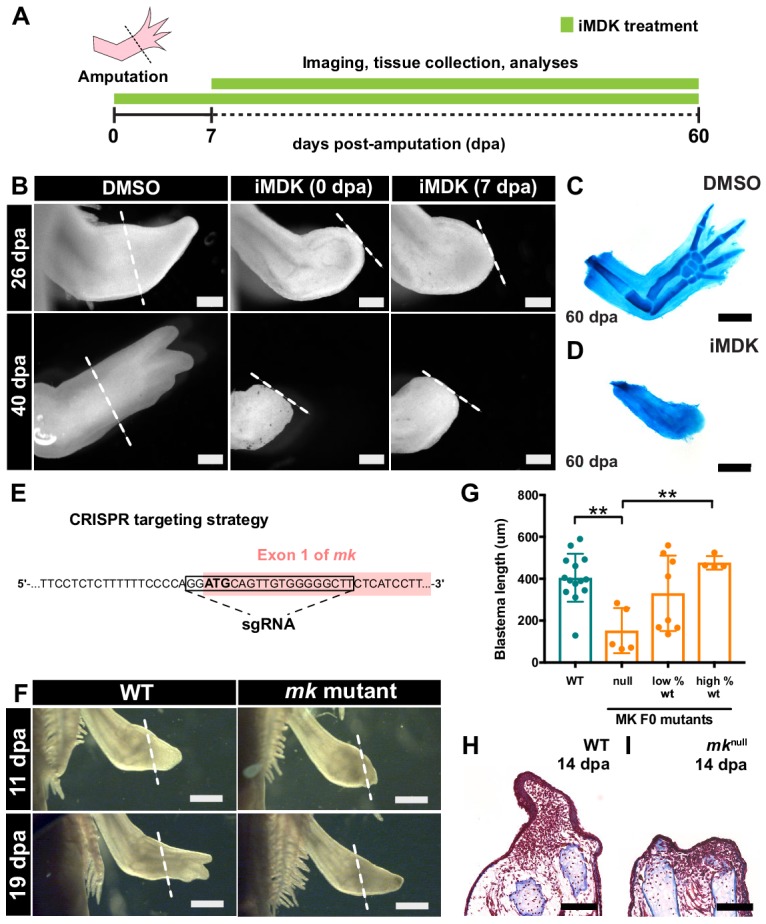
Figure 3—figure supplement 1. CRISPR generation of mk mosaic null mutants.
Mk loss-of-function leads to impaired AEC development and persistent inflammation
To elucidate the cellular mechanism of the regeneration defect, we further characterized both iMDK-treated and mk null (mknull) mutant regenerating limbs (Figure 4). Interestingly, iMDK-treated regenerating limbs had significantly lower percentages of total EdU+ cells during late (14 dpa), but not early stages of limb regeneration (5 dpa) (Figure 4A–B). Furthermore, blastemal cells accumulated at the amputation plane in iMDK-treated limbs even at 40 dpa (Figure 4—figure supplement 1), however blastemal outgrowth never occurred, altogether suggesting that mk is involved in maintenance, but not induction of cellular proliferation. In addition, we observed a significantly thinner wound epidermis in both iMDK-treated and mknull mutant limbs during later stages of regeneration compared to controls (Figure 4C–H). These dual observations indicated a defect in wound epidermis expansion and function during AEC development.
Figure 4. Mk perturbations result in defective AEC development.
(A–B) EdU staining (A) and quantification (B) of the total percentage of EdU+ nuclei of DMSO/iMDK-treated limbs at 5 dpa (N = 6 DMSO, 6 iMDK) and 14 dpa (N = 4 DMSO, 5 iMDK). White arrowheads denote the amputation plane in A. (C–D’) Picro-mallory stained sections from regenerating limbs from DMSO/iMDK-treated animals. Higher magnification images of the AEC are shown in C’-D’. (E) Quantification of wound epidermis (WE) thickness in DMSO/iMDK-treated regenerating limbs at 14 dpa (N = 4 DMSO, 5 iMDK). (F–G’) Picro-mallory stained sections from regenerating limbs from regenerating limbs of control WT (mkWT) or mknull animals. Higher magnification images of the developing AEC at 10 dpa are shown in F’-G’. (H) Quantification of WE thickness in regenerating limbs of mkWT or mknull animals at 14 dpa (N = 8 mkWT, 8 mknull). Picro-mallory staining of a 40 dpa iMDK-treated limb is shown in Figure 4—figure supplement 1. Each N represents a limb from a different animal. Two-tailed unpaired student’s t-test was used for statistical analysis. Graphs are mean ± SD. *p<0.05, **p<0.005, ***p<0.001. Scale bars, A, C-D, F-G: 200 µm, C’-D’, F’-G’: 100 µm. bl, blastema, dpa, days post-amputation.
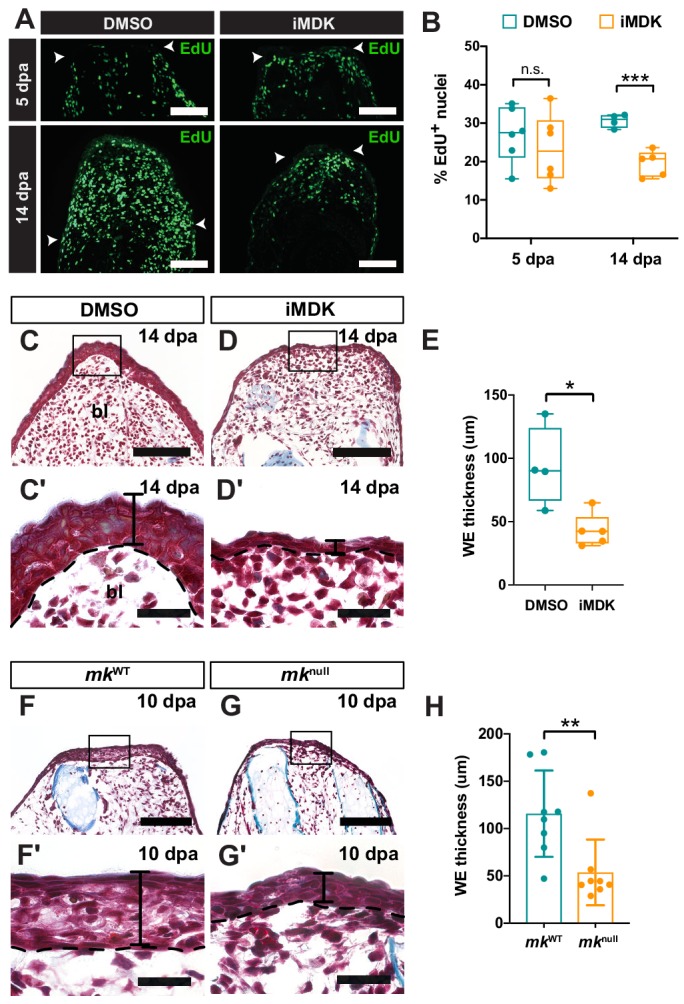
Figure 4—figure supplement 1. iMDK-treated limbs do not regenerate.

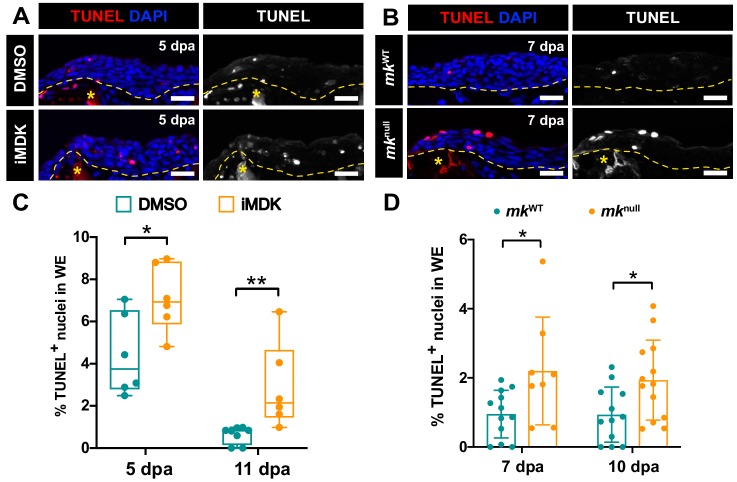
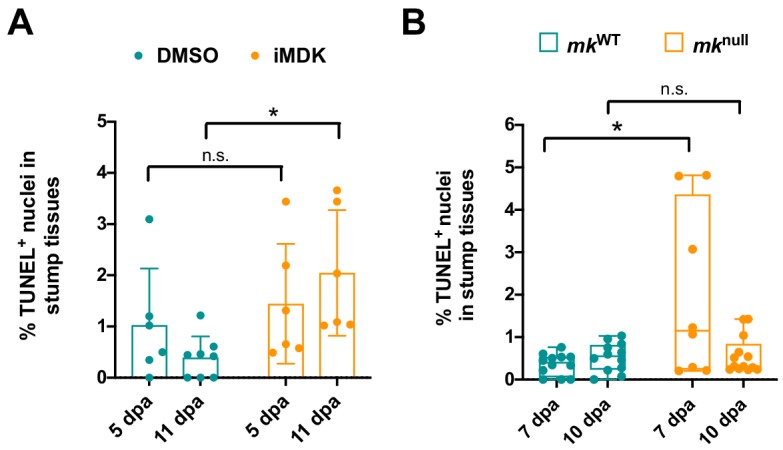
Further analysis of TUNEL staining in both mknull mutant and iMDK-treated regenerating limbs revealed that the defect in AEC development was primarily attributed to higher levels of cell death in the wound epidermis in both contexts (Figure 5). Altogether, these data indicate that mk mainly regulates early wound epidermis expansion by acting as a survival factor during the early wound epidermis transition to the AEC. In addition, while iMDK-treated limbs exhibited persistently high levels of cell death in blastemal cells, mknull mutants had higher levels of cell death during early stages that eventually resolved to that of wildtype limbs, suggesting mk is not required for blastemal cell survival (Figure 5—figure supplement 1).
Figure 5. Mk acts as a critical survival factor during AEC development.
(A) Representative TUNEL-stained sections from limbs of DMSO and iMDK-treated limbs. (B) Representative TUNEL-stained sections from limbs of regenerating mkWT control or mknull mutants. (C) Quantification of the % TUNEL+ nuclei in the wound epidermis of DMSO or iMDK-treated limbs limbs at 5 dpa (N = 6 DMSO, 6 iMDK) or 11 dpa (N = 8 DMSO, 6 iMDK). (D) Quantification of the percentage of TUNEL+ nuclei in mkWT control and mknull mutants at 7 dpa (N = 12 mkWT, 8 mknull) and 10 dpa (N = 12 mkWT, 13 mknull). Asterisks mark auto-fluorescent bone. Each N represents a limb from a different animal. Quantification of levels of blastemal cell death in DMSO/iMDK-treated and mkWT/mknull regenerating limbs can be found in Figure 5—figure supplement 1. Two-tailed unpaired student’s t-tests were used for statistical analysis. Graphs are mean ± SD. *p<0.05, **p<0.005. Scale bars: 100 µm. dpa, days post-amputation.
Figure 5—figure supplement 1. Mk is not required for blastemal cell survival.
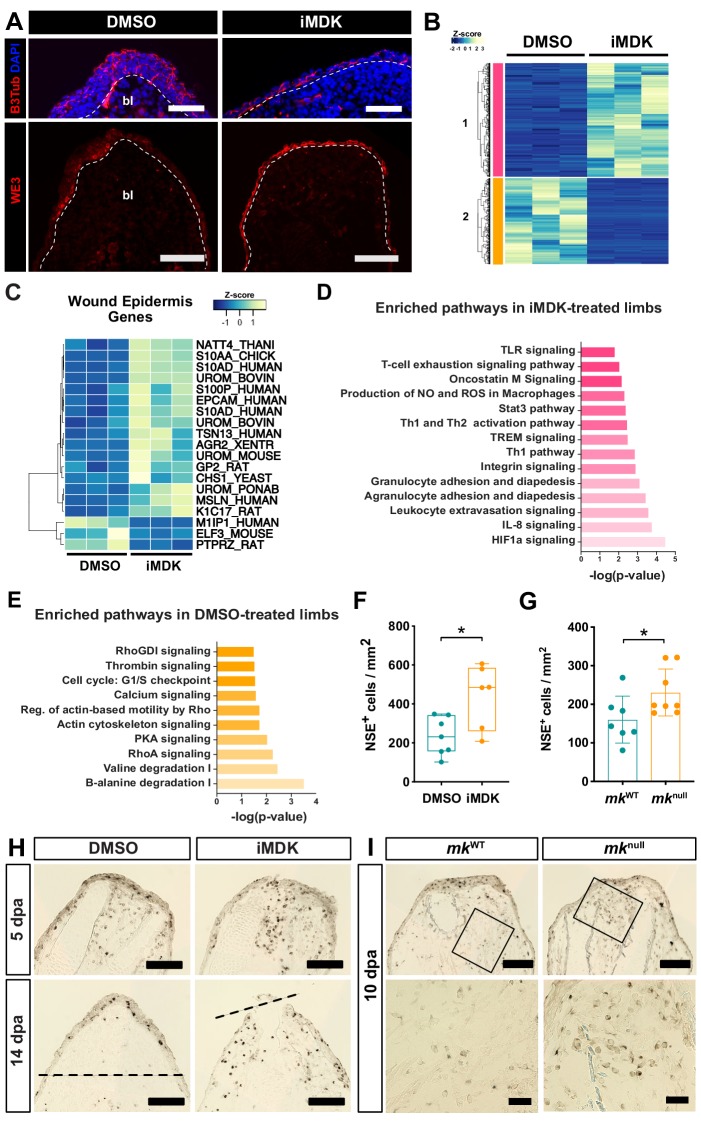
To determine the extent of defective AEC development, we also examined other aspects of the wound epidermis including innervation, which has been shown to be important for the AEC transition (Satoh et al., 2008; Satoh et al., 2012). Interestingly, iMDK-treatment did not impact wound epidermis innervation (Figure 6A) and data from a published microarray dataset showed similar induction of mk expression in innervated vs. denervated regenerating limbs (Monaghan et al., 2009). These results collectively indicate that mk operates in a nerve-independent manner. However, while expression of the known wound epidermis/AEC marker WE3 was observed (Figure 6A), sequencing of DMSO- and iMDK-treated limbs at 11 dpa revealed transcriptional dysregulation of many wound epidermis genes, suggestive of impaired functionality (Figure 6B–C, Supplementary file 4) (Campbell et al., 2011; Gerber et al., 2018; Leigh et al., 2018).
Figure 6. iMDK-treated and mk mutant regenerating limbs display dysregulated wound epidermis gene expression and persistent inflammation.
(A) Beta-III tubulin staining or WE3 staining of DMSO- or iMDK-treated limbs. White dotted line marks boundary of wound epidermis/AEC-blastema. (B) Heatmap of annotated differentially expressed transcripts in DMSO- and iMDK-treated limbs (N = 3 each) reveals two main clusters (colored pink and orange) of transcripts either enriched in DMSO or iMDK treatments. Transcript expression was normalized per row and plotted as a Z-score. Differentially expressed transcripts can be found in Supplementary file 4. (C) Heatmap of normalized TPM expression levels of wound epidermis genes in DMSO- or iMDK-treated regenerating limbs at 11 dpa. (D) Plot of enriched pathways in iMDK-treated limbs. (E) Plot of enriched pathways in DMSO-treated limbs. (F) Quantification of NSE+ monocytes at 5 dpa in DMSO/iMDK-treated limbs at 5 dpa (N = 7 DMSO, 6 iMDK). (G) Quantification of the density of NSE+ monocytes in mkWT control and mknull regenerating limbs at 10 dpa (N = 7 mkWT, 8 mknull). (H) NSE staining of DMSO- and iMDK-treated limbs at 5 dpa and 14 dpa. Dotted lines demarcate the amputation plane. (I) Representative NSE stained sections from regenerating limbs of mkWT control and mknull mutants at 10 dpa. Higher magnification insets are shown in bottom two panels. Each N represents a limb from a different animal. Data demonstrating rescue of mknull phenotypes via overexpression of mk during regeneration in mutant limbs as well as electroporation efficiency metrics can be found in Figure 6—figure supplements 1, 2 and 3. A two-tailed unpaired student’s t-test was used for statistical analysis. Graphs are mean ± SD. *p<0.05. Scale bars, A (top): 100 µm, A (bottom), H-I (top): 200 µm, I (bottom): 50 µm. bl, blastema, dpa, days post-amputation.
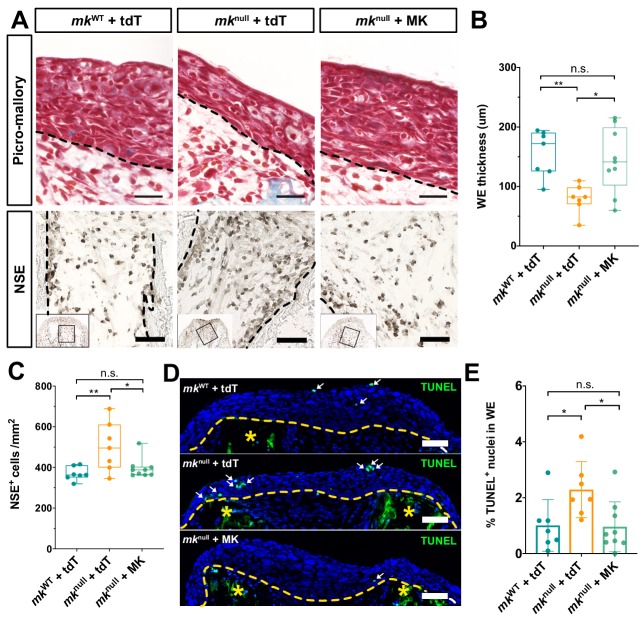
Figure 6—figure supplement 1. Mk overexpression in mknull regenerating limbs rescues delayed regeneration.
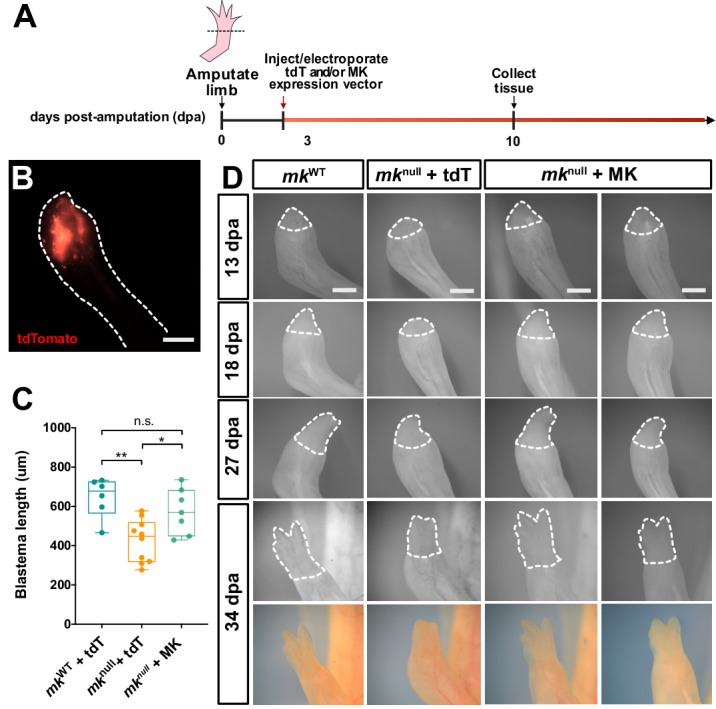
Figure 6—figure supplement 2. Mk overexpression in mknull regenerating limbs rescues mutant wound epidermis and monocyte density phenotypes.
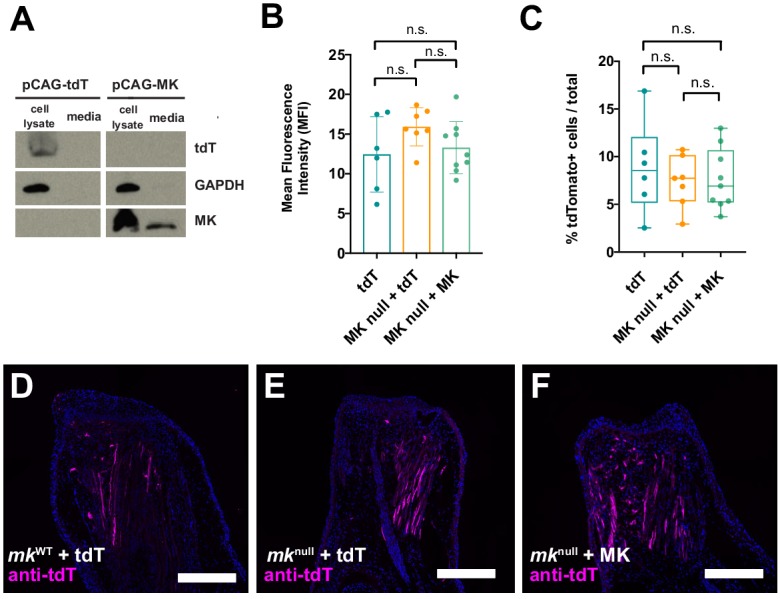
Figure 6—figure supplement 3. Electroporation efficiencies are similar between pCAG-tdTomato and pCAG-MK injected mknull mutant and mkWT regenerating limbs.
Notably, many pro-inflammatory pathways were enriched in inhibitor treated limbs (Figure 6D–E) and both mknull mutant and iMDK-treated regenerating limbs exhibited higher monocyte densities (Figure 6F–I), suggesting that functional perturbation of mk likely leads to prolonged inflammation. As we uncovered an immunomodulatory role for the early wound epidermis from our transcriptional data, it is possible that the persistent inflammation in mk loss-of-function contexts could be indirectly caused by impaired wound epidermis functionality from defective AEC development.
Most importantly, overexpression of mk in mknull mutant regenerating limbs rescued the delayed blastema formation and regeneration phenotype as well as the wound epidermis defects and high monocyte density (Figure 6—figure supplements 1–3), confirming that mk is indeed a regulator of both AEC development and the resolution of inflammation. As electroporation of constructs into limb epidermis is challenging and only occurs at a very low efficiency (Fei et al., 2016), the majority of mk was expressed and secreted from regenerating stump tissues. Therefore, these data further suggested that mk did not necessarily need to be produced in the wound epidermis to rescue AEC development.
Mk is sufficient to control proliferation and inflammation in regenerating limbs
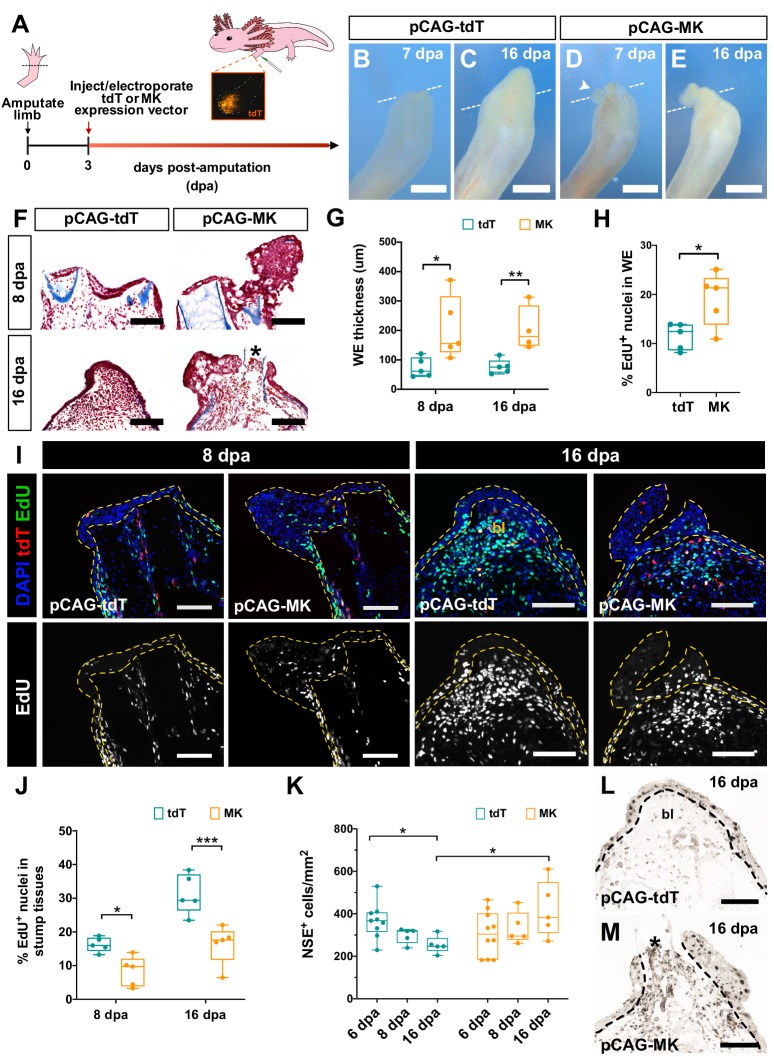
As the AEC forms via epithelial cell proliferation in peripheral skin and subsequent migration to the amputation plane (Satoh et al., 2012), we also investigated whether perturbing mk affected proliferation in the wound epidermis. Interestingly, iMDK-treated limbs displayed both consistently lower levels of wound epidermis proliferation as well as defects in the maintenance, but not induction, of blastemal cell proliferation in stump tissues (Figure 7—figure supplement 1A,C). However, we unexpectedly did not observe any differences in the proliferation of the wound epidermis or blastemal cells of mk mutants, suggesting mk is not required for proliferation (Figure 7—figure supplement 1B,D). These phenotypic differences in addition to the earlier presented blastemal cell death data were likely the main contributing causes to the observed variance in the severity in phenotype between iMDK-treated and mknull regenerating limbs and suggested that genetic compensatory mechanisms may be at play in the mk mutants (El-Brolosy et al., 2019). Nevertheless, overexpression of mk in regenerating limbs beginning at 3 dpa induced visible dramatic overgrowth of the wound epidermis by as early as 7 dpa (Figure 7A–G, Figure 7—figure supplement 2). In some mk-overexpressing limbs, the bone also protruded through the expanded wound epidermis, indicating that the tissue integrity of the wound epidermis was likely compromised. Further analysis confirmed that this expansion was attributed to a significant increase in the percentage of EdU+ cells within the wound epidermis (Figure 7H–I), strongly suggesting that mk is sufficient, but not required, to induce proliferation in the wound epidermis.
Figure 7. Overexpression of mk in regenerating limbs leads to wound epidermis expansion, decreased blastemal cell proliferation, and improper resolution of inflammation.
(A) Schematic of mk overexpression experiment. (B–E) Representative brightfield images of tdTomato (tdT)- (B–C) or mk-overexpressing (D–E) regenerating limbs at 7 and 16 dpa. White dotted line denotes amputation planes and white arrowhead in D denotes small aberrant skin growth. (F) Representative picro-mallory stained sections of tdT- or mk-overexpressing regenerating limbs. Asterisk marks protruding bone. (G) Quantification of wound epidermis thickness in tdTomato- or mk-overexpressing limbs at 8 dpa (N = 5 each) and 16 dpa (N = 5 tdT, N = 4 MK). (H) Quantification of the percentage of EdU+ nuclei in the wound epidermis of tdTomato- or mk-overexpressing limbs at 8 dpa (N = 5 each). (I) Representative images of EdU-stained sections of tdTomato- or mk-overexpressing limbs. Wound epidermis/AEC are outlined with yellow dotted lines. (J) Quantification of the percentage of EdU+ nuclei in regenerating stump tissues of tdT- or mk-overexpressing limbs. (K) Quantification of the density of NSE+ monocytes in tdT- or mk-overexpressing limbs. (L–M) Representative images of NSE-stained sections in tdT- or mk-overexpressing limbs. Black dotted line denotes wound epidermis boundary. Asterisk denotes protruding bone. Quantification of EdU+ cells in the wound epidermis and blastemal cells of DMSO/iMDK-treated and WT/mknull regenerating limbs is shown in Figure 7—figure supplement 1. Electroporation efficiency data can be found in Figure 7—figure supplement 2. Data showing that mk-overexpressing regenerating limbs display delayed regeneration is shown in Figure 7—figure supplement 3. Data demonstrating that overexpression of mk in non-regenerating intact limbs does not affect cellular proliferation or monocyte density can be found in Figure 7—figure supplement 4. Graphs are mean ± SD. Two-tailed unpaired student’s t-tests were used for statistical analysis. *p<0.05, **p<0.01, ***p<0.005. Scale bars, B-E: 500 µm, F, I, L-M: 200 µm. bl, blastema, dpa, days post-amputation.
Figure 7—figure supplement 1. Mk is not required for proliferation in the wound epidermis or blastemal cells.
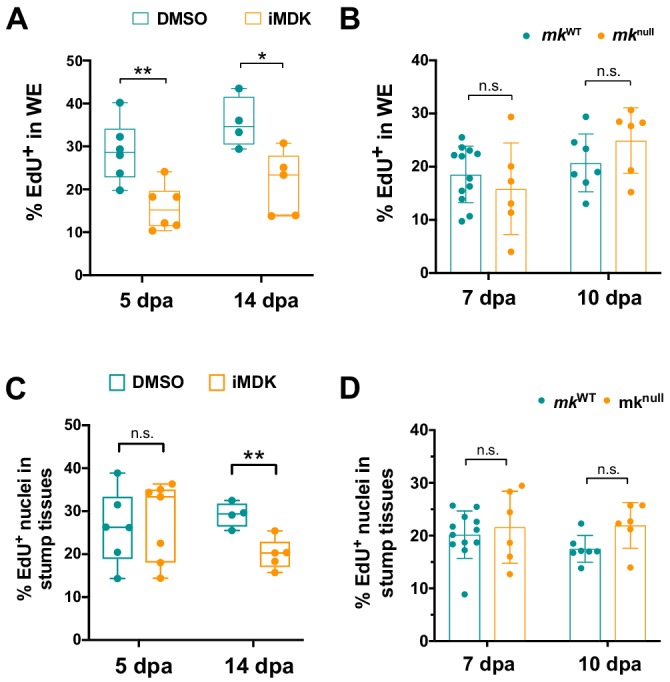
Figure 7—figure supplement 2. Electroporation efficiencies of pCAG-tdT and pCAG-MK injected regenerating limbs are similar.
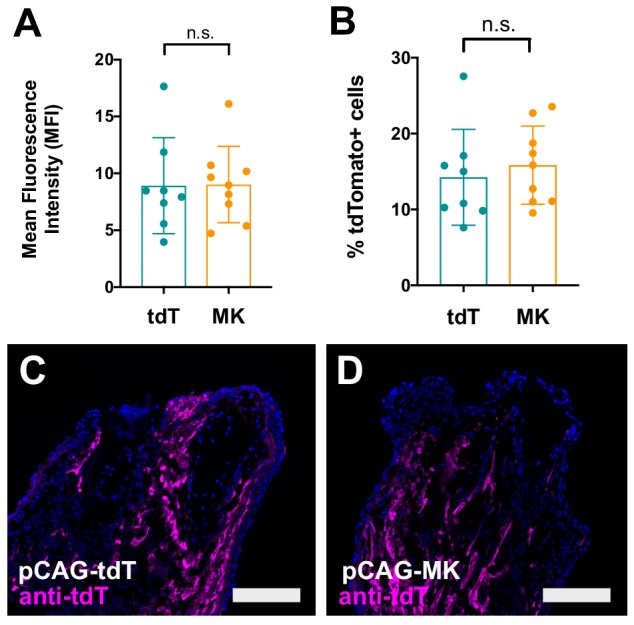
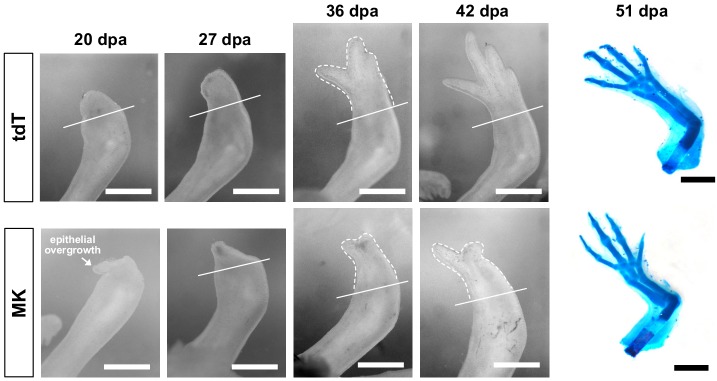
Figure 7—figure supplement 3. Mk-overexpressing limbs resolve epithelial overgrowth and undergo delayed regeneration.
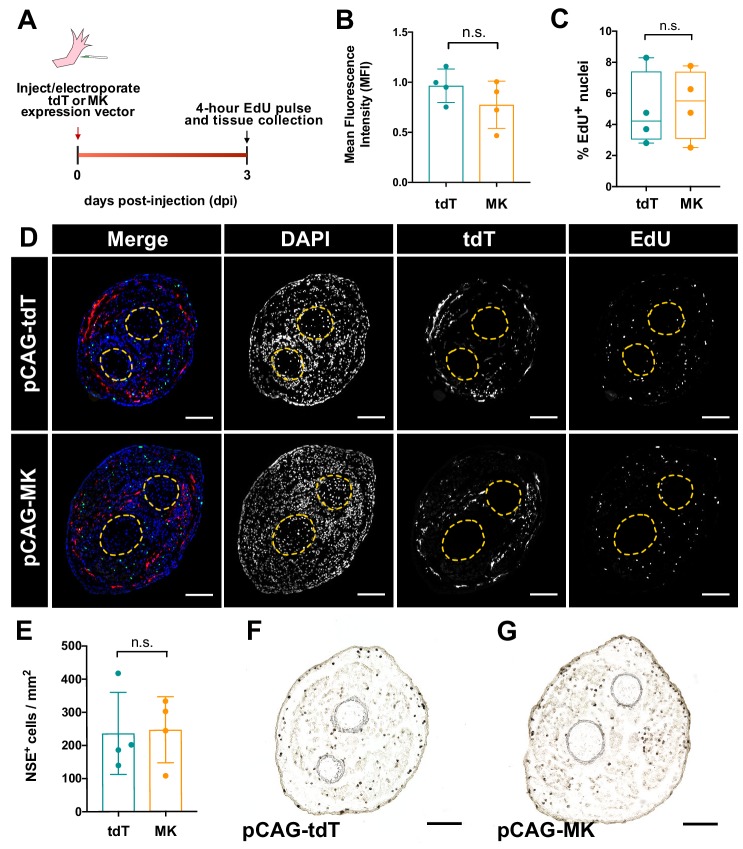
Figure 7—figure supplement 4. Overexpression of mk in non-regenerating intact limbs does not affect cellular proliferation or monocyte density.
We also observed lower levels of proliferation in blastemal cells of mk-overexpressing limbs (Figure 7J), indicating that high levels of mk during regeneration can have a suppressive effect on blastemal cell division. Furthermore, while control tdTomato-expressing limbs exhibited a significant decrease in monocyte density throughout regeneration, mk-overexpressing limbs displayed a persistently high monocyte density (Figure K-M), suggestive of prolonged inflammation. Together with our loss-of-function data, these results collectively suggest that mk levels must be tightly regulated during regeneration for proper resolution of inflammatory responses. Surprisingly, mk-overexpressing limbs eventually regenerated with a substantial delay, gradually resolving the overgrown wound epidermis/AEC and forming a blastema (Figure 7—figure supplement 3). Finally, in contrast to regeneration, overexpression of mk in intact limbs was not sufficient to affect cell proliferation or monocyte density (Figure 7—figure supplement 4), suggesting that the ability for mk to regulate these processes may be restricted to regenerative contexts.
Mk receptors are expressed throughout regenerating limb tissues
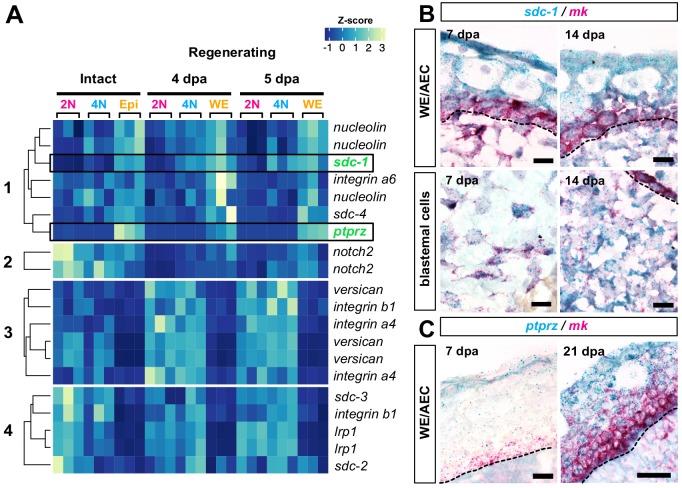
The strong multi-tissue expression pattern of mk in regenerating limbs led us to investigate whether mk may likely be acting through autocrine or paracrine signaling. Due to mk’s pleiotropic functional nature, mk has been shown to bind to many different receptors depending on the biological context, none of which have been shown to exclusively bind mk (Xu et al., 2014). To determine the expression levels and patterns of mk receptors, we first examined our dataset. We found that transcriptional isoforms of several known receptors were expressed in both intact and/or early regenerating tissues (Figure 8A). Clustering of normalized transcript levels revealed four main clusters of expression patterns: receptors that were enriched in both the intact skin and regenerating wound epidermis/AEC, and receptors that were either downregulated, up-regulated, or maintained expression in regenerating stump tissues during regeneration.
Figure 8. Mk receptors are expressed throughout regenerating tissues.
(A) Heatmap of normalized transcript levels for expressed mk receptors in intact and regenerating subpopulations from our RNA-seq dataset revealed four general patterns of expression (Figure 8—source data 1). (B–C) Double RNAscope in situ hybridization of mk (bright red puncta) with its cognate receptors sdc-1 (B) or ptprz (C) (dark blue puncta). Dotted black line denotes wound epidermis/AEC boundary. Since ptprz expression was low during early stages of regeneration, ptprz/mk in situs were performed without a hematoxylin counterstain to ease visualization of dark blue puncta in the wound epidermis at 7 dpa. Scale bars, B and C (left panel): 25 µm for 63x magnification, C (right panel): 50 µm for 40x magnification. WE, wound epidermis, AEC, apical epithelial cap, bl, blastema, dpa, days post-amputation.
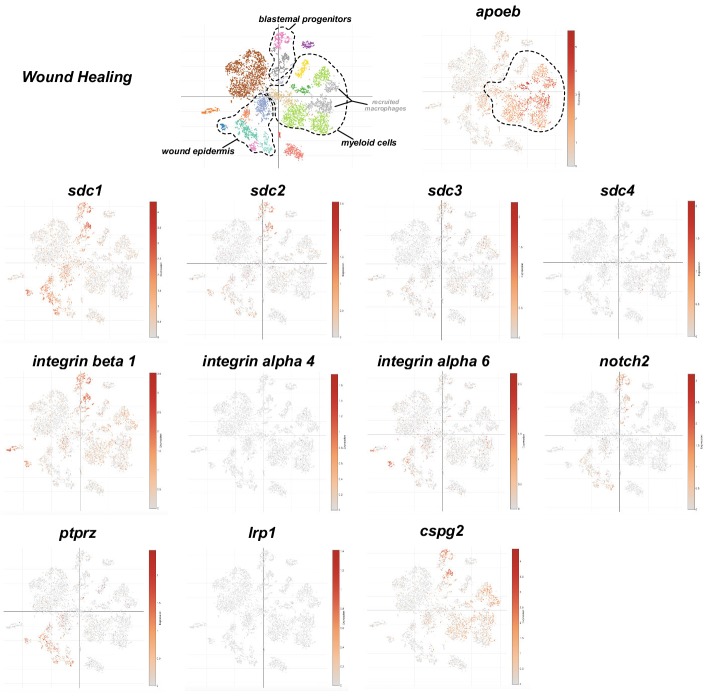
Figure 8—figure supplement 1. Mk receptors are not highly expressed in myeloid cells during wound healing.
We also examined data from published single cell limb regeneration studies and found that several of these receptors were identified as markers for various different cell types throughout homeostasis and regeneration (Gerber et al., 2018; Leigh et al., 2018). For example, syndecans (sdc-1,–2, −3,–4) and integrins (integrin alpha a4, a6, and b1) were detected at high levels in blastemal cells, basal and outer layers of the wound epidermis/AEC, endothelial cells, and schwann cells, indicating that mk likely directly signals to these cells. Surprisingly, most mk receptors were detected at low levels or not detected at all in myeloid cells, with the exception of integrin b1, which was moderately expressed (Figure 8—figure supplement 1). Although not a receptor, the extracellular chondroitin sulfate proteoglycan versican (cspg2) was also detected at moderate levels in myeloid cells. Versican is a high affinity binding partner of mk that can serve as an intermediary between mk and target receptors, potentially on immune cells (Zou et al., 2000; Muramatsu, 2014; Wight et al., 2014). Given the low expression of mk receptors in myeloid cells including monocytes/macrophages, these data altogether signify that mk may either directly signal through these or unidentified receptors, indirectly modulate inflammation through other tissues, or both.
Interestingly, one mk receptor, sdc-1 (Mitsiadis et al., 1995b), was expressed in both the blastemal cells and wound epidermis during early and later stages of regeneration, a pattern that seemed to overlap with that of mk. Double in situ hybridization of mk with sdc-1 indeed validated the co-expression of sdc-1 in mk+ blastemal and basal wound epidermal/AEC cells at 7 and 14 dpa (Figure 8B). Furthermore, sdc-1+ single positive cells were found in the outer layers of the wound epidermis/AEC, suggesting mk is capable of autocrine and paracrine signaling.
While sdc-1 exhibited a multi-cellular expression pattern throughout regenerating tissue, we noticed that the mk receptor ptprz exhibited restricted expression in the wound epidermis/AEC (Figure 8C, Figure 8—figure supplement 1). Double in situ hybridization of mk and ptprz revealed low levels of ptprz expression largely restricted to the outer layers of the wound epidermis at 7 dpa. By 21 dpa, ptprz expression increased in the AEC and overlapped with mk expression in the basal layers. Furthermore, we noticed that ptprz failed to be up-regulated in iMDK-treated limbs, suggesting ptprz expression may rely on a mk-dependent feedback mechanism (Figure 6C). As signaling through ptprz has been shown to be important for cell survival (Xia et al., 2019), its restricted epithelial expression suggests that it may be a key mk receptor for wound epidermis-specific signaling, potentially acting in conjunction with other mk receptors that are also expressed in the wound epidermis. In all, these expression data demonstrate the complexity of mk signaling in a wide range of cell types in both limb regeneration and homeostasis.
Discussion
While it is well known that the wound epidermis is a specialized epithelium required for the initiation of blastema formation and limb regeneration (Goss, 1956a; Thornton, 1957; Thornton, 1958; Mescher, 1976; Tassava and Garling, 1979), its roles during early stages of regeneration as well as the molecular mechanisms mediating its transition into the AEC remain largely unknown. Our study has revealed the functional roles of the early wound epithelium by examining how the separate transcriptional programs of blastemal progenitors and the surrounding tissues change in its absence. Here, we identified both early wound epidermis-dependent transcriptional programs as well as wound epithelial gene expression programs likely to be dependent on signaling from regenerating stump tissues. Importantly, our approach enabled us to identify mk as dual regulator of wound epidermis expansion during AEC development and the resolution of inflammation - two events that are essential for proper initiation of blastema formation.
The global transcriptional profiles of dividing progenitors are largely wound epidermis-independent during the initial stages of regeneration. Rather, we find that the transcriptional profiles of the surrounding tissues are more heavily affected when wound epidermis formation is prevented. A majority of these dysregulated transcripts are predominantly involved in inflammation, ECM remodeling, and tissue histolysis, indicating that the wound epidermis heavily modulates these processes during early stages of regeneration. Many characteristics of the initial wound healing stages post-amputation mirror those of mammals including activation of canonical inflammatory pathways (Godwin et al., 2013). Yet while most mammals activate a fibrotic scarring response post-wound healing, salamanders initiate blastema formation, suggesting there exist key differences during wound healing resolution. It is well known that differences in immune responses and ECM composition can tip the balance between scarring and regeneration (Godwin et al., 2013; Godwin et al., 2014). In light of our data, we propose that the early wound epidermis orchestrates the proper pro-regenerative niche during wound healing that is necessary for blastema formation by regulating early inflammation, ECM remodeling, and tissue histolysis.
Given that the initial cell cycle re-entry of progenitors during regeneration is independent of the wound epidermis (Mescher, 1976; Tassava and Garling, 1979; Johnson et al., 2018), our data provide transcriptional evidence to support the notion that the onset of progenitor activation is stimulated by processes inherent to the injury itself such as blood clotting (Tanaka and Brockes, 1998; Tanaka et al., 1999; Wagner et al., 2017). However, even though the overall transcriptional profile of early dividing progenitors is relatively wound epidermis-independent, we find that transcriptional activation of HIPPO, Wnt, and TGF-beta signaling in dividing progenitors is dependent on the wound epidermis. The loss of activation of not one, but all three pathways, when wound epidermis formation was prevented strongly suggests potential common regulation and synergism between these pathways, similar to other developmental and tumorigenic contexts (McNeill and Woodgett, 2010; Attisano and Wrana, 2013). As the wound epidermis/AEC is known to maintain blastemal cell proliferation (Mescher, 1976; Tassava and Garling, 1979), it is therefore possible that it does so by inducing and/or sustaining activation of HIPPO, canonical Wnt, and TGF-beta signaling in dividing progenitors. Thus, dissecting the functional interplay between these three pathways will likely elucidate the mechanisms that control early blastemal cell behaviors critical for the initial phases of blastema formation.
Common down-regulation of the macrophage marker mpeg1 in both the non-dividing cells and epithelia of full skin flap sutured limbs further indicates that the wound epidermis may play a role in macrophage recruitment during early stages of regeneration. Macrophage depletion during early stages of limb regeneration inhibits blastema formation and results in abnormal ECM deposition reminiscent of fibrosis (Godwin et al., 2013), suggesting a tight interplay between the immune system and scarring. We find that many critical regenerative ECM remodeling enzymes and components, including various matrix metalloproteinases (Vinarsky et al., 2005) and tenascin, fail to be strongly induced in regenerating stump tissues in the absence of the wound epidermis. Our data therefore suggest the possibility that the wound epidermis may mediate ECM remodeling indirectly through immune cells including macrophages. Interestingly, cross-talk between the wound epithelium and macrophages through interleukin one beta (il1b) signaling is important for early stages of fish fin fold regeneration (Hasegawa et al., 2017). Our data also indicate that contact between epithelial cells and underlying regenerating stump tissues is important for proper regulation of inflammation and ECM remodeling during salamander limb regeneration. In addition, we found signatures reminiscent of defects in bone resorption, indicating that the wound epidermis may play a role in mediating early skeletal remodeling. As this area has been understudied during limb regeneration, it will be interesting to explore the potential involvement of the wound epidermis in the regulation of this process in future studies.
Notably, we identify the pleotropic growth factor cytokine mk as a dual regulator of AEC development and the resolution of early inflammation. Mk is an important mediator of a variety of processes including development, tumorigenesis, and inflammation (Muramatsu, 2010; Weckbach et al., 2011; Sorrelle et al., 2017). We demonstrate that mk controls early wound epidermis expansion during AEC development by acting as a cell survival signal. Increased cell death in the wound epidermis of both iMDK-treated and mknull regenerating limbs leads to a thin wound epithelium that exhibits aberrant gene expression and likely impaired functionality. Mis-expression of wound epidermis genes including chs1, which is normally only expressed during wound healing (Leigh et al., 2018), also demonstrates a lack of normal wound epidermis progression from early to blastemal stages. Others have shown the role of mk as a survival factor in several contexts including neural crest cell development, immune cell maintenance, and tumorigenesis (Muramatsu, 2010; Cohen et al., 2012; Vieceli and Bronner, 2018), suggesting that it may be a conserved function. In fact, mk signaling through one of its most well-studied receptors ptprz regulates cell survival (Xu et al., 2014). Based on our data, it is likely that mk-dependent ptprz signaling similarly modulates cell survival during AEC development in limb regeneration given its wound epidermis-specific expression pattern. Furthermore, mk induced anti-apoptotic signaling has been linked to activation of the PI3K/AKT signaling cascade in B-cells (Cohen et al., 2012), neurons (Owada et al., 1999), and cancer (Dai et al., 2006). As the inhibitor iMDK also inhibits PI3K phosphorylation (Masui et al., 2016), it is possible that PI3K/AKT signaling acts downstream of mk in AEC development. Furthermore, ptprz modulates endogenous phosphorylated ß-catenin levels through its phosphatase activity, which becomes inactive upon ligand binding (Meng et al., 2000). Since Wnt signaling is essential for limb regeneration and Wnt ligands are also expressed in the wound epidermis (Kawakami et al., 2006; Yokoyama et al., 2007; Ghosh et al., 2008; Knapp et al., 2013), mk may also act in concert with and/or up-stream of Wnt signaling to regulate wound epidermis development.
In a variety of other contexts, mk also acts as a mitogen to stimulate tumor growth, tissue growth during organogenesis, and tissue repair/regeneration (Calinescu et al., 2009; Gramage et al., 2015; Masui et al., 2016; Karaman and Alitalo, 2017; Nagashima et al., 2019). For example, mk has been shown to induce proliferation of Müller glia in an autocrine manner during zebrafish photoreceptor regeneration (Gramage et al., 2015; Nagashima et al., 2019). Therefore, we were surprised to find that mknull mutants did not display differences in wound epidermis or blastemal cell proliferation, despite observing lower levels in iMDK-treated limbs. We also observed higher cell death in blastemal cells that eventually resolved in mknull mutants, but not iMDK-treated limbs. In all, these data suggested that genetic compensation may be occurring in the mutant and mk is likely not required for general proliferation or blastemal cell survival. While it is tempting to assume compensation is the sole cause for these differences, we cannot discount the possibility that they may have been caused by non-specific effects of the inhibitor given the exact inhibitory mechanism and direct target of iMDK remains unknown (Masui et al., 2016). Nevertheless, while not required for proliferation, mk was sufficient to affect proliferation at high levels. Most notably, overexpression during regeneration induced a striking ectopic expansion of the wound epidermis. Interestingly, innervation of the wound epidermis has also been shown to regulate keratinocyte proliferation in the AEC (Satoh et al., 2012). While our data suggest mk does not affect innervation, mk may act in parallel with nerve-derived factors to finely control cell death/proliferation dynamics during early wound epidermis expansion. Unexpectedly, overexpression of mk decreased blastemal cell proliferation. Despite ample evidence of the role of mk as a mitogen, mk has been shown to have anti-proliferative activities in some contexts. For example, MK exerts an anti-proliferative effect on dental mesenchyme during mouse tooth development, yet a proliferative effect if combined with FGF-2 (Mitsiadis et al., 1995a), indicating that MK activity can differ depending on the presence of other growth factors in the micro-environment. Hence, mk may act cooperatively with other secreted molecules that modulate blastema formation and growth (Kumar et al., 2007; Currie et al., 2016; Farkas et al., 2016; Sugiura et al., 2016; Tanaka, 2016; Bryant et al., 2017b) to exert stage- and cell-type specific effects during regeneration.
Notably, both loss- and gain-of-function of mk additionally led to signs of persistent inflammation including higher monocyte density, altogether suggesting that mk levels need to be tightly controlled for proper regulation of inflammation during early regeneration. Many relatively recent studies have focused on the pro-inflammatory role of mk, driving chronic inflammatory diseases including rheumatoid arthritis and atherosclerosis (Weckbach et al., 2011; Sorrelle et al., 2017). Removing or reducing mk activity in mouse models in these contexts led to less inflammation attributed to decreased leukocyte recruitment. In contrast, our data suggest that mk acts as an anti-inflammatory cytokine to resolve injury-induced inflammation during the initiation of limb regeneration. While inflammation is required for regeneration (Kyritsis et al., 2012; Hasegawa et al., 2017; Tsarouchas et al., 2018), prolonged inflammation can be detrimental. Recent studies have shown that macrophages play a role in resolving inflammation in both zebrafish and salamanders, by acting as a source of anti-inflammatory factors (Godwin et al., 2013; Hasegawa et al., 2017; Tsarouchas et al., 2018). Our data further suggest that both blastemal progenitors and the wound epidermis may be additional sources of anti-inflammatory molecules including mk that aid in resolving inflammation during early stages of limb regeneration. Moreover, it is possible that mk expression may be induced by macrophage-derived factors, an interesting future avenue of investigation. These findings thus highlight the need to advance our understanding of the complex molecular cross-talk between the immune system and various regenerating tissues during wound healing to gain insight into what leads to regenerative versus non-regenerative outcomes.
Finally, both overlapping and non-overlapping expression of mk and its receptors indicates that mk is likely acting through both autocrine and paracrine signaling in limb regeneration. The epithelial-mesenchymal dual expression of mk has been known for decades in the development of various organs including the lungs, gut, and limb buds of mice (Mitsiadis et al., 1995b). Although the expression pattern is suggestive of inductive signaling between the epithelium and developing mesenchyme, the exact functional nature of mk in these contexts remains largely unknown. While we did not examine mk expression in developing axolotl limbs, it is possible that mk exhibits a similar expression pattern in both the limb bud ectoderm and mesenchyme. As such, the up-regulation of mk in blastemal progenitors and the wound epidermis may signify the re-induction of a developmental program during the initiation of limb regeneration. Additionally, mk is first expressed in blastemal progenitors prior to the wound epidermis and failed to be strongly up-regulated when wound epidermis formation was prevented. Therefore, mk up-regulation during the initiation of limb regeneration may depend on positive feedback signaling between mk+ blastemal progenitors and the wound epidermis. Moreover, mk receptors are expressed in intact epithelial cells and our rescue data suggest the wound epidermis is responsive to mk from non-epithelial sources. Hence, it is possible that mk secreted from blastemal progenitors may first induce mk expression in the wound epidermis through paracrine signaling, which is then self-sustained through autocrine signaling.
While our mk receptor expression data suggest that mk likely directly signals to the wound epidermis to regulate AEC development, how mk regulates inflammation remains less clear. To the best of our knowledge, besides moderate levels of integrin b1, we were not able to identify robust expression of mk receptors on populations of immune cells in our data or published single cell data. Thus, it is possible that mk may regulate monocyte activity directly through the low expression of these or as of yet unidentified receptor(s), indirectly through other tissues, or both. Given the data, it is likely that both regulatory mechanisms are at play. As mk receptors are more highly expressed throughout many other cell types and tissues, it is possible that mk indirectly regulates the resolution of inflammation by stimulating the secretion of anti-inflammatory factors in these tissues that then signal directly to monocytes. One likely example of such a tissue may be the wound epidermis. As we have shown that the early wound epidermis is immunomodulatory, it is conceivable that the prolonged inflammation we observed in both mk loss- and gain-of-function contexts was indirectly or partially caused by an impaired wound epidermis/AEC, opening up the possibility that mk may regulate the resolution of inflammation in an indirect manner. In all, the widespread expression of mk receptors throughout regenerating limb tissues underscores the complexity of mk function and makes it clear that future studies using tissue-specific inducible mk and receptor knockout models will be necessary to deconvolute the exact nature of cell-type specific mk signaling during regeneration.
To conclude, we have revealed the functional roles of the early wound epidermis during the initial stages of limb regeneration and further identified mk as a key regulator of both AEC development and the resolution of inflammation. The early expression of mk in both blastemal progenitors and the basal wound epidermis, key cell types that coordinate limb regeneration, suggests its expression may be linked with the ability to regenerate. As mk is highly conserved in other vertebrates, examining whether it plays similar roles in other appendage regeneration models and if it is differentially activated in non-regenerative systems will be important future lines of questioning. In all, we believe the findings presented here provide a solid foundation to further elucidate the molecular underpinnings of wound epidermis biology in appendage regeneration.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Ambystoma mexicanum) |
Midkine (mk) | |||
| Strain, strain background (Ambystoma mexicanum) |
Axolotl, White (d/d) | Ambystoma Genetic Stock Center (AGSC) (RRID:SCR_006372) |
Cat. #AGSC_1015 | Subadults and juveniles, used for FSF transcriptional analysis and functional experiments |
| Strain, strain background (Ambystoma mexicanum) |
Axolotl, Albino (a/a) |
Ambystoma Genetic Stock Center (AGSC) (RRID:SCR_006372) |
Cat. #AGSC_1025 | Subadults, used only for FSF transcriptional analysis |
| Genetic reagent (Ambystoma mexicanum) | Mknull mutants | This paper | Generated in d/d strain | |
| Cell line (Homo sapiens) |
293T HEK Cells | ATCC (RRID:SCR_001672) |
Cat. #CRL-3216 (RRID:CVCL_0063) |
For validation of MK overexpression construct |
| Antibody | Polyclonal goat anti-tdTomato | LS Bio (RRID:SCR_013414) |
Cat. #LS-C340696 (RRID:AB_2819022) |
WB: 1:200 |
| Antibody | Polyclonal chick anti-GAPDH | Millipore (RRID:SCR_008983) |
Cat. #AB2302 (RRID:AB_10615768) |
WB: 1:2000 |
| Antibody | Polyclonal rabbit anti-midkine (axolotl-specific) | This paper, NEPeptide | IF: 1:500, WB: 1:2000 | |
| Antibody | Monoclonal mouse Anti-WE3 | DSHB (RRID:SCR_013527) |
Cat. #WE3 (RRID:AB_531902) |
IF: 1:10 |
| Antibody | Monoclonal mouse Anti-Beta III Tubulin | Sigma Aldrich (RRID:SCR_008988) |
Cat. #T8578 (RRID:AB_1841228) |
IF: 1:200 |
| Recombinant DNA reagent | pCAG-tdTomato | Pathania et al., 2012 | Addgene: Cat. #83029 | Injected and electroporated at 500 ng/uL |
| Recombinant DNA reagent | pCAG-MK | This paper | Injected and electroporated at 500 ng/uL | |
| Sequenced-based reagent | Custom RNAscope axolotl prrx-1 probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl csf1r probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl pax7 probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl pecam probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl midkine probe (C2) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl ptprz probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl sdc1 probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Alt-R CRISPR-Cas9 crRNA (mk specific): 5’AAGCCCCCACAACTGCATCC −3’ |
This paper | Midkine specific short guide RNA sequence | |
| Sequenced-based reagent | Alt-R CRISPR-Cas9 tracrRNA | Integrated DNA Technologies (IDT) | Cat. #1072534 | |
| Sequenced-based reagent |
Mk forward genotyping primer: 5’-TTGCTTATTCCTTGTGATCATGC-3’ |
This paper | Genotyping primer | |
| Sequenced-based reagent |
Mk reverse genotyping primer: 5’- GGCACATTATTACACAGAAAGCTC-3’ |
This paper | Genotyping primer | |
| Sequenced-based reagent |
Mk nested PCR forward primer: 5’- tctttccctacacgacgctcttccgatctGAGGTTTGATTGGACCCTGA-3’ |
This paper | Genotyping primer | |
| Sequenced-based reagent |
Mk nested PCR reverse primer: 5’-tggagttcagacgtgtgctcttccgatctGGCACATTATTACACAGAAAGCTC-3’ |
This paper | Genotyping primer | |
| Peptide, recombinant protein | Axolotl Midkine blocking peptide (amino acids 126 to 142) | This paper, NEPeptide | Used to validate custom polyclonal axolotl MK antibody | |
| Chemical compound, drug | iMDK (Midkine inhibitor) | Tocris Bio (RRID:SCR_003689) |
Cat. #5126 | Used at 10 uM |
| Chemical compound, drug | EdU | Thermofisher Scientific (RRID:SCR_008452) |
Cat. #A10044 | Used at 8 mg/mL concentration |
| Commercial assay or kit | In Situ Cell Death Detection Kit, TMR Red | Roche (RRID:SCR_001326) |
Cat. #12156792910 | |
| Commercial assay or kit | In Situ Cell Death Detection Kit, Fluorescein | Roche (RRID:SCR_001326) |
Cat. #11684795910 |
|
| Commercial assay or kit | Click-iT EdU Cell Proliferation Kit for Imaging | Thermofisher Scientific (RRID:SCR_008452) |
Cat. #C10337 | |
| Commercial assay or kit | α-Naphthyl Acetate Esterase (NSE) Kit | Sigma Aldrich (RRID:SCR_008988) |
Cat. #91A | |
| Commercial assay or kit | RNAScope 2.5 HD Duplex Assay | ACD Bio | Cat. #322430 | |
| Commercial assay or kit | Miseq Reagent Nano Kit v2 (300-cycle) | Illumina (RRID:SCR_010233) |
Cat. #MS-102–2002 | |
| Commercial assay or kit | Illumina Nextera XT DNA Library Prep Kit | Illumina (RRID:SCR_010233) |
Cat. #FC-131–1024 | |
| Commercial assay or kit | Ovation RNA-seq System V2 | Integrated Sciences | Cat. #7102–32 | |
| Commercial assay or kit | PrepX ILM 32i DNA Library Prep Kit | Takara Bio | Cat. #400076 | |
| Software, algorithm | Trinity | Grabherr et al., 2011 | https://github.com/trinityrnaseq/trinityrnaseq/wiki | |
| Software, algorithm | Kallisto | Bray et al., 2016 | https://pachterlab.github.io/kallisto/ | |
| Software, algorithm | Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic | |
| Software, algorithm | DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | |
| Software, algorithm | WEB Gestalt | Wang et al., 2017 | http://www.webgestalt.org/ | |
| Software, algorithm | CRISPResso | Pinello et al., 2016 | http://crispresso.pinellolab.partners.org/ | |
| Software, algorithm | Complex Heatmaps | Gu et al., 2016 | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html | |
| Software, algorithm | ImageJ | Schindelin et al., 2012 | https://imagej.net/Fiji |
Experimental model and subject details
Axolotl (Ambystoma mexicanum) husbandry and surgeries were performed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) and Institutional Animal Care and Use Committee (IACUC) guidelines at Harvard University. Sub-adult white and albino axolotls (15–18 cm) provided by the Ambystoma Genetic Stock Center (AGSC, University of Kentucky, KY) were used for the RNA-sequencing experiment of full skin flap sutured regenerating limbs. Juvenile white axolotls (3–6 cm animals) were utilized for the midkine inhibitor (iMDK) (Tocris Bio) experiments and mk overexpression experiments. Mk mutants were generated using CRISPR/Cas9 in the white background and genotyped/analyzed as juveniles as well.
Animal surgeries and drug treatments
For amputations, animals were anesthetized in 0.1% tricaine (Sigma Aldrich) until unresponsive to a tail pinch. The limb was then amputated at the mid-radius/ulna level using sterilized dissection scissors. All of the limb amputations were performed on naïve regenerating animals that is animals that had not regenerated before. The amputations for all functional experiments were performed at the mid-radius/ulna level of axolotl forelimbs. For the transcriptional profiling experiment of full skin flap sutured limbs, amputations were performed at the mid-zeugopodial level (radius/ulna or –tibia/fibula) and full skin flap surgeries were performed on all four limbs of nine animals immediately post-amputation as described in Mescher (1976). Briefly, limbs were amputated in the distal zeugopodial region and skin was carefully peeled back. The exposed underlying non-epithelial tissues were then amputated at the mid-zeugopodial level and the excess skin was sutured directly over the amputation plane using 9–0 sutures (Ethicon).
For iMDK experiments, limbs of animals were amputated and the animals were immersed in DMSO or 10 uM iMDK solution beginning at either 0 or 7 dpa. Drug solution was changed daily for the duration of the inhibitor experiments and animals were fed normally every other day.
RNA isolation and sequencing
For RNA-sequencing of full skin flap sutured limbs, approximately 2–3 mm of tissue directly proximal to the amputation plane was collected at 5 dpa. Twelve limbs (6 forelimbs and 6 hindlimbs) were pooled for each replicate per timepoint and the experiment was performed in triplicate. Pooling was necessary to collect enough cells for RNA isolation.. Dissociation, DAPI cell cycle analysis, and RNA isolation from the dividing and non-dividing cells in the non-epithelial stump tissues as well as the full thickness skin flap was performed as described in Tsai et al. (2019). Briefly, the wound epidermis or full skin flap at the distal amputation plane was carefully micro-dissected off of regenerating limb tissue and dissociated in 0.25% Trypsin-EDTA, while the remaining non-epithelial stump tissues along with some intact skin proximal to the amputation plane were dissociated in a mixture of collagenase, dispase, and glucose in 80% PBS. Both dissociated populations underwent DAPI staining and FACS was performed on the stained non-epithelial stump tissue fraction to sort the population into 2N and 4N fractions. RNA was isolated from the 3 DAPI stained cellular populations of wound/full skin flap epithelial cells, dividing cells (4N) and non-dividing cells (2N) from regenerating non-epithelial stump tissues. cDNA was synthesized using the NuGEN Ovation RNAseq System V2 protocol (Integrated Sciences) using 10 ng of starting material and cDNA libraries were generated using the Wafergen PrepX Complete ILMN DNA Library kit (Takara Bio). Library concentrations were measured using the Qubit dsDNA HS Assay kit (Thermofisher Scientific) and Kapa Illumina Library Quantification Kit (Kapa Biosystems). The samples (regenerating and full skin flap samples) were pooled altogether and sequenced on an Illumina Hiseq 2500 system (Illumina) (125 bp reads) at the Harvard Bauer Core Sequencing Facility. The regenerating dataset (data from 0 and 5 dpa) presented in this study used for comparison was previously published in Tsai et al. (2019), while the full skin flap samples are newly presented in this work.
Raw reads were filtered and trimmed using Trimmomatic (Bolger et al., 2014) and aligned to the axolotl transcriptome (Bryant et al., 2017a) using Kallisto (Bray et al., 2016). Raw TPM values from each sample were analyzed using DESeq2 (Love et al., 2014). Differentially expressed transcripts were filtered for fold change >2 and adjusted p-value<0.05. For pathway analysis, Uniprot IDs of blastx hits from different species for all differentially expressed transcripts were converted to human Uniprot IDs using HUGO Gene Nomenclature Committee (HGNC) and/or Uniprot ID conversion tools online. Ingenuity pathway analysis (IPA) analysis was performed on differentially expressed transcripts to determine growth factor signaling pathway activation or inhibition for the following comparisons: 0 dpa 2N vs 5 dpa 2N, 0 dpa 4N vs 5 dpa 4N, 0 dpa epi vs 5 dpa wound epidermis (WE), 0 dpa 2N vs 5 dpa FSF 2N, 0 dpa 4N vs 5 dpa FSF 4N, and 0 dpa epi vs 5 dpa FSF epi. Heatmaps of normalized TPM levels for differentially expressed transcripts in each fraction (2N, 4N, or epidermis) were generated in RStudio utilizing the ComplexHeatmap (Gu et al., 2016) or gplots packages (Warnes et al., 2015). Clusters of differentially expressed transcripts (5 dpa regenerating vs. 5dpa FSF) for each cellular fraction were analyzed using WebGestalt (Wang et al., 2017).
For sequencing of DMSO- or iMDK-treated limbs, RNA was isolated from DMSO or iMDK-treated regenerating limbs at 11 dpa. Tissue was homogenized in Trizol and RNA was purified using the Qiagen RNeasy mini kit. We collected the samples in biological triplicate for each condition. Quality and quantity of RNA were assessed using the Agilent RNA 6000 pico kit and run on a BioAnalyzer 2100 (Agilent). Sequencing libraries were then synthesized using the Illumina Nextera XT kit (Illumina) and sequenced on a Nextseq 500 at the Biopolymers Facility at Harvard Medical School. Fastq data were filtered, trimmed, and aligned as described above. Differential expression analysis was performed with DESeq2 (Love et al., 2014). For the differential expression analysis, the cutoff was set at an adjusted p-value<0.05 and fold change >2. K-means clustering of the transcripts was performed using the Complex Heatmap package (Gu et al., 2016) and pathway analysis of DMSO- or iMDK-enriched transcripts was performed on differentially expressed transcripts using Ingenuity Pathway Analysis (IPA) (Qiagen) software.
Generation of mk mutants and genotyping
In order to generate the mk mutants, CRISPR-cas9 technology was utilized and the start codon of the mk locus was targeted with a single guide RNA (sgRNA) (ATG start is bolded): 5’AAGCCCCCACAACTGCATCC −3’. This sequence was also unique within the axolotl genome. The mk crRNA and generic tracrRNA were ordered from IDT and reconstituted according to their instructions. One-cell stage embryos were injected with cas9 ribonucleoprotein (cas9 RNP) complexes to generate mutants. To generate the cas9 RNP complexes, mk crRNA and tracrRNA were mixed and annealed to form the mk sgRNA. Mk sgRNA (final concentration 200–400 ng/µL) was then mixed with concentrated cas9 (final concentration 500 ng/µL) (PNA Bio Inc) and incubated for 10 min at 37°C to generate the cas9 ribonucleoprotein (RNP) complex. For each fertilized egg, 2–3 nl of injection mix was injected. Control embryos were injected with cas9 RNPs that were complexed with tracrRNA only.
For genotyping, genomic DNA was collected from the original limbs that were amputated from regenerating tracrRNA controls and mk mutants. Tissue was incubated in 50 mM NaOH for 20 min. at 94°C and neutralized with TE buffer pH 7.5. The supernatant was then PCR purified using the Qiagen PCR purification kit according to the manufacturer’s instructions. Genomic PCR was performed to amplify the area surrounding the cut site with the following primers:
For 5’-TTGCTTATTCCTTGTGATCATGC-3
Rev 5’- GGCACATTATTACACAGAAAGCTC-3’
Next, two rounds of additional PCRs were performed to generate DNA sequencing libraries as described in Gagnon et al. (2014). Briefly, one round of nested PCR was performed using the following gene specific primers with universal overhangs:
For 5’- tctttccctacacgacgctcttccgatctGAGGTTTGATTGGACCCTGA-3’
Rev 5’- tggagttcagacgtgtgctcttccgatctGGCACATTATTACACAGAAAGCTC-3’
The next round of PCR was performed to add on i7 indices and P5/P7 sequences to generate DNA sequencing libraries. Up to 24 libraries were pooled together and sequenced on a department owned Miseq using the Miseq reagent nano kit v2 (300 cycle) (Illumina). Fastq data were then analyzed utilizing CRISPResso (Pinello et al., 2016). For the genotyping analysis, mutants in which less than 1% of wildtype alleles were detected were deemed null (most had mosaic null genotypes), with up to 10–15% wildtype alleles deemed low % wildtype, and greater than 15% deemed high % wildtype (most of these animals were actually closer to 80–90% wildtype). Only mosaic null mk mutants were considered for the downstream characterization.
RNAscope in situ hybridization and expression analysis
Tissue from intact or regenerating limbs was collected and fixed in 4% paraformaldehyde overnight, brought up a sucrose gradient, embedded in OCT, and cryo-sectioned at 12 μm. Chromogenic RNAscope in situ hybridization was performed utilizing the RNAscope 2.5HD Duplex Assay Kit (ACD Bio) according to the manufacturer’s instructions using custom axolotl RNAscope probes generated for mk in the C1 channel and pax7, prrx-1, pecam, ptprz, sdc1, and csf1r in the C2 channel.
For the quantification of the breakdown of cell types expressing mk, sections were quantified within the distal most 500 μm from the amputation plane. The total numbers of single positive mk+, pax7+, prrx-1+, and pecam+ cells as well as double positive cells were used to calculate the percentage of mk+ cells expressing each respective marker at 0, 3, and seven dpa. Although mk is lowly expressed in the blastema as well, the densely packed nuclei precluded accurate quantification of co-expression. For co-expression analysis of mk in csf1r+ monocytes, single and double positive cells were counted and the respective breakdown out of total counted cells was shown.
Histological characterization of mk mutants
Brightfield images of regenerating limbs were taken of tracrRNA controls and mk mutants during regeneration using an Olympus SZX16 dissecting microscope. Tissue was collected at 10 and 14 dpa for analyses. For the blastema length measurement analysis, picro-mallory staining was performed on sections and measured the length at 14 dpa. For wound epidermis thickness, the thickest part of the wound epidermis was measured and averaged across at least two sections from regenerating limbs of tracrRNA and mk mutants at 10 dpa.
α-Naphthyl acetate esterase (NSE) staining and analysis
NSE staining was performed using the α-Naphthyl Acetate Esterase staining kit (Sigma) according to the manufacturer’s instructions, except a 10 min fixation step was performed instead of 30 s to 1 min. The number of NSE+ monocytes/unit area mm2 within 500 µm of the amputation plane were quantified in DMSO/iMDK-treated, mkWT and mknull, as well as tdTomato- or mk-overexpressing intact and regenerating limbs.
TUNEL staining and analysis
TUNEL staining was performed using the In Situ Cell Death Detection Kit (Roche) according to the manufacturer’s instructions except for tissue permeabilization which was performed by incubation at −20°C for 5 min in 1:2 glacial acetic acid/ethanol solution. To quantify apoptotic cells in the wound epidermis, the percentage of TUNEL+ nuclei out of total DAPI+ cells was calculated in the wound epidermis within 500 µm of the amputation plane for each condition. To quantify the total percentage of apoptotic cells, the percentage of TUNEL+ nuclei out of total DAPI+ cells was calculated within 500 µm of the amputation plane.
Immunostaining and EdU analysis
Limb tissue was collected and fixed overnight in 4% paraformaldehyde, brought up a sucrose gradient, and embedded in OCT. Tissue was cryo-sectioned at a thickness of 12 μm. For MK immunostaining, a custom polyclonal rabbit antibody against the C-terminus (amino acids 125–142) of axolotl MK (New England Peptide) was generated and validated with a peptide blocking assay. Western blots were performed on 20 µg of 10 dpa protein extracts and blocked with increasing concentrations of the peptide used to generate the antibody to validate a depletion of signal from the anti-MK staining (1:2000) (Figure 2—figure supplement 1B). MK immunostaining was performed at a dilution of 1:500. For other stains, the following antibodies were used: Beta-III tubulin (Sigma, 1:200), WE3 (DSHB, 1:10), GAPDH (Millipore, 1:2000), tdTomato (LS Bio, 1:200). Sections were blocked for 1 hr at room temperature (0.1% triton-X, 0.3%BSA, 8% donkey serum in PBS), incubated in primary antibodies at 4°C overnight, and stained with secondary Alexafluor antibodies.
For EdU analysis, animals were injected with 100–200 µL of an 8 mg/mL solution of EdU (Thermofisher Scientific) and the Click-it EdU A488 Imaging Kit was utilized (Thermofisher Scientific) according to the manufacturer’s instructions. To quantify the proliferating cells in the wound epidermis, the percentage of EdU+ cells out of total DAPI+ cells in the wound epidermis was quantified within 500 µm of the amputation plane for each condition. For total EdU quantification, the percentage of EdU+ cells out of total DAPI+ cells across all tissues within 500 µm of the amputation plane was calculated.
Overexpression of mk in regenerating limbs
To generate the mk overexpression vector, the open reading frame of mk was cloned into the pCAG-tdTomato backbone using EcoRI and NotI (Addgene plasmid #83029) to generate a pCAG-MK overexpression vector. To validate the expression and secretion of mk, 293 T cells were transfected with either pCAG-tdTomato or pCAG-MK vectors. Transfected cells and media were collected at 72 hr post-transfection (hpt). Protein lysates from the transfected 293 T cells were collected using RIPA buffer and western blots were performed on each of the protein lysates and media samples using anti-tdTomato (1:1000) (LS Bio LS-C340696), our custom generated anti-MK (1:2000), and anti-GAPDH (1:2000) (Millipore AB2302) antibodies. The presence of axolotl mk was detected in both pCAG-MK transfected 293 T cell lysates and media (Figure 6—figure supplement 3A).
Limbs of regenerating animals were injected and electroporated at 3 dpa with either the pCAG-MK overexpression vector and/or the pCAG-tdTomato vector for both the mk rescue experiment in mknull mutants and the general mk overexpression experiments. The electroporation settings were: 3 × 5 ms poring pulses at 150 V with a pulse interval of 50 ms (+ polarity, 0% decay) and 5 × 50 ms transfer pulses at 50 V with a pulse interval of 999 ms (+ polarity, 0% decay). Approximately 1 μg total was injected into each limb. Comparison of electroporation efficiency between conditions was quantified both via calculation of the mean fluorescence intensity (MFI) and quantification of the percentage of tdTomato+ cells in tissues 500 µm proximal of the amputation plane. Brightfield images of regenerating limbs were taken using an Olympus SZX16 dissecting microscope. For quantification of proliferating cells in the wound epidermis, EdU+ cells were quantified out of total DAPI+ cells in the wound epidermis within 200 µm of the amputation plane.
Alcian staining
Regenerated limbs of DMSO/iMDK-treated limbs or tdT/mk-overexpressing limbs were collected at 60 dpa or 51 dpa, respectively, and fixed in 95% ethanol overnight. Limbs were then placed in acetone overnight and subsequently stained for 3–5 days at 37°C in Alcian blue/Alizarin red staining solution: 5% glacial acetic acid, 5% Alcian blue stock solution, and 5% Alizarin red stock solution in 70% ethanol. The tissue was then digested and cleared through a 1% KOH/glycerol gradient with increasing concentrations of glycerol.
Mk receptor single cell RNA-seq data analysis
The single cell RNA-seq data browser was utilized from Leigh et al. (2018), to identify expression patterns of mk receptors in different cell types during homeostasis and regeneration (https://singlecell.broadinstitute.org/single_cell/study/SCP422/transcriptomic-landscape-of-the-blastema-niche-in-regenerating-adult-axolotl-limbs-at-single-cell-resolution-intact-limb).
Quantification and statistical analysis
All of the imaging analysis in each functional experiment was conducted randomized and blinded to prevent bias. For statistical analysis in each experiment, an unpaired two-tailed t-test was conducted unless otherwise stated. Besides the initial full skin flap sequencing experiment, every sample (N) in each experiment was an independent biological replicate that is a limb from a different animal. All of the data were represented as the mean ± standard deviation. For quantification, the value from each biological replicate was an average of values from 2 to 3 sections from the same regions of the limb (using the radius and ulna as histological landmarks across different samples) to capture variability across the tissue.
Acknowledgements
We thank S Ionescu and J Lavecchio in the FACS core at the Harvard Department of Stem Cell and Regenerative Biology (HSCRB) for helping with the cell sorting, I Adatto, L Krug, and Harvard Office of Animal Resources (OAR) for their dedicated animal care, as well as both the Biopolymers Facility and Bauer Sequencing Core at Harvard for their sequencing services. We are also grateful to all of the members of the Melton lab, N Leigh, J Whited, C Tabin, and C Extavour for comments and suggestions on the project and manuscript. DAM is an investigator of the Howard Hughes Medical Institute (HHMI). This research was performed using resources and/or funding from the Harvard Stem Cell Institute (HSCI) and HHMI.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Douglas A Melton, Email: dmelton@harvard.edu.
Phillip A Newmark, Morgridge Institute for Research, United States.
Anna Akhmanova, Utrecht University, Netherlands.
Funding Information
This paper was supported by the following grant:
Howard Hughes Medical Institute to Douglas A Melton.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing - original draft, Writing - review and editing.
Conceptualization, Data curation, Formal analysis, Validation, Investigation, Writing - review and editing.
Conceptualization, Resources, Supervision, Funding acquisition, Methodology, Writing - review and editing.
Ethics
Animal experimentation: Axolotl (Ambystoma mexicanum) husbandry and surgeries were performed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) and Institutional Animal Care and Use Committee (IACUC) guidelines at Harvard University. All procedures conducted under the axolotl animal protocol #11-32 were approved by Harvard IACUC. All surgeries were performed under tricaine-induced anesthesia and every effort was made to minimize suffering.
Additional files
This excel table contains the list of differentially expressed transcripts in dividing cells (enriched for progenitors) in both conditions at 5 dpa. The respective fold change, blastx hit, and adjusted p-values are listed for each hit.
This excel table contains the list of differentially expressed transcripts in non-dividing cells in stump tissues in both conditions at 5 dpa. The respective fold change, blastx hit, and adjusted p-values are listed for each hit.
This excel table contains the list of differentially expressed transcripts in epithelial cells of full thickness skin vs. wound epithelial cells at 5 dpa. The respective fold change, blastx hit, and adjusted p-values are listed for each hit.
This excel table contains the list of differentially expressed transcripts in DMSO vs. iMDK-treated limbs at 11 dpa with the respective fold change, blastx hit, and adjusted p-values for each hit.
Data availability
The raw reads and normalized TPM values for each RNA-sequencing dataset are deposited on GEO at accession numbers GSE132325 for the full skin flap dataset and accession number GSE132317 for the iMDK dataset.
The following datasets were generated:
Stephanie L Tsai, Clara Baselga-Garriga, Douglas A Melton. 2020. Wound epidermis-dependent transcriptional programs. NCBI Gene Expression Omnibus. GSE132317
Stephanie L Tsai, Clara Baselga-Garriga, Douglas A Melton. 2020. Sequencing of iMDK-treated regenerating limbs. NCBI Gene Expression Omnibus. GSE132325
The following previously published dataset was used:
Tsai SL, Baselga-Garriga C, Melton DA. 2019. Blastemal progenitors modulate immune signaling during early limb regeneration. NCBI Gene Expression Omnibus. GSE111213
References
- Attisano L, Wrana JL. Signal integration in TGF-β, WNT, and hippo pathways. F1000Prime Reports. 2013;5:17. doi: 10.12703/P5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boilly B, Albert P. In vitro control of blastema cell proliferation by extracts from epidermal cap and mesenchyme of regenerating limbs of axolotls. Roux's Archives of Developmental Biology. 1990;198:443–447. doi: 10.1007/BF00399054. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo TH, Davis FG, Bateman J, Bryant S, Guzikowski AR, Tsai SL, Coyne S, Ye WW, Freeman RM, Peshkin L, Tabin CJ, Regev A, Haas BJ, Whited JL. A Tissue-Mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Reports. 2017a;18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Sousounis K, Payzin-Dogru D, Bryant S, Sandoval AGW, Martinez Fernandez J, Mariano R, Oshiro R, Wong AY, Leigh ND, Johnson K, Whited JL. Identification of regenerative roadblocks via repeat deployment of limb regeneration in axolotls. Npj Regenerative Medicine. 2017b;2:30. doi: 10.1038/s41536-017-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calinescu AA, Vihtelic TS, Hyde DR, Hitchcock PF. Cellular expression of midkine-a and midkine-b during retinal development and photoreceptor regeneration in zebrafish. The Journal of Comparative Neurology. 2009;514:1–10. doi: 10.1002/cne.21999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LJ, Suárez-Castillo EC, Ortiz-Zuazaga H, Knapp D, Tanaka EM, Crews CM. Gene expression profile of the regeneration epithelium during axolotl limb regeneration. Developmental Dynamics. 2011;240:1826–1840. doi: 10.1002/dvdy.22669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LJ, Crews CM. Wound epidermis formation and function in urodele amphibian limb regeneration. Cellular and Molecular Life Sciences : CMLS. 2008;65:73–79. doi: 10.1007/s00018-007-7433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen RN, Weinstein M, Tassava RA. Expression of fibroblast growth factors 4, 8, and 10 in limbs, flanks, and blastemas ofAmbystoma. Developmental Dynamics. 2002;223:193–203. doi: 10.1002/dvdy.10049. [DOI] [PubMed] [Google Scholar]
- Christensen RN, Tassava RA. Apical epithelial cap morphology and fibronectin gene expression in regenerating axolotl limbs. Developmental Dynamics. 2000;217:216–224. doi: 10.1002/(SICI)1097-0177(200002)217:2<216::AID-DVDY8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Cohen S, Shoshana OY, Zelman-Toister E, Maharshak N, Binsky-Ehrenreich I, Gordin M, Hazan-Halevy I, Herishanu Y, Shvidel L, Haran M, Leng L, Bucala R, Harroch S, Shachar I. The cytokine midkine and its receptor rptpζ regulate B cell survival in a pathway induced by CD74. The Journal of Immunology. 2012;188:259–269. doi: 10.4049/jimmunol.1101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, Tanaka EM. Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Developmental Cell. 2016;39:411–423. doi: 10.1016/j.devcel.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai LC, Wang X, Yao X, Lu YL, Ping JL, He JF. Antisense oligonucleotides targeting midkine induced apoptosis and increased chemosensitivity in hepatocellular carcinoma cells. Acta Pharmacologica Sinica. 2006;27:1630–1636. doi: 10.1111/j.1745-7254.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, Fukuda R, Gerri C, Giraldez AJ, Stainier DYR. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568:193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas JE, Freitas PD, Bryant DM, Whited JL, Monaghan JR. Neuregulin-1 signaling is essential for nerve-dependent axolotl limb regeneration. Development. 2016;143:2724–2731. doi: 10.1242/dev.133363. [DOI] [PubMed] [Google Scholar]
- Fei JF, Knapp D, Schuez M, Murawala P, Zou Y, Pal Singh S, Drechsel D, Tanaka EM. Tissue- and time-directed electroporation of CAS9 protein-gRNA complexes in vivo yields efficient multigene knockout for studying gene function in regeneration. Npj Regenerative Medicine. 2016;1:16002. doi: 10.1038/npjregenmed.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei JF, Schuez M, Knapp D, Taniguchi Y, Drechsel DN, Tanaka EM. Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration. PNAS. 2017;114:12501–12506. doi: 10.1073/pnas.1706855114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei JF, Lou WP, Knapp D, Murawala P, Gerber T, Taniguchi Y, Nowoshilow S, Khattak S, Tanaka EM. Application and optimization of CRISPR-Cas9-mediated genome engineering in axolotl (Ambystoma mexicanum) Nature Protocols. 2018;13:2908–2943. doi: 10.1038/s41596-018-0071-0. [DOI] [PubMed] [Google Scholar]
- Fischman DA, Hay ED. Origin of osteoclasts from mononuclear leucocytes in regenerating newt limbs. The Anatomical Record. 1962;143:329–337. doi: 10.1002/ar.1091430402. [DOI] [PubMed] [Google Scholar]
- Flowers GP, Sanor LD, Crews CM. Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. eLife. 2017;6:e25726. doi: 10.7554/eLife.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 Protein-Mediated oligonucleotide insertion and Large-Scale assessment of Single-Guide RNAs. PLOS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac-Santel M, Nowoshilow S, Kageyama J, Khattak S, Currie JD, Camp JG, Tanaka EM, Treutlein B. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 2018;362:eaaq0681. doi: 10.1126/science.aaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Roy S, Séguin C, Bryant SV, Gardiner DM. Analysis of the expression and function of Wnt-5a and Wnt-5b in developing and regenerating axolotl (Ambystoma mexicanum) limbs. Development, Growth & Differentiation. 2008;50:289–297. doi: 10.1111/j.1440-169X.2008.01000.x. [DOI] [PubMed] [Google Scholar]
- Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. PNAS. 2013;110:9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin J, Kuraitis D, Rosenthal N. Extracellular matrix considerations for scar-free repair and regeneration: insights from regenerative diversity among vertebrates. The International Journal of Biochemistry & Cell Biology. 2014;56:47–55. doi: 10.1016/j.biocel.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Goss RJ. Regenerative inhibition following limb amputation and immediate insertion into the body cavity. The Anatomical Record. 1956a;126:15–27. doi: 10.1002/ar.1091260103. [DOI] [PubMed] [Google Scholar]
- Goss RJ. The regenerative responses of amputated limbs to delayed insertion into the body cavity. The Anatomical Record. 1956b;126:283–297. doi: 10.1002/ar.1091260303. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramage E, D’Cruz T, Taylor S, Thummel R, Hitchcock PF. Midkine-a protein localization in the developing and adult retina of the zebrafish and its function during photoreceptor regeneration. PLOS ONE. 2015;10:e0121789. doi: 10.1371/journal.pone.0121789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Edholm ES, Robert J. Mechanisms of amphibian macrophage development: characterization of the Xenopus laevis colony-stimulating factor-1 receptor. The International Journal of Developmental Biology. 2014;58:757–766. doi: 10.1387/ijdb.140271jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Han MJ, An JY, Kim WS. Expression patterns of Fgf-8 during development and limb regeneration of the axolotl. Developmental Dynamics. 2001;220:40–48. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1085>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Hall CJ, Crosier PS, Abe G, Kawakami K, Kudo A, Kawakami A. Transient inflammatory response mediated by interleukin-1β is required for proper regeneration in zebrafish fin fold. eLife. 2017;6:e22176. doi: 10.7554/eLife.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED, Fischman DA. Origin of the blastema in regenerating limbs of the newt Triturus viridescens. An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Developmental Biology. 1961;3:26–59. doi: 10.1016/0012-1606(61)90009-4. [DOI] [PubMed] [Google Scholar]
- Johnson K, Bateman J, DiTommaso T, Wong AY, Whited JL. Systemic cell cycle activation is induced following complex tissue injury in axolotl. Developmental Biology. 2018;433:461–472. doi: 10.1016/j.ydbio.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman S, Alitalo K. Midkine and melanoma metastasis: a malevolent mix. Developmental Cell. 2017;42:205–207. doi: 10.1016/j.devcel.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Martí M, Dubova I, Izpisúa Belmonte JC. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes & Development. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp D, Schulz H, Rascon CA, Volkmer M, Scholz J, Nacu E, Le M, Novozhilov S, Tazaki A, Protze S, Jacob T, Hubner N, Habermann B, Tanaka EM. Comparative transcriptional profiling of the axolotl limb identifies a tripartite Regeneration-Specific gene program. PLOS ONE. 2013;8:e61352. doi: 10.1371/journal.pone.0061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyritsis N, Kizil C, Zocher S, Kroehne V, Kaslin J, Freudenreich D, Iltzsche A, Brand M. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012;338:1353–1356. doi: 10.1126/science.1228773. [DOI] [PubMed] [Google Scholar]
- Leigh ND, Dunlap GS, Johnson K, Mariano R, Oshiro R, Wong AY, Bryant DM, Miller BM, Ratner A, Chen A, Ye WW, Haas BJ, Whited JL. Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nature Communications. 2018;9:5153. doi: 10.1038/s41467-018-07604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui M, Okui T, Shimo T, Takabatake K, Fukazawa T, Matsumoto K, Kurio N, Ibaragi S, Naomoto Y, Nagatsuka H, Sasaki A. Novel midkine inhibitor iMDK inhibits tumor growth and angiogenesis in oral squamous cell carcinoma. Anticancer Research. 2016;36:2775–2781. [PubMed] [Google Scholar]
- McCusker C, Bryant SV, Gardiner DM. The axolotl limb blastema: cellular and molecular mechanisms driving blastema formation and limb regeneration in tetrapods. Regeneration. 2015;2:54–71. doi: 10.1002/reg2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Woodgett JR. When pathways collide: collaboration and connivance among signalling proteins in development. Nature Reviews Molecular Cell Biology. 2010;11:404–413. doi: 10.1038/nrm2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng K, Rodriguez-Peña A, Dimitrov T, Chen W, Yamin M, Noda M, Deuel TF. Pleiotrophin signals increased tyrosine phosphorylation of beta beta-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase beta/zeta. PNAS. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher AL. Effects on adult newt limb regeneration of partial and complete skin flaps over the amputation surface. Journal of Experimental Zoology. 1976;195:117–127. doi: 10.1002/jez.1401950111. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Muramatsu T, Muramatsu H, Thesleff I. Midkine (MK), a heparin-binding growth/differentiation factor, is regulated by retinoic acid and epithelial-mesenchymal interactions in the developing mouse tooth, and affects cell proliferation and morphogenesis. The Journal of Cell Biology. 1995a;129:267–281. doi: 10.1083/jcb.129.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis TA, Salmivirta M, Muramatsu T, Muramatsu H, Rauvala H, Lehtonen E, Jalkanen M, Thesleff I. Expression of the heparin-binding cytokines, midkine (MK) and HB-GAM (pleiotrophin) is associated with epithelial-mesenchymal interactions during fetal development and organogenesis. Development. 1995b;121:37–51. doi: 10.1242/dev.121.1.37. [DOI] [PubMed] [Google Scholar]
- Monaghan JR, Epp LG, Putta S, Page RB, Walker JA, Beachy CK, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biology. 2009;7:1. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T. Midkine, a heparin-binding cytokine with multiple roles in development, repair and diseases. Proceedings of the Japan Academy, Series B. 2010;86:410–425. doi: 10.2183/pjab.86.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T. Structure and function of midkine as the basis of its pharmacological effects. British Journal of Pharmacology. 2014;171:814–826. doi: 10.1111/bph.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima M, D’Cruz TS, Hesse D, Sifuentes CJ, Raymond PA, Hitchcock PF. Midkine-a is required for cell cycle progression of müller Glia during neuronal regeneration. bioRxiv. 2019 doi: 10.1101/668210. [DOI] [PMC free article] [PubMed]
- Owada K, Sanjo N, Kobayashi T, Mizusawa H, Muramatsu H, Muramatsu T, Michikawa M. Midkine inhibits caspase-dependent apoptosis via the activation of mitogen-activated protein kinase and phosphatidylinositol 3-kinase in cultured neurons. Journal of Neurochemistry. 1999;73:2084–2092. [PubMed] [Google Scholar]
- Pathania M, Torres-Reveron J, Yan L, Kimura T, Lin TV, Gordon V, Teng ZQ, Zhao X, Fulga TA, Van Vactor D, Bordey A. miR-132 enhances dendritic morphogenesis, spine density, synaptic integration, and survival of newborn olfactory bulb neurons. PLOS ONE. 2012;7:e38174. doi: 10.1371/journal.pone.0038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, Yuan GC. Analyzing CRISPR genome-editing experiments with CRISPResso. Nature Biotechnology. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repesh LA, Oberpriller JC. Scanning electron microscopy of epidermal cell migration in wound healing during limb regeneration in the adult newt, Notophthalmus viridescens. American Journal of Anatomy. 1978;151:539–555. doi: 10.1002/aja.1001510408. [DOI] [PubMed] [Google Scholar]
- Sandoval-Guzmán T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, Simon A. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Satoh A, Graham GM, Bryant SV, Gardiner DM. Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum) Developmental Biology. 2008;319:321–335. doi: 10.1016/j.ydbio.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Satoh A, Bryant SV, Gardiner DM. Nerve signaling regulates basal keratinocyte proliferation in the blastema apical epithelial cap in the axolotl (Ambystoma mexicanum) Developmental Biology. 2012;366:374–381. doi: 10.1016/j.ydbio.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrelle N, Dominguez ATA, Brekken RA. From top to bottom: midkine and pleiotrophin as emerging players in immune regulation. Journal of Leukocyte Biology. 2017;102:277–286. doi: 10.1189/jlb.3MR1116-475R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocum DL, Dearlove GE. Epidermal-mesodermal interaction during morphogenesis of the limb regeneration blastema in larval salamanders. Journal of Experimental Zoology. 1972;181:49–61. doi: 10.1002/jez.1401810106. [DOI] [Google Scholar]
- Sugiura T, Wang H, Barsacchi R, Simon A, Tanaka EM. MARCKS-like protein is an initiating molecule in axolotl appendage regeneration. Nature. 2016;531:237–240. doi: 10.1038/nature16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Drechsel DN, Brockes JP. Thrombin regulates S-phase re-entry by cultured newt myotubes. Current Biology. 1999;9:792–799. doi: 10.1016/S0960-9822(99)80362-5. [DOI] [PubMed] [Google Scholar]
- Tanaka EM. The molecular and cellular choreography of appendage regeneration. Cell. 2016;165:1598–1608. doi: 10.1016/j.cell.2016.05.038. [DOI] [PubMed] [Google Scholar]
- Tanaka EM, Brockes JP. A target of thrombin activation promotes cell cycle re-entry by urodele muscle cells. Wound Repair and Regeneration. 1998;6:S-371–S-370. doi: 10.1046/j.1524-475X.1998.60413.x. [DOI] [PubMed] [Google Scholar]
- Tassava RA, Garling DJ. Regenerative responses in larval axolotl limbs with skin grafts over the amputation surface. Journal of Experimental Zoology. 1979;208:97–109. doi: 10.1002/jez.1402080111. [DOI] [PubMed] [Google Scholar]
- Tassava RA, Loyd RM. Injury requirement for initiation of regeneration of newt limbs which have whole skin grafts. Nature. 1977;268:49–50. doi: 10.1038/268049a0. [DOI] [PubMed] [Google Scholar]
- Thornton CS. The effect of apical cap removal on limb regeneration in amblystoma larvae. Journal of Experimental Zoology. 1957;134:357–381. doi: 10.1002/jez.1401340209. [DOI] [PubMed] [Google Scholar]
- Thornton CS. The inhibition of limb regeneration in Urodele larvae by localized irradiation with ultraviolet light. Journal of Experimental Zoology. 1958;137:153–179. doi: 10.1002/jez.1401370108. [DOI] [PubMed] [Google Scholar]
- Tsai SL, Baselga-Garriga C, Melton DA. Blastemal progenitors modulate immune signaling during early limb regeneration. Development. 2019;146:dev169128. doi: 10.1242/dev.169128. [DOI] [PubMed] [Google Scholar]
- Tsarouchas TM, Wehner D, Cavone L, Munir T, Keatinge M, Lambertus M, Underhill A, Barrett T, Kassapis E, Ogryzko N, Feng Y, van Ham TJ, Becker T, Becker CG. Dynamic control of proinflammatory cytokines Il-1β and Tnf-α by macrophages in zebrafish spinal cord regeneration. Nature Communications. 2018;9:4670. doi: 10.1038/s41467-018-07036-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieceli FM, Bronner ME. Leukocyte receptor tyrosine kinase interacts with secreted midkine to promote survival of migrating neural crest cells. Development. 2018;145:dev164046. doi: 10.1242/dev.164046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinarsky V, Atkinson DL, Stevenson TJ, Keating MT, Odelberg SJ. Normal newt limb regeneration requires matrix metalloproteinase function. Developmental Biology. 2005;279:86–98. doi: 10.1016/j.ydbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wagner I, Wang H, Weissert PM, Straube WL, Shevchenko A, Gentzel M, Brito G, Tazaki A, Oliveira C, Sugiura T, Shevchenko A, Simon A, Drechsel DN, Tanaka EM. Serum proteases potentiate BMP-Induced cell cycle Re-entry of dedifferentiating muscle cells during newt limb regeneration. Developmental Cell. 2017;40:608–617. doi: 10.1016/j.devcel.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Research. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. Gplots: Various R Programming Tools for Plotting Data 2015
- Weckbach LT, Muramatsu T, Walzog B. Midkine in inflammation. The Scientific World JOURNAL. 2011;11:2491–2505. doi: 10.1100/2011/517152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited JL, Tsai SL, Beier KT, White JN, Piekarski N, Hanken J, Cepko CL, Tabin CJ. Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development. 2013;140:1137–1146. doi: 10.1242/dev.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight TN, Kang I, Merrilees MJ. Versican and the control of inflammation. Matrix Biology. 2014;35:152–161. doi: 10.1016/j.matbio.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Ouyang D, Li Q, Li M, Zou Q, Li L, Yi W, Zhou E. The expression, functions, interactions and prognostic values of PTPRZ1: a review and bioinformatic analysis. Journal of Cancer. 2019;10:1663–1674. doi: 10.7150/jca.28231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhu S, Wu M, Han W, Yu Y. Functional receptors and intracellular signal pathways of midkine (MK) and pleiotrophin (PTN) Biological and Pharmaceutical Bulletin. 2014;37:511–520. doi: 10.1248/bpb.b13-00845. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Ogino H, Stoick-Cooper CL, Grainger RM, Moon RT. Wnt/beta-catenin signaling has an essential role in the initiation of limb regeneration. Developmental Biology. 2007;306:170–178. doi: 10.1016/j.ydbio.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K, Muramatsu H, Ikematsu S, Sakuma S, Salama RH, Shinomura T, Kimata K, Muramatsu T. A heparin-binding growth factor, midkine, binds to a chondroitin sulfate proteoglycan, PG-M/versican. European Journal of Biochemistry. 2000;267:4046–4053. doi: 10.1046/j.1432-1327.2000.01440.x. [DOI] [PubMed] [Google Scholar]