Abstract
Myosin 1s have critical roles in linking membranes to the actin cytoskeleton via direct binding to acidic lipids. Lipid binding may occur through PIP3/PIP2-specific PH domains or nonspecific ionic interactions involving basic-hydrophobic (BH) sites but the mechanism of myosin 1s distinctive lipid targeting is poorly understood. Now we show that PH domains occur in all Dictyostelium myosin 1s and that the BH sites of Myo1A, B, C, D, and F are in conserved positions near the β3/β4 loops of their PH domains. In spite of these shared lipid-binding sites, we observe significant differences in myosin 1s highly dynamic localizations. All myosin 1s except Myo1A are present in macropinocytic structures but only Myo1B and Myo1C are enriched at the edges of macropinocytic cups and associate with the actin in actin waves. In contrast, Myo1D, E, and F are enclosed by the actin wave. Mutations of BH sites affect localization of all Dictyostelium myosin 1s. Notably, mutation of the BH site located within the PH domains of PIP3-specific Myo1D and Myo1F completely eradicates membrane binding. Thus, BH sites are important determinants of motor targeting and may have a similar role in the localization of other myosin 1s.
INTRODUCTION
Class-I myosins, the first-discovered unconventional myosins (Pollard and Korn, 1973), are one of the most widely expressed and ancient members of the myosin superfamily (Pollard and Korn, 1973; Richards and Cavalier-Smith, 2005; Odronitz and Kollmar, 2007; Kollmar and Muhlhausen, 2017). These ubiquitous motors have a wide range of cellular functions (McConnell and Tyska, 2010; Maxeiner et al., 2015; Kittelberger et al., 2016; McIntosh and Ostap, 2016; Ouderkirk-Pecone et al., 2016; Visuttijai et al., 2016; Caridi et al., 2018; Juan et al., 2018; McIntosh et al., 2018; Nevzorov et al., 2018). All myosin 1s are monomeric and have a single heavy chain with an N-terminal head (motor domain), a neck with one or more IQ sites that bind light chains, and a basic tail homology-1 (TH1) region. Long-tail myosin 1s have, in addition, a Pro-rich region and a C-terminal SH3 domain (see Figure 1; Pollard et al., 1991; Sellers, 2000; Berg et al., 2001). Most organisms harbor multiple isoforms of myosin 1s. For example, Dictyostelium discoideum has three short-tail (DdMyo1A, DdMyo1E, and DdMyo1F), and three long-tail (DdMyo1B, DdMyo1C, and DdMyo1D) myosin 1s, while Homo sapiens have six short-tail (HsMyo1A, HsMyo1B, HsMyo1C, HsMyo1D, HsMyo1G, and HsMyo1H) and two long-tail (HsMyo1E and HsMyo1F) myosin 1s.
FIGURE 1:
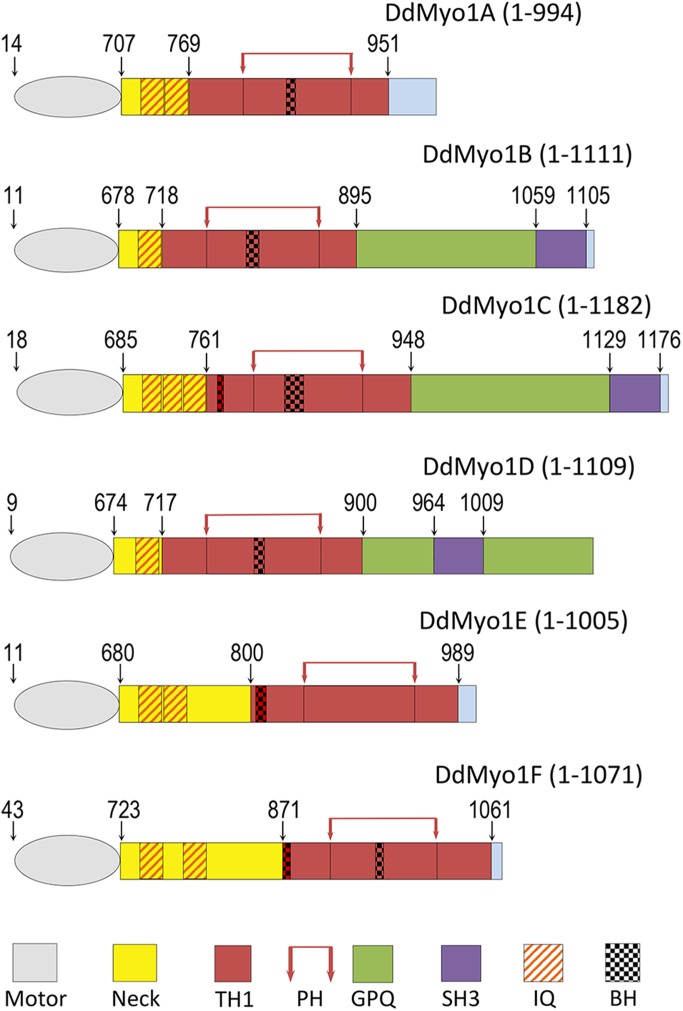
Dictyostelium myosin 1s. Schematic representation of Dictyostelium Myo1A-Myo1F. The heads (motors), necks, and regions within the tails are color coded, and their boundaries are marked on the top according to Cymobase (Odronitz and Kollmar, 2006; Kollmar and Muhlhausen, 2017). The tails (but not the motors) are drawn in proportion. The BH sites are marked according to BH search (Brzeska et al., 2010). Positions of PH domains are additionally marked by red arrows on the top. Light-chain binding sites (IQ motifs) are marked according to Crawley et al. (2011). The exact locations that are not marked in the figure by numbers are as follows: MIA: IQ1: 725–746; IQ2: 747–768; PH: 820–923; BH: 858–865, MIB: IQ: 696–717; PH: 765–865; BH: 802–811, MIC: IQ1: 703–720; IQ2: 721–738; IQ3: 739–760; BH-N: 775–780; PH: 809–907; BH: 837–854, MID: IQ: 692–713; PH: 760–864; BH: 804–814, MIE: IQ1: 698–719; IQ2: 720–741; BH: 804–815; PH: 850–952, MIF: IQ1: 741–762; IQ2: 781–802; BH-N: 871–878; PH: 912–1010; BH: 947–955.
Dictyostelium myosin 1 isoforms may have unique as well as redundant functions because removal of multiple isoforms usually causes stronger phenotypic defects than removal of any single one (Novak et al., 1995; Jung et al., 1996; Ostap and Pollard, 1996; Falk et al., 2003; Rivero, 2008; Chen et al., 2012). Most of the known functions of myosin 1s require binding to membranes via acidic phospholipids. Mapping of the critical regions required for membrane targeting of each myosin 1 is needed for a full understanding of their individual and shared functions and how their lipid-binding sequences dictate their specific intracellular targeting and function.
The binding of myosin 1s to lipids occurs through the basic TH1 domain either by a PIP3/PIP2-specific PH domain (Hokanson and Ostap, 2006; Komaba and Coluccio, 2010) or by non–lipid-specific ionic interactions (Brzeska et al., 2008, 2010, 2012, 2016; Feeser et al., 2010; Mazerik and Tyska, 2012). Three Dictyostelium myosin 1s (Myo1D, Myo1E, and Myo1F) bind PIP3 with high specificity, whereas the other three (Myo1A, Myo1B, and Myo1C) do not (Chen et al., 2012). Dictyostelium Myo1B and Myo1A bind to acidic phospholipids principally through a short basic-hydrophobic (BH) region located within the TH1 domain of their tails (Brzeska et al., 2010, 2012, 2014, 2016) as shown originally for Acanthamoeba myosin 1C (Brzeska et al., 2008).These BH sites all have a high content of basic and hydrophobic residues but no sequence homology (Brzeska et al., 2010).
PH domains have low sequence homology, similar to BH sites, but a highly conserved structure (seven β-sheets and a C-terminal α-helix). One of the major functions of PH domains is lipid binding (Lemmon et al., 2002; Scheffzek and Welti, 2012; Feng et al., 2018); however, most PH domains do not display any lipid specificity (Lemmon et al., 2002). A high-resolution structure of HsMyo1C identified all the conserved structural elements of its PH domain (Lu et al., 2015), and the presence of PH domains was predicted for other human myosin 1s (Hokanson et al., 2006; Lu et al., 2015).
Dictyostelium cells are extremely motile; they undergo fast and dramatic morphological changes, form impressive actin waves, and macropinocytose robustly, making them an excellent model for studying mechanisms of myosin 1s targeting. The cellular localization of long-tail DdMyo1B (Brzeska et al., 2012, 2014) and short-tail DdMyo1A, DdMyo1E, and DdMyo1F (Brzeska et al., 2016) have been previously shown for cells in different motile states. To directly compare the localization of all myosin 1 isoforms in a single cell type, the dynamic localizations of long-tail DdMyo1C and DdMyo1D have now been characterized. We also investigated the importance of the BH sites for localization of Dictyostelium myosins 1C, 1D, 1E, and 1F.
RESULTS
Identification of BH sites within the PH domains of Dictyostelium and human myosin 1s
Myosin 1s bind to membranes via their TH1 domain. The recently published crystal structure of the tail region of human Myo1C (Lu et al., 2015) revealed the secondary structure of its TH1 region and of the PH domain within it. We aligned the sequences of Dictyostelium and human myosin 1s (Edgar, 2004) and superimposed the structural elements from the crystal structure of human HsMyo1C tail (Lu et al., 2015) and identified the positions of the BH sites (Supplemental Figures S1 and S2, and Table 1). BH sites are defined as amino acid residues with a BH score above 0.6 and the BH search identifies BH sites by including nine residues before and after the BH site (Brzeska et al., 2010). Thus, a designated BH site can be a single amino acid, as for HsMyo1A (Figures 1 and 2A and Table 1). Figure 1 shows the schematic representations of full-length Dictyostelium myosin 1s with BH sites marked. The boundaries of head, neck, TH1, GPQ, and SH3 domains are marked according to Cymobase (Odronitz and Kollmar, 2006; Kollmar and Muhlhausen, 2017) and the IQ sites are marked according to Crawley et al. (2011).
TABLE 1:
Localization of BH peaks in the tails of Dictyostelium and human myosin 1s.
| Myosin | Tail length | Name of BH peak | Location | Sequence |
|---|---|---|---|---|
| Dictyostelium | ||||
| DdMyo1A | Short | BH | β3/β4 | (858) KGDSWFAI (865) |
| DdMyo1B | Long | BH | β3/β4 | (802) KKVLVHTLIR (811) |
| DdMyo1C | Long | BH | β3/β4 | (837) TKQAIYLIKLKQKKNLAT (854) |
| BH-N | α4/α5 | (775) NRFSMI (780) | ||
| DdMyo1D | Long | BH | β3/β4 | (804) KRPWIYVQKRR (814) |
| DdMyo1E | Short | BH-N | α4/α5 | (804) YDIFHGKKKWDF (815) |
| DdMyo1F | Short | BH-N | α4/α5 | (871) RKKEWDCR (878) |
| BH | β3/β4 | (947) KYTQKKVGL (955) | ||
| Human | ||||
| HsMyo1A | Short | BH-N | α3 | (830) KRF (832) |
| BH | β1/β2 | (909) T | ||
| HsMyo1B | Short | BH-N | α3 | (915) KRIFHLWRCKKYRDQ (929) |
| HsMyo1C | Short | BH | β1/β2 | (894) DRKGYKPR (901) |
| HsMyo1D | Short | BH | β1 | (856) NVLFSCH (862) |
| HsMyo1E | Long | β1/β2 | (777) FKGVKRDLLLT (787) | |
| HsMyo1F | Long | BH | β1/β2 | (773) RRFKPIKRDLI (783) |
| BH-C | ? | (927) RKGMAKGKPR (936) | ||
| HsMyo1G | Short | BH | β1/β2 | (876) RKVNRFHKIR (885) |
| HsMyo1H | Short | BH | β1/β2 | (887) KYDRKGFKARQ (899) |
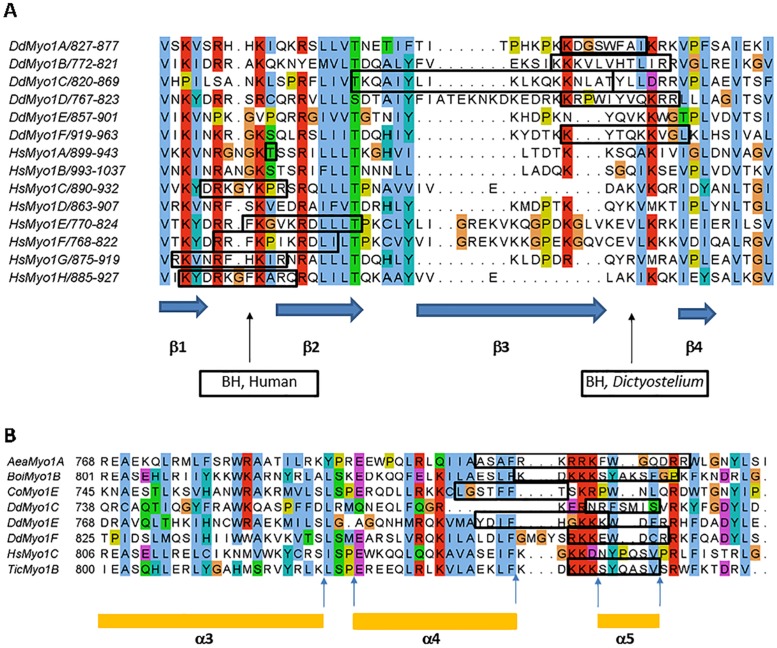
The positions of BH peaks were defined by comparing BH plots of myosins (Supplemental Figures S1 and S2) to their sequence alignments (Figures 1 and 2 and Supplemental Figure S3). BH sites are defined as amino acid residues with BH score higher than 0.6 but nine additional residues on both sides contribute to their appearance (Brzeska et al., 2010).
FIGURE 2:
BH sites of Dictyostelium myosin 1s are located in conserved positions. (A) BH sites of Dictyostelium and human myosin 1s localized within the β2/β3 and β3/β4 loops of their PH domains. Note that most BH sites are located in the vicinity of loops between β1 and β2 for human (Hs) myosin 1s and between β3 and β4 for Dictyostelium (Dd) myosin 1s. Dictyostelium Myo1E and human Myo1B and Myo1D do not have BH sites in this region. The blue arrows at the bottom of the figure mark the positions of β1, β2, β3, and β4 for human Myo1C as defined from its crystal structure (Lu et al., 2015). (B) BH sites of Dictyostelium and other myosin 1s located N-terminal to the PH domain. Secondary structures are marked according to the crystal structure of HsMyo1C (Lu et al., 2015) that does not have a BH site in this region but is included in the alignment as a reference sequence. Alignments were done with MUSCLE (Edgar, 2004). BH sites were defined by BH search (Brzeska et al., 2010) and are boxed. For locations of other BH sites, see Table 1. The sequences are from Cymobase (Odronitz and Kollmar, 2006; Kollmar and Muhlhausen, 2017) and their accession names are shown in the figure. In Figure 2B, sequences from the additional following species are presented: mosquito (Aea, Aedes aegypti), bumble bee (Boi, Bombus impatiens), single-celled eukaryote (Co, Capsaspora owczarzaki), and red flour beetle (Tic, Tribolium castaneum).
The alignments of the human and Dictyostelium TH1 domain sequences unexpectedly reveal that the BH sites of all Dictyostelium myosin 1s except Myo1E are in or near the β3/β4 loop of their PH domains (Figure 2A). The only BH site of Myo1E and the second BH sites of Myo1C and Myo1F are N-terminal to their PH domains but still within the TH1 tail segment (Figure 2B and BH-Ns in Table 1). Based on the homology to human myosin 1C, these BH-N sites are located in the vicinity of the α4/α5 loop of their TH1 domains. We found only four phylogenetically diverse myosin 1s with BH sites in homologous positions (Figure 2B). Interestingly, the BH sites of all human myosin 1s except HsMyo1B are instead found in the vicinity of the β1/β2 loop of their PH domains (Figure 2A), that is, in a region of the PH domain N-terminal to that harboring the BH sites of Dictyostelium myosin 1s. The single BH site of HsMyo1B and a second BH site of HsMyo1A are in the C-terminus of the neck region (BH-N in Table 1). BH sites in this location are also present in myosin 1s from diverse species (Supplemental Figure S3). The long-tailed HsMyo1F has a second BH site in the proline-rich region of the tail (BH-C in Table 1).
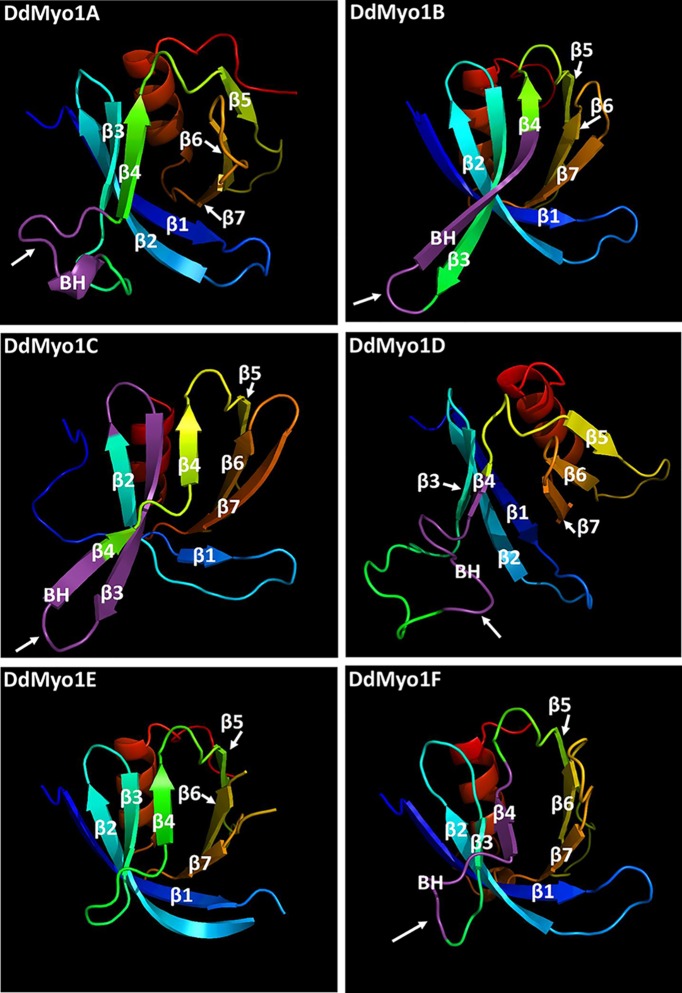
Identification of the BH sites of Dictyostelium myosin 1s in or near the unstructured β3/β4 loop of PH domains was supported by an analysis of predicted protein folding using the PHYRE2 Protein Fold Recognition Server (Kelley et al., 2015). All PH domains of Dictyostelium myosin 1s matched the human Myo1C tail crystal structure (PDB 4R8G) with 96–100% confidence, higher than the confidence of the match with all other PH domains in the Protein Data Bank. The predicted secondary and tertiary structures of the putative Dictyostelium myosin 1 PH domains agree with the secondary structure of the PH domain of human Myo1C and consist of seven β-sheets and a C-terminal α-helix (Figure 3). The exact boundaries and lengths of these structures for the individual myosin 1s are slightly different in the Phyre2 model (see Supplemental Figure S4) than in the sequence alignment shown in Figure 2 but in both alignments the BH sites are in the vicinity of the β3/β4 loop. The other difference is that Phyre2 introduces a short α-helix in the β3/β4 loop of Myo1A and split β4 of Myo1C into two segments (Figure 3). Our three-dimensional (3D) models show that the β3/β4 loops of all Dictyostelium myosin 1s, like the β1/β2 loops of human myosin 1s (Lu et al., 2015), protrude from the PH domain (Figure 3) making them excellent candidates for mediating intermolecular interactions.
FIGURE 3:
3D models of the PH domains of Dictyostelium myosin 1s. 3D models of the PH domains of Dictyostelium Myo1A, Myo1B, Myo1C, Myo1D, and Myo1F were created in Phyre2 (Kelley et al., 2015) using the crystal structure of the PH domain of human Myo1C (Lu et al., 2015) as a template. The arrows point to the BH sites (marked in purple) that are located in the vicinity of the β3/β4 loop of Dictyostelium myosin 1s. DdMyo1E does not have a BH site at this region. The BH sites of human myosin 1s (unpublished data) are located in the vicinity of the β1/β2 loop.
In vivo imaging of myosin 1s and their mutants
Dictyostelium cells were transfected with the plasmids encoding GFP-Myo1s listed in Table 2 and RFP-LifeAct, and the localization of the expressed proteins monitored by fluorescence microscopy. The expression and stability of each GFP-myosin fusion was examined by immunoblotting of SDS–PAGE of cell extracts. GFP-myosin heavy-chain bands were observed at the expected molecular weights and no lower molecular weight bands were present in the blots (Supplemental Figure S5). The blotting results along with the absence of fluorescent autophagosomes in microscopy images argues against any significant proteolysis and therefore also against unfolding of expressed proteins. Although the expression of the GFP-myosin 1s, as seen in total cell extracts, varied between cell lines expressing different myosins and mutants (Supplemental Figure S5), the level of expression in individual cells within the same cell line also varied. Therefore, there was always a considerable pool of cells with similar expression level for comparison (Supplemental Figure S6).
TABLE 2:
Myosins and mutants used in this study.
| Protein name | Description | Mutation |
|---|---|---|
| Myo1C | Full length, 1–1182 | None |
| Myo1C-BH-Ala | BH site mutated | K838A, K845A, K847A |
| Myo1C-BH-N-Ala | BH-N site mutated | R776A, R783A, K784A |
| Myo1C-2BH-Ala | Both BH sites mutated | R776A, R783A, K784A, K838A, K845A, K847A |
| Myo1D | Full length, 1–1109 | None |
| Myo1D-BH-Ala | BH site mutated | K804A, R805A, K812A, R831A |
| Myo1E | Full length, 1–1005 | None |
| Myo1E-BH-N-Ala | BH-N site mutated | K810A, K811A, K812A |
| Myo1F | Full length, 1–1071 | None |
| Myo1F-BH-Ala | BH site mutated | K946A, K947A, K951A, K952A |
| Myo1F-BH-N-Ala | BH-N site mutated | R871A, K872A, K873A, R878A, R879A, K880A |
| Myo1B-BH-Myo1D | BH sites swapped | KKKVLVHTLIRR/RKKRPWIYVQKRR |
In Myo1B-BH-Myo1D the BH site in Myo1B (801)KKKVLVHTLIRR(812) was replaced with the BH site of Myo1D (802)RKKRPWIYVQKRR(814).
The localization of each mutant myosin was quantified in at least 100 cells in two independent transfections in parallel with wild-type protein, and the images shown are representative of a large population of cells for a given cell line.
Localization of PIP3-specific long-tail Myo1D
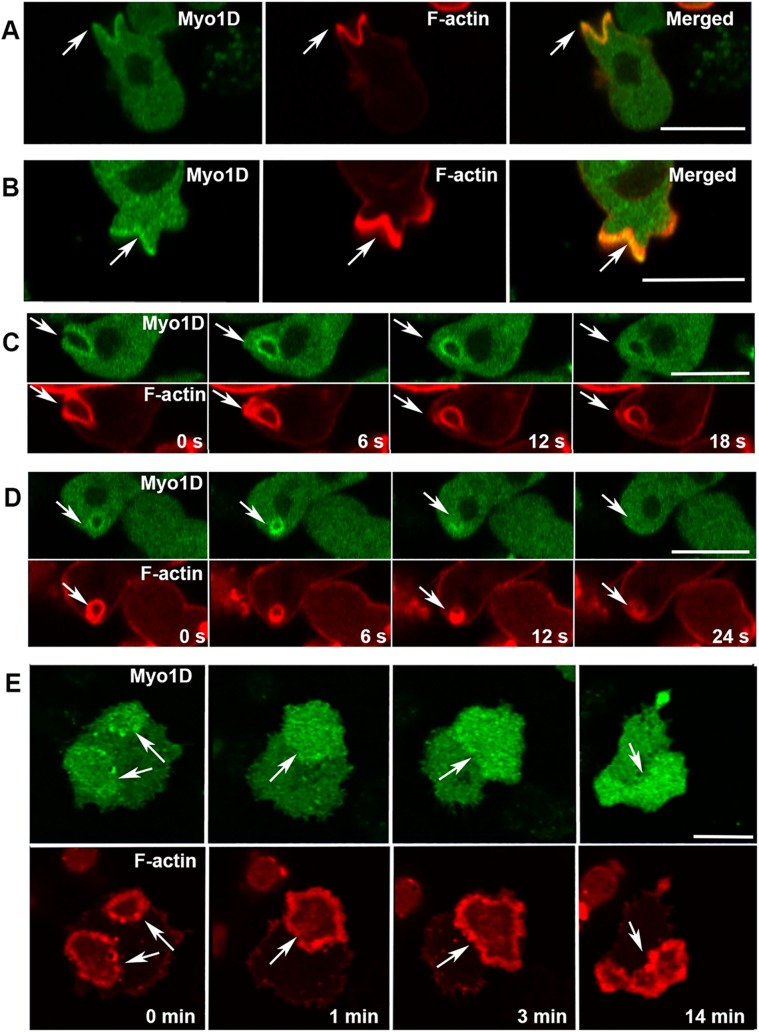
Three Dictyostelium myosin 1s display PIP3-binding; two short-tailed myosins, Myo1E and Myo1F, and the long-tailed Myo1D that has a GPQ domain that targets Myo1B to actin waves. It is not known whether these three myosins have identical cellular localizations and whether the BH site plays a role in their targeting. The dynamic localization of Myo1D largely resembles that of both Myo1E and Myo1F. Expressed Myo1D localized sharply and uniformly to the plasma membrane of nonmotile cells (Figure 4A), diffusely at the front of chemotaxing cells (Figure 4F), as seen for all other Dictyostelium myosin 1s (Brzeska et al., 2012, 2014, 2016, and see below), and localized to cell–cell contacts (Figure 4B).
FIGURE 4:
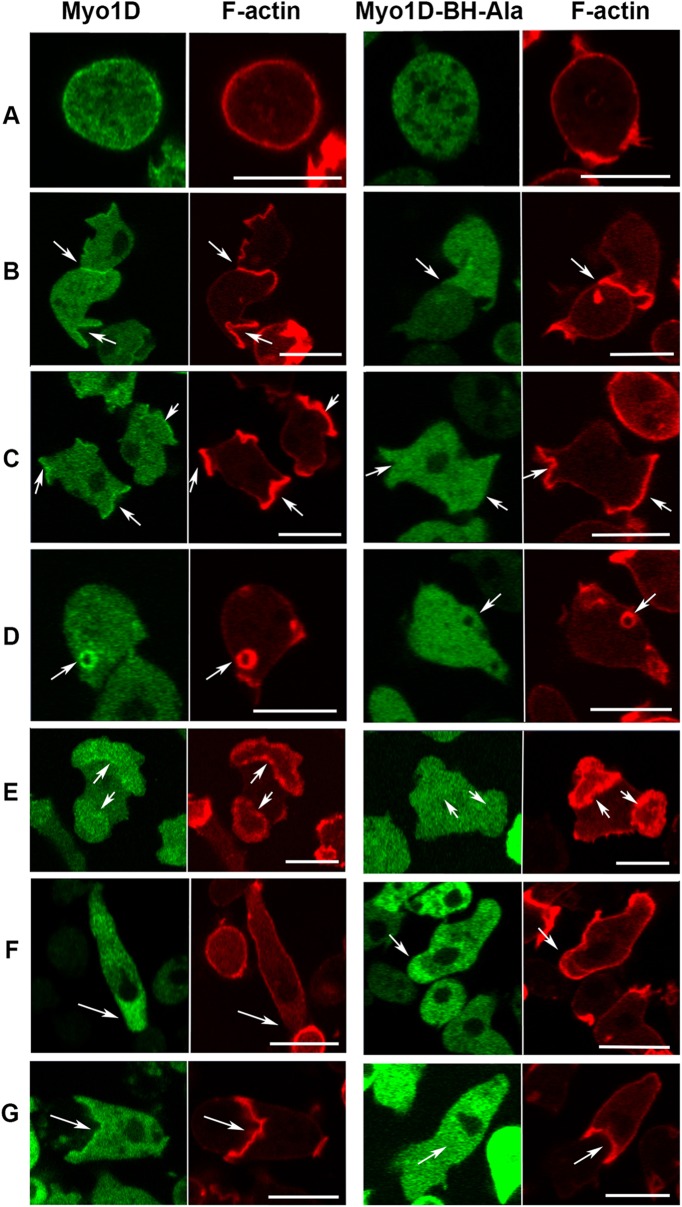
Localization of Myo1D and the effect of BH-Ala mutation. Myo1D (left panels) localizes uniformly on the plasma membrane in nonmotile cells (A), at cell–cell contacts (B), in macropinocytotic protrusions (C), at micropinocytic vesicles (D), in the region encircled by actin wave (E), at the front of chemotaxing cells (F), and in engulfing mouth (G). Myo1D-BH-Ala mutant (right panels) is absent from all these locations (A–E, G) except the front of chemotaxing cells (F). Cells in row A were fixed and live cells are shown in all other rows. Arrows point to regions of interest. Experiments were done in AX2 cells. The localization of MID was the same in Myo1B null cells (unpublished data). Bars are 10 μm. The number of independent transfections was four for Myo1D and two for Myo1D-BH-Ala. At least two independent experiments were performed for each transfection under each condition. The images are representative of more than 100 cells observed under each condition.
A striking difference between Myo1D and the other long-tailed (but not PIP3 specific) Myo1B localization in actin waves was observed (Figures 4E and 5E). Myo1D localized, like short-tail Myo1E and Myo1F, in the PIP3-rich region encircled by the actin wave (Figure 4E), with only occasional enrichment in the actin wave itself (see 0 and 14 min in Figure 5E), whereas Myo1B colocalizes only with the actin (Brzeska et al., 2012, 2014). Thus, in spite of the presence of an actin-binding domain (the GPQ region), the Myo1D PIP3-binding site in the TH1 region plays a dominant role in Myo1D localization.
FIGURE 5:
Myo1D localization in macropinocytic structures and actin waves. (A, B) Myo1D is present but not enriched at the edges of macropinocytic cups. (C) Myo1D dissociates from macropinocytic vesicle before actin. (D) Myo1D fluorescence around the pinosome transiently intensifies after ingestion (see 6 s). (E) Myo1D localizes in the region surrounded by the actin wave and is only occasionally and temporarily enriched in waves (see 0 min). Arrows point to the regions of interest. Experiments were done in AX2 cells. All images are of live cells. Bars are 10 μm. Four independent Myo1D transfections were done and at least two independent experiments were performed for each transfection. The images are representative of more than 100 cells observed under each condition.
All myosin 1s except Myo1A are associated with macropinosomes during their formation and internalization (Brzeska et al., 2016, and see below). Myo1D is also found in pinocytic protrusions and cups, and freshly internalized vesicles (Figure 4, C, D, and G). It is localized along the entire cup surface (Figure 5, A and B), and, after cup closure, along the entire vesicle surface (Figure 5, C and D). In contrast to Myo1B, Myo1D is not enriched at the sites of cup closure (Figures 5C, 6 s, and 6D, 0 s). Myo1D is seen at cup edges before closure, although it is not highly enriched, in contrast to Myo1E and Myo1F that are largely absent from this location (compare Myo1D in Figure 5, A and B, with Myo1F in Supplemental Figure S7B, 0 s, and with Myo1F and Myo1E in Brzeska et al., 2016). Myo1D dissociated from macropinocytic vesicles before actin (Figure 5, C, 18 s, and D, 24 s).
FIGURE 6:
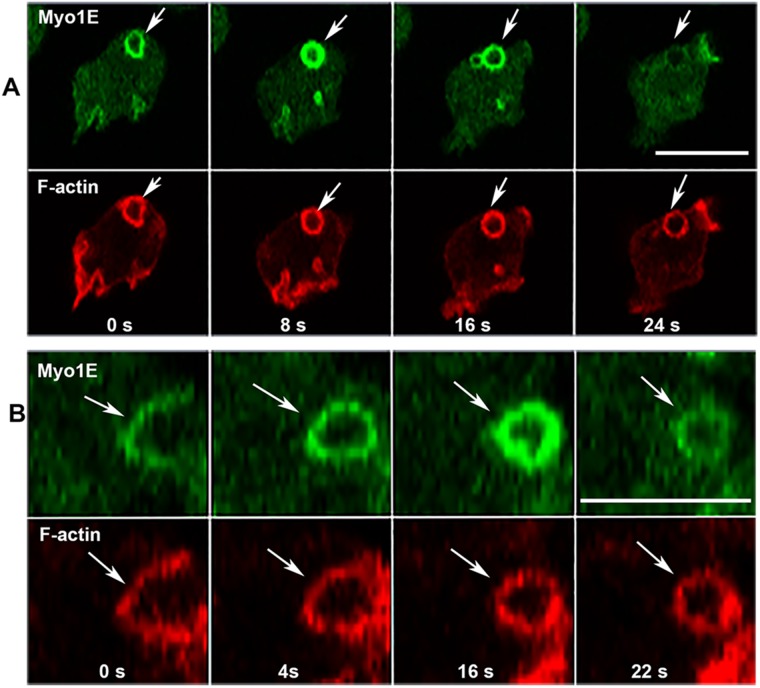
Changes in Myo1E association with macropinocytic vesicles. (A) Myo1E dissociates from vesicle before F-actin. (B) Myo1E association with vesicle intensifies after vesicle internalization (16 s); images show high magnification of a single vesicle. Arrows point to the regions of interest in both A and B. Images are of live AX2 cells. Bars are 10 μm. Two independent Myo1E transfections were done and two independent experiments were performed for each transfection. The images are representative of more than 100 cells observed.
Differential association of Myo1E and Myo1F with macropinocytic vesicles
Myo1E and Myo1F both have PIP3-specific PH domains (Chen et al., 2012) and localize to the plasma membrane of nonmotile cells, to cell–cell contacts and macropinocytic structures, the PIP3-rich region encircled by actin waves, and the engulfing mouth and front of chemotaxing cells (Chen et al., 2012; Brzeska et al., 2016, and see below). The localizations of both Myo1E and Myo1F resemble that of Myo1D shown in Figures 4 and 5 except that Myo1E and Myo1F are excluded from the edges of pinocytic cups (Brzeska et al., 2016). The localizations of Myo1E and Myo1F during the process of micropinocytosis were examined in more detail to determine whether they were identical.
Myo1E has previously been shown to remain associated with pinocytic vesicles for a longer time than Myo1B (Brzeska et al., 2016). We now show that Myo1E dissociates from macropinocytic vesicles before actin (Figure 6, A, 24 s, and B, 22 s), that is, earlier than Myo1F that remains associated with pinocytic vesicles after actin dissociation (Brzeska et al., 2016, and see later in this article). Thus, while the overall localization of Myo1F and Myo1E during macropinocytosis is similar, Myo1F stays associated with internalized vesicle longer than Myo1E. Thus far, this is the only observed difference in the localizations of Myo1E and Myo1F.
PIP3-specific myosin 1s associate strongly with freshly ingested macropinosome
All myosin 1s except Myo1A associated with macropinosomes during their formation and internalization (Brzeska et al., 2012, 2016, and this article). While the levels of actin remained constant during this process, the fluorescence of PIP3-specific Myo1D, Myo1E, and Myo1F increased transiently before dissociating from the early macropinosome (Figure 7; see also Figures 5D, 6 s, and 6B, 16 s). The long-tailed Myo1B is similarly localized to the early macropinosomes and dissociates before F-actin, but its levels do not change during macropinosome internalization (Brzeska et al., 2016).
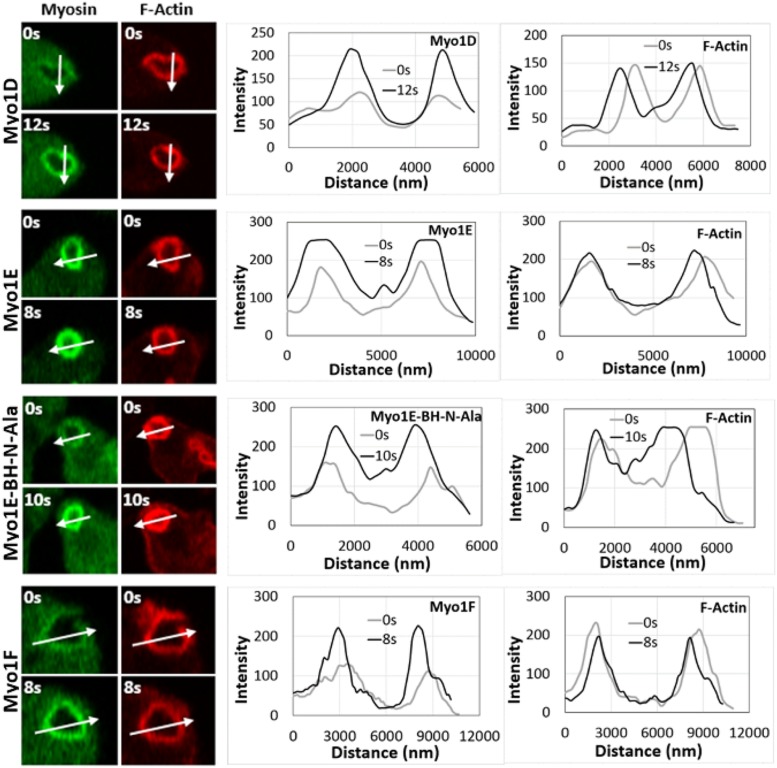
FIGURE 7:
Fluorescence of myosin 1s intensifies after vesicle incorporation, whereas fluorescence of F-actin remains stable. The images of cells taken at indicated time points are shown in the left panels (Myo1D, Myo1E, Myo1E-BH-Ala, and Myo1F, top to bottom). Arrows show the direction of line scans for myosins and F-actin shown in the right panels, and 0 distance is the beginning of the arrow. The images of each Myo1 are representative of more than 100 cells observed in two independent experiments.
Mutation of BH site located in PIP3-specific PH domain of Myo1D abolishes membrane association
BH sites are essential for in vivo membrane association of Myo1B (Brzeska et al., 2012) and Myo1A (Brzeska et al., 2016) and for in vitro lipid binding of Acanthamoeba Myo1C (Brzeska et al., 2008). Peptides corresponding to BH sites at this position in Myo1D, Myo1B, and Myo1C bind to acidic lipid vesicles in vitro and scrambling the peptide sequence does not affect its affinity for lipids (Brzeska et al., 2010). Dictyostelium Myo1D has a single BH site located in the β3/β4 loop of its PIP3-specific PH domain (Figures 1 and 8 and Table 1). Therefore, it seemed reasonable to consider that the BH site of Myo1D contributes to its in vivo membrane association by direct binding to acidic membrane lipids.
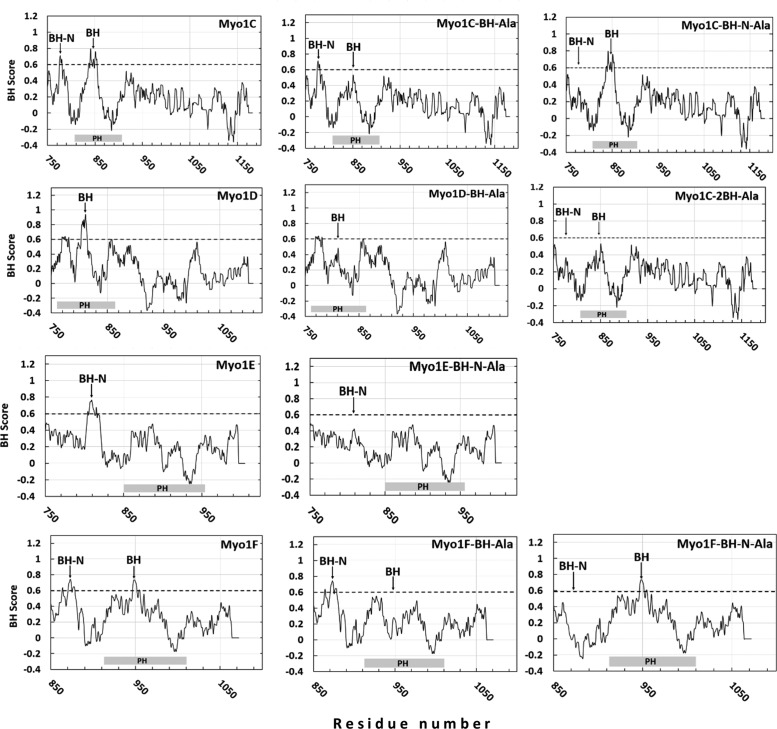
FIGURE 8:
BH plots of the tail regions of Dictyostelium myosin 1s and their mutants. Myosins are shown in the left column and mutants in which the BH sites were mutated are in the center and the right columns. Positions of BH peaks are marked with arrows and positions of PH domains are marked at the bottom of each panel as gray rectangles.
Mutation of Arg and Lys residues in the BH site to Ala, which eliminates its single BH peak (Figure 8 and Table 2), completely abolished the association of Myo1D with the plasma membrane of nonmotile cells, cell–cell contacts, macropinocytic structures (Figure 4, A–D), the PIP3-rich membrane region encircled by actin waves (Figure 4E), and the engulfing mouth of chemotaxing cells (Figure 4G). However, Myo1D-BH-Ala still localized diffusely at the front of chemotaxing cells (Figure 4F), consistent with this localization being due to myosin association with actin by the myosin head, as shown earlier for Myo1B and Myo1A (Brzeska et al., 2012, 2016). These results reveal for the first time the importance of a BH site located within a PIP3-specific domain for membrane-lipid binding.
Role of the N-terminal BH site in myosin 1E membrane association
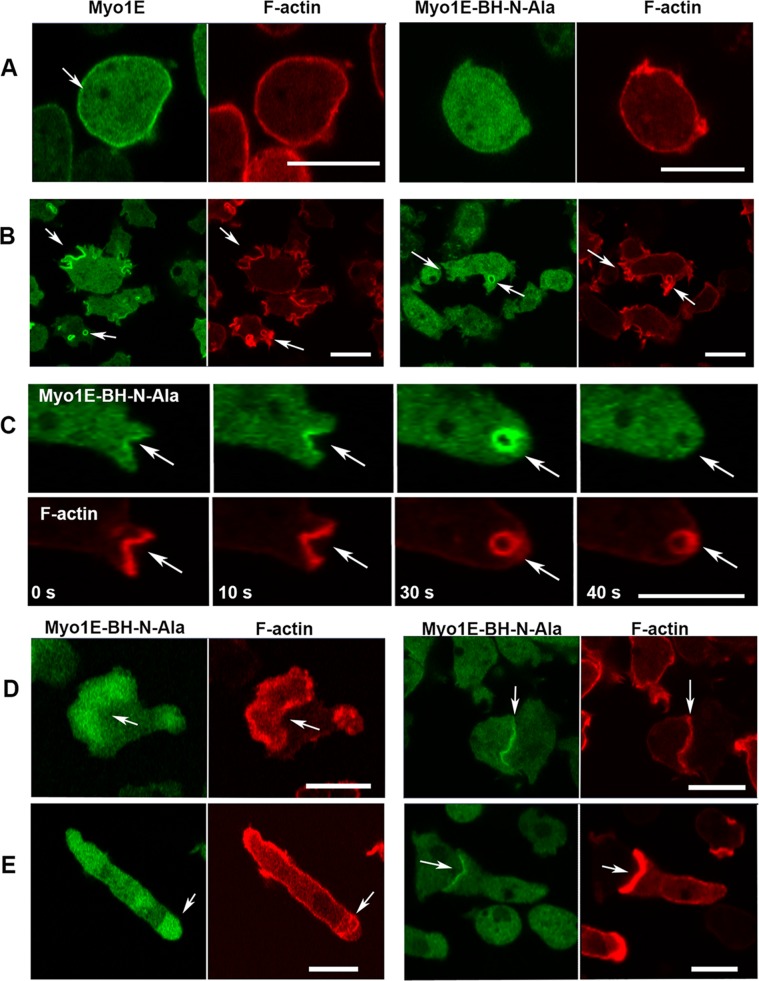
Myo1E is the only Dictyostelium myosin 1 with a single BH site at the N-terminus of its TH1 region and none in its PH domain (Figure 1 and Table 1). Mutation of this BH site (Table 2 and Figure 8) strongly inhibited Myo1E localization to the plasma membrane of nonmotile cells (Figure 9A) but the effect of mutation was not substantial when cells became motile. Myo1E-BH-N-Ala associated with macropinocytic protrusions more weakly than Myo1E (especially at the early stages of starvation, Figure 9B) but it associated strongly with macropinocytic vesicles (Figure 9, B and C) and localized to the region encircled by actin waves, cell–cell contacts of randomly moving cells, and the front and engulfing mouth of chemotaxing cells in a manner indistinguishable from Myo1E (Figure 9, D and E).
FIGURE 9:
Localization of Myo1E-BH-N-Ala. (A, B) Myo1E-BH-N-Ala is shown in right panels and control MIE is shown in left panels. (A) Myo1E-BH-N-Ala does not associate with the plasma membrane of nonmotile cells. (B) Myo1E-BH-N-Ala associates with early protrusions weaker than Myo1E but it associates strongly with vesicles. (C) Time course of vesicle ingestion. Myo1E-BH-N-Ala is not enriched at macropinocytic cup edges (0 s, 10 s), is transiently enriched on ingested vesicle (30 s), and dissociates from vesicles before actin (40 s). (D) Myo1E-BH-N-Ala localizes to the region encircled by actin waves (left panels) and to the cell–cell contacts (right panels). (E) Myo1E-BH-N-Ala localizes diffusely to the front (left panels) and sharply to the engulfing mouth (right panels) of chemotaxing cells. Localization was the same in AX2 cells (A, B) and in Myo1B null cells (C–E). Fixed cells are shown in panel A and live cells are shown in other panels. Arrows point to the regions of interest. Bars are 10 μm. The number of independent transfections was two for Myo1E and three for Myo1E-BH-N-Ala. At least two independent experiments were performed for each transfection under each condition. The images are representative of more than 100 cells observed under each condition.
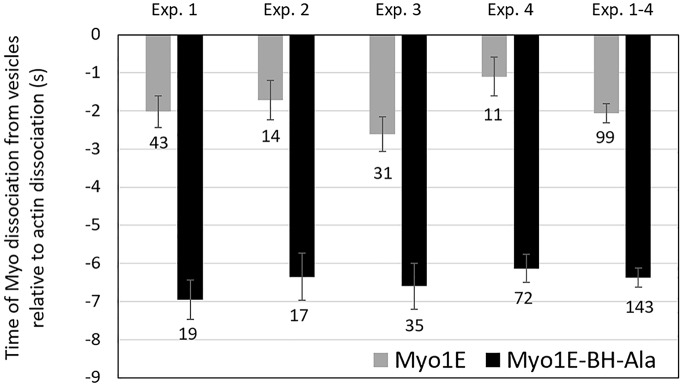
The Myo1E-BH-N-Ala mutant was transiently enriched on pinocytic vesicles after their ingestion (Figures 9C, 30 s, and 7), and dissociated from pinocytic vesicles before actin (Figure 9C, 40 s) as seen for the wild-type Myo1E (Figure 6A). However, the BH-N-Ala mutation decreased the time that Myo1E remained associated with pinocytic vesicles (Figure 10). On average, Myo1E dissociated from vesicles ∼2 s before actin, whereas Myo1E-BH-N-Ala dissociated ∼6 s before actin, indicating that the Ala mutant associated with pinocytic vesicles more weakly than wild-type Myo1E. Thus, the single BH site of Myo1E, located outside the PH domain, contributes to Myo1E localization, especially in nonmotile cells. However, in contrast to Dictyostelium Myo1D, it is not the main determinant of Myo1E intracellular localization.
FIGURE 10:
Time of the dissociation of Myo1E and Myo1E-BH-N-Ala from vesicles relative to the dissociation of F-actin. A time value of −2 s means that myosin dissociated from vesicle 2 s before F-actin. Four independent experiments are shown with SEM marked and the number of cells indicated at the bottom of each bar. The last set of bars is an average of all experiments. Experiments were done in AX2 cells.
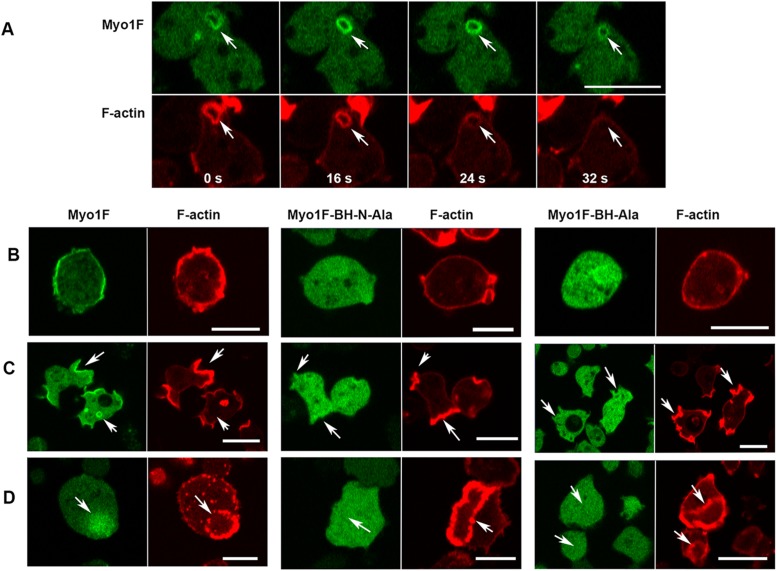
The two BH sites of Myo1F are required for membrane association
Myo1F is the only myosin 1 that stays associated with pinocytic vesicles longer than F-actin (Figure 11A). Myo1F has two BH sites: one located in the β3/β4 loop of its PH domain and a second one outside the PH domain at the N-terminus of TH1 region, in the same region as the sole Myo1E BH site (Figures 1 and 8 and Table 1). The contribution of the two BH sites of Myo1F to its localization was tested by mutating each separately (see Tables 1 and 2 and Figure 8 for mutations). When the BH site in the PH domain was mutated, Myo1F-BH-Ala did not localize to the plasma membranes of nonmotile cells, or to macropinocytic structures, or to regions encircled by actin waves (see Myo1F-BH-Ala in Figure 11, B–D). This effect was very similar to the effect of mutation of the homologous site in Myo1D, emphasizing the importance of the BH site in the PH domain for membrane interactions. Mutation of the BH site at the N-terminus of the TH1 region, Myo1F-BH-N-Ala, also completely abolished membrane association of Myo1F, and its binding to the interior of actin waves (see Myo1F-BH-N-Ala in Figure 11, B–D), a much stronger effect than mutation of the homologous site in Myo1E. These results show that each of the two BH sites of Myo1F make significant contributions to the association of Myo1F with membranes.
FIGURE 11:
Localization of Myo1F and its two BH-Ala mutants. (A) Myo1F localization during vesicle ingestion. MIF is temporarily enriched on freshly ingested vesicles (16 s) and stays on vesicles longer than F-actin (32 s). (B–D) Both the Myo1F-BH-Ala and the Myo1F-BH-N-Ala mutations abolished association of Myo1F with the plasma membrane of nonmotile cells (B), association with macropinocytic structures (C), and localization to the regions encircled by actin waves (D). Experiments were done in Myo1B null cells. Panel B shows fixed cells; other panels show live cells. Arrows point to the regions of interest. Bars are 10 μm. The number of independent transfections was four for Myo1F, and two for Myo1F-BH-Ala and Myo1F-BH-N-Ala. At least two independent experiments were performed for each transfection under each condition. The images are representative of more than 100 cells observed under each condition.
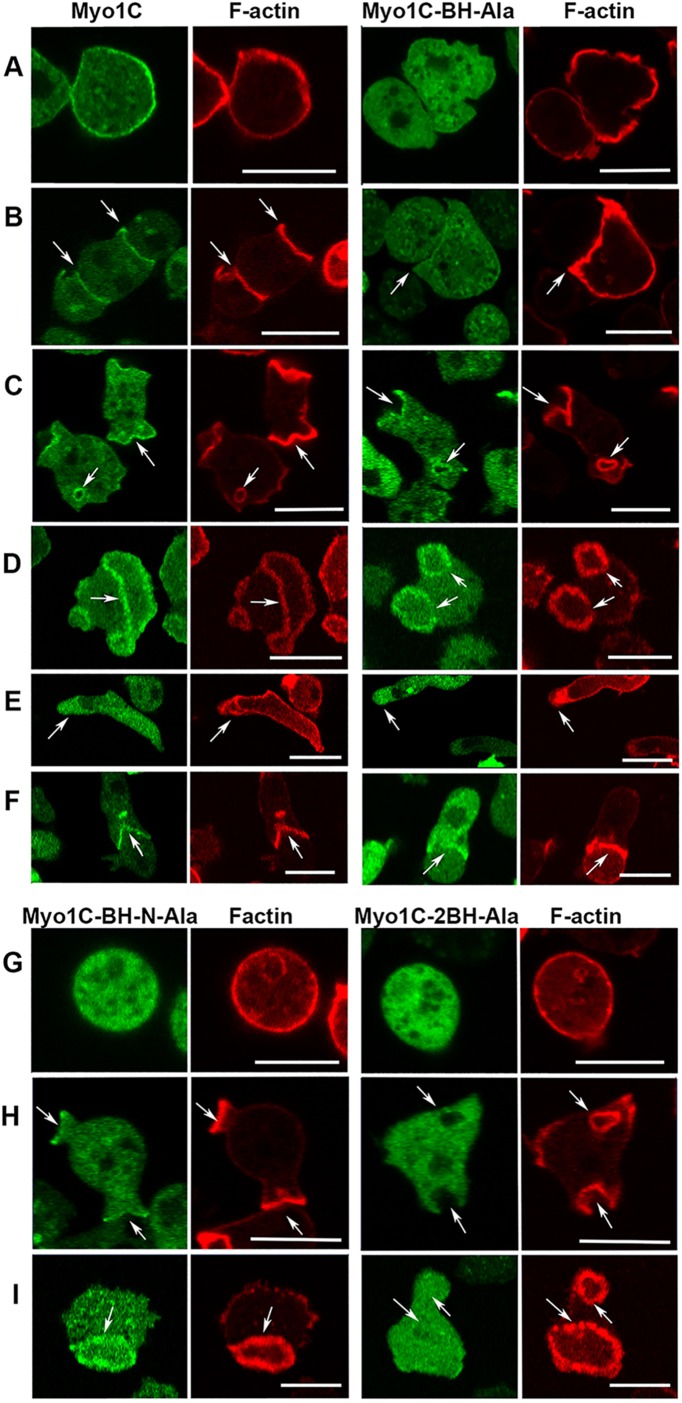
Cellular localization of long-tail Myo1C is very similar to the localization of long-tail Myo1B
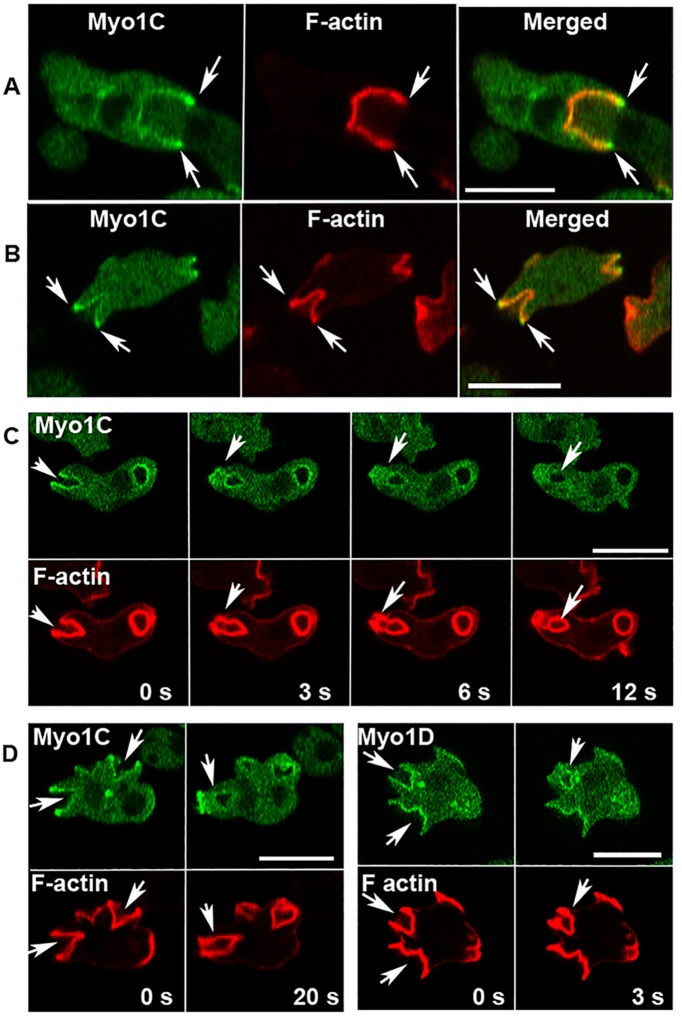
The finding that localization of long-tail Myo1D and Myo1B in actin waves and macropinosomes differs raised the question of whether the third long-tail Dictyostelium myosin 1, Myo1C, which does not display PIP3 specificity (Chen et al., 2012), behaves similarly to Myo1D or to Myo1B. The localization of Myo1C was found to be quite similar to that of Myo1B (Brzeska et al., 2012, 2014, 2016); it is distributed sharply and uniformly along the plasma membrane of nonmotile cells, and tightly localized to cell–cell contact sites, macropinocytic structures of randomly moving cells, and the “mouth” of a cell engulfing another cell during chemotaxis (Figure 12, A–C and F). Like all other Dictyostelium myosin 1s, Myo1C is localized diffusely at the front of chemotaxing cells (Figure 12E). Myo1C is associated with F-actin in actin waves (Figure 12D), similar to Myo1B.
FIGURE 12:
Localization of Myo1C and its BH mutants. (A–F) Localization of Myo1C and Myo1C-BH-Ala. Myo1C localizes sharply on the plasma membrane in nonmotile cells (A), in cell–cell contacts (B), in macropinocytic protrusions and vesicles (C), together with actin in actin waves (D), diffusely at the front of elongated cells (E), and sharply at the mouth of chemotaxing cells (F). Myo1C-BH-Ala is barely detectable in plasma membrane of nonmotile cells (A) and at cell–cell contacts (B) but it is clearly present in macropinocytic protrusions and vesicles (C), in actin waves (D) at the front of chemotaxing cells (E), and it is detectable in the mouth of streaming cells (F). (G, H) Localization of Myo1C-BH-N-Ala and Myo1C-2BH-Ala mutants on the plasma membrane of nonmotile cells (G), in macropinocytic protrusions (H), and in actin waves (I). Note that mutation of both BH sites together (Myo1C-2BH-Ala) abolished membrane association of Myo1C, whereas mutations of single sites had only a partial effect. Arrows point to the regions of interest. Cells in A and G were fixed; all others are images of live cells. The same localization was observed in AX2 and Myo1B null cells. Images shown are of AX2 cells. Bars are 10 μm. The number of independent transfections was eight for Myo1C, four for Myo1C-BH-Ala, and two for Myo1C-BH-N-Ala and Myo1C-2BH-Ala. At least two independent experiments were performed for each transfection under each condition. The images are representative of more than 100 cells observed under each condition.
Myo1C is significantly enriched in the edges of macropinocytic cups (Figure 13, A and B), similar to Myo1B (Brzeska et al., 2016) and in contrast to Myo1D (Figure 13D). Following cup closure, Myo1C remained associated with newly internalized macropinocytic vesicles for a short period of time but dissociated from these vesicles before actin (Figure 13C, 12 s). Myo1C was also diffusely enriched in a “closing patch” after vesicle ingestion (Figure 13C, 3 and 6 s), and often found sharply localized to the plasma membrane at the site of previous vesicle ingestion, which sometimes converted to a new macropinocytic cup (Figure 11D, 20 s, and Supplemental Figure S7A). Interestingly, vesicle enclosure at the plasma membrane site was often first detected by the accumulated fluorescence of GFP-labeled Myo1C followed by enclosure by F-actin (Figure 13C, 3 and 6 s). Similar phenomena were also observed for expressed Myo1F (Supplemental Figure S7B). The localization of expressed Myo1C was the same in wild-type AX2 cells and Myo1B null cells (Figure 13, A and B), in agreement with our earlier data showing that the presence or absence of endogenous myosin 1s did not affect the localization of expressed myosin 1s (Brzeska et al., 2016).
FIGURE 13:
Localization of Myo1C in macropinocytic structures. (A, B) Myo1C is enriched at cup edges in Myo1B null cells (A) and in AX2 cells (B). (C) Myo1C dissociates from the vesicle before actin and Myo1C is diffused in closing patches (12 s). Note that Myo1C fully surrounds the vesicle before it is fully enclosed by F-actin (3 s). (D) Comparison of Myo1C (left panels) and Myo1D (right panels). Myo1C is enriched at the edge of the macropinocytic cup where Myo1D is also present but not enriched (0 s). After vesicle ingestion Myo1C localizes sharply to the site of vesicle closure (20 s). Both myosins seal the vesicle entrance site before actin does (20 s for Myo1C and 3 s for Myo1D). All images are of live cells; Myo1B null cells are shown in row A and AX2 cells in other rows. Bars are 10 μm. Eight Myo1C independent transfections were done and two independent experiments were performed for each transfection. The images are representative of more than 100 cells observed.
Mutation of each of the two BH sites weakens, and mutation of both BH sites abolishes membrane association of Myo1C
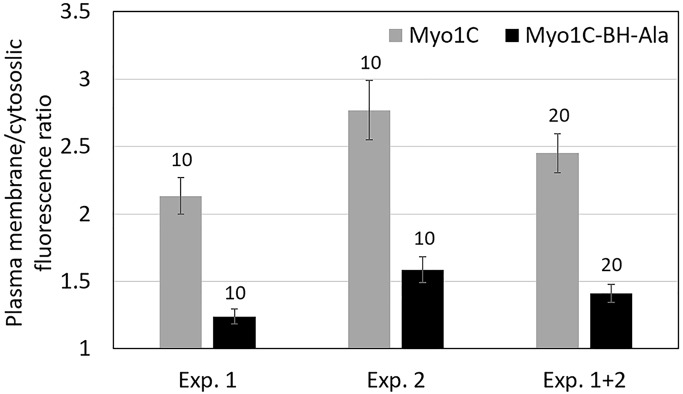
Long-tail Myo1C has two BH sites: one located within the PH domain, like the single BH site of Myo1B and Myo1D, and one at the N-terminus of the TH1 region (Figures 1 and 8 and Table 1). Mutation of the BH site within the PH domain (Myo1C-BH-Ala in Figure 8 and Table 2) significantly weakened, but did not completely abolish, Myo1C association with membranes (Figure 12, A–F), the effect being most pronounced in nonmotile cells (Figure 12A). We quantified the difference by measuring the ratio of the maximum fluorescence intensity on the plasma membrane to the average fluorescence intensity in the cytoplasm of Myo1C and Myo1C-BH-Ala images in two independent experiments (Figure 14). The ratios were 2.45 ± 0.146 SEM for Myo1C and 1.41 ± 0.067 SEM for Myo1C-BH-Ala, clearly demonstrating that the BH-Ala mutation strongly affected plasma membrane association in these cells.
FIGURE 14:
Ratio of the plasma membrane vs. cytoplasmic fluorescence of expressed GFP-labeled Myo1C and Myo1C-BH-Ala. The fluorescence ratio was determined by comparing the maximum fluorescence intensity of the plasma membrane to the average fluorescence intensity in the cytoplasm from line scans of nonmotile cells. The results of two independent experiments are shown. The number of examined cells is on the top of bars and standard errors are marked. Experiments were done in AX2 cells.
The differences between Myo1C and Myo1C-BH-Ala were less pronounced in motile cells. Myo1C-BH-Ala was detected in cell–cell contacts, although the signal was very weak (Figure 10B). The BH-Ala mutant clearly associated with macropinocytic protrusions and vesicles (Figure 12C), although less robustly than the wild-type Myo1C and was detectable in the engulfing mouth of chemotaxing cells (Figure 12F). Myo1C-BH-Ala and Myo1C associated equally well with actin waves (Figure 12D). Myo1C-BH-Ala also localized diffusely at the front of chemotaxing cells (Figure 12E), not surprisingly as diffuse associations with the front had been shown to be independent of the BH site of other myosin 1s (Brzeska et al., 2012, 2016).
The weak effect of the Ala mutation of the BH site in the PH domain on Myo1C membrane association might be explained by the presence of the second BH site N-terminal to the PH domain (BH-N-Ala site in Table 1). We tested the importance of this BH site by mutating it separately or together with the first site (Myo1C-2BH-Ala; Figures 1 and 8 and Tables 1 and 2). The effect of mutating the BH-N site alone (Myo1C-BH-N-Ala in Figures 1 and 8, and Tables 1 and 2) was similar to the effect of mutating only the BH site within the PH domain abolishing association of Myo1C with the plasma membrane of nonmotile cells but not its association with macropinocytic protrusions and actin waves (Figure 12, G–I). However, when both BH sites were mutated (Myo1C-2BH-Ala) association with membranes was completely abolished in nonmotile cells, macropinocytic protrusions, and in actin waves (Figure 10, G–I); that is, both BH sites contribute to Myo1C membrane targeting, whereas either BH site may be sufficient to localize the Myo1C to actin waves, presumably due to the presence of the actin-binding GPQ domain.
A stronger BH site increases Myo1B membrane association but not its localization
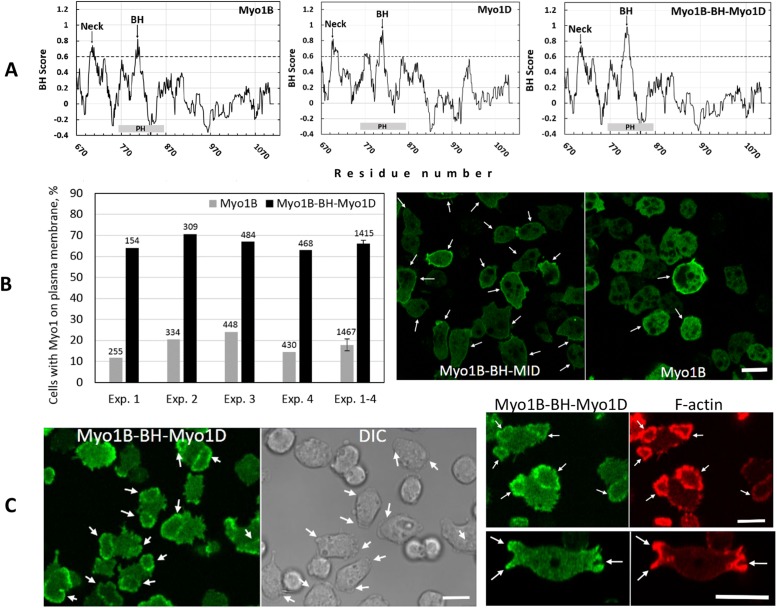
Our earlier in vitro data showed that BH sites bind membrane lipids by nonspecific interactions whose overall strength is proportional to the positive charge density of the BH site (Brzeska et al., 2008) and independent of its sequence (Brzeska et al., 2010). We replaced the BH site of non–PIP3-specific Myo1B with the stronger BH site of PIP3-specific Myo1D (Figure 15A). The BH peak of the resulting Myo1B-BH-Myo1D mutant is significantly stronger than that of Myo1B (Figure 15A). It is also stronger than the BH peak of Myo1D because amino acids on both sides of a BH site affect the BH peak size. We compared the plasma membrane association of Myo1B and of the Myo1B-BH-Myo1D mutant in cells after starvation for 10 min. Myo1B-BH-Myo1D was localized uniformly to the plasma membrane in 66% of cells but Myo1B in only 18% (Figure 15B), demonstrating that strengthening the BH site of Myo1B causes stronger membrane association in vivo consistent with our in vitro data that lipid binding by BH sites depends on their amino acid composition rather than their sequence (Brzeska et al., 2010). However, Myo1B-BH-Myo1D mutant colocalized with actin in actin waves and was enriched at the macropinocytic cup edges (Figure 15C) similarly to Myo1B and not like Myo1D. This demonstrates that the BH site of Myo1D does not carry any lipid specificity by itself.
FIGURE 15:
Localization of Myo1B-BH-Myo1D mutant. The BH site in Myo1B was replaced by the BH site of Myo1D. (A) BH plots of Myo1B, Myo1D, and Myo1B-BH-Myo1D mutant. The BH site of Myo1D is stronger than that of Myo1B, and the BH site of Myo1B-BH-Myo1D is stronger than both. (B) Left panel, Percentage of cells with wild-type or mutant Myo1 localized on plasma membrane in nonmotile cells. Four independent experiments are shown with the number of cells scored in each experiment listed at the top of bars. The last set of bars is an average of the four experiments with standard error marked. Right panel, Examples of scored cells. Cells were starved for 10 min and fixed. The arrows point to cells with Myo1 on plasma membrane and the bar is 10 μM. (C) Localization of Myo1B-BH-Myo1D mutant in actin and at the edges of pinocytic cups. The left panel shows a field of cells making waves. The right top panel shows colocalization of Myo1B-BH-MID with F-actin in waves. The right bottom panel shows the localization of MIB-BH-Ala in the edges of macropinocytic cups. Images are of live cells. Arrows point to waves and cups. Experiments were done in Myo1B null cells. Bars are 10 μm. Three independent transfections of Myo1B-BH-Myo1D were done and two independent experiments were performed for each transfection. The images are representative of more than 100 cells observed for each condition.
DISCUSSION
Cells typically express more than one myosin 1 with both shared and unique functions (McConnell and Tyska, 2010; McIntosh and Ostap, 2016).The localization of a particular myosin 1 dictates its cellular role and this is largely determined by membrane-binding sequences in the common TH1 domain. Comparison of the localizations of all of Dictyostelium myosin 1s and identification of sequences critical for the membrane targeting (Tables 3 and 4) now provides a more complete understanding of the distribution of myosin I isoforms in a single cell and insight into the molecular determinants that tune their intracellular localizations.
TABLE 3:
Summary of in vivo localization of Dictyostelium myosin 1s.
| Localization | Myosin 1s present | Myosin 1s absent |
|---|---|---|
| Plasma membrane of nonmotile cells | Myo1A, Myo1B, Myo1C, Myo1D, Myo1E, Myo1F | None |
| Cell–cell contacts of randomly moving and chemotaxing cells | Myo1A, Myo1B, Myo1C, Myo1D, Myo1E, Myo1F | None |
| Front of polarized cells | Myo1A, Myo1B, Myo1C, Myo1D, Myo1E, Myo1F | None |
| Actin waves | Myo1B, Myo1C | Myo1A, Myo1D, Myo1E, Myo1F |
| Region encircled by actin wave | Myo1D, Myo1E, Myo1F | Myo1A, Myo1B, Myo1C |
| Macropinocytic structures (protrusions, cups, and vesicles) | Myo1B, Myo1C, Myo1D, Myo1E, Myo1F | Myo1A |
| Enriched at cup edges | Myo1B, Myo1C | Myo1A, Myo1D, Myo1E, Myo1F |
| Dissociates from vesicles before actin | Myo1B, Myo1C, Myo1D, Myo1E | Myo1F |
| Dissociates from vesicles after actin | Myo1F | Myo1B, Myo1C, Myo1D, Myo1E, |
| Transiently enriched at vesicles | Myo1D, Myo1E, Myo1F | Myo1A, Myo1B, Myo1C |
The sources of the data in the table are as follows: MIA, Brzeska et al., 2016; MIB, Brzeska et al., 2012, 2014, 2016; MIC, this article; MID, this article; MIE, Brzeska et al., 2016, and this article; MIF, Brzeska et al., 2016, and this article.
TABLE 4:
Localization of BH sites in Dictyostelium myosin 1s and effect of their mutation on membrane association.
| Myosin | Tail length | Mutated BH site | BH site location within TH1 domain | Mutation of BH site affects membrane association |
|---|---|---|---|---|
| Myo1A | Short | BH | β3/β4 | + |
| Myo1B | Long | BH | β3/β4 | + |
| Myo1C | Long | BH | β3/β4 | +/− |
| BH-N | β3/β4 | +/− | ||
| BH and BH-N | β3/β4 and α4/α5 | + | ||
| Myo1D | Long | BH | β3/β4 | + |
| Myo1E | Short | BH-N | α4/α5 | +/− |
| Myo1F | Short | BH | β3/β4 | + |
| BH-N | α4/α5 | + |
The sources of the data on the effect of BH sites mutations are as follows: MIA, Brzeska et al., 2016; MIB, Brzeska et al., 2012, 2014, 2016; MIC, this article; MID, this article; MIE, this article; MIF, this article.
Differential localizations of Dictyostelium myosin 1s
All six Dictyostelium myosin 1s localize sharply to the plasma membrane of nonmotile cells and diffusely at the front of chemotaxing cells (Table 3). The tails of the myosins are responsible for the former and the heads for the latter. However, the dynamic localizations of the six myosin 1s in actin waves and macropinocytic structures are different, most strikingly for actin waves (Table 3).
Myo1B, Myo1C, Myo1D, Myo1E, and Myo1F, but not Myo1A, associate with macropinocytic structures (protrusions, cups, and vesicles), consistent with their involvement in endocytosis (Novak et al., 1995; Neuhaus and Soldati, 2000; Durrwang et al., 2006; Chen et al., 2012). The overall pattern of myosin 1s localizations suggests that the isoforms are likely to have different roles in macropinocytosis. Myo1B and Myo1C are localized to the leading edges of pinocytic cups (Table 3) and concentrated at the site of membrane sealing on the newly internalized vesicle indicating that these myosins may be involved in the closure of macropinocytic cups. Myo1B, Myo1C, and F-actin levels remain constant on the forming macropinosome; however, shortly after vesicle ingestion, there is a transient enhancement of Myo1D, Myo1E, and Myo1F on the vesicle membrane. This likely reflects an enhancement of PIP3, to which Myo1D, Myo1E, and Myo1F bind specifically. The myosin 1s associate with pinocytic vesicles for different lengths of time. Myo1B, Myo1C, and Myo1D first dissociate from the newly formed macropinosome, followed by Myo1E, then F-actin. Myo1F remains on these endocytic vesicles after actin dissociation (Brzeska et al., 2016, and this article) and is more likely to be involved in later stages of macropinocytosis.
The dynamic localization of myosin 1s differs prominently in actin waves (Table 3).
Actin waves have been compared with large macropinocytic cups (Bretschneider et al., 2009; Gerisch, 2010), the common features being PIP2 and actin in the periphery, and PIP3 enrichment in the center (Gerisch et al., 2012; Veltman et al., 2016). The PIP3-specific short-tail Myo1E and Myo1F, and long-tail Myo1D, localize to the membrane region encircled by the actin wave (see Table 3), consistent with the presence of PIP3 in these structures. These three myosins may contribute to stabilizing PIP3-rich patches. In contrast, Myo1B and Myo1C localize to regions of the waves containing PIP2 and enriched in actin. Myo1B and Myo1C fully colocalize with dynamic actin in the wave, and both the lipid-binding BH site and the actin-binding GPQ region are required for this localization (Brzeska et al., 2014). Myo1B and Myo1C may be involved in the recruitment of actin to these waves. It is of interest to note that Myo1D, that has both PIP3-binding specificity and actin-binding GPQ domain in the tail, is predominantly associated with PIP3-containing membranes, indicative of a higher affinity for PIP3 than for actin. Thus, the ability to bind PIP3 directs a subset of myosin 1s to the membrane encircled by the actin wave while myosin 1s that have the GPQ region and do not show PIP3 specificity target the actin wave itself. Interestingly, myosin 1s that colocalize with actin in actin waves also contain an SH3 domain that enables CARMIL-mediated interaction between myosin 1 and Arp 2/3 (Jung et al., 2001; Bretschneider et al., 2009).
BH sites and membrane binding of Dictyostelium myosin 1s
All of the Dictyostelium myosin 1s have a BH site and a putative PH domain in the membrane-binding TH1 domain of their tails. Interestingly, the BH site of five myosin 1s, Myo1A, Myo1B, Myo1C, Myo1D, and Myo1F, is contained within their PH domains. These BH sites are located near the β3/β4 loop of the PH domain that in our models of the PH domains of Dictyostelium myosin 1s, as well as in a crystal structure of human Myo1C (Lu et al., 2015), protrude from the PH domain, making them perfect candidates for intramolecular interactions. The number and the location of BH sites can vary. The sole BH site of Myo1E and the second BH sites of Myo1C and Myo1F lie outside the canonical PH domain but near the α4/α5 loop of the extended PH domain (Lu et al., 2015) at the N-terminus of the TH1 region.
Elimination of each of the tail-located BH sites abolished or weakened membrane association of all Dictyostelium myosin 1s (Table 4). The basic amino acid/Ala mutations of the BH sites in the PH domains abolished all membrane binding of PIP3-specific Myo1D and Myo1F (this article) and of non–PIP3-specific Myo1A and Myo1B (Brzeska et al., 2012, 2016) while weakening membrane binding of non–PIP3-specific Myo1C (this article). Mutation of the N-terminal BH sites of Myo1E, Myo1C, and Myo1F that are within the basic TH1 tail segment abolished the association of Myo1F with membranes and weakened the plasma membrane association of Myo1E and Myo1C. The greater effect of mutation of the BH sites of Myo1C and Myo1E in nonmotile cells suggests that the plasma membrane of these cells may have a lower negative charge density than specific regions of the plasma membrane in motile cells.
BH sites are defined by their amino acid composition and lack a consensus sequence homology (Brzeska et al., 2010). Therefore the BH sites themselves are unlikely to provide a conserved steric specificity required for selective binding to certain phospholipids. Indeed, replacing the BH site of non–PIP3-specific Myo1B with the BH site of PIP3-specific Myo1D resulted in a mutant that was not PIP3 specific and localized like the parent Myo1B.
Although increasing the size of the single BH peak in Myo1B strengthened its membrane association, there are factors other than the BH site that may affect myosin 1s localizations and membrane associations. These may include a functional PH domain, more than one lipid-binding site, different degrees of BH site exposure, and the interaction of the myosin with other proteins. For example, we know that the heads of Myo1B (Brzeska et al., 2012) and Myo1A (Brzeska et al., 2016) are required for the relocation of myosin from the plasma membrane of nonmotile cells to motile protrusions and the front of polarized cells. However, the fact that the Myo1B-BH-Myo1D mutant associates with the same membrane sites as Myo1B argues against the BH site being involved in any sequence-specific inter- or intramolecular protein interactions.
BH sites and PH domains in other proteins
BH sites are present within and outside the PH domains of several myosin 1s, other than those in Dictyostelium, as well as in other lipid-binding proteins (Brzeska et al., 2010), but their roles in membrane binding have not been systematically studied. For example, 67 of 77 Dictyostelium proteins containing PH domains listed in Park et al. (2008) have BH peaks located within their PH domains.
BH sites might provide initial nonspecific binding to membranes required for PIP3-specific binding by other residues within the PH domain. Such a nonspecific interaction is required for Myo1D binding to PIP3-enriched regions in vivo. Conversely, because the interaction of BH sites with membranes is proportional to the negative charge density of the membrane, PIP2/PIP3-enriched regions would enhance BH binding because of their higher charge.
To our knowledge there are only two reports describing mutations of BH sites in mammalian myosin 1s. Mutations of each of the two BH sites of mammalian Myo1A abolished its targeting to microvilli (Mazerik and Tyska, 2012), suggesting that the role of the BH sites in mammalian Myo1A is similar to their role in Dictyostelium myosin 1s. Interestingly, the BH sites of mammalian Myo1A are located outside the PH domain and mutation of conserved residues of the PH domain did not eliminate targeting of mammalian Myo1A. The mutation of the BH site located within the PH domain of mammalian Myo1A has not been tested. Mutation of residues within the BH site of mammalian Myo1E did not abolish lipid binding (Feeser et al., 2010) but this mutation also did not abolish the BH peak so this result is not conclusive and further studies on the role of the Myo1E BH site are needed.
In general, mutations of conserved residues within the PH domains of myosin 1s have given mixed results, abolishing membrane association in some cases but not in others. Mutation of a conserved residue within the PH domain of Acanthamoeba Myo1C had very little effect on its non–PIP3-specific lipid binding driven by the BH site (Brzeska et al., 2008). In contrast, mutation of conserved residues in the PH domain of Dictyostelium Myo1E, that does not have a BH site in its PH domain, caused the loss of lipid binding (Chen et al., 2012), while mutation of the BH site located N-terminal to the PH domain weakened membrane association (this article). Mutation of conserved residues abolished lipid binding of mammalian Myo1B (Komaba and Coluccio, 2010) and Myo1F (Chen et al., 2012) but had no effect on Myo1E (Feeser et al., 2010). The mutation of conserved residues within the PH domain also abolished lipid binding for mammalian Myo1C (Hokanson et al., 2006), but this myosin may interact with lipids by nonspecific ionic interactions (McKenna and Ostap, 2009). Similarly, mutation of a conserved residue within the PH domain abolished membrane association of mammalian Myo1G, but this myosin also engages regions other than the PH domain in lipid binding (Patino-Lopez et al., 2010). Taken together, these results suggest that nonspecific ionic interactions may play a role in the association with membranes of other myosin 1s with functional PH domains, similarly as shown for Dictyostelium myosin 1s.
It is important to keep in mind, however, that the BH search identifies only potential lipid-binding sites. The basic-hydrophobic composition of BH sites also makes them good candidates for inter- or intramolecular interactions with acidic-hydrophobic regions of other proteins, and only some of the BH sites that have been tested are involved in lipid binding (Lanier et al., 2016). Additionally, many lipid-binding proteins do not have clearly defined BH peaks.
The BH search identified known lipid-binding sites in proteins other than myosin 1s (Brzeska et al., 2010) and previously unknown lipid-binding sites in varied proteins including kinases (Barbosa et al., 2016; Lee et al., 2018; Gerganova et al., 2019), phosphatases (Itoh et al., 2014), ion channels (Bernier et al., 2013; Giblin et al., 2019), CARMIL (Lanier et al., 2016), and proteins of PAR polarity complex (Bailey and Prehoda, 2015; Ramanujam et al., 2018). In addition, the basic region located within the β3/β4 loop of the PH domain of the metastatic factor, P-Rex-1, was recently found to serve as a nonspecific membrane localization element (Cash et al., 2016, 2019) and this region corresponds to a strong BH peak in the BH profile of P-Rex-1 (unpublished data). BH sites may prove to be good mutagenic targets for some other lipid-binding proteins as well as for other than Dictyostelium myosin 1s because elimination of BH site(s) abolishes lipid binding independently of the myosin 1s lipid specificity. This may be especially useful for proteins for which mutation of conserved residues in putative PH domains has no effect.
MATERIALS AND METHODS
Cell lines, cell culturing, and cell imaging conditions
Cells were grown, transfected, and prepared for microscopy as described earlier (Brzeska et al., 2014). Briefly, GFP-Myo1 expression plasmid and the RFP-LifeAct plasmid were cotransfected into AX2 or Myo1Bnull cells (Brzeska et al., 2014) by electroporation (Gaudet et al., 2007). We have shown earlier that the localizations of expressed myosin 1s are not affected by the presence or absence of other endogenous myosin 1 isoforms (Brzeska et al., 2016). Cells were grown in 10-cm Petri dishes in HL5 media (Formedium; HLG0101; Sussman, 1987) in the presence of appropriate selective antibiotics at the following concentrations: G418 sulfate (Mediatech) at 12 μg/ml for expressing myosin 1s and their mutants, and hygromycin (Invitrogen) at 50 μg/ml for expressing LifeAct (Riedl et al., 2008). Myo1B null cells were grown in the presence of blasticidin S HCl (Invitrogen) at 7 μg/ml. For microscopy, cells were transferred to chambered cover glass (Nalge Nunc International; 1555383) and allowed to attach for 30 min. HL5 media was then exchanged for starvation solution (20 mM phosphate buffer, pH 6.2, 0.2 mM CaCl2, 2 mM MgCl2). For monitoring of nonmotile cells, attachment time was shortened to 20 min and cells were fixed immediately after solution exchange. Random movement and macropinocytosis were observed in live cells between 0.5 and 4 h of starvation and chemotaxis after overnight incubation at 4°C. Actin waves were induced by adding 1 μM latrunculin (latrunculin A; Sigma) into the cells that were starved for 30 min. Waves persisted for at least 3 h in the presence of latrunculin and were recorded in live cells.
Image recording and processing
Cells were viewed with a Zeiss 780 confocal microscope with a 63× plan Achromat 1.4 objective. Image recording and profile scanning were done using Zeiss Zen software and final illustrations were prepared using Zeiss Zen software, Photoshop, and Excel.
Protein sequence alignments and modeling
Sequences of myosin 1s were aligned using MUSCLE (multiple sequence comparison by log-expectation) (Edgar, 2004) available on the PIR web site (https://pir.georgetown.edu/pirwww/search/multialn.shtml) or the EMBL-EBI web site (www.ebi.ac.uk/Tools/msa/muscle/), the latter with applying Person/Fasta parameters. The positions of BH sites were determined using BH search (Brzeska et al., 2010; https://hpcwebapps.cit.nih.gov/bhsearch/). The 3D modeling was done using the PHYRE2 Protein Fold Recognition Server (Kelley et al., 2015; www.sbg.bio.ic.ac.uk/phyre2/html/). The locations of other myosin 1 domains were according to Cymobase (Odronitz and Kollmar, 2006, 2007; Kollmar and Muhlhausen, 2017; www.cymobase.org/cymobase).
Amino acid sequences of all myosin 1s were obtained from Cymobase and their accession names are listed in Figure 2 and Supplemental Figure S3.
DNA constructs
The myosins and their mutants used in this study are shown in Table 1. Base clones for each myosin 1 gene (myoC, myoE, myoD, and myoF ) were generated by TA cloning (Strataclone system; Agilent) of a full-length genomic fragment generated by PCR using AX2 genomic DNA as a template. Q5 mutagenesis (New England Biolabs) was then used to introduce mutations into the region of each gene encoding one or both of the BH regions (see Supplemental Table S1). The sequence of all PCR-generated clones was verified (University of Minnesota BioMedical Genomics Center). The wild-type or mutant full-length genes were then restriction enzyme cloned into a low copy number origin of replication extrachromosomal expression plasmid carrying a G418 resistance cassette, pTX-GFP (Levi et al., 2000). The resulting plasmid encoded each of the myosin 1s with GFP fused at its N-terminus. The plasmid carrying RFP-LifeAct and hygromycin resistance was described earlier (Riedl et al., 2008; Veltman et al., 2009; Brzeska et al., 2014).
Immunoblotting
Cells from each strain were collected and dissolved in 100 µl of urea-containing Laemmli sample buffer (Novak et al., 1995) to a final concentration of (3–4) × 107 cells/ml and run on an 8–16% TGX separating gel (Bio-Rad Laboratories), transferred to nitrocellulose membrane (LI-COR Biosciences), and then probed simultaneously with anti-GFP (BioLegend; catalogue #902601, clone B4; used at 1:4000) and anti-Dictyostelium actin (Westphal et al., 1997; used at 1:1000) monoclonal antibodies. The blots were incubated in goat anti-mouse Alexa 800 (LI-COR Biosciences; catalogue #926-32210; used at 1:4,000) and then scanned using an Odyssey infrared imaging system (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We acknowledge the support of the Light Microscopy Core of the Intramural Research Program of the National Heart, Lung, and Blood Institute. We thank Livia Songster for help with transformations and the initial screen of expressed proteins and Michael Bagnoli for help with cell culturing and data processing. This work was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute and by National Institutes of Health Grant no. R01GM-122917 to M.A.T. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- BH
basic-hydrophobic site
- PH
pleckstrin homology domain
- PIP2
PI(4,5)P2 phosphatidylinositol 4,5-bisphosphate
- PIP3
PI(3,4,5)P3 phosphatidylinositol 3,4,5-triphosphate
- SH3
src homology domain 3
- TH1
tail homology-1 domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-08-0475) on November 27, 2019.
REFERENCES
- Bailey MJ, Prehoda KE. (2015). Establishment of Par-polarized cortical domains via phosphoregulated membrane motifs. Dev Cell , 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa IC, Shikata H, Zourelidou M, Heilmann M, Heilmann I, Schwechheimer C. (2016). Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development , 4687–4700. [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. (2001). A millennial myosin census. Mol Biol Cell , 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier LP, Ase AR, Seguela P. (2013). Post-translational regulation of P2X receptor channels: modulation by phospholipids. Front Cell Neurosci , 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T, Anderson K, Ecke M, Muller-Taubenberger A, Schroth-Diez B, Ishikawa-Ankerhold HC, Gerisch G. (2009). The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys J , 2888–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Guag J, Preston GM, Titus MA, Korn ED. (2012). Molecular basis of dynamic relocalization of Dictyostelium myosin IB. J Biol Chem , 14923–14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Guag J, Remmert K, Chacko S, Korn ED. (2010). An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J Biol Chem , 5738–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Hwang KJ, Korn ED. (2008). Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J Biol Chem , 32014–32023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Koech H, Pridham KJ, Korn ED, Titus MA. (2016). Selective localization of myosin-I proteins in macropinosomes and actin waves. Cytoskeleton (Hoboken) , 68–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Pridham K, Chery G, Titus MA, Korn ED. (2014). The association of myosin IB with actin waves in dictyostelium requires both the plasma membrane-binding site and actin-binding region in the myosin tail. PLoS One , e94306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, D’Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I. (2018). Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature , 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash JN, Davis EM, Tesmer JJG. (2016). Structural and biochemical characterization of the catalytic core of the metastatic factor P-Rex1 and its regulation by PtdIns(3,4,5)P3. Structure , 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash JN, Sharma PV, Tesmer JJG. (2019). Structural and biochemical characterization of the plecstrin homology domain of the RhoGEF P-Rex2 and its regulation by PIP3. J Struct Biol X , 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Wang Y, Sesaki H, Iijima M. (2012). Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci Signal , ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Liburd J, Shaw K, Jung Y, Smith SP, Cote GP. (2011). Identification of calmodulin and MlcC as light chains for Dictyostelium myosin-I isozymes. Biochemistry , 6579–6588. [DOI] [PubMed] [Google Scholar]
- Durrwang U, Fujita-Becker S, Erent M, Kull FJ, Tsiavaliaris G, Geeves MA, Manstein DJ. (2006). Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J Cell Sci , 550–558. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res , 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk DL, Wessels D, Jenkins L, Pham T, Kuhl S, Titus MA, Soll DR. (2003). Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J Cell Sci , 3985–3999. [DOI] [PubMed] [Google Scholar]
- Feeser EA, Ignacio CM, Krendel M, Ostap EM. (2010). Myo1e binds anionic phospholipids with high affinity. Biochemistry , 9353–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, He L, Li Y, Xiao F, Hu G. (2018). Modeling of PH domains and phosphoinositides interactions and beyond. Adv Exp Med Biol, 10.1007/5584_2018_236. [DOI] [PubMed] [Google Scholar]
- Gaudet P, Pilcher KE, Fey P, Chisholm RL. (2007). Transformation of Dictyosteliumdiscoideum with plasmid DNA. Nat Protoc , 1317–1324. [DOI] [PubMed] [Google Scholar]
- Gerganova V, Floderer C, Archetti A, Michon L, Carlini L, Reichler T, Manley S, Martin SG. (2019). Multi-phosphorylation reaction and clustering tune Pom1 gradient mid-cell levels according to cell size. Elife , 10.7554/eLife.45983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch G. (2010). Self-organizing actin waves that simulate phagocytic cup structures. PMC Biophys , 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch G, Schroth-Diez B, Muller-Taubenberger A, Ecke M. (2012). PIP3 waves and PTEN dynamics in the emergence of cell polarity. Biophys J , 1170–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giblin JP, Etayo I, Castellanos A, Andres-Bilbe A, Gasull X. (2019). Anionic phospholipids bind to and modulate the activity of human TRESK background K(+) channel. Mol Neurobiol , 2524–2541. [DOI] [PubMed] [Google Scholar]
- Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. (2006). Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell , 4856–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Ostap EM. (2006). Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc Natl Acad Sci USA , 3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh A, Uchiyama A, Taniguchi S, Sagara J. (2014). Phactr3/scapinin, a member of protein phosphatase 1 and actin regulator (phactr) family, interacts with the plasma membrane via basic and hydrophobic residues in the N-terminus. PLoS One , e113289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan T, Geminard C, Coutelis JB, Cerezo D, Poles S, Noselli S, Furthauer M. (2018). Myosin1D is an evolutionarily conserved regulator of animal left-right asymmetry. Nat Commun , 1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA., 3rd (2001). The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol , 1479–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Wu X, Hammer JA., 3rd (1996). Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol , 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc , 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelberger N, Breunig M, Martin R, Knolker HJ, Miklavc P. (2016). The role of myosin 1c and myosin 1b in surfactant exocytosis. J Cell Sci , 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M, Muhlhausen S. (2017). Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol Biol , 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaba S, Coluccio LM. (2010). Localization of myosin 1b to actin protrusions requires phosphoinositide binding. J Biol Chem , 27686–27693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier MH, McConnell P, Cooper JA. (2016). Cell migration and invadopodia formation require a membrane-binding domain of CARMIL2. J Biol Chem , 1076–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Weber ZT, Zourelidou M, Hofmeister BT, Schmitz RJ, Schwechheimer C, Dobritsa AA. (2018). Arabidopsis protein kinase D6PKL3 is involved in the formation of distinct plasma membrane aperture domains on the pollen surface. Plant Cell , 2038–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS. (2002). Pleckstrin homology domains and the cytoskeleton. FEBS Lett , 71–76. [DOI] [PubMed] [Google Scholar]
- Levi S, Polyakov M, Egelhoff TT. (2000). Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid , 231–238. [DOI] [PubMed] [Google Scholar]
- Lu Q, Li J, Ye F, Zhang M. (2015). Structure of myosin-1c tail bound to calmodulin provides insights into calcium-mediated conformational coupling. Nat Struct Mol Biol , 81–88. [DOI] [PubMed] [Google Scholar]
- Maxeiner S, Shi N, Schalla C, Aydin G, Hoss M, Vogel S, Zenke M, Sechi AS. (2015). Crucial role for the LSP1-myosin1e bimolecular complex in the regulation of Fcγ receptor-driven phagocytosis. Mol Biol Cell , 1652–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazerik JN, Tyska MJ. (2012). Myosin-1A targets to microvilli using multiple membrane binding motifs in the tail homology 1 (TH1) domain. J Biol Chem , 13104–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. (2010). Leveraging the membrane-cytoskeleton interface with myosin-1. Trends Cell Biol , 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh BB, Ostap EM. (2016). Myosin-I molecular motors at a glance. J Cell Sci , 2689–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh BB, Pyrpassopoulos S, Holzbaur ELF, Ostap EM. (2018). Opposing kinesin and myosin-I motors drive membrane deformation and tubulation along engineered cytoskeletal networks. Curr Biol , 236–48.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna JM, Ostap EM. (2009). Kinetics of the interaction of myo1c with phosphoinositides. J Biol Chem , 28650–28659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus EM, Soldati T. (2000). A myosin I is involved in membrane recycling from early endosomes. J Cell Biol , 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevzorov I, Sidorenko E, Wang W, Zhao H, Vartiainen MK. (2018). Myosin-1C uses a novel phosphoinositide-dependent pathway for nuclear localization. EMBO Rep , 290–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Peterson MD, Reedy MC, Titus MA. (1995). Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J Cell Biol , 1205–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. (2006). Pfarao: a web application for protein family analysis customized for cytoskeletal and motor proteins (CyMoBase). BMC Genomics , 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. (2007). Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol , R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap EM, Pollard TD. (1996). Overlapping functions of myosin-I isoforms? J Cell Biol , 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouderkirk-Pecone JL, Goreczny GJ, Chase SE, Tatum AH, Turner CE, Krendel M. (2016). Myosin 1e promotes breast cancer malignancy by enhancing tumor cell proliferation and stimulating tumor cell de-differentiation. Oncotarget , 46419–46432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WS, Heo WD, Whalen JH, O’Rourke NA, Bryan HM, Meyer T, Teruel MN. (2008). Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell , 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino-Lopez G, Aravind L, Dong XY, Kruhlak MJ, Ostap EM, Shaw S. (2010). Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent Pleckstrin homology (PH) domain (Myo1PH). J Biol Chem , 8675–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Doberstein SK, Zot HG. (1991). Myosin-I. Annu Rev Physiol , 653–681. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Korn ED. (1973). Acanthamoeba myosin. I. Isolation from Acanthamoebacastellanii of an enzyme similar to muscle myosin. J Biol Chem , 4682–4690. [PubMed] [Google Scholar]
- Ramanujam R, Han Z, Zhang Z, Kanchanawong P, Motegi F. (2018). Establishment of the PAR-1 cortical gradient by the aPKC-PRBH circuit. Nat Chem Biol , 917–927. [DOI] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. (2005). Myosin domain evolution and the primary divergence of eukaryotes. Nature , 1113–1118. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods , 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F. (2008). Endocytosis and the actin cytoskeleton in Dictyosteliumdiscoideum. Int Rev Cell Mol Biol , 343–397. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Welti S. (2012). Pleckstrin homology (PH) like domains—versatile modules in protein–protein interaction platforms. FEBS Lett , 2662–2673. [DOI] [PubMed] [Google Scholar]
- Sellers JR. (2000). Myosins: a diverse superfamily. Biochim Biophys Acta , 3–22. [DOI] [PubMed] [Google Scholar]
- Sussman M. (1987). Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol , 9–29. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Akar G, Bosgraaf L, Van Haastert PJ. (2009). A new set of small, extrachromosomal expression vectors for Dictyosteliumdiscoideum. Plasmid , 110–118. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Williams TD, Bloomfield G, Chen BC, Betzig E, Insall RH, Kay RR. (2016). A plasma membrane template for macropinocytic cups. Elife , 10.7554/eLife.20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visuttijai K, Pettersson J, Mehrbani Azar Y, van den Bout I, Orndal C, Marcickiewicz J, Nilsson S, Hornquist M, Olsson B, Ejeskar K, Behboudi A. (2016). Lowered expression of tumor suppressor candidate MYO1C stimulates cell proliferation, suppresses cell adhesion and activates AKT. PLoS One , e0164063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Muhlbauer B, Heizer C, Schwartz JM, Marriott G, Gerisch G. (1997). Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol , 176–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.