Abstract
Targeting definite genomic locations using CRISPR-Cas systems requires a set of enzymes with unique protospacer adjacent motif (PAM) compatibilities. To expand this repertoire, we engineered nucleases, cytosine base editors, and adenine base editors from the archetypal Streptococcus thermophilus CRISPR1-Cas9 (St1Cas9) system. We found that St1Cas9 strain variants enable targeting to five distinct A-rich PAMs and provide a structural basis for their specificities. The small size of this ortholog enables expression of the holoenzyme from a single adeno-associated viral vector for in vivo editing applications. Delivery of St1Cas9 to the neonatal liver efficiently rewired metabolic pathways, leading to phenotypic rescue in a mouse model of hereditary tyrosinemia. These robust enzymes expand and complement current editing platforms available for tailoring mammalian genomes.
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins form a prokaryotic adaptive immune system, and some of its components have been harnessed for robust genome editing (Komor et al. 2017). Type II–based editing tools rely on a large multidomain endonuclease, Cas9, guided to its DNA target by an engineered single-guide RNA (sgRNA) chimera (Jinek et al. 2012; for a classification of CRISPR-Cas systems, see Koonin et al. 2017; Shmakov et al. 2017; Makarova et al. 2018). The Cas9–sgRNA binary complex finds its target through the recognition of a short sequence called the protospacer adjacent motif (PAM), and the subsequent base-pairing between the guide RNA and DNA leads to a double-strand break (DSB) (Komor et al. 2017; Hille et al. 2018). Although Streptococcus pyogenes (SpCas9) remains the most widely used Cas9 ortholog for genome engineering, the diversity of naturally occurring RNA-guided nucleases is astonishing (Shmakov et al. 2017). Hence, Cas9 enzymes from different microbial species can contribute to the expansion of the CRISPR toolset by increasing targeting density, improving activity and specificity, as well as easing delivery (Esvelt et al. 2013; Komor et al. 2017).
In principle, engineering complementary CRISPR-Cas systems from distinct bacterial species should be relatively straightforward, as they have been minimized to only two components. However, many such enzymes were found inactive in human cells despite being accurately reprogrammed for DNA binding and cleavage in vitro (Ran et al. 2015; Zetsche et al. 2015; Chen et al. 2017a). Nevertheless, an example of the value of alternative Cas9 enzymes is the implementation of the type II-A Cas9 from Staphylococcus aureus (SaCas9) for in vivo editing using a single recombinant adeno-associated virus (AAV) vector (Ran et al. 2015; Maeder et al. 2019). More recently, Campylobacter jejuni and Neisseria meningitidis Cas9s from the type II-C (Mir et al. 2018) CRISPR-Cas systems have been added to this repertoire (Kim et al. 2017; Ibraheim et al. 2018; Edraki et al. 2019). In vivo editing offers the possibility to generate phenotypes in animal models in order to better recapitulate the interactions between cell types and organs. In addition, it can be envisioned as a novel class of human therapeutics that enables precise molecular correction of genetic defects underlying diseases. Therefore, further development of robust and wide-ranging CRISPR-based technologies for in vivo editing may help to decipher disease mechanisms and offer novel therapeutic options (Lau and Suh 2017; Schneller et al. 2017).
Here we revisited the properties of Streptococcus thermophilus type II-A CRISPR1-Cas9, a model system central to the discovery of CRISPR and its function (Barrangou and Horvath 2017; Hille et al. 2018). We aimed to engineer potent RNA-guided nucleases and base editors with distinctive PAM sequences for both in vitro and in vivo applications.
Results
Robust and potent DNA cleavage by St1Cas9 in human cells
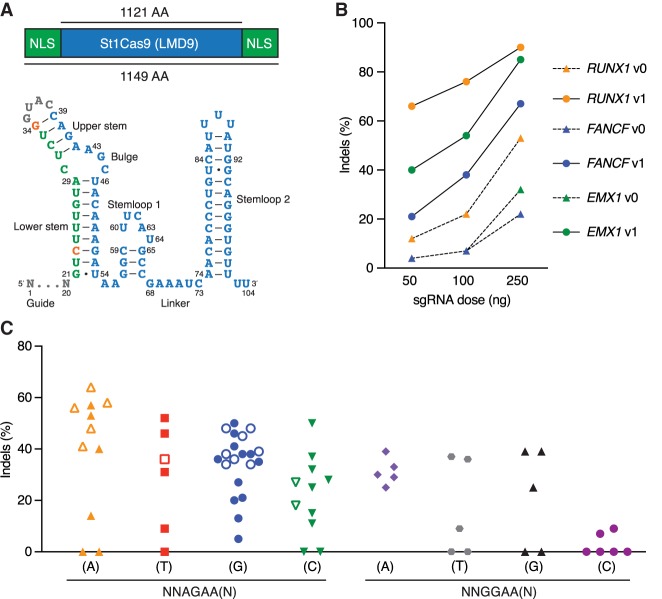
While characterizing the interplay between St1Cas9 and anti-CRISPR proteins isolated from phages infecting S. thermophilus (Hynes et al. 2018), we noticed the substantial levels of editing achieved in human cells, an observation contrasting with previous reports (Chari et al. 2015; Ran et al. 2015). We thus attempted to optimize its activity. First, we flanked the human codon-optimized ORF (Kleinstiver et al. 2015b) with nuclear localization signals (NLSs) (Fig. 1A). Second, we customized the sgRNA sequence to maximize nuclease activity and tested our constructs at three endogenous loci (Fig. 1A,B; Supplemental Fig. S1A–F). The best-performing sgRNA architecture (v1) was engineered by truncating the repeat:antirepeat region (Briner et al. 2014) and substituting a wobble base pair present in the lower stem for a canonical Watson–Crick base pair (Fig. 1A). These modifications also markedly improved transcriptional activation using dSt1Cas9-VPR (Supplemental Fig. S2; Chavez et al. 2015). This analysis revealed that high gene disruption rates could be obtained under standard conditions using St1Cas9 in human cells.
Figure 1.
Functional PAM sequences for robust and potent DNA cleavage by St1Cas9 LMD9 in mammalian cells. (A) Schematic representations of St1Cas9 LMD9 flanked by nuclear localization signals (NLSs) and its engineered sgRNA (v1). Nucleotide sequence and functional modules are depicted; crRNA (green), loop (gray), tracrRNA (blue), and mutated nucleotides (orange). (B) K562 cells stably expressing St1Cas9 were transfected with indicated sgRNA expression vectors at increasing doses, and TIDE assays were performed 3 d later to determine the frequency of indels. An expression vector encoding EGFP (−) was used as a negative control. The experiment was performed twice and yielded equivalent results; only one is shown. (C) Screening for guides targeting St1Cas9 LMD9 to various PAMs was performed by transient transfections in K562 (solid shapes) and Neuro-2a (open shapes) cells using single-vector constructs driving the expression of St1Cas9 and its sgRNA. Surveyor assays were performed 3 d later to determine the frequency of indels. An expression vector encoding EGFP (−) was used as a negative control. See also Supplemental Figure S1.
Functional PAM sequences for St1Cas9 LMD9 in mammalian cells
Cas9 orthologs rely on different PAMs as the first step in target recognition, and the consensus PAM for St1Cas9 (LMD9 and DGCC7710 S. thermophilus strains that differ by only 2 aa within their N terminus) was originally defined as NNAGAAW (where W is A or T) (Deveau et al. 2008). However, sequences closely related to the consensus can be functional in test tubes or when transplanted in Escherichia coli for St1Cas9 LMD9, the strain variant first engineered for genome editing (Cong et al. 2013; Esvelt et al. 2013; Kleinstiver et al. 2015b; Leenay et al. 2016). We thus explored its PAM preference by targeting endogenous loci in human and mouse cells. This analysis revealed that St1Cas9 LMD9 functions efficiently at both NNAGAA and NNGGAA PAMs (Fig. 1C). Although a C is tolerated at position 7, there is a trend for these guides to be less efficacious. This bias was also observed in bacterial cells (Leenay et al. 2016). Thus, the functional core PAM sequence is constituted of four specific base pairs and defined as NNRGAA (where R is A or G). The optimal PAM sequence to regularly achieve high levels of editing is NNRGAAD (where D is A or G or T). The length of the nonconserved PAM linker (NN) has also been shown to be flexible, and an extension from two to three bases can be tolerated in bacterial cells (Briner et al. 2014; Chen et al. 2014), but we failed to reproduce this observation in human cells, suggesting a higher stringency of the system (Supplemental Fig. S1G). We also explored the impact of varying guide length on activity and observed no obvious correlation, confirming previous observations (Supplemental Fig. S1H; Kleinstiver et al. 2015b). As 20-bp guides are markedly less tolerant of mismatches than longer ones for SaCas9 (type II-A SaCas9 and St1Cas9 share 37% identity), we favor the use of 20-bp guides (Tycko et al. 2018). Hence, the flexibility of PAM recognition by St1Cas9 LMD9 enhances its targeting capabilities. Although recognition of an A-rich PAM may facilitate targeting A/T-rich regions of genomes, the targeting range of St1Cas9 in mammalian cells is less constrained than originally thought.
Engineering St1Cas9 variants to expand its targeting range
Although not formally defined as the PAM at the time, the presence of a degenerate consensus sequence situated downstream from protospacers has been observed in strains of S. thermophilus ∼15 yr ago (Bolotin et al. 2005). For example, inferred consensus PAM sequences for St1Cas9 from strains CNRZ1066 and LMG18311 are NNACAA(W) and NNGYAA(A) (where Y is C or T), respectively (Bolotin et al. 2005). Accordingly, the CRISPR1-Cas system of S. thermophilus strain LMG18311 transplanted in E. coli or reconstituted from purified components has been shown to target DNA using a NNGCAAA PAM (Chen et al. 2014).
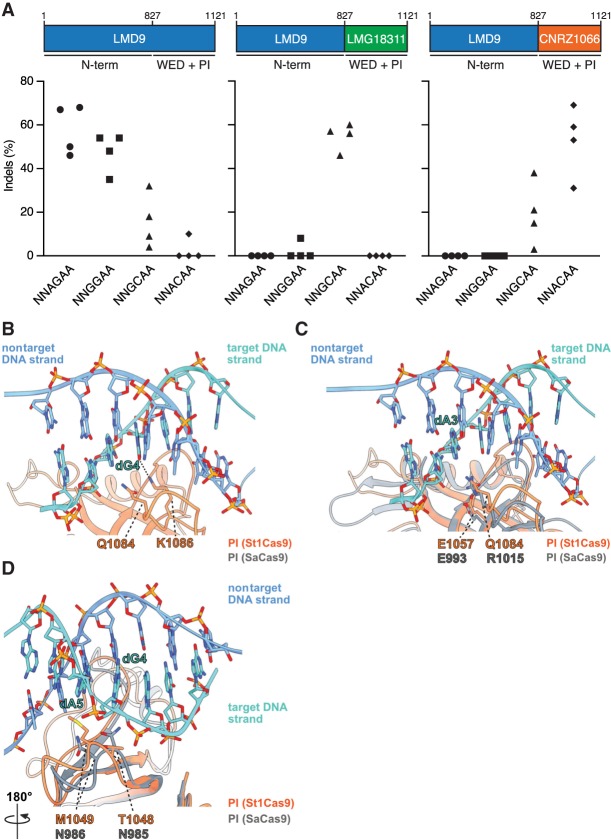
At the protein level, the sequence of those St1Cas9 strain variants diverges mostly within the C-terminal wedge (WED) and PAM-interacting (PI) domains, implying that they have evolved to recognize distinct PAM sequences (Supplemental Fig. S3A,D). Because the PAM duplex is sandwiched between them (Fuchsbauer et al. 2019), we tested whether swapping the WED and PI domains of St1Cas9 LMD9 with the ones from LMG18311 and CNRZ1066 could reprogram PAM specificity (Fig. 2A). The same sgRNA architecture was used with all St1Cas9 variants for these experiments. Whereas St1Cas9 LMD9 targeted NNAGAA and NNGGAA PAMs, the hybrid constructs targeted with high efficacy NNGCAA and NNACAA PAMs, respectively (Fig. 2A). We observed minimal levels of cross-reactivity when swapping sgRNAs between nucleases, indicating that these variants have distinct PAM requirements for high cleavage efficacy (Fig. 2A). LMD9- and CNRZ1066-based variants recognized the noncognate NNGCAA PAM with some sgRNAs, albeit cleavage efficacy was diminished (Fig. 2A).
Figure 2.
Structural basis for PAM specificity of engineered St1Cas9 variants with expanded targeting range. (A) Schematic representation of St1Cas9 hybrid proteins containing the N terminus of LMD9 and the C-terminal domains (WED + PI) of LMG18311 or CNRZ1066. To determine the activity of St1Cas9 variants programmed with sgRNAs compatible with different PAMs, K562 cells were transiently transfected with single-vector constructs driving expression of St1Cas9 and its sgRNA. For each PAM and nuclease combination, four different sgRNAs (targets) were tested. Surveyor assays were performed 3 d later to determine the frequency of indels. An expression vector encoding EGFP (−) was used as a negative control. The experiment was performed twice and yielded equivalent results; only one is shown. (B) Close-up view of the 5′-GCAGAAA-3′ PAM bound to the St1Cas9 (DGCC7010) PI domain (PDB: 6RJD). The target (turquoise) and nontarget (blue) strands are shown as sticks (the phosphate–sugar backbones are also shown as ribbons). The ribbon representation of the PI domain is orange. The hydrogen bonds between the side chain of St1Cas9 K1086 and the nucleobase of dG4 is shown as a dashed line. (C,D) The PI domains of St1Cas9 and SaCas9 (PDB: 5CZZ, gray ribbon) are superimposed. (C) The St1Cas9 Q1084 and SaCas9 R1015 occupy the same positions relative to the PAM (dA3). The St1Cas9 E1057 and SaCas9 E993 occupy the same positions relative to St1Cas9 Q1084 and SaCas9 R1015, respectively. (D) St1Cas9 T1048 and M1049 (substituted for N1048 and D1049 in some variants) superimpose onto SaCas9 N985 and N986 that specify purines in positions 4 and 5 of the PAM. See also Supplemental Figure S3.
Sequence database mining using the “search for PAMs by alignment of targets” (SPAMALOT) tool (Chatterjee et al. 2018) predicts that even more diversity exists within CRISPR1-StCas9 systems, and two additional groups represented by strains TH1477 and MTH17CL396 potentially target NNGAAA and NNAAAA PAMs, respectively (Supplemental Fig. S3B). By using the strategy described above, we were able to construct highly active nucleases targeting NNGAAA and NNAAAA PAMs (Supplemental Fig. S3C). These variants also displayed a high level of specificity for their cognate PAMs despite differing by only two residues (see below) (Supplemental Fig. S3C,D).
These data highlight the modularity inherent to Cas9 enzymes and a simple strategy to further expand the targeting range of St1Cas9s. Currently, this set of nucleases based on the St1Cas9 backbone can target five unique A-rich PAMs (LMD9, NNRGAA; LMG18311, NNGCAA; CNRZ1066, NNACAA; TH1477, NNGAAA; and MTH17CL396, NNAAAA). Tapping into the natural diversity found within S. thermophilus strains results in true reprograming toward a distinct PAM as opposed to relaxing specificity. In addition, despite their sequence and structural conservation (Nishimasu et al. 2015; Fuchsbauer et al. 2019) St1Cas9 variants could not cleave at SaCas9 PAMs (NNGRRT; where R is A or G) in human cells, further highlighting their specificity (Supplemental Fig. S4A–D). These orthologs also differ in their sensitivity to anti-CRISPR proteins as St1Cas9 is inhibited by both AcrIIA5 and AcrIIA6, whereas SaCas9 can only be blocked by AcrIIA5 (Supplemental Fig. S4E,F; Hynes et al. 2018; Fuchsbauer et al. 2019; Garcia et al. 2019). This comparison between St1Cas9 and SaCas9 suggests that they function orthogonally and could be used in a combinatorial manner.
Structural basis for St1Cas9s PAM specificity
We recently determined the structure of St1Cas9 (DGCC7710) bound to its sgRNA and to a target DNA containing a PAM to an overall resolution of 3.3 Å using single-particle cryo-electron microscopy (Fuchsbauer et al. 2019). In this structure, the 5′-GCAGAAA-3′-containing PAM duplex is formed by seven Watson–Crick base pairs, and its major and minor grooves are sandwiched between the WED and PI domains. Although the resolution of this structure prevents us from mapping all amino acid contacts with the PAM, the side chain of K1086 in the PI domain hydrogen bonds with the guanine at position 4 (NNAG4AA) (Fig. 2B). Accordingly, St1Cas9 variants predicted by SPAMALOT (Chatterjee et al. 2018) to specify a guanine at this position contain K1086 (Supplemental Fig. S3B,D). There is only one type of substitution at that position, where K1086 is replaced by I1086 (Supplemental Fig. S3D). This set of variants, which includes LMG18311 (NNGC4AA), CNRZ1066 (NNAC4AA), TH1477 (NNGA4AA), and MTH17CL396 (NNAA4AA), have lost the specificity for a guanine at position 4. At position 1084, the same type of analysis reveals that substitution of Q1084 for R1084 leads to the recognition of a guanine at position 3, as directly observed for LMG18311 (NNG3CAA) and TH1477 (NNG3AAA) (Fig. 2A; Supplemental Fig. S3C). Note that K1086 and R1084 are mutually exclusive, and their co-occurrence would result in a steric clash (Supplemental Fig. S3D). In the structure of SaCas9 bound to 5′-TTG3AAT-3′ PAM, the guanine at position 3 is recognized by R1015 (Nishimasu et al. 2015). Structural comparison reveals that SaCas9 R1015 and St1Cas9 Q1084 occupy the same position relative to their PAMs (Fig. 2C). In addition, SaCas9 R1015 is anchored via salt bridges to E993, a position equivalent to E1057 in St1Cas9 (Fig. 2C). Thus, St1Cas9 variants with R1084 likely recognize guanine at position 3 in an analogous manner as SaCas9 does. Finally, a distinct set of amino acids surrounding positions 1048–1052 likely specify an adenine at position 4 in some variants as it is the case for TH1477 (NNGA4AA) and MTH17CL396 (NNAA4AA) (Supplemental Fig. S3D). T1048 and M1049 are replaced by N1048 and D1049 in those St1Cas9 variants and are predicted to occupy the same positions as N985 and N986 in SaCas9, the residues that specify purines at positions 4 and 5 in the NNGR4R5T PAM (where R is A or G) (Fig. 2D; Supplemental Fig. S3D; Nishimasu et al. 2015). Structural comparison predicts that N1048 could directly contact the adenine in position 4 and that N1048 and D1049 would contact the adenine in positions 4 and 5, potentially via water-mediated hydrogen bonds as observed in SaCas9 (Fig. 2D; Nishimasu et al. 2015). Taken together, these observations provide a first glimpse at PAM recognition by St1Cas9 variants.
Broadening the targeting scope of base editors using St1Cas9 variants
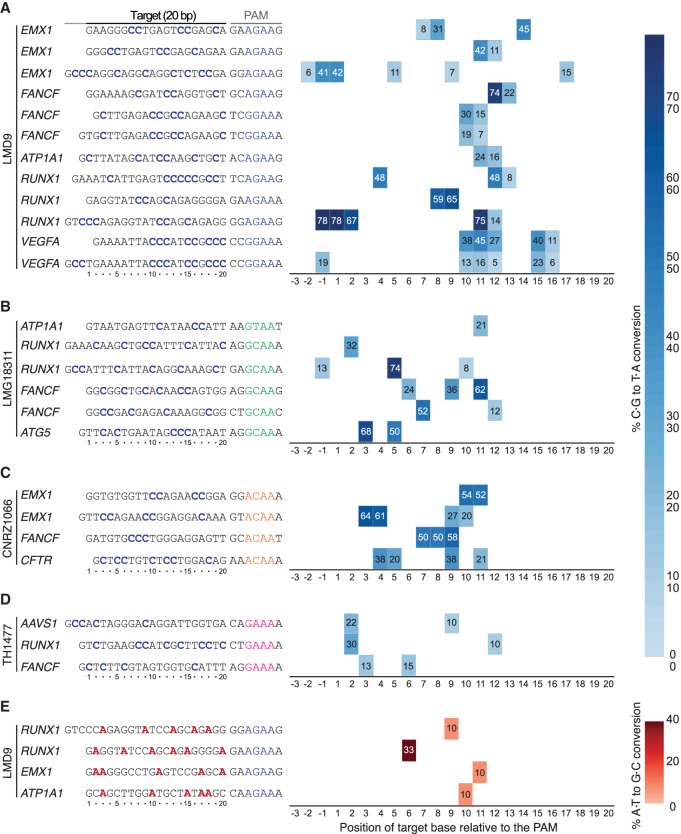
DNA base editors comprise fusions between a catalytically impaired Cas nuclease and a base modification enzyme that operates on single-stranded DNA (ssDNA) (Rees and Liu 2018). Cytosine base editors (CBEs) convert a C•G base pair into a T•A using the APOBEC1 cytidine deaminase. Fusion of APOBEC1 to a S. pyogenes Cas9 (SpCas9) D10A mutant (nickase) and two copies of the uracil DNA glycosylase inhibitor (UGI), resulted in the creation of the SpBE4max enzyme (Koblan et al. 2018). A limitation of the current base editing technology is that the PAM must be appropriately positioned relative to the target base to ensure efficient editing (Rees and Liu 2018). Thus, there is a need to develop base editors with additional PAM compatibilities to increase the number of targetable bases in a genome. As such, SaCas9 has also been converted into a base editor to create SaBE4 (Rees and Liu 2018). In an analogous manner, we have created St1BE4max by exchanging SpCas9 D10A for St1Cas9 LMD9 D9A into the SpBE4max construct (Koblan et al. 2018). This created a potent CBE with novel targeting specificity owing to the unique PAM of St1Cas9 (Fig. 3A). Our data indicate that St1BE4max has an activity window similar to SaBE4, which is wider than SpBE4max, and sometimes extends upstream of the guide (Fig. 3A; Supplemental Data; Rees and Liu 2018).
Figure 3.
Broadening the targeting scope of base editors using St1Cas9 variants. (A) K562 cells were transiently transfected with single-vector constructs driving expression of St1BE4max LMD9 and its sgRNA. Genomic DNA was harvested 3 d later, and quantification of base editing was performed on PCR amplified target sites using EditR. The target sequence was defined as the 20 bases upstream of the PAM and are numbered in decreasing order from the PAM. Sequence of the guides and related PAMs are shown with target cytosine highlighted in blue. An expression vector encoding EGFP (−) was used as a negative control. (B–D) Same as A but using St1BE4max LMG18311, CNRZ1066, and TH1477 chimeric proteins. (E) Same as A but using St1ABEmax LMD9. Target adenines are highlighted in red. Most sgRNAs were tested at least twice; only one experiment is shown. See also Supplemental Figure S5.
We then proceeded to test if St1Cas9 strain variants that display unique PAM preferences are also functional as CBEs. Indeed, LMG18311-, CNRZ1066-, and TH1477-based St1BE4max are efficient base editors at NNGCAA, NNACAA, and NNGAAA PAMs, respectively (Fig. 3B–D). St1BE4max variants were inactive at noncognate PAMs, indicating that they function in an orthogonal manner (Supplemental Fig. S5). We also generated an adenine base editor (St1ABEmax LMD9) to mediate the conversion of A•T to G•C in genomic DNA. We observed moderate editing efficiencies of St1ABEmax, a phenomenon also observed for SaABEmax, indicating that the ABEmax architecture is not fully compatible with these shorter Cas9s (Fig. 3E; Huang et al. 2019). Nevertheless, these architectures can serve as a starting point for further improvements. Taken together, these data further show that St1Cas9 variants can be used as a scaffold to expand the targeting range of base editors.
In vivo genome editing using St1Cas9
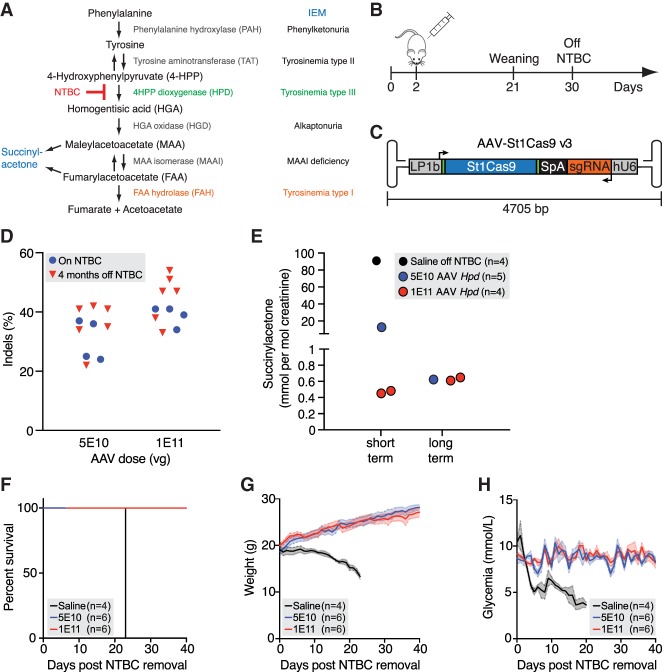
The small size of St1Cas9 makes it potentially permissive for packaging holo-St1Cas9 (St1Cas9 + sgRNA) into AAV vectors for in vivo delivery. To test the cleavage activity of St1Cas9 in vivo, we used the hereditary tyrosinemia type I (HT-I) mouse model, a disease caused by a deficiency of fumarylacetoacetate hydrolase (FAH), the last enzyme of the tyrosine catabolic pathway (OMIM 276700; Orphanet ORPHA:882) (Fig. 4A). Fah−/− mutant mice die as neonates with severe hepatic dysfunction and kidney damage owing to the accumulation of toxic metabolites unless treated with nitisone (NTBC), a drug that inhibits 4-hydroxyphenylpyruvate dioxygenase (HPD) upstream in the pathway (Fig. 4A; Grompe 2017). Because genetic ablation of Hpd in mice can also prevent liver damage and lethality by creating a much milder HT-III phenotype (Endo et al. 1997; Pankowicz et al. 2016), we attempted to inactivate Hpd in our studies using St1Cas9.
Figure 4.
In vivo genome editing using St1Cas9. (A) The tyrosine degradation pathway and associated inborn errors of metabolism (IEMs). (B) Experimental design. Neonatal (2-d-old) Fah−/− mice were injected with AAV8-St1Cas9 or saline into the retro-orbital sinus and weaned at 21 d, and NTBC was removed at 30 d of age. Mice off NTBC were sacrificed when they lost 20% of their body weight. (C) Schematic representation of the AAV-St1Cas9 v3 vector. Annotated are the liver-specific promoter (LP1b), synthetic polyadenylation sequence (SpA), and hU6 promoter. Arrows indicate the direction of transcriptional unit. (D) Neonatal Fah−/− mice were injected with either 5 × 1010 or 1 × 1011 vector genomes (vg) of AAV8-St1Cas9 v3 targeting Hpd exon 13 and sacrificed 28 d following injection or kept alive for phenotypic and metabolic studies for 4 mo post NTBC removal. Genomic DNA was extracted from whole-liver samples, and the Surveyor assay was used to determine the frequency of indels. Each dot represents a different mouse. A mouse injected with saline (−) was used as a negative control. (E) SUAC levels in urine from treated mice were determined 15 d (short term) or 4 mo (long term) following NTBC removal. Samples were collected from the indicated treatment groups over a 24-h period using metabolic cages. Number of mice per group/metabolic cage (n) and AAV doses (vg) is indicated. SUAC levels are undetectable in C57BL/6N (wild-type) mice. (F–H) Survival analysis, body weight, and glycemia following NTBC removal in treated mice. Body weight was measured daily, and glycemia was monitored in nonfasted mice. Solid lines designate the mean; and error bars are represented by shaded areas and denote SEM. See also Supplemental Figure S6.
To deliver holo-St1Cas9 to the liver, we generated a first set of AAV plasmids (AAV-St1Cas9 v1 and v2) containing a liver-specific promoter and sgRNA expression cassettes in opposite orientations and produced hepatotropic AAV serotype 8 (AAV8) vectors (Supplemental Fig. S6A,B; Colella et al. 2018). We injected NTBC-treated Fah−/− mice at day 2 of life into the retro-orbital sinus with these vectors and isolated total liver DNA at day 28 post injection in treated mice (Fig. 4B; Supplemental Fig. S6A,B). The titration showed that the degree of target editing at two different exons of Hpd was substantial and dependent on the dose of AAV8-St1Cas9 (Supplemental Fig. S6A,B). We then evaluated if alternative AAV-St1Cas9 expression cassettes could further improve cleavage efficacy in vivo while minimizing vector size. In our best-performing design (AAV-St1Cas9 v3), we engineered a liver-specific promoter (LP1b) by combining elements from the human apolipoprotein E/C-I gene locus control region (ApoE-HCR), a modified human α1 antitrypsin promoter (hAAT), and an SV40 intron and used a synthetic polyadenylation signal element (Fig. 4C; Nathwani et al. 2006; McIntosh et al. 2013). These modifications increased cleavage efficacy markedly, especially at a low AAV8 dose, and led to the creation of a vector of ∼4.7 kb in size, which is optimal for viral particle packaging (Fig. 4C,D; Colella et al. 2018). It is worth noting that as the genomic DNA was extracted from pieces of total livers, the effective activity is likely to be underestimated because hepatocytes make up 70% of the liver's mass (Palaschak et al. 2019). Under the same experimental conditions, the levels of in vivo editing achieved with AAV-St1Cas9 v3 were comparable to the ones obtained using the gold standard AAV-SaCas9 system (Supplemental Fig. S6C; Ran et al. 2015). Of note, because modifications made to AAV plasmids can occasionally result in loss of potency of the recombinant AAV, we did not alter the structure of the published SaCas9 vector (Ran et al. 2015) for this comparison. Nevertheless, AAV8-mediated delivery into neonatal mice results in transient expression (see Discussion), which constitutes a stringent test of potency for the two nuclease systems.
To test if AAV8-St1Cas9 v3 can achieve phenotypic correction in vivo, NTBC was withdrawn shortly after weaning in the remaining subset of treated mice. Systemic delivery via a single neonatal injection normalized the levels of excretion of succinylacetone (SUAC), a toxic metabolite and a diagnostic marker for HT-I (Fig. 4E; Grompe 2017). Even at the lower vector dose (5 × 1010), we observed delayed but near complete elimination of SUAC secretion 4 mo following NTBC removal, which is likely owing to the potent selective growth advantage of targeted hepatocytes that can extensively repopulate the diseased organ (Fig. 4E; Grompe 2017). This is also reflected by the increased levels of indels detected in liver samples over the same period (Fig. 4D). Consequently, treatment rescued lethality in all mice, whereas saline-treated animals had to be sacrificed after ∼3 wk as they met the weight loss criterion (Fig. 4F). Likewise, glycemia and weight loss were normalized in the treatment groups (Fig. 4G,H). Therefore, AAV8-mediated delivery of St1Cas9 in neonatal mice can result in efficient DNA cleavage, stable genetic modification, and phenotypic correction by rewiring a metabolic pathway through gene inactivation.
To corroborate these findings, we targeted Pck1, the gene encoding phosphoenolpyruvate carboxykinase 1, cytosolic, which plays a broad role in integrating hepatic energy metabolism and gluconeogenesis (Yang et al. 2009). Mice with a liver-specific deletion of the gene are viable but display an impaired response to fasting (She et al. 2000). Neonatal 2-d-old C57BL/6N (wild-type) pups were injected with AAV8-St1Cas9 v3 targeting Pck1, and at 6 wk of age, they were fasted for 24 h and sacrificed for metabolic profiling and evaluation of gene disruption efficacy (Supplemental Fig. S6D,E). Systemic delivery via a single neonatal injection resulted in substantial hepatic Pck1 gene disruption (Supplemental Fig. S6D). Plasma and hepatic triglyceride content were also markedly increased (Supplemental Fig. S6E). However, we found no change in circulating free fatty acid levels, and hepatic glycogen stores were not depleted, suggesting that the observed phenotype may be intermediary to the one described in a prenatal hepatic knockout model (Supplemental Fig. S6E; She et al. 2000). We speculate that normal PCK1 in any nontargeted hepatocytes can partially compensate for the loss of function resulting from in vivo editing. Nevertheless, AAV8-mediated delivery of St1Cas9 in neonatal mice can efficiently disrupt the function of a key metabolic enzyme, leading to clear and substantial phenotype in vivo. Collectively, these data support the notion that St1Cas9 can be engineered as a powerful tool for in vivo genome editing.
Discussion
Here we report that St1Cas9 can be harnessed for robust and efficient genome editing in vitro and in vivo, thereby expanding the CRISPR-Cas toolbox. We optimized this system to create potent nucleases, transcription activators, and base editors. We further validated its use in mice by showing efficient rewiring, rescue, and creation of metabolic defects using all-in-one AAV vectors. Our work offers a comprehensive analysis and highlights novel fields of application for this CRISPR-Cas9 platform in mammalian cells (Kleinstiver et al. 2015b; Müller et al. 2016). St1Cas9 also functions efficiently for the labeling of chromosomal loci in human cells and in mouse zygotes to create animal models (Ma et al. 2015; Fujii et al. 2016). In other systems, such as mycobacteria and the plant Arabidopsis thaliana, St1Cas9 is at least comparable to, and can even outperform, SpCas9 (Steinert et al. 2015; Rock et al. 2017).
Structure-guided and random mutagenesis have been combined to successfully reprogram the PI domain of Cas9s to alter its specificity toward a distinct sequence but also to relax its specificity (e.g., NNGRRT would become NNNRRT or NGG would become NGN) (Kleinstiver et al. 2015a,b; Hu et al. 2018; Nishimasu et al. 2018). As expected, relaxed PAM recognition typically decreases genome-wide specificity by increasing the number of off-targets (Kleinstiver et al. 2015a, 2019; Nishimasu et al. 2018). As a corollary, off-target sites are generally low in number for Cas9s with longer PAMs versus with shorter ones (Kleinstiver et al. 2015b; Tsai et al. 2015; Kleinstiver et al. 2019). This highlights an emerging connection between the length and complexity of the PAM versus the absolute specificity of a Cas protein that warrants further exploration (Müller et al. 2016). In this work, we engineered St1Cas9 variants with distinct PAM requirements and observed limited cross-reactivity toward noncognate PAMs. Although the complete characterization of PAM preference for each of these variants remains to be performed, these data suggest a stringent PAM requirement for St1Cas9s. Some level of flexibility in PAM recognition has been observed and is to be expected for CRISPR-Cas systems in human cells. For example, wild-type SpCas9 (consensus NGG) can also recognize NAG, NGA, NGT, and NGC with some, but not all, sgRNAs (Tsai et al. 2015; Nishimasu et al. 2018). Wild-type SaCas9 (consensus NNGRRT) can cleave at noncanonical NNARRT PAMs (Kleinstiver et al. 2015a). Finally, wild-type AsCas12a (consensus TTTV) can recognize noncanonical GTTV and GCTV PAMs (Jacobsen et al. 2019). Although this flexibility in PAM recognition is a concern regarding potential off-target activity (Tsai et al. 2015), DNA cleavage activity is typically much lower at these sites. Nevertheless, a comprehensive genome-wide profiling of off-target cleavage remains to be performed for St1Cas9 variants. The recently described structure of St1Cas9 should facilitate the creation of high-fidelity St1Cas9s (Slaymaker et al. 2016; Chen et al. 2017b; Vakulskas et al. 2018; Fuchsbauer et al. 2019).
The engineering of CRISPR-Cas systems with unique PAM sequences is of utmost importance and should not be guided uniquely by the absolute targeting range (the total number of PAMs present in a genome) as the exact location of binding is most often key for genome editing. Applications such as disruption of small genetic elements, allele-specific targeting, seamless gene correction via recombination, base editing, or gene correction via microhomology-mediated end joining require highly precise targeting (Canver et al. 2015; Rees and Liu 2018; Gyorgy et al. 2019; Iyer et al. 2019). To achieve single-nucleotide precision in targeting, a plethora of Cas9 orthologs harboring both wild-type and altered PAM specificities will be needed. To illustrate the utility of St1Cas9 variants in such contexts, we identified several disease-causing mutations that could potentially be targeted in an allele-specific manner (Supplemental Fig. S7A). We also identified highly active St1Cas9 nucleases targeting narrow regions within the 5′ UTR and first intron of the mouse albumin gene (Supplemental Fig. S7B,C). These sites are of particular interest because the albumin gene has been described as a safe-harbor locus for targeted integration of therapeutic transgenes and liver-directed protein replacement therapies (Sharma et al. 2015). Base editors using St1Cas9 variants could also be relevant to correct metabolic diseases (Supplemental Fig. S8).
Recombinant AAV vectors are prime in vivo gene delivery vectors for nonproliferative tissues. However, a limitation in the therapeutic use of AAV is the loss of episomal vector genomes from actively dividing cells, resulting in transient expression of therapeutic transgenes. Hence, the combination of genome editing technology with AAV-mediated delivery could lead to permanent genome modification and positive therapeutic outcome in young patients when tissues, such as the liver and retina, are still growing (Li et al. 2011; Yang et al. 2016). As a side benefit, the elimination of vector genomes would lead to transient nuclease expression in proliferating tissues that likely prevents accumulation of mutations at off-target sites (Li et al. 2011; Yang et al. 2016). In this perspective, the development of alternative in vivo genome editing platforms based on orthologous CRISPR-Cas systems would further increase the options available for therapeutic interventions.
Methods
Cell culture and transfection
K562 were obtained from the ATCC (CCL-243) and maintained at 37°C under 5% CO2 in RPMI medium supplemented with 10% FBS, penicillin–streptomycin, and GlutaMAX. Neuro-2a were obtained from the ATCC and maintained at 37°C under 5% CO2 in DMEM medium supplemented with 10% FBS, penicillin–streptomycin, and GlutaMAX. All cell lines are tested for absence of mycoplasma contamination. Cells (2 × 105 per transfection) were transfected using the Amaxa 4D-Nucleofector (Lonza) per the manufacturer's recommendations. Unless otherwise specified, 0.5 µg and 1 µg of single-vector constructs driving the expression of both the sgRNA and St1Cas9 variants (nucleases or base editors) were used for transient transfections in Neuro-2a and K562 cells, respectively. K562 cell lines expressing St1Cas9 from the AAVS1 safe-harbor locus were generated as previously described (Dalvai et al. 2015; Agudelo et al. 2017). Briefly, simultaneous selection and cloning were performed for 10 d in methylcellulose-based semisolid RPMI medium supplemented with 0.5 µg/mL puromycin starting 3 d post-transfection. Clones were picked and expanded in 96 wells for 3 d and transferred to 12-well plates for another 3 d before cells were harvested for western blot analysis.
St1Cas9 strain variants
Sequences of St1Cas9 variants were retrieved from NCBI's Identical Protein Groups (IPG) resource, which contained 29 unique protein sequences at the time of analysis. Predicted PAM sequences for St1Cas9 LMD9, LMG18311, and CNRZ1066 were previously published (Bolotin et al. 2005; Deveau et al. 2008; Chen et al. 2014). Predictions for St1Cas9s related to TH1477 and MTH17CL396 originated from https://github.com/mitmedialab/SPAMALOT (Chatterjee et al. 2018). Alignments were performed using Clustal Omega (Sievers et al. 2011) in combination with the sequence alignment renderer ESPript 3 (Robert and Gouet 2014).
Structural analysis of PAM specificity
Coot and PISA were used to analyze the 3D structures (Krissinel and Henrick 2007). UCSF ChimeraX was used to prepare the figures (Goddard et al. 2018).
Genome editing vectors
Vectors for in vitro and in vivo genome editing with the CRISPR1-Cas9 (St1Cas9) system of S. thermophilus generated in this study are available from Addgene (Supplemental Fig. S9). Protein and DNA sequences for all St1Cas9 ORFs are available in Supplemental Tables S1 through S5. The mammalian expression vector for St1Cas9 (LMD9) fused to SV40 NLS sequences at the N and C terminus (MSP1594_2x_NLS; Addgene plasmid 110625) was constructed from MSP1594 (Addgene plasmid 65775, a gift from Keith Joung) (Kleinstiver et al. 2015b). The U6-driven sgRNA expression cassettes for St1Cas9 (LMD9; v1, v2, v3; St1Cas9_LMD-9_sgRNA_pUC19; Addgene plasmid 110627) were synthesized as gBlock gene fragments (Integrated DNA Technologies) and cloned into pUC19 (Supplemental Table S11). BPK2301 (v0; Addgene plasmid 65778, a gift from Keith Joung) (Kleinstiver et al. 2015b) was used to compare St1Cas9 sgRNA architectures. The single-vector mammalian expression system containing a CAG promoter-driven St1Cas9 LMD9 and its U6-driven sgRNA (U6_sgRNA_CAG_hSt1Cas9_LMD9; Addgene plasmid 110626) was built from the above-described plasmids. LMG18311, CNRZ1066, TH1477, and MTH17CL396 C-terminal sequences were synthesized as gBlock gene fragments (Integrated DNA Technologies) and subcloned into U6_sgRNA_CAG_hSt1Cas9_LMD9 to produce the chimeric vectors (Addgene plasmids 136653, 136651, 136655, 136656).
Base editors were constructed into U6_sgRNA_CAG_hSt1Cas9_LMD9 (or the chimeric variants) using fragments derived from pCMV_BE4max_3xHA and pCMV_ABEmax_3xHA (Addgene plasmids 112096 and 112098, a gift from David Liu) (Koblan et al. 2018). Protein and DNA sequences for all St1Cas9 base editors are available in Supplemental Tables S6 through S10. St1Cas9BE4max (Addgene plasmids 136652, 136654, 136657, 136659) and ABEmax (Addgene plasmid 136660) variants are available for distribution.
The single-vector rAAV-St1Cas9 LMD9 systems containing liver-specific promoters were assembled from the above-described components into a derivative of pX602 (Addgene plasmid 61593, a gift from Feng Zhang) (Ran et al. 2015) containing a deletion within the backbone to eliminate BsmBI restriction sites. The LP1b promoter was engineered by combining elements from previously described AAV expression cassettes (Supplemental Table S12; Nathwani et al. 2006; McIntosh et al. 2013). We deposited the most active version of this vector (v3) (pAAV_LP1B_St1Cas9_LMD-9_SpA_U6_sgRNA; Addgene plasmid 110624).
To establish clonal K562 cell lines constitutively expressing C-terminally tagged St1Cas9 under the control of an hPGK1 promoter, the Cas9 ORF from MSP1594_2x_NLS was subcloned into AAVS1_Puro_PGK1_3xFLAG_Twin_Strep (Addgene plasmid 68375) (Dalvai et al. 2015).
The CRISPOR (Haeussler et al. 2016) web tool was used to design guide sequences against mouse and human targets for St1Cas9 LMD9. For St1Cas9 variants, the guides were identified by manual inspection of target sequences. Guide sequences are available in Supplemental Tables S13 through S18.
Surveyor nuclease, TIDE, and base editing assays
Genomic DNA from 2.5 × 105 cells was extracted with 250 µL of QuickExtract DNA extraction solution (Lucigen) per the manufacturer's recommendations. The various loci were amplified by 30 cycles of PCR using the primers described in Supplemental Table S19. Assays were performed with the Surveyor mutation detection kit (Transgenomics) as previously described (Guschin et al. 2010; Agudelo et al. 2017). Samples were separated on 10% PAGE gels in TBE buffer. Gels were imaged using a ChemiDoc MP (Bio-Rad) system, and quantifications were performed using the Image laboratory software (Bio-Rad). TIDE analysis was performed using a significance cut-off value for decomposition of P < 0.001 (Brinkman et al. 2014). EditR (Kluesner et al. 2018) was used to quantify base editing from Sanger sequencing reads with the P-value cutoff set to 0.01. Under these settings, any editing levels ≤5% is considered background. All chromatograms are available as Supplemental Data.
Recombinant AAV production
Production of recombinant adeno-associated viral vectors was performed by the triple plasmid transfection method essentially as previously described (Gray et al. 2011). Briefly, HEK293T17 cells were transfected using polyethylenimine (PEI; Polysciences) with helper plasmid pxx-680 (a gift from R.J. Samulski), the rep/cap hybrid plasmid pAAV2/8 (a gift from James Wilson), and the rAAV vector plasmid. Twenty-four hours post transfection, media was replaced with growth media without FBS, and cells were harvested 24 h later. rAAV particles were extracted from cell extracts by freeze/thaw cycles and purified on a discontinuous iodixanol gradient. Virus were resuspended in PBS 320 mM NaCl + 5% D-sorbitol + 0.001% pluronic acid (F-68), aliquoted, and stored at −80°C. rAAVs were titrated by qPCR (Roche) using SYBR Green and ITR primers as previously described (Aurnhammer et al. 2012). The yields for all vectors varied between 1 × 1013 and 2 × 1013 vg/mL. The purity of the viral preparations was determined by SDS-PAGE analysis on a 10% stain free gel (Bio-Rad) in Tris-glycine-SDS buffer (Supplemental Fig. S10). ITR integrity was assessed following a BssHII digestion of the AAV plasmid. The vector core facility at the Canadian neurophotonics platform (molecular tools) produced the rAAV8s.
Animal experiments (Fah−/− mouse model)
Fah−/− mice (Grompe et al. 1993) on a C57BL/6 genetic background were group-housed and fed a standard chow diet (Harlan 2018SX) with free access to food and water. Fah−/− mice drinking water was supplemented with 7.5 mg (2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione) (NTBC)/L, and pH was adjusted to 7.0. Mice were exposed to a 12-h:12-h dark–light cycle and kept under an ambient temperature of 23 ± 1°C. Animals were cared for and handled according to the Canadian Guide for the Care and Use of Laboratory Animals. The Université Laval animal care and use committee approved the procedures.
Two-day-old neonatal mice were injected intravenously in the retro-orbital sinus (Yardeni et al. 2011) with different doses of rAAV8 or saline in a total volume of 20 µL. Mice were weaned at 21 d of age, and NTBC was removed 7 d later. Body weight and glycemia were monitored daily following NTBC removal. Mice were not fasted for measurement of glycemia; data collection occurred between 9:00 and 10:00 a.m. Animals were sacrificed by cardiac puncture under anesthesia at predetermined time points or when weight loss reached 20% of body weight. Livers were snap frozen for downstream applications.
Urine collection and SUAC quantification
Urine from groups of three to four mice was collected overnight in metabolic cages (Tecniplast) 15 d and 4 mo after NTBC removal. Urine was centrifuged at 2000 rpm for 5 min, aliquoted, and frozen at −80°C. SUAC was quantified in urine samples by a sensitive method using gas chromatography–mass spectrometry (GC-MS) as previously described (Cyr et al. 2006). The biochemical genetics laboratory at the Centre Hospitalier Universitaire de Sherbrooke performed the analyses.
Additional method details
Methods related to transcription activation (Supplemental Fig. S2) and experiments performed in C57BL/6N mice (Supplemental Fig. S6) are described in the Supplemental Methods section.
Data access
All vectors generated in this study have been submitted to Addgene (https://www.addgene.org). All raw Sanger sequencing data generated in this study are available as Supplemental Data.
Competing interest statement
An international patent application has been filed in relation to this work. Y.D. is named as an inventor.
Supplementary Material
Acknowledgments
This study was supported by grants from the Canadian Institutes of Health Research (CIHR) to M.L. and Y.D. and by the Banting Research Foundation to Y.D. S.M. acknowledges funding from NSERC. S.M. holds a Tier 1 Canada Research Chair in Bacteriophages. A.G. acknowledges funding from the French National Research Agency (ANR-18-CE11-0016-01). UCSF ChimeraX that was used for molecular graphics and analyses is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, and receives support from the National Institutes of Health (NIH) R01-GM129325 and P41-GM103311. Salary support was provided by the Fonds de la recherche du Québec-Santé (FRQS) to M.L. and Y.D. D.A. holds a Vanier Canada graduate scholarship. S.L. holds a Frederick Banting and Charles Best Canada graduate scholarship. A.D. and J-F.R. hold graduate training awards from the Fonds de la recherche du Québec-Santé (FRQS). J-F.R. is also supported by the Fondation du Grand défi Pierre Lavoie. We thank Marie-Ève Paquet and the skilled vector core facility staff at the Canadian neurophotonics platform for AAV8 production. Robert Tanguay provided the mouse model of HT-I, nitisone, expertise, and support.
Author contributions: Conceptualization was done by D.A., S.C., M.V., A.D., J-F.R., S.M., A.G., and Y.D. Methodology was performed by D.A., S.C., M.V., A.D., J-F.R., S.L., J.L., M.M., D.C., P.J.W., M.L., A.G., and Y.D. Investigation was performed by D.A., S.C., M.V., A.D., J-F.R., S.L., J.L., M.M., and D.C. Writing of the original draft was done by Y.D. Review and editing were done by all authors; supervision was done by P.J.W., M.L., and Y.D. Funding acquisition was by M.L., S.M., A.G., and Y.D. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.255414.119.
References
- Agudelo D, Duringer A, Bozoyan L, Huard CC, Carter S, Loehr J, Synodinou D, Drouin M, Salsman J, Dellaire G, et al. 2017. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat Methods 14: 615–620. 10.1038/nmeth.4265 [DOI] [PubMed] [Google Scholar]
- Aurnhammer C, Haase M, Muether N, Hausl M, Rauschhuber C, Huber I, Nitschko H, Busch U, Sing A, Ehrhardt A, et al. 2012. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum Gene Ther Methods 23: 18–28. 10.1089/hgtb.2011.034 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Horvath P. 2017. A decade of discovery: CRISPR functions and applications. Nat Microbiol 2: 17092 10.1038/nmicrobiol.2017.92 [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. 2014. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell 56: 333–339. 10.1016/j.molcel.2014.09.019 [DOI] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, van Steensel B. 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42: e168 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. 2015. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527: 192–197. 10.1038/nature15521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R, Mali P, Moosburner M, Church GM. 2015. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 12: 823–826. 10.1038/nmeth.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P, Jakimo N, Jacobson JM. 2018. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv 4: eaau0766 10.1126/sciadv.aau0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. 2015. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12: 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Choi J, Bailey S. 2014. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J Biol Chem 289: 13284–13294. 10.1074/jbc.M113.539726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Ding X, Feng Y, Seebeck T, Jiang Y, Davis GD. 2017a. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat Commun 8: 14958 10.1038/ncomms14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. 2017b. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550: 407–410. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella P, Ronzitti G, Mingozzi F. 2018. Emerging issues in AAV-mediated in vivo gene therapy. Mol Ther Methods Clin Dev 8: 87–104. 10.1016/j.omtm.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr D, Giguère R, Villain G, Lemieux B, Drouin R. 2006. A GC/MS validated method for the nanomolar range determination of succinylacetone in amniotic fluid and plasma: an analytical tool for tyrosinemia type I. J Chromatogr B Analyt Technol Biomed Life Sci 832: 24–29. 10.1016/j.jchromb.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P, Côté J, Doyon Y. 2015. A scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell Rep 13: 621–633. 10.1016/j.celrep.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400. 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edraki A, Mir A, Ibraheim R, Gainetdinov I, Yoon Y, Song CQ, Cao Y, Gallant J, Xue W, Rivera-Perez JA, et al. 2019. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol Cell 73: 714–726.e4. 10.1016/j.molcel.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo F, Kubo S, Awata H, Kiwaki K, Katoh H, Kanegae Y, Saito I, Miyazaki J, Yamamoto T, Jakobs C, et al. 1997. Complete rescue of lethal albino c14CoS mice by null mutation of 4-hydroxyphenylpyruvate dioxygenase and induction of apoptosis of hepatocytes in these mice by in vivo retrieval of the tyrosine catabolic pathway. J Biol Chem 272: 24426–24432. 10.1074/jbc.272.39.24426 [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. 2013. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10: 1116–1121. 10.1038/nmeth.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsbauer O, Swuec P, Zimberger C, Amigues B, Levesque S, Agudelo D, Duringer A, Chaves-Sanjuan A, Spinelli S, Rousseau GM, et al. 2019. Cas9 allosteric inhibition by the anti-CRISPR protein AcrIIA6. Mol Cell 76: 922–937. 10.1016/j.molcel.2019.09.012 [DOI] [PubMed] [Google Scholar]
- Fujii W, Kakuta S, Yoshioka S, Kyuwa S, Sugiura K, Naito K. 2016. Zygote-mediated generation of genome-modified mice using Streptococcus thermophilus 1–derived CRISPR/Cas system. Biochem Biophys Res Commun 477: 473–476. 10.1016/j.bbrc.2016.06.070 [DOI] [PubMed] [Google Scholar]
- Garcia B, Lee J, Edraki A, Hidalgo-Reyes Y, Erwood S, Mir A, Trost CN, Seroussi U, Stanley SY, Cohn RD, et al. 2019. Anti-CRISPR AcrIIA5 potently inhibits all Cas9 homologs used for genome editing. Cell Rep 29: 1739–1746.e5. 10.1016/j.celrep.2019.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SJ, Choi VW, Asokan A, Haberman RA, McCown TJ, Samulski RJ. 2011. Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration. Curr Protoc Neurosci. Chapter 4: Unit 4 17 10.1002/0471142301.ns0417s57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M. 2017. Fah knockout animals as models for therapeutic liver repopulation. Adv Exp Med Biol 959: 215–230. 10.1007/978-3-319-55780-9_20 [DOI] [PubMed] [Google Scholar]
- Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG, Soriano P. 1993. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev 7: 2298–2307. 10.1101/gad.7.12a.2298 [DOI] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649: 247–256. 10.1007/978-1-60761-753-2_15 [DOI] [PubMed] [Google Scholar]
- Gyorgy B, Nist-Lund C, Pan B, Asai Y, Karavitaki KD, Kleinstiver BP, Garcia SP, Zaborowski MP, Solanes P, Spataro S, et al. 2019. Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat Med 25: 1123–1130. 10.1038/s41591-019-0500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17: 148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E. 2018. The biology of CRISPR-Cas: backward and forward. Cell 172: 1239–1259. 10.1016/j.cell.2017.11.032 [DOI] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, et al. 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556: 57–63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TP, Zhao KT, Miller SM, Gaudelli NM, Oakes BL, Fellmann C, Savage DF, Liu DR. 2019. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat Biotechnol 37: 626–631. 10.1038/s41587-019-0134-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes AP, Rousseau GM, Agudelo D, Goulet A, Amigues B, Loehr J, Romero DA, Fremaux C, Horvath P, Doyon Y, et al. 2018. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat Commun 9: 2919 10.1038/s41467-018-05092-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheim R, Song CQ, Mir A, Amrani N, Xue W, Sontheimer EJ. 2018. All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol 19: 137 10.1186/s13059-018-1515-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Suresh S, Guo D, Daman K, Chen JCJ, Liu P, Zieger M, Luk K, Roscoe BP, Mueller C, et al. 2019. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568: 561–565. 10.1038/s41586-019-1076-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen T, Liao C, Beisel CL. 2019. The Acidaminococcus sp. Cas12a nuclease recognizes GTTV and GCTV as non-canonical PAMs. FEMS Microbiol Lett 366: fnz085 10.1093/femsle/fnz085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, et al. 2017. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8: 14500 10.1038/ncomms14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. 2015a. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 33: 1293–1298. 10.1038/nbt.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. 2015b. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I, et al. 2019. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 37: 276–282. 10.1038/s41587-018-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluesner MG, Nedveck DA, Lahr WS, Garbe JR, Abrahante JE, Webber BR, Moriarity BS. 2018. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J 1: 239–250. 10.1089/crispr.2018.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, Maianti JP, Raguram A, Liu DR. 2018. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol 36: 843–846. 10.1038/nbt.4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. 2017. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 169: 559 10.1016/j.cell.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37: 67–78. 10.1016/j.mib.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Lau CH, Suh Y. 2017. In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Res 6: 2153 10.12688/f1000research.11243.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. 2016. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell 62: 137–147. 10.1016/j.molcel.2016.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. 2011. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221. 10.1038/nature10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. 2015. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci 112: 3002–3007. 10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, Chao H, Ciulla DM, DaSilva JA, et al. 2019. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med 25: 229–233. 10.1038/s41591-018-0327-9 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. 2018. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J 1: 325–336. 10.1089/crispr.2018.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J, Lenting PJ, Rosales C, Lee D, Rabbanian S, Raj D, Patel N, Tuddenham EG, Christophe OD, McVey JH, et al. 2013. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood 121: 3335–3344. 10.1182/blood-2012-10-462200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A, Edraki A, Lee J, Sontheimer EJ. 2018. Type II-C CRISPR-Cas9 biology, mechanism, and application. ACS Chem Biol 13: 357–365. 10.1021/acschembio.7b00855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. 2016. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Ther 24: 636–644. 10.1038/mt.2015.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Gray JT, Ng CY, Zhou J, Spence Y, Waddington SN, Tuddenham EG, Kemball-Cook G, McIntosh J, Boon-Spijker M, et al. 2006. Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver. Blood 107: 2653–2661. 10.1182/blood-2005-10-4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. 2015. Crystal structure of Staphylococcus aureus Cas9. Cell 162: 1113–1126. 10.1016/j.cell.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, et al. 2018. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361: 1259–1262. 10.1126/science.aas9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaschak B, Herzog RW, Markusic DM. 2019. AAV-mediated gene delivery to the liver: overview of current technologies and methods. Methods Mol Biol 1950: 333–360. 10.1007/978-1-4939-9139-6_20 [DOI] [PubMed] [Google Scholar]
- Pankowicz FP, Barzi M, Legras X, Hubert L, Mi T, Tomolonis JA, Ravishankar M, Sun Q, Yang D, Borowiak M, et al. 2016. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat Commun 7: 12642 10.1038/ncomms12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Liu DR. 2018. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19: 770–788. 10.1038/s41576-018-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, et al. 2017. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol 2: 16274 10.1038/nmicrobiol.2016.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneller JL, Lee CM, Bao G, Venditti CP. 2017. Genome editing for inborn errors of metabolism: advancing towards the clinic. BMC Med 15: 43 10.1186/s12916-017-0798-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Anguela XM, Doyon Y, Wechsler T, DeKelver RC, Sproul S, Paschon DE, Miller JC, Davidson RJ, Shivak D, et al. 2015. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126: 1777–1784. 10.1182/blood-2014-12-615492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P, Shiota M, Shelton KD, Chalkley R, Postic C, Magnuson MA. 2000. Phosphoenolpyruvate carboxykinase is necessary for the integration of hepatic energy metabolism. Mol Cell Biol 20: 6508–6517. 10.1128/MCB.20.17.6508-6517.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. 2017. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol 15: 169–182. 10.1038/nrmicro.2016.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert J, Schiml S, Fauser F, Puchta H. 2015. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84: 1295–1305. 10.1111/tpj.13078 [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. 2015. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33: 187–197. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko J, Barrera LA, Huston NC, Friedland AE, Wu X, Gootenberg JS, Abudayyeh OO, Myer VE, Wilson CJ, Hsu PD. 2018. Pairwise library screen systematically interrogates Staphylococcus aureus Cas9 specificity in human cells. Nat Commun 9: 2962 10.1038/s41467-018-05391-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, Bode NM, McNeill MS, Yan S, Camarena J, et al. 2018. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med 24: 1216–1224. 10.1038/s41591-018-0137-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kalhan SC, Hanson RW. 2009. What is the metabolic role of phosphoenolpyruvate carboxykinase? J Biol Chem 284: 27025–27029. 10.1074/jbc.R109.040543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, et al. 2016. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 34: 334–338. 10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardeni T, Eckhaus M, Morris HD, Huizing M, Hoogstraten-Miller S. 2011. Retro-orbital injections in mice. Lab Anim (NY) 40: 155–160. 10.1038/laban0511-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.