Abstract
CBL interacting protein kinases play important roles in adaptation to stress conditions. In the present study, we isolated a CBL-interacting protein kinase homolog (AdCIPK5) from a wild peanut (Arachis diogoi) with similarity to AtCIPK5 of Arabidopsis. Expression analyses in leaves of the wild peanut showed AdCIPK5 induction by exogenous signaling molecules including salicylic acid, abscisic acid and ethylene or abiotic stress factors like salt, PEG and sorbitol. The recombinant AdCIPK5-GFP protein was found to be localized to the nucleus, plasma membrane and cytoplasm. We overexpressed AdCIPK5 in tobacco plants and checked their level of tolerance to biotic and abiotic stresses. While wild type and transgenic plants displayed no significant differences to the treatment with the phytopathogen, Phytophthora parasitica pv nicotianae, the expression of AdCIPK5 increased salt and osmotic tolerance in transgenic plants. Analysis of different physiological parameters revealed that the transgenic plants maintained higher chlorophyll content and catalase activity with lower levels of H2O2 and MDA content during the abiotic stress conditions. AdCIPK5 overexpression also contributed to the maintenance of a higher the K+/Na+ ratio under salt stress. The enhanced tolerance of transgenic plants was associated with elevated expression of stress-related marker genes; NtERD10C, NtERD10D, NtNCED1, NtSus1, NtCAT and NtSOS1. Taken together, these results indicate that AdCIPK5 is a positive regulator of salt and osmotic stress tolerance.
Subject terms: Plant molecular biology, Plant stress responses
Introduction
Plants have developed an elaborate network of signaling pathways to counter the challenges posed by various stressful environmental conditions. In general, Ca2+ serves as an ubiquitous secondary messenger that is reported to be involved in several biological processes including abiotic stress responses, pathogen defense and ion homeostasis adjustment1. Studies on Ca2+ dynamics have indicated stimulus-specific elevations in cytosolic Ca2+ concentration, termed as ‘calcium signatures’. To decode each signature, cells retain precise tools and mechanisms that include Ca2+ sensors and their downstream target proteins2. The sensor proteins exhibit a Ca2+ binding site in their helix-loop-helix region3. These proteins are classified into two categories as sensor responders and sensor relays4. Sensor responders such as Ca2+ dependent protein kinases (CDPKs) contain both Ca2+ binding and kinase activities, while sensor relays like calmodulin (CaM) and calmodulin-like proteins (CML) do not exhibit kinase activity. However, after binding with Ca2+, they interact with other protein kinases to regulate their activities5.
Compared to other organisms, the Ca2+ signaling mechanism in plants is more complex with the acquisition of several specific Ca2+ sensor proteins. The calcineurin B-like protein (CBL) family is one of these specific sensor proteins. Arabidopsis CBL4/SOS3 is the first CBL protein reported from the plant system6. Since then, several CBL proteins have been identified from different plants7. These proteins act as sensor relays and target a family of Ser/Thr kinases known as CBL-interacting protein kinases (CIPKs). Like CBLs, CIPKs are unique to the plant system8. Genome-wide studies have identified 26 CIPKs in Arabidopsis, 31 in rice, 27 in poplar, and 32 in sorghum7. The overall structure of CIPKs includes a catalytic N-terminal domain and a regulatory C-terminal domain1,9. Within the regulatory domain, a highly conserved stretch of 24 amino acids has been designated as NAF domain. It is believed to be responsible for mediating the CIPK interaction with CBL proteins10.
Many CIPKs have been functionally characterized in the recent past. For example, Arabidopsis AtCIPK24/SOS2 was initially identified as an AtCBL4 target protein that participated in salt tolerance mechanism by activating the Na+/H+ antiporter located on the root plasma membrane11,12, while AtCBL10-AtCIPK24 system was shown to be involved in the maintenance of Na+ homeostasis in shoots and leaves13. Moreover, reports suggest that AtCIPK24 might also be involved in activating the H+/Ca+ antiporter to maintain the intracellular Ca2+ levels14. The role of AtCBL1-AtCIPK1 complex was identified in ABA-dependent stress responses, whereas AtCBL9-AtCIPK1 complex appeared to modulate ABA-independent stress responses15. AtCBL9-AtCIPK3 complex was shown to be a negative regulator of ABA responses during seed germination16. CIPKs were also thoroughly studied in other plant species such as maize, rice, soybean and sorghum7. The transcript level analysis of ZmCIPK16 in maize seedlings showed its strong upregulation by Polyethylene glycol (PEG), ABA, NaCl, dehydration, heat, and drought17. The expression of PsCIPK gene in Pisum sativum was induced in various abiotic and biotic stresses but not by dehydration and ABA treatments18. A CaCIPK25 gene from chickpea was reported to enhance root growth along with dehydration and salt stress tolerance in transgenic tobacco plants19.
Peanut (Arachis hypogaea) is an important oil crop grown worldwide for its oil and protein. Its productivity is greatly influenced by various environmental stresses. However, studies have shown that its wild relatives exhibit the high level of tolerance to various biotic and abiotic stress conditions20 and Arachis diogoi is one of them. In comparison to others, very little is known about the CIPKs from Arachis species. In the present study, using the available partial cDNA sequence of AdDR-5 (NCBI Accession No. EF371923) identified in a differential gene expression study of Arachis diogoi treated with the fungal pathogen, Phaeoisariopsis personata21, a full length cDNA was cloned and named as AdCIPK5. Its transcript levels were analyzed in Arachis diogoi during various treatments. Tobacco transgenic plants overexpressing AdCIPK5 were checked for tolerance against various stress treatments. These observations are reported in this communication.
Results
Isolation of full length cDNA and sequence analysis
A full length cDNA of 2,031 bp was isolated by using the RACE-PCR approach. Sequence analysis of this revealed the presence of 1,386 bp long ORF region along with 408 bp of 5′ and 237 bp of 3′UTRs in the cDNA (Fig. S1). The ORF encodes a 461 amino acid polypeptide. The encoded protein had a predicted molecular mass of ~52 kDa with the isoelectric point value of 8.75. The SignalP analysis did not show any signal peptide in the deduced amino acid sequence. The amino acid sequence exhibited 62% and 99% similarity with CIPK5 protein from Arabidopsis thaliana and Arachis duranensis, respectively. Hence, this cDNA was designated as AdCIPK5. A Blast analysis revealed the presence of conserved N-terminal activation loop and C-terminal NAF domain in the predicted protein sequence (Fig. S2). Like other CIPKs, the activation loop of AdCIPK5 harbors three conserved amino acids, Serine (S), Threonine (T) and Tyrosine (Y) as possible target sites for phosphorylation by other protein kinases. Phylogenetic analysis showed the presence of two subgroups as intron-rich and intron-less in the phylogenetic tree (Fig. S3). AdCIPK5 was present in intron-less subgroup along with AduCIPK5, GmCIPK25, AtCIPK5, AtCIPK25 and AtCIPK16 as the closest ones.
AdCIPK5 expression analysis
The expression of AdCIPK5 was analyzed in A. diogoi leaves in response to different phytohormones and stress treatments at various time point (Fig. 1). We used four basic phytohormones; Ssalicylic acid (SA), Methyl-jasmonate (MJ), Abscisic acid (ABA) and Ethephon (ET) in this study. These phytohormones are essential for plant growth and development and also mimic various stress conditions22,23. Our analyses showed that SA treatment caused early induction (at 3 h) of AdCIPK5 but the level of expression (around 2-fold) was almost consistent between 3 to 12 h of treatment while it got downregulated later to the ground level at 24 h. It has been known that SA and MJ/ET associated molecular pathways work antagonistically to each other24. Therefore, we also checked the AdCIPK5 expression during both MJ and ET treatments. MJ did not cause any induction whereas some upregulation was observed at 3 h of ET treatment. Interestingly, downregulation in AdCIPK5 expression was observed at later stages of MJ treatment (12 and 24 hr). ABA is a major signaling molecule associated with abiotic stress responses. We observed that ABA treatment caused an early induction of AdCIPK5 expression, which was nearly 8-fold higher at 6 h post-treatment. Even at the later time points (12 and 24 h), the expression remained around 2 to 3-fold higher to the basal level of expression. Compared to the marginal expression caused by other phytohormones, strong upregulation observed during ABA treatment suggests that ABA acts a major regulator of AdCIPK5 expression.
Figure 1.
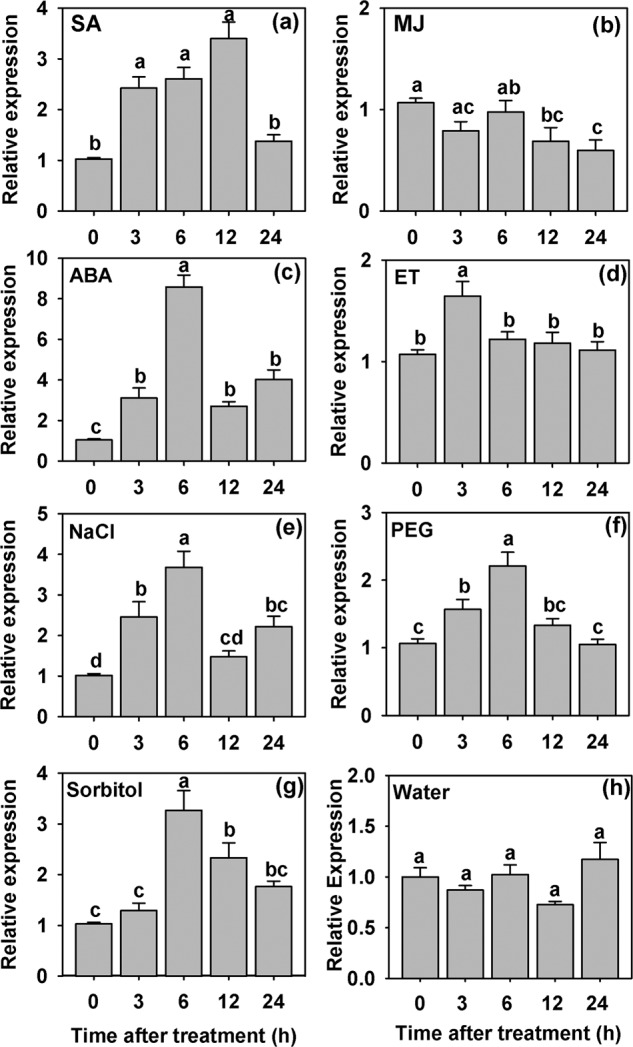
Expression analysis of AdCIPK5 in mature leaf samples of Arachis diogoi under different treatments by qRT-PCR. (a) 500 μM Salicylic acid treatment; (b) 100 μM Methyl jasmonate treatment; (c) 100 μM, Abscisic acid; (d) 250 μM Ethephon treatment; (e) 200 mM NaCl treatment; (f) 10% Polyethylene glycol treatment; (g) 300 mM Sorbitol treatment; (h) water treatment. Error bar represents means ± SE (n = 3), and each replicate comprises 5 leaves. Different letters indicate significant difference (P ≤ 0.05) with each other.
Significant increase in AdCIPK5 expression levels were also observed at different time points under NaCl, PEG and Sorbitol stress treatments. Under salt stress caused by NaCl, the expression level of this gene reached a maximum of around 3-fold higher compared to the initial level whereas PEG and Sorbitol enhanced its expression around 2-fold higher. Interestingly, the maximum expression levels were obtained at 6 h post-treatment, which was similar for all the treatments with the stressor molecules like the ABA treatment. The basal expression level of AdCIPK5 was almost constant at different time points during the control (water) treatment (Fig. 1h).
Subcellular distribution of AdCIPK5
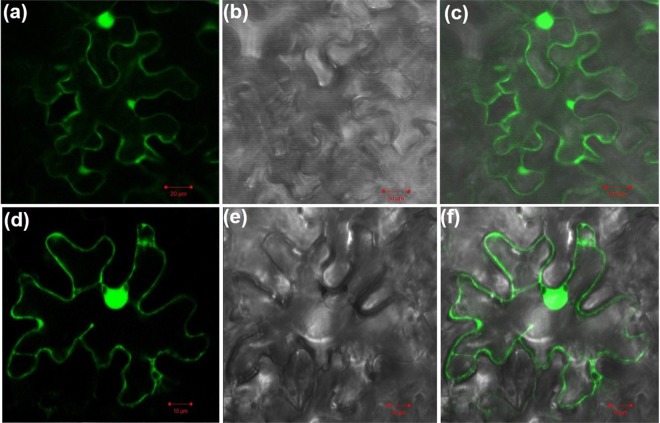
The subcellular localization of AdCIPK5 was studied by constructing a C-terminal translational fusion of green fluorescent protein (GFP) with AdCIPK5 and expressing the fusion gene transiently in Nicotiana benthamiana leaves through Agroinfiltration method. A control vector overexpressing GFP only (35S:GFP) was used as a control. After 48 h incubation at room temperature, the localization of the fusion protein was observed under confocal microscopy. When GFP alone was transiently expressed, the GFP fluorescence was visualized throughout the cell, while the recombinant AdCIPK5:GFP protein was appeared to be localized mainly in nucleus and on the plasma membrane with some fluorescence being observed in cytoplasm as well (Fig. 2).
Figure 2.
Subcellular localization of AdCIPK5. N. benthamiana leaves were infiltrated with Agrobacterium culture harboring empty vector pCAMBIA1302 and recombinant pCAMBIA1302-AdCIPK5 constructs for studying the subcellular localization of AdCIPK5. Leaf cells were visualized under a Confocal Laser Scanning Microscope. (a–c) Control pCAMBIA1302 vector showing GFP throughout the cell. (d–f) pCAMBIA1302-AdCIPK5 recombinant vector showing GFP expression in nucleus and plasma membrane with some presence in cytoplasm.
PCR and RT-PCR analysis of putative transgenic plants
Nine independent primary transgenic tobacco plants showed the expected amplification of 700 bp of nptII and 1383 bp of AdCIPK5, respectively in PCR analysis (Fig. S4). In a semi-quantitative RT-PCR, the putative transgenic plants 1 and 6 showed highest expression level of AdCIPK5 while the transgenic plant 2 was with lowest expression level (Fig. S5a). Actin amplification was used as the internal control. The high expression plants (1 and 6) exhibited the expected 3:1 segregation for resistance to the selection antibiotic (Kanamycin) in T1 generation. Homozygosity of the transgenic lines was checked by 100% seed germination on kanamycin selection medium in T2 generation. Based on these results, line 1 and 6 (high expression) and line 2 (low expression) plants were used for subsequent analyses. The transcript level AdCIPK5 in these three lines and wild type (WT) was also determined in T2 generation (Fig. S5b). In the present work, line 1, 6 and 2 were represented as H1, H2 and Low respectively.
Biotic stress analysis
WT and transgenic T2 plants were used in detached leaf treatments with the plant pathogen Phytophthora parasitica pv nicotianae. This pathogen causes severe damage to tobacco plants and has been used in plant resistance analysis in various studies25,26. We observed the infection symptoms after 2 days post inoculation (dpi) and after 5 dpi, and necrotic areas covered almost complete surface of WT and transgenic leaves (Fig. S6a). The level of resistance was measured by calculating the percentage diseased leaf area (DLA) and by measuring the cell death in leaves caused due to pathogen treatment. Our analysis revealed that around 90% of all the tested leaves were damaged after 5 dpi (Fig. S6b). Further, the Evans blue staining of infected leaves also showed similar cell death in WT and transgenic lines (Fig. S6c) suggesting that the level of AdCIPK5 expression in transgenic tobacco had no significant effect on imparting resistance to the test pathogen in tobacco.
Seed germination analysis
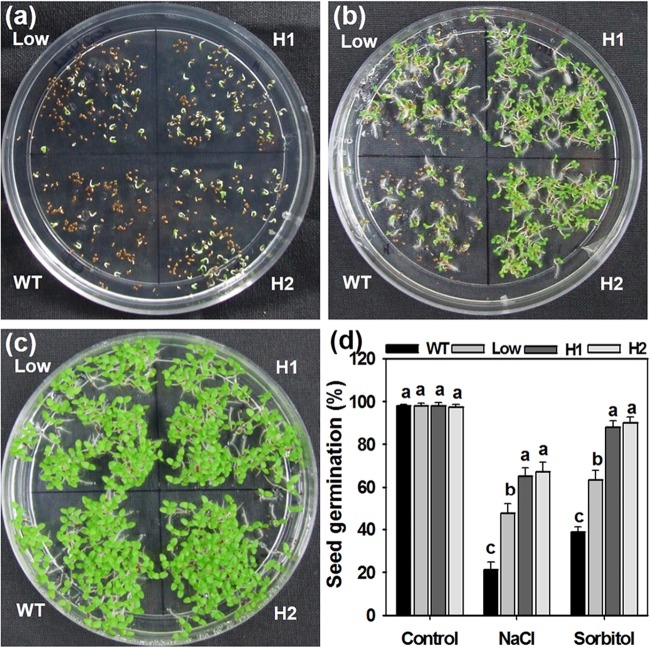
Seeds of the WT and transgenic plants were surface sterilized and plated on half strength MS medium containing 200 mM NaCl and 300 mM Sorbitol (Fig. 3). Medium lacking the stressor agents served as the control. After germination for 7 d, the seeds that developed green cotyledons were scored on the stress media. Out of 150 seeds taken for analysis, almost 98 to 100% seeds germinated on the control plates from all the lines. However, the germination percentage on the medium containing 200 mM NaCl was around 20% for WT while the low expression line exhibited 50% and it was 70% for both the high expression lines H1 and H2. Similarly, 40–45% of WT, 70% of low and almost 90–95% seeds of H1 and H2 germinated on 300 mM Sorbitol containing medium after 7 d treatment. These results demonstrated a higher level of tolerance in transgenic lines against abiotic stress conditions like salt and osmotic stress during the seed germination stage.
Figure 3.
Seed germination assay under salt and osmotic stress. The germination rate of WT and transgenic seeds was detected on a medium supplemented with (a) 200 mM NaCl and (b) 300 mM Sorbitol for 7 d. (c) Control plate without any stress treatment was maintained for the same period of time. (d) Graphical representation of percentage seed germination after 7 d NaCl and osmotic stress treatments. H1 and H2 represent high expression lines whereas Low represents low expression line seedlings. Values are means ± SE (n = 3), and each replicate comprises 150 seeds. Different letters indicate significant difference (P ≤ 0.05) with each other.
Salt stress tolerance at the seedling stage
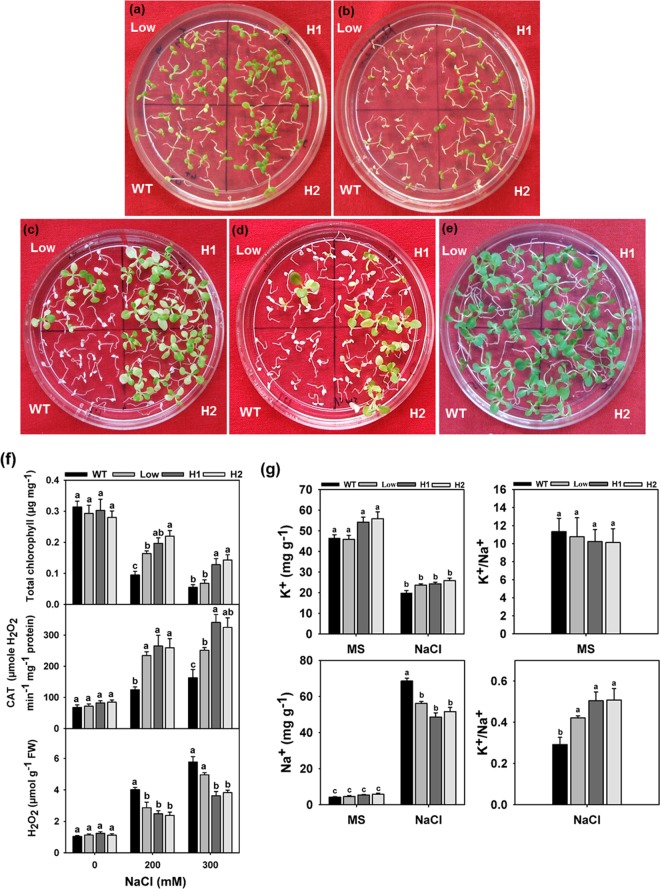
The 10 d old seedlings of the WT and transgenic lines germinated on stress free medium were transferred to different concentrations (200 and 300 mM) of NaCl. After 10 d treatment, WT seedlings from 200 mM NaCl medium displayed higher chlorosis and retarded root elongation compared to the transgenic lines. Chlorosis was also observed in low expression line seedlings, whereas both H1 and H2 developed normally without significant chlorosis (Fig. 4a). After 7 d treatment with 300 mM NaCl, the WT and low expression line seedlings were severely affected (Fig. 4b). However, the high expression lines showed better growth with mild chlorosis. The ability of treated seedlings to recover from stress condition was also checked by transferring them from salt to salt free medium. Considering the severity of damage to the seedlings due to the stress condition, we chose to transfer the seedling after 6 d of treatment from both 200 mM and 300 mM NaCl media to the recovery plates and then observed their recovery for 15 d (Fig. 4c,d). Control seedlings were also maintained simultaneously (Fig. 4e). We observed complete bleaching of WT seedlings on both the recovery plates, whereas the high expression lines from 200 mM media were able to recover with true leaf and root formation while the 300 mM concentration of NaCl appeared to be too high even for the high expression transgenic lines, which was evident from only 30 to 40 percent recovery of H1 and H2 lines.
Figure 4.
Effect of NaCl stress on seedlings growth. The 10 d old seedlings were treated with NaCl. Clear differences were observed between WT and transgenic seedlings after 10 d of 200 mM (a) and 7 d of 300 mM (b) NaCl treatment. Seedlings on NaCl-free medium; recovery (for 15 d) after 6 d treatment with 200 mM and 300 mM NaCl (c,d). (e) Unstressed seedlings were regularly sub-cultured along with the NaCl-treated seedlings. (f) Total chlorophyll content, CAT activity and H2O2 generation was measured after 6 d treatment. Values are means ± SE (n = 3) and each replicate comprises a minimum of 10 seedlings. (g) Measurement of K+ and Na+ content under NaCl treatment after 6 d. The whole seedlings of the WT and transgenic lines were sampled to detect K+ and Na+ contents, and then the ratio of K+ and Na+ was calculated. Values are means ± SE (n = 3), and each replicate comprises minimum 20 seedlings. Different letters indicate significant difference (P ≤ 0.05) with each other. H1 and H2 represent high expression lines whereas Low represents low expression line seedlings.
Further, the estimation of various physiological parameters revealed that the transgenic seedlings retained higher chlorophyll content and catalase (CAT) activity with lower H2O2 generation compared to WT on 200 mM NaCl containing medium. On 300 mM medium, only the high expression lines retained significantly higher chlorophyll content and CAT activity with lower H2O2 generation (Fig. 4f). In addition, the effect of AdCIPK5 overexpression on Na+ and K+ accumulation in the transgenic plants was also estimated during 200 mM salt treatment (Fig. 4g). In the absence of any stress, all the samples showed almost similar Na+ and K+ levels whereas transgenic seedlings from NaCl treatment showed significantly lower Na+ levels compared to WT with no apparent change in the K+ levels. Hence, the transgenic seedlings were able to maintain higher K+/ Na+ ratio on NaCl treatment compared to the control.
Osmotic stress tolerance
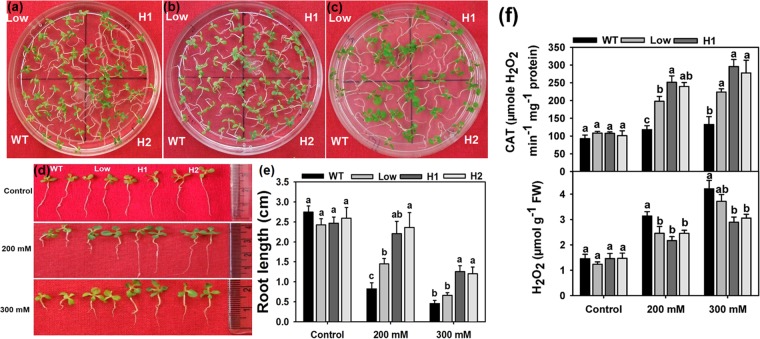
To check the osmotic stress response of the transgenics at seedling stage, the 10 d old WT and transgenic seedlings were transferred to media containing 200 and 300 mM Sorbitol and observations were made after 15 d of treatments with appropriate control (Fig. 5a–c). We observed that the growth of transgenic lines on both 200- and 300-mM sorbitol plates was near normal and better than the WT seedlings. The differences in growth patterns were clearly depicted in Fig. 5d. Further, the measurement and graphical representation of root length of the treated seedlings of all the lines clearly showed that transgenic lines performed better with higher root length compared to the WT under two levels of osmotic stress (Fig. 5e). However, on 300 mM sorbitol, the root length of the low expression line seedlings was also suppressed similar to the WT. Further on, analysis of CAT activity and H2O2 generation revealed that the transgenic seedlings exhibited higher CAT activity with lower H2O2 generation on stress media (Fig. 5f).
Figure 5.
Effect of osmotic stress on seedling growth. The 10 d old WT and transgenic seedlings were transferred to 200 mM (a) and 300 mM (b) Sorbitol medium for 15 days. (c) Seedlings not subjected to stress. (d) Phenotypic difference in the growth of seedlings after 15 d of treatment. (e) Graphical representation of root length measurements. Values are means ± SE (n = 3), and each replicate comprises 15 seedlings. (f) Measurement of CAT activity and H2O2 content after 10 d of stress treatment. Values are means ± SE (n = 3), and each replicate comprises minimum 10 seedlings. Different letters indicate significant difference (P ≤ 0.05) between each other. H1 and H2 represent high expression lines whereas Low represents low expression line seedlings.
Leaf Disc assay for salt and osmotic stress
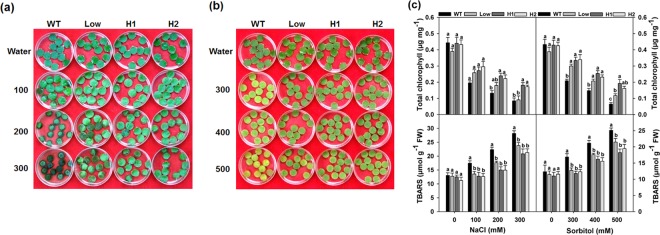
To assess the stress tolerance at mature leaf stage, a leaf disc assay was performed using different concentrations of NaCl and Sorbitol. Leaf discs from 7-week-old plants were used in both the treatments. NaCl tolerance in the discs of transgenic plants was observed after 3 d treatment (Fig. 6a). Chlorophyll estimation revealed a dose dependent loss of total chlorophylls in the leaf discs of the WT plants compared with those from the transgenic plants, which were able to retain significantly higher total chlorophyll content (Fig. 6c). However, a non-significant difference was observed between WT and low line plants on the 300 mM medium. Similarly, the levels of Thiobarbituric acid reactive substances (TBARS) increased in the WT compared to the transgenic lines. In case of Sorbitol treatment, chlorosis started appearing after 3 d treatment (data not shown), which became more prominent after 5 d treatment in WT plants with increasing concentration of Sorbitol, whereas the leaf discs of transgenic plants showed reduced chlorosis (Fig. 6b). Leaf discs from distilled water treatment remained green in both WT and transgenic plants. Further, the chlorophyll content and lipid peroxidation levels (as evidenced by TBARS) of the leaf discs after 4 d treatment confirmed the observed phenotypic differences (Fig. 6c).
Figure 6.
Leaf disc assay. (a) Stress tolerance exhibited by the leaf discs of WT and transgenic lines at various concentrations of NaCl (100, 200 and 300 mM) and (b) sorbitol 300, 400 and 500 mM treatments. Photographs were taken post 3 d NaCl and 5 d Sorbitol treatments. (c) Total chlorophyll and TBARS were measured in WT and transgenic leaf discs after 2 d NaCl treatment and after 4 d Sorbitol treatment. Values are means ± SE (n = 3), and each replicate comprises minimum 15 leaf discs. Different letters indicate significant difference (P ≤ 0.05) between each other. H1 and H2 represent high expression lines whereas Low represents low expression line seedlings.
ROS analysis
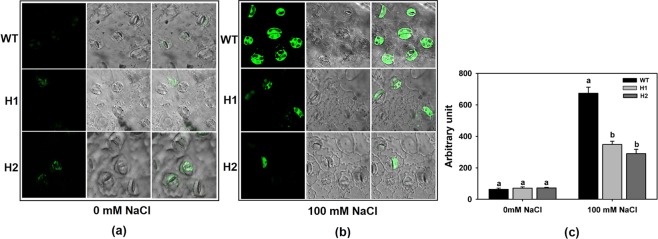
Both the high expression lines H1 and H2 along with the WT were used in the analysis of ROS generation (Fig. 7). In untreated control samples (without NaCl), the observed fluorescence due to the formation of Reactive Oxygen Species (ROS) as evidenced by the staining with 2ʹ,7ʹ-Dihydrodichlorofluorescein diacetate (H2DCFDA) in stomatal guard cells was minimal and more or less similar in WT and transgenic tobacco plants. Increased fluorescence as indicative of enhanced H2O2 formation was observed in guard cells of WT and transgenic plants in NaCl (100 mM) treatment. However, the fluorescence displayed by the transgenic plants was significantly less compared to the WT plants, which implied that there was reduced ROS formation in transgenic guard cells under sodium chloride stress.
Figure 7.
ROS detection in leaf epidermal guard cells using Confocal Microscopy. Fluorescence levels in WT and high expression line plants (H1 and H2) were observed after (a) control and (b) 100 mM NaCl treatments by staining with H2DCFDA. WT samples showed higher ROS accumulation during NaCl stress. Bright field images were also displayed. (c) Quantification of ROS production by using Image-J software. Values are means ± SE (n = 3), and each replicate comprises 100 stomata. Different letters indicate significant difference (P ≤ 0.05) between each other.
Transcript level analysis of stress related genes
To understand further the relative effect of AdCIPK5 overexpression in salt and osmotic stress tolerance, the expression of different stress-related genes was analyzed in WT and the transgenic lines (Low and H1) with and without stress treatment (Fig. 8). The transcript level of six genes (NtCAT, NtERD10C, NtERD10D, NtNCED1, NtSus1 and NtSOS1), which were reported to be involved in response to abiotic stress were analyzed. The 10 d old WT and transgenic seedlings grown in Petri dishes were exposed to salt (200 mM NaCl) and Sorbitol (300 mM) treatments for 3 d followed by a real time analysis of the expression of the above mentioned genes. The results showed that all stress-responsive genes analyzed were significantly induced in the high expression line compared to the control plants when exposed to salt and sorbitol treatments suggesting that AdCIPK5 overexpression in tobacco has been associated with the upregulated expression of stress-related genes under salt and osmotic stress conditions.
Figure 8.
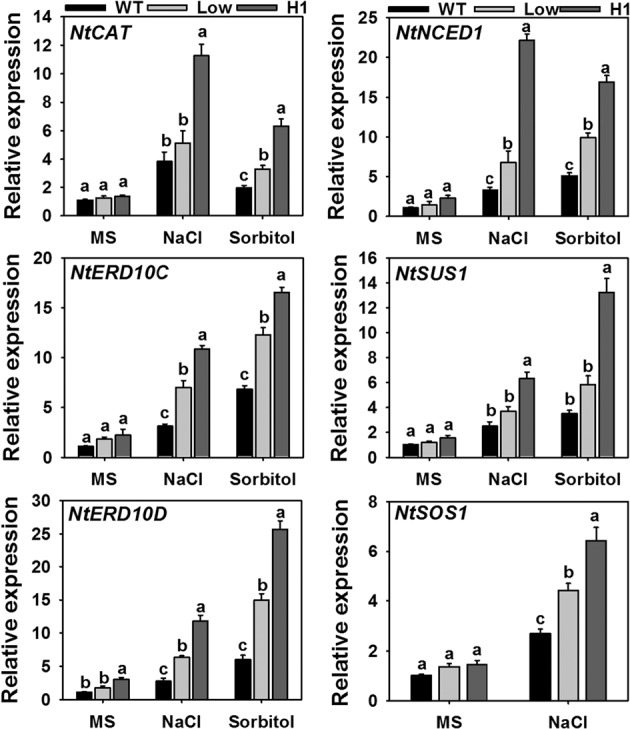
Transcript level analysis of stress-related genes in plants under NaCl and osmotic stress conditions. The 10 d old seedlings of WT and transgenic lines (low and H1) were exposed to NaCl treatment (200 mM NaCl) and osmotic stress treatment (300 mM Sorbitol) for 3 d followed by real time expression analysis of stress related genes. Values are means ± SE (n = 3), and each replicate comprises 5 seedlings. Different letters indicate significant difference (P ≤ 0.05) between each other.
Discussion
In the present investigation, a full-length cDNA of AdCIPK5 gene that was identified in differential gene expression analysis was cloned using the sequence of a partial cDNA from a wild peanut, Arachis diogoi21 using RACE-PCR approach. The analysis of deduced AdCIPK5 protein sequence showed the presence of highly conserved motifs such as the activation loop and NAF domain, which are typical of the CIPK family and are important for their plant function1,10. Phylogenetic analysis revealed the close homology of AdCIPK5 with AduCIPK5 of Arachis duranensis, GmCIPK5 of Glycine max and AtCIPK5, AtCIPK25, and AtCIPK16 of Arabidopsis thaliana. The Arabidopsis CIPKs exhibited roles in the plant developmental processes, salt stress responses and fungal infection27–29.
In the present study, we checked the quantitative expression of AdCIPK5 in treatment with different phytohormones and stress inducers. The analysis showed that, the transcript levels were upregulated differentially at least at some point during the SA, ABA and ET treatments in A. diogoi leaves except MJ treatment. These hormones are regarded as essential components of plant adaptation system, but their responses are governed by complex interlinked mechanisms22,23. For example, during the pathogen attack, SA is associated with strong immune responses while JA/ET pathways act antagonistically to SA24,30. Similarly, ABA has long been known for its role in abiotic stress, but recent studies suggest that it can also regulate plant immunity in association with other hormones31. Since these phytohormones regulate the different sets of mechanisms, AdCIPK5 induction can be assumed to be associated with multiple signaling pathways in the plant. Previous findings on CIPKs have confirmed their potential role in the abiotic stress tolerance7. Therefore, under stress conditions such as NaCl, Sorbitol and PEG, we examined the expression profile of AdCIPK5. During the treatments, the enhanced expression of AdCIPK5 was observed in all the stress treatments, which suggested that the gene is positively involved in abiotic stress tolerance mechanisms.
CIPKs could be located at different sites within the cell9. For example, wheat proteins TaCIPK14 and TaCIPK29 have been observed across cells32,33. AtCIPK1 from Arabidopsis was found to be localized to the plasma membrane and to some extent also to the nucleus and cytosol, which was further recruited to the plasma membrane after interacting with AtCBL1 and AtCBL915. Similarly, the AtCIPK5 was shown to be dynamically translocated from cytoplasm to tonoplast34. The analysis of the amino acid sequence revealed that the AdCIPK5 had no recognizable localization signal that was consistent with previous CIPKs findings1. However, with some presence in cytoplasm, the AdCIPK5:GFP fusion protein appeared to be located mainly in the nucleus and plasma membrane. Hence, it could be speculated that the role of AdCIPK5 is not limited to a specific organelle and that localization may depend on specific interacting partners.
As an active participant of Ca+2 signaling pathways, CIPKs play crucial roles in plant responses during the microbe and other pathogen attacks. An earlier study on tomato CIPK6/CBL10 complex reported its involvement in immune responses by generating ROS and ultimately leading to the programmed cell death in the plant35. Similarly, a TaCBL4/TaCIPK5 complex is involved in modulation of fungal resistance in wheat36. As elaborated in introduction, the partial cDNA sequence of AdCIPK5 was initially identified during the fungal infection21. Further, in this study, the observed induction of AdCIPK5 expression in response to SA, ABA, and ET suggested that this gene could be involved in plant defense mechanisms. Therefore, the transgenic tobacco plants overexpressing AdCIPK5 were raised and their resistance was analyzed along with the WT in treatment with an oomycetes pathogen, P. nicotianae. Detached leaves were inoculated with the pathogen. The level of resistance in WT and transgenic leaves was determined by calculating the percentage diseased leaf area and by measuring the cell death caused by the P. nicotianae infection. No significant differences were observed in our analyses between WT and transgenic leaves. These findings suggested that the expression of AdCIPK5 did not enhance the pathogen resistance in tobacco transgenic plants. It is possible that the AdCIPK5 role is limited in pathogen resistance mechanisms with some unknown function. Likewise, rice OsCIPK14/15 also displayed a wide range of defence responses in cultured cells, but the adult plant showed no resistance to blast fungus37.
The functions of CIPKs in abiotic stress conditions have been widely studied. In Arabidopsis several CIPKs have been shown for their roles in regulating the stress responses2,5. In rice, overexpression of OsCIPK3, OsCIPK12 and OsCIPK15 significantly improved cold, drought and salt tolerance respectively38. Transgenic tobacco plants overexpressing TaCIPK14 and TaCIPK29 from wheat also conferred single or multiple stress tolerance32,33.
In the present investigation, we observed that the AdCIPK5 transcript levels were upregulated during the abiotic stress treatments in the wild peanut. Therefore, using NaCl and Sorbitol, the WT and transgenic plants were tested for salt and osmotic stresses. The initial analysis was carried out at the stage of seed germination, which is considered to be the most crucial phase for the establishment of the plant and its final yield. Significantly, higher percentages of transgenic seed germination were recorded on both 200 mM of NaCl and 300 mM of Sorbitol media compared to WT indicating enhanced tolerance to both salt and osmotic stresses in tobacco transgenic plants expressing AdCIPK5. These transgenic tobacco lines were also able to perform better on both 200 and 300 mM NaCl containing medium at the seedling stage compared to the WT plants. However, the concentration of 300 mM of NaCl seemed to be not tolerable even for the transgenic plants. It was also confirmed on a medium of recovery where only 30–40 percent of high-expression seedlings could recover from NaCl stress of 300 mM, whereas most of them could recover from 200 mM level of stress. Similarly, during osmotic stress treatment, transgenic lines exhibited better growth patterns compared to their non-transformed WT counterparts. The transgenic seedlings showed higher root length than the WT on both 200 and 300 mM of Sorbitol stress media. However, a non-significant difference was observed between seedlings of WT and low-expression line on 300 mM medium.
Various physiological parameters were recorded to confirm the enhanced levels of abiotic stress tolerance exhibited by the transgenic plants ectopically expressing AdCIPK5. Both NaCl and osmotic stresses affect nearly all plant growth and developmental stages resulting in the generation of excessive reactive oxygen species (ROS) that can cause damage by oxidizing various vital cell components such as proteins, lipids and DNA39. To survive under the NaCl stress, plants must sequester to an appropriate cellular compartment or expel the excessive Na+ and therefore, maintain a higher K+ level, which ultimately helps in scavenging higher ROS levels40,41. In comparison to the WT under NaCl stress, the AdCIPK5 transgenic seedlings showed better ability to retain higher K+/Na+ ratio. This observation was supported by other parameters like higher chlorophyll content, higher CAT activity with reduced levels of H2O2 in the transgenic plants. These parameters are important in assessing the ability of the plant to sustain itself against the impending stresses using the antioxidant defence mechanisms42. Similarly, with increased CAT activity and lower H2O2 content, the AdCIPK5 transgenic seedlings maintained better growth during the osmotic stress conditions also. Furthermore, the WT and transgenic plants were also evaluated against stress conditions at a mature stage. For this purpose, leaf discs from seven week-old pot grown plants were treated with different NaCl and Sorbitol concentrations. Total chlorophyll content and TBARS values were analyzed in all the treated samples. Analysis of TBARS is used to assess the ROS-mediated membrane damage43. Transgenic lines were able to retain higher chlorophyll content compared to WT plants during both the treatments with significantly reduced TBARS levels. This clearly indicated reduced lipid peroxidation and membrane damage in AdCIPK5 overexpression lines that encountered stress treatments. Direct ROS level measurement is used as an effective index to evaluate multiple stress tolerance in plants42. For example, higher ROS levels were recorded under salt stress in Arabidopsis Atcipk24 mutant while ZmCIPK21 overexpressing lines in maize exhibited lower levels of ROS compared to the control plants44,45. In our study, staining with a ROS sensitive fluorescent dye, H2DCFDA was used during NaCl treatment to monitor the visible differences in ROS levels in stomatal guard cells. The results showed that there was significantly reduced fluorescence in tobacco transgenic plants, which could be associated with the constitutive expression of AdCIPK5. No major differences in control samples were observed. Taken together, these observations clearly indicate that AdCIPK5 functions in a pathway that controls ROS detoxification in response to NaCl and osmotic stresses.
To understand the function of AdCIPK5 in regulation of gene expression under NaCl and osmotic stresses, the transcript levels of several stress-related marker genes were evaluated in transgenic plants expressing AdCIPK5 ectopically. Results clearly showed that the expression of genes NtERD10C, NtERD10D, NtNCED1, NtSus1, NtCAT and NtSOS1 was significantly higher in the high expression line plants compared with the WT under NaCl stress. These genes have been reported earlier to contribute to mechanisms of plant stress tolerance. Both NtERD10C and NtERD10D encode LEA proteins that act as macromolecule stabilizers and membrane protectants while NtNECD1 is an important gene of the ABA biosynthesis pathway46,47. In stress conditions, SUS1 plays a crucial role in osmotic maintenance48. CAT is a catalase that assists the cell in scavenging excessive H2O249. SOS1 encodes a putative Na+/H+ antiporter that modulates the ion movement across the membrane and is involved in the mechanism of salt tolerance12. Similarly, the higher expressions of NtERD10C, NtERD10D, NtNCED1, NtSus1 and NtCAT were also observed during osmotic stress conditions. These findings suggest that by modulating the expression of stress-related genes, AdCIPK5 maintains the cellular homeostasis in NaCl and osmotic stress conditions. Further studies are required for further insights into the mechanism by which AdCIPK5 regulates the associated genes with stress responses.
In conclusion, AdCIPK5 is clearly identified as a stress-responsive gene by our results. The gene was reported during the fungal infection but our analysis showed its involvement in abiotic stress tolerance. This can be explained on the basis of previous findings suggesting that CIPKs are not very specific to their role. By forming different sets of CBL-CIPK complex, they can participate in different stress tolerance mechanisms5,50. Despite extensive studies of the CBL/CIPK network in Arabidopsis and other plants, their identities and roles in Arachis diogoi are largely unknown. Therefore, the identification of AdCIPK5 target proteins will provide more insights in understanding its regulatory mechanism in response to NaCl and osmotic stresses. The high similarities between AdCIPK5 and the CIPK5 of Arachis duranensis whose genome has been sequenced recently51 would help unravel the AdCIPK5 interacting partners in peanuts in future investigations. Taken together, AdCIPK5 overexpression conferred tolerance to NaCl and osmotic stresses at various stages of growth in transgenic tobacco. To our best knowledge, this is the first report of the functional characterization of any CIPK gene from Arachis spp. We believe that our results would help in understanding the role that CIPK family proteins play in plant stress management.
Materials and Methods
Plant materials and treatments
Wild peanut (Arachis diogoi, ICG8962) and tobacco (Nicotiana tabacum var Samsun) plants were maintained in the green house. Detached leaves of A. diogoi were utilized for different treatments, and the experiments were performed essentially as described earlier52. The leaves were used in various treatments, 500 μM salicylic acid (SA), 100 μM methyl jasmonate (MeJA), 100 μM abscisic acid (ABA), 250 μM ethephon, 200 mM NaCl, 300 mM Sorbitol and 10% PEG, while the treatment with water served as a control. Samples were collected at regular intervals, quick-frozen in liquid nitrogen, and stored at −80 °C.
RACE and isolation of full length cDNA
Rapid amplification of cDNA ends (RACE) reaction was performed to obtain full length cDNA of AdCIPK5 from the partial sequence identified earlier21 by using SMARTer™ rapid amplification of cDNA ends (RACE) Kit (Clontech, USA) following the manufacturer’s instructions. All the reactions were performed using hot-start DNA polymerase provided along with the kit. The gene specific primers (GSPs) used for 5′/3′ RACE-PCR reaction were provided in Table S1. The full-length cDNA sequence of AdCIPK5 was obtained by aligning 5′/3′ RACE products and the partial AdDR-13 cDNA sequences. The open reading frame (ORF) of the AdCIPK5 sequence was amplified with ORF-F1 and ORF-R1 primers (Table S1) by using Phusion™ high fidelity DNA polymerase (Finnzymes, NEB, UK). The amplified PCR products were cloned in pTZ57R/T vector and sequenced commercially for the confirmation.
Analysis of cDNA and protein sequence
Basic sequences were analyzed and compared by using BLASTn and BLASTp similarity searches. ExPASy tools were used for nucleotide translations, isoelectric point prediction and molecular mass calculations. Multiple sequence alignment and phylogenetic tree construction were performed using ClustalW and MEGA 6 software respectively. Signal peptide prediction was done using SignalP 4.1.
Real time gene expression analysis
The qRT-PCR reaction was performed essentially as described earlier50. Alcohol dehydrogenase class III (adh3) and Ubiqutin (Ubq) genes were used as internal controls for Arachis diogoi and tobacco samples respectively53,54. The ΔΔCT method was used for the estimation of relative fold change in RNA expression. Primers used in this study were provided in Table S2.
AdCIPK5 subcellular localization analysis
The AdCIPK5 ORF was reamplified by using ORF-F1 and ORF-R1 primers with NcoI and SpeI restriction sites respectively. The amplified product was digested and cloned in binary vector pCAMBIA 1302, digested with the same set of enzymes. Further, the confirmed recombinant vector was mobilized into Agrobacterium strain, EHA105 using the standard freeze thaw method. Similarly, the empty 1302 vector was also mobilized into the same Agrobacterium strain. Nicotiana benthamiana leaves were used for agroinfiltration21. The GFP expression was visualized in infiltrated leaves using laser scanning confocal microscopy (Leica TCS SP2 with Leica DM6000 microscope).
Construct preparation and tobacco transformation
The AdCIPK5 ORF was reamplified with ORF-F2 and ORF-R2 primers harboring ApaI and KpnI restriction sites respectively and cloned in pTZ57R/T vector. Digested fragments were cloned in pRT100 plant expression vector at corresponding sites. After confirmation, the expression cassette of AdCIPK5 was isolated using HindIII enzyme and cloned in pCAMBIA2300 vector. The confirmed binary vectors were mobilized into Agrobacterium strain EHA105. Tobacco (Nicotiana tabacum cv Samsun) was transformed through the standard leaf disc method.
Molecular analysis of transgenic plants
Transformants were raised on half strength MS media supplemented with 125 mg/l Kanamycin. Plantlets with well-developed root system were shifted to soil cups for acclimatization and later allowed to grow in green house for maturation and seed collection. Putative transgenic plants were screened by PCR for the presence of AdCIPK5 and nptII transgenes using gene-specific primers. The transcript level of AdCIPK5 was checked in different lines by using semi-quantitative RT-PCR reactions. Mendelian segregation analysis was performed for Kanamycin resistance and sensitive seedlings at seed germination stage.
Evaluation of fungal resistance in transgenic plants
The fully expanded detached leaves from transgenic and WT control plants were used for the anti-fungal bioassay in T2 generation. Fungal resistance was checked against the pathogen, Phytophthora parasitica pv nicotianae, which causes black shank disease on tobacco. Abrasions were made on the adaxial surface of the leaves and actively growing fungus along with potato dextrose agar block (0.5 cm2) was inoculated on abraded areas. The lesions were observed regularly, photographed after 5 days post-inoculation (dpi) and the percentage diseased leaf areas (DLA) were calculated as described earlier55. Additionally, cell death was quantified using the Evans blue dye56. In brief, equal size of the leaf discs (1 cm2) were cut out from WT and transgenic leaves post 5 dpi and submerged into 0.25% (w/v) Evans blue solution for 30 mints. The stained discs were washed with Mili-Q water to remove the unbound dye. Further, the discs were ground in 1% SDS solution and centrifuged at 12000 rpm for 10 min. The supernatant was collected and optical density (OD) was measured at 600 nm of wavelength for cell death measurement.
Seed germination assay
Mature WT and transgenic seeds were surface sterilized with 4% sodium hypochlorite solution and washed properly with sterile distilled water. From each T2 generation transgenic lines, around 150 seeds were taken and transferred to two separate sets of half strength MS media without organics supplemented with 200 mM NaCl and 300 mM Sorbitol separately. Germination was observed regularly and graphs were plotted after 7 d in both treatments. Control plates without any stress agent were also maintained simultaneously.
Seedling assay
Surface sterilized transgenic seeds were grown on 125 mg/l Kanamycin for 10 d. Simultaneously WT seeds were grown on medium without Kanamycin and maintained at 27 ± 1 °C with a photoperiod of 16 h light and 8 h dark. The seedlings from each of the transgenic plants along with the WT were used in different stress treatments, 200 and 300 mM each of NaCl and sorbitol.
Leaf disc assay
Leaf discs from the leaves of 7-week-old potted WT and transgenic tobacco plants were used in stress treatments with 100, 200 and 300 mM concentrations of NaCl and 300, 400 and 500 mM concentration of Sorbitol. The treatments were carried out in continuous white light at 27 ± 1 °C until visible differences were observed among the lines.
Catalase (CAT) and H2O2 measurement
The 10 d old WT and transgenic seedlings were transferred to media with different concentrations of NaCl and Sorbitol. In the case of NaCl treatment of the transgenics, both CAT and H2O2 were measured after 6 d, while these measurements were made after 10 d of treatment in the case of Sorbitol. The CAT activity was measured spectrophotometrically by following the oxidation of H2O2 at 240 nm57. The reaction mixture contained 50 mM sodium phosphate buffer (pH 7.0), 20 mM H2O2 and enzyme extract equivalent to 10 µg protein in a final volume of 1 ml. Δε for H2O2 at 240 nm was 43.6 mM−1 cm−1.
For H2O2 measurement, 150 mg seedlings samples (with and without NaCl treatment) were homogenized in 5 ml cold acetone and centrifuged at 1,250 g. Activated carbon was used for the adsorption of chlorophyll contents. The supernatant (200 µl) was added to 1 ml reaction buffer (0.25 mM FeSO4, 0.25 mM (NH4)2SO4, 25 mM H2SO4, 1.25 mM Xylenol orange, and 1 mM Sorbitol) at room temperature for 1 h. The H2O2 levels were quantified at the 560 nm absorbance as described earlier58.
Quantification of Na+ and K+ ion
Na+ and K+ ions were quantified by referring to the methods described by Deng et al.33. and ion estimation was performed using an inductively coupled Plasma Atomic Emission Spectrometer. The 10 d old WT and transgenic seedlings were transferred to a medium supplemented with 200 mM NaCl for 6 d. A NaCl-free control plate was maintained simultaneously.
Total chlorophyll and TBARS measurement
Total chlorophyll content and lipid peroxidation were measured by following Arnon59 and, Heath and Packer60 respectively. In seedling experiments, a 50 mg of sample was used from each line for chlorophyll estimation, while 100 mg tissue was used for both chlorophyll and TBARS analysis in leaf discs experiments.
ROS detection
By using the epidermal peels from the abaxial surface of fully expanded WT and transgenic leaves, ROS levels were measured using the fluorescence substrate H2DCFDA and confocal microscopy (kex-488 nm and kem-530 nm). The experiment was repeated thrice with at least n = 100 stomata. Fluorescence quantification was performed using Image-J 1.42 software by selecting appropriate pigmentation areas.
Statistical analysis
Statistical analyses were conducted using SIGMASTAT version 11.0. One-way ANOVA (Duncan’s Multiple Range Test) was performed to confirm the variability of results and to determine significant differences between treatment groups. P ≤ 0.05 was considered significantly different with each other and were represented by different letters.
Supplementary information
Acknowledgements
The authors are thankful to DBT-CREBB, DST-FIST, UGC-SAP, for extending the facilities to the Department of Plant Sciences, University of Hyderabad. The.pngt of the Arachis diogoi seed sample from the International Crops Research Institute for Semi-Arid Tropics (ICRISAT), Patancheru, India is also acknowledged.
Author contributions
Conceived and designed the experiments: Naveen Kumar Singh and P.B. Kirti. Performed the experiments: Naveen Kumar Singh. Analyzed the data: Naveen Kumar Singh and Pawan Shukla. Contributed reagents/materials/analysis tools: Naveen Kumar Singh and Pawan Shukla. Wrote the manuscript: Naveen Kumar Singh and P.B. Kirti.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-57383-x.
References
- 1.Kolukisaoglu Ü, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant physiology. 2004;134:43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luan S. The CBL–CIPK network in plant calcium signaling. Trends in Plant Science. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. Biochemical Journal. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 4.Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. The Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R, et al. Functions and mechanisms of the CBL–CIPK signaling system in plant response to abiotic stress. Progress in Natural Science. 2009;19:667–676. doi: 10.1016/j.pnsc.2008.06.030. [DOI] [Google Scholar]
- 6.Liu J, Zhu J-K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 7.Yu Q, An L, Li W. The CBL–CIPK network mediates different signaling pathways in plants. Plant cell reports. 2014;33:203–214. doi: 10.1007/s00299-013-1507-1. [DOI] [PubMed] [Google Scholar]
- 8.Sanyal SK, Pandey A, Pandey GK. The CBL–CIPK signaling module in plants: a mechanistic perspective. Physiologia plantarum. 2015;155:89–108. doi: 10.1111/ppl.12344. [DOI] [PubMed] [Google Scholar]
- 9.Batistic O, Kudla J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta. 2004;219:915–924. doi: 10.1007/s00425-004-1333-3. [DOI] [PubMed] [Google Scholar]
- 10.Albrecht V, Ritz O, Linder S, Harter K, Kudla J. The NAF domain defines a novel protein–protein interaction module conserved in Ca2+−regulated kinases. The EMBO journal. 2001;20:1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halfter U., Ishitani M., Zhu J.-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proceedings of the National Academy of Sciences. 2000;97(7):3735–3740. doi: 10.1073/pnas.97.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H., Ishitani M., Kim C., Zhu J.-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences. 2000;97(12):6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BG, et al. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. The Plant Journal. 2007;52:473–484. doi: 10.1111/j.1365-313X.2007.03249.x. [DOI] [PubMed] [Google Scholar]
- 14.Cheng N-H, Pittman JK, Zhu J-K, Hirschi KD. The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. Journal of Biological Chemistry. 2004;279:2922–2926. doi: 10.1074/jbc.M309084200. [DOI] [PubMed] [Google Scholar]
- 15.D’Angelo C, et al. Alternative complex formation of the Ca2+−regulated protein kinase CIPK1 controls abscisic acid‐dependent and independent stress responses in Arabidopsis. The Plant Journal. 2006;48:857–872. doi: 10.1111/j.1365-313X.2006.02921.x. [DOI] [PubMed] [Google Scholar]
- 16.Pandey GK, Grant JJ, Cheong YH, Kim B-G, Luan S. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Molecular Plant. 2008;1:238–248. doi: 10.1093/mp/ssn003. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, et al. Cloning and characterization of a novel CBL-interacting protein kinase from maize. Plant molecular biology. 2009;69:661–674. doi: 10.1007/s11103-008-9445-y. [DOI] [PubMed] [Google Scholar]
- 18.Mahajan S, Sopory SK, Tuteja N. Cloning and characterization of CBL‐CIPK signalling components from a legume (Pisum sativum) The FEBS journal. 2006;273:907–925. doi: 10.1111/j.1742-4658.2006.05111.x. [DOI] [PubMed] [Google Scholar]
- 19.Meena, M. K., Ghawana, S., Dwivedi, V., Roy, A. & Chattopadhyay, D. Expression of chickpea CIPK25 enhances root growth and tolerance to dehydration and salt stress in transgenic tobacco. Frontiers in plant science6 (2015). [DOI] [PMC free article] [PubMed]
- 20.Pande S, Rao JN. Resistance of wild Arachis species to late leaf spot and rust in greenhouse trials. Plant Disease. 2001;85:851–855. doi: 10.1094/PDIS.2001.85.8.851. [DOI] [PubMed] [Google Scholar]
- 21.Kumar KRR, Kirti PB. Differential gene expression in Arachis diogoi upon interaction with peanut late leaf spot pathogen, Phaeoisariopsis personata and characterization of a pathogen induced cyclophilin. Plant molecular biology. 2011;75:497–513. doi: 10.1007/s11103-011-9747-3. [DOI] [PubMed] [Google Scholar]
- 22.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant molecular biology. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 23.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Current opinion in plant biology. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Dong X. SA, JA, ethylene, and disease resistance in plants. Current opinion in plant biology. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 25.Li Jb, Luan Ys, Liu Z. Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiologia plantarum. 2015;155:248–266. doi: 10.1111/ppl.12315. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, et al. Stomatal closure and SA-, JA/ET-signaling pathways are essential for Bacillus amyloliquefaciens FZB42 to restrict leaf disease caused by Phytophthora nicotianae in Nicotiana benthamiana. Frontiers in microbiology. 2018;9:847. doi: 10.3389/fmicb.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huibers RP, de Jong M, Dekter RW, Van den Ackerveken G. Disease-specific expression of host genes during downy mildew infection of Arabidopsis. Molecular plant-microbe interactions: MPMI. 2009;22:1104–1115. doi: 10.1094/mpmi-22-9-1104. [DOI] [PubMed] [Google Scholar]
- 28.Schmid Markus, Davison Timothy S, Henz Stefan R, Pape Utz J, Demar Monika, Vingron Martin, Schölkopf Bernhard, Weigel Detlef, Lohmann Jan U. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 29.Roy SJ, et al. A novel protein kinase involved in Na+ exclusion revealed from positional cloning. Plant, cell & environment. 2013;36:553–568. doi: 10.1111/j.1365-3040.2012.02595.x. [DOI] [PubMed] [Google Scholar]
- 30.Zander M, Chen S, Imkampe J, Thurow C, Gatz C. Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Molecular Plant. 2012;5:831–840. doi: 10.1093/mp/ssr113. [DOI] [PubMed] [Google Scholar]
- 31.Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends in Plant Science. 2009;14:310–317. doi: 10.1016/j.tplants.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Deng X, et al. TaCIPK29, a CBL-interacting protein kinase gene from wheat, confers salt stress tolerance in transgenic tobacco. PloS one. 2013;8:e69881. doi: 10.1371/journal.pone.0069881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng X, et al. Ectopic expression of wheat TaCIPK14, encoding a calcineurin B‐like protein‐interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiologia plantarum. 2013;149:367–377. doi: 10.1111/ppl.12046. [DOI] [PubMed] [Google Scholar]
- 34.Schlücking K, et al. A New β-Estradiol-Inducible Vector Set that Facilitates Easy Construction and Efficient Expression of Transgenes Reveals CBL3-Dependent Cytoplasm to Tonoplast Translocation of CIPK5. Molecular Plant. 2013;6:1814–1829. doi: 10.1093/mp/sst065. [DOI] [PubMed] [Google Scholar]
- 35.de la Torre F, et al. The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. The Plant Cell Online. 2013;25:2748–2764. doi: 10.1105/tpc.113.113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, P. et al. The Calcium Sensor TaCBL4 and its Interacting Protein TaCIPK5 are Required for Wheat Resistance to Stripe Rust Fungus. Journal of experimental botany (2018). [DOI] [PubMed]
- 37.Kurusu T, et al. Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant physiology. 2010;153:678–692. doi: 10.1104/pp.109.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, Huang Y, Xiong L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant physiology. 2007;144:1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mittler Ron, Vanderauwera Sandy, Gollery Martin, Van Breusegem Frank. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9(10):490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Archives of biochemistry and biophysics. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 41.Zhu, J.-K. In Trends in Plant Science Vol. 6 66–71 (2001). [DOI] [PubMed]
- 42.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant physiology and biochemistry. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Iturbe-Ormaetxe I, Escuredo PR, Arrese-Igor C, Becana M. Oxidative damage in pea plants exposed to water deficit or paraquat. Plant physiology. 1998;116:173–181. doi: 10.1104/pp.116.1.173. [DOI] [Google Scholar]
- 44.Zhu J, et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Molecular and cellular biology. 2007;27:5214–5224. doi: 10.1128/MCB.01989-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, et al. ZmCIPK21, a maize CBL-interacting kinase, enhances salt stress tolerance in Arabidopsis thaliana. International journal of molecular sciences. 2014;15:14819–14834. doi: 10.3390/ijms150814819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amara I, et al. Insights into late embryogenesis abundant (LEA) proteins in plants: from structure to the functions. American Journal of Plant Sciences. 2014;5:3440. doi: 10.4236/ajps.2014.522360. [DOI] [Google Scholar]
- 47.Taylor IB, Burbidge A, Thompson AJ. Control of abscisic acid synthesis. Journal of experimental botany. 2000;51:1563–1574. doi: 10.1093/jexbot/51.350.1563. [DOI] [PubMed] [Google Scholar]
- 48.Rosa M, et al. Soluble sugars: Metabolism, sensing and abiotic stress: A complex network in the life of plants. Plant signaling & behavior. 2009;4:388–393. doi: 10.4161/psb.4.5.8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma, P., Jha, A. B., Dubey, R. S. & Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. Journal of Botany2012 (2012).
- 50.Chen X-f, GU Z-M, Feng L, Zhang H-S. Molecular analysis of rice CIPKs involved in both biotic and abiotic stress responses. Rice Science. 2011;18:1–9. doi: 10.1016/S1672-6308(11)60001-2. [DOI] [Google Scholar]
- 51.Bertioli, D. J. et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nature genetics (2016). [DOI] [PubMed]
- 52.Singh NK, Kumar KRR, Kumar D, Shukla P, Kirti P. Characterization of a pathogen induced thaumatin-like protein gene AdTLP from Arachis diogoi, a wild peanut. PloS one. 2013;8:e83963. doi: 10.1371/journal.pone.0083963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar D, Rampuria S, Singh NK, Shukla P, Kirti P. Characterization of a vacuolar processing enzyme expressed in Arachis diogoi in resistance responses against late leaf spot pathogen, Phaeoisariopsis personata. Plant molecular biology. 2015;88:177–191. doi: 10.1007/s11103-015-0318-x. [DOI] [PubMed] [Google Scholar]
- 54.Sharma A, et al. Ectopic Expression of an Atypical Hydrophobic Group 5 LEA Protein from Wild Peanut, Arachis diogoi Confers Abiotic Stress Tolerance in Tobacco. PloS one. 2016;11:e0150609. doi: 10.1371/journal.pone.0150609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vijayan S, Singh N, Shukla P, Kirti P. Defensin (TvD1) from Tephrosia villosa exhibited strong anti-insect and anti-fungal activities in transgenic tobacco plants. Journal of pest science. 2013;86:337–344. doi: 10.1007/s10340-012-0467-5. [DOI] [Google Scholar]
- 56.Baker CJ, Mock NM. An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell, Tissue and Organ Culture. 1994;39:7–12. doi: 10.1007/BF00037585. [DOI] [Google Scholar]
- 57.Patterson BD, Payne LA, Chen Y-Z, Graham D. An inhibitor of catalase induced by cold in chilling-sensitive plants. Plant physiology. 1984;76:1014–1018. doi: 10.1104/pp.76.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue T, et al. Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. Journal of experimental botany. 2009;60:339–349. doi: 10.1093/jxb/ern291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant physiology. 1949;24:1. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heath RL, Packer L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Archives of biochemistry and biophysics. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.