Abstract
Background
Genome-wide association studies (GWAS) have significantly contributed to the association of many clinical conditions and phenotypic characteristics with genomic variants. The majority of these genomic findings have been deposited to the GWAS catalog. So far, findings uncovering associations of single nucleotide polymorphisms (SNPs) with treatment efficacy in mood disorders are encouraging, but not adequate.
Methods
Statistical, genomic, and literature information was retrieved from EBI’s GWAS catalog, while we also searched for potential clinical information/clinical guidelines in well-established pharmacogenomics databases regarding the assessed drug-SNP correlations of the present study.
Results
Here, we provide an overview of significant genome-wide associations of SNPs with the response to commonly prescribed antipsychotics and antidepressants. Up to date, this is the first study providing novel insight in previously reported pharmacogenomics associations for antipsychotic/antidepressant treatment. We also show that although there are published CPIC guidelines for antidepressant agents, as well as the FDA labels include genome-based drug prescription information for both antipsychotic and antidepressant treatments, there are no specific clinical guidelines for the assessed drug-SNP correlations of this study.
Conclusions
Our present findings suggest that more effort should be implemented towards identifying GWA-significant antipsychotic and antidepressant pharmacogenomics correlations. Moreover, additional functional studies are required in order to characterise the potential role of the assessed SNPs as biomarkers for the response of patients to antipsychotic/antidepressant treatment.
Keywords: Antipsychotics, Antidepressants, Pharmacogenomics, Statistical assessment, GWAS catalog, GWAS findings, Clinically approved guidelines
Background
Pharmacogenomics refers to the use of genomic biomarkers to predict an individual’s response to drug efficacy and toxicity. Pharmacogenomics has also seen a raise in terms of research findings, which span over the last decade. Although additional clinical factors such as disease severity, diet, and concurrent medications clearly contribute to the variability in response to drug therapy, inherited differences in the metabolism and action of drugs at their target sites or in the pharmacokinetics and pharmacodynamics of a drug harbor a predominant effect in the therapy outcome [1, 2]. The complexity of drug response can be multifactorial and variable over time, since subjective clinical scales are usually implemented, thus making it challenging to identify genetic variants that robustly predict drug response.
Over the last decade, genome-wide association studies (GWAS) have widely focused on tailoring the genetic background of psychiatric diseases [3, 4]. However, most of these findings usually fail to be replicated in subsequent genetic studies. Candidate gene studies have also been performed as an alternative approach, although the sample sizes have been quite small in some instances [5]. Regardless of each study’s design limitations, genetic studies have provided useful insight in delineating associations of genetic variation with psychiatric disease [6–8] and new ideas about disease etiology. Interesting psychiatric genetic findings include specific SNPs, which were characterised as genome-wide significant for both bipolar disorder and schizophrenia. These SNPs were identified within the following genes: CACNA1CS, ANK3, and ITIH3-ITIH4. In contrast, SNPs within MHC, ODZ4, TCF4, and other genetic loci were genome-wide significant for either disorder separately but not for both [9].
GWAS for delineating drug treatment response and toxicity for psychiatric disorders have also been performed in order to tailor antipsychotic or antidepressant treatment. It is worth noting that most of the identified associations have not been individually replicated, thus leaving the pharmacogenomics background of commonly prescribed antipsychotic or antidepressant drugs quite vague. To this end, Allen and Bishop performed a systematic review of the existing literature for GWAS findings for antipsychotic treatment response. In this review, 15 genome-wide significant loci were identified (CNTNAP5, GRID2, GRM7, 8q24 (KCNK9), PCDH7, SLC1A1 and TNIK), seven of which were replicated in other antipsychotic genome-wide studies [10]. However, further validation of these findings is needed in order to demonstrate the clinical utility of these pharmacogenomics markers.
The United States Food and Drug Administration (FDA; http://www.fda.gov) started incorporating pharmacogenomics information on its labels especially after 2005 and the completion of the Human Genome Project (“www.fda.gov”, [11]). Almost 15 years later, more than 200 drug labels are accompanied by pharmacogenomics information highlighting the progressive acknowledgment of the major regulatory body regarding this field of precision medicine [12]. Such information is gathered and represented in detail in the FDA’s Table of Pharmacogenomics Biomarkers in Drug Labels [13] bearing variable levels of significance, from informational to necessary guidance.
The unmet need, for translating the voluminous literature into the association pairs that could serve as possible biomarkers in a real-time clinical setting, was significantly catalyzed by the formation of the renowned online resource, the Clinical Pharmacogenetics Implementation Consortium (CPIC; http://www.cpicpgx.org) in late 2009 [49, 50]. So far, the CPIC has published 23 peer-reviewed and evidence-based guidelines for over 60 drug–gene pairs in an effort to bridge the gap between the available pharmacogenomics test results and their usefulness in genome-informed prescribing [15]. As for the drugs implicated in antidepressant treatment, two guidelines regarding the selective serotonin reuptake inhibitors (SSRI’s) and the tricyclic antidepressants (TCA’s) and their association with CYP2D6 and CYP2C19 were published in August 2015 and December 2016, respectively [54, 55].
In this study, we aimed to identify and assess genetic associations either with GWA or with nominal significance for commonly prescribed antipsychotic and antidepressant treatment, for which clinical guidelines exist. For this purpose, we assessed the pharmacogenomics associations of commonly prescribed antidepressant/antipsychotic drugs as these were reported in publications deposited in the EBI-GWAS catalog.
Results
Assessing the GWAS catalog data for identification of psychiatric pharmacogenomics findings
First, we began by downloading the pharmacogenomics GWA findings from 20 research studies and by excluding review papers (Additional file 1: Table S1). We extracted the following information from each study: the assessed drug (type of antidepressant or antipsychotic compound), the SNP associated with drug response, the gene within which we identified the specific SNP, the PubMed ID of the study, as well as the web link.
Subsequently, we performed literature search in order to annotate the study type of each one of the 20 research studies. The study findings could be either GWAS findings, or (the findings) could constitute a combination of findings from GWAS and (genome-wide) cell assays. We also identified SNPs, which were associated with response to more than one drug related to antipsychotic or antidepressant treatment (Additional file 1: Table S1). More precisely, 555 drug–SNP correlations were identified in total, of which 462 drug–SNP correlations were characterised as unique (Additional file 1: Table S2) upon removal of any duplicate drug–SNP correlations.
Manhattan plots were also created in order to highlight the significance levels of the SNP associations with antipsychotic or antidepressant drug response. Interestingly, one of these variants had a GWA p value exceeding the level of GWA significance (p = 9 × 10-66). This variant was rs10023464 SNP (AC093720.1 - AC021146.8 intergenic region) that was characterised as a locus associated with plasma clozapine–norclozapine ratio in treatment-resistant schizophrenia patients. Amongst SNPs, which exceeded the GWA significance level, are rs11725502 (AC021146.8 - UGT2B10 intergenic region; p = 5 × 10-15), rs12767583 (CYP2C19; p = 5 × 10-14), rs7668556 (AC111000.6 - AC111000.1 intergenic region; p = 4 × 10-13), rs2814778 (ACKR1; p = 4 × 10-21), and rs117752187 (MIR100HG; p = 6 × 10-11). These SNPs were associated with clozapine response, except for rs117752187, which was associated with paliperidone response (Fig. 1).
Fig. 1.
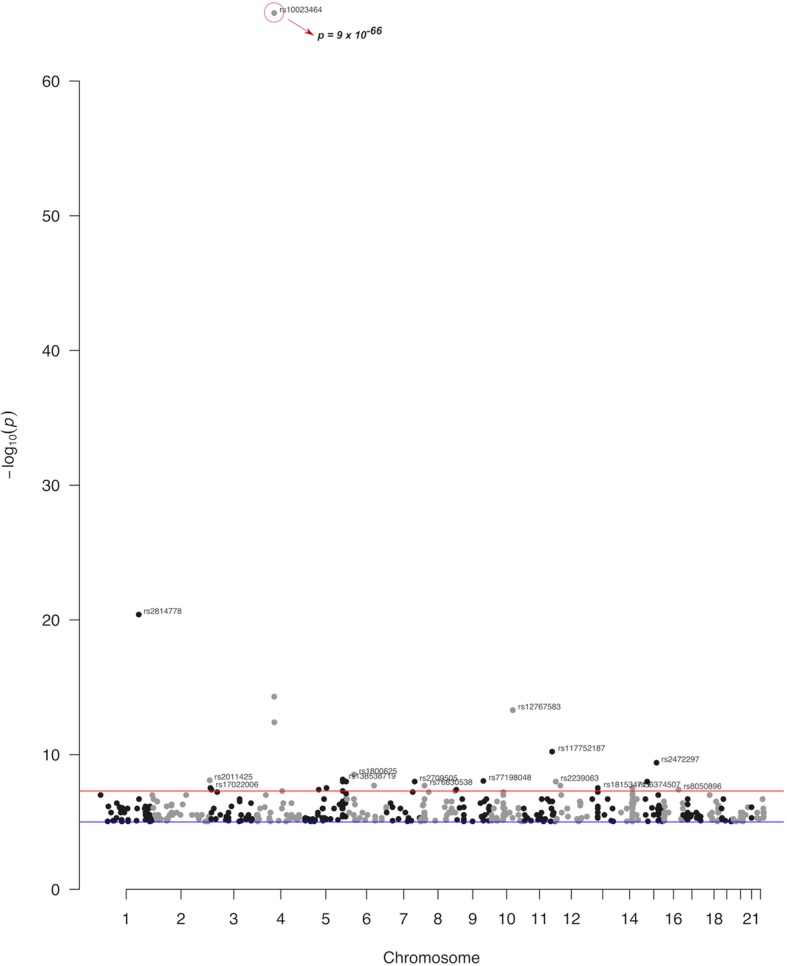
Manhattan plot of the significant associations of pharmacogenomics variants as identified in the GWAS studies deposited in the GWAS catalog. These variants have been associated with response to antipsychotic treatment. The red line denotes the genome-wide threshold of significance (p = 5× 10-8) and the blue line the suggestive threshold of significance (p = 10-5)
Another interesting observation from the Manhattan plots is the higher number of drug–SNP correlations for antipsychotic drugs compared to antidepressant treatment (Fig. 2). However, this observation could be explained by the overall limited number of the assessed GWAS studies.
Fig. 2.
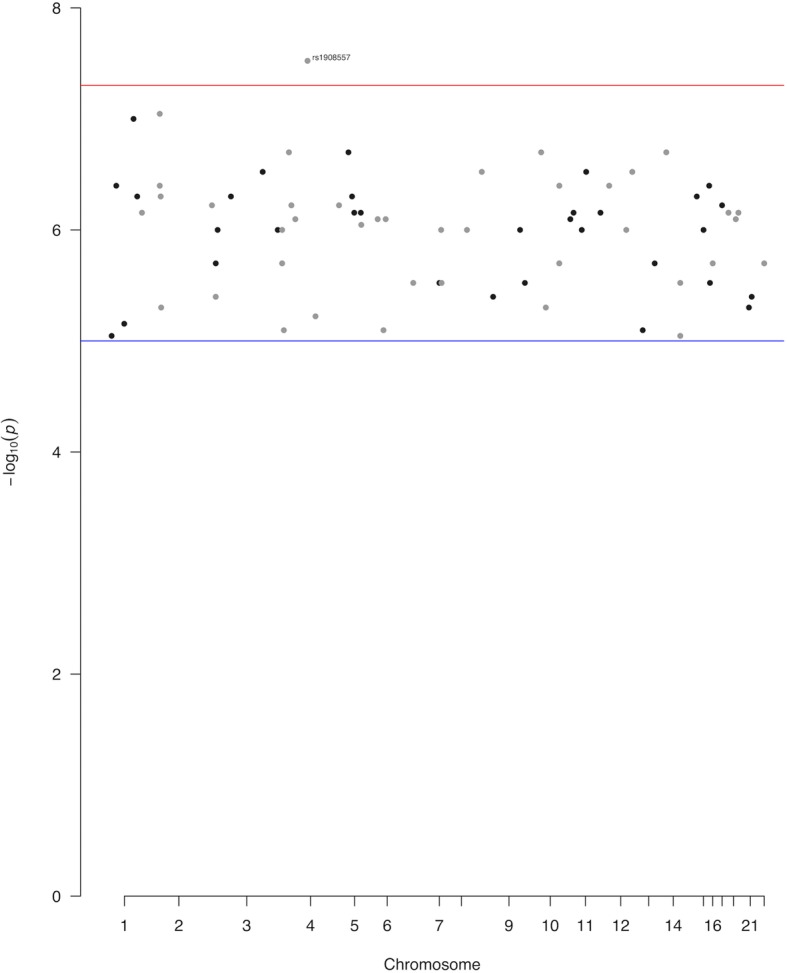
Manhattan plot of the significant associations of pharmacogenomics variants as identified in the GWAS studies deposited in the GWAS catalog. These variants have been associated with response to antidepressant treatment. The red line denotes the genome-wide threshold of significance (p = 5 × 10-8) and the blue line the suggestive threshold of significance (p = 10-5)
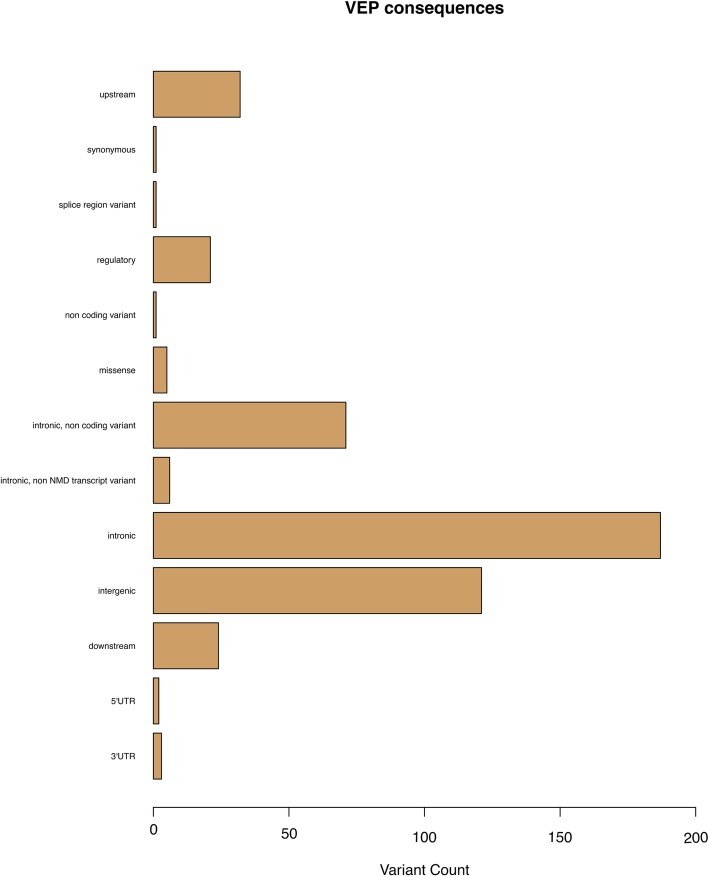
Regarding the SNPs’ functional protein consequence, more than 90% of the identified GWA-significant SNPs were characterised as intronic or intergenic variants, whilst only 5 were characterised as missense: rs2236295, rs2307441, rs17815774, rs17727261, and rs41314643 (Table 1; Fig. 3). rs2236295 (ADO) and rs2307441 (POLG) were identified within the first genome-wide association study (GWAS) in Generalized Anxiety Disorder (GAD), and they were characterised as potential predictors of venlafaxine extended release (XR) treatment outcome. Moreover, rs17815774 (TRPM1) and rs17727261 (CNTNAP5) were identified in a genome-wide pharmacogenomics study and they were associated with the response to treatment with risperidone. Lastly, in another GWAS study, rs41314643 (NMNAT2) was associated with the risk of clozapine-induced agranulocytosis/granulocytopenia.
Table 1.
Functional annotation of the five identified missense variants within the psychiatric pharmacogenomics GWAS studies of interest in this study
| Variant type | rsID | Gene | Drug correlation | Protein damaging pred. (SIFT*, Polyphen2#) |
|---|---|---|---|---|
| missense | rs4314643 | NMNAT2 | Clozapine | Benign (0.13, 0.232) |
| missense | rs2236295 | ADO | Venflaxine | Benign (0.74, 0.007) |
| missense | rs17815774 | TRPM1 | Risperidone | Benign (0.07, 0.191) |
| missense | rs2307441 | POLG | Venlafaxine | Benign (0.16, 0.334) |
| missense | rs17727261 | CNTNAP5 | Risperidone | Benign (0.06, 0.006) |
SIFT and Polyphen2 are in silico tools used to assess the protein damaging effect of missense variants
*SIFT score for protein damaging prediction (< 0.05; deleterious)
#Polyphen2 score for deleterious variants ( > 0.908; probably damaging, 0.446 < score ≤ 0.908; possibly damaging)
Fig. 3.
Plot of the number of pharmacogenomics variants assessed in the present study against their Variant Effect Prediction (VEP) consequence. NMD, nonsense mediated decay; 5’UTR, variant in the 5’UTR region; 3’UTR, variant in the 3’UTR region
Μoreover, we assessed the number of drug–SNP correlations, after removing any duplicate associations, and we observed a high number of common SNPs associated with antidepressant or antipsychotic efficacy and toxicity (Table 2). An almost equal number of low frequency, rare, and SNPs with no 1000 Genomes MAF was also observed, whilst the lowest number of drug-SNP correlations was found within SNPs with intermediate frequency (Table 2). This observation indicates that a couple of the identified drug-common SNP associations could be correlated with potential clinical actionability of these SNPs.
Table 2.
Number of common (MAF > 0.10), intermediate frequency (MAF 0.05–0.10), low frequency (0.01–0.05) and rare variants (MAF < 0.01) for each assessed drug
| Drug | Common (N genes or SNPs) | Interm (N genes or SNPs) | Low freq (N genes or SNPs) | Rare (N genes or SNPs) | No1000 G MAF (N genes or SNPs) | Total (per drug) |
|---|---|---|---|---|---|---|
| Aripiprazole | 2 | - | - | - | - | 2 |
| Bupropion | 6 | - | 5 | 8 | 2 | 21 |
| Clozapine | 35 | 7 | 16 | 11 | 8 | 77 |
| Citalopram | 7 | 2 | 7 | 8 | 7 | 31 |
| Duloxetine | 1 | - | - | - | 1 | 2 |
| Escitalopram | 3 | 1 | 1 | 8 | 4 | 17 |
| Haloperidol | 6 | 1 | - | - | 1 | 8 |
| Iloperidone | 5 | 1 | - | - | - | 6 |
| Ketamine | 13 | 2 | - | - | 4 | 19 |
| Lurasidone | 11 | - | - | - | 2 | 13 |
| Olanzapine | 10 | 3 | - | - | 2 | 15 |
| Oxcarbazepine | 4 | 1 | 2 | - | - | 7 |
| Paliperidone | 56 | 22 | 34 | 56 | 24 | 192 |
| Perphenazine | 11 | 1 | 3 | - | 1 | 16 |
| Quetiapine | 14 | 1 | 1 | - | 2 | 18 |
| Risperidone | 15 | - | 1 | - | 4 | 20 |
| Sertraline | - | 1 | 1 | - | - | 2 |
| Venlafaxine | 11 | 1 | 2 | 1 | 3 | 18 |
| Ziprasidone | 8 | 1 | - | 1 | - | 10 |
| Total (per MAF) | 218 | 45 | 73 | 93 | 65 |
The number of SNPs counted is equal to the number of genes as duplicates of the same drug–SNP correlations were removed
interm intermediate, low freq low frequency, MAF minor allele frequency
Measuring the association strength of the identified SNPs based on their allele frequency
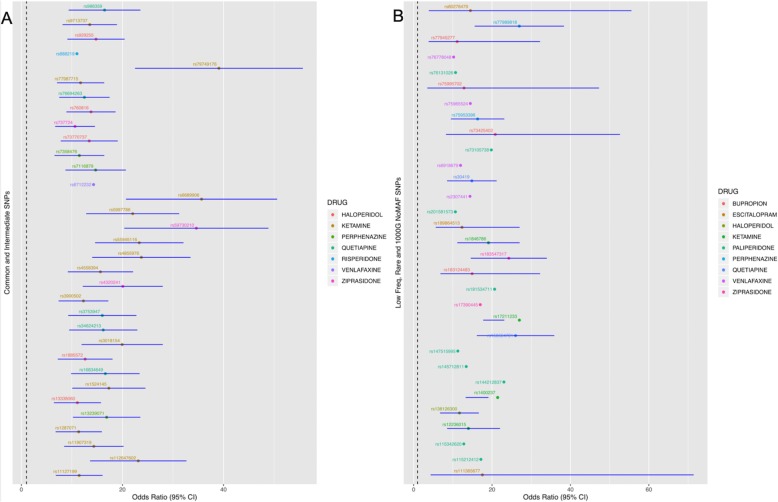
As explained in the Methods section, odds ratio values (ORs) and confidence interval values (CIs) were retrieved and assessed for the identified SNPs (Fig. 4). These values provide an estimation for the protective or risk effect of these SNPs and can be directly associated with the potential clinical outcome. More precisely, in this section, we aim to delineate the SNP effect on the risk for showing adverse drug reactions (ADRs) by analysing odds ratio values and their association with ADR risk. The drug–SNP associations with OR higher than 1 were associated with an increased risk for developing ADRs, whilst drug–SNP associations with OR less than 1 were associated with a lower risk for developing ADRs.
Fig. 4.
a Odds ratio values of pharmacogenomics (PGx) variants of common (MAF > 0.10) or intermediate (0.05 ≤ MAF ≤ 0.10) frequency are plotted alongside with the 95% of their confidence intervals. The PGx variants are colored based on the antipsychotic or antidepressant drug of the association. b Odds ratio values of pharmacogenomics (PGx) variants of low frequency (0.01 ≤ MAF < 0.05); rare (MAF < 0.01) frequency are plotted alongside with the 95% of their confidence intervals. Moreover, the odds ratio values and the confidence intervals of pharmacogenomics variants that were not found within the 1000 Genomes project are also plotted. The PGx variants are colored based on the antipsychotic or antidepressant drug association
Additional data (Additional file 2: Figsure S1–S10) show the ORs and CIs plotted for all identified SNPs and color-coded based on the drug compound. Two graphs, one with OR less than 1 and one with OR bigger than 1, were created for the 5 different classes of MAFs: common (1000 Genomes MAF higher than 10%), intermediate (1000 Genomes MAF between 5 and 10%), low-frequency (1000 Genomes MAF between 1 and 5%), rare (1000 Genomes MAF less than 1%), and finally variants for which there was no MAF in the 1000 Genomes Project.
We observed that there were an equally high number of common SNPs that had ORs either below 1 or above 1, thus hinting at a decreased or increased possibility with drug toxicity, respectively. Similar observations were also applied for GWA-significant drug—intermediate frequency SNP correlations.
In contrast, most of the rare and low-frequency variants had high ORs, usually higher than 5, thus suggesting that these variants can be potentially used for further clinical tests. Our findings indicate that the majority of SNPs with frequency less than 5% have high ORs, thus hinting that these SNPs may be associated with increased drug toxicity. SNPs that have an OR less than 1 indicate an inversed association, which is basically defined as protection against the drug response (Additional file 2: Figures S5–S10). Interestingly, all low-frequency and rare SNPs as well as SNPs that were not annotated in 1000 Genomes, with ORs between 0 and 1, have been associated with reduced probability for drug toxicity.
Amongst the SNPs with high ORs were rs117375960 and rs188843168, which were excluded from the forest plots in order to allow a clearer presentation of the findings. rs117375960 was identified within NT5C2 and was associated with the response to citalopram and escitalopram (p = 2 × 10-6, OR = 142.86). Similarly, rs188843168 (ATP5MD) was also associated with the response to citalopram and escitalopram (p = 4 × 10-7, OR = 449.83). Both of these SNPs are associated with an increased risk for drug toxicity induced from citalopram or escitalopram treatment.
Forest plots were also created for variants that had ORs higher than 10. As also stated in the Methods section, this OR cut-off value was selected since the majority of the clinical studies suggest an OR greater than 10, as sufficient enough for decision-making. However, since the ORs for most of these variants were quite high, it was difficult to provide an accurate estimate of the confidence intervals for these variants. The high ORs combined with the statistically significant association of the identified SNPs provide evidence for the potential role of these variants as biomarkers for antipsychotic drug toxicity.
Lack of CPIC guidelines for the presented antipsychotic drug–SNP correlations
In an effort to mine valuable pharmacogenomics information from the CPIC online resource, we found out that the commonly prescribed antipsychotic agents, namely clozapine, risperidone, haloperidol, perphenazine, aripiprazole, iloperidone, and olanzapine, are not yet accompanied by a corresponding pharmacogenomics (PGx) guideline. However, all the aforementioned drugs are associated with CYP2D6 bearing a CPIC level B or C, while their corresponding CPIC guideline will be compiled in the future. Of note, the US regulatory body already includes pharmacogenomics information regarding CYP2D6 in the respective drug labels, either vaguely commenting on the pharmacokinetics of the administered agent (as in the case of risperidone-CYP2D6) or proposing specific genome-informed dose adjustment (as in the case of aripiprazole-CYP2D6). Two SNRI’s, venlafaxine and duloxetine, are deemed to be accompanied by a CPIC guideline in the future regarding their association with CYP2D6. As for the SSRI’s, citalopram, escitalopram, and sertraline, a published CPIC guideline proposes specific therapeutic strategies based on CYP2C19 genotype results. In parallel, the US regulatory body refers to the pharmacogenomics information related to the aforementioned drug–gene association pairs in the corresponding drug inserts. It is noteworthy that there is no mention regarding the variants discussed in our paper that are deemed ambitious as for their clinical actionability in either two sources.
Discussion
There is no doubt that GWAS research has yielded many discoveries in the field of neuropsychiatric and neurodevelopmental diseases. However, constant research and improvements in “big data” parsing methods are essential for improvements in the discovery of pharmacogenomics findings. GWAS studies can be useful in unraveling potential mechanisms and pathways that underlie human characteristics, diseases, and drug response.
For example, researchers in the Schizophrenia Working Group of the Psychiatric Genetics Consortium (PGC) performed a GWAS and identified 108 genome-wide significant loci [18], thus showing that increased sample sizes can boost the discovery in schizophrenia genetics research and psychiatric pharmacogenomics research.
As with most GWAS studies, those focusing on identification of drug-genomic variant correlations may be characterised by heterogeneity issues across the different study sample datasets. Any discrepancies between individual studies must be taken into consideration as confounding variables. For example, studies included a broad range of sample sizes, some of which only included a single racial or ethnic group; these studies should be replicated within different populations [51].
Nonetheless, PGx GWAS have resulted in the identification of several actionable genetic variants that have been genotyped and used to inform drug selection and dosage. However, the most significant PGx GWAS achievements have been associated with the identification of non-HLA markers, such as NUDT15, which is associated with thiopurine-induced leukopenia [19].
So far, only a few psychiatric clinical pharmacogenomics and psychiatric PGx GWAS studies have been published thus pointing to a need for properly designed psychiatric pharmacogenomics. Two characteristic examples of clinical psychiatric PGx studies, which are currently on-going, are the following: one is the “PREPARE” by the Ubiquitous Pharmacogenomics (U-PGx; http://www.upgx.eu) Consortium and the other one is an effort run at the Karolinska Institute [20, 21]. The “PREPARE” study aims to perform pre-emptive genotyping of a panel of clinically relevant PGx-markers, for which guidelines are available, whilst being implemented across healthcare institutions in seven European countries. Amongst the individuals recruited are individuals diagnosed with a variety of psychiatric disorders and the upper study goal is the identification of biomarkers associated with antipsychotic/antidepressant drug responses [21]. Regarding the other effort, as run by the Karolinska Institute, findings from this study indicate that CYP2D6 pre-emptive genotyping would be valuable for individualising risperidone and aripiprazole dosing, thus leading to treatment optimisation [20].
It is worth to note that only about half of the psychiatric genome-wide significant associations have been validated in subsequent studies, and more precisely either in an independent study or in a replication sample. Because of the possibility of false discovery, the likelihood of a GWAS signal being a true marker of the tested phenotype holds fairly limited promise prior to replication. Overall, findings from both of these efforts highlight the need for improving and boosting discovery of clinically relevant psychiatric pharmacogenomics biomarkers [22].
As of submission of this manuscript, this is the first study focusing on 20 published research articles, as deposited in the GWAS catalog, and which assesses GWA-significant antipsychotic/antidepressant drug–SNP correlations. Interestingly, only two SNPs were identified within or between clinically approved pharmacogenes, (rs2472297 between CYP1A1-CYP1A2 and rs12767583 within CYP2C19) and they were both associated with the response to clozapine. No clinical guidelines either from CPIC or from FDA exist for the rest of the identified drug–SNP correlations.
Undoubtedly, implementation of PGx research findings in the clinic requires lots of time and effort owing to the ever-increasing need for functional validation studies, regulatory clearance and development of the appropriate translational tools. To this end, our findings can be proven useful for unravelling the background for identification of pharmacogenomics biomarkers for psychiatric drug treatment. These findings may be proven meaningful for informing the findings from pharmacogenomics clinical studies (i.e., Ubiquitous Pharmacogenomics—U-PGx), since lots of the assessed antipsychotic/antidepressant compounds are included in the U-PGx clinical study. Therefore, the identified drug–SNP correlations may be of particular interest in future pharmacogenomics clinical studies and they may be in linkage disequilibrium with other known psychiatric pharmacogenomics biomarkers.
Conclusions
To our knowledge, this is the first study that summarizes and provides novel insight in previously identified and GWA-significant pharmacogenomics associations for antipsychotic/antidepressant treatment. We showed that common pharmacogenomics association for antipsychotic/antidepressant drugs with genomic biomarkers might harbour large effect sizes, thus hinting at a potential clinical utility. We also demonstrate that there are multiple drug–SNP correlations, for which little or no clinical information is available and for which no approved clinical guidelines exist. So far, little is known about the functional impact of variants on inter-individual variability in drug response, which are identified in genomic loci apart from well-characterised pharmacogenes. Το this end, more effort should be placed towards the identification of pharmacogenomics biomarkers for antipsychotic treatments. This can be achieved by leveraging the information from databases, such as the GWAS catalog, as well as by updating and renewing the freely available information regarding clinically approved guidelines (i.e. CPIC, FDA).
Methods
Data collection
The data of this study include all studies recorded by the GWAS catalog [23] with the date of the datasets download being the 27th of August 2019. Statistical, genomic, and literature information were retrieved from 20 research articles (Additional file 1: Table S1) [4, 24–46] as deposited in the GWAS catalog of EBI. The information of the datasets includes literature sources, phenotype information, p values, and identified SNPs, ORs and CIs.
The selection of antipsychotic or antidepressant drugs was based on examples of antidepressant or antipsychotic medications, which are the gold-standard treatment options for schizophrenia and other common mental conditions. The query criteria in the GWAS catalog database were the name of the assessed antipsychotic or antidepressant compounds.
Statistical analysis
Odds ratio values, chi-square p values, and 95% of confidence intervals were used in order to statistically examine the association strength of the GWA significant SNPs. These statistical measurements were also directly retrieved from the GWAS catalog. Moreover, drug–SNP correlations were examined in order to identify potential associations, which were identified in more than one study. The results were presented either as Forest plots or Manhattan plots and they were created in R software (“https://www.r-project.org” [46–48]). Forest plots were built in the ‘ggplot2’ R package, while Manhattan plots were created by implementing the ‘qqman’ R package [16, 17]. Moreover, forest plots were also created separately for SNPs with ORs higher than 10, since most studies suggest 10 as a threshold sufficient enough for decision-making [52].
Variant annotation
Functional annotation of the identified SNPs and MAF annotation from the 1000 Genomes Project was performed using the Variant Effect Predictor (VEP) freely available from ENSEMBL [53].
PGx information mining
Moreover, specific pharmacogenomics information was retrieved from the FDA’s Table of Pharmacogenomics Biomarkers in Drug Labels regarding the pharmaceutical agents currently used in antidepressant and antipsychotic treatment, as well as from the respective published CPIC guidelines and their supplementary information [54, 55].
Supplementary information
Additional file 1: Table S1. Summary information for the identified drug SNP-correlations. Table S2. List of the antipsychotic/antidepressant drug-SNP correlations that have been reported once (duplicates removed) in the assessed 20 studies deposited in the GWAS catalog.
Additional file 2: Figures S1-S10. showing the odds ratio values split in two categories (between 0-1 and between 1-60) for commonfrequency (MAF > 0.10), intermediate frequency (0.05 <= MAF <= 0.10), low frequency (0.01 <= MAF < 0.05), rare frequency (MAF < 0.01) variants as well as for variants not found in 1000Genomes Project. The PGx variants are colored based on the antipsychotic or antidepressant drug of the association.
Acknowledgements
Not applicable
Abbreviations
- ADRs
Adverse drug reactions
- CI
Confidence interval
- CPIC
Clinical Pharmacogenetics Implementation Consortium
- EBI
European Bioinformatics Institute
- FDA
Food and Drug Administration
- GAD
Generalised anxiety disorder
- GWA
Genome-wide association
- GWAS
Genome-wide association study
- MAF
Minor allele frequency
- OR
Odds ratio
- PGC
Psychiatric Genetics Consortium
- PGx
Pharmacogenomics
- SNP
Single-nucleotide polymorphism
- SNRI
Serotonin norepinephrine reuptake inhibitor
- SSRI
Selective serotonin reuptake inhibitor
- U-PGx
Ubiquitous pharmacogenomics
- VEP
Variant effect predictor
- XR
Extended release
Authors’ contributions
MK conceived the study. MK and SK performed the analysis. SK assessed the clinical guidelines from the FDA and CPIC. All authors have compiled, read and approved the final manuscript.
Funding
SK is a recipient of an Onassis foundation scholarship. This work was partly funded by grants from the European Commission [(H2020-668353; U-PGx (www.upgx.eu) and H2020-860895; TranSYS)] to GPP.
Availability of data and materials
All datasets are publicly available specified in the manuscript. The initial data, as retrieved from the GWAS catalog, can be found in the Additional file 1.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare no conflict of interest. GPP is Full Member and National Representative at the European Medicines Agency, Committee for Human Medicinal Products (CHMP)–Pharmacogenomics working Party in Amsterdam, the Netherlands.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40246-019-0254-y.
References
- 1.Sweeney GD. Variability in the human drug response. Thromb Res Suppl. 1983;4:3–15. doi: 10.1016/0049-3848(83)90353-5. [DOI] [PubMed] [Google Scholar]
- 2.Westervelt P, Cho K, Bright DR, Kisor DF. Drug-gene interactions: inherent variability in drug maintenance dose requirements. P T. 2014;39(9):630–637. [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H, Yan H, Wang L, Li J, Tan L, Deng W, et al. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry. 2018;5(4):327–338. doi: 10.1016/S2215-0366(18)30049-X. [DOI] [PubMed] [Google Scholar]
- 5.Patnala R, Clements J, Batra J. Candidate gene association studies: a comprehensive guide to useful in silico tools. BMC Genet. 2013;14:39. doi: 10.1186/1471-2156-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasi G, Selvaggi P, Fazio L, Antonucci LA, Taurisano P, Masellis R, et al. Variation in dopamine D2 and serotonin 5-HT2A receptor genes is associated with working memory processing and response to treatment with antipsychotics. Neuropsychopharmacology. 2015;40(7):1600–1608. doi: 10.1038/npp.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang R, Tiwari AK, Zai CC, Felsky D, Remington E, Wallace T, et al. Dopamine D4 and D5 receptor gene variant effects on clozapine response in schizophrenia: replication and exploration. Prog Neuropsychopharmacol Biol Psychiatry. 2012;37(1):62–75. doi: 10.1016/j.pnpbp.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 8.Pouget JG, Goncalves VF, Nurmi EL, Laughlin CP, Mallya KS, Mc Cracken JT, et al. Investigation of TSPO variants in schizophrenia and antipsychotic treatment outcomes. Pharmacogenomics. 2015;16(1):5–22. doi: 10.2217/pgs.14.158. [DOI] [PubMed] [Google Scholar]
- 9.Psychosis Endophenotypes International C, Wellcome Trust Case-Control C. Bramon E, Pirinen M, Strange A, Lin K, et al. A genome-wide association analysis of a broad psychosis phenotype identifies three loci for further investigation. Biol Psychiatry. 2014;75(5):386–397. doi: 10.1016/j.biopsych.2013.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen JD, Bishop JR. A systematic review of genome-wide association studies of antipsychotic response. Pharmacogenomics. 2019;20(4):291–306. doi: 10.2217/pgs-2018-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Food and Drug Administration. 2019. http://www.fda.gov. Accessed 25 Oct 2019.
- 12.Cheng Y, He YJ, Tang J, Zhou HH. Translating pharmacogenetics and pharmacogenomics: the last 60 years and the rise of collective innovation as a force multiplier for personalized medicine. Curr Pharmacogenomics Person Med. 2014;12:15–31. doi: 10.2174/1875692111666131223235548. [DOI] [Google Scholar]
- 13.Lauschke VM, Ingelman-Sundberg M. How to consider rare genetic variants in personalized drug therapy. Clin Pharmacol Ther. 2018;103(5):745–748. doi: 10.1002/cpt.976. [DOI] [PubMed] [Google Scholar]
- 14.Food and Drug Administration. Table of pharmacogenic biomarkers in drug labeling. In: Science and and research drugs. Food and Drug Administration. 2019. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling. Accessed 25 Oct 2019.
- 15.Clinical Pharmacogenetics Implementation Consortium. (2019). https://cpicpgx.org. Accessed 25 Oct 2019.
- 16.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 17.Turner, S.D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. J Open Source Softw. 2018;3(25):731.
- 18.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung WH, Chang WC, Lee YS, Wu YY, Yang CH, Ho HC, et al. Genetic variants associated with phenytoin-related severe cutaneous adverse reactions. JAMA. 2014;312(5):525–534. doi: 10.1001/jama.2014.7859. [DOI] [PubMed] [Google Scholar]
- 20.Jukic MM, Smith RL, Haslemo T, Molden E, Ingelman-Sundberg M. Effect of CYP2D6 genotype on exposure and efficacy of risperidone and aripiprazole: a retrospective, cohort study. Lancet Psychiatry. 2019;6(5):418–426. doi: 10.1016/S2215-0366(19)30088-4. [DOI] [PubMed] [Google Scholar]
- 21.van der Wouden CH, Cambon-Thomsen A, Cecchin E, Cheung KC, Davila-Fajardo CL, Deneer VH, et al. CORRIGENDUM: implementing pharmacogenomics in Europe: design and implementation strategy of the ubiquitous pharmacogenomics consortium. Clin Pharmacol Ther. 2017;102(1):152. doi: 10.1002/cpt.725. [DOI] [PubMed] [Google Scholar]
- 22.Tam V, Patel N, Turcotte M, Bosse Y, Pare G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20(8):467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 23.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(Database issue):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Loebel A, Meltzer HY. Identifying the genetic risk factors for treatment response to lurasidone by genome-wide association study: a meta-analysis of samples from three independent clinical trials. Schizophr Res. 2018;199:203–213. doi: 10.1016/j.schres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Volpi S, Heaton C, Mack K, Hamilton JB, Lannan R, Wolfgang CD, et al. Whole genome association study identifies polymorphisms associated with QT prolongation during iloperidone treatment of schizophrenia. Mol Psychiatry. 2009;14(11):1024–1031. doi: 10.1038/mp.2008.52. [DOI] [PubMed] [Google Scholar]
- 26.McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, et al. Genome-wide pharmacogenomics analysis of response to treatment with antipsychotics. Mol Psychiatry. 2011;16(1):76–85. doi: 10.1038/mp.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adkins DE, Aberg K, McClay JL, Bukszár J, Zhao Z, Jia P, et al. Genomewide pharmacogenomics study of metabolic side effects to antipsychotic drugs. Mol Psychiatry. 2011;16(3):321–332. doi: 10.1038/mp.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aberg K, Adkins DE, Liu Y, McClay JL, Bukszár J, Jia P, et al. Genome-wide association study of antipsychotic-induced QTc interval prolongation. Pharmacogenomics J. 2012;12(2):165–172. doi: 10.1038/tpj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark SL, Adkins DE, Aberg K, Hettema JM, McClay JL, Souza RP, et al. Pharmacogenomics study of side-effects for antidepressant treatment options in STAR*D. Psychol Med. 2012;42(6):1151–1162. doi: 10.1017/S003329171100239X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark SL, Souza RP, Adkins DE, Aberg K, Bukszár J, McClay JL, et al. Genome-wide association study of patient-rated and clinician-rated global impression of severity during antipsychotic treatment. Pharmacogenet Genomics. 2013;23(2):69–77. doi: 10.1097/FPC.0b013e32835ca260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drago A, Giegling I, Schäfer M, Hartmann AM, Konte B, Friedl M, et al. Genome-wide association study supports the role of the immunological system and of the neurodevelopmental processes in response to haloperidol treatment. Pharmacogenet Genomics. 2014;24(6):314–319. doi: 10.1097/FPC.0000000000000052. [DOI] [PubMed] [Google Scholar]
- 32.Goldstein JI, Jarskog LF, Hilliard C, Alfirevic A, Duncan L, Fourches D, et al. Clozapine-induced agranulocytosis is associated with rare HLA-DQB1 and HLA-B alleles. Nat Commun. 2014;5:4757. doi: 10.1038/ncomms5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Athanasiu L, Smorr LL, Tesli M, Røssberg JI, Sønderby IE, Spigset O, et al. Genome-wide association study identifies common variants associated with pharmacokinetics of psychotropic drugs. J Psychopharmacol. 2015;29(8):884–891. doi: 10.1177/0269881115584469. [DOI] [PubMed] [Google Scholar]
- 34.Sacchetti E, Magri C, Minelli A, Valsecchi P, Traversa M, Calza S, et al. The GRM7 gene, early response to risperidone, and schizophrenia: a genome-wide association study and a confirmatory pharmacogenetic analysis. Pharmacogenomics J. 2017;17(2):146–154. doi: 10.1038/tpj.2015.90. [DOI] [PubMed] [Google Scholar]
- 35.Saito T, Ikeda M, Mushiroda T, Ozeki T, Kondo K, Shimasaki A, et al. Pharmacogenomics study of clozapine-induced agranulocytosis/granulocytopenia in a Japanese population. Biol Psychiatry. 2016;80(8):636–642. doi: 10.1016/j.biopsych.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Legge SE, Hamshere ML, Ripke S, Pardinas AF, Goldstein JI, Rees E, et al. Genome-wide common and rare variant analysis provides novel insights into clozapine-associated neutropenia. Mol Psychiatry. 2017;22(10):1502–1508. doi: 10.1038/mp.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodds KN, Beckett EA, Evans SF, Grace PM, Watkins LR, Hutchinson MR. Glial contributions to visceral pain: implications for disease etiology and the female predominance of persistent pain. Transl Psychiatry. 2016;6(9):e888. doi: 10.1038/tp.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li QS, Tian C, Seabrook GR, Drevets WC, Narayan VA. Analysis of 23andMe antidepressant efficacy survey data: implication of circadian rhythm and neuroplasticity in bupropion response. Transl Psychiatry. 2016;6(9):e889. doi: 10.1038/tp.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Wineinger NE, Fu DJ, Libiger O, Alphs L, Savitz A. Genome-wide association study of paliperidone efficacy. Pharmacogenet Genomics. 2017;27(1):7–18. doi: 10.1097/FPC.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung J, Tawa EA, Muench C, Rosen AD, Rickels K, Lohoff FW. Genome-wide association study of treatment response to venlafaxine XR in generalized anxiety disorder. Psychiatry Res. 2017;254:8–11. doi: 10.1016/j.psychres.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maciukiewicz M, Marshe VS, Tiwari AK, Fonseka TM, Freeman N, Kennedy JL, et al. Genome-wide association studies of placebo and duloxetine response in major depressive disorder. Pharmacogenomics J. 2018;18(3):406–412. doi: 10.1038/tpj.2017.29. [DOI] [PubMed] [Google Scholar]
- 42.McCoy TH, Castro VM, Cagan A, Snapper L, Roberson A, Perlis RH. Cytochrome P450 interactions are common and consequential in Massachusetts hospital discharges. Pharmacogenomics J. 2018;18(2):347–350. doi: 10.1038/tpj.2017.30. [DOI] [PubMed] [Google Scholar]
- 43.Guo W, Machado-Vieira R, Mathew S, Murrough JW, Charney DS, Grunebaum M, et al. Exploratory genome-wide association analysis of response to ketamine and a polygenic analysis of response to scopolamine in depression. Transl Psychiatry. 2018;8(1):280. doi: 10.1038/s41398-018-0311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legge SE, Pardiñas AF, Helthuis M, Jansen JA, Jollie K, Knapper S, et al. A genome-wide association study in individuals of African ancestry reveals the importance of the Duffy-null genotype in the assessment of clozapine-related neutropenia. Mol Psychiatry. 2019;24(3):328–337. doi: 10.1038/s41380-018-0335-7. [DOI] [PubMed] [Google Scholar]
- 45.Berghuis B, Stapleton C, Sonsma ACM, Hulst J, de Haan GJ, Lindhout D, et al. A genome-wide association study of sodium levels and drug metabolism in an epilepsy cohort treated with carbamazepine and oxcarbazepine. Epilepsia Open. 2019;4(1):102–109. doi: 10.1002/epi4.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardiñas AF, Nalmpanti M, Pocklington AJ, Legge SE, Medway C, King A, et al. Pharmacogenomics variants and drug interactions identified through the genetic analysis of clozapine metabolism. Am J Psychiatry. 2019;176(6):477–486. doi: 10.1176/appi.ajp.2019.18050589. [DOI] [PubMed] [Google Scholar]
- 47.Core Team R. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 48.R Project. 2019. https://www.r-project.org. Accessed 25 Oct 2019.
- 49.Caudle KE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Relling MV, Klein TE. Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am J Health Syst Pharm. 2016;73(23):1977–1985. doi: 10.2146/ajhp150977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526(7573):343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakobsen JC, Gluud C, Winkel P, Lange T, Wetterslev J. The thresholds for statistical and clinical significance - a five-step procedure for evaluation of intervention effects in randomised clinical trials. BMC Med Res Methodol. 2014;14:34. doi: 10.1186/1471-2288-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ensembl GRCh37. 2019. http://grch37.ensembl.org/Homo_sapiens/Tools/VEP?db=core;tl=j2bBNIwPINafz22S-5364081. Accessed 25 Oct 2019.
- 54.Hicks JK, Bishop JR, Sangkuhl K, Müller DJ, Ji Y, Leckband SG, Leeder JS, Graham RL, Chiulli DL, LLerena A, Skaar TC, Scott SA, Stingl JC, Klein TE, Caudle KE, Gaedigk A. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline forCYP2D6andCYP2C19Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clinical Pharmacology & Therapeutics. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Müller DJ, Shimoda K, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37–44. doi: 10.1002/cpt.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Summary information for the identified drug SNP-correlations. Table S2. List of the antipsychotic/antidepressant drug-SNP correlations that have been reported once (duplicates removed) in the assessed 20 studies deposited in the GWAS catalog.
Additional file 2: Figures S1-S10. showing the odds ratio values split in two categories (between 0-1 and between 1-60) for commonfrequency (MAF > 0.10), intermediate frequency (0.05 <= MAF <= 0.10), low frequency (0.01 <= MAF < 0.05), rare frequency (MAF < 0.01) variants as well as for variants not found in 1000Genomes Project. The PGx variants are colored based on the antipsychotic or antidepressant drug of the association.
Data Availability Statement
All datasets are publicly available specified in the manuscript. The initial data, as retrieved from the GWAS catalog, can be found in the Additional file 1.