Abstract
Glioblastoma is the most common malignant primary brain tumor among adults and one of the most lethal cancers. It is characterized by the deregulation of signaling pathways involving proliferation, growth, survival, and other factors. MicroRNAs (miRNAs) play a role in the regulation of genes by affecting the 3′ untranslated region (UTR) of mRNA and affect many cell functions. The present study showed that miR-129 decreased the expression of retinoblastoma and p53 signaling pathways’ genes, including CDK4, CDK6, and MDM2. The real-time PCR data indicated that expression of CDK4 in U251 and U87 cell lines declined by 69.8% and 47% (p < 0.05), respectively, and expression of CDK6 and MDM2 in U251 cells decreased by 55.3% (p < 0.0001) and 34.7% (p < 0.05), respectively. Luciferase assays confirmed that overexpression of miR-129 decreased the expression of the CDK4 gene by 58.9% (p < 0.01), CDK6 by 35.7% (p < 0.0001), and MDM2 by 49% (p < 0.001). Moreover, cell cycle assays showed a decrease of the G2-phase population to 10% and pre-G2 arrest in U87 cells (p < 0.05). Additionally, wound healing assays indicated that miR-129 overexpression inhibits cell growth of glioblastoma cells. These findings introduced novel targets for miR-129 in glioblastoma cells.
Keywords: glioblastoma, cell cycle, miR-129, CDK4, CDK6, MDM2
Introduction
Grade IV astrocytoma or glioblastoma multiforme (GBM) is a kind of tumor that stems from glial (supportive cells) tissue of the brain and spinal cord in the CNS (central nervous system) and contains cells that grow fast and look very different in comparison with normal cells. This cancer is more common in adults than children and affects the brain more often than the spinal cord.1 The median survival for GBM patients is less than 15–20 months.2 The incidence of GBM in European and US populations is less than 2 and about 3 per 100,000, respectively, per year, and it tends to be higher in more developed countries. It is 1.5-fold more common in men.3
Numerous factors contribute to a tragic outcome for GBM patients. (1) Surgery and radiation therapy have limitations because of the proximity of critical functional structures in the brain. (2) The tumor margin is hardly defined, since cancerous cells tend to be highly invasive, and they migrate into the brain parenchyma and through blood vessels and white matter tracks. Therefore, recurrence typically occurs within a few centimeters. (3) The existence of cancer stem cells (CSCs) that are tumor initiating and propagating, and in some cases radiotherapy and chemotherapy resistant, provides the chance of recurrence. (4) Targeting specific oncogenic pathways because of heterogeneous genotype, histology, and pathology is complex. (5) The blood-brain barrier (BBB) limits therapeutic delivery. (6) The immune-privileged status of the brain and immune-suppressive features of the tumor limit systemic immune responses. As a result, there is an essential need for new therapeutic strategies that can overcome these barriers to treatment, and gene therapy may hold promise for the treatment of GBM.4 The most common strategies to treat GBM via gene therapy include suicide gene therapy, immune response activation, oncolytic viruses, and reprogramming.5
Advances in the understanding of signaling pathways that underlie glioblastoma pathogenesis have resulted in the development of new therapeutic approaches targeting multiple oncogenic signaling aberrations associated with GBM. The signaling pathways that are physiologically and pathologically relevant to GBM consist of p53, retinoblastoma (RB), phosphatidylinositol 3-kinase (PI3K)/phosphatase and tensin homolog (PTEN)/Akt/mammalian target of rapamycin (mTOR), RAS/mitogen-activated protein kinase (MAPK), and signal transducer and activator of transcription 3 (STAT3)/ZIP4.6 Based on The Cancer Genome Atlas (TCGA) research, the RB signaling pathway was altered in 78% of glioblastoma samples, consisting of 52% mutation or homozygous deletion of CDKN2A (P16/INK4A), 47% homozygous deletion of CDKN2B, 2% homozygous deletion of CDKN2C, and amplification of CDK4 and CDK6 to 18% and 1%, respectively.7,8 The tumor suppressor RB (pRB) has a crucial role in inhibiting cell cycle progression by binding and inhibiting E2F family transcription factors. In brief, in the G1 phase, pRB is normally inactivated by Cyclin D/CDK4/CDK6-induced phosphorylation, which leads to the release of pRB from E2F and the subsequent stimulation of cell progression into the S phase. CDKN2B, which is a CDK inhibitor, forms a complex with CDK4 or CDK6, thereby preventing the activation of CDKs. The downstream effect of this inhibition is preventing cell growth and the cell cycle through G1 progression, by maintaining the activation state of pRB. Amplification of CDK4 and CDK6 in glioblastomas is common, and they both play pivotal roles in astrocytic tumorigenesis and glioma progression. Since the pRB pathway is inactivated by the kinase activity of the CDK4/CDK6/Cyclin D complex, inhibition of CDK4 and CDK6 may be a chemotherapeutic treatment strategy in GBM patients with aberrantly expressed pRB.6 Amplification of both or either CDK4 or CDK6 could be one of the important events that provide a growth advantage to astrocytic tumor.9
In addition, TCGA research reveals that the p53 signaling pathway was altered in 87% of glioblastoma samples and consisted of 49% mutation or homozygous deletion of CDKN2A (ARF), 35% homozygous deletion or mutation of TP53, and amplification of MDM2 and MDM4 to 14% and 7%, respectively.10,11 MDM2 is an E3 ubiquitin ligase and important negative regulator of the p53 tumor suppressor. It negatively regulates p53 in two ways: direct binding and transcriptional inhibition, and degradation through its E3 ligase activity.12 Amplification of MDM2 only occurred in tumors without a p53 mutation, suggesting that MDM2 overexpression may provide alternative means for tumors to inactivate p53-regulated growth control without having to alter p53 itself.13
MicroRNAs (miRNAs) are single-stranded RNAs (ssRNAs) of ∼22 nt in length, and they are generated from endogenous hairpin-shaped transcripts. miRNA molecules play a guiding role in post-transcriptional gene regulation by base pairing with the target mRNAs, usually in the 3′ UTR (untranslated region). miRNA and target mRNA binding typically leads to translational repression and exonucleolytic mRNA decay, although highly complementary targets can be cleaved endonucleolytically. Other types of regulation, such as translational activation and heterochromatin formation, have also been described. It is predicated that more than one-third of human genes are directly targeted by miRNAs, and the unique combination of miRNAs in each cell type determines the use of thousands of mRNAs.14 Precise control of miRNA levels is crucial to maintain normal cellular functions, and there is a relationship between deregulated miRNAs and a variety of cancers, such as glioblastoma and medulloblastoma.15 There is some evidence that implicates miRNAs as having a role in the control of cyclin expression and, consequently, cell cycle progression. For instance, let-7 regulates cyclin D2, and it is poorly expressed in lung tumors and lung cancer cell lines.16 For another example, miR-122 was downregulated in hepatic tumors. Gramantieri et al.17 showed that miR-122a downregulates cyclin G1 expression in a hepatocellular carcinoma (HCC)-derived cell line.
Taking all of these examples into account, miRNA-mediated suppression of upregulated genes that are involved in cell cycle signaling and progression seems a promising strategy to inhibit the proliferation and invasiveness of cancer cells. In our study, we chose miR-129, which targets CDK4, CDK6, and MDM2 genes based on bioinformatics databases, and it potentially can inhibit cancer proliferation. The goal of our study was to investigate the effect of overexpression of miR-129 on the expression of CDK4, MDM2, and CDK6 in glioma cells and provide proof of a relationship between miR-129 and CDK4/CDK6/MDM2 expression.
Results
miRNA and Gene Selection
According to the KEGG database and published documents, CKD4, CKD6, and MDM2 were selected as upregulated genes that play a role in RB and p53 signaling pathways. Based on the Expression Atlas, the expression of CDK4 in U-251 and U-87 cell lines is medium (142 TPM [transcripts per million] and 218 TPM, respectively). Additionally, the expression of CDK6 is medium in both of the cell lines (77 TPM for U251 and 18 TPM for U87), and the expression of MDM2 is medium (23 TPM for U251 and 26 TPM for U87). Thus, we were able to inhibit RB and p53 signaling pathways in glioblastoma via downregulation of these genes in these cell lines. Afterward, we used TargetScan, RNA22, and miRanda algorithms to choose miRNAs that can potentially target desirable genes. A candidate miRNA, hsa-miR-129-5p, was selected according to the table score (Table 1).
Table 1.
miR-129-5p Binding Sites of CDK4, CDK6, and MDM2 3′ UTRs Predicted by Bioinformatics Databases
| TargetScan |
miRanda |
RNA22 |
||
|---|---|---|---|---|
| Seed of miR-129-5p | Site Type | Contex++ Score | mirSVR Score | Folding Energy (kcal/mol) |
| poorly conserved 7-mer-1A | −0.06 | ---- | −11.30 | |
| poorly conserved 7-mer-m8 | −0.17 | −0.0049 | −13.70 | |
| poorly conserved 7-mer-1A | −0.02 | −0.0021 | −16.20 | |
Induction of miR-129 Expression in Glioblastoma Cells
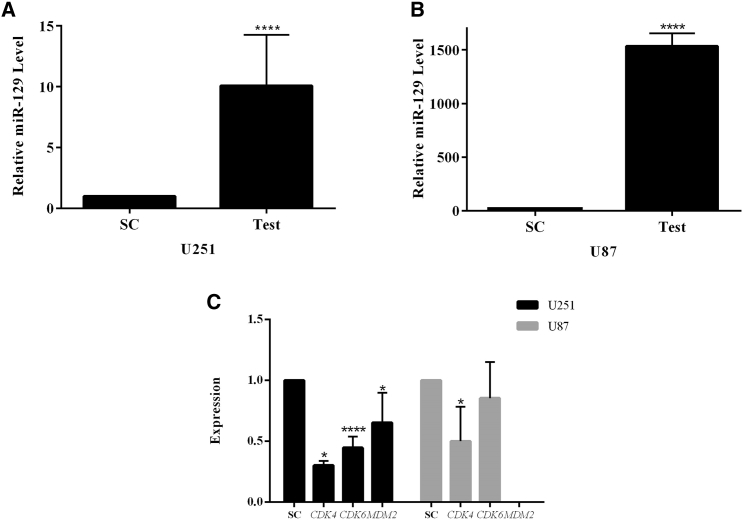
Lentiviral particles that contain miR-129 (LV-miR-129) were used for transduction of glioblastoma cell lines U251 (Figure S1A) and U87 (Figure S1B). Consequently, the expression level of miR-129 in U251 and U87 cell lines was quantified via real-time PCR. The results show that the miR-129 expression increased to 10.08- and 1,533.99-fold (p < 0.0001) in U251 and U87 cells, respectively, in comparison with the control group (Figures 1A and 1B).
Figure 1.
Upregulation of miR-129 in GBM Cells Can Affect Genes' Expression
(A and B) Upregulation of miR-129 in (A) U251 and (B) U87 cells compared with the control group. (C) Downregulation of CDK4, CDK6, and MDM2 in transduced cells with LV-miR-129 compared with the control group. *p < 0.05, ****p < 0.0001. SC, scrambled control.
Expression of CDK4, CDK6, and MDM2 in Glioblastoma Cell Lines
Following the proof of significant overexpression of miR-129 in transduced cell lines, the expressions of CDK4, CDK6, and MDM2 genes were analyzed by real-time PCR. We analyzed the total mRNA of these cells and determined that expression of CDK4 in U251 and U87 cell lines declined by 69.8% and 47% (p < 0.05), respectively, and expression of CDK6 and MDM2 in U251 cells decreased by 55.3% (p < 0.0001) and 34.7% (p < 0.05), respectively (Figure 1C). Our bioinformatics studies suggested that miR-129 targets these genes and may proceed to inhibit cell signaling. Hence, the real-time PCR data suggested that the prediction could be trustworthy.
miR-129 Targets CDK4, CDK6, and MDM2 Dramatically
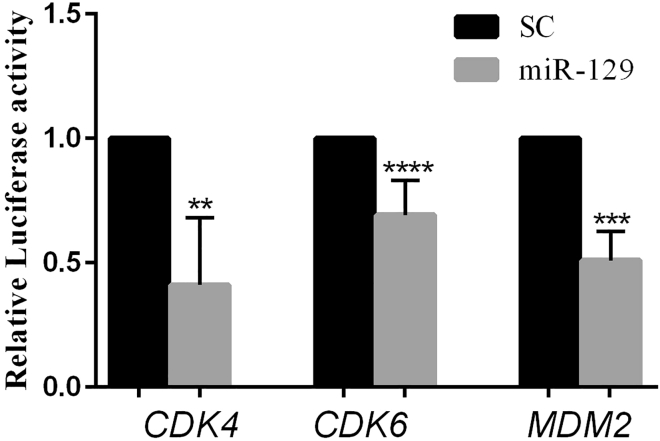
A luciferase reporter assay was taken into account to prove the significant interaction between miR-129 and the predicted binding site in the 3′ UTR of the understudied genes. The results indicated that overexpression of miR-129 can decrease the expression of the CDK4 gene by 58.9% (p < 0.01) and reduce the CDK6 expression by 35.7% (p < 0.0001) in HEK293 cells. The MDM2 expression was reduced by 49% (p < 0.001) as well (Figure 2). As a result, we can claim that miR-129 significantly targets and inhibits CDK4, CDK6, and MDM2 expression significantly.
Figure 2.
CDK4, CDK6, and MDM2 Are Targeted by miR-129
Analysis of luciferase assay in HEK293 cells co-transfected with miR-129 and CDK4-3′ UTR or CDK6-3′ UTR, or MDM2-3′ UTR in comparison with control groups that are HEK293 cells co-transfected with scramble control and 3′ UTR vectors. **p < 0.01, ***p < 0.001, ****p < 0.0001. SC, scrambled control. n = 3.
Overexpression of miR-129 Inhibits Cell Cycle and Growth in Glioblastoma Cells
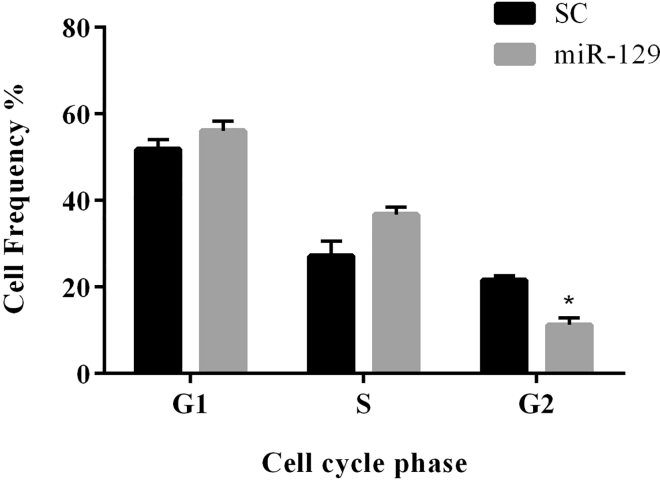
To investigate whether miR-129 affects glioblastoma cell growth via regulation of CDK4, MDM2, and CDK6, we performed a cell cycle analysis. First, we transduced the glioblastoma cells with LV-miR-129 and then analyzed the cell cycle by the flow cytometry method. We found that miR-129 arrests U87 cells in pre-G2 phase, and the population of cells in the G2 phase decreased significantly to 10% (p < 0.05) (Figure 3; Figure S2). Cell cycle analysis of U251 cells showed no significant changes after miR-129 overexpression.
Figure 3.
Flow Cytometry Analysis of Cell Cycle
The pre-G2 arrest of U87 cell line after overexpression of miR-129 compared with U87 cells transduced with scramble control. *p < 0.05. SC, scrambled control. n = 3.
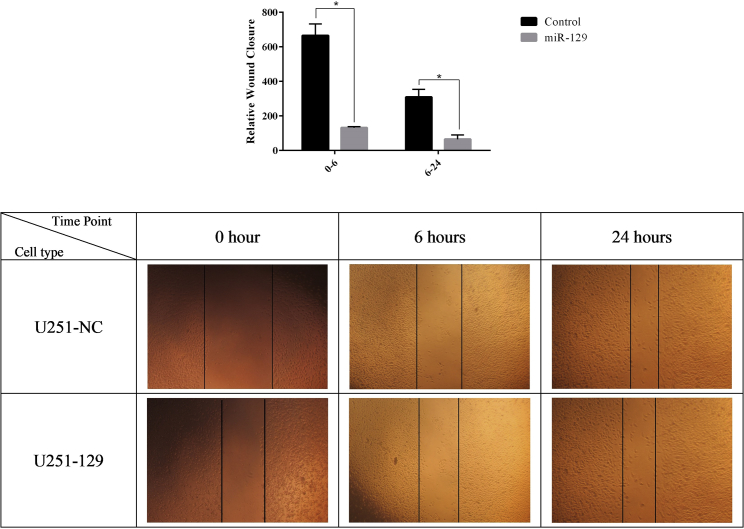
The wound healing assay results demonstrated that miR-129-5p-transduced cells had a significantly lower wound closure rate than that of the control group (Figure 4). Inhibition of the U87 cell line was so strong that we could not expand U87-transduced cells for this assay, although we cultured them for more than 2 weeks. These results suggested that overexpression of miR-129 correlated with inhibition of cell cycle progression in glioblastoma cells, and that miR-129 may serve as a tumor suppressor in glioma.
Figure 4.
U251 Transduced Cells by LV-miR-129 Demonstrated a Lower Wound Closure Rate Than Did the Control Group
*p < 0.05. n = 3.
Discussion
Glioblastoma is one of the most lethal forms of human cancer. Despite improvements in understanding GBM molecular mechanisms, certain treatments are not available and require further studies. Previously, it was shown that miR-129 is downregulated in human glioma cancer.18 miRNAs play roles in almost all biological pathways, such as cell proliferation, apoptosis, and tumorigenesis. Dysregulation of miRNAs can lead to different kinds of diseases, such as cancer. They have the potential of being tumorigenic or tumor suppressive at the beginning of cancer and affect its progression, metastasis, and response to treatment.19
Previously, it was shown that extrinsic miR-129 expression functions as a tumor suppressor in colon20 and papillary thyroid cancer.21 Additionally, some targets and functions have been recognized for miR-129 through its induced overexpression in glioma cell lines. In brief, in 2017 it was shown that miR-129-5p could target FNDC3B by binding to the 3′ UTR. The overexpression of miR-129-5p and the inhibition of FNDC3B can both inhibit U87 cell viability, proliferation, migration, and invasion while promoting cell apoptosis.22 Xiong et al.23 indicated that miR-129 inhibits the expression of SOX2 and suppresses glioma stem cell proliferation. Alternatively, MALAT1 as a long non-coding RNA (lncRNA) promotes the expression of SOX2 and boosts the viability and proliferation of glioma stem cells. The effects of MALAT1 and miR-129 on the glioma tumor in a xenograft mouse model indicated that MALAT1 promotes glioma tumor growth by regulating miR-129 and SOX2. In another study, it has been shown that miR-129-2 is downregulated in human glioma cancer, partially due to its DNA promoter hypermethylation. Overexpression of miR-129-2 suppressed cell growth, migration, and invasion and promoted cell apoptosis in U373 and U87 glioma cell lines through targeting the oncogene HMGB1 (high-mobility group box 1), which has been found to be upregulated and involved in the pathogenesis of many kinds of human cancers, and inhibiting its expression in glioma cells.18 Moreover, it has been shown that miR-129 is a new inducer of autophagy both through mTOR signaling and upregulation of Beclin-1 by targetedly suppressing Notch-1 in U251 and U87 cells. In fact, miR-129-5p triggered autophagy partially by the Notch-1/E2F7/Beclin-1 axis.24
In our study, two cell lines were used to compare the results in different mutation patterns and to confirm the reproducibility of our tests. Both U251 and U87 cell lines are mutant in the PTEN gene and have a methylated MGMT promoter, while the p53 gene is mutant in the U251 cell line as well.25 The present study indicated that induced expression of miR-129 negatively correlated with expression of CKD4, CDK6, and MDM2 in real-time PCR. To confirm the significant correlation between miR-129 and genes, a luciferase assay was used. Analysis of assays demonstrated that miR-129 dramatically targets the 3′ UTR of CDK4, CDK6, and MDM2. Targeted genes have function in RB and p53 signaling pathways and are upregulated in glioma. Therefore, their suppression via miR-129 can potentially inhibit cell signaling and promote cell cycle arrest. As we expected, cell cycle analysis verified pre-G2 arrest, and wound healing assays showed significant proliferation inhibition in comparison with the control group.
Conclusion
In summary, the findings of the present study identified CDK4, CDK6, and MDM2 as new targets of miR-129 in glioma cells, and these new findings can be of help in future clinical and GBM signaling-based research.
Materials and Methods
Bioinformatics Analysis
The KEGG database was used to investigate signaling pathways, and upregulated genes were identified via published documents based on TCGA project. Additionally, the high-throughput data from the Expression Atlas database was used to assess the expression of genes in glioma cell lines. To select miRNAs that target desirable upregulated genes, we used the following bioinformatics databases: TargetScan with a context++ score lower than −0.01, RNA22 with maximum folding energy for a heteroduplex of −10 kcal/mol, and the miRanda algorithm with a mirSVR score lower than −0.001 thresholds.
Cell Lines and Culture
Studied cell lines consisted of U87MG (IBRC C10982) and HEK293 cells (IBRC C10139), which were purchased from the Iranian Biological Resource Center (IBRC, Tehran, Iran). U251 was gifted from the Stem Cell Technology Research Center (Tehran, Iran), and short tandem repeat (STR) DNA profiling analysis verified cell authentication. All of the cell lines were cultured in high-glucose DMEM (4.5 g/L) (Bio Idea, Tehran, Iran) supplemented with 10% fetal bovine serum (FBS) (Gibco) and antibiotics (100 U/mL penicillin,100 mg/mL streptomycin) in a 5% CO2 incubator at 37°C. All cells were passaged at 2- or 3-day intervals.
Cloning of Genes and miRNA
The mir-129 gene was amplified by PCR using Taq DNA polymerase (cinnaGen) and specific primers (Table S1). Then, it was cloned into a pCDH-CMV-MCS-EF1-cGFP-T2A-puro vector (System Biosciences, USA). The full-length 3′ UTR of the CDK4 gene (1,060th nt to the end of mRNA NM_000075.4) and partial lengths of CDK6 3′ UTR (6,998–11,580 nt of transcript variant 1 mRNA NM_001259.8) and MDM2 3′ UTR (1,593–6,048 nt of transcript variant 3 mRNA NM_001145337.3) were amplified by PCR on genomic DNA using Takara SpeedSTAR HS DNA polymerase (catalog no. RR070A/B) and specific primers (Table S1). PCR fragments were cloned into the psiCHECK-2 Dual-Luciferase reporter plasmid separately (Promega, Madison, WI, USA) downstream of the Renilla luciferase gene. The pLenti-III-eGFP scramble vector was used as a negative control for miRNA.
RNA Isolation and Quantitative Real-Time RT-PCR
Total RNA was extracted from transduced glioma cell lines after 72 h using a GeneAll Hybrid-R kit (catalog no. 305-101), according to the manufacturer’s instructions. The quality and concentration of total RNA were examined using spectrophotometry. Extracted total RNA was converted to cDNA by using Moloney murine leukemia virus (M-MLV) reverse transcription (RT) enzyme (Thermo Fisher, catalog no. 28025013) and a specific RT primer for miR-129 and random hexamer for CDK4, CDK6, and MDM2 expression.
For qPCR, miR-129 expression was quantitated by using specific primers and a TaqMan probe relative to SNORD47 as an endogenous control. The CDK4, CDK6, and MDM2 genes’ expressions were calculated relative to the β2M gene using SYBR Green. The primers in this study used for cDNA synthesis and real-time PCR are as shown in Tables S2 and S3. Relative expression was determined using the 2−ΔΔCt method.
Virus Production
Recombinant lentiviruses were produced by transfection of one T25 flask of HEK293 cells at 60%–80% confluency with 2 μg of lentiviral vector (carrying hsa-miR-129), 2 μg of psPAX2 (gag/pol), and 1 μg of pMD2.G (VSVg) using the QIAGEN PolyFect transfection reagent (QIAGEN, Hilden, Germany, catalog no. 301107). psPAX2 and pMD2.G virus packaging helper vectors were purchased from the Stem Cell Technology Research Center (Tehran, Iran). Virus supernatant was collected in the 3 following days. For in vitro assays, glioblastoma cell lines were cultured in a T25 flask at 60%–80% confluency and were transduced by using hexadimethrine bromide (also known as Polybrene) as a transduction enhancer.
Luciferase Reporter Assay
In brief, for the luciferase reporter assay, miR-129 or scramble vector was transfected into cells, along with CDK4-3′ UTR, CDK6-3′ UTR, or MDM2-3′ UTR, using Lipofectamine 2000 transfection reagent (Invitrogen), according to the manufacturer’s protocol. Luciferase activity was evaluated 48 h after transfection using a Dual-Luciferase reporter assay system (Promega), according to the manufacturer’s instructions. Firefly luciferase activity was used for normalization.
Flow Cytometry Analysis
Cell cycle was evaluated 72 h after transduction. Transduced cells were trypsinized and washed in ice-cold PBS. Afterward, cells were fixed with cold paraformaldehyde for 1 h at 4°C. Then, we permeabilized cells by adding 70% ethanol at −20°C drop-wise to the cell pellet with the tube sitting on the vortex mixer and incubated the cell suspension overnight at −20°C. Subsequently, we stained cells with propidium iodide (PI) working solution, which consisted of PI, PBS, Triton X-100, and RNase A, and incubated them at 37°C in the dark for 30 min. Then, we analyzed the samples on a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Statistical Analysis
The data in this study are presented as mean ± SD. Data were compared using Student’s t test with GraphPad Prism 6.0 software. Real-time data were statistically analyzed by REST 2009 software, which is under QIAGEN supervision. Differences at p ≤ 0.05 were considered significant.
Wound-Healing Assay
Cell migration was analyzed using a wound-healing assay. Briefly, miR-129 or non-transduced cells were cultured to 80% confluency in 24-well plates. Then, an artificial wound was made, after which the cells were further incubated (0 h). Migration to the scratched area was imaged at 6 and 24 h after wounding. The wound closure was measured by ImageJ software relative to the total area of wounding.
Author Contributions
The study was conceptualized by E.A.; The methodology was given by E.A. and M.S.; A.M. carried out the experiments and analyzed the data. R.R.V. and S.G. contributed to designing microRNA and luciferase vectors constructs and assisted with gene cloning. F.J.A. assisted with lentiviral vector packaging and manipulation. The manuscript is written by A.M. and edited by E.A.
Acknowledgments
The authors would like to thank the National Institute for Medical Research Development (NIMAD) for the financial support of this research (grant 942974).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.033.
Supplemental Information
References
- 1.National Cancer Institute What You Need To Know About Brain Tumors. 2011. https://abc2.org/sites/default/files/NCI%20What%20You%20Need%20To%20Know%20About%20Brain%20Tumors_0.pdf
- 2.Grossman S.A., Ye X., Piantadosi S., Desideri S., Nabors L.B., Rosenfeld M., Fisher J., NABTT CNS Consortium Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin. Cancer Res. 2010;16:2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandes A.A., Tosoni A., Franceschi E., Reni M., Gatta G., Vecht C. Glioblastoma in adults. Crit. Rev. Oncol. Hematol. 2008;67:139–152. doi: 10.1016/j.critrevonc.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Murphy A.M., Rabkin S.D. Current status of gene therapy for brain tumors. Transl. Res. 2013;161:339–354. doi: 10.1016/j.trsl.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwiatkowska A., Nandhu M.S., Behera P., Chiocca E.A., Viapiano M.S. Strategies in gene therapy for glioblastoma. Cancers (Basel) 2013;5:1271–1305. doi: 10.3390/cancers5041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao H., Lebrun D.G., Yang J., Zhu V.F., Li M. Deregulated signaling pathways in glioblastoma multiforme: molecular mechanisms and therapeutic targets. Cancer Invest. 2012;30:48–56. doi: 10.3109/07357907.2011.630050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahir B.K., Ozer H., Engelhard H.H., Lakka S.S. MicroRNAs in glioblastoma pathogenesis and therapy: A comprehensive review. Crit. Rev. Oncol. Hematol. 2017;120:22–33. doi: 10.1016/j.critrevonc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Dunn G.P., Rinne M.L., Wykosky J., Genovese G., Quayle S.N., Dunn I.F., Agarwalla P.K., Chheda M.G., Campos B., Wang A. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam P.Y., Di Tomaso E., Ng H.K., Pang J.C., Roussel M.F., Hjelm N.M. Expression of p19INK4d, CDK4, CDK6 in glioblastoma multiforme. Br. J. Neurosurg. 2000;14:28–32. doi: 10.1080/02688690042870. [DOI] [PubMed] [Google Scholar]
- 10.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itahana K., Mao H., Jin A., Itahana Y., Clegg H.V., Lindström M.S., Bhat K.P., Godfrey V.L., Evan G.I., Zhang Y. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Reifenberger G., Ichimura K., Reifenberger J., Elkahloun A.G., Meltzer P.S., Collins V.P. Refined mapping of 12q13–q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets. Cancer. 1996;56:5141–5145. [PubMed] [Google Scholar]
- 14.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 17.Gramantieri L., Ferracin M., Fornari F., Veronese A., Sabbioni S., Liu C.G., Calin G.A., Giovannini C., Ferrazzi E., Grazi G.L. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y., Huang J.Q., Zhang X., Shen L.F. miR-129-2 functions as a tumor suppressor in glioma cells by targeting HMGB1 and is down-regulated by DNA methylation. Mol. Cell. Biochem. 2015;404:229–239. doi: 10.1007/s11010-015-2382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romano G., Veneziano D., Acunzo M., Croce C.M. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485–491. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q., Meng W.Y., Jie Y., Zhao H. lncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J. Cell. Physiol. 2018;233:6750–6757. doi: 10.1002/jcp.26383. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Cai Y., Zheng L., Zhang Z., Lin X., Jiang N. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J. Cell. Physiol. 2018;233:6638–6648. doi: 10.1002/jcp.26425. [DOI] [PubMed] [Google Scholar]
- 22.Xu H., Hu Y., Qiu W. Potential mechanisms of microRNA-129-5p in inhibiting cell processes including viability, proliferation, migration and invasiveness of glioblastoma cells U87 through targeting FNDC3B. Biomed. Pharmacother. 2017;87:405–411. doi: 10.1016/j.biopha.2016.12.100. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Z., Wang L., Wang Q., Yuan Y. lncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J. Cell. Mol. Med. 2018;22:3929–3940. doi: 10.1111/jcmm.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Zhang Y., Shi Y., Lian H., Tu H., Han S., Yin J., Peng B., Zhou B., He X., Liu W. miR-129 triggers autophagic flux by regulating a novel Notch-1/ E2F7/Beclin-1 axis to impair the viability of human malignant glioma cells. Oncotarget. 2016;7:9222–9235. doi: 10.18632/oncotarget.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sesen J., Dahan P., Scotland S.J., Saland E., Dang V.T., Lemarié A., Tyler B.M., Brem H., Toulas C., Cohen-Jonathan Moyal E. Metformin inhibits growth of human glioblastoma cells and enhances therapeutic response. PLoS ONE. 2015;10:e0123721. doi: 10.1371/journal.pone.0123721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.